Abstract
Phosphoinositide-dependent protein kinase 1 (PDK1) plays a central role in cellular signaling by phosphorylating members of the AGC family of kinases. This family includes protein kinase C (PKC), protein kinase B (PKB), p70/p90 ribosomal S6 kinases (RSK and S6K), and the catalytic subunit of cAMP-dependent protein kinase (PKA). Although PDK1 phosphorylates and activates PKC, PKB, and RSK in vivo, PDK1 regulation of PKA remains controversial. We isolated ksg1, the fission yeast ortholog of mammalian PDK1, as a suppressor of growth defects caused by loss of the stress-activated MAP kinase, Spc1. Here, we demonstrate that Ksg1 is required for activation of PKA. Cells containing the ksg1.12 thermolabile allele exhibit pleiotropic phenotypes, including the failure to arrest in G1 and an inability to conjugate. The ksg1.12 allele strongly suppresses defects associated with unregulated PKA. Pka1, the catalytic subunit of cAMP-dependent protein kinase, is phosphorylated in vivo at Thr-356, which is located in the activation loop of the kinase and corresponds to Thr-197 in mammalian PKA. Phosphorylation of Thr-356 is required for in vivo activation of Pka1 and is dependent upon Ksg1. These data provide experimental evidence that PKA is a physiological substrate for PDK1.
A recurrent theme in biological systems is regulation of signal transduction cascades by protein phosphorylation. Diverse mechanisms regulate kinase activity and protein kinases are themselves often controlled by phosphorylation.
All eukaryotic protein kinases are related through a conserved catalytic core that has a bilobal composition. The smaller amino-terminal lobe is responsible for nucleotide binding; the large lobe participates in substrate binding and catalysis. The large lobe contains an activation segment. Kinase activity is often regulated by an upstream kinase that phosphorylates specific amino acid residues in the activation segment. Phosphorylation likely positions the loop for substrate recognition and for catalysis (Hagopian et al. 2001; Prowse and Lew 2001).
cAMP-dependent protein kinase (PKA) is the structural prototype for members of the kinase family (Taylor et al. 1993). The PKA catalytic subunit is an active enzyme held in an inactive state by association with a regulatory subunit. cAMP binding to the regulatory subunit frees and activates the catalytic subunit. The PKA catalytic subunit prepared from animal tissues is phosphorylated on both Thr-197 and Ser-338 (Shoji et al. 1979, 1983; Yonemoto et al. 1993). Ser-338 phosphorylation does not contribute to catalytic activity but full activation depends upon phosphorylation of Thr-197 (Steinberg et al. 1993; Adams et al. 1995). The phosphothreonine-deficient form of the catalytic subunit has reduced affinities for ATP analogs and for its inhibitor peptide; it also has elevated Kms for both ATP and peptide substrates (Steinberg et al. 1993). The kinase responsible for phosphorylation of Thr-197 is controversial but may be PDK1 (Cauthron et al. 1998; Cheng et al. 1998; Moore et al. 2002).
PDK1 activates members of the AGC family of serine/threonine kinases by activation loop phosphorylation (Belham et al. 1999; Peterson and Schreiber 1999; Toker and Newton 2000; Alessi 2001). The AGC family includes all isoforms of protein kinase C (PKC), protein kinase B (PKB), p70/p90 ribosomal S6 kinases (S6K and RSK, respectively), and the catalytic subunit of PKA. Members of this family are structurally related and contain a highly conserved activation loop phosphorylation site, known as the T-loop residue (Hanks and Hunter 1995). For PKA, the T-loop residue corresponds to Thr-197.
In vitro phosphorylation studies identified PDK1 as a kinase capable of phosphorylating and activating PKB in the presence of insulin and other growth factors (Alessi et al. 1996, 1997). Subsequent studies revealed that a number of kinases are in vitro substrates for PDK1, including PKC (Dutil et al. 1998), S6 kinase (Pullen et al. 1998), and PKA (Cheng et al. 1998). Thus, PDK1 has broad substrate specificity for the threonine in the activation loop of members of the AGC family of protein kinases.
The functional significance of PDK1 phosphorylation was investigated using mouse embryonal stem (ES) cells devoid of PDK1 (Williams et al. 2000). Surprisingly, the cells were viable and grew with rates similar to wild-type cells, even though S6K, RSK, and PKB were not activated. PKA was active and contained a phosphorylated T-loop residue in PKD1−/− cells. PDK1 knockout mice revealed that PDK1 is required for embryonic development and viability (Lawlor et al. 2002). Consistent with findings in ES cells, these studies showed that PKB, S6K, and RSK were neither phosphorylated at the conserved threonine nor activated. Phosphorylation and activation of PKA was not affected by the lack of PDK1. These studies have cast doubt on the physiological significance of PDK1-dependent phosphorylation of PKA. PKA differs from other members of the AGC kinase family in that it autophosphorylates its T-loop residue (Shoji et al. 1979, 1983; Yonemoto et al. 1993). In vitro, this phosphorylation is inefficient and may be caused by the high concentrations of kinase used in the in vitro reactions. An activity capable of phosphorylating the catalytic subunit on Thr-197 was purified from protein kinase A-deficient lymphoma cells (Cauthron et al. 1998). This, together with the finding that PDK1 expressed in mouse cells phosphorylates recombinant PKA (Cheng et al. 1998), suggests that a heterologous kinase phosphorylates Thr-197 in vivo and is likely PDK1. However, the significance of PDK1 phosphorylation of PKA remains unclear.
Elements of the mammalian PKA pathway are conserved in Schizosaccharomyces pombe (Young et al. 1989; Devoti et al. 1991; Isshiki et al. 1992; Maeda et al. 1994; Fujita and Yamamoto 1998). In this yeast, PKA has a role in growth, stationary phase, meiosis, and gluconeogenesis. In general, conditions that cause unregulated PKA repress meiosis (Devoti et al. 1991; Mochizuki and Yamamoto 1992) and gluconeogenesis (Hoffman and Winston 1991) and cause cell death upon entry into the G0 stationary phase (Costello et al. 1986; Devoti et al. 1991). Regulation of yeast PKA is similar to that in higher eukaryotes. Its association with the regulatory subunit inactivates the catalytically active C subunit. cAMP binds the regulatory subunit causing dissociation from and activation of the catalytic subunit (Devoti et al. 1991).
The ksg1 gene encodes the fission yeast ortholog of mammalian PDK1 (Niederberger and Schweingruber 1999). We previously isolated a temperature-sensitive allele of ksg1 (ksg1.12) in a screen to identify genetic suppressors of the gluconate growth defect of spc1 mutants (our unpublished data). Spc1 is a fission yeast stress-activated protein kinase (Millar et al. 1995; Shiozaki and Russell 1995). Pka1 is also involved in gluconate utilization. In this report, we present genetic and biochemical evidence indicating that the fission yeast ortholog of PDK1 activates PKA in vivo through phosphorylation of its T loop residue, Thr-356.
MATERIALS AND METHODS
Strains, media, and plasmids:
S. pombe strains are listed in Table 1. The pka1::pka1-3XFLAG ura4+ alleles were derived by integration of ura4 plasmids containing the 3′ coding region of pka1 fused in-frame with a C-terminal 3× FLAG tag. T356A, T356D, and T356E are single amino acid substitutions in the Pka1 protein.
