Abstract
The aim of this study was to develop and evaluate multiplex and nested PCR-reverse line blot (RLB) hybridization assays for detection and serovar identification of Chlamydia trachomatis. Two sets of primers targeting the VD2 region of the omp1 gene and one set targeting the cryptic plasmid were designed for use in multiplex (both targets) and nested PCR (omp1 only). For the RLB assay, labeled omp1 amplicons were hybridized to a membrane containing probes specific for 15 C. trachomatis serovars. The assays were used to test 429 clinical specimens, which had been previously tested for C. trachomatis using the COBAS AMPLICOR system. Specimens were tested without knowledge of the COBAS AMPLICOR result. Of 205 specimens that were positive by COBAS AMPLICOR, 201 (98%) were positive by multiplex PCR-RLB and 188 (92%) were also positive by omp1 nested PCR-RLB. In addition, three of 224 COBAS AMPLICOR-negative specimens were positive by omp1 nested PCR-RLB. One hundred sixty-six of 191 (87%) specimens in which C. trachomatis serovars were identified contained only one serovar and 25 (13%) contained two or three serovars. Serovars D, E, and F were found in 31 (16%), 83 (43%), and 51 (27%) specimens, respectively. Serovar E (41%) was the most commonly identified single serovar. Serovars J and K were found alone uncommonly (<2% each), but 18 of 25 (72%) specimens with multiple C. trachomatis serovars contained one or both (10 specimens) of these serovars. The nested (ompI) PCR-RLB is a specific and sensitive method for simultaneous detection and serovar identification of C. trachomatis, which can reliably identify mixed C. trachomatis serovars. It is suitable for use in epidemiological studies.
Chlamydia trachomatis is one of the most common sexually transmissible pathogens. An estimated 92 million new cases occur worldwide each year (19). This is probably an underestimate, because C. trachomatis infection in men and women is often asymptomatic, and unrecognized infections are a reservoir for sexual transmission.
C. trachomatis infections can be diagnosed by cell culture, immunofluorescence (IF), enzyme immunoassay (EIA), direct DNA hybridization, and PCR. Laboratory diagnosis of chlamydial infection by culture is limited by the fact that collection of urethral swabs is unacceptable to many asymptomatic men. PCR, using various gene targets, including the cryptic plasmid, omp1 (which encodes the major outer membrane protein, MOMP), and rRNA genes, is more sensitive than culture, EIA, or IF (4, 7). Moreover, urine specimens can be used for PCR, which are more convenient to collect and more acceptable to patients.
Serotyping of C. trachomatis is unnecessary to make a clinical diagnosis of chlamydial infection. However, it is useful for epidemiologic research, investigation of person-to-person transmission, and study of differences in clinical manifestations or responses to treatment between serovars. While most C. trachomatis infections are due to single serovars, up to 15% of infections have been reported to involve two or more (1-3, 7,13-16). IF and EIA are commonly used for serotyping and detection of multiple serotypes in C. trachomatis cultures. PCR, plus restriction fragment length polymorphism (RFLP) analysis or DNA sequencing of omp1 amplicons, is needed to identify serovars directly from clinical specimens (2), but neither can reliably detect mixed infections (6, 7, 17, 22). Recently the combination of PCR with reverse dot blot or reverse line blot (RLB) assays has been described (12, 17). In this study, we modified these methods to detect 15 C. trachomatis serovars in a variety of clinical specimens from men and women.
MATERIALS AND METHODS
Reference strains.
The following reference strains were used in this study: C. trachomatis serotype H ATCC UR-898, Chlamydia pneumoniae TWAR strain TW-183, Chlamydia psittaci serotype ATCC VR-628, Ureaplasma urealyticum serovar 1 ATCC 27813, Ureaplasma urealyticum serovar 4 ATCC 27816, Mycoplasma hominis ATCC 23114, Mycoplasma genitalium ATCC 33530 (G37), Gardnerella vaginalis ATCC 14018, and Neisseria gonorrhoeae WHO A.
Clinical specimens.
Four hundred twenty-nine specimens, which had been referred to the Centre for Infectious Diseases and Microbiology for diagnosis of C. trachomatis infection between January 2004 and February 2005, were selected (by staff from the diagnostic laboratory) so as to ensure a fairly even distribution of C. trachomatis-positive and -negative specimens. They consisted of first voided urine (309 specimens), rectal swabs (2 specimens), and cervical swabs (118 specimens).
