Abstract
Mutation in SGS1, which encodes the yeast homolog of the human Bloom helicase, or in mismatch repair (MMR) genes confers defects in the suppression of mitotic recombination between similar but nonidentical (homeologous) sequences. Mutational analysis of SGS1 suggests that the helicase activity is required for the suppression of both homologous and homeologous recombination and that the C-terminal 200 amino acids may be required specifically for the suppression of homeologous recombination. To clarify the mechanism by which the Sgs1 helicase enforces the fidelity of recombination, we examined the phenotypes associated with SGS1 deletion in MMR-defective and recombination-defective backgrounds. Deletion of SGS1 caused no additional loss of recombination fidelity above that associated with MMR defects, indicating that the suppression of homeologous recombination by Sgs1 may be dependent on MMR. However, the phenotype of the sgs1 rad51 mutant suggests a MMR-independent role of Sgs1 in the suppression of RAD51-independent recombination. While homologous recombination levels increase in sgs1Δ and in srs2Δ strains, the suppression of homeologous recombination was not relaxed in the srs2 mutant. Thus, although both Sgs1 and Srs2 limit the overall level of mitotic recombination, there are distinct differences in the roles of these helicases with respect to enforcement of recombination fidelity.
MITOTIC recombination is critical for the repair of DNA double-strand breaks and is therefore an important mechanism for maintaining genome integrity. The identical (homologous) duplexes of sister chromatids are preferentially used as templates for recombinational repair (Kadyk and Hartwell 1992), as use of nonidentical (homeologous) sequences can lead to sequence changes and chromosomal rearrangements. The mismatch repair (MMR) system, well known for its mismatch detection and repair functions during DNA replication, also plays a key role in the suppression of homeologous recombination (Harfe and Jinks-Robertson 2000a). The genome instability resulting from defects in MMR has been implicated in enhanced cancer susceptibility; specifically, mutations in mismatch repair factors are often seen in tumor cells and are the cause of the cancer syndrome hereditary nonpolyposis colorectal cancer (Fishel and Kolodner 1995; Balogh et al. 2003; Chen et al. 2003).
In yeast, recognition of mismatches in newly replicated DNA is carried out by MutS-like Msh2:Msh3 or Msh2:Msh6 heterodimers (Johnson et al. 1996; Marsischky et al. 1996). Then interaction of the MutL-like Mlh1:Pms1 heterodimer with a Msh2 complex leads to the removal of the nascent strand containing the mismatch (Harfe and Jinks-Robertson 2000b). The relative contributions of these MMR factors to the suppression of homeologous recombination have been examined in detail, and differences between their replication and recombination functions have been noted. Msh2, for example, has greater antirecombination activity than does Mlh1 (Chen and Jinks-Robertson 1999; Nicholson et al. 2000), but the effect of each protein on mutation avoidance appears to be the same. In addition, the Msh2:Msh3 complex, which is not involved in repairing base-base mismatches during DNA replication, has an antirecombination role when the interacting sequences contain only potential base-base mismatches (Nicholson et al. 2000). Finally, mutations in PMS1 that partially uncouple the replication and recombination roles have been identified, suggesting that the steps downstream of mismatch recognition may differ (Welz-Voegele et al. 2002). Although binding of MMR proteins to mismatches present in heteroduplex recombination intermediates may be sufficient to block the progression of recombination between nonidentical substrates, additional factors such as helicases also may be required to actively reject or abort mismatch-containing recombination intermediates (Alani et al. 1994; Chen and Jinks-Robertson 1998). To identify additional factors that enforce the fidelity of the recombination process in yeast, we undertook both a genetic screen and a candidate gene approach. The only non-MMR protein identified in the genetic screen was the helicase Sgs1, which has previously been described as being important in the suppression of homeologous recombination (Myung et al. 2001).
The yeast Sgs1 helicase, the homolog of the Escherichia coli RecQ helicase, has been studied by both biochemical and genetic approaches. Loss of Sgs1 causes sensitivity to DNA-damaging agents, hyperrecombination, and premature aging (Oakley and Hickson 2002). These phenotypes in yeast reflect the syndromes caused by defects in the human RecQ homologs: Bloom (BLM), Werner (WRN), and Rothmund-Thomson (RecQL4) syndromes (Ellis et al. 1995; Yu et al. 1996; Kitao et al. 1998). Although the relevant proteins may play several roles in humans, loss of any one of these proteins is associated with genomic instability (Nakayama 2002). In vitro, Sgs1 binds at the junction of single-strand and double-strand DNA, unwinding the duplex with 3′–5′ polarity with respect to the 3′ single-strand tail (Bennett et al. 1998). Sgs1 can also unwind G4-DNA (four-strand structures stabilized by hydrogen bonds between quartets of guanines; Sun et al. 1998) and DNA:RNA hybrids and can facilitate branch migration of synthetic Holliday junctions (Karow et al. 2000). Finally, physical interactions of Sgs1 have been identified with many proteins, including all three yeast topoisomerases (Top1, Top2, and Top3), as well as with the nucleotide excision repair protein Rad16, the recombination protein Rad51, and the MMR proteins, Mlh1, Msh2, and Msh6 (Gangloff et al. 1994; Duno et al. 2000; Saffi et al. 2000; Pedrazzi et al. 2001; Wu et al. 2001; Gavin et al. 2002). A complex of human Bloom and topoisomerase IIIα proteins can resolve artificial double Holliday junctions in vitro, leading to the suggestion that a similar activity could suppress crossing over in vivo (Wu and Hickson 2003).
In addition to the physical interactions, genetic interactions between SGS1 and several genes involved in DNA metabolism have been demonstrated. Indeed, Sgs1 was originally identified by the ability of sgs1 mutations to suppress the slow growth phenotype associated with topoisomerase III (top3) mutations (Gangloff et al. 1994). In contrast to the improvement of growth of top3 mutants, however, loss of SGS1 causes synthetic growth defects in topoisomerase I (top1) mutants and in mutants of another helicase gene, SRS2 (Lu et al. 1996; Gangloff et al. 2000). Although small effects of TOP3 deletion on homeologous recombination have been reported (Myung et al. 2001), the role of TOP1 in this regulation has not been studied. Srs2, like Sgs1, is a helicase with 3′–5′ polarity whose loss confers a mitotic hyperrecombination phenotype. Srs2 has some homology to E. coli UvrD, but no mammalian homolog has been identified (Rong and Klein 1993). The overlap in the function of Sgs1 and Srs2 that causes the severe growth defect in double mutants is dependent on recombination; the growth defect of the sgs1 srs2 double mutant is rescued when recombination is blocked by mutation of RAD51, RAD52, RAD55, or RAD57 (Gangloff et al. 2000). Biochemical analyses have demonstrated the ability of Srs2 to remove the strand-exchange protein Rad51 from nucleoprotein filaments (Krejci et al. 2003; Veaute et al. 2003). Such an activity not only might serve to limit homologous recombination but also could be involved more specifically in the rejection of homeologous recombination intermediates. Finally, recent genetic studies suggest that both Sgs1 and Srs2 play a role in the suppression of mitotic crossover events (Ira et al. 2003).