TABLE 1.
S. pombe strains
| Strains | Genotype | Sourcea |
|---|---|---|
| SPB173 | h+ | Lab stock |
| SPB193 | h− | Lab stock |
| SPB229 | h− leu1-32 pck2::Leu2 | T. Toda |
| SPB231 | h− leu1-32 ura4-D18 pck1:: ura4+ | T. Toda |
| SPB54 | h−90 leu1-32 ade6M-216 git6-261 | C. S. Hoffman |
| SPB56 | h− leu1-32 ade6M-210 his7-336 git6-261 | C. S. Hoffman |
| SPB330 | h− ura4-D18 cgs1:: ura4+ | |
| SP870a | h90 ade6-M216 leu1-32 ura4-D18 pka1::pka1-3XFLAG ura4+ | |
| SP870b | h90 ade6-M216 leu1-32 ura4-D18 pka1::pka1(T356A)-3XFLAG ura4+ | |
| SP870c | h90 ade6-M216 leu1-32 ura4-D18 pka1::pka1(T356E)-3XFLAG ura4+ | |
| SP870d | h90 ade6-M216 leu1-32 ura4-D18 pka1::pka1(T356D)-3XFLAG ura4+ | |
| S12.4 | h+ ksg1.12 | |
| S12.5 | h− ksg1.12 | |
| S12.8 | h+ ade6-M216 ksg1.12 | |
| S12.17 | h− ade6-M210 ksg1.12 | |
| S12.32 | h− ura4-D18 ksg1.12 cgs1:: ura4+ |
Except those indicated, all strains were constructed in this study.
Yeast cells were cultured in rich medium (YEA) or minimal medium (EMM) containing 2% glucose (Alfa et al. 1993). Staurosporine sensitivity was tested by growing cells on EMM agar supplemented with the indicated amounts of staurosporine. When required, 2% gluconate was substituted for glucose in EMM agar. SPA medium was used to measure conjugation and sporulation (Moreno et al. 1991).
Flow cytometery:
Wild-type (SPB193) or ksg1.12 cells (S12.5) were grown at 25° in EMM medium to 1 × 107 cells/ml and shifted to EMM-N medium for the indicated times. A total of 1 × 107 cells were fixed and stained in 2 μg/ml propidium iodide (Moreno et al. 1991).
Cell viability in stationary phase:
Cells were cultured at 25° to a density of 1 × 108. Incubation was continued for 6 days. Cell viability was determined by plating a portion of each culture on YEA agar, incubating for 25° for 3 days, and counting colonies.
Conjugation and sporulation:
A total of 1 × 107 cells of opposite mating type (S12.4 and S12.5) were mixed in 100 μl water. Twenty-microliter cells were spotted onto SPA medium. Incubation at 25° was continued for 2 days. Conjugation was scored as “total conjugating cells = 2X zygotes + 1/2 spores.” Sporulation was examined in diploid cells formed by mating S12.8 and S12.17. Sporulation was scored as “total asci forming cells = 2X zygotes with asci + 1/2 spores.” To measure conjugation and sporulation of cells expressing either wild-type or mutant pka1 alleles, SP870a, SP870b, SP870c, and SP870d cells were cultured in YEA to the indicated densities.
Spore germination:
Individual spores were isolated using glusulase (Sigma, St. Louis) treatment. Following extensive washes the spores were counted using a hemocytometer and plated on rich medium (YEA).
Quantitative RT-PCR:
RNA was isolated using glass bead breakage and an RNeasy kit (QIAGEN, Valencia, CA). Total RNA was purified using the DNA-free kit (Ambion, Austin, TX). Quantitative PCR reactions were carried out as described (Peng et al. 2003).
Mutagenesis:
The pka1 alleles T356A, T356D, and T356E were created using a Quickchange site-directed mutagenesis kit (Strategene, La Jolla, CA). Primers were: T356A, 5′ CGTCTCTACTAGCAACTGTTGCGCGCTTTGTGGTACCCCC 3′; T356D, 5′ CGTCTCTACTAGCAACTGTTGTGATCTTTGTGGTACCCCC 3′; T356E, 5′ CGTCTCTACTAGCAACTGTTGTGAGCTCTGTGGTACCCCC 3′.
Cell extract preparation, immunoprecipitation, and Western blot:
Total cell protein was extracted from 2 × 108 cells in a denaturing buffer (10 mm Tris HCl, 0.1 m NaH2PO4, 8 m urea, pH 8.0) using glass bead breakage. Total cell extract (1 mg) was incubated with 20 μl ANTI-FLAG M2 agarose affinity gel (Sigma) for immunoprecipitation of FLAG-tagged Pka1. For some experiments, immunoprecipitates were treated with 0.2 units threonine/serine phosphatase PP1 (New England Biolabs, Beverly, MA). Immunoprecipitates were examined by a Western blot developed with anti-FLAG M2 antibody (Sigma).
RESULTS
Identification of PKA and PKC as potential substrates for Ksg1:
ksg1.12 was identified in a screen designed to obtain suppressors of the inability of spc1::ura4 cells to grow on medium containing gluconate as a sole carbon source. This analysis identified 12 suppressors defining four complementation groups. ksg1.12 was initially chosen for further study because it displayed a temperature-sensitive phenotype. Genetic analysis of the remaining suppressors will be presented elsewhere.
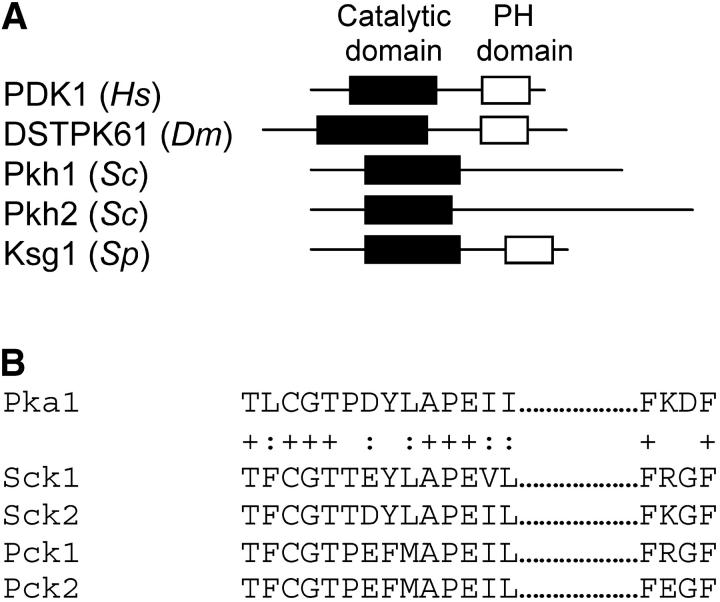
ksg1 encodes the fission yeast ortholog of mammalian PDK1 (Niederberger and Schweingruber 1999). Fission yeast Ksg1 exhibits 46% overall identity with human PDK1. It contains an N-terminal kinase domain that displays 56% identity with the kinase domain of mammalian PDK1. In common with its mammalian and Drosophilia counterparts, fission yeast Ksg1 contains a predicted C-terminal PH domain (Niederberger and Schweingruber 1999; see Figure 1A).
