Abstract
Resistance of soybean against the oomycete pathogen Phytophthora sojae is conferred by a series of Rps genes. We have characterized a disease resistance gene-like sequence NBSRps4/6 that was introgressed into soybean lines along with Rps4 or Rps6. High-resolution genetic mapping established that NBSRps4/6 cosegregates with Rps4. Two mutants, M1 and M2, showing rearrangements in the NBSRps4/6 region were identified from analyses of 82 F1's and 201 selfed HARO4272 plants containing Rps4. Fingerprints of these mutants are identical to those of HARO4272 for 176 SSR markers representing the whole genome except the NBSRps4/6 region. Both mutants showed a gain of race specificities, distinct from the one encoded by Rps4. To investigate the possible mechanism of gain of Phytophthora resistance in M1, the novel race specificity was mapped. Surprisingly, the gene encoding this resistance mapped to the Rps3 region, indicating that this gene could be either allelic or linked to Rps3. Recombinant analyses have shown that deletion of NBSRps4/6 in M1 is associated with the loss of Rps4 function. The NBSRps4/6 sequence is highly transcribed in etiolated hypocotyls expressing the Phytophthora resistance. It is most likely that a copy of the NBSRps4/6 sequence is the Rps4 gene. Possible mechanisms of the deletion in the NBSRps4/6 region and introgression of two unlinked Rps genes into Harosoy are discussed.
OVER 40 disease-resistance genes (R) have been cloned in plants (Hulbert et al. 2001; Martin et al. 2003). It has been observed that the proteins encoded by most of the resistance genes are structurally very similar although they provide resistance against a wide range of phytopathogens and pests, such as viruses, bacteria, fungi, nematodes, and aphids. Broadly, cloned disease-resistance genes can be classified into eight classes (Hulbert et al. 2001). The majority of the resistance genes encode nucleotide-binding sites (NBS) and leucine-rich repeat regions (LRR). This class includes genes from dicotyledons as well as monocotyledons. Some members of this class contain an N-terminal coiled coil domain and others contain a toll-interleukin receptor-like domain (Dangl and Jones 2001; Mondragon-Palomino et al. 2002). Overall sequence homology among members of the class is low but small domains are well conserved (Dangl and Jones 2001). R-gene diversity is largely controlled by LRR domain, which goes through adaptive selection to generate new race-specificities (Dangl and Jones 2001). Resistance genes are evolved at a fast rate to protect plants against rapidly evolving new pathogenic races or isolates.
Soybean [Glycine max L. (Merrill)] is an important oilseed crop and the United States is a major soybean producer. An average 71 million tons of soybean valued at up to ∼16 billion dollars is produced in the United States annually (Wrather et al. 2001). Soybean yields per hectare are significantly reduced due to diseases. Soybean suffers annual yield losses worth nearly 2.7 billion dollars from all diseases together (Wrather et al. 2001). Phytophthora stem and root rot disease caused by Phytophthora sojae alone results in annual yield losses worth up to ∼300 million dollars (Wrather et al. 2001). The disease can occur at any growth stages. Application of chemicals is expensive and sometimes ineffective in controlling the pathogen. Monogenic resistance encoded by Rps (resistance to Phytophthora sojae) genes has provided reasonable protection for soybean against this pathogen for the last 4 decades. Over 50 physiological races of this oomycete pathogen have been reported, and the number of races is increasing rapidly (Leitz et al. 2000). Mutation and rare outcrossing between races and isolates are the main mechanisms of evolution of novel races in this pathogen (Förster et al. 1994). Due to the highly variable nature of P. sojae it is difficult to control this pathogen by the use of most Rps genes (Schmitthenner et al. 1994). Identification of new useful Rps genes is a constant need for protecting soybean against newly evolved races of the pathogen. To date, 14 Rps genes have been mapped to eight genomic loci: Rps1, Rps2, Rps3, Rps4, Rps5, Rps6, Rps7, and Rps8. Rps1 carries five Rps genes, viz. Rps1-a, -b, -c, -d, and -k, and was mapped to the molecular linkage group (MLG) N. Rps2, Rps3, Rps4, Rps5, Rps6, Rps7, and Rps8 were mapped to MLG J, F, G, G, G, N, and A2, respectively. Three functional Rps genes, viz. Rps3-a, -b, and -c, were mapped to the Rps3 locus (Diers et al. 1992; Lohnes and Schmitthenner 1997; Demirbas et al. 2001; Weng et al. 2001; Burnham et al. 2003). Map location of Rps6 has not been consistent in previous studies (Athow and Laviolette 1982; Demirbas et al. 2001). Demirbas et al. (2001) putatively mapped Rps4 and Rps6 to a similar region in the linkage group G. Furthermore, both genes encode resistance against almost the same set of P. sojae races (Buzzell et al. 1987).
We are interested in investigating the molecular differences that make the two genes distinct in their race specificities. Here we have shown that Rps4 and Rps6 are either allelic or tightly linked genes. We have isolated an NBS-LRR-type disease-resistance sequence from the Rps4/6 region that was introgressed into soybean isolines. Deletion of a few copies of this sequence is associated with the loss of Rps4 function. In this investigation we have also shown that additional Rps genes were introgressed into isolines during incorporation of Rps4 through backcrossing. Possible mechanisms of the deletion in the NBSRps4/6 region and introgression of two unlinked Rps genes into the cultivar Harosoy are discussed.
MATERIALS AND METHODS
Plant materials:
The near-isogenic lines carrying different Rps genes [Harosoy lines: HARO14 (Rps1-c), HARO1472 (Rps1-c, 7), HARO13 (Rps1-b), HARO15 (Rps1-k), HARO(1-7) (rps1-7), HARO1372 (Rps1-b, 7), HARO16 (Rps1-d), HARO1572 (Rps1-k, 7), L-70-6494 (Rps2, 7), HARO3272 (Rps3, 7), HARO5272 (Rps5, 7), HARO4272 (Rps4, 7), HARO6272 (Rps6, 7), HARO72 (rps1, Rps7); Williams lines: Williams (rps), L-75-6141 (Rps1-a), L-77-1863 (Rps1-b), L-75-3735 (Rps1-k), Williams 82 (Rps1-k), L-76-1988 (Rps2), L-83-570 (Rps3), L-85-2352 (Rps4), L-85-3059 (Rps5), L-81-4352 (Rps1-c, 2), Williams 79 (Rps1-c), L-88-1479 (Rps3-b), L-89-1581 (Rps6)] were used to study the organization of the NBS sequence isolated in a previous study (M. K. Bhattacharyya, unpublished results). For mapping the Rps4 gene, 17 F1's were generated from crosses made between Williams (rps4) and HARO4272 (Rps4) at the Agronomy Research Center, Ames, Iowa, during 2000. Additional 65 F1's were generated from crosses between 20 Williams and 20 HARO4272 plants at the Iowa State University soybean research site at the Isabela substation of the University of Puerto Rico during the winter of 2000–2001. The resulting F1 plants were selfed to produce F2 populations. Crosses were also made between HARO4272 (Rps4) and L89-1581 (Rps6) and an F2 population was developed to determine the segregation ratio of Rps4 and Rps6 and also to map the Rps6 gene.
Disease evaluation:
The pathogen races were grown for 6 days in dark at 22° and zoospores were produced by following published methods (Ward et al. 1979). F2 plants were screened by inoculating detached leaves (Bhattacharyya and Ward 1986). Two unifoliate leaves of 14-day-old F2 plants were detached and placed in moist petri plates. Fifteen microliters of P. sojae race 1 zoospore droplets (105 spores/ml) were placed on each half of the leaf. Disease reactions were scored 3 and 5 days following inoculation. For wounded green hypocotyl inoculation, mycelia from a 6-day-old culture were used. Hypocotyls of 7-day-old light-grown seedlings were slit 1.0 cm open and fungal mycelia grown in 1/4 V8 medium were introduced into the slits (Schmitthenner et al. 1994). Resistant and susceptible seedlings were scored at a 24-hr interval up to 1 week following inoculation.