The identification of sgs1 mutants in our screen for factors involved in regulating recombination highlights the importance of Sgs1 in genome stability. In the current study, we have explored which domains of Sgs1 are required for its role in suppressing homeologous recombination and find that deletion of the C terminus of Sgs1 causes an increase in homeologous recombination but not homologous recombination. In addition, the relationship between MMR proteins and Sgs1 in the regulation of homeologous recombination was examined in a recombination-proficient RAD background, as well as in strains defective in the strand-exchange protein Rad51 or in the strand-annealing protein Rad59. Results demonstrate that the relative roles of MMR proteins and Sgs1 vary for the different recombination pathways, suggesting that Sgs1 may affect the fidelity of recombination in both MMR-dependent and MMR-independent manners. Furthermore, although mutation of SRS2 or TOP1 generally causes hyperrecombination similar to that seen in sgs1 mutants, neither relieved the suppression of homeologous recombination. These results provide novel insight into the regulation of recombination fidelity and reveal a complex interplay between MMR proteins and non-MMR factors in this process.
MATERIALS AND METHODS
Media and growth conditions:
Yeast strains were grown nonselectively in YEP medium (1% yeast extract, 2% Bacto-peptone, 250 mg/liter adenine; 2% agar for plates) supplemented with either 2% dextrose (YEPD) or 2% glycerol and 2% ethanol (YEPGE). Selective growth was done on synthetic complete (SC) media lacking the appropriate nutrient (Sherman 1991) and supplemented with 2% dextrose (SCD) or 2% galactose, 2% glycerol, and 2% ethanol (SCGGE). Strains mutant at TRP5 were grown on media supplemented with 30 mg/liter tryptophan. Ura− derivatives were identified on medium containing 5-fluoroorotic acid (5-FOA; Boeke et al. 1987). Geneticin- and hygromycin-resistant transformants were isolated on YEPD supplemented with 200 mg/liter geneticin (G418) or 300 mg/liter hygromycin B, respectively. Mutator phenotype was assessed by forward mutation at CAN1 on SC-arginine containing 60 mg/liter canavanine. All incubations were done at 30°.
Strains and plasmids:
A complete list of yeast strains used in this study is given in Table 1. All strains were derived from the congenic strains SJR1486 [MATα ade2-101oc his3Δ200 ura3(Nhe)-(HIS3::intron::cβ2/cβ2)-ura3 lys2ΔRV::hisG leu2K-(lys2Δ5′-lys2Δ3′)-LEU2] and SJR1487 [MATα ade2-101oc his3Δ200 ura3(Nhe)-(HIS3::intron::cβ2/cβ7)-ura3 lys2ΔRV::hisG leu2K-(lys2Δ5′-lys2Δ3′)-LEU2], containing cβ2/cβ2 100%- or cβ2/cβ7 91%-identical inverted repeats, respectively, inserted at the URA3 locus on chromosome V (Spell and Jinks-Robertson 2003). Both strains also contained the 100%-identical lys2Δ5′-lys2Δ3′ inverted repeats, inserted at the LEU2 locus on chromosome XV. The size of the inverted repeats in the HIS3 and in the LYS2 assays are similar (783 vs. 916 bp). Standard genetic techniques were used to disrupt relevant genes. SGS1, SRS2, and TOP1 were disrupted by transformation with PCR-generated kanMX2 cassettes (Wach et al. 1994), while RAD59 was disrupted in sgs1Δ background using a PCR-generated hygMX2 cassette (Goldstein and McCusker 1999). Other disruptions were performed as described (Spell and Jinks-Robertson 2003). To create the msh2 sgs1 strain, MSH2 was disrupted first, and then the single mutant was transformed with pSR505 (obtained from G. Crouse, Emory University), a CEN plasmid containing the wild-type MSH2 gene. Following disruption of SGS1, loss of the plasmid was selected on 5-FOA. This method was employed to prevent the accumulation of suppressor mutations in the background of the double msh2 sgs1 mutant. The presence of each targeted disruption was confirmed by appropriate phenotypic tests and by PCR.
TABLE 1.
S. cerevisiae strains used in recombination rate analysis
| Strain
|
|||
|---|---|---|---|
| Relevant genotype | 100%-identical substrates |
91%-identical substrates |
Source |
| Wild type | SJR1486 | SJR1487 | Spell and Jinks-Robertson (2003) |
| sgs1Δ::kan | SJR1694 | SJR1695 | This study |
| srs2Δ::kan | SJR1704 | SJR1705 | This study |
| top1Δ::kan | SJR1667 | SJR1668 | This study |
| msh2Δ::hisG | SJR1652 | SJR1653 | Spell and Jinks-Robertson (2003) |
| mlh1Δ::hyg | SJR1946 | SJR1947 | Spell and Jinks-Robertson (2003) |
| rad51Δ::URA3 | SJR1551 | SJR1552 | Spell and Jinks-Robertson (2003) |
| rad59Δ::kan | SJR1670 | SJR1554 | Spell and Jinks-Robertson (2003) |
| sgs1Δ::kan rad51Δ::URA3 | SJR1784 | SJR1785 | This study |
| sgs1Δ::kan rad59Δ::hyg | SJR1786 | SJR1787 | This study |
| sgs1Δ::kan msh2Δ::hisG | SJR1724 | SJR1697 | This study |
| srs2Δ::kan msh2Δ::hisG | SJR1708 | SJR1707 | This study |
| sgs1Δ::kan with pRS402 | SJR1716 | This study | |
| sgs1Δ::kan with pRS402-SGS1 | SJR1717 | This study | |
| sgs1Δ::kan with pRS402-sgs1-hd | SJR1718 | This study | |
| sgs1Δ::kan with pRS402-sgs1-ΔN158 | SJR1741 | This study | |
| sgs1Δ::kan with pRS402-sgs1-ΔN158hd | SJR1742 | This study | |
| sgs1Δ::kan with pRS402-sgs1-ΔN322 | SJR1719 | This study | |
| sgs1Δ::kan with pRS402-sgs1-ΔN644 | SJR1720 | This study | |
| sgs1Δ::kan with pRS402-sgs1-ΔC200 | SJR1721 | This study | |
| sgs1Δ::kan with pRS402-sgs1-ΔC300 | SJR1722 | This study | |
| sgs1Δ::kan with pRS402-sgs1-ΔC795 | SJR1723 | This study | |
All strains were derived from the congenic strains SJR1486 and SJR1487 containing cβ2/cβ2 100%- and cβ2/cβ7 91%-identical inverted repeats fused to HIS3, respectively. Both strains also contain the lys2Δ5′-lys2Δ3′ 100%-identical inverted repeats.