Figure 1.—
(A) A schematic of PDK1-related proteins. The solid box indicates the conserved catalytic domain sequences. The open box shows conserved PH domain sequences. (B) Sequence alignment of the activation loop of putative Ksg1 substrates in fission yeast. (+) identical amino acids; (:) similar amino acids. The accession numbers for the indicated proteins are Pka1, BAA04891 (Maeda et al. 1994); Sck1, S55694 (Jin et al. 1995); Sck2, CAA93901 (Fujita and Yamamoto 1998); Pck1, P36582 (Yonemoto et al. 1993); and Pck2, P36583 (Yonemoto et al. 1993).
All substrates for mammalian PDK1 have two highly conserved regions (Cheng et al. 1998; Moore et al. 2002). One region, within the activation loop, contains the phosphoacceptor threonine. The other is a short, hydrophobic F-X-X-F motif, found at the C terminus of PDK1 substrates (Figure 1B). Substitutions in the F-X-X-F motif abolish substrate function (Moore et al. 2002). To identify potential substrates for Ksg1, we interrogated the fission yeast genome, which has been entirely sequenced (Wood et al. 2002), using both conserved motifs. This analysis identified five potential substrate proteins (Figure 1B). Each is a known kinase. Three (Pka1, Sck1, and Sck2) are highly related to protein kinase A (Fujita and Yamamoto 1998). The remaining two (Pck1 and Pck2) are isoforms of protein kinase C (Toda et al. 1993). Recently, it was reported that Gad8 kinase is a member of the AGC family and an in vitro substrate for Ksg1 (Matsuo et al. 2003). Inspection of Gad8 shows that the activation loop is conserved with respect to PDK1 substrates but the protein does not contain an FXXF motif. It is not known if Gad8 is a physiological substrate for Ksg1.
Characterization of the ksg1 temperature-sensitive allele:
A previous study demonstrated that ksg1 is essential and that cells containing conditional ksg1 alleles display defects in growth; stationary phase induced cell cycle arrest and the ability to conjugate and sporulate (Niederberger and Schweingruber 1999). We conducted a detailed examination of cells containing ksg1.12 with respect to those and other phenotypes. This analysis revealed previously undescribed phenotypes.
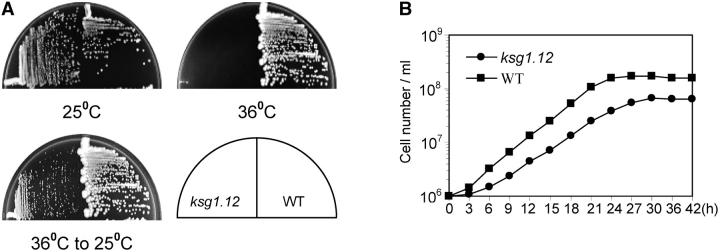
ksg1.12 cells fail to form colonies when incubated at 35°. The cells do not die at the high temperature, but return to growth when the culture is shifted to 25° (Figure 2A). Twenty-five degrees is only a semipermissive temperature for ksg1.12 because, although the cells grow at this temperature, they display mutant phenotypes. At 25°, ksg1.12 cells grow more slowly than wild-type cells (doubling times of 4.5 and 3.0 hr, respectively). Also, ksg1.12 cells consistently enter G0 at a lower density than wild-type cells (Figure 2B).
Figure 2.—
ksg1.12 cells exhibit pleiotropic phenotypes. (A) ksg1.12 cells are sensitive to high temperature. Cells were plated for single colonies and incubated at 25° or at 36° for 3 days. At that time, the plate incubated at 36°, which showed no growth, was transferred to 25°. Incubation was continued for 3 days prior to photography. (B) A growth curve of wild-type (WT; SPB173) and ksg1.12 (S12.4) cells grown in complete medium at 25°.
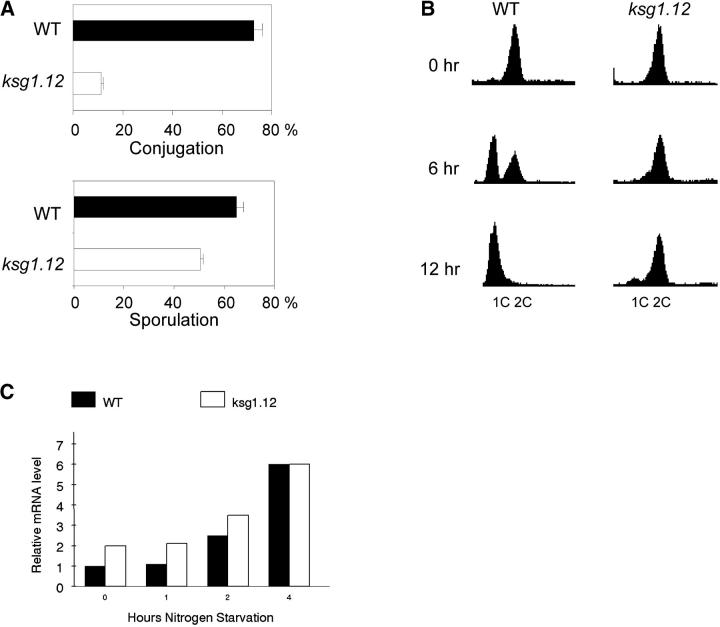
In fission yeast, meiosis is preceded by conjugation and, together, the two processes are considered sexual differentiation. Differentiation requires limiting nutrients and pheromone signaling. Loss of pka1 bypasses the starvation requirement but not the need for pheromone signaling (Maeda et al. 1994). In contrast, unregulated PKA activity prevents conjugation and sporulation (Devoti et al. 1991). As previously reported (Niederberger and Schweingruber 1999), ksg1 is required for efficient conjugation (Figure 3A, top). Interestingly, and in contrast with the results of others (Niederberger and Schweingruber 1999), ksg1.12 diploid cells sporulate nearly as efficiently as wild-type cells (∼50% vs. ∼65%, Figure 3A, bottom). The ksg1 alleles used in this and the published study were isolated independently of one another and the difference in sporulation capability may be allele specific.
Figure 3.—
Sexual development phenotypes of ksg1.12 cells. (A) Conjugation and sporulation frequencies for wild-type (WT) and ksg1.12 cells. Cells were examined microscopically following incubation at 25°. Conjugation was scored by examination of h90 ksg1.12 cells following incubation of SPA medium to determine the number that had formed zygotes or asci. Sporulation was measured by counting asci formation in h+ ksg1.12/h− ksg1.12 diploid cells incubated on SPA medium. (B) Flow cytometery of nitrogen-starved cells at 25°. (C) Quantitative PCR analysis of mei2 mRNA from nitrogen-starved cells. Strains are WT (SPB193) and ksg1.12 (S12.5). Cells were grown at 25° to midlog growth and then shifted to nitrogen-free medium and incubated for the indicated times.
