Abstract
Quantitative detection of hepatitis B virus (HBV) in serum or plasma has become the most direct and reliable method for monitoring chronic hepatitis B. Here, we report the performance characteristics of a real-time PCR hepatitis B DNA quantitative assay, the COBAS TaqMan (CTM) HBV test (Roche Diagnostics, Meylan, France), in combination with an automated DNA extraction on the COBAS AmpliPrep (CAP) instrument using the total nucleic acid isolation kit (TNAI kit), a generic reagent for nucleic acid isolation (both from Roche Diagnostics). The linearity, accuracy, and specificity of the CAP-TNAI-CTM HBV test were evaluated using various reference panels and standards (HBV panel 2004 from Quality Control for Molecular Diagnostics, OptiQuant HBV panel from AcroMetrix, WHO International Standard for HBV, and Teragenix hepatitis B genotype panel). Quantitative results show that the CAP-TNAI-CTM HBV test performed well with respect to linearity, accuracy, and reproducibility from at least 100 to 500,000 HBV DNA IU/ml. Based on the log10 IU of HBV DNA/ml measured, the intra-assay variation ranged from 2.49% to 8.46% and the interassay variation ranged from 1.88% to 7.83%. The test was extremely sensitive and could detect samples containing HBV DNA below the reported quantification threshold (<30 IU/ml). All HBV genotypes were correctly amplified, and no cross-contamination occurred during the automated sample preparation. In addition, 402 human serum samples were tested comparatively to the VERSANT HBV DNA 3.0 assay (bDNA; Bayer Diagnostics, Puteaux, France). The viral load results of the CAP-TNAI-CTM test and bDNA were significantly correlated, but the agreement between the two tests was poor, with large differences between results for individual samples. The hands-on time was estimated to be reduced from 2.30 h with bDNA to 45 min with the CAP-TNAI-CTM test, and up to 84 samples were completely processed within a working day. Overall, the performance characteristics of the CAP-TNAI-CTM test demonstrated that it provides a high-throughput sensitive and reliable method for quantitation of HBV DNA levels in the routine molecular laboratory.
Hepatitis B virus (HBV) is a major causative agent of chronic hepatitis. Approximately 350 million to 400 million people worldwide are chronically infected. Chronic HBV infection can lead to liver cirrhosis and hepatocellular carcinoma (13). Detection and quantification of HBV DNA in serum or plasma are now considered important indicators for managing disease in HBV-infected patients and predicting and monitoring the efficiency of antiviral treatment as well as identifying the emergence of drug resistance by detecting HBV DNA breakthrough (4, 24). Several commercial molecular tests for the quantitation of HBV DNA in serum or plasma have been developed and are routinely used in diagnostic virology laboratories (10, 29). These molecular tests are based on target amplification (HBV Monitor test; Roche Diagnostics, Meylan, France), branched-DNA signal amplification (VERSANT HBV DNA 3.0 assay; Bayer Diagnostics, Puteaux, France), or hybridization of a chemiluminescent probe (Digene Corporation, Gaithersburg, Md). They differ in their performance characteristics (limits of detection and dynamic ranges) and their requirements for sample processing, which imply limitations in the screening of large numbers of samples (21, 38).
Real-time PCR technology has recently been introduced as a new molecular method for the detection and quantitation of PCR products and is increasingly being used as a diagnostic test for infectious diseases (2, 12, 25). Different studies have evaluated the quantitation of HBV DNA by real-time PCR assays and reported a higher sensitivity, a broader dynamic range, and an accurate quantitation of HBV DNA compared to those of the existing commercial assays (1, 8, 28, 32, 35).
The COBAS TaqMan HBV test (CTM HBV test; Roche Diagnostics, Meylan, France) is a recent commercial nucleic acid amplification test for HBV DNA viral load determination based on TaqMan PCR chemistry (16, 23, 37). Due to an increasing demand for molecular tests and for taking advantage of real-time PCR, a fully automated extraction system is considered a prerequisite (18, 19, 36). Several solutions for nucleic acid (NA) extraction on automated platforms have recently been evaluated for the diagnosis of viral infections (9, 11, 15, 17, 20, 22, 26, 27). The COBAS AmpliPrep system (CAP; Roche Diagnostics) is a new instrument designed for rapid high-throughput purification of DNAs and RNAs from biological fluids (such as serum and plasma) (5, 14, 19, 31, 33, 34). The automated NA extraction with the CAP is based on either a specific probe capture or a generic extraction method with magnetic glass particle technology. Clinical samples are loaded into the instrument, which then automatically releases NAs using ready-to-use reagents.
The aim of this study was to evaluate the clinical performance of the CAP using the total nucleic acid isolation (TNAI) kit (Roche Diagnostics) for generic nucleic acid extraction combined with the CTM HBV test for the quantitative detection of HBV DNA in the routine diagnostic laboratory. Analytical performance parameters, including specificity, reproducibility, linearity, limits of detection, and quantification, were determined by using different panels of HBV DNA standards and routine clinical samples. The results were compared to those obtained with the VERSANT HBV DNA 3.0 assay (bDNA; Bayer Diagnostics).
MATERIALS AND METHODS
QCMD HBV panel 2004.
The Quality Control for Molecular Diagnostics (QCMD) 2004 Hepatitis B Virus Proficiency Program was tested. It was prepared by Boston Biomedica, Inc. (Massachusetts) and consisted of eight coded serum samples. Among them, seven HBV positive samples were supplied in a concentration range of 38 to 190,114 IU/ml (or 200 to 106 copies/ml). Two genotypes, A and D, were included. One sample was HBV negative (see Table 1). The HBV DNA concentration for each sample of the QCMD HBV panel 2004 was expressed as copies/ml and obtained by Boston Biomedica, Inc. from release testing of 3× replicates using the COBAS Amplicor HBV Monitor test (Roche Diagnostics). The conversion in IU/ml (1 IU is equivalent to 5.26 HBV DNA copies) was made according to the manufacturer's instructions. In our assay, each sample of the QCMD HBV panel 2004 was tested once in one run, both with bDNA and the CAP-TNAI-CTM HBV test.
TABLE 1.