The ApaI-SacI fragments containing the SGS1 gene, the helicase-defective allele sgs1-hd (K706A), sgs1-ΔN158, sgs1-ΔN158-hd, sgs1-ΔN322, sgs1-ΔN644, sgs1-ΔC200, sgs1-ΔC300, and sgs1-ΔC795 were isolated from pJM526, pJM511, pJM527, pSM102-hd, pJM528, pJM530, pJM512, pRL5, and pRL1, respectively (Mullen et al. 2000). These fragments were inserted at the ApaI-SacI sites of pRS402, a yeast-integrating plasmid containing ADE2 (Brachmann et al. 1998), creating pSR791, pSR792, pSR803, pSR804, pSR793, pSR794, pSR795, pSR796, and pSR805, respectively. Each plasmid was targeted to the ADE2 locus of SJR1695 (sgs1Δ::kan) by digestion with PflMI, and integration was confirmed by PCR. Correct expression at this locus was inferred by complete rescue of the sgs1Δ MMS-sensitive phenotype upon integration of pSR791.
Screen for mutations that relax the fidelity of recombination:
A genetic screen for loss of suppression of homeologous recombination was performed with strains containing homeologous (his3) and homologous (lys2) recombination substrates, SJR984 or SJR1392 (URA3 trp5 and trp5 relatives of SJR1487, respectively). UV mutagenesis was performed as follows. Cultures were grown overnight in 1 ml of YEPD, washed, plated at 105 cells/plate on YEPD plates, and exposed to UV light for 45 sec. Plates were wrapped in foil and incubated overnight. Cells were then washed off each plate and frozen as independent pools. EMS mutagenesis was performed as follows. Two cultures were grown overnight in 5 ml YEPD, washed with 10 ml sterile water, and resuspended in 10 ml sterile water. Two milliliters of cells were exposed to 3% EMS for 30 min, the EMS was neutralized with an equal volume of 10% sodium thiosulfate, and cells were washed twice with sterile water. Appropriate dilutions were plated on YEPD and SCD-arginine + canavanine to determine viability and the induction of forward mutation at CAN1, respectively. The EMS mutagenesis achieved ∼50–75% killing and a 100-fold increase in mutation frequency. Aliquots from the independent pools of UV- or EMS-mutagenized cells were plated to 100–300 colonies per plate. Colonies were patched to YEPD and grown for 2 days and then replicated to SCGGE-His. Papillation frequency, reflecting recombination between the 91%-identical inverted repeats that resulted in His+ prototrophy, was assessed after 5 days growth on SCGGE-His. Candidates with an elevated number of His+ recombinants were purified from the original YEPD plate and repatched and replica plated to SCGGE-His and SCD-Lys to compare homeologous and homologous recombination frequencies, respectively. Candidates with a qualitative increase in the number of His+ colonies relative to the number of Lys+ colonies were crossed to a bank of tester strains containing mutations in genes shown previously to be involved in the inhibition of homeologous recombination: MSH2, MSH3, MSH6, MLH1, PMS1, RAD1, and SGS1. Failure to complement the elevated homeologous recombination phenotype indicated allelism with the relevant mutant gene. On the basis of the mutation frequency of the CAN1 gene (3.4 × 10−4) and the number of colonies tested (17,413), approximately six candidates for each gene were expected. Mutation of the SGS1 gene in sgs1 candidates was confirmed by sequencing.
Determination of recombination rates:
Cultures inoculated with individual yeast colonies were grown nonselectively in YEPGE and then plated on YEPD, SCD-Lys, and SCGGE-His to determine the total number of viable cells and number of recombinants per culture. A minimum of six cultures of at least two isolates of each strain was tested. Recombination rates (recombinants/cell/generation) were calculated using the method of the median (Lea and Coulson 1949), and the 95% confidence interval for each rate was determined as described previously (Spell and Jinks-Robertson 2004).
RESULTS
Genetic screen for factors that suppress homeologous recombination:
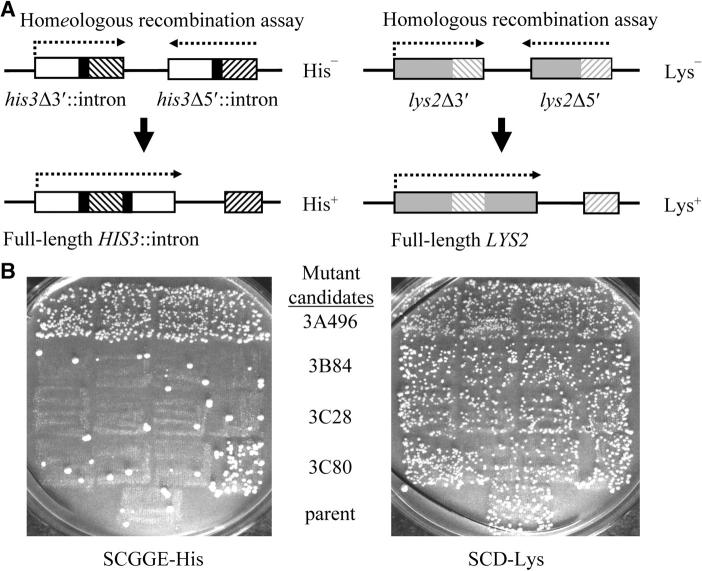
An intron-based inverted repeat assay was used in a genetic screen to identify factors important for the inhibition of homeologous recombination between 91%-identical inverted repeat (IR) substrates fused to HIS3 sequences (Figure 1A). Recombination that reorients the region between the homeologous IR substrates leads to His+ prototrophy. Mutant candidates with elevated homeologous recombination were retested (as represented in Figure 1B) for homologous as well as homeologous recombination to eliminate genes with nonspecific effects on general homologous recombination. The level of homologous recombination was assessed using 100%-identical inverted repeats of the LYS2 gene, with recombination leading to Lys+ prototrophy. The identities of the genes mutated in the candidates with a specific homeologous recombination phenotype were determined by complementation analysis with strains mutant in genes known to be involved in the regulation of homeologous recombination. Expected mutator and UV- and MMS-sensitive phenotypes were confirmed for each identified mutant candidate. The screen was successful in detecting the expected number of mutations in genes known to play a role in the regulation of homeologous recombination; 33 mutants (27 of them known to be independent) were identified, representing six different genes (Table 2). In addition to multiple identifications of the MMR proteins known to be involved in suppressing homeologous recombination, two mutant candidates failed to complement the recombination phenotype of an sgs1Δ strain. Sequencing confirmed the presence of a mutation in SGS1 in each candidate (sgs1-Q325Stop and sgs1-L1150Stop).