To define the ksg1-mediated pathway that regulates conjugation, the cell cycle distribution of starved ksg1.12 cells was examined. Growing cells have a G2 DNA content but arrest in G1 when deprived of nitrogen (Nurse and Bissett 1981). We found that ksg1 is required for efficient G1 arrest induced by nitrogen starvation (Figure 3B). Conjugation requires G1 arrest as well as transcription of a set of developmental genes that are regulated by an HMG-box protein, Ste11p (Sugimoto et al. 1991). Transcription of ste11 (data not shown) and its direct target, mei2 (Figure 3C), is derepressed by nitrogen starvation in ksg1.12 cells. Thus, the inability of ksg1.12 cells to conjugate is not likely caused by a failure to activate developmental gene expression but by a failure to accumulate in G1 in nutrient-limited medium. Cells containing activated PKA are unable to activate expression of ste11 (Sugimoto et al. 1991). These results indicate that, although loss of ksg1 and activation of pka1 share phenotypes related to differentiation, ksg1.12 only superficially mimics activated PKA. The molecular mechanisms used by each to regulate sexual development are different.
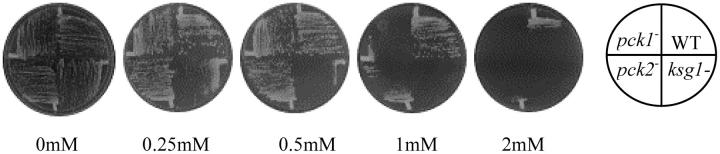
Fission yeast contains two protein kinase C-related genes, pck1 and pck2, which share an overlapping essential function (Toda et al. 1993). Two-hybrid assays indicate that Ksg1 interacts with PCK1 (Graub et al. 2003). Thus, we examined growth of ksg1.12 cells on staurosporine, a potent inhibitor of protein kinase C. ksg1.12 cells are more sensitive to staurosporine than cells containing mutations in either pck1 or pck2 alone (Figure 4).
Figure 4.—
ksg1.12 cells are hypersensitive to staurosporine. WT (SPB173), ksg1.12 (S12.4) pck1− (SPB231), and pck2− (SPB229) cells were plated with the indicated amounts of staurosporine. Photographs were taken after 5 days incubation.
The above observations indicate that ksg1 has functions in common with both the PKA and the PKC pathways. The pleiotropic phenotypes displayed by ksg1.12 cells are consistent with the hypothesis that Ksg1 has multiple molecular targets that regulate growth and differentiation. Absence of PKA or PKC activity causes cells to become very sick or to die. Thus, the extent to which ksg1.12 cells mimic the phenotype of cells devoid of PKA or PKC activity cannot be determined.
Loss of Ksg1 reverses defects caused by unregulated PKA activity:
Clearly, phosphorylation is critical for activation of PKA and the PKA catalytic subunit is an efficient in vitro substrate for PDK1 kinase. However, there are no reports that definitively identify PDK1 as the in vivo activating kinase for PKA. In addition to activation loop phosphorylation, PKA is regulated by association with a regulatory subunit. The Pka1 regulatory subunit in fission yeast is cgs1. In the absence of cgs1, Pka1 activity is not regulated by cAMP and cells display severe defects in sexual differentiation and in survival during G0 (Devoti et al. 1991). We reasoned that, if Ksg1 is required to fully activate Pka1, then defects caused by loss of cgs1 ought to require Ksg1. To investigate this, a ksg.112 cgs1::ura4 strain was constructed. Phenotypes displayed by the double mutant were examined with respect to those displayed by cells carrying single mutations in either gene.
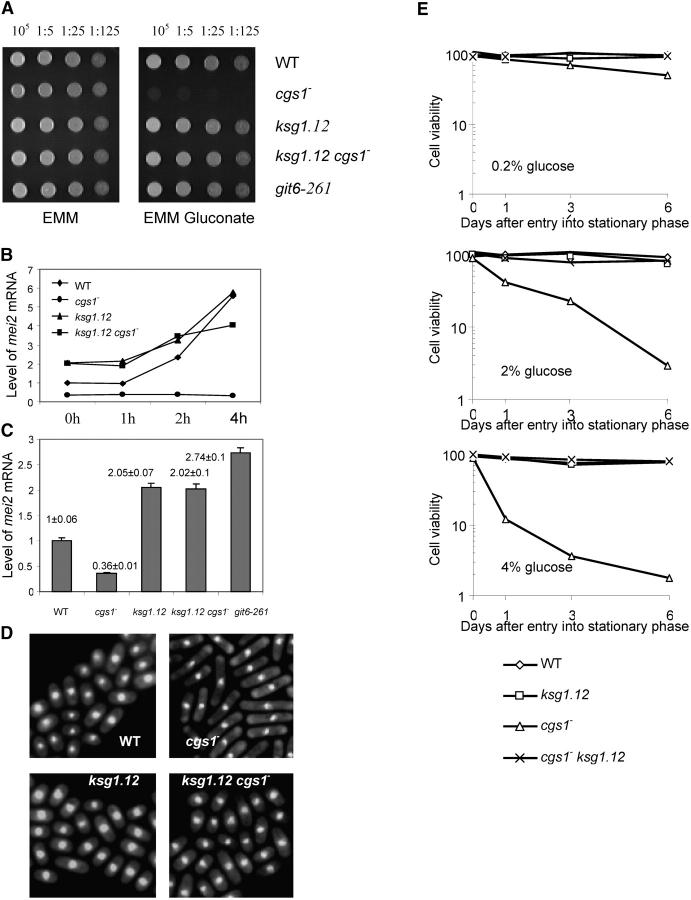
High Pka1 activity has a negative role in transcription of genes required for gluconeogenesis or for sexual development (Hoffman and Winston 1991). Thus, cgs1 cells fail to derepress gluconeogenic enzymes and are unable to utilize gluconate as a carbon source (Caspari 1997). As shown in Figure 5A, both ksg1.12 cells and the pka1− mutant, git6-261 (Byrne and Hoffman 1993; Jin et al. 1995), utilize gluconate as well as wild-type cells. In contrast, cgs1::ura4 cells fail to grow on gluconate. Significantly, loss of ksg1 reverses this phenotype and cgs1::ura4− ksg1.12 cells grow well on gluconate.
Figure 5.—
Loss of ksg1 reverses phenotypes imparted by loss of the PKA regulatory subunit. (A) Photograph of WT (SPB193), cgs1::ura4 (SPB330), ksg1.12 (S12.17), ksg1.12 cgs1::ura4 (S12.32), or pka1− (git6.261;SPB54) cells following growth at 25° on EMM/glucose or EMM/gluconate medium. (B and C) Quantitative PCR analysis of mei2 mRNA from nitrogen-starved (top) or growing (bottom) cells. Strains and conditions correspond to those used in A. (D) Photomicrographs of the indicated cells (described in A) grown to late log phase and stained with Hoechst 33324. (E) Cells were grown to stationary phase (1 × 108 cells/ml) at 25°. Incubation was continued for the indicated number of days, and at each time point, a portion of the culture was removed, counted, and plated onto complete medium to determine the number of viable cells.