Comparative results obtained for eight members of the QCMD HBV panel 2004 with the CAP-TNAI-CTM HBV test and bDNAa
| Panel member | Genotype | No. of copies/mlb | No. of IU/ml | No. of log10 IU/ml | Result by CAP-TNAI-CTM HBV test
|
Result by bDNA
|
Difference between bDNA and CAP-TNAI-CTM values (log10 IU/ml) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of IU/mlc | No. of log10 IU/ml | Difference between measured and expected values (log10 IU/ml) | No. of IU/mld | No. of log10 IU/ml | ||||||
| HBV-09 | D | 100,000 | 19,011 | 4.3 | 43,400 | 4.6 | +0.3 | 53,973 | 4.7 | +0.1 |
| HBV-10 | A | 1,000 | 190 | 2.3 | 556 | 2.7 | +0.4 | 731 | 2.9 | +0.2 |
| HBV-11 | A | 100,000 | 19,011 | 4.3 | 55,500 | 4.7 | +0.4 | 60,542 | 4.8 | +0.1 |
| HBV-12 | D | 1,000 | 190 | 2.3 | 909 | 3.0 | +0.7 | 766 | 2.9 | −0.1 |
| HBV-13 | Negative | 0 | ND | <357 | <2.79 | |||||
| HBV-14 | A | 10,000 | 1,901 | 3.3 | 5,930 | 3.8 | +0.5 | 3,775 | 3.6 | −0.2 |
| HBV-15 | A | 1,000,000 | 190,114 | 5.3 | 313,000 | 5.5 | +0.2 | 430,381 | 5.6 | +0.1 |
| HBV-16 | A | 200 | 38 | 1.6 | 96.8 | 2.0 | +0.4 | <357 | <2.79 | |
One replicate of each sample was tested.
The mean values obtained by the production laboratory (Boston Biomedica, Inc.) were found by testing 3× replicates using the Roche COBAS Amplicor Monitor, version 2.0. The conversion factor between HBV copies/ml and HBV IU/ml is 5.26 copies/IU for the Roche COBAS Amplicor Monitor.
The conversion factor between HBV copies/ml and HBV IU/ml is 5.82 copies/IU for the CTM HBV test. ND, target not detected.
The conversion factor between HBV copies/ml and HBV IU/ml is 5.60 copies/IU for bDNA.
OptiQuant standard.
A commercially available standard containing HBV DNA (genotype A) at 2 × 107 IU/ml (OptiQuant panel 94-2012, NAP-HBV006; AcroMetrix, Inc., Benicia, CA, purchased from Biocentric, Bandol, France) was used. Serial dilutions of this reference standard were prepared to obtain four different dilutions with viral concentrations of 100,000, 10,000, 1,000 and 100 IU/ml. Dilutions were made in human serum (reference no. D119-00-0100; Rockland Immunochemicals, Inc., Gilbertsville, PA) purchased from Tebu-Bio SA (Le Perray en Yvelines, France) and then stored at −30°C prior to analysis using the CAP-TNAI-CTM HBV system. Each dilution was tested 18 times in a single run. Linearity of quantitation was evaluated as the mean log10 titer of HBV DNA (IU/ml) compared to the assigned log10 value (IU/ml), and the intra-assay variation was determined.
WHO standard.
To evaluate the sensitivity and the interassay variation of the CAP-TNAI-CTM HBV test, the World Health Organization (WHO) International Standard for HBV DNA quantification (National Institute for Biological Standards and Control, code 97/746) was used. This standard was established by a collaborative study (30). Each vial contained 500,000 IU of HBV DNA in lyophilized form (genotype A, subtype adw, 5 × 105 IU). An aliquot of the WHO International Standard was reconstituted with 0.5 ml of sterile water and then serially diluted to HBV DNA concentrations of 400, 200, 100, 50, 25, 12.5, and 6.25 IU/ml in human serum as described above. Each concentration was tested in duplicate daily over a total of 4 days.
HBV genotype panel.
To assess the genotype inclusivity of the CAP-TNAI-CTM HBV test, an international reference serum panel was obtained from Teragenix Corporation (Fort Lauderdale, FL). This panel consisted of 15 members of clinical HBV DNA-positive specimens representing genotypes A to G. The viral load values (IU/ml) assigned by Teragenix for each sample of this panel were determined with Roche COBAS TaqMan 48 at 1:10 dilution. Viral load values were calculated from this dilution, and genotype results were generated utilizing the INNO-LiPA HBV genotyping assay (Innogenetics N.V, Ghent, Belgium).
Clinical specimens.
A total of 402 human serum samples previously analyzed by bDNA at the L.C.L. Laboratory were selected for the study. The clinical serum samples were divided into two groups: the first group (n = 104) was below the limit of detection of bDNA (below 357 IU/ml), and the second group consisted of 298 samples with viral loads of >357 IU/ml with bDNA. All samples were stored at −30°C until use for this study.
AmpliPrep sample processing and COBAS TaqMan 48 HBV test.
HBV DNA was extracted from clinical specimens and various standards with the CAP instrument, using the TNAI kit according to the manufacturer's instructions. The protocol for isolation of NAs was essentially based on the method developed by Boom et al. (7). Extraction, amplification, and detection steps were performed in batches of 24 samples (21 clinical specimens and 3 controls) without the user's intervention. Briefly, 1 ml of the clinical sample or control was manually transferred into bar code-labeled sample input tubes (S-tubes) and then placed into sample racks in the CAP instrument. Note that there were three sample rack positions, each rack holding 24 S-tubes for an onboard capacity of 72 samples. Once treated, sample racks can be removed and new racks loaded while the CAP is running for continuous operation. All CAP reagents were contained into TNAI kit bar-coded cassettes with all necessary reagents, including the HBV quantitation standard (QS) DNA adjusted in an appropriate dilution with the TNAI/QS diluent. Sample preparation occurred in a disposable sample processing unit. After the preparation process ended, eluted NAs were transferred into output S-tubes that were sealed. Then, 50 μl of eluates was manually transferred into kinetic reaction tubes (K-tubes) together with 50 μl of TaqMan HBV working master mix containing primer pairs and probes specific for both HBV DNA and HBV QS DNA at a known copy number. K-tubes were placed in a K-tube rack (K-carrier, 24 tubes each) and then loaded onto the CTM analyzer for real-time PCR according to the manufacturer's instructions. The detection of amplified DNA was performed using target-specific and QS-specific dual-labeled oligonucleotide probes, allowing an independent identification of HBV and HBV QS amplicons.