Figure 1.—
Inverted repeat assay. (A) Homeologous recombination was assayed using 91% identical cβ2/cβ7 inverted repeats (hatching) fused to intron splice sites (solid boxes) and placed next to the 5′ and 3′ halves of the HIS3 gene (open boxes). Recombination between the repeats that leads to reorientation of the intervening sequence reconstitutes a full-length HIS3 gene and results in a His+ phenotype. Homologous recombination was assayed using overlapping 5′ and 3′ portions of the LYS2 gene (shaded boxes), thus generating 100%-identical repeats in an inverted orientation (shaded hatching). Recombination between the repeats that leads to reorientation of the intervening sequence reconstitutes a full-length LYS2 gene and results in a Lys+ phenotype. Quantitative measurements of homologous recombination could also be measured in strains containing 100%-identical cβ2/cβ2 repeats fused to HIS3 sequences. Similar effects on homologous recombination are seen with the 100% identical LYS2 substrates and the 100% identical HIS3 substrates. (B) Screen for mutants defective in the regulation of homeologous recombination. Representative plates from retests of mutant candidates are shown. The unmutagenized parent (bottom square) and four purified isolates of each mutant candidate were patched (row of four squares) onto YEPD and then replica plated to SDGGE-His (left) and SCD-Lys (right). Prototrophic papillae represent recombinants. Whereas homeologous recombination (His+) is suppressed relative to homologous recombination (Lys+) in the parent strain, only candidate 3A496 exhibited a consistent elevation in homeologous recombination relative to homologous recombination and was later determined to contain a mutation in MSH6.
TABLE 2.
Genes identified in the screen for mutants with elevated homeologous recombination
| Gene | No. of candidates identified |
Additional phenotype |
|---|---|---|
| MSH6 | 9 | Mutator |
| MSH2 | 8 | Mutator |
| PMS1 | 7 | Mutator |
| MLH1 | 6 | Mutator |
| SGS1 | 2 | MMS sensitive |
| RAD1 | 1 | UV sensitive |
Domains of Sgs1 involved in the regulation of homeologous and homologous recombination:
Sgs1 is involved in several processes that impact replication and recombination and that may require interaction with many different proteins (Oakley and Hickson 2002). To determine which domains of Sgs1 are required for the suppression of homeologous recombination and homologous recombination, we quantified the rate of recombination in strains containing a helicase-defective allele (sgs1-hd) or N- and C-terminal deletions of SGS1 (Mullen et al. 2000). The production of protein from sgs1 mutant constructs was previously confirmed by Western analysis (Mullen et al. 2000). For a given type of substrate (homeologous vs. homologous), the recombination rates of the mutants were compared to that of the wild-type strain; this will be referred to throughout as the relative recombination rate. In the sgs1 null mutant (sgs1Δ::kan), the homeologous recombination rate increased more than the homologous recombination rate (71-fold vs. 11-fold, respectively; Table 3). Integration of the wild-type allele (sgs1Δ::kan + SGS1) rescued the recombination and MMS-sensitivity phenotypes of the null mutant (Table 3 and data not shown).
TABLE 3.
Homologous and homeologous recombination rates insgs1 mutants
| Homologous recombination
|
Homeologous recombination
|
|||
|---|---|---|---|---|
| Relevant genotype | Lys+ rate × 10−8 | Relative rate |
His+ rate × 10−8 | Relative rate |
| Wild type | 230 (210–270) | 1.0 | 4.2a (4.0–4.7) | 1.0 |
| sgs1Δ | 2500 (2100–2700) | 11 | 300 (270–310) | 71 |
| sgs1Δ + vector | 1900 (1300–2500) | 8.1 | 170 (130–260) | 41 |
| sgs1Δ + SGS1 | 190 (150–230) | 0.84 | 3.0 (2.4–3.5) | 0.71 |
| sgs1Δ + sgs1-hd | 1500 (1300–1900) | 6.6 | 130 (110–170) | 31 |
| sgs1Δ + sgs1-ΔN158 | 2700 (1400–3400) | 12 | 230 (140–260) | 55 |
| sgs1Δ + sgs1-ΔN158hd | 1900 (1600–2600) | 8.4 | 190 (130–220) | 46 |
| sgs1Δ + sgs1-ΔN322 | 2200 (1600–2800) | 9.7 | 130 (64–260) | 32 |
| sgs1Δ + sgs1-ΔN644 | 1100 (910–1500) | 4.7 | 50 (36–68) | 12 |
| sgs1Δ + sgs1-ΔC200 | 170 (130–190) | 0.72 | 13 (8.5–18) | 3.2 |
| sgs1Δ + sgs1-ΔC300 | 1300 (950–1800) | 5.8 | 75 (61–84) | 18 |
| sgs1Δ + sgs1-ΔC795 | 1000 (870–1100) | 4.3 | 91 (76–110) | 22 |
Homologous recombination (Lys+) and homeologous recombination (His+) in null sgs1Δ::kan mutants (sgs1Δ) containing a wild-type or mutant version of SGS1 integrated at the ADE2 locus were compared to that of the wild-type SGS1 parent. The mutant alleles of SGS1 (full-length 1447 amino acids) contain the helicase-defective allele (K706A) and/or terminal deletions of the indicated number of amino acids from the amino (N) or the carboxy (C) terminus (Mullen et al. 2000). Confidence intervals of 95% are indicated within parentheses. The relative rate was calculated by dividing the mutant rate by the wild-type rate.
Rate is from Spell and Jinks-Robertson (2003).
The relative rates of homologous and homeologous recombination in a strain containing the helicase-defective allele (sgs1-hd) were indistinguishable from those of a null mutant, indicating that the helicase activity of Sgs1 is required for its role in maintaining the fidelity of recombination (Table 3). Each of the deletion constructs caused a loss of the fidelity of recombination similar to that seen in the null mutant, as indicated by a greater increase in the relative level of homeologous recombination (His+) than in the relative level of homologous recombination (Lys+). Interestingly, one deletion mutant (sgs1-ΔC200) displayed an unusual phenotype of an increase in homeologous recombination (3.2-fold relative to wild type) but not of homologous recombination (0.72-fold relative to wild type). Thus, the sgs1-ΔC200 mutant displays a loss of recombination fidelity without a general hyperrecombination phenotype.