To determine if ksg1.12 suppresses other transcription defects of cgs1::ura4 cells, we examined nitrogen-starvation-induced mei2 expression. As shown in Figure 3, ksg1.12 cells fail to conjugate but undergo meiosis in response to nitrogen starvation. Meiosis requires expression of mei2, which is transcriptionally activated by starvation through inactivation of the cAMP-PKA pathway (Watanabe et al. 1988). cgs1::ura4 cells fail to express mei2 when starved (Devoti et al. 1991). ksg1.12 restores mei2 expression to cgs1::ura4 cells (Figure 5, B and C). Significantly, the basal level of mei2 mRNA in pka1− (git6-261), ksg1.12, and cgs1::ura4 ksg1.12 cells was equivalent to that measured in wild-type cells starved of nitrogen for 2 hr. Together, these results demonstrate that, in the absence of the regulatory subunit, activation of Pka1 requires ksg1 with respect to its role in regulation of gene expression.
cgs1::ura4 has only a minor effect on cell growth until nutrients become limiting and cells enter stationary phase. Compared with ksg1.12 or wild-type cells, cgs1::ura4 cells become highly elongated as they approach stationary phase (Devoti et al. 1991). Loss of ksg1 reverses the elongated cgs1-phenotype (Figure 5D). G0 cells are characterized by long-term survivability (Costello et al. 1986). G0 survival of double-mutant cells (ksg1.12, cgs1::ura4) was compared with that of cgs1::ura4 cells. To accomplish this, cells were cultured to stationary phase in three different media, which varied in the amount of carbon source (glucose). At daily intervals, the viability of cells from each culture was determined by plating a portion of each onto fresh medium. Consistent with previous results (Devoti et al. 1991), the long-term survivability of cgs1::ura4 cells decreases as cells remain at G0 to an extent that depends on the composition of the medium. In contrast, continuing survivability of both ksg1.12 and cgs1::ura4 ksg1.12 was excellent in all three media tested (Figure 5E). Taken together, these results indicate that ksg1 is epistatic to cgs1. Formally, then, ksg1 functions either downstream of cgs1 or on a separate, convergent pathway. These data do not establish the epistatic relationship between ksg1 and pka1. However, studies of the mammalian PDK1 show that optimal activation of PKA requires phosphorylation of Thr-197 and this may be accomplished by PDK1. This, together with the above genetic data, is consistent with the hypothesis that Ksg1 is upstream of Pka1.
Phosphorylation of Pka1-Thr-356 is required for its in vivo activation and is dependent on Ksg1:
To determine if activation loop phosphorylation might be a mechanism by which Pka1 activity is downregulated in ksg1.12 cells, we used a Western blot to examine the phosphorylation state of Pka1 and Pka1 variants. Thr-356 in fission yeast Ksg1 corresponds to Thr-197 in mammalian PKA (Figure 1). Strains were constructed to express Pka1 proteins containing substitutions of Thr-356 with alanine (to mimic a nonphosphorylatable site) or aspartic acid or glutamic acid (to mimic constitutive phosphorylation). To detect the Pka1 proteins, each was C-terminally tagged with a triple FLAG epitope.
The density of cells growing in defined medium was measured at various times (Table 2). Cells containing pka1-T36E (SP870c) or pka1-T356D (SP870d) alleles have a doubling time similar to wild-type cells (SP870a). In contrast, the doubling time of cells containing the pka1-T356A allele (SP870b; doubling time = 5.1 hr) is more similar to that of cells containing a loss-of function pka1 allele (git6-261; doubling time = 4.8 hr). Extracts were prepared from the cells and used for immunoprecipitation of the FLAG-tagged Pka1 proteins. An immunoblot of the precipitated material was developed with anti-FLAG M2 antibody. Two Pka1-FLAG bands were detected (Figure 6A, lane 2). This pattern resembles that of mammalian PKA on SDS-PAGE. Phosphorylation of mammalian PKA1 at Thr-197 causes a characteristic reduction in the mobility of the protein in SDS-PAGE (Steinberg et al. 1993; Yonemoto et al. 1993). To determine if the shift represents phosphorylated Pka1, the immunoprecipitated material was treated with phosphatase prior to SDS-PAGE. The upper band disappeared after PP1 treatment and the density of the lower band increased (Figure 6A and data not shown). Thus, the slowly migrating protein is likely modified by phosphorylation. The band shift is dependent on Ksg1 because it is not observed when Pka1 is immunoprecipitated from ksg1.12 cells (Figure 6A; compare lane 7 with lanes 2 and 3). The upper band was also not detected when extract was prepared from cells containing pka1-T36E, pka1-T356D, or pka1-T356A alleles (Figure 6A lanes 4–6). Thus, the band shift is dependent on both ksg1 and the presence of threonine at position 356. These findings suggest that Ksg1 regulates Pka1 through phosphorylation of Thr-356 in vivo.
TABLE 2.
Cell growth rate analysis
| Strain | Relevant genotype | Doubling time (hr) |
|---|---|---|
| SP870a | h90 ade6-M216 leu1-32 ura4-D18 | 3.9 ± 0.1 |
| SP870c | h90 ade6-M216 leu1-32 ura4-D18 pka1::pka1(T356E)-3XFLAG ura4+ | 4.1 ± 0.4 |
| SP870d | h90 ade6-M216 leu1-32 ura4-D18 pka1::pka1(T356D)-3XFLAG ura4+ | 4.2 ± 0.2 |
| SP870b | h90 ade6-M216 leu1-32 ura4-D18 pka1::pka1(T356A)-3XFLAG ura4+ | 5.1 ± 0.2 |
| SPB54 | h90 ade6M-216 leu1-32 git6-261 | 4.8 ± 0.2 |
Cells were grown in minimal medium to a density of 2 × 106/ml and counted every 3 hr for 12 hr. Values are means ±SE for the doubling time of two independent cultures per strain.