The HBV DNA concentration was automatically calculated by comparing the HBV signal to the HBV QS for each specimen and control. Runs were validated if low positive, high positive, and negative controls were in an acceptable range. As stated by the manufacturer, the limit of detection was 6 IU/ml, and the limit of quantitation was reported to be 30 IU/ml. The linear dynamic range of the CAP-TNAI-CTM HBV test was 30 HBV IU/ml to 1.1 × 108 HBV IU/ml. For all values of ≥1.1 × 108 HBV IU/ml, it was recommended to retest the sample after dilution if required. The AmpliLink software (Roche Molecular Systems) reported the results as follows: a positive result was considered to be any quantitative value, regardless of the value, or any result of less than 6 IU/ml. Results in which the target was not detected were considered a negative result. Although a numeric titer below 30 IU/ml was considered a positive result, the relation between the calculated value based upon the HBV DNA and HBV QS DNA cycle threshold values was not totally linear (coefficient of variation [CV], >40%), showing the difference between the limit of detection and the limit of quantitation. According to the manufacturer's insert, 1 IU is equivalent to 5.82 HBV DNA copies.
bDNA.
bDNA, a sandwich nucleic acid hybridization test with signal amplification, was performed according to the manufacturer's instructions using the Q340 analyzer. The test used 50 μl of serum or plasma. Samples were processed in two 96-microwell plates per run and had lower and upper detection limits of 357 and 1.78 × 107 IU/ml (or 2.0 × 103 and 1.8 × 108 copies/ml), respectively. According to the manufacturer's insert, 1 IU is equivalent to 5.60 HBV DNA copies.
Comparative analysis.
Regression lines and their characteristics were calculated using Microsoft Excel (Microsoft Office 2000; Microsoft Corp., Redmond, Wash.). For comparison of quantitative results from the overlapping dynamic range of the CAP-TNAI-CTM test and bDNA after log10 transformation of IU/ml values, scatter plots and Spearman's coefficient were determined, and the Bland-Altman (6) and Passing-Bablock (MedCalc software) (3) methods were used to assess the agreement between the values obtained with the two assays.
RESULTS
Evaluation with the QCMD HBV panel 2004 and correlation with the VERSANT HBV DNA 3.0 assay.
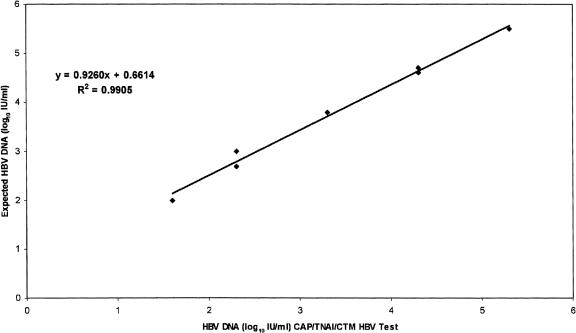
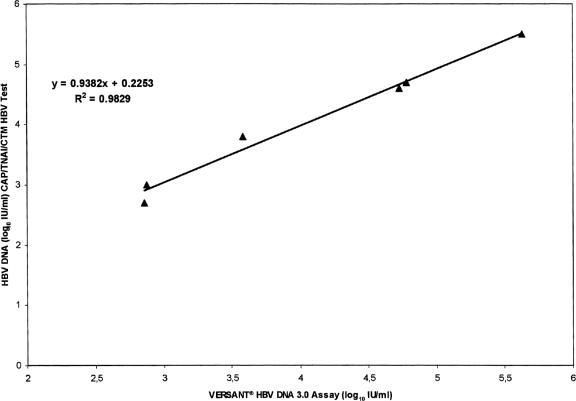
The accuracy of the CAP-TNAI-CTM HBV test was evaluated using eight samples from the QCMD HBV panel 2004. The results obtained were in acceptable agreement with the reported values for each sample (Table 1) but were systematically higher. The mean log10 IU/ml difference between measured and expected values was +0.41 (range, +0.2 to +0.7). The linear regression plot had a slope of 0.92, with a correlation coefficient (R2) of 0.99 (Fig. 1). The HBV-16 sample, containing a low number of HBV DNA (38 IU/ml, equivalent to 200 copies/ml), was correctly quantified, and the negative sample (HBV-13) was correctly detected. We then compared these results with those obtained by bDNA. The viral load measured with the two tests showed a high correlation (slope = 0.93; R2 = 0.98) (Fig. 2). The mean difference between HBV DNA levels obtained with bDNA and the CAP-TNAI-CTM HBV test was 0.13 log10 (range, −0.2 to +0.2).
FIG. 1.
Linearity of the CAP-TNAI-CTM 48 HBV test using the eight members of the QCMD HBV panel 2004. HBV DNA log10 IU/ml measured with the CAP-TNAI-CTM HBV test were plotted against log10 expected concentration. One replicate of each member of the QCMD HBV panel 2004 was tested.
FIG. 2.
Correlation of log10 quantitative HBV DNA concentrations of samples obtained from the QCMD HBV panel 2004 and measured with the CAP-TNAI-CTM HBV test and the VERSANT HBV DNA 3.0 assay. HBV DNA log10 IU/ml measured with the CAP-TNAI-CTM HBV test were plotted against log10 HBV DNA IU/ml obtained with bDNA. One replicate of each member of the QCMD HBV panel 2004 was tested.
Linearity and intra-assay variation.
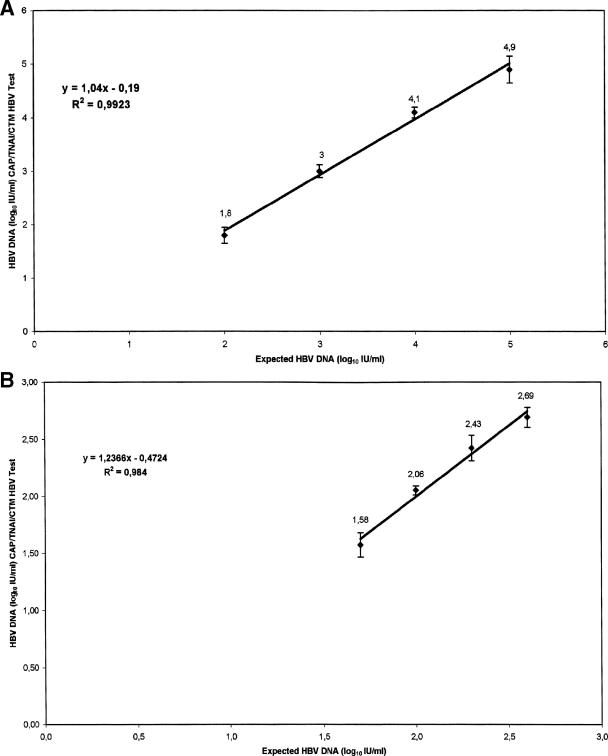
Table 2 shows the mean log10 (IU/ml) viral loads, standard deviations, and CVs of serially diluted samples from the OptiQuant standard. HBV DNA was detected in all the replicates, and viral load values from the CAP-TNAI-CTM test were highly correlated to expected values (slope = 1.04; R2 = 0.99) and very reproducible over the various dilutions, with a CV ranging from 2.49% to 8.46%. The measured versus expected values are plotted in Fig. 3A.