COBAS AMPLICOR testing.
Specimens were kept at −20°C until testing and then thawed, resuspended in 500 μl wash buffer, vortexed vigorously, and incubated at 37°C for 30 min. After centrifugation at 13,000 × g for 15 min, the supernatant was discarded, 250 μl C. trachomatis lysis buffer was added, and after another 15-min incubation at 37°C, 250 μl C. trachomatis specimen diluent was added to the lysate. The contents of the tubes were mixed by vortexing, centrifuged at 13,000 × g for 10 min, and incubated at 95°C for 10 min. All specimens were tested for C. trachomatis using the COBAS AMPLICOR (Amplicor; Roche Diagnostics Australia Pty. Limited Systems, Castle Hill, Australia), as described previously (9-11) and according to the manufacturer's instructions. DNA extracts were frozen at −20°C until required for further testing in this study, in which they were tested without knowledge of the COBAS AMPLICOR result.
Probe and primer design.
Two sets of primers targeting the VD2 region of C. trachomatis and one set targeting the cryptic plasmid were designed or modified from previous publications. For each of 15 serovars, serovar-specific oligonucleotide probes, based on the published VD2 region of omp1 sequences, and two probes based on published cryptic plasmid sequences of C. trachomatis, were designed or modified (Table 1). All probes and primers were checked for specificity against all sequences in GenBank by using QueryBD, WebANGIS GCG, SeqSearch, and the Browse code in the Australian National Genomic Information Services (ANGIS) programs (http://www1.angis.org.au/WebANGIS/WebFM). Probes were designed to have similar melting temperatures (Tm) of more than 59°C, and their lengths varied from 19 to 27 bp. Oligonucleotide probes were labeled with an amine group at the 5′ end, and primers were labeled with biotin at the 5′ end (Sigma-Aldrich, St. Louis, Mo.), respectively.
TABLE 1.
Primers and probes used for C. trachomatis PCR-RLB and DNA sequencing
| Primer/probea | Tm (°C)b | Target | GenBank accession no. | Sequencec | Source or reference |
|---|---|---|---|---|---|
| CP24b | 67.33 | Cryptic plasmid | X06707 | 840GGGATTCCTGTAACAACAAGTCAGG864 | 22 |
| CTS1p | 63.59 | omp1 | X06707 | 865TTGCGCATAATTTTAGGCTTG885 | This study |
| CTA2p | 62.57 | omp1 | X06707 | 1021ACACTTTGTCTCGATGAAAGACA999 | This study |
| CP27b | 67.38 | Cryptic plasmid | X06707 | 1047CCTCTTCCCCAGAACAATAAGAACAC1022 | 22 |
| CTSb | 61.96 | omp1 | J03813 | 556AATATYTGGGATCGYTTTGATGT578 | This study |
| CTSNb | 62.55 | omp1 | J03813 | 572TTGATGTATTYTGTACAYTRGGAGC596 | 13 (modified) |
| CTSp | 61.86 | omp1 | J03813 | 613AAAGGAAAYTCHGCWTCYTTCAA635 | 13 (modified) |
| CTANb | 62.92 | omp1 | J03813 | 774GCTGCDCGAGCDCCNACRCT757 | 13 (modified) |
| CTAb | 60.99 | omp1 | J03813 | 791CCRCAYTCCCASARAGCTGC772 | 13 (modified) |
| CMP1 | 64.98 | omp1 | J03813 | 333bTGACGCTATCAGCATGCG349 | 10 |
| CMP6AS | 61.85 | omp1 | J03813 | 808TGAGCRTATTGGAAWGAAGC827 | 10 (modified) |
| Ap | 69.4 | Serovar A omp1 | J03813 | 664CAATCTTCTGGCTTTGATACAGCGAAT690 | 17 |
| B/Bap | 61.57 | Serovar B/Ba omp1 | AF063208 | 394TCAAATGGTACGTTTGTACCAA415 | 12 (modified) |
| Cp | 61.44 | Serovar C omp1 | M17343 | 978TCTAGCTTTAATACAGCGAAGCTTAT1003 | This study |
| D/Dap | 67.08 | Serovar D/Da omp1 | AF063195 | 610AAAAAACGGTCAAAGCGGAGTC631 | This study |
| Ep | 59.89 | Serovar E omp1 | AF063198 | 448ACAGATACTGCCTTCTCTTGG468 | This study |
| Fp | 61.01 | Serovar F omp1 | AF063212 | 388ACGAAACCTGCTGCAGATA406 | 12, 17 |