Pathogen used:
P. sojae races 1 and 4 and isolates I-CC5A, I-CC5C, I-CC8, I-CC9A, I-CC10D, I-CE5, I-CE10, I-CE20, I-CW1, I-LEE1, I-MARIONCO1-2, I-MARSHALL1-1, I-MARSHALL2-1, I-NW1, I-NW5A, I-NW8A, I-NW8B, I-NW9B, I-NW10, I-POLK1-1, I-POLK3-3, I-SC1A, I-SC4A, I-SC10, I-SE10, I-SE10A, I-SE11A, I-SW1, I-SW11A, and I-SW11B were used. Isolates were identified in a previous investigation (S. Cianzio and X. B. Yang, unpublished results).
DNA analysis:
Genomic DNA was prepared according to Anderson et al. (1992) for DNA gel blot and PCR analyses. Ten micrograms of genomic DNA was digested with appropriate restriction enzymes and electrophoretically separated on 0.8% agarose gels (Kasuga et al. 1997). DNA was blotted from the gel onto nylon membrane (Zeta Probe, Bio-Rad Laboratories, Hercules, CA) using capillary action of 0.4 m NaOH overnight at room temperature.
Probe preparation, hybridization, and autoradiography:
About 80 ng DNA was labeled with 50 μCi of [α32P]dATP (Feinberg and Vogelstein 1983). Hybridization was performed in 10 ml of hybridization buffer (50% formamide, 1% SDS, 1 m NaCl, 5× Denhardt's, 100 μg/ml herring sperm DNA), incubated at 42° for 16–18 hr in a hybridization rotisserie oven (Hybaid). Blots were washed with 2× SSC for 5 min at 42° followed by 65° in 0.2× SSC, 0.1% SDS solution for 45 min, and then once more at 65° in 0.1× SSC, 0.1% SDS for 45 min. Blots were exposed to X-ray films for 3–7 days.
Marker analysis:
For SSR analysis 30 ng genomic DNA was used as template in 10-μl reaction mixtures containing 1× buffer (10 mm Tris-HCl, 50 mm KCl, pH 8.3), 2.0 mm MgCl2, 0.25 μm of each primer, 200 μm dNTPs, and 0.5 units of Biolase DNA polymerase (Bioline). The PCR conditions were as follows: initial 2 min at 94° followed by 35 cycles consisting of denaturation at 94° for 30 sec, primer annealing at 58° for 30 sec, and extension at 72° for 1 min. A single 8-min period for extension was provided at the end of the amplification reactions. The amplification products were size separated on a 4% agarose gel.
Mapmaker 2.0 program was used for determining map distances and developing genetic maps (Lander et al. 1987). Marker orders were determined at a LOD score of 3.0.
Physical mapping:
A Williams 82 soybean BAC library representing 10 soybean haploid genomes was screened with the NBSRps4/6 probe (M. K. Bhattacharyya, unpublished results). BAC160N2 containing the NBSRps4/6 region was identified, and both ends of this clone were sequenced. The primers for one end of the BAC clone were designed on the basis of the open reading frame sequence for the putative protein phosphatase 2 gene, and the PCR product for this end was named 160N2FEP. Similarly, primers designed on the basis of the putative importin gene sequence from the other BAC160N2 end were used to PCR amplify the 160N2REP probe. Both probes were used to screen the soybean BAC library and identify a clone that overlapped with the 160N2REP end. No clones were obtained for the 160N2FEP end. To identify clones for this end, the Faribault soybean BAC library constructed at the University of Minnesota was screened, and three clones that overlapped with the 160N2FEP end were identified (Danesh et al. 1998). Each end of the four new BAC clones was sequenced to develop PCR primers, which were subsequently used to generate the BAC contig of the Rps4 region.
cDNA cloning:
It has been shown that the upper one-third portion of the etiolated hypocotyls of 7-day-old dark-grown seedlings expresses gene-specific Phytophthora resistance (Lazarovits et al. 1981). We therefore used the upper one-third portion of the etiolated hypocotyls of 7-day-old dark-grown HARO4272 seedlings (Rps4) for generating a cDNA library in the Uni-ZAP XR λ-vector (Stratagene, La Jolla, CA; Bhattacharyya 2001). The unamplified library was screened using the NBSRps4/6 probe. Positive clones were purified and excised. The cDNA clones that hybridized to all four NBSRps4/6-specific HindIII fragments of BAC160N2 were sequenced to confirm their identities. Sequencing was carried out in an ABI 3100 automated DNA sequencer at the DNA facility, Iowa State University. Sequence of a representative cDNA clone has been deposited in GenBank (accession no. AY258630).
RESULTS
Cosegregation of the disease-resistance-gene-like sequence NBSRps4/6 with the Phytophthora resistance genes Rps4 and Rps6:
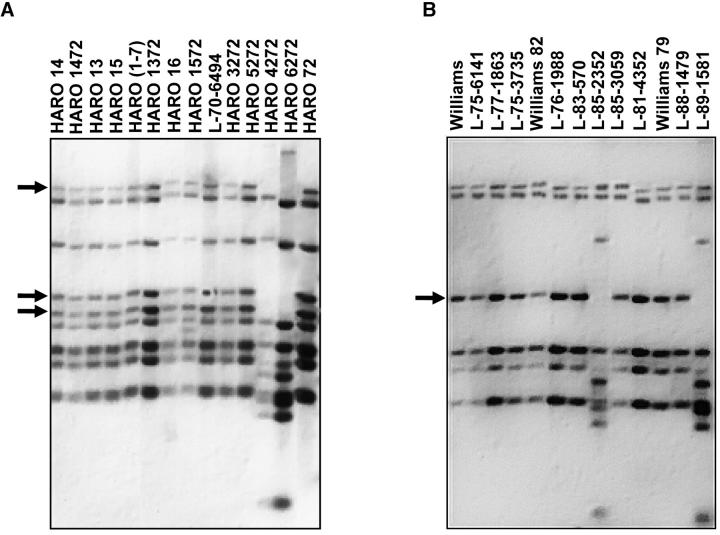
A pulsed-field gel-purified DNA fraction containing molecular markers that flank both sides of the Rps1 locus, and primers synthesized for the conserved sequences of the NBS domain of cloned disease-resistance genes were used in PCR experiments (Yu et al. 1996; Bhattacharyya et al. 2001). The amplified product showed high sequence identity to the NBS domain of cloned NBS-LRR-type disease-resistance genes and hybridized to several HindIII DNA fragments in DNA gel blot analyses (Figure 1, A and B). This sequence showed polymorphisms between near-isogenic lines differing at the Rps4 and Rps6 regions, but not among near-isogenic lines that differ for other Rps genes (Figure 1, A and B).
Figure 1.—
Genomic organization of NBSRps4/6 among near-isogenic soybean lines. (A) Harosoy isolines (details for isolines in materials and methods) for Rps genes. (B) Williams isolines for Rps genes. Arrows indicate NBS-specific HindIII fragments of the recurrent parent that are linked to Rps4 and Rps6 in the repulsion phase.
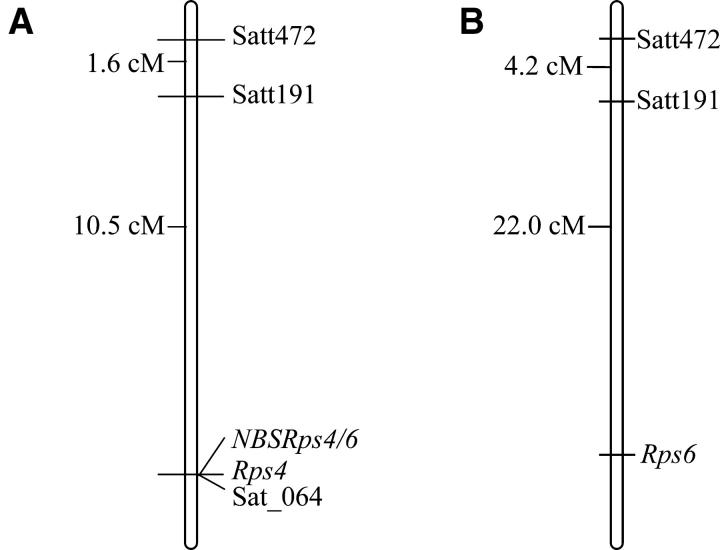
To determine the extent of linkage between NBSRps4/6 and Rps4, an F2 population developed by crossing Williams with HARO4272 was investigated. Williams is susceptible to P. sojae and does not contain any known Rps genes. Detached leaves of F2 plants were inoculated with P. sojae race 1 zoospores for determining the host responses (Bhattacharyya and Ward 1986). On the basis of previous information (Demirbas et al. 2001) three SSR markers, Sat_064, Satt191, and Satt472, were selected for mapping Rps4. A total of 1295 F2 plants were screened, and 254 of these plants showing susceptibility to P. sojae race 1 were selected for mapping the Rps4 region using these three SSR markers and NBSRps4/6. Sat_064 and NBSRps4/6 cosegregated with Rps4 (Figure 2A).