TABLE 2.
Linearity and intra-assay variation of the CAP-TNAI-CTM HBV test
| OptiQuant standard (expected HBV DNA level [log10 IU/ml]) | No. of replicates tested | Mean level (log10 IU/ml) of HBV DNA measured | SD (log10 IU/ml) | CV (%) |
|---|---|---|---|---|
| 5 | 18 | 4.9 | 0.25 | 5.17 |
| 4 | 18 | 4.1 | 0.10 | 2.49 |
| 3 | 18 | 3 | 0.12 | 4.03 |
| 2 | 18 | 1.8 | 0.15 | 8.46 |
FIG. 3.
(A) Plot of mean log10 (IU/ml) measured against expected log10 (IU/ml) of HBV DNA for serial dilutions (100,000, 10,000, 1,000, and 100 IU/ml) prepared from the OptiQuant panel. Each point represents the mean log10 value (IU/ml) at each dilution for a total of 18 replicates. (B) Plot of mean log10 (IU/ml) measured against expected log10 (IU/ml) of HBV DNA for serial dilutions (400, 200, 100, and 50 IU/ml) prepared from the WHO standard for HBV DNA (NIBSC 97/746). Each dilution was tested in duplicate in four separate runs with the COBAS AmpliPrep coupled with the COBAS TaqMan HBV test. Each point represents the mean log10 value (IU/ml) of HBV DNA at each dilution for a total of eight replicates.
Sensitivity and interassay variation.
Assays were performed on two replicates of serially diluted samples from the WHO HBV standard (6.25 to 400 IU/ml) tested daily over 4 days. One hundred percent of replicates were detected at all the seven levels of dilution (Table 3). Although HBV DNA was detected in all replicates in dilutions containing 50 IU/ml (mean value, 38.5 IU/ml; range, 21 to 59 IU/ml; CV, 6.85%), it was inconsistently quantified within the lower limit of the linear dynamic range of the CTM HBV test, with two replicates giving values below the lower quantification limit of 30 IU/ml (21 and 29 IU/ml). The interassay CVs of the log10 HBV DNA IU/ml measured with the CAP-TNAI-CTM HBV test ranged from 1.88% to 7.83%, and a linear relationship (slope = 1.23; R2 = 0.98) was obtained between the measured and expected values, ranging from 50 to 400 IU/ml (Fig. 3B).
TABLE 3.
Analytical sensitivity of the CAP-TNAI-CTM HBV test and results of interassay testinga
| WHO HBV standard dilution [IU/ml (log10 IU/ml)] | No. of IU/ml forb:
|
Mean (log10 IU/ml) | SD (log10 IU/ml) | CV (%) | |||
|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | Run 4 | ||||
| 400 (2.60) | 495-502 | 530-367 | 405-417 | 728-549 | 2.69 | 0.09 | 3.21 |
| 200 (2.30) | 219-271 | 271-203 | 237-208 | 378-400 | 2.43 | 0.11 | 4.59 |
| 100 (2.00) | 117-100 | 111-114 | 141-114 | 121-87 | 2.06 | 0.04 | 1.88 |
| 50 (1.70) | 44-42 | 21-39 | 29-32 | 59-42 | 1.58 | 0.11 | 6.85 |
| 25 (1.40) | 22-23 | 16-13 | 17-18 | 19-30 | 1.29 | 0.10 | 7.83 |
| 12.5 (1.10) | 12-<6 | 9-8 | 8-<6 | <6 | |||
| 6.25 (0.80) | <6 | <6 | <6 | <6 | |||
One duplicate of each dilution was tested daily over 4 days.
Results below the limit of quantitation (30 IU/ml) of the assay are indicated in italics.
Evaluation of HBV genotype inclusivity.
Using the HBV 15-member genotype panel (genotypes A, B, C, D, E, F, and G) from Teragenix, 10 samples tested were quantified within the dynamic range of the CAP-TNAI-CTM HBV test (Table 4). Two samples (members 5 and 9) were above the upper limit of quantification (>110,000,000 IU/ml or 8 log10 IU/ml), and for three samples (members 4, 10, and 14), the values obtained by the CAP-TNAI-CTM HBV test were near or below the limit of quantitation (30 IU/ml). The expected and measured HBV DNA concentrations were significantly correlated (R2 = 0.98).
TABLE 4.
Evaluation of HBV genotype panel from Teragenix using the CAP-TNAI-CTM HBV testa
| Member | Genotype | Origin | Expected viral load
|
Result by CAP-TNAI-CTM HBV test
|
||
|---|---|---|---|---|---|---|
| No. of IU/ml | No. of log10 IU/ml | No. of IU/ml | No. of log10 IU/ml | |||
| 1 | A | United States | 182,000.0 | 5.3 | 123,000.0 | 5.1 |
| 2 | F | Venezuela | 542.0 | 2.7 | 670.0 | 2.8 |
| 3 | C | Indonesia | 309.0 | 2.5 | 291.0 | 2.5 |
| 4 | E | Ivory Coast | 61.8 | 1.8 | 31.7 | 1.5 |
| 5 | B | Indonesia | 512,000,000.0 | 8.7 | >110,000,000 | >8 |
| 6 | D | United States | ND | ND | ||
| 7 | A | United States | 20,000.0 | 4.3 | 18,800.0 | 4.3 |
| 8 | A | United States | 984.0 | 3.0 | 1,140.0 | 3.1 |
| 9 | E | Ivory Coast | 195,000,000.0 | 8.3 | >110,000,000 | >8 |
| 10 | F | Venezuela | 128.0 | 2.1 | 25.9 | 1.4 |
| 11 | C | Venezuela | 1,490.0 | 3.2 | 1,590.0 | 3.2 |
| 12 | G | United States | 37,700,000.0 | 7.6 | 40,500,000.0 | 7.6 |
| 13 | B | Indonesia | 145,000.0 | 5.2 | 117,000.0 | 5.1 |
| 14 | B | Indonesia | 20.7 | 1.3 | 6.3 | 0.8 |
| 15 | C/E | Ivory Coast | 496.0 | 2.7 | 775.0 | 2.9 |
The regression equation comparing HBV DNA (log10 IU/ml) measured by the CAP-TNAI-CTM HBV test versus expected values (log10 IU/ml) is as follows: y = 1.0484x − 0.2847; R2 = 0.9827. ND, target not detected.