| G/Gap | 77.99 | Serovar G/Ga omp1 | AF063199 | 496GCCACGCAGCCTGCTGCAACA516 | 12 |
| Hp | 59.54 | Serovar H omp1 | X16007 | 661ACAAAATCTTCTGATTTTAATACAGC686 | 12 (modified) |
| Ip | 60.87 | Serovar I omp1 | AF063200 | 494CACAATCTTCTAACTTTAATACAGCG519 | 12 |
| Jp | 60.51 | Serovar J omp1 | AF063202 | 519GAATCTTTTTCCTAACACTGCTTT542 | 12 (modified) |
| Jap | 60.51 | Serovar Ja omp1 | AF063203 | 519GAAGCTTATTCCTAACACTGCTTT542 | This study |
| Kp | 70.34 | Serovar K omp1 | AF063207 | 421AACACTGCTTTGGATCGAGCTGTG444 | 17 |
| L1p | 63.35 | Serovar L1 omp1 | M36533 | 700GGTCAAAAAGGATGCTGTCC720 | This study |
| L2p | 59.32 | Serovar L2 omp1 | M14738 | 975GTTTCAGATAGTAAGCTTGTACCAA999 | This study |
| L3p/Jap | 60.1 | Serovar L3 omp1 | X55700 | 541TTGAATCAAGCTGTAGTTGAGC562 | This study |
p indicates biotin-labeled probe, and b indicates biotin-labeled primer.
Melting temperatures provided by manufacturer.
Numbers relate to positions in GenBank sequences. Underlined sequences indicate modifications compared with published sequences.
Multiplex PCR amplification.
Amplification was performed using primers CP24b/CP27b for the cryptic plasmid and CTSb/CTAb and CTSNb/CTANb for omp1. The amplifications were performed using a 25-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 200 μM (each) deoxynucleoside triphosphate (dNTP), 25 pmol of each primer, 1 U of HotStart DNA polymerase (QIAGEN, Pty. Ltd., Doncaster, Australia), and 10 μl of DNA. The thermal profile involved initial denaturation for 15 min at 96°C and 35 cycles with the following steps: 0.5 min of denaturation at 94°C, 0.5 min of annealing at 60°C, and 1 min of extension at 72°C, with a final extension for 10 min at 72°C.
Nested PCR amplification.
Amplification of VD2 omp1 was performed with outer primers CTSb and CTAb in a 25-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 200 μM (each) dNTP, 25 pmol of each primer, 1 U of DNA polymerase (Promega, Pty. Ltd., Annandale, Australia), and 10 μl of DNA. The thermal profile involved initial denaturation for 15 min at 96°C and 35 cycles with the following steps: 0.5 min of denaturation at 94°C, 0.5 min of annealing at 60°C, and 1 min of extension at 72°C, with a final extension for 10 min at 72°C. Ten microliters of the primary PCR product was used for a secondary PCR, which was prepared and run using the same reagents and conditions as the primary PCR, except that the (inner) primers used were CTSNb and CTANb. Strict procedures to avoid specimen contamination and carryover were followed when performing nested PCRs. All DNA extractions were done in a room dedicated to PCR use.
PCR products were electrophoresed on a 1.5% agarose gel and analyzed using SRBR Safe DNA gel stain (Molecular Probes Europe BV, Leiden, The Netherlands).
RLB assay.
The RLB assay was performed using a system that has been described previously (21). Briefly, slots (Miniblotter 45; Immunetics) were filled with 150 μl of two to three different concentrations of each probe solution (0.3125, 0.625, and 1.25 pmol). Each PCR product was denatured and immediately chilled on ice. Hybridization was performed at 60°C for 60 min. The washed membrane was incubated in peroxidase-labeled streptavidin conjugate (Roche, Mannheim, Germany) at 42°C for 45 min. Then the washed membrane was incubated in chemiluminescence blotting substrate (ECL Direct System; Roche) for 1 min, and the membrane was covered with Hyperfilm X-ray film (Amersham) for detection of chemiluminescence. The film was exposed for 7 min.
Sequencing and sequence searching.