Figure 2.—
Genetic linkage maps of the Rps4 and Rps6 regions. (A) High-resolution genetic linkage map of the Rps4 region. (B) Genetic linkage map showing the location of Rps6.
NBSRps4/6 detected several similar introgressed HindIII DNA fragments in near-isogenic lines containing either Rps4 or Rps6 (Figure 1, A and B). Introgression of these fragments was also accompanied by loss of HindIII fragments that were presumably linked tightly to the Rps4 and Rps6 loci in repulsion (Figure 1, A and B). These data strongly indicate that Rps4 and Rps6 are linked genes. However, earlier reports suggested that Rps4 and Rps6 segregated independently (Athow and Laviolette 1982). We therefore reinvestigated the possible linkage between these two Rps genes. An F2 population developed from the cross between HARO4272 (Rps4) and L89-1581 (Rps6) was used to map these two Rps genes. A total of 120 plants were leaf inoculated with P. sojae race 1. Both genes confer resistance against race 1. All 120 F2 plants were resistant to race 1. No double-recessive susceptible recombinants were identified in this study. If these genes were to assort independently, ∼8 susceptible plants (rps4rps6) should have been identified. These results suggest that two genes are linked with a χ2 probability of 0.0046.
To distinguish the race specificity of Rps4 and Rps6, we infected HARO4272 (Rps4) and L89-1581 (Rps6) with five selected P. sojae isolates, viz. I-CC5C, I-CC8, I-CE5A, I-NW8A, and POLK3-3 that were collected from soybean fields of Iowa. Isolate I-NW8a was found to be virulent to HARO4272 (Rps4) and avirulent to L89-1581 (Rps6). This isolate was used to screen 38 F2:3 families generated from the cross between HARO4272 (Rps4) and L89-1581 (Rps6). The families segregated in a 1:2:1 ratio with a χ2 probability of 0.08. Relative positions of polymorphic SSR markers Satt191 and Satt472 in relation to Rps6 suggest that Rps4 and Rps6 are most likely allelic or tightly linked genes (Figure 2B).
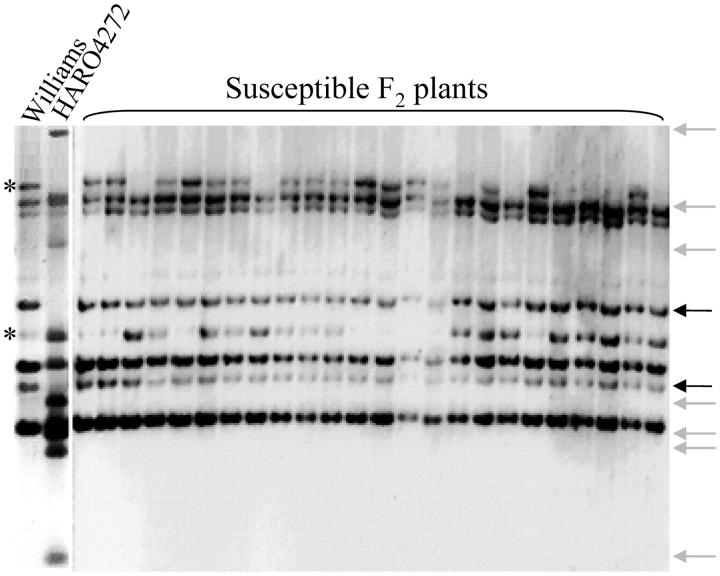
It was observed that seven NBSRps4/6-specific HindIII fragments of the resistant parent HARO4272 were missing among all susceptible F2 plants (Figure 3). These fragments were mapped to the Rps4 region. One HindIII fragment specific to HARO4272 and one to Williams segregated independently of Rps4. Presumably, these sequences mapped to a second homeologous locus (shown by asterisks in Figure 3). Four fragments were not polymorphic between two parents, and therefore map positions of these fragments could not be predicted. These results suggest that the NBSRps4/6 sequence mapped to at least two loci, one of which cosegregates with Rps4.
Figure 3.—
Cosegregation of NBSRps4/6 with the Rps4 locus. Solid arrows represent HindIII DNA fragments that cosegregate with the rps4 allele; shaded arrows represent HindIII fragments that cosegregate with Rps4, which are absent in all susceptible F2 plants. (*) HindIII fragments that segregate independently of the Rps4 locus.
Identification of rearrangements at the NBSRps4/6 region:
Seventeen F1's were generated by crossing Williams with HARO4272 for developing a mapping population that segregated for the Rps4 region. In one F1, all except one HARO4272-specific NBSRps4/6 HindIII fragment were lost (data not shown). One novel HindIII fragment was evolved in this F1, indicating the presence of a deletion in the NBSRps4/6 region. We were therefore interested in studying (i) if this locus is unstable and (ii) if deletion of this sequence resulted in the loss of Rps4 function.
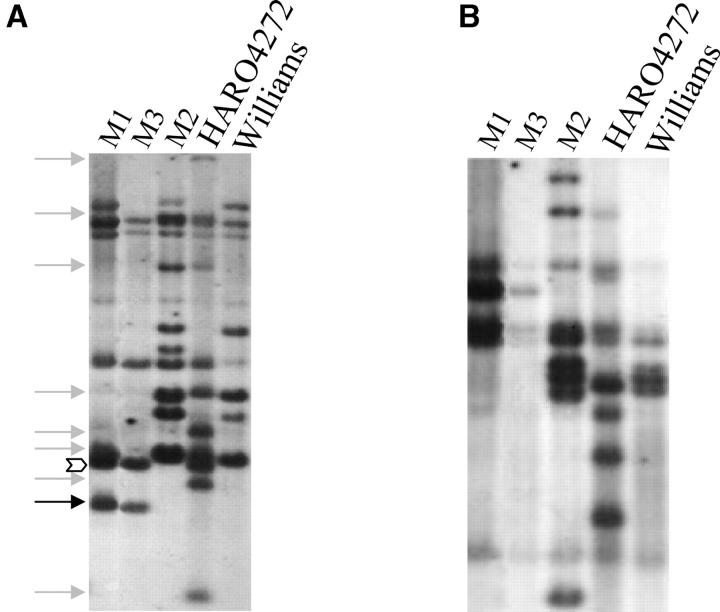
Identification of a loss-of-function mutant could establish the NBSRps4/6 sequence as a potential candidate for the Rps4 gene. This mutant will then facilitate the cloning of Rps4. We analyzed 65 additional F1's and 201 selfed HARO4272 plants for detecting any possible rearrangements in the NBSRps4/6 sequence and identified five additional mutants. These five mutant plants were classified into two classes on the basis of their NBSRps4/6-specific HindIII fingerprints (data not shown). In total we identified three classes of mutations. The mutant originally identified from analysis of 17 F1's is termed M1, and the other two classes of mutants identified from selfed HARO4272 lines are M2 and M3, respectively. Of the four rearranged plants of the M2 class, only one was in homozygous condition; the NBSRps4/6-specific HindIII fingerprint in this mutant is identical to that of the recurrent parent Harosoy (data not shown). M3 was found in homozygous condition, and its Rps4-linked NBSRps4/6-specific HindIII fingerprint is comparable to that of M1 (Figure 4A).
Figure 4.—
Genomic organization of NBSRps4/6 among the mutants. (A) DNA digested with HindIII. Loss of eight HindIII fragments (shaded arrows) and gain of a novel fragment (solid arrow) are shown. Both M1 and M3 retained a unique NBSRps4/6-specific HindIII fragment shown by an open arrowhead. (B) DNA digested with DraI.