Clinical performance characteristics of the CAP-TNAI-CTM HBV test.
A total of 402 clinical samples previously tested with bDNA were evaluated with the CAP-TNAI-CTM HBV test. Samples were divided into two groups according to the viral load previously measured by bDNA.
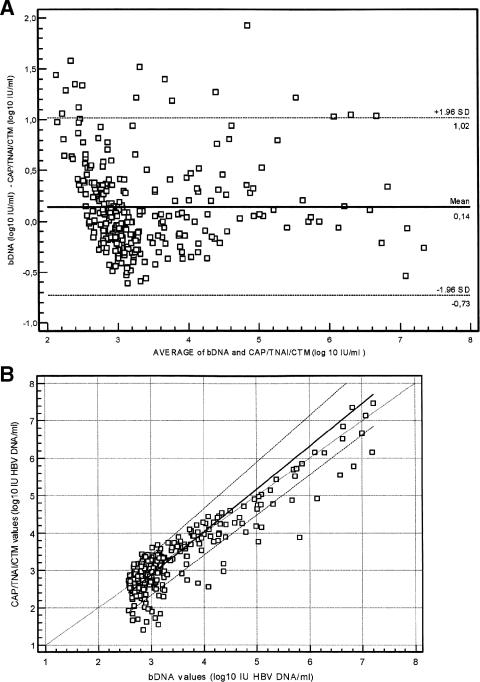
In the first group (n = 104), the bDNA HBV load was <357 IU/ml (<2.55 log10 IU/ml), and in the second group, the viral load was >357 IU/ml (>2.55 log10 IU/ml). Among the first group, 40 samples did not detect HBV DNA and 64 samples (62%) yielded a positive result with the CAP-TNAI-CTM HBV test (Table 5). Among them, 29 samples were found to contain less than 357 IU/ml (mean, 244 IU/ml), and 9 samples demonstrated values above 357 IU/ml (mean, 616 IU/ml; range, 402 to 1,000 IU/ml) with the CAP-TNAI-CTM HBV test. For the remaining 26 samples (41%), HBV DNA was detected but not quantified by the CAP-TNAI-CTM HBV test (<30 IU/ml). In the second group, of 298 samples with values above 357 IU/ml with bDNA, 283 (94.9%) samples were found with a viral load above 357 IU/ml with the CAP-TNAI-CTM HBV test (Table 6). Six samples had titers above the linear range of both bDNA (7.25 log10 IU/ml) and the CAP-TNAI-CTM HBV test (8.04 log10 IU/ml). For 12 (4%) samples positive with bDNA (>357 IU/ml), HBV DNA was detected below 30 IU/ml with the CAP-TNAI-CTM HBV test. The average titer of these samples with bDNA was 681 IU/ml (2.83 log10). Three (1%) samples positive with bDNA (453, 448, and 446 IU/ml) were reported not detectable by the CAP-TNAI-CTM HBV test. Based on these data, a comparison between the two assays was performed from 277 samples with HBV DNA values included in the linear dynamic range of both assays. Spearman's coefficient showed a significant correlation between the two tests (R = 0.768; 95% confidence interval, 0.715 to 0.812; P < 0.0001). A plot of the differences between the log10 IU/ml values reported by both assays for the 277 samples versus the average log10 IU/ml for each specimen was established (Bland-Altman analysis) (Fig. 4A). The mean log10 IU/ml difference between bDNA and the CAP-TNAI-CTM HBV test was 0.14 (means, 3.46 versus 3.31, respectively; P = 0.0781).
TABLE 5.
Results for 104 serum samples with a viral load below the lower limit of the VERSANT HBV DNA 3.0 assay analyzed by the CAP-TNAI-CTM HBV test
| Result by CAP-TNAI-CTM HBV test (n = 104) | No. of samples by VERSANT HBV DNA 3.0 assay (<357 IU of HBV DNA/ml) |
|---|---|
| HBV DNA not detected | 40 |
| 6 IU/ml < x < 30 IU/ml (HBV DNA detected but not within the linear range)a | 26 |
| >30 IU/ml | 38 |
x, HVB DNA level.
TABLE 6.
Results for 298 serum samples with a viral load above the upper limit of the VERSANT HBV DNA 3.0 assay analyzed by the CAP-TNAI-CTM HBV testa
| Result by CAP-TNAI-CTM HBV test (n = 298) | No. of samples by VERSANT HBV DNA 3.0 assay
|
|
|---|---|---|
| 357 IU/ml < x < 1.78 × 107 IU/ml | >1.78 × 107 IU/ml | |
| HBV DNA not detected | 3 | |
| 6 IU/ml < x < 30 IU/ml (HBV DNA detected but not within the linear range) | 12 | |
| >30 IU/ml | 277 | |
| >1.1 × 108 IU/ml | 6 | |
x, HVB DNA level.
FIG. 4.
(A) Bland-Altman comparison of bDNA and the CAP-TNAI-COBAS TaqMan HBV test on 277 samples initially quantified above 357 IU/ml with bDNA. For each specimen, the difference between the log10 IU/ml values is plotted against the mean of the bDNA and the CAP-TNAI-CTM log10 IU values for each paired result. (B) Passing-Bablock agreement between viral loads detected by bDNA and the CAP-TNAI-COBAS TaqMan HBV test. Assay correlation was determined by processing 277 clinical specimens with the CAP-TNAI-COBAS TaqMan HBV test, which were initially quantified above 357 IU/ml with bDNA. Slope, 1.1429 (95% CI, 1.06 to 1.23); and intercept, −0.5471 (95% CI, −0.86 to −0.30). The solid diagonal line represents theoretical, perfect agreement between viral loads measured with the CAP-TNAI-COBAS TaqMan HBV test and bDNA.
Values with bDNA were more often greater than with the CAP-TNAI-CTM HBV test. As shown in Fig. 4A, 93.8% (260/277) of HBV DNA values were within the 95% confidence interval (±1.96 standard deviation), but they were not homogeneously distributed, since all off-limits values were above +1.96 standard deviation. Furthermore, 18% of paired results differed by more than 0.5 log10 IU/ml, which is a deviation regarded as clinically important in consecutive viral load determinations. The comparative analysis performed using Passing-Bablock agreement showed a slope of 1.142 and a y intercept of −0.547, demonstrating a significant deviation from linearity (P < 0.01) (Fig. 4B).