C. trachomatis omp1 amplicons, generated by nested PCR, were sequenced as described previously (21), using primers CMP1 and CMP6AS (Table 1) and Applied Biosystems (Foster City, Calif.) BigDye terminator chemistry on an ABI Prism 373 DNA sequencer. Sequences were identified using the FastA program group accessed through WebANGIS.
RESULTS
Sensitivities of multiplex and nested PCR-RLB.
The results of multiplex and nested PCR—in which amplicons were identified by RLB—are shown in Table 2. Three specimens that had been negative in the COBAS AMPLICOR assay were positive in the nested PCR-RLB and identified by hybridization with the corresponding probes as serovar E. Assuming that all 208 positive PCR results of all three assays are true positives, the relative sensitivities of COBAS AMPLICOR, multiplex PCR-RLB, and nested PCR-RLB for detection of C. trachomatis were 98.6%, 96.6%, and 91.8%, respectively.
TABLE 2.
Multiplex and nested PCR results in 429 clinical specimens previously tested for Chlamydia trachomatis by COBAS Amplicor assay
| No. with COBAS Amplicor cp result | Result bya:
|
|||
|---|---|---|---|---|
| Multiplex PCR-RLB (cp and omp1)
|
Nested PCR-RLB (omp1)
|
|||
| Positiveb | Negative | Positive | Negative | |
| Positive, 205 | 201 | 4 | 188 | 17 |
| Negative, 224 | 0 | 224 | 3 | 237 |
PCR targets are shown in parentheses: cp, cryptic plasmid; omp1, outer membrane protein gene.
Both the cryptic plasmid and omp1 were amplified from 149 specimens; the cryptic plasmid only was amplified from 52 specimens.
In the multiplex PCR-RLB, the cryptic plasmid was more likely to be amplified than omp1 (Table 2; Fig. 1). The serovar was identified by multiplex PCR-RLB in only 149 of 201 (74%) C. trachomatis-positive specimens. However, the omp1 nested PCR-RLB, using probes for each serovar, amplified 188 of 201 (94%) specimens positive by multiplex PCR as well as 3 of 224 that were negative by COBAS AMPLICOR and multiplex PCR (Table 2; Fig. 2). It was more sensitive than multiplex PCR-RLB for serovar identification and more convenient than sequencing.
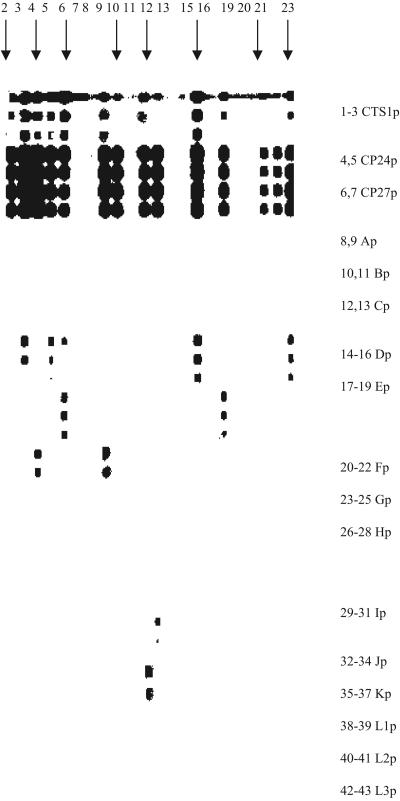
FIG. 1.
Hybridization, with 18 probes, of multiplex PCR products from a representative sample of 23 C. trachomatis-positive specimens. From top to bottom, probes are as shown in numbered rows. The probe concentrations in each set of three rows (top to bottom) are 0.3125, 0.625, and 1.25 pmol (or 0.625 and 1.25 pmol when there are only two rows per probe). Lanes 1, 7, 8, 11, 15, 17, 19, and 20 are negative; lanes 2 to 6, 9 to 10, 12 to 13, 16, 18, and 21 to 23 are cryptic plasmid positive; lanes 21 and 22 are omp1 (CTS1p) negative and so cannot be subtyped; lanes 2 and 10 are omp1 positive but cannot be subtyped; results for the remaining lanes are as follows: 3, D; 4, F; 5, D; 6, D and E; 9, F; 12, J; 13, I;16, D; 18, E; and 23, D.