We investigated if the observed rearrangements were due to methylation. It has been considered that HindIII is sensitive to cytosine methylation (Kessler and Manta 1990). To confirm that the differences in restriction patterns observed among mutants were not due to differences in DNA methylation, we compared NBSRps4/6 fingerprints of HARO4272 and mutants using methylation-insensitive restriction enzyme DraI (Figure 4B). DraI-digested DNA samples of all three mutants resulted in NBSRps4/6-specific fingerprints that are distinct from the one for HARO4272. These results suggested that the rearrangements were not due to DNA methylation.
Rearrangements at the Rps4 region were not generated from seed or pollen contamination:
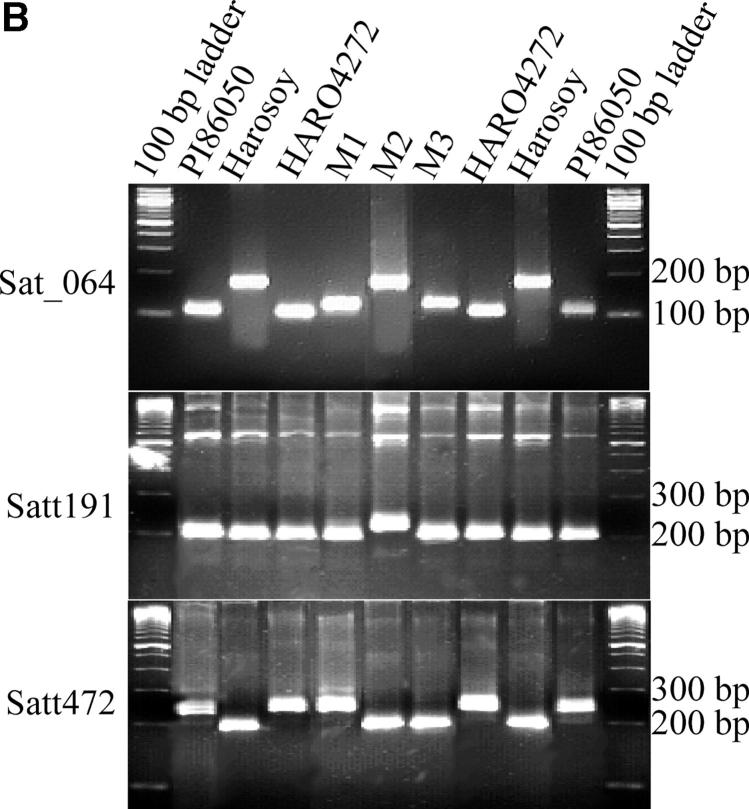
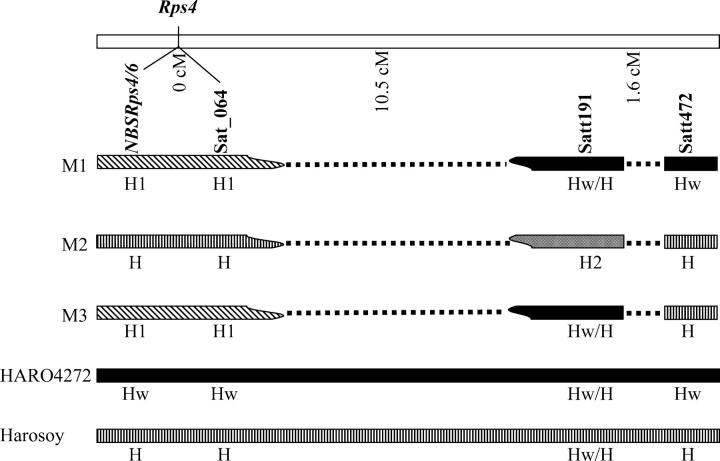
To investigate if the mutations originated from mechanical seed mixture, cross-fertilization, or through recombination, we genotyped all three mutants and HARO4272 using 180 SSR markers representing all 20 soybean chromosomes (see supplemental Table S1 at http://www.genetics.org/supplemental/). Mutants were identical to HARO4272 for all SSR markers except for a small region of MLG G that contains Rps4 (Figure 5). Haplotypes for M1 and M3 are very comparable for the Rps4 region. M1 and M3 differ only for Satt472. This SSR marker showed the HARO4272-type pattern in M1 and the Harosoy-type pattern in M3 haplotype (Figure 5B and Figure 6). These results indicate the occurrence of a recombination event between M1 and Harosoy haplotypes for generation of the M3 haplotype. M2 and Harosoy haplotypes are similar for the whole Rps4 region except Satt191 (Figure 5B and Figure 6). On the basis of the variations in linked SSR markers and NBSRps4/6 we concluded that there are at least four haplotypes for the Rps4 region in the cultivar HARO4272 (Table 1 and Figure 6).
Figure 5.—
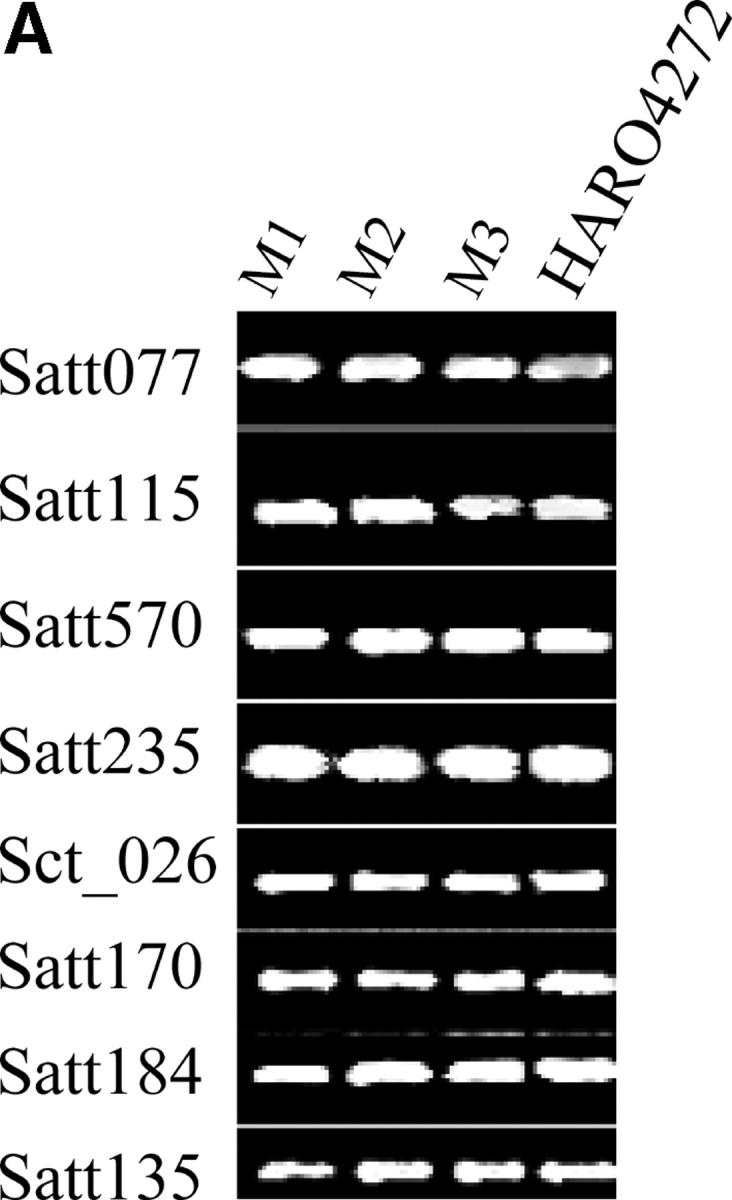
Genome-wide simple sequence repeat analyses. (A) Identical simple sequence repeat phenotypes among mutants and HARO4272. Amplification products of a few selected SSR markers are shown. (B) Simple sequence repeat sequences depicting differences of haplotypes at the Rps4 region. PI86050, donor parent; Harosoy, recurrent parent; HARO4272, the Rps4 line; M1, M2, and M3 are three mutants.