In our hands, the time required to prepare and manually load both specimens into bar-coded sample racks, reagent tips, cassettes, sample processing units, and S-tubes and K-tubes prior to automated processing was approximately 30 min. The total time required for the extraction process with the CAP instrument was 2.0 h for 21 samples plus 3 controls without any hands-on work. The real-time PCR using the TaqMan 48 analyzer takes approximately 2.15 h. Finally, the total duration of the process from extraction to final data output was approximately 6 h.
DISCUSSION
Molecular techniques have proven to be of great utility in the diagnosis and management of HBV infection. Because HBV DNA is detectable in serum prior to biochemical evidence of hepatitis and persists at variable levels throughout the course of chronic disease, sensitive and accurate HBV DNA tests are required. Currently, molecular diagnostic laboratories are faced with an increasing demand to improve the efficiency of and time to reportable results. Real-time PCR is considered a powerful and rapid technique for nucleic acid testing, combining both high sensitivity and high specificity. Because amplification and detection are performed in a single vessel, they offer time and labor savings while reducing the risk of amplicon carryover contamination. Furthermore, automated extraction systems have recently been introduced for minimizing labor-intensive nucleic acid isolation from the sample. In the present study, we evaluated the performance of a fully automated system for the hepatitis B quantitative assay combining the COBAS AmpliPrep analyzer, an automated nucleic acid extractor, using the TNAI kit (CAP-TNAI), with that of the CTM HBV test, a recent automated HBV quantitative assay based on real-time PCR. The performance characteristics were established using standardized panels and clinical samples and compared to those of bDNA.
While our study did not evaluate the entire reportable dynamic range of the COBAS TaqMan HBV test (30 IU/ml to 1.1 × 108 IU/ml, as stated in the technical insert), our data from the QCMD HBV panel 2004 and OptiQuant panel testing showed that the CAP-TNAI-CTM test demonstrated an accurate and linear quantification of HBV DNA from 100 to 500,000 IU/ml, with a good intra-assay variation ranging from 2.49 to 8.46%.
The results obtained with the QCMD panel were found to be within ±0.5 log10 of the expected viral load (mean, +0.4 log10) for each panel member except HBV-12 (+0.7 log10 difference). For this sample, the log10 difference between the CAP-TNAI-CTM HBV test and bDNA was very weak (−0.1 log10). In addition, in the QCMD 2004 HBV final report, viral loads obtained for HBV-12 from five reference laboratories ranged from 136 to 875 IU/ml. The overestimation of the viral load for this sole sample should be interpreted with caution and might be due to an isolated technical problem. When various dilutions of the WHO International Standard for HBV DNA were tested, 100% of repeats with 100 IU/ml (2 log10 IU/ml) were accurately quantified (mean, 2.06 log10 IU/ml; CV, 1.88%). At the 50-IU/ml (1.7 log10 IU/ml) dilution, the mean value was 1.58 log10 IU/ml, and six out of eight repeats demonstrated HBV DNA values above the quantification threshold. Since the quantified values for the two remaining repeats were near 30 IU/ml (21 and 29 IU/ml), the limit of quantification of the test could be estimated between 30 and 50 IU/ml. HBV DNA was consistently detected from all repeats at 6.25 IU/ml. These results confirmed the limit of quantification and the limit of detection claimed by the manufacturer, 30 and 6 IU/ml, respectively. When the Teragenix genotype panel was tested, the CAP-TNAI-CTM HBV test appeared to measure viral load regardless of the infecting genotype (A through G). However, the viral loads from two samples (genotypes B and F) were underestimated (−0.5 to −0.7 log10 IU/ml) against the expected values. Since the expected viral loads for these samples ranged between 1.3 log10 and 2.1 log10 IU/ml, one explanation might be a reduced effectiveness of the CAP-TNAI-CTM HBV test at low HBV DNA concentration. However, another possibility might be an extrapolation error, considering that viral loads were calculated by Teragenix from a 1:10 dilution to get final results. The HBV genotype panel consisted of a 1-ml volume per vial; thus, insufficient material was available to control our results, and too few samples were tested to conclude that a potential for genotype-dependent bias in HBV DNA quantification might exist. In addition, no significant genotype bias was observed in recent reports using the COBAS TaqMan HBV test (16, 23, 37).
From the results of our clinical study, it first appeared that the CAP-TNAI-CTM HBV test was highly sensitive compared to bDNA. Samples (62%) demonstrating bDNA results below the lower limit of the quantification (<357 IU/ml) yielded a positive result with the CTM HBV test. Of these, 59% were quantified within the dynamic range of the CTM HBV test, with nine samples having viral loads above 357 IU/ml, and for the remaining 41%, HBV DNA levels were found detectable but not quantified within the linear range of the CTM HBV test. The increased sensitivity of the COBAS TaqMan HBV test (range, 30 IU/ml to 1.1 × 108 IU/ml) could improve a clinician's ability to predict virological relapse or treatment failure in patients undergoing anti-HBV therapy (8). The high sensitivity of the CAP-TNAI-CTM HBV test might be due in part to the large sample volume (1 ml) required for DNA extraction. This could be an inconvenience when only limited volumes of patient sample are available.
Two hundred ninety-eight clinical samples previously positive (>357 IU/ml) with bDNA were also quantifiable with the CAP-TNAI-CTM HBV test. A correlation was calculated that included 277 samples with viral load found inside the linear range in two assays. The results of the two assays showed a significant correlation (Spearman's coefficient = 0.768; P < 0.0001). The bDNA values were more often greater than those of the CAP-TNAI-CTM HBV test (mean difference, 0.14 log10 IU/ml). However, the two tests showed poor agreement, with a significant deviation from linearity.
This could be explained by the extraction method, the input specimen volume used (1 ml versus 200 μl for bDNA), or the assay format (real-time PCR versus signal amplification methodology). Since 18% of paired results differed by more than a 0.5-log10 IU/ml deviation, which is by consensus considered clinically important, our data indicated that the two tests could not be used interchangeably during the patient's monitoring.
Discrepant results were noted for 15 samples. Among them, 12 (average titer with bDNA, 681 IU/ml or 2.83 log10 IU/ml) were detectable but under the quantification threshold with the CAP-TNAI-CTM test. In addition, three samples with respective titers of 453 IU/ml, 448 IU/ml, and 446 IU/ml (2.65 log10 IU/ml) with bDNA were found not detectable with the CAP-TNAI-CTM test, in contrast to the above-mentioned higher sensitivity of this test. Although a technical error could not be excluded, a false positivity of bDNA results at values near the positive cutoff was considered likely, as previously noted in a multicenter evaluation of the VERSANT HBV DNA 3.0 assay (38). Unfortunately, sufficient material was not available for further studies of these discrepant samples.