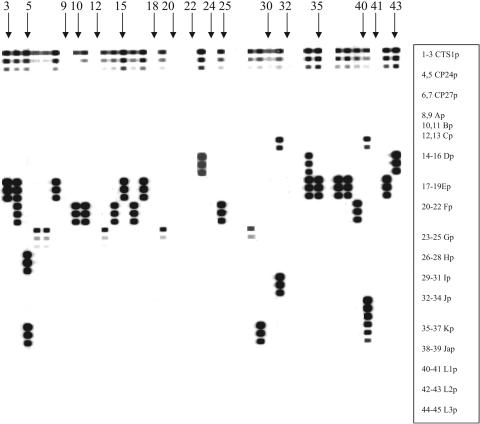
FIG. 2.
Hybridization, with 18 probes, of nested PCR products from a representative sample of 23 C.trachomatis-positive specimens. From top to bottom are probes CTS1p (concentrations 1, 2, and 3),CP24p (concentrations 1 and 2), CP27p (concentrations 1 and 2), Ap (concentrations 1 and 2), Bp (concentrations 1 and 2),Cp (concentrations 1 and 2), Dp (concentrations 1, 2, and 3), Ep (concentrations 1, 2, and 3), Fp (concentrations 1, 2, and 3), Gp (concentrations 1, 2, and 3), Hp (concentrations 1, 2, and 3), Ip (concentrations 1, 2, and 3), Jp (concentrations 1, 2, and 3), Kp (concentrations 1, 2, and 3), Ja (concentrations 1 and 2), L1 (concentrations 1 and 2), L2 (concentrations 1 and 2), and L3 (concentrations 1 and 2). Concentrations 1, 2, and 3 represent probe concentrations 0.325, 0.625, and 1.25pmol, respectively. Results for lanes are as follows: 3, E; 4, E and F; 5, H and K; 6, G; 7, G; 8, E; 10, F; 11, F; 13, G; 14, F; 15, E;16, F; 17, E; 19, G; 23, D; 25, F; 28, G; 29, K; 31, C and I; 34, D and E; 35, E; 37, E; 38, E; 39, F; 40, C, J, and K; 42, E; and 43, D.
Specificity of C. trachomatis typing assay.
There were no cross-reactions between C. trachomatis probes and DNA extracts from the panel of other potential urogenital pathogens (see Materials and Methods). The specificities of both the mutiplex and nested PCR-RLB assays were further evaluated against individual C. trachomatis serovars (C, D, F, E, G, H, I, J, and K)—confirmed by sequencing (see below)—all of which hybridized with the corresponding probe with no cross-reaction with other probes.
DNA sequencing was performed on omp1 amplicons from 23 selected specimens, of which 10 contained single serovars and 11 contained multiple serovars (Table 3). DNA sequencing confirmed the results of PCR-RLB for all specimens in which a single serovar was identified but was uninterpretable for specimens with multiple serovars. Eleven of 12 amplicons from single serovars had omp1 sequences identical to the corresponding sequences in GenBank; the other was a variant serovar C sequence, with 5 nucleotide differences compared with the GenBank sequence.
TABLE 3.
Results of C. trachomatis PCR-RLB assay and DNA sequencing in 23 selected omp1-positive specimens
| RLB results (n) | No. of specimens | DNA sequencing result(s) |
|---|---|---|
| Single infections | 12 | |
| 1 each of C, D, E (3), F, G, H, I, J (2), K | Results corresponded with those of RLB; 1 serovar C specimen was sequence variant | |
| Multiple infections | 11 | |
| 2 serovars: C/I, D/E, J/K (4), H/K | 7 | Uninterpretable |
| 3 serovars: A+C+J, F/J/K, C/I/K, D/E/H | 4 | Uninterpretable |
Typing of C. trachomatis.
Of 208 C. trachomatis-positive specimens, 191 (92%) were successfully typed by omp1 nested PCR-RLB (Table 4). Single C. trachomatis serovars were detected in 166 (87%) and two or three serovars in 25 (13%) positive specimens. Overall, serovars D, E, and F were found in 15%, 41%, and 26% of specimens (including mixed), respectively. Serovar E was the serovar most commonly identified alone (39%). Serovar H, J, or K was identified in only 6 of 166 (3.6%) specimens with single serovars (or 3% of all positive specimens), but these serovars were found together or with other serovars in 19 of 25 (76%) specimens with mixed serovars (or 10% of all positive specimens). This discrepancy was most marked for serovar K, which was found alone in only 1 of 166 (0.6%) specimens with single serovars compared with 15 of 25 (60%) of those with mixed serovars (P < 0.001) (or 1 versus 15 of 191 positive specimens; P < 0.001). The equivalent figures for serovar J were 3 of 166 (1.8%) versus 11 of 25 (44%) (P < 0.001) (or 3 versus 11 of 191; P = 0.03).