Figure 6.—
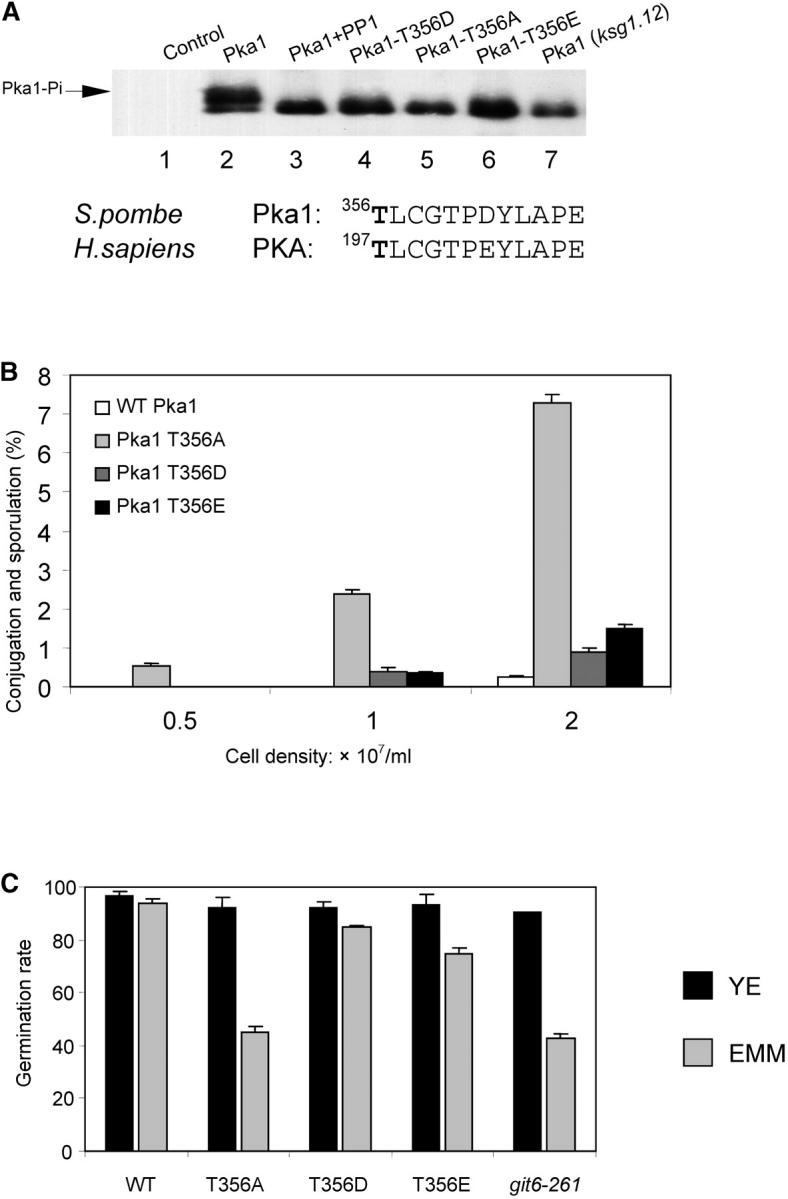
Activation loop phosphorylation of PKA is dependent on Ksg1. (A) Cell extracts were prepared from control cells (lane 1) or cells containing FLAG-tagged Pka1 (lanes 2 and 3), FLAG-Pka-T356D (lane 4), FLAG-Pka-T356A (lane 5), FLAG-Pka-T356E (lane 6), or FLAG-tagged Pka1 and ksg1.12 (lane 7). Cells were grown at 25°. The immunoprecipitated sample shown in lane 3 was treated with protein phosphatase I prior to SDS-PAGE. Epitope-tagged Pka1 proteins were visualized using a Western blot developed with anti-FLAG antibodies. The partial amino acid sequence of fission yeast Pka1 and human PKA is shown below to indicate the threonine phosphorylated by Ksg1/PDK. (B) Conjugation and sporulation was measured in wild-type h90 cells or h90 cells containing alanine (A), aspartic acid (D), or glutamic acid (E) substitutions for the threonine at position 356. Cells were grown to the indicated density in YEA and conjugating pairs or asci were quantitated using a microscope. (C) Spore germination was determined as described in materials and methods. The relevant yeast strains are WT (SP870), Pka1− (SPB56), Pka1 T356A (SP870b), Pka1 T356D (SP870d), and Pka1 T356E (SP870c).
Next, the physiological consequence of Pka1-T356 phosphorylation was examined. Unlike wild-type cells, cells lacking Pka1 conjugate in rich media and the frequency of conjugation and sporulation is cell density dependent (Maeda et al. 1994; Fujita and Yamamoto 1998). Conjugation and sporulation of cells containing pka1-T36A, pka1-T356D, or pka1-T356E alleles were compared with wild-type cells (Figure 6B). This experiment revealed that the pka1-T356A allele caused cells to conjugate and sporulate in a density-dependent manner on rich medium. In contrast, few conjugating or sporulating cells were observed in the wild-type culture (<0.2%). Cells containing pka1-T356D or pka1-T356E alleles displayed a marginally elevated level of differentiating cells compared with wild-type cells. These data demonstrate that Thr-356 is essential for in vivo activation of Pka1 and that substitution of this residue with an alanine inactivates Pka1. In contrast, pka1-T356D and pka1-T356E are partially functional. This is consistent with findings from an in vitro study showing that the catalytic efficiency of mammalian PKA1-T197D and PKA1-T197A mutants is reduced 53-fold and 490-fold, respectively (Adams et al. 1995).
Spore germination is a transition from a quiescent state to a proliferative one and requires Pka activity (Hatanaka and Shimoda 2001). We compared spore germination of cells containing substitutions of Thr-356 with that of either wild-type cells or cells containing a loss-of-function pka1− (git6.261) allele (Figure 6C). In complete, rich medium (YEA), spore germination is high (between 85 and 95%) in wild-type cells, pka1− cells, and all three pka1-Thr-356 substituted alleles. However, in minimal defined medium (EMM) differences between the strains becomes apparent. Compared with wild-type spores, germination of spores containing either pka1.261 or pka1-T356A alleles is reduced by >50%. Significantly, spores derived from cells containing mutations that mimic constitutive phosphorylation of T356 (pka1-T356E and pka1-T356D) have rates of spore germination similar to spores derived from wild-type cells.
These results establish the physiological significance of phosphorylation of Pka1 at Thr-356 within the activation loop of the kinase. In addition, they support the identification of Ksg1 as the kinase that phosphorylates and activates Pka1 in vivo. These data provide a foundation for establishing a mechanistic role for the Thr-356 phosphorylation in activation of PKA.
DISCUSSION
The major finding reported here is that ksg1, the fission yeast ortholog of mammalian PDK1, is a component of the cAMP-regulated pathway. ksg1 is an essential gene and all mutant alleles isolated thus far are thermolabile. Cells containing the ksg1.12 allele exhibit pleiotropic phenotypes, including the failure to arrest in G1 and an inability to conjugate. However, diploid ksg1.12/ksg1.12 cells sporulate, indicating that only some requirements for sexual differentiation are inoperative in ksg1.12 cells. ksg1.12 cells do not die when incubated for periods of time at the restrictive temperature but resume growth when downshifted to a temperature permissive for growth. Phenotypes associated with the ksg1.12 allele differ somewhat from those isolated and characterized by the Schweingruber group. In those studies, cells containing a thermolabile ksg1 allele lyse and die at the restrictive temperature. Additionally, the cells are unable to form spores (Niederberger and Schweingruber 1999; Graub et al. 2003). These differences may be allele specific. Alternatively, they may be due to differences under the experimental growth conditions employed by both studies.
An important phenotype exhibited by cells containing ksg1.12 is that they enter a quiescent state when raised to 36°. At the restrictive temperature, the cells do not grow or divide but remain viable. This state superficially resembles the G0 stationary phase. Fission yeast cells exit the cell cycle and enter G0 when deprived of nutrients. The cells acquire long-term viability, become resistant to various stresses, and express G0-specific genes (Costello et al. 1986; Dimitrov and Sazer 1998). Whether quiescence induced by inactivation of ksg1 is accompanied by these characteristics remains to be determined.
The cAMP-regulated pathway has a major role in the G0 stationary phase. Unregulated PKA activity, such as is measured in cgs1::ura4 cells (cgs1 encodes the regulatory subunit of PKA), causes cells to die in G0. The cells are healthy and viable until they approach stationary phase. At that point, the cells cease division but continue to grow. Since fission yeast grows by length extension, the cells are highly elongated. Eventually, the cells die and are unable to form colonies when plated on fresh medium (Devoti et al. 1991). This indicates a role for the cAMP-regulated pathway in fission yeast life span. In budding and fission yeasts, life span is defined by the methods used to measure it. Replicative life span quantifies the total number of daughter cells generated by a mother cell (Mortimer and Johnston 1959; Barker and Walmsley 1999). Chronological life span, on the other hand, is measured by the ability of stationary cultures to maintain viability over time (Fabrizio et al. 2001). Significantly, ksg1.12 reverses the inability of cgs1::ura4 cells to survive for periods of time at G0. Additionally, the elongated growth phenotype of cgs1::ura4 cells is suppressed by ksg1.12. These results indicate a role for ksg1 in the chronological life span of fission yeast.