In our study, no cross-contamination was observed during the whole process, and none of the human serum samples exhibited evidence of containing PCR inhibitors (data not shown). However, we noticed some problems or failures of sample processing associated with the CAP instrument during routine operations. The turnaround time for the bDNA assay (semiautomated assay) is approximately 18 h for a total number of 168 tests in one run being performed in a 96-well format (a maximum of 84 samples can be processed per plate).
In our routine experience, since the CAP instrument was able to work continuously once the run was started (samples, reagents, and consumables being replaced during the extraction process), 84 samples were completely processed (from extraction to final data output) within 9 h using one CAP coupled with two CTM 48 analyzers (note that the COBAS TaqMan 96 analyzer is not presently available in France). Combining multiple TaqMan analyzers could be an attractive option for laboratories handling large numbers of samples. When we performed this study, processed samples and controls were manually transferred into the prealiquoted master mixes in PCR K-tubes. This inconvenience is now overcome in the last version of the CAP instrument's software, which allows the automated distribution of eluates and PCR master mix in K-tubes by the CAP itself. This could further decrease the total time required while avoiding human pipetting error. Because the automated sample preparation by the CAP instrument is performed without any hands-on work, it allows the operator time to perform other laboratory duties in contrast to the manual processing steps with bDNA. Moreover, the benefits of the real-time PCR format include a reduction of the hands-on time by a complete abolishment of postamplification processing and a reduction of the carryover risk compared to conventional quantitative HBV PCR tests. From our experience, the hands-on time is estimated to be reduced from 2.30 h for the manual method with bDNA to 45 min with the CAP-TNAI-CTM HBV test. In summary, the results of this study show that the automated CAP-TNAI-CTM HBV test may be considered an attractive high-throughput and sensitive tool for the quantitative determination of HBV DNA load.
Because the sample treatment is a key component of nucleic acid detection, as it affects both the reliability and the reproducibility of target amplification, one of the major advantages of automating the extraction is the ability to provide a standardized process among samples. However, it is important to take some precautions before using the CAP instrument, particularly when installing consumables and loading samples and reagents. The combination with the CTM 48 analyzer significantly reduces hands-on work time and labor intensity while reducing the risk of contamination and human error, despite the present incapability of the CAP instrument to proceed directly from bar-coded primary sample tubes. Overall, the CAP-TNAI-CTM test is a sensitive, accurate, reproducible, and labor-saving automation system and may be considered a valuable step toward complete automation for the determination of HBV loads in the routine molecular laboratory.
Acknowledgments
We thank Roche Diagnostics, Meylan, France, for supplying the TNAI kits and COBAS TaqMan 48 HBV test reagents.
We thank Magali Fucina for her help and her excellent technical assistance.
REFERENCES
- 1.Abe, A., K. Inoue, T. Tanaka, J. Kato, N. Kajiyama, R. Kawaguchi, S. Tanaka, M. Yoshiba, and M. Kohara. 1999. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J. Clin. Microbiol. 37:2899-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arya, M., I. S. Shergill, M. Williamson, L. Gommersall, N. Arya, and H. R. Patel. 2005. Basic principles of real-time quantitative PCR. Expert Rev. Mol. Diagn. 5:209-219. [DOI] [PubMed] [Google Scholar]
- 3.Bablok, W., H. Passing, R. Bender, and B. Schneider. 1988. A general regression procedure for method transformation. J. Clin. Chem. Clin. Biochem. 26:783-790. [DOI] [PubMed] [Google Scholar]
- 4.Berger, A., W. Preiser, and H. W. Doerr. 2001. The role of viral load determination for the management of human immunodeficiency virus, hepatitis B virus and hepatitis C virus infection. J. Clin. Virol. 20:23-30. [DOI] [PubMed] [Google Scholar]
- 5.Berger, A., L. Scherzed, M. Sturmer, W. Preiser, H. W. Doerr, and H. F. Rabenau. 2002. Evaluation of the COBAS AmpliPrep/COBAS Amplicor HIV-1 Monitor ultrasensitive test: comparison with the COBAS Amplicor HIV-1 Monitor test (manual specimen preparation). J. Clin. Virol. 25(Suppl. 3):S103-S107. [DOI] [PubMed] [Google Scholar]
- 6.Bland, J. M., and D. G. Altman. 1995. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet 346:1085-1087. [DOI] [PubMed] [Google Scholar]
- 7.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, R. W., H. Piiparinen, M. Seppanen, P. Koskela, S. Sarna, and M. Lappalainen. 2001. Real-time PCR for detection and quantitation of hepatitis B virus DNA. J. Med. Virol. 65:250-256. [DOI] [PubMed] [Google Scholar]
- 9.Cook, L., K.-W. Ng, A. Bagabag, L. Corey, and K. R. Jerome. 2004. Use of the MagNA Pure LC automated nucleic acid extraction system followed by real-time reverse transcription-PCR for ultrasensitive quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 42:4130-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Franchis, R., A. Hadengue, G. Lau, D. Lavanchy, N. McIntyre, A. Mele, G. Paumgartner, A. Pietrangelo, J. Rodes, W. Rosenberg, D. Valla, and the EASL Jury. 2003. EASL International Consensus Conference on Hepatitis B. J. Hepatol. 39(Suppl. 1):S3-S25. [PubMed] [Google Scholar]
- 11.Fiebelkorn, K. R., B. G. Lee, C. E. Hill, A. M. Caliendo, and F. S. Nolte. 2002. Clinical evaluation of an automated nucleic acid isolation system. Clin. Chem. 48:1613-1615. [PubMed] [Google Scholar]
- 12.Foy, C. A., and H. C. Parkes. 2001. Emerging homogeneous DNA-based technologies in the clinical laboratory. Clin. Chem. 47:990-1000. [PubMed] [Google Scholar]
- 13.Ganem, D., and A. M. Prince. 2004. Hepatitis B virus infection-natural history and clinical consequences. N. Engl. J. Med. 350:1118-1129. [DOI] [PubMed] [Google Scholar]