Epistasis relationship between SGS1 and mismatch repair genes:
To characterize the relationship between MMR and Sgs1 in the regulation of homeologous recombination, we tested the effect of Sgs1 loss in strains disrupted for the MutS homolog, MSH2, or the MutL homolog MLH1 (Table 4). To compare recombination between homeologous and homologous substrates that differ only in sequence identity, we compared the recombination rates for strains containing 91%-identical substrates fused to HIS3 sequences to strains containing 100%-identical substrates fused to HIS3 sequences, respectively. The differential effect on the inhibition of homeologous recombination is best described by the ratio of the relative increase in homeologous recombination vs. the relative increase in homologous recombination; this will be referred to throughout as the relative homeologous/homologous ratio. The relative homeologous/homologous ratio increased 11-fold upon loss of Sgs1 (Table 4). The stimulation of recombination between homologous substrates in the sgs1 msh2 and sgs1 mlh1 strains (6.5- and 7.6-fold increases in the relative homologous recombination rate, respectively) was similar to that observed in the sgs1 single mutant (6.5-fold increase). Homeologous recombination increased dramatically in the msh2 sgs1 double mutant (230-fold relative to the wild-type strain), but the loss of Sgs1 conferred no additional increase in the relative homeologous/homologous ratio above that conferred by the loss of msh2 (e.g., a 35-fold increase in msh2 sgs1 vs. a 34-fold increase in msh2). Although loss of Msh2 caused a larger increase in the homeologous/homologous ratio than did loss of Mlh1 (Chen and Jinks-Robertson 1999; Nicholson et al. 2000), the relationship of an MMR defect and Sgs1 loss was maintained; there was an 18-fold increase in the homeologous/homologous ratio in the mlh1 sgs1 double mutant vs. a 19-fold increase in the mlh1 single mutant. The similarity in the increase in the relative homeologous/homologous ratio in the MMR-defective sgs1 double-mutant strains and the single MMR mutants suggests that the MMR defect is epistatic to sgs1, although additivity cannot be statistically excluded.
TABLE 4.
Homologous and homeologous recombination rates insgs1 mutants defective in MMR
| Homologous recombination (HR)
|
Homeologous recombination (HER)
|
||||
|---|---|---|---|---|---|
| Relevant genotype | His+ rate × 10−8 | Relative rate |
His+ rate × 10−8 | Relative rate |
Relative HER/HR ratio |
| Wild typea | 170 (150–210) | 1.0 | 4.2 (4.0–4.7) | 1.0 | 1.0 |
| sgs1Δ | 1100 (960–1200) | 6.5 | 300 (270–310) | 71 | 11 |
| msh2Δ | 320 (220–470) | 1.9 | 270 (240–320) | 64 | 34 |
| msh2Δ sgs1Δ | 1100 (890–1900) | 6.5 | 960 (860–1400) | 230 | 35 |
| mlh1Δ | 250 (190–290) | 1.5 | 120 (100–130) | 29 | 19 |
| mlh1Δ sgs1Δ | 1300 (1000–1300) | 7.6 | 570 (450–790) | 140 | 18 |
Confidence intervals of 95% are indicated within parentheses. The relative rate was calculated by dividing the mutant rate by the wild-type rate. HR, homologous recombination; HER, homeologous recombination.
Rates are from Spell and Jinks-Robertson (2003).
Relationship between SGS1 and MMR proteins in the RAD51- and RAD59-independent recombination pathways:
Recombination in Saccharomyces cerevisiae is completely dependent on RAD52, but different pathways can be defined by their dependence on RAD51 and RAD59 (Symington 2002). Our previous studies of the regulation of homeologous recombination in the RAD51-independent and RAD59-independent recombination pathways (defined here as recombination occurring in rad51 and rad59 mutants, respectively) have demonstrated different levels of suppression of homeologous recombination in the two pathways (Spell and Jinks-Robertson 2003). RAD59-independent recombination shows more stringent regulation of homeologous recombination than does RAD51-independent recombination, with the relative homeologous/homologous ratio thus decreasing in a rad59 mutant and increasing in a rad51 mutant (Spell and Jinks-Robertson 2003). In addition, the MMR system (specifically Msh2) plays a larger role in regulating homeologous recombination in the RAD59-independent pathway than in the RAD51-independent pathway. Thus, elimination of Msh2 elevates the homeologous/homologous ratio 32-fold in a rad59 background, but only 2.5-fold in a rad51 background.
To understand the importance of Sgs1 for regulating the fidelity of recombination in the different pathways, the suppression of homeologous recombination by Sgs1 was examined in rad51 and rad59 mutant backgrounds. Although homologous recombination increased in both recombination-defective backgrounds when Sgs1 was removed, homeologous recombination increased to a much greater extent than homologous recombination (Table 5). The loss of Sgs1 caused a 53-fold increase in the homeologous/homologous ratio of the RAD59-independent pathway and a 12-fold increase in the homeologous/homologous ratio of the RAD51-independent pathway. In each of the pathways, it should be noted that elimination of Sgs1 elevated homeologous recombination to a level that is statistically equivalent to the level of homologous recombination. In the RAD59-independent pathway the effect of Sgs1 loss was slightly greater than that of Msh2 loss (53- vs. 32-fold, respectively), suggesting some residual regulation of homeologous recombination in a msh2 mutant. In the RAD51-independent pathway, however, elimination of Sgs1 increased the homeologous/homologous ratio to a much greater extent than did elimination of Msh2 (12- vs. 2.5-fold, respectively). This is the reverse of the pattern observed in a wild-type background, where the increase in the homeologous/homologous ratio was greater for the msh2 mutant than for the sgs1 mutant.
TABLE 5.
Homologous and homeologous recombination rates inrad mutants defective forMSH2 vs. SGS1
| Homologous recombination (HR) |
Homeologous recombination (HER) |
||||
|---|---|---|---|---|---|
| Relevant genotype | Rate × 10−8 | Relative rate |
Rate × 10−8 | Relative rate |
Relative HER/HR ratio |
| RADa | 170 (150–210) | 1.0 | 4.2 (4.0–4.7) | 1.0 | 1.0 |
| RAD msh2Δa | 320 (220–470) | 1.9 | 270 (240–320) | 64 | 34 |
| RAD sgs1Δ | 1100 (960–1200) | 6.5 | 300 (270–310) | 71 | 11 |
| rad51Δa | 53 (46–62) | 1.0 | 5 (4.5–5.4) | 1.0 | 1.0 |
| rad51Δ msh2Δa | 77 (45–81) | 1.5 | 18 (15–23) | 3.6 | 2.5 |
| rad51Δ sgs1Δ | 130 (120–180) | 2.5 | 120 (110–130) | 29 | 12 |
| rad59Δa | 76 (64–85) | 1.0 | 1.1 (0.91–1.3) | 1.0 | 1.0 |
| rad59Δ msh2Δa | 64 (57–110) | 0.84 | 30 (25–37) | 27 | 32 |
| rad59Δ sgs1Δ | 430 (320–500) | 5.7 | 330 (270–410) | 300 | 53 |
Confidence intervals of 95% are indicated within parentheses. The relative rate was calculated by dividing the mutant rate by the wild-type rate. HR, homologous recombination; HER, homeologous recombination.
Rates are from Spell and Jinks-Robertson (2003).