Figure 6.—
Identification of four haplotypes at the Rps4 region in HARO4272. Dotted lines represent genomic region for which no information is available. M1, M2, and M3 are mutants; H1, haplotype for M1; H2, haplotype for M2; H, Harosoy haplotype; Hw, HARO4272 haplotype. M1 and M3 are different only for Satt472.
TABLE 1.
Identification of multiple haplotypes at theRps4 region
| Line | Proportion in population |
NBSRps4/6 | Sat_064 | Rps4/rps4 | Satt191 | Satt472 |
|---|---|---|---|---|---|---|
| M1 | 0.059 | H1 | H1 | NK | Hw/H | Hw |
| M2 | 0.020 | H | H | NK | H2 | H |
| M3 | 0.005 | H1 | H1 | NK | Hw/H | H |
| HARO4272 | NA | Hw | Hw | Rps4 | Hw/H | Hw |
| Harosoy | NA | H | H | rps4 | Hw/H | H |
M1, mutant 1; M2, mutant 2; M3, mutant 3; Hw, wild-type haplotype (HARO4272); H, Harosoy-type haplotype; Hw/H, not polymorphic between HARO4272 and Harosoy; H1, M1-type haplotype; H2, M2-type haplotype; NA, not applicable; NK, not known.
All four haplotypes are, however, identical for all 176 SSR markers, unlinked to the Rps4 region. PI86050 was the donor parent of HARO4272. As expected, HARO4272 and PI86050 are identical for Sat_064, Satt191, Satt472, and NBSRps4/6. Twenty individual PI86050 plants were evaluated for Sat_064 and Satt472. All 20 plants showed identical banding patterns for these two markers (data not shown). It is most unlikely that four novel haplotypes identified in HARO4272 were introgressed from the donor parent. Selection for Phytophthora resistance after each backcrossing should have allowed the introgression of a specific genomic region from the donor parent, because a single Rps gene should have been enough to confer resistance against race 1 in each backcross generation during development of HARO4272. Second, only a single resistant plant was selected for the next backcrossing. Therefore, it is very unlikely that the introgression of more than one haplotype was possible during introgression of Rps4. These results strongly suggest that the additional three haplotypes varying at the Rps4 region evolved from rearrangements through recombination events rather than from mechanical seed mixture or pollen contamination.
Loss of NBSRps4/6 is associated with the loss of Rps4 function in M1:
We hypothesized that if a copy of NBSRps4/6 is the Rps4 gene sequence, then deletion of this sequence in M1 should have resulted in the loss of Rps4 function. F2 individuals of the cross between Williams and M1 were segregating for resistance and susceptibility when leaves were inoculated with race 1, suggesting the intactness of Phytophthora resistance function in this mutant.
Although leaves of HARO4272 are resistant to P. sojae race 1 (Figure 2A), the cultivar is highly susceptible to race 1 when wounded hypocotyls of light-grown plants are inoculated with the race. Responses of leaves and hypocotyls to the race were consistent throughout the investigation. On the other hand, wounded hypocotyls of mutants were highly resistant to this race (see supplemental Figure S1 at http://www.genetics.org/supplemental/). Thirty P. sojae isolates (see materials and methods) collected from different soybean fields of Iowa were used to determine if there were any differences between Phytophthora resistance in mutants and the one encoded by Rps4 in HARO4272. Four of these isolates and race 1 can distinguish mutants and HARO4272 (Table 2). M1 and M3 showed similar disease reactions against all 30 isolates. Furthermore, except for the loosely linked SSR marker Satt472, both mutants showed the same allelic constitutions for all molecular loci of the Rps4 region (Figure 6). Therefore, most likely M1 and M3 were evolved from the same original mutation. M2 showed the race specificity, which is distinct from those of HARO4272, M1, M3, and Harosoy. These results suggest that there are at least two distinct race specificities among three mutants.
TABLE 2.
Differential responses of mutants againstP. sojae isolates
|
P. sojae isolate
|
|||||
|---|---|---|---|---|---|
| Soybean line | I-CC5C | I-CC8 | I-CE5 | I-POLK3-3 | Race 1 |
| PI86050 (Rps4) | R | R | R | R | R |
| HARO4272 (Rps4) | R | S | S | I | S |
| M1 | R | R | R | R | R |
| M2 | R | S | S | R | I |
| M3 | R | R | R | R | R |
| Harosoy (Rps7) | S | S | S | S | S |
R, resistant, i.e., >80% plants survived; I, intermediate, i.e., 50–80% plants survived; S, susceptible, i.e., <50% plants survived.
P. sojae race 1 has been reported to be avirulent against soybean lines containing Rps4 (Buzzell et al. 1987). In our investigation, HAR04272 containing Rps4 was susceptible to race 1 when wounded hypocotyls were inoculated, but resistant when leaves were inoculated. We investigated if race 1 used in this study still maintains its original race specificity against various Rps genes. Differential lines carrying all 14 Rps genes were evaluated for their responses to race 1. HARO4272 and PI91160 carrying Rps4 and Rps5, respectively, were susceptible to this race (Table 3). P. sojae race 1 is expected to be avirulent to all differential lines carrying any Rps genes except Rps7 (Table 3; Buzzell et al. 1987). It appears that race 1 used in this study has lost its avirulence function against two known Rps genes.
TABLE 3.
Responses of differentials againstP. sojae race 1
| Rps gene | Line | Expected reaction |
Observed reaction |
|---|---|---|---|
| 1a | Mukden/L75-6141 | R | R |
| 1b | Sanga | R | R |
| 1c | Mack | R | R |
| 1d | PI103091 Wu An | R | R |
| 1k | Kingwa | R | R |
| 2 | CNS | R | I |
| 3a | PI171442 | R | R |
| 3b | PI172901 | R | R |
| 3c | PI380046 | R | R |
| 4 | PI86050 Rasuto San | R | R |
| 5 | T240/PI91160 | R | S |
| 6 | Altona | R | R |
| 7 | Harosoy | S | S |
| 8 | PI399073 | — | R |
| 4, 7 | HARO4272 | R | S |
| — | Williams | S | S |
R, resistant; I, intermediate; S, susceptible; —, data not available.
We were interested in understanding the mechanism of gain of Phytophthora resistance function associated with deletion of the NBSRps4/6 sequence. As a first step, we investigated the inheritance of resistance in M1. Wounded green hypocotyls of 96 F2:3 families obtained from the cross between Williams and M1 were infected with race 1. As a control, 48 F2:3 families derived from the cross between Williams and HARO4272 were similarly inoculated with race 1. Each family consisted of 20 progeny plants. All F2:3 families obtained from the cross between Williams and HARO4272 were susceptible to race 1 (see supplemental Figure S2 at http://www.genetics.org/supplemental/). On the other hand, F2:3 families of the cross between Williams and M1 segregated in a 1:2:1 ratio for a single dominant gene with a χ2 probability of 0.573. These data suggest that a single major gene controls the Phytophthora resistance expressed in wounded hypocotyls of M1 against race 1.