In Caenorhabditis elegans, PDK1 is a central element of the insulin/IGF-1-like pathway. This pathway controls metabolism and determines whether animals grow reproductively or hibernate. Hibernation (the dauer diapause state) is induced by pheromone and is accompanied by many morphological changes: growth is arrested, metabolism is shifted to fat storage, and life span increases. Isolation and characterization of long-lived worm mutants show that those related to dauer formation have a marked ability to live longer. Consequently, mutations in pdk-1 result in increased life span (Paradis et al. 1999). Thus, PDK1/Ksg1 may be structurally and functionally conserved between diverse species.
The relationship between ksg1 and the cAMP-regulated pathway extends to other cellular processes regulated by Pka1 activity. Inactivation of cgs1 inhibits conjugation and sporulation, at least in part by preventing expression of meiosis-specific genes (Devoti et al. 1991; Maeda et al. 1994; see Stiefel et al. 2004 for a discussion of cgs1− phenotypes). cgs1::ura4 ksg1.12 cells conjugate and express mei2, a gene that facilitates conjugation and is essential for meiosis. Also, although cgs1::ura4 cells fail to grow on gluconate as a sole carbon source (Caspari and Urlinger 1996), cgs1::ura4 ksg1.12 cells are fully capable of growth on gluconate. These data indicate that cgs1::ura4 and ksg1 are on the same genetic pathway. This, along with the well-established observation that mammalian PKA1 is an in vitro substrate for PDK1 (Cheng et al. 1998), led us to examine phosphorylation of Pka1 as a function of Ksg1 activity.
Mammalian PKA1 is phosphorylated at Thr-197 in its activation loop. Phosphorylation of PKA1 by PDK1 produces a characteristic PKA1 band shift on SDS-PAGE (Cauthron et al. 1998). Using a Western blot, we observed a band-shifted version of Pka1 in wild-type cells that was absent in ksg1.12 cells grown at 25°. The band shift is likely the result of phosphorylation because treatment of cell lysate with phosphatase causes Pka1 to migrate as a single band on SDS-PAGE. PKA1 Thr-197 corresponds to Thr-365 in fission yeast Pka1. Substitution of Thr-365 with phosphomimetic or inactivating mutations results in proteins that do not display the upper-band-shifted version of Pka1. These results indicate that fission yeast Pka1 is phosphorylated on Thr-365 in a manner that is dependent on Ksg1.
In addition to PKA1, mammalian PDK1 activates other members of the AGC family of serine/threonine kinases by activation loop phosphorylation (Belham et al. 1999; Peterson and Schreiber 1999; Toker and Newton 2000; Alessi 2001). Other AGC family includes PKC, AKT/PKB, and p70/p90 ribosomal S6 kinases (S6K and RSK, respectively). The functional significance of the in vivo role of PDK1 was investigated in PDK1 knockout mouse embryonic cells. PDK1−/PDK1− cells contain PKC and PKB that is not phosphorylated. However, these studies showed that PKA was phosphorylated at Thr-365. Thus, the role of PDK1 in phosphorylation of PKA is controversial. To assess the physiological function of Pka1 phosphorylation at its activation loop in fission yeast, we integrated the Thr-365 phosphomimetic and inactivating mutations into the endogenous pka1 locus. Previous studies showed that loss of pka1 has minor effects on cell growth. However, pka1− cells are distinguished from wild-type cells by their density-dependent ability to conjugate. Also, pka1 defective spores germinate poorly on defined media. Pka1-T365A, but not Pka1-T365E, cells mimic pka1− cells. These data indicate that substitution of activation loop Thr-365 does not uniformly lead to reduced Pka1 activity because the phenotypes observed are dependent on the nature of the pka1 substitution. Taken together, these experiments provide strong genetic evidence that Pka1 is a physiological substrate for Ksg1. In addition, they indicate a role for Pka1 and Ksg1 in regulation of chronological aging.
Acknowledgments
We thank C. S. Hoffman (Boston College), M. E. Schweingruber (University of Berne), and T. Toda (London Research Institute) for providing strains. Kun Cai is thanked for help with quantitative PCR. Members of the McLeod laboratory, past and present, are acknowledged for helpful suggestions and discussion. This work was supported by National Institutes Health grant 5RO1GM56875.
References
- Adams, J. A., M. L. McGlone, R. Gibson and S. S. Taylor, 1995. Phosphorylation modulates catalytic function and regulation in the cAMP-dependent protein kinase. Biochemistry 34: 2447–2454. [DOI] [PubMed] [Google Scholar]
- Alessi, D. R., 2001. Discovery of PDK1, one of the missing links in insulin signal transduction. Biochem. Soc. Trans. 29: 1–14. [DOI] [PubMed] [Google Scholar]
- Alessi, D. R., M. Andjelkovic, B. Caudwell, P. Cron, N. Morrice et al., 1996. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15: 6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Alessi, D. R., S. R. James, C. P. Downes, A. B. Holmes, P. R. Gaffney et al., 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase B alpha. Curr. Biol. 7: 261–269. [DOI] [PubMed] [Google Scholar]
- Alfa, C., P. Fantes, J. Hyams, M. McLeod and E. Warbrick, 1993 Experiments With Fission Yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Barker, M. G., and R. M. Walmsley, 1999. Replicative ageing in the fission yeast Schizosaccharomyces pombe. Yeast 15: 1511–1518. [DOI] [PubMed] [Google Scholar]
- Belham, C., S. Wu and J. Avruch, 1999. Intracellular signalling: PDK1—a kinase at the hub of things. Curr. Biol. 9: R93–R96. [DOI] [PubMed] [Google Scholar]
- Byrne, S. M., and C. S. Hoffman, 1993. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 105: 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari, T., 1997. Onset of gluconate-H+ symport in Schizosaccharomyces pombe is regulated by the kinases Wis1 and Pka1, and requires the gti1+ gene product. J. Cell Sci. 110: 2599–2608. [DOI] [PubMed] [Google Scholar]
- Caspari, T., and S. Urlinger, 1996. The activity of the gluconate-H+ symporter of Schizosaccharomyces pombe cells is down-regulated by D-glucose and exogenous cAMP. FEBS Lett. 395: 272–276. [DOI] [PubMed] [Google Scholar]
- Cauthron, R. D., K. B. Carter, S. Liauw and R. A. Steinberg, 1998. Physiological phosphorylation of protein kinase A at Thr-197 is by a protein kinase A kinase. Mol. Cell. Biol. 18: 1416–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X., Y. Ma, M. Moore, B. A. Hemmings and S. S. Taylor, 1998. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc. Natl. Acad. Sci. USA 95: 9849–9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello, G., L. Rodgers and D. Beach, 1986. Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr. Genet. 11: 119–125. [Google Scholar]
- Devoti, J., G. Seydoux, D. Beach and M. McLeod, 1991. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 10: 3759–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov, K., and S. Sazer, 1998. The role of fnx1, a fission yeast multidrug resistance protein, in the transition of cells to a quiescent G0 state. Mol. Cell. Biol. 18: 5239–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutil, E. M., A. Toker and A. C. Newton, 1998. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr. Biol. 8: 1366–1375. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., F. Pozza, S. D. Pletcher, C. M. Gendron and V. D. Longo, 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292: 288–290. [DOI] [PubMed] [Google Scholar]