Topoisomerase I and Srs2 helicase do not play a role in maintenance of recombination fidelity:
To determine if the genetic interactions found between TOP1 and SGS1 reflect a role for Top1 in the regulation of recombination, we examined the phenotype of top1 strains with respect to homologous and homeologous recombination (Table 6). Deletion of TOP1 had no significant effect on homologous recombination and only a marginal effect on homeologous recombination (1.4- and 2.0-fold increases, respectively). These data suggest a very minor role, if any, of Top1 in the suppression of homeologous recombination.
TABLE 6.
TOP1 andSRS2 are not involved in the suppression of homeologous recombination
| Homologous recombination (HR) |
Homeologous recombination (HER) |
||||
|---|---|---|---|---|---|
| Relevant genotype | Rate × 10−8 | Relative rate |
Rate × 10−8 | Relative rate |
Relative HER/HR ratio |
| Wild typea | 170 (150–210) | 1.0 | 4.2 (4.0–4.7) | 1.0 | 1.0 |
| srs2Δ | 1900 (1600–2200) | 11 | 8.7 (7.8–12) | 2.1 | 0.19 |
| msh2Δa | 320 (220–470) | 1.9 | 270 (240–320) | 64 | 34 |
| msh2Δ srs2Δ | 1100 (670–1600) | 6.5 | 410 (370–560) | 98 | 15 |
| top1Δ | 230 (200–390) | 1.4 | 8.3 (5.2–11) | 2.0 | 1.4 |
Confidence intervals of 95% are indicated within parentheses. The relative rate was calculated by dividing the mutant rate by the wild-type rate. HR, homologous recombination; HER, homeologous recombination.
Rates are from Spell and Jinks-Robertson (2003).
Genetic interactions between SGS1 and SRS2 suggest that both helicases act to prevent recombination (Gangloff et al. 2000). To determine whether Srs2, like Sgs1, acts to suppress homeologous recombination, we measured homeologous and homologous recombination in an srs2 mutant (Table 6). As expected, homologous recombination increased 11-fold when SRS2 was deleted. Unexpectedly, there was a much smaller (2-fold) effect of Srs2 loss on homeologous recombination. Thus, the relative ratio of homeologous/homologous recombination was reduced 5-fold in the srs2 mutant relative to a wild-type strain. In an srs2 background, loss of Msh2 increased the relative homeologous/homologous ratio 79-fold, which is greater than the increase observed upon loss of Msh2 in a wild-type strain (34-fold). This suggests that much of the decrease in the homeologous/homologous ratio observed in an srs2 mutant derives from activity of the MMR system.
DISCUSSION
During double-strand break repair, a single strand from the broken chromosome invades an intact duplex to form heteroduplex DNA. The presence of mismatches in the heteroduplex recombination intermediates either initiates a repair process (gene conversion) or prevents the recombination event from going to completion (antirecombination). Although both outcomes are dependent on mismatch recognition by the MMR machinery, the downstream steps that complete the processes are likely to be different. The helicase Sgs1, for example, has been shown to suppress mitotic recombination between nonidentical (homeologous) sequences (Myung et al. 2001), and yet mutations in this gene have no reported effect on the removal of DNA replication errors. In the experiments reported here, we have characterized the role of Sgs1 in regulating recombination not only in MMR-competent and MMR-defective cells, but also in recombination-defective cells lacking the Rad51 strand-exchange protein or the Rad59 strand-annealing protein.
In an otherwise wild-type strain, the effect of Sgs1 loss on homeologous recombination was stronger than the effect on homologous recombination, resulting in an 11-fold increase in the homeologous/homologous ratio relative to that in a wild-type strain. A helicase-defective allele of Sgs1 produced a phenotype similar to that of a null allele, indicating that the helicase activity of Sgs1 is required not only for the general suppression of recombination (Mullen et al. 2000), but also for the specific regulation of homeologous recombination. However, the loss of the last 200 amino acids of Sgs1 (sgs1-ΔC200) caused an increase in homeologous recombination but not in homologous recombination or MMS sensitivity, suggesting that this allele may represent a separation-of-function allele that distinguishes different potential roles of Sgs1 in replication and the regulation of recombination vs. the specific regulation of homeologous recombination. Recent studies in yeast and humans suggest that this domain may be important for interaction of Sgs1 or BLM with the MMR factor Mlh1 (Langland et al. 2001; Pedrazzi et al. 2001; Gellon et al. 2002). Alternatively, the less severe phenotype of the sgs1-ΔC200 allele could result from an intermediate level of function rather than from a disruption of a specific function. Additional mutational analysis of SGS1 and MMR repair factors will be necessary to uncover the importance of the interaction of these proteins in the regulation of recombination.
Both msh2 and mlh1 appeared to be epistatic to sgs1, with the double mutants exhibiting relative homeologous/homologous ratios higher than that in the sgs1 single mutant but indistinguishable from those in the corresponding msh2 and mlh1 single mutants. In contrast to the epistasis observed in our studies, Myung et al. (2001) previously reported a synergistic effect of Msh2 and Sgs1 loss on the homeologous/homologous recombination rate ratio. The earlier study reported an unexpected decrease in the rate of homologous recombination in the msh2 sgs1 mutant, such that the homeologous recombination rate was actually higher than the homologous rate. This decrease is surprising because both msh2 and sgs1 single mutants display an increase in homologous recombination levels. The unexpected decrease of homologous recombination resulted in an elevated homeologous/homologous ratio that was interpreted as a synergistic increase, leading to the suggestion of independent contributions by Msh2 and Sgs1 to the regulation of homeologous recombination. We have never seen the rate of homeologous recombination exceed that of homologous recombination in any mutant background and suggest that the anomalous homologous recombination rate in the msh2 sgs1 double mutant may explain the discrepancy between the two studies. If one just considers the homeologous recombination rate data of Myung et al. (2001), their results are consistent with an epistatic or additive effect of simultaneous Msh2 and Sgs1 loss on homeologous recombination.
Our data are consistent with a model in which MMR and Sgs1 act in the same pathway to suppress homeologous recombination, and we suggest that the helicase activity of Sgs1 acts downstream of the MMR system, perhaps to unwind heteroduplex recombination intermediates that contain recognized mismatches. Previous studies of the genetic requirements of the regulation of homeologous recombination have demonstrated that both homeologous and homologous recombination are dependent on RAD52 and on either RAD51 or RAD59, suggesting that the recombination mechanism is the same for homeologous recombination as for homologous recombination (Spell and Jinks-Robertson 2003). Although the mechanism is the same, it is possible that the recombination intermediates that form in the presence of Sgs1 are different from those that arise in its absence, with the latter being affected less by the potential mismatches. It should be noted, however, that loss of either Msh2 or Mlh1 elevates the relative homeologous/homologous ratio to a higher level than does loss of Sgs1 alone (Table 4), indicating that some of the MMR-dependent inhibition of homeologous recombination occurs independently of Sgs1. The residual inhibition could involve unwinding by another helicase or could occur by a completely different mechanism. It has been demonstrated, for example, that the E. coli MutS and MutL proteins can block RecA-mediated strand exchange in vitro (Worth et al. 1994). Regardless of whether the effect of Sgs1 on homeologous recombination is direct or indirect, it clearly has an important role in promoting genome stability by preventing inappropriate recombination.