TABLE 4.
Serovar distribution in 191 C. trachomatis omp1-positive urogenital specimens
| Serovar | No. of specimens containing serovar(s)
|
%a | ||
|---|---|---|---|---|
| Men (n = 94) | Women (n = 97) | Total (n = 191) | ||
| Single | 82 | 84 | 166 | 87 |
| C | 1 | 1 | 2 | 1 |
| D | 15 | 13 | 28 | 15 |
| E | 34 | 40 | 74 | 39 |
| F | 25 | 21 | 46 | 24 |
| G | 4 | 4 | 8 | 4 |
| H | 0 | 1 | 1 | 0.5 |
| I | 0 | 2 | 2 | 1 |
| J | 2 | 1 | 3 | 2 |
| K | 1 | 1 | 2 | 1 |
| Multiple | 12 | 13 | 25 | 13 |
| C+I | 0 | 1 | 1 | 0.5 |
| D+E | 1 | 1 | 2 | 1 |
| E+F | 1 | 1 | 2 | 1 |
| H+K | 3 | 2 | 5 | 3 |
| J+K | 5 | 4 | 9 | 5 |
| A+C+J | 1 | 1 | 2 | 1 |
| C+J+K | 1 | 0 | 1 | 0.5 |
| D+E+H | 0 | 1 | 1 | 0.5 |
| F+G+K | 0 | 1 | 1 | 0.5 |
Values are percentages of total numbers of C. trachomatis-positive specimens (n = 191).
DISCUSSION
We have developed a PCR-RLB hybridization assay for simultaneous detection and genotyping of C. trachomatis which is faster and more convenient than other serotyping methods such as PCR sequencing, PCR-RFLP, and culture-based methods using antisera. First we used multiplex PCR to amplify omp1 and the cryptic plasmid and then hybridized biotin-labeled amplicons to serotype-specific omp1 and plasmid probes, respectively. When multiplex PCR was negative or C. trachomatis could not be typed, we used nested PCR to amplify omp1 and hybridized biotin-labeled amplicons to the serotype-specific omp1 probes.
Nucleic acid amplification is now considered to be the most sensitive and specific method for screening and diagnosis of C. trachomatis infections. The sensitivity of PCR varies, depending on the target and, in particular, whether there are one or more copies. Mahony et al. compared the sensitivities of five PCR assays—including two targeting the cryptic plasmid (of which there are 10 copies, on average), two targeting omp1 (single copy), and one targeting rRNA genes (multiple copies)—by testing serial dilutions of C. trachomatis DNA and genitourinary tract specimens (9-11). The plasmid PCRs can detect as little as 0.1 fg of C. trachomatis plasmid DNA and 10 fg of total cellular DNA (which is equivalent to ∼0.01 inclusion-forming unit [IFU], whereas the two omp1 PCRs and one rRNA PCR could only detect the equivalent of 1, 100, and 10 IFU or 0.1 pg, 10 pg, and 1 pg, respectively, of cellular DNA.
Our multiplex PCR, in which cryptic plasmid and omp1 DNA were amplified from 98% and 78%, respectively, of 205 COBAS AMPLICOR-positive samples, is relatively insensitive for serovar identification, using RLB to detect serovar-specific omp1 sequences. The nested format increased the sensitivity of the omp1 PCR to a level close to that of plasmid PCR; it amplified DNA from 92% of COBAS AMPLICOR-positive samples, as well as 3 of 224 negative samples. This latter finding is consistent with previous reports of C. trachomatis strains which do not have cryptic plasmids (9-12, 18). Molano et al. (12) reported that nested omp1 PCR-RLB identified C. trachomatis in 94% of specimens in which it had been detected by a cryptic plasmid PCR.
Serovars B, L1, L2, and L3 were not identified in our study—as would be expected in the population from which the specimens were derived (mainly women and heterosexual men). However, theoretically, our PCR-RLB typing method should be able to detect and identify these serovars and, if so, could be used for the diagnosis of lymphogranuloma venereum as well as genital chlamydial and other infections due to the trachoma serovars. However, further evaluation is required to confirm this.