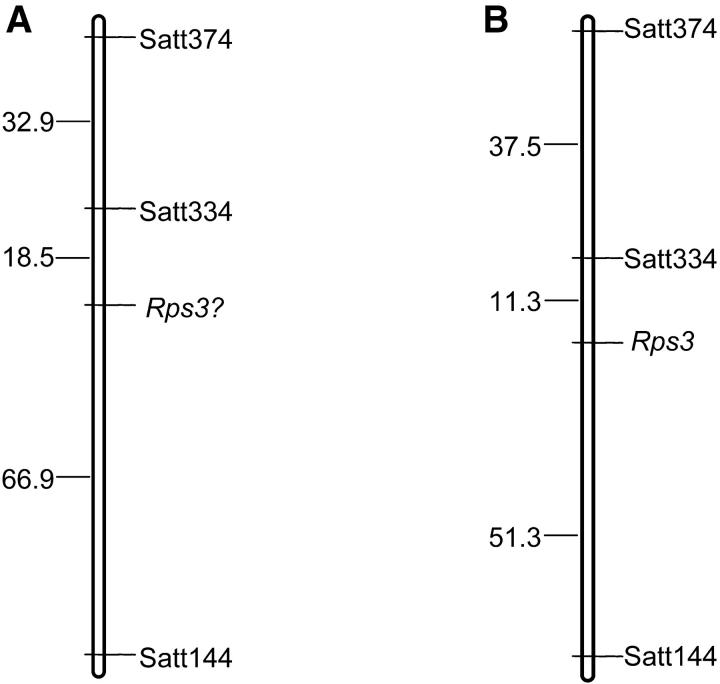
During backcrossing usually only a single genomic region carrying a desirable trait is introgressed into recurrent parent. Therefore, except for the introgressed region, the rest of the genome in the near-isogenic line is expected to be identical to that of the recurrent parent. Gain of Phytophthora resistance in this mutant indicated that deletion in the NBSRps4/6 region is responsible for expression of this novel race specificity. Loss of a genetic factor or element from the NBSRps4/6 region could be responsible for gain of the novel race specificity in M1. To test this hypothesis, SSR markers were used to map the novel Phytophthora resistance in M1. Bulk segregant analysis was applied to map the new Phytophthora resistance gene (Michelmore et al. 1991). Resistant and susceptible bulks were prepared from 8 homozygous-resistant and 8 homozygous-susceptible F3 families, respectively. These families were selected from 96 F2:3 families obtained from the cross between Williams and M1. A total of 379 SSR markers representing the whole soybean genome were used to PCR amplify from both bulks. Satt334 showed polymorphism between resistant and susceptible bulks. Satt334 and polymorphic SSR markers close to Satt334 were used to map the unknown gene by using 31 selected F2:3 families generated from the cross between Williams and M1. Map position of the unknown gene is shown in Figure 7. The Rps gene identified in this mutant mapped closely to the Rps3 locus that contains three functional alleles. We conclude that the Rps gene identified in M1 could be either allelic or tightly linked to Rps3. We tentatively named this new Rps gene “Rps3?.”
Figure 7.—
The gene encoding Phytophthora resistance in M1 mapped to the Rps3 region. (A) Because the gene encoding Phytophthora resistance in M1 mapped to the Rps3 region, it could be allelic to Rps3 and is termed as Rps3?. (B) Map shows the Rps3 region in the soybean composite genetic linkage map for MLG F (http://soybase.ncgr.org/cgi-bin/ace/generic/pic/soybase?name=F-Composite&class=Map; Song et al. 2004).
Analysis of the segregating F2:3 families and identification of the map location of the gene Rps3? in M1 allowed us to investigate whether deletion in the NBSRps4/6 (ΔNBSRps4/6) region played any role in the expression of Rps3?. If a negative regulator in the NBSRps4/6 region were controlling the expression of the Phytophthora resistance encoded by Rps3?, then lines carrying ΔNBSRps4/6 should always be resistant. Both Rps3? and ΔNBSRps4/6 segregated independently, and we were able to identify susceptible genotypes homozygous for ΔNBSRps4/6 and rps3? alleles (e.g., families Ax17931-1-85, Ax17931-1-103). Therefore, the deletion unlikely resulted in the expression of Rps3?.
Generation of recombinant F2:3 families homozygous for ΔNBSRps4/6 and rps3? alleles also allowed us to investigate if deletion of any NBSRps4/6 copies in M1 resulted in loss of the Rps4 function. Earlier, leaf inoculation of F2 plants of the cross between Williams and M1 showed segregation of resistance and susceptibility in a 3:1 ratio, suggesting that there was no loss of Phytophthora resistance (data not shown). Identification of families such as Ax17931-1-85 and Ax17931-1-103 carrying ΔNBSRps4/6 and rps3? alleles in homozygous condition allowed us to investigate whether the leaf-specific Rps4 function against race 1 is intact in the haplotype containing the ΔNBSRps4/6 allele. Results of such a leaf-inoculation study are presented in Figure 8. It appears that progenies of the families homozygous for ΔNBSRps4/6 and rps3? alleles were susceptible to race 1. This observation strongly supported that Rps4, which cosegregates with NBSRps4/6 (Figure 2A), must have been deleted in M1. We hypothesize that a copy of NBSRps4/6 is the Rps4 gene.
Figure 8.—

Recombinants showing the association of deletion at the NBSRps4/6 region with the loss of Rps4 function. Disease reactions of mutants, recombinants, and control genotypes are shown. Rps3?, Rps gene mapped to the Rps3 region; Rps?, Rps gene that has not been characterized; ΔNBSRps4/6, deletion in the NBSRps4/6 region; nbsrps4/6w, NBSRps4/6 allele from Williams; nbsrps4/6h, NBSRps4/6 allele from Harosoy. In this experiment P. sojae race 1 zoospores were used. In leaves Rps4 confers resistance against this race. (A) Williams (nbsrps4/6w, rps3?). (B) 17931-1-85 (ΔNBSRps4/6, rps3?). (C) 17931-1-103 (ΔNBSRps4/6, rps3?). (D) Harosoy (nbsrps4/6h, rps3?). (E) HARO4272 (NBSRps4/6, rps3?). (F) 17931-1-81 (ΔNBSRps4/6, Rps3?). (G) 17931-1-102 (nbsrps4/6w, Rps3?). (H) 17931-1-113 (nbsrps4/6w, Rps3?). (I) M2 (nbsrps4/6h, Rps?). (J) M3 (ΔNBSRps4/6, Rps3?). (K) PI86050 (NBSRps4/6, Rps3?).
Physical mapping of the Rps4 region:
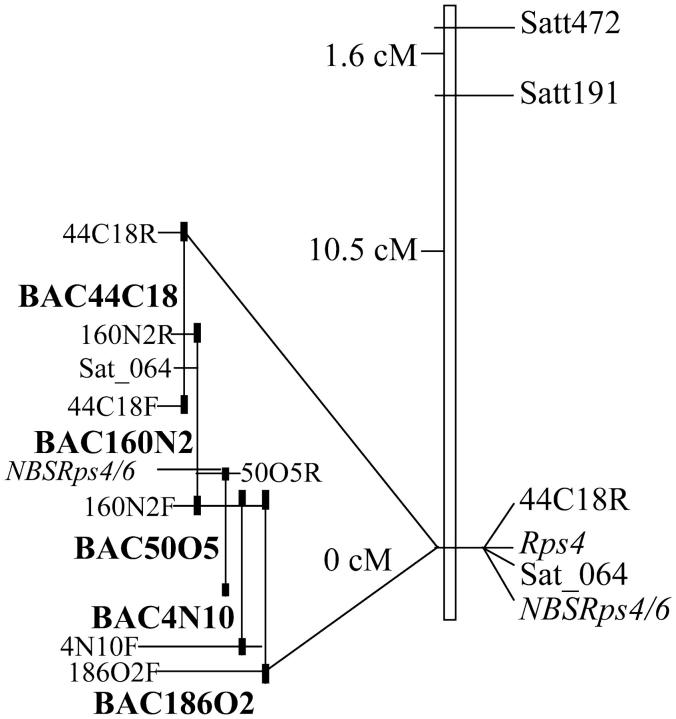
To generate a physical map for the region, the NBSRps4/6 probe was used to screen a Williams 82 BAC library (M. K. Bhattacharyya, unpublished results). Screening of the library resulted in identification of BAC160N2 containing all four NBSRps4/6-specific HindIII restriction fragments of the susceptible rps4 haplotype of Williams 82 (Figure 1). Ends of this BAC were sequenced to develop end-specific primers. Open reading frames identified in each end were used to generate primers. Amplified sequences were used to isolate BAC44C18 that overlaps with the BAC160N2R end. The BAC160N2F end was used to identify three clones, BAC50O5, 4N10, and 186O2, from a separate BAC library constructed in the Young Laboratory (Danesh et al. 1998). Ends of these BACs were sequenced to develop BAC-end-specific probes for developing a BAC contig of the Rps4 region (Figure 9). The contig most likely includes the NBSRps4/6 region, because BAC160N2 contains all four HindIII NBSRps4/6 fragments specific to the susceptible Williams haplotype. Marker 44C18R was mapped to the Rps4 region by using an F2 population of the cross between Williams and HARO4272 (Figure 9). BAC160N2 and BAC44C18 carry the Sat_064 marker that cosegregates with Rps4 (Figure 9).