In the presence of the MMR machinery, the homeologous/homologous ratio increased in a rad51 mutant and decreased in a rad59 mutant (Spell and Jinks-Robertson 2003 and Table 5). This suggests that RAD51-independent recombination has less stringent and that RAD59-independent recombination has more stringent identity requirements than does recombination that occurs in the presence of both Rad51 and Rad59. Multiple mechanisms of recombination have been proposed for inverted repeat assays such as the one used here (for a review, see Symington 2002), and this could account for differences observed between wild-type, rad51, and rad59 strains. In a RAD background, the predominant mechanism likely initiates with a Rad51-dependent strand invasion step, followed by an annealing step that may involve Rad59. In a rad51 mutant, a canonical strand invasion reaction cannot take place, and “invasion” of a duplex would presumably occur by annealing between single strands. On the basis of current recombination models, it seems likely that synthesis-dependent strand annealing and/or gene conversion predominate in a RAD strain, while a less efficient version of either or both might occur in the absence of Rad59. In the absence of Rad51, recombination may involve break-induced replication coupled with single-strand annealing.
The rates of homologous and homeologous recombination were equivalent in either the rad59 sgs1 or the rad51 sgs1 background, indicating the complete loss of the suppression of recombination between substrates that are not identical. In these backgrounds, therefore, mismatches seem to exert no detectable negative effect on recombination. While the effects of Sgs1 or Msh2 loss on RAD59-independent homeologous recombination were similar, Msh2 had a much weaker suppressive effect than did Sgs1 on RAD51-independent homeologous recombination, which is in contrast to the stronger effect of Msh2 in the RAD background. These data suggest that the strand invasion recombination intermediates that form in the presence of Rad51 (i.e., in RAD or rad59 strains) are more susceptible to the recombination-editing activity of MMR than are the presumptive strand-annealing intermediates that form in absence of Rad51 (i.e. in rad51 strains). It is possible, for example, that the presence of the third strand in the invasion intermediate favors MMR-initiated reversal of the heteroduplex DNA by Sgs1 helicase activity.
On the basis of the results presented here, we suggest that mismatch binding by a Msh2-containing complex is necessary to suppress homeologous recombination when both Rad51 and Rad59 are present and that part of the suppression is mediated through the helicase activity of Sgs1. Sgs1 seems to be similarly involved in the MMR-dependent suppression of HO-initiated single-strand annealing when the interacting sequences contain potential mismatches (Sugawara et al. 2004). In contrast, Sgs1 can regulate homeologous recombination in a partially Msh2-independent manner when recombination is compromised by loss of either Rad51 or Rad59. Loss of Rad51 or Rad59 could lead to the formation of different recombination intermediates (see above) or may simply retard an otherwise normal recombination process. If, for example, heteroduplex DNA is shorter or forms more slowly in the absence of Rad51 or Rad59, mismatches might have a greater destabilizing effect and could make the heteroduplex a better target for the helicase action of Sgs1. In a wild-type strain, recombination might proceed at such a pace that the MMR machinery is required to efficiently target Sgs1 to reversible recombination intermediates.
Although the Srs2 helicase, like the Sgs1 helicase, exerts a general suppressive effect on mitotic recombination in yeast, studies suggest that recombination-suppressing roles of these two helicases are functionally distinct. It has been proposed that Sgs1 may limit the accumulation of recombination-initiating lesions/structures by promoting replication fork progression (Fabre et al. 2002), while in vitro data suggest that Srs2 may suppress homologous recombination more directly by disassembling Rad51 nucleoprotein filaments (Krejci et al. 2003; Veaute et al. 2003). In addition, both proteins are speculated to have additional roles in processing/resolving recombination intermediates (Fabre et al. 2002; Ira et al. 2003). The data presented here demonstrate that Sgs1 and Srs2 have opposing effects on the regulation of homeologous recombination. Whereas the suppression of recombination between homeologous sequences was reduced in sgs1 mutants, the suppression was enhanced in srs2 mutants. As expected, a srs2 mutant had strong increases in homologous recombination, but had surprisingly weak increases in homeologous recombination, resulting in a fivefold decrease in the homeologous/homologous ratio relative to that in a wild-type strain. There are several potential explanations for the difference in the increases in homologous vs. homeologous recombination in srs2 mutants. First, it is possible that recombination intermediates containing mismatches may require Srs2 to complete the recombination process. Alternatively, the type of recombination that is normally prevented by Srs2 action may be subsequently blocked by other factors that specifically suppress homeologous recombination, such as MMR proteins or Sgs1. In vitro experiments have implicated Srs2 specifically in the prevention of Rad51-dependent recombination, and as noted above, homology requirements for recombination that occurs in the presence of Rad51 are more stringent than those that occur in its absence (Spell and Jinks-Robertson 2003 and Table 5). In the absence of Srs2, RAD51-dependent recombination presumably would be favored, resulting in more stringent homology requirements and, therefore, the observed decrease in the homeologous/homologous ratio. Our results are thus consistent with in vitro observations and provide in vivo support for a role of Srs2 in the disruption of Rad51 nucleoprotein filaments. In addition, the phenotype of msh2 srs2 mutants suggests that much of the prevention of hyperrecombination between homeologous recombination substrates is dependent on mismatch recognition, which also would be expected for RAD51-dependent recombination. The synthetic growth phenotype of sgs1 srs2 double mutants precludes testing of the role of Sgs1 in regulating homeologous recombination in an srs2 mutant background.
In summary, the experiments reported here reveal distinct roles of the Sgs1 and Srs2 helicases in the regulation of recombination fidelity in yeast. We suggest that Sgs1 directly suppresses recombination between homeologous sequences by using its helicase activity to unwind mismatch-containing heteroduplex DNA, although the effect may be more indirect. Data suggest that the suppression of homeologous recombination by Sgs1 can be MMR dependent or MMR independent, depending on the presence of the Rad51 strand exchange and/or Rad59 strand-annealing protein. While a separation-of-function mutant of SGS1 suggests that the specific suppressive effect of Sgs1 on homeologous recombination is distinct from its more general suppressive effect on mitotic recombination, the increased fidelity of recombination in the srs2 mutant likely results from the same activity (i.e., removal of Rad51 from nucleoprotein filaments) that is thought to suppress general recombination.