- 14.Gärtner, B. C., J. M. Fischinger, A. Litwicki, K. Roemer, and N. Mueller-Lantzsch. 2004. Evaluation of a new automated, standardized generic nucleic acid extraction method (total nucleic acid isolation kit) used in combination with cytomegalovirus DNA quantification by COBAS AMPLICOR CMV MONITOR. J. Clin. Microbiol. 42:3881-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Germer, J. J., M. M. Lins, M. E. Jensen, W. S. Harmsen, D. M. Ilstrup, P. S. Mitchell, F. R. Cockerill III, and R. Patel. 2003. Evaluation of the MagNA Pure LC instrument for extraction of hepatitis C virus RNA for the COBAS AMPLICOR Hepatitis C Virus Test, version 2.0. J. Clin. Microbiol. 41:3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordillo, R. M., J. Gutierrez, and M. Casal. 2005. Evaluation of the COBAS TaqMan 48 real-time PCR system for quantitation of hepatitis B virus DNA. J. Clin. Microbiol. 43:3504-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, P. R., C. M. Sims, F. Krieg-Schneider, E. M. Love, R. Eglin, and R. S. Tedder. 2002. Automated screening of blood donations for hepatitis C virus RNA using the Qiagen BioRobot 9604 and the Roche COBAS HCV Amplicor assay. Vox Sang. 82:169-176. [DOI] [PubMed] [Google Scholar]
- 18.Ivnitski, D., D. J. O'Neil, A. Gattuso, R. Schlicht, M. Calidonna, and R. Fisher. 2003. Nucleic acid approaches for detection and identification of biological warfare and infectious disease agents. BioTechniques 35:862-869. [DOI] [PubMed] [Google Scholar]
- 19.Jungkind, D. 2001. Automation of laboratory testing for infectious diseases using the polymerase chain reaction-our past, our present, our future. J. Clin. Virol. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 20.Kessler, H. H., G. Muhlbauer, E. Stelzl, E. Daghofer, B. I. Santner, and E. Marth. 2001. Fully automated nucleic acid extraction: MagNA Pure LC. Clin. Chem. 47:1124-1126. [PubMed] [Google Scholar]
- 21.Konnick, E. Q., M. Erali, E. R. Ashwood, and D. R. Hillyard. 2005. Evaluation of the COBAS Amplicor HBV Monitor assay and comparison with the Ultrasensitive HBV Hybrid Capture 2 assay for quantification of hepatitis B virus DNA. J. Clin. Microbiol. 43:596-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leb, V., M. Stocher, E. Valentine-Thon, G. Holzl, H. Kessler, H. Stekel, and J. Berg. 2004. Fully automated, internally controlled quantification of hepatitis B virus DNA by real-time PCR by use of the MagNA Pure LC and LightCycler instruments. J. Clin. Microbiol. 42:585-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindh, M., and C. Hannoun. 2005. Dynamic range and reproducibility of hepatitis B virus (HBV) DNA detection and quantification by Cobas Taqman HBV, a real-time semiautomated assay. J. Clin. Microbiol. 43:4251-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lok, A. S., and B. J. McMahon. 2004. Practice Guidelines Committee, American Association for the Study of Liver Diseases (AASLD). Chronic hepatitis B: update of recommendations. Hepatology 39:857-861. [DOI] [PubMed] [Google Scholar]
- 25.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng, Q., C. Wong, A. Rangachari, S. Tamatsukuri, M. Sasaki, E. Fiss, L. Cheng, T. Ramankutty, D. Clarke, H. Yawata, Y. Sakakura, T. Hirose, and C. Impraim. 2001. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 39:2937-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsunaga, S., K. Fujimura, C. Matsumoto, R. Shiozawa, S. Hirakawa, K. Nakajima, K. Tadokoro, and T. Juji. 2002. High-throughput HBV DNA and HCV RNA detection system using a nucleic acid purification robot and real-time detection PCR: its application to analysis of posttransfusion hepatitis. Transfusion 42:100-106. [DOI] [PubMed] [Google Scholar]
- 28.Pas, S. D., E. Fries, R. A. De Man, A. D. Osterhaus, and H. G. Niesters. 2000. Development of a quantitative real-time detection assay for hepatitis B virus DNA and comparison with two commercial assays. J. Clin. Microbiol. 38:2897-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pawlotsky, J. M. 2003. Hepatitis B virus (HBV) DNA assays (methods and practical use) and viral kinetics. J. Hepatol. 39(Suppl. 1):S31-S35. [DOI] [PubMed] [Google Scholar]
- 30.Saldanha, J., W. Gerlich, N. Lelie, P. Dawson, K. Heermann, and A. Heath. 2001. An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sang. 80:63-71. [DOI] [PubMed] [Google Scholar]
- 31.Schorling. S., G. Schalasta, G. Enders, and M. Zauke. 2004. Quantification of parvovirus B19 DNA using COBAS AmpliPrep automated sample preparation and LightCycler real-time PCR. J. Mol. Diagn. 6:37-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shyamala, V., P. Arcangel, J. Cottrell, D. Coit, A. Medina-Selby, C. McCoin, D. Madriaga, D. Chien, and B. Phelps. 2004. Assessment of the target-capture PCR hepatitis B virus (HBV) DNA quantitative assay and comparison with commercial HBV DNA quantitative assays. J. Clin. Microbiol. 42:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stelzl, E., A. Kormann-Klement, J. Haas, E. Daghofer, B. I. Santner, E. Marth, and H. H. Kessler. 2002. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J. Clin. Microbiol. 40:1447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stelzl, E., Z. Muller, E. Marth, and H. H. Kessler. 2004. Rapid quantification of hepatitis B virus DNA by automated sample preparation and real-time PCR. J. Clin. Microbiol. 42:2445-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sum, S. S., D. K. Wong, M. F. Yuen, H. J. Yuan, J. Yu, C. L. Lai, D. Ho, and L. Zhang. 2004. Real-time PCR assay using molecular beacon for quantitation of hepatitis B virus DNA. J. Clin. Microbiol. 42:3438-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernet, G. 2004. Molecular diagnostics in virology. J. Clin. Virol. 31:239-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss, J., H. Wu, B. Farrenkopf, T. Schultz, G. Song, S. Shah, and J. Siegel. 2004. Real time TaqMan PCR detection and quantitation of HBV genotypes A-G with the use of an internal quantitation standard. J. Clin. Virol. 30:86-93. [DOI] [PubMed] [Google Scholar]
- 38.Yao, J. D., M. G. Beld, L. L. Oon, C. H. Sherlock, J. Germer, S. Menting, S. Y. Se Thoe, L. Merrick, R. Ziermann, J. Surtihadi, and H. J. Hnatyszyn. 2004. Multicenter evaluation of the VERSANT hepatitis B virus DNA 3.0 Assay. J. Clin. Microbiol. 42:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]