The most commonly identified serovars in our study, overall, were D (15%), E (41%), and F (26%) (Table 3). These results are consistent with those in studies of urogenital chlamydial infection elsewhere, although there are some differences in relative proportions in different geographic areas. For example, in Sweden (7) serovar E (47%) and in Thailand (2) serovars F (25%) and D (23%) were the most commonly identified; in Colombia (12) serovars D, F, G, and E together accounted for 74% of isolates, compared with 85% in our study.
The distribution of serovars in our study was similar between specimens from women and (predominantly) heterosexual men. A recent study in Melbourne, Australia (8), showed significant differences in serovar distribution between women and men who have sex with men, especially in the relative distribution of the two most common serovars, E (17% and 3%, respectively) and D (7% and 54%, respectively). Serovar G was present in higher proportions of specimens (17% and 26% in women and men who have sex with men, respectively) than in our study (4%). The number of specimens tested in the Melbourne study was relatively small, and mixed serovars were not detected (serovars were identified by sequencing). Although difficult to interpret, these differences suggest that further investigation of the relative distribution of serovars in different patient groups would be useful.
In our study, 13% of C. trachomatis-positive specimens contained two or three serovars. Using identical probes, Molano et al. reported that 9% of specimens from women in Colombia contained more than one C. trachomatis serovar (12). Many other studies from different parts of the world (12-15, 20-22) have shown small proportions of specimens with mixed serovars (2 to 15%) (1, 5, 6, 13, 14). Using a reverse dot blot assay, Stothard (17) was able to identify both C. trachomatis serovars in eight artificially mixed specimens and seven of eight clinically mixed specimens (each containing two serovars), as identified by immunofluorescence. However, as in our study, individual serovars were poorly recognized by DNA sequencing, which may depend on the relative proportions of each serovar present. In one study, when DNA sequencing identified mixed infection, reanalysis of the RFLP pattern showed minor bands that had been overlooked but that could be interpreted as representing multiple serovars (17, 18).
The infrequent occurrence of serovars J and K in specimens with single serovars is consistent with other studies. However, as far as we know, the relatively high frequency of these serovars together or with others has not been previously reported. The explanation for this phenomenon is not clear. Perhaps serovars K and J are relatively low-grade pathogens, which can only persist in the presence of, or act synergistically with, other serovars. Further investigation is required to confirm these findings, which were only possible because of the ability of the PCR-RLB method to identify mixed serovars.
Our RLB assay facilitates the detection of multiple serovars that are difficult to identify by the more commonly used PCR-RFLP and sequencing methods. This is important to advance our knowledge of the natural history of C. trachomatis infections and to determine whether there are significant differences in infectivity, immune response, treatment response, or reinfection rates between C. trachomatis serovars (2). Our omp1 nested PCR-RLB method will be useful in large epidemiological studies. Compared with sequencing or RFLP, it can analyze large numbers of clinical samples relatively rapidly and inexpensively without specialized instruments.
Acknowledgments
We thank Susan Alderson, Terry Flood, and Beverley Horne for COBAS AMPLICOR testing for C. trachomatis and selection of specimens. We thank Ilya Henner for help in DNA sequencing.