Acknowledgments
We thank Steven Brill for generously providing sgs1 mutant plasmids; Gray Crouse for MSH2 plasmid; Stacey Jeffries, Zareen Gauhar, and Greg Rosen for technical assistance; and members of the S.J.R. lab for critical reading of this manuscript. This work was supported by a National Research Service Award grant GM20753 (to R. M. Spell) and grant GM38464 (to S. Jinks-Robertson) from the National Institutes of Health.
References
- Alani, E., R. A. G. Reenan and R. D. Kolodner, 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogh, G. A., I. H. Russo and J. Russo, 2003. Mutations in mismatch repair genes are involved in the neoplastic transformation of human breast epithelial cells. Int. J. Oncol. 23: 411–419. [PubMed] [Google Scholar]
- Bennett, R. J., J. A. Sharp and J. C. Wang, 1998. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem. 273: 9644–9650. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., J. Trueheart, G. Natsoulis and G. R. Fink, 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175. [DOI] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Chen, W., and S. Jinks-Robertson, 1998. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol. 18: 6525–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., and S. Jinks-Robertson, 1999. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics 151: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., J. Wang, M. M. Fraig, K. Henderson, N. K. Bissada et al., 2003. Alterations in PMS2, MSH2 and MLH1 expression in human prostate cancer. Int. J. Oncol. 22: 1033–1043. [PubMed] [Google Scholar]
- Duno, M., B. Thomsen, O. Westergaard, L. Krejci and C. Bendixen, 2000. Genetic analysis of the Saccharomyces cerevisiae Sgs1 helicase defines an essential function for the Sgs1-Top3 complex in the absence of SRS2 or TOP1. Mol. Gen. Genet. 264: 89–97. [DOI] [PubMed] [Google Scholar]
- Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon et al., 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83: 655–666. [DOI] [PubMed] [Google Scholar]
- Fabre, F., A. Chan, W. D. Heyer and S. Gangloff, 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99: 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel, R., and R. D. Kolodner, 1995. Identification of mismatch repair genes and their role in the development of cancer. Curr. Opin. Genet. Dev. 5: 382–395. [DOI] [PubMed] [Google Scholar]
- Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur and R. Rothstein, 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff, S., C. Soustelle and F. Fabre, 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25: 192–194. [DOI] [PubMed] [Google Scholar]
- Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch et al., 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gellon, L., M. Werner and S. Boiteux, 2002. Ntg2p, a Saccharomyces cerevisiae DNA N-glycosylase/apurinic or apyrimidinic lyase involved in base excision repair of oxidative DNA damage, interacts with the DNA mismatch repair protein Mlh1p. Identification of a Mlh1p binding motif. J. Biol. Chem. 277: 29963–29972. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. a DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34: 359–399. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. b Mismatch repair proteins and mitotic genome stability. Mutat. Res. 451: 151–167. [DOI] [PubMed] [Google Scholar]
- Ira, G., A. Malkova, G. Liberi, M. Foiani and J. E. Haber, 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. E., G. K. Kovvali, L. Prakash and S. Prakash, 1996. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J. Biol. Chem. 271: 7285–7288. [DOI] [PubMed] [Google Scholar]
- Kadyk, L. C., and L. H. Hartwell, 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow, J. K., A. Constantinou, J. L. Li, S. C. West and I. D. Hickson, 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97: 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao, S., I. Ohsugi, K. Ichikawa, M. Goto, Y. Furuichi et al., 1998. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics 54: 443–452. [DOI] [PubMed] [Google Scholar]
- Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy et al., 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Langland, G., J. Kordich, J. Creaney, K. H. Goss, K. Lillard-Wetherell et al., 2001. The Bloom's syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J. Biol. Chem. 276: 30031–30035. [DOI] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264–285. [DOI] [PubMed] [Google Scholar]
- Lu, J., J. R. Mullen, S. J. Brill, S. Kleff, A. M. Romeo et al., 1996. Human homologues of yeast helicase. Nature 383: 678–679. [DOI] [PubMed] [Google Scholar]
- Marsischky, G. T., N. Filosi, M. F. Kane and R. Kolodner, 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10: 407–420. [DOI] [PubMed] [Google Scholar]
- Mullen, J. R., V. Kaliraman and S. J. Brill, 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K., A. Datta, C. Chen and R. D. Kolodner, 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27: 1–4. [DOI] [PubMed] [Google Scholar]
- Nakayama, H., 2002. RecQ family helicases: roles as tumor suppressor proteins. Oncogene 21: 9008–9021. [DOI] [PubMed] [Google Scholar]
- Nicholson, A., M. Hendrix, S. Jinks-Robertson and G. F. Crouse, 2000. Regulation of mitotic homeologous recombination in yeast: functions of mismatch repair and nucleotide excision repair genes. Genetics 154: 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, T. J., and I. D. Hickson, 2002. Defending genome integrity during S-phase: putative roles for RecQ helicases and topoisomerase III. DNA Repair 1: 175–207. [DOI] [PubMed] [Google Scholar]
- Pedrazzi, G., C. Perrera, H. Blaser, P. Kuster, G. Marra et al., 2001. Direct association of Bloom's syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 29: 4378–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, L., and H. L. Klein, 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268: 1252–1259. [PubMed] [Google Scholar]
- Saffi, J., V. R. Pereira and J. A. Henriques, 2000. Importance of the Sgs1 helicase activity in DNA repair of Saccharomyces cerevisiae. Curr. Genet. 37: 75–78. [DOI] [PubMed] [Google Scholar]
- Sherman, F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–20. [DOI] [PubMed] [Google Scholar]
- Spell, R. M., and S. Jinks-Robertson, 2003. Role of mismatch repair in the fidelity of RAD51- and RAD59-dependent recombination in Saccharomyces cerevisiae. Genetics 165: 1733–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell, R. M., and S. Jinks-Robertson, 2004. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 262: 3–12. [DOI] [PubMed] [Google Scholar]
- Sugawara, N., T. Goldfarb, B. Studamire, E. Alani and J. E. Haber, 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101: 9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., J. K. Karow, I. D. Hickson and N. Maizels, 1998. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273: 27587–27592. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam et al., 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Welz-Voegele, C., J. E. Stone, P. T. Tran, H. M. Kearney, R. M. Liskay et al., 2002. Alleles of the yeast PMS1 mismatch-repair gene that differentially affect recombination- and replication-related processes. Genetics 162: 1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth, L. J., S. Clark, M. Radman and P. Modrich, 1994. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc. Natl. Acad. Sci. USA 91: 3238–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., and I. D. Hickson, 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu, L., S. L. Davies, N. C. Levitt and I. D. Hickson, 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276: 19375–19381. [DOI] [PubMed] [Google Scholar]
- Yu, C. E., J. Oshima, Y. H. Fu, E. M. Wijsman, F. Hisama et al., 1996. Positional cloning of the Werner's syndrome gene. Science 272: 258–262. [DOI] [PubMed] [Google Scholar]