Figure 9.—
Physical map of the Rps4 region. A BAC contig is shown in parallel to the linkage map. BAC160N2, which is ∼70 kb, contains all four NBSRps4/6 HindIII fragments specific to the Williams haplotype and the Sat_064 marker.
The candidate NBSRps4/6 sequence is transcribed in tissues that confer Rps4-specific Phytophthora resistance:
High-resolution genetic and physical mapping data and loss of Rps4-encoded Phytophthora resistance in M1 indicate physical association of NBSRps4/6 with Rps4. Random sequencing of BAC160N2 indicated that NBSRps4/6 may be the only class of disease-resistance-gene-like sequence in the Rps4 region (H. Gao, unpublished results). We therefore investigated if the NBSRps4/6 sequence is transcribed. The NBSRps4/6 probe was used to screen a cDNA library generated from HARO4272 (Rps4) etiolated hypocotyls. The upper one-third portion of etiolated hypocotyls expressing Rps4-specific resistance was used to generate the cDNA library (Bhattacharyya 2001). About 1.5 × 106 plaque-forming units were screened for the NBSRps4/6 sequence. Thirty-two positive clones were identified from this screening. Identity of individual clones was verified by gel blot and sequence analyses. Blast search of a partial cDNA sequence (GenBank accession no. AY258630) revealed 43% amino acid identity of NBSRps4/6 with the Arabidopsis thaliana RPP13 gene. Search for conserved domains using Reverse Position Specific BLAST (RPBLAST of NCBI) revealed an NB-ARC domain. No similarity with any known domains was found for the truncated N-terminal region.
DISCUSSION
In this study, we have identified the disease-resistance-gene-like sequence NBSRps4/6 that was introgressed into a soybean line along with Phytophthora resistance genes Rps4 and Rps6 from PI86050 and Altona, respectively. Earlier Athow and Laviolette (1982) reported independent assortment between Rps4 and Rps6 genes. However, Demirbas et al. (2001) identified a SSR marker that mapped closely to both Rps4 and Rps6. Loose linkages of RFLP and SSR markers with Rps4 have also been reported (Diers et al. 1992; Demirbas et al. 2001). High-resolution genetic mapping data showed that the SSR marker Sat_064 cosegregates with Rps4 and is physically linked to NBSRps4/6. Rps4 and Rps6 are reported to encode resistance against very comparable sets of P. sojae races (Buzzell et al. 1987). Therefore, most likely these two genes are related. Rps6 and Rps4 isolines also exhibit similar DNA fingerprints for NBSRps4/6 (Figure 1). Mapping data have shown that Rps4 and Rps6 are either allelic or clustered genes (Figure 2).
In this investigation we have discovered several rearrangements at the NBSRps4/6 region in the cultivar HARO4272 (Table 1). On the basis of race specificities, mutants can be classified into two classes: (i) M1 and M3 in class I and (ii) M2 in class II. However, molecular marker data showed that mutants M1 and M3, although they have the same race specificity, are different for Satt472, possibly because of a recombination event (Figure 5B and Figure 6).
A significant number of polymorphisms can be detected in soybean with the aid of SSR markers. A genetic diversity study of 131 Asian soybean accessions revealed that the average probability of detecting SSR-based polymorphisms between any two lines is 0.78. A minimum probability of 0.05 for detecting polymorphisms was observed between highly related Japanese soybean lines originating from the same cultivar group (Abe et al. 2003). In our study, even if we consider the lower probability value for detecting polymorphism by any SSR marker, the use of 176 SSR markers representing the whole genome, except the Rps4 region, should have detected at least nine polymorphisms if mutants were to originate from mechanical seed mixture or cross-pollination rather than from recombination events. These data, therefore, suggest that genetic recombination at the Rps4 region was the mechanism for generation of novel Phytophthora race specificities in HARO4272 (Table 2).
Investigation of the Phytophthora resistance encoded in M1 led to the identification of a novel gene Rps3?, which could be either allelic or linked to Rps3 in MLG F. Eight loci have already been assigned to accommodate 14 Rps genes (Diers et al. 1992; Lohnes and Schmitthenner 1997; Demirbas et al. 2001; Weng et al. 2001; Burnham et al. 2003). Rps3? was most likely introgressed along with Rps4 into the Harosoy background. HARO4272 was developed in Harrow, Ontario, during 1980s. The initial cross was made between L62-904 (white-flowered Harosoy) and PI86050 (Rps4). Selection for the Phytophthora resistance gene Rps4 was accomplished by inoculating each backcrossing generation with P. sojae race 4 and race 5. Rps4 encodes resistance against race 4 but not race 5 (Buzzell et al. 1987). Nine BC6F1 plants were selfed to generate nine segregating F2 populations that were evaluated for resistance against P. sojae race 1. Homozygous-resistant families were subsequently bulked and named HARO4272 (Buzzell et al. 1987; D. Buzzell, personal communication). Our data established that HARO4272 is composed of several haplotypes and carried multiple Rps genes. This can be explained if we assume that race 4 used for selection of Rps4 was a mixture of isolates because a mixture of distinct isolates can provide selection pressure for two or more independent resistance genes. For example, one isolate carried the corresponding Avr gene for Rps4, while the other one carried the corresponding Avr gene for Rps3?. As a result of the use of a mixture of two isolates, simultaneous selection for both Rps4 and Rps3? was possible. M2 showed a distinct race specificity (Table 2) and contains an unknown Rps gene that confers resistance in leaves against race 1 (Figure 9). The presence of a third isolate in the race mixture can be considered for selection of this unknown Rps? gene detected in M2. Introgression of all three Rps genes was therefore accomplished in each backcross generation with the use of an isolate mixture. Use of single-spore-derived races was not in practice until the 1990s and consideration of an isolate mixture as a race was quite possible in the 1980s (Bhat et al. 1993; T. Anderson, personal communication).
Recombination most likely has taken place at the NBSRps4/6 region between Harosoy and the donor parent PI86050 during selfing generations of nine BC6F1 plants. Occurrence of such a recombination process during backcrossing generations was very unlikely because only a single resistant plant was selected for crossing with the recurrent parent after each backcrossing generation, allowing introgression of only one haplotype into Harosoy. Instead, we have documented four haplotypes at the Rps4 region in varying proportions in the cultivar HARO4272 (Table 1). We hypothesize that M1 was derived from a single unequal recombination event during selfing generations following the completion of backcrosses. This recombination event led to a deletion in the NBSRps4/6 region. M3 is most likely derived from M1 through a subsequent recombination event between NBSRps4/6 and Satt472 (Figure 6).
Unequal crossing over is considered as one of the most important mechanisms for evolution of disease-resistance genes. For example, in maize unequal crossing over is the main mechanism of meiotic instability at the Rp1 region (Sudupak et al. 1993). In the Arabidopsis ecotype Columbia, the nonfunctional chimeric rpp8 gene was most likely evolved from unequal crossing over between the functional oomycete resistance gene RPP8 and its homolog RPH8A (McDowell et al. 1998). It has been proposed that the HRT gene encoding viral resistance was evolved by unequal crossing over between progenitor genes related to RPP8 and RPH8A (Cooley et al. 2000). Unequal crossing over has also been considered to play an important role in the generation of alleles at the Rsv1 locus in soybean (Hayes et al. 2004). The NBSRps4/6 HindIII fingerprints show that the sequence has many more paralogous copies in lines carrying either Rps4 or Rps6 than in those carrying the corresponding recessive alleles (Figure 1). Therefore, this region is highly diverse between the resistant donor and the susceptible recurrent parents, and a recombination at this region most likely will be an unequal one, which can cause loss of sequences. For example, nine deletions in the maize Rp1-D gene family have been considered to generate from unequal crossing over (Collins et al. 1999). The unequal crossing over between donor and recurrent parents at the NBSRps4/6 region most likely led to deletion of Rps4 in M1.