- Fujita, M., and M. Yamamoto, 1998. S. pombe sck2+, a second homologue of S. cerevisiae SCH9 in fission yeast, encodes a putative protein kinase closely related to PKA in function. Curr. Genet. 33: 248–254. [DOI] [PubMed] [Google Scholar]
- Graub, R., N. Hilti, C. Niederberger and M. E. Schweingruber, 2003. Ksg1, a homologue of the phosphoinositide-dependent protein kinase 1, controls cell wall integrity in Schizosaccharomyces pombe. J. Basic Microbiol. 43: 473–482. [DOI] [PubMed] [Google Scholar]
- Hagopian, J. C., M. P. Kirtley, L. M. Stevenson, R. M. Gergis, A. A. Russo et al., 2001. Kinetic basis for activation of CDK2/cyclin A by phosphorylation. J. Biol. Chem. 276: 275–280. [DOI] [PubMed] [Google Scholar]
- Hanks, S. K., and T. Hunter, 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9: 576–596. [PubMed] [Google Scholar]
- Hatanaka, M., and C. Shimoda, 2001. The cyclic AMP/PKA signal pathway is required for initiation of spore germination in Schizosaccharomyces pombe. Yeast 18: 207–217. [DOI] [PubMed] [Google Scholar]
- Hoffman, C. S., and F. Winston, 1991. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 5: 561–571. [DOI] [PubMed] [Google Scholar]
- Isshiki, T., N. Mochizuki, T. Maeda and M. Yamamoto, 1992. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 6: 2455–2462. [DOI] [PubMed] [Google Scholar]
- Jin, M., M. Fujita, B. M. Culley, E. Apolinario, M. Yamamoto et al., 1995. sck1, a high copy number suppressor of defects in the cAMP-dependent protein kinase pathway in fission yeast, encodes a protein homologous to the Saccharomyces cerevisiae SCH9 kinase. Genetics 140: 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor, M. A., A. Mora, P. R. Ashby, M. R. Williams, V. Murray-Tait et al., 2002. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 21: 3728–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, T., Y. Watanabe, H. Kunitomo and M. Yamamoto, 1994. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J. Biol. Chem. 269: 9632–9637. [PubMed] [Google Scholar]
- Matsuo, T., Y. Kubo, Y. Watanabe and M. Yamamoto, 2003. Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22: 3073–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, J. B., V. Buck and M. G. Wilkinson, 1995. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 9: 2117–2130. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N., and M. Yamamoto, 1992. Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol. Gen. Genet. 233: 17–24. [DOI] [PubMed] [Google Scholar]
- Moore, M. J., J. R. Kanter, K. C. Jones and S. S. Taylor, 2002. Phosphorylation of the catalytic subunit of protein kinase A. Autophosphorylation versus phosphorylation by phosphoinositide-dependent kinase-1. J. Biol. Chem. 277: 47878–47884. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Mortimer, R. K., and J. R. Johnston, 1959. Life span of individual yeast cells. Nature 183: 1751–1752. [DOI] [PubMed] [Google Scholar]
- Niederberger, C., and M. E. Schweingruber, 1999. A Schizosaccharomyces pombe gene, ksg1, that shows structural homology to the human phosphoinositide-dependent protein kinase PDK1, is essential for growth, mating and sporulation. Mol. Gen. Genet. 261: 177–183. [DOI] [PubMed] [Google Scholar]
- Nurse, P., and Y. Bissett, 1981. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature 292: 558–560. [DOI] [PubMed] [Google Scholar]
- Paradis, S., M. Ailion, A. Toker, J. H. Thomas and G. Ruvkun, 1999. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 13: 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., W. Wang, A. Schettino, B. Leung and M. McLeod, 2003. Inactivation of Ran1/Pat1 kinase bypasses the requirement for high-level expression of mei2 during fission yeast meiosis. Curr. Genet. 43: 178–185. [DOI] [PubMed] [Google Scholar]
- Peterson, R. T., and S. L. Schreiber, 1999. Kinase phosphorylation: keeping it all in the family. Curr. Biol. 9: R521–R524. [DOI] [PubMed] [Google Scholar]
- Prowse, C. N., and J. Lew, 2001. Mechanism of activation of ERK2 by dual phosphorylation. J. Biol. Chem. 276: 99–103. [DOI] [PubMed] [Google Scholar]
- Pullen, N., P. B. Dennis, M. Andjelkovic, A. Dufner, S. C. Kozma et al., 1998. Phosphorylation and activation of p70s6k by PDK1. Science 279: 707–710. [DOI] [PubMed] [Google Scholar]
- Shiozaki, K., and P. Russell, 1995. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature 378: 739–743. [DOI] [PubMed] [Google Scholar]
- Shoji, S., K. Titani, J. G. Demaille and E. H. Fischer, 1979. Sequence of two phosphorylated sites in the catalytic subunit of bovine cardiac muscle adenosine 3′:5′-monophosphate-dependent protein kinase. J. Biol. Chem. 254: 6211–6214. [PubMed] [Google Scholar]
- Shoji, S., L. H. Ericsson, K. A. Walsh, E. H. Fischer and K. Titani, 1983. Amino acid sequence of the catalytic subunit of bovine type II adenosine cyclic 3′,5′-phosphate dependent protein kinase. Biochemistry 22: 3702–3709. [DOI] [PubMed] [Google Scholar]
- Steinberg, R. A., R. D. Cauthron, M. M. Symcox and H. Shuntoh, 1993. Autoactivation of catalytic (C alpha) subunit of cyclic AMP-dependent protein kinase by phosphorylation of threonine 197. Mol. Cell. Biol. 13: 2332–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiefel, J., L. Wang, D. A. Kelly, R. T. Janoo, J. Seitz et al., 2004. Suppressors of an adenylate cyclase deletion in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 3: 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, A., Y. Iino, T. Maeda, Y. Watanabe and M. Yamamoto, 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5: 1990–1999. [DOI] [PubMed] [Google Scholar]
- Taylor, S. S., E. Radzio-Andzelm, D. R. Knighton, L. F. Ten Eyck, J. M. Sowadski et al., 1993. Crystal structures of the catalytic subunit of cAMP-dependent protein kinase reveal general features of the protein kinase family. Receptor 3: 165–172. [PubMed] [Google Scholar]
- Toda, T., M. Shimanuki and M. Yanagida, 1993. Two novel protein kinase C-related genes of fission yeast are essential for cell viability and implicated in cell shape control. EMBO J. 12: 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker, A., and A. C. Newton, 2000. Cellular signaling: pivoting around PDK-1. Cell 103: 185–188. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., Y. Lino, K. Furuhata, C. Shimoda and M. Yamamoto, 1988. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 7: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M. R., J. S. Arthur, A. Balendran, J. Van Der Kaay, V. Poli et al., 2000. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 10: 439–448. [DOI] [PubMed] [Google Scholar]
- Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne et al., 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Yonemoto, W., S. M. Garrod, S. M. Bell and S. S. Taylor, 1993. Identification of phosphorylation sites in the recombinant catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 268: 18626–18632. [PubMed] [Google Scholar]
- Young, D., M. Riggs, J. Field, A. Vojtek, D. Broek et al., 1989. The adenylyl cyclase gene from Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 86: 7989–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]