Abstract
It is well established that sequence divergence has an inhibitory effect on homologous recombination. However, a detailed analysis of this relationship is missing for most higher eukaryotes. We have measured the rate of somatic recombination between direct repeats as a function of the number, type, and position of divergent nucleotides in Arabidopsis. We show that a minor divergence level of 0.16% (one mutation in otherwise identical 618 bp) has a profound effect, decreasing the recombination rate approximately threefold. A further increase in the divergence level affects the recombination rate to a smaller extent until a “divergence saturation” effect is reached at relatively low levels of divergence (∼0.5%). The type of mismatched nucleotide does not affect recombination rates. The decrease in the rate of recombination caused by a single mismatch was not affected by the position of the mismatch along the repeat. This suggests that most recombination intermediate tracts contain a mismatch and thus are as long as the full length of the 618-bp repeats. Finally, we could deduce an antirecombination efficiency of ∼66% for the first mismatch in the repeat. Altogether, this work shows some degree of conservation across kingdoms when compared to previous reports in yeast; it also provides new insight into the effect of sequence divergence on homologous recombination.
HOMOLOGOUS recombination (HR) plays a major role in promoting genetic diversity. Ironically, it is also essential for maintaining genome stability in various ways. It ensures proper chromosome segregation by forming a physical link between homologs during meiosis. It enables the accurate repair of potentially lethal DNA double-strand breaks using a homologous donor sequence as template. It also plays a crucial role in controlling the choice of partners during the recombination process. HR between wrongly chosen partner sequences poses a threat to the organism by ways of genome rearrangements. This is an especially challenging problem in higher eukaryotes, which often have repeat-rich genomes. Plant genomes, for example, contain a high proportion of repetitive elements and are often polyploid, containing two or more divergent (homeologous) genomes. Two important physical factors that affect the rate of HR between DNA fragments are sequence length and divergence. The rate of HR was found to increase with the increase in length of the recombining homologous sequences in several organisms (Rubnitz and Subramani 1984; Shen and Huang 1986; Liskay et al. 1987; Ahn et al. 1988; Puchta and Hohn 1991; Deng and Capecchi 1992; Jinks-Robertson et al. 1993; Bell and McCulloch 2003). This increase was often, but not always, linearly dependent on the length.
In general, the rate of HR is lower between divergent sequences than between identical sequences (see review in Modrich and Lahue 1996). A single nucleotide heterology was shown to inhibit recombination in bacteria (Claverys and Lacks 1986), in yeast (Datta et al. 1997), and in mammalian cells (Lukacsovich and Waldman 1999). In most studies in yeast and bacteria, the relationship between recombination and divergence was log linear (Zawadzki et al. 1995; Datta et al. 1997; Vulic et al. 1997). Interestingly, in yeast the first mismatches were shown to have a much stronger inhibitory effect than the additional mismatches (Datta et al. 1997), a phenomenon termed as the “rapid drop-off” of recombination. The mechanism responsible for this rapid drop-off effect is the mismatch repair (MMR) machinery. It was shown that in the absence of MMR activities the DNA of distant species could recombine (Rayssiguier et al. 1989). Similarly, in yeast the rapid drop-off was abolished in MMR mutants (Datta et al. 1997).
In plants, most of our knowledge regarding the effects of sequence divergence on HR rates comes from studies on meiotic recombination between chromosomes or chromosomal segments. For example, homeologous chromosomes in wheat do not normally pair at meiosis except in some pairing mutants (Sears 1976). Such homeologous pairing can give rise to multivalents and to a high proportion of sterile gametes. Studies on interspecific tomato hybrids between Lycopersicon esculentum, the edible tomato, and its wild relatives have also provided evidence for suppression of HR between homeologous chromosomal segments. For example, recombination between chromosomes from L. esculentum and L. pennellii is suppressed in F1 hybrids (Rick 1969). Similarly, when Solanum lycopersicoides (the most distant relative of tomato that can still cross-hybridize) is crossed with L. esculentum, there is an overall reduction in recombination in the hybrid that can reach a 200-fold suppression for some chromosomal segments (Chetelat et al. 2000). The MMR machinery was shown to affect microsattellite stability in Arabidopsis (Leonard et al. 2003) but its possible antirecombination role has not yet been studied.
We devised an intrachromosomal recombination assay in Arabidopsis similar to that designed in yeast (Datta et al. 1997). This enables the first study of intrachromosomal recombination between diverged repeats in plants and also provides an interkingdom comparison of this process. We have, in a systematic manner, studied the effect that the location within the repeat and the type and frequency of nucleotide divergence between repeats has on the rate of somatic recombination. Comparisons to the yeast data suggest a strong conservation in the recombination-divergence relationship between the two kingdoms, as well as several significant differences. Moreover, our analysis estimated that most recombination intermediates were as long as the size of the repeat (618 bp) used to monitor homologous recombination. Finally, we could predict the efficiency of the antirecombination machinery.
MATERIALS AND METHODS
Primers used in the study:
The primers used in this study to amplify various DNA segments of the recombination assay are described in Table 1. This includes the primers used for site-directed mutagenesis.
TABLE 1.
Primers used in this study
| Primer name | Primer sequence (5′ → 3′) |
|---|---|
| 51321 | AGCCGCTCGAGGTCCTGTAGAAACCCC |
| 51322 | GGGGTACCGCGGCCGCATTCCGATCTAGTAACAT |
| 51323 | GCTCTAGAGAGTCAAAGATTCAAATAGAG |
| 51324 | GCTCTAGAGAATTCTGGCCACCACCTGCCAGTCA |
| 51325 | CCGCTCGAGACTGTCTGCTTACATAAACAG |
| 51326 | CCATCGATGGTCATGAGATTATC |
| 51327 | GCGTTGAACTGCGTGATG |
| 51328 | TCAGCAAGCGCACTTACA |
| 51329 | GTGGAATCGATCAGCGTTGGT |
| 51330 | ACCAACGCTGATCGATTCCAC |
| 51331 | GGGTCAACAATCAGGAAGTGAT |
| 51332 | ATCACTTCCTGATTGTTGACCC |
| 51333 | CGCCGAACACGTGGGTGGACGA |
| 51334 | TCGTCCACCCACGTGTTCGGCG |
| 51335 | GTGAACAACGAGCTCAACTGGC |
| 51336 | GCCAGTTGAGCTCGTTGTTCAC |
| 51337 | AAGCGCCTTACAAGAAAGCCGGGCGATTGC |
| 51338 | GCAATCGCCCGGCTTTCTTGTAAGGCGCTT |
| 51339 | GGAATTCATCGCAGCGTAATGCTATACACC |
| 51340 | GGTGTATAGCATTACGCTGCGATGAATTCC |
| 51341 | AGCCGCTCGAGGTCCTGTAGAAACCCC |
| 51342 | GGGGTACCGCGGCCGCATTCCGATCTAGTAACAT |
| 51343 | GCTCTAGAGAGTCAAAGATTCAAATAGAG |
| 51344 | GCTCTAGAGAATTCTGGCCACCACCTGCCAGTCA |
| 51345 | CCGCTCGAGACTGTCTGCTTACATAAACAG |
| 51346 | CCATCGATGGTCATGAGATTATC |
| 51347 | GCGTTGAACTGCGTGATG |
| 51348 | TCAGCAAGCGCACTTACA |
| 51349 | GTGGAATCGATCAGCGTTGGT |
| 51350 | ACCAACGCTGATCGATTCCAC |
| 51351 | GGGTCAACAATCAGGAAGTGAT |
| 51352 | ATCACTTCCTGATTGTTGACCC |
| Dya1 | CGCCGAACACGTGGGTGGACGA |
| Dya2 | TCGTCCACCCACGTGTTCGGCG |
| Dya3 | GTGAACAACGAGCTCAACTGGC |
| Dya4 | GCCAGTTGAGCTCGTTGTTCAC |
Cloning of the intrachromosomal recombination assay constructs:
The experimental construct is similar to that designed by Swoboda et al. (1994). The 1304-bp 5′ GU part (Figure 1) of the uidA gene encoding for the β-glucuronidase enzyme (GUS) was amplified by PCR using PFU DNA polymerase (Promega, Madison, WI) from vector pCHN1DC4B1 (kindly provided by Puchta Holger) using primers 51323 and 51324 (see Table 1). This part of the gene contains the 35S promoter as well as a peptide “leader” sequence, enhancing the activity of the enzyme (part G; Swoboda et al. 1994). It also contains the 618-bp repeat (or “U”) that codes for the 5′ region of the GUS ORF. The 2163-bp 3′ part (see US in Figure 1) of the gene was amplified by PCR from vector pJD330 (kindly provided by Virginia Walbot) using primers 51321 and 51322. This part of the gene contains the 618-bp U repeat, followed by the rest of the GUS gene, and a Nopaline Synthase terminator. Both fragments were purified using a QIAGEN (Valencia, CA) PCR purification kit. Fragment GU was digested by EcoRI and XbaI, run on a gel, extracted, and ligated to a pBluescript II KS plasmid in the same sites, thus creating clone KS-GU. Fragment US was similarly cloned into pBluescript KS II by digestion of insert and vector with KpnI and XhoI, creating clone KS-US. As a spacer between the two repeats, a 965-bp fragment containing the nptII gene (neomycin phosphotransferase), was amplified from plasmid pGREEN 0029 (Hellens et al. 2000) using primers 51325 and 51326. This fragment was purified, digested with XhoI and ClaI, and cloned into the same sites of a Bluescript II KS vector. This resulted in creation of KS-npt. Next, KS-npt was digested with XhoI and HindIII. The resulting 971-bp nptII-containing fragment was gel-purified and ligated into the same sites of clones KS-GU and KS-US. The resulting clones were called KS-GUnpt and KS-nptUS respectively. Next, KS-US was digested with KpnI and XhoI, and the ∼2160-bp fragment containing US was purified and cloned into the corresponding restriction sites of KS-GUnpt. The resulting construct was called KS-GUnptUS. Vector KS-GUnptUS contains two MscI restriction sites (one in the U region of GU and the other in the U region of US). After digestion of the vector with MscI, it was gel-purified and self-ligated, resulting in creation of vector KS-GUS.