REFERENCES
- 1.Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunham, R., C. Yang, I. Maclean, J. Kimani, G. Maitha, and F. Plummer. 1994. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Investig. 94:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunham, R. C., M. Laga, J. N. Simonsen, D. W. Cameron, R. Peeling, J. McDowell, H. Pamba, J. O. Ndinya-Achola, G. Maitha, and F. A. Plummer. 1990. The prevalence of Chlamydia trachomatis infection among mothers of children with trachoma. Am. J. Epidemiol. 132:946-952. [DOI] [PubMed] [Google Scholar]
- 4.Claas, H. C., W. J. Melchers, I. H. de Bruijn, M. de Graaf, W. C. van Dijk, J. Lindeman, and W. G. Quint. 1990. Detection of Chlamydia trachomatis in clinical specimens by the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 9:864-868. [DOI] [PubMed] [Google Scholar]
- 5.Dean, D., and K. Millman. 1997. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J. Clin. Investig. 99:475-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean, D., E. Oudens, G. Bolan, N. Padian, and J. Schachter. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J. Infect. Dis. 172:1013-1022. [DOI] [PubMed] [Google Scholar]
- 7.Jurstrand, M., L. Falk, H. Fredlund, M. Lindberg, P. Olcén, S. Andersson, K. Persson, J. Albert, and A. Bäckman. 2001. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 39:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister, N. A., S. N. Tabrizi, C. K. Fairley, A. Smith, P. H. Janssen, and S. Garland. 2004. Variability of the Chlamydia trachomatis omp1 gene detected in samples from men tested in male-only saunas in Melbourne, Australia. J. Clin. Microbiol. 42:2596-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahony, J. B., K. E. Luinstra, M. Tyndall, J. W. Sellors, J. Krepel, and M. Chernesky. 1995. Multiplex PCR for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in genitourinary specimens. J. Clin. Microbiol. 33:3049-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahony, J. B., K. E. Luinstra, J. Waner, G. McNab, H. Hobranzska, D. Gregson, J. W. Sellors, and M. A. Chernesky. 1994. Interlaboratory agreement study of a double set of PCR plasmid primers for detection of Chlamydia trachomatis in a variety of genitourinary specimens. J. Clin. Microbiol. 32:87-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahony, J. B., X. Song, S. Chong, M. Faught, T. Salonga, and J. Kapala. 2001. Evaluation of the NucliSens Basic Kit for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in genital tract specimens using nucleic acid sequence-based amplification of 16S rRNA. J. Clin. Microbiol. 39:1429-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molano, M., C. J. L. M. Meijer, S. A. Morré, R. Pol, and A. J. C. van den Brule. 2004. Combination of PCR targeting the VD2 of omp1 and reverse line blot analysis for typing of urogenital Chlamydia trachomatis serovars in cervical scrape specimens. J. Clin. Microbiol. 42:2935-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morré, S. A., R. Moes, I. Van Valkengoed, J. P. Boeke, J. T. M. van Eijk, C. J. L. M. Meijer, and A. J. C. van den Brule. 1998. Genotyping of Chlamydia trachomatis in urine specimens will facilitate large epidemiological studies. J. Clin. Microbiol. 36:3077-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morré, S. A., J. M. Ossewaarde, J. Lan, G. J. J. van Doornum, J. M. M. Walboomers, D. M. MacLaren, C. J. L. M. Meijer, and A. J. C. van den Brule. 1998. Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis. J. Clin. Microbiol. 36:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morré, S. A., J. M. Ossewaarde, P. H. M. Savelkoul, J. Stoof, C. J. L. M. Meijer, and A. J. C. van den Brule. 2000. Analysis of genetic heterogeneity in Chlamydia trachomatis clinical isolates of serovars D, E, and F by amplified fragment length polymorphism. J. Clin. Microbiol. 38:3463-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morré, S. A., L. Rozendaal, I. G. M. van Valkengoed, A. J. P. Boeke, P. C. Voorst Vader, J. Schirm, S. de Blok, J. A. R. van Den Hoek, G. J. J. van Doornum, C. J. L. M. Meijer, and A. J. C. van den Brule. 2000. Urogenital Chlamydia trachomatis serovars in men and women with a symptomatic or asymptomatic infection: an association with clinical manifestations? J. Clin. Microbiol. 38:2292-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stothard, D. R. 2001. Use of a reverse dot blot procedure to identify the presence of multiple serovars in Chlamydia trachomatis urogenital infection. J. Clin. Microbiol. 39:2655-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stothard, D. R., J. A. Williams, B. Van Der Pol, and R. B. Jones. 1998. Identification of a Chlamydia trachomatis serovar E urogenital isolate which lacks the cryptic plasmid. Infect. Immun. 66:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor-Robinson, D. 2000. Genitourinary chlamydial infections. Int. J. STD AIDS 11:272. [DOI] [PubMed] [Google Scholar]
- 20.van de Laar, M. J., Y. T. van Duynhoven, J. S. Fennema, J. M. Ossewaarde, A. J. van den Brule, G. J. van Doornum, R. A. Coutinho, and J. A. van Den Hoek. 1996. Differences in clinical manifestations of genital chlamydial infections related to serovars. Genitourin. Med. 72:261-265. [PMC free article] [PubMed] [Google Scholar]
- 21.van den Brule, A. J. C., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. L. M. Meijer, and P. J. F. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, C. L., I. Maclean, and R. C. Brunham. 1993. DNA sequence polymorphism of the Chlamydia trachomatis omp1 gene. J. Infect. Dis. 168:1225-1230. [DOI] [PubMed] [Google Scholar]