We have shown through analysis of segregants that leaves of recombinant lines homozygous for ΔNBSRps4/6 and rps3? alleles are susceptible to race 1 (Figure 8, B and C). Thus, M1 carrying ΔNBSRps4/6 does not contain Rps4. M2 also does not contain Rps4 (Table 2; and slight spread in Figure 8I). Loss of Rps4, presumably through unequal crossing over, led to selection pressure for Rps3? in M1 or M3. Earlier studies have shown that most Rps genes confer resistance against race 1 (Buzzell et al. 1987). Therefore, lines carrying any of these three genes should be selected during the selfing generations, because race 1 but not race 4 was used to inoculate the segregating population (D. Buzzell, personal communication). If we assume that Rps4 provided the strongest resistant response as compared to that of the other two genes against race 1, then use of race 1 instead of race 4 during selfing generations most likely relieved the selection pressure for Rps3? and Rps?. Lack of selection pressure for either Rps3? or Rps? among individuals carrying Rps4 most likely led to partial loss of Rps3? and Rps? from the HARO4272 population (see supplementary Figure S1 at http://www.genetics.org/supplemental/).
High-resolution genetic mapping data have shown that Rps4 and NBSRps4/6 cosegregate. NBSRps4/6 is also transcribed and most likely is the only candidate disease-resistance-gene-like sequence at the NBSRps4/6 region (H. Gao, unpublished results). Loss of a few copies of the NBSRps4/6 gene family is also associated with the loss of Rps4-specific resistance, suggesting that a copy of the NBSRps4/6 gene family could be the Rps4 gene.
Acknowledgments
We are grateful to Peter Peterson and Randy Shoemaker for reviewing this manuscript. We are also grateful to Nieves Rivera-Velez and Grace Welke for generating the F1's. This work was funded by the Iowa Soybean Promotion Board.
References
- Abe, J., D. H. Xu, Y. Suzuki, A. Kanazawa and Y. Shimamoto, 2003. Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor. Appl. Genet. 106: 445–453. [DOI] [PubMed] [Google Scholar]
- Anderson, J. A., Y. Ogihara, M. E. Sorrells and S. D. Tanksley, 1992. Development of a chromosomal arm map for wheat based on RFLP markers. Theor. Appl. Genet. 83: 1035–1043. [DOI] [PubMed] [Google Scholar]
- Athow, K. L., and F. A. Laviolette, 1982. Rps6, a major gene for resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathology 72: 1564–1567. [Google Scholar]
- Bhat, R. G., B. A. McBlain and A. F. Schmitthenner, 1993. Development of pure lines of Phytophthora sojae races. Phytopathology 83: 473–477. [Google Scholar]
- Bhattacharyya, M. K., 2001 Construction of cDNA libraries, pp. 42–62 in Essential Molecular Biology: A Practical Approach, edited by T. A. Brown. Oxford University Press, London/New York/Oxford.
- Bhattacharyya, M. K., and E. W. B. Ward, 1986. Resistance, susceptibility and accumulation of glyceollins I–III in soybean organs inoculated with Phytophthora megasperma f.sp. glycinea. Physiol. Mol. Plant Pathol. 29: 227–237. [Google Scholar]
- Bhattacharyya, M. K., B. G. Espinosa, T. Kasuga, S. S. Salimath, M. Gijzen et al., 2001 Towards understanding the recognition and signal transduction processes in the soybean-Phytophthora sojae interaction, pp. 5–1043 in Symposium on Plant Signal Transduction, October 4–6, 1999, edited by S. K. Sopory, R. Oelmuller and S. C. Maheshwari. Kluwer Academic/Plenum, International Center for Genetic Engineering and Biotechnology, New Delhi.
- Burnham, K. D., A. E. Dorrance, D. M. Francis, R. J. Fioritto and S. K. St. Martin, 2003. Rps8, a new locus in soybean for resistance to Phytophthora sojae. Crop Sci. 43: 101–105. [Google Scholar]
- Buzzell, R. I., T. R. Anderson and B. D. Rennie, 1987. Harosoy Rps isolines. Soyb. Genet. Newsl. 14: 79–81. [Google Scholar]
- Collins, N., J. Drake, M. Ayliffe, Q. Sun, J. Ellis et al., 1999. Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutant. Plant Cell 11: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, M. B., S. Pathirana, H. J. Wu, P. Kachroo and D. F. Klessig, 2000. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12: 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh, D., S. Penuela and J. Mudge, 1998. A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistance gene. Theor. Appl. Genet. 96: 196–202. [Google Scholar]
- Dangl, J. L., and J. D. Jones, 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Demirbas, A., B. G. Rector, D. G. Lohnes, R. J. Fioritto, G. L. Graef et al., 2001. Simple sequence repeat markers linked to the soybean Rps genes for Phytophthora resistance. Crop Sci. 41: 1220–1227. [Google Scholar]
- Diers, B. W., L. Mansur, J. Imsande and R. C. Shoemaker, 1992. Mapping Phytophthora resistance loci in soybean with restriction fragment length polymorphism markers. Crop Sci. 32: 377–383. [Google Scholar]
- Feinberg, A. P., and B. Vogelstein, 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6–13. [DOI] [PubMed] [Google Scholar]
- Förster, H., B. M. Tyler and M. D. Coffey, 1994. Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. Mol. Plant Microbe Interact. 7: 780–791. [Google Scholar]
- Hayes, A. J., S. C. Jeong, M. A. Gore, Y. G. Yu, G. R. Buss et al., 2004. Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 166: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S. H., C. A. Webb, S. M. Smith and Q. Sun, 2001. Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39: 285–312. [DOI] [PubMed] [Google Scholar]
- Kasuga, T., S. S. Salimath, J. Shi, M. Gijzen, R. I. Buzzell et al., 1997. High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. Mol. Plant Microbe Interact. 10: 1035–1044. [Google Scholar]
- Kessler, C., and V. Manta, 1990. Specificity of restriction endonucleases and DNA modification methyltransferases a review (edition 3). Gene 92: 1–248. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lazarovits, G., P. Stössel and E. W. B. Ward, 1981. Age-related changes in specificity and glyceollin production in the hypocotyl reaction of soybeans to Phytophthora megasperma var. sojae. Phytopathology 71: 94–97. [Google Scholar]
- Leitz, R. A., G. L. Hartman, W. L. Pedersen and C. D. Nickell, 2000. Races of Phytophthora sojae on soybean in Illinois. Plant Dis. 84: 487. [DOI] [PubMed] [Google Scholar]
- Lohnes, D. G., and A. F. Schmitthenner, 1997. Position of the Phytophthora resistance gene Rps7 on the soybean molecular map. Crop Sci. 37: 555–556. [Google Scholar]
- Martin, G. B., A. J. Bogdanove and G. Sessa, 2003. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61. [DOI] [PubMed] [Google Scholar]
- McDowell, J. M., M. Dhandaydham, T. A. Long, M. G. Aarts, S. Goff et al., 1998. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10: 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R. W., I. Paran and R. V. Kesseli, 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., B. C. Meyers, R. W. Michelmore and B. S. Gaut, 2002. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12: 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitthenner, A. F., M. Hobe and R. G. Bhat, 1994. Phytophthora sojae races in Ohio over a 10-year interval. Plant Dis. 78: 269–276. [Google Scholar]
- Song, Q. J., L. F. Marek, R. C. Shoemaker, K. G. Lark, V. C. Concibido et al., 2004. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109: 122–128. [DOI] [PubMed] [Google Scholar]
- Sudupak, M. A., J. L. Bennetzen and S. H. Hulbert, 1993. Unequal exchange and meiotic instability of disease-resistance genes in the Rp1 region of maize. Genetics 133: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E. W. B., G. Lazarovits, C. H. Unwin and R. I. Buzzell, 1979. Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megasperma var. sojae. Phytopathology 69: 951–955. [Google Scholar]
- Weng, C., K. Yu, T. R. Anderson and V. Poysa, 2001. Mapping genes conferring resistance to Phytophthora root rot of soybean, Rps1a and Rps7. J. Hered. 92: 442–446. [DOI] [PubMed] [Google Scholar]
- Wrather, J. A., W. C. Stienstra and S. R. Koenning, 2001. Soybean disease loss estimates for the United States from 1996 to 1998. Can. J. Plant Pathol. 23: 122–131. [Google Scholar]
- Yu, Y. G., G. R. Buss and M. A. Saghai Maroof, 1996. Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc. Natl. Acad. Sci. USA 93: 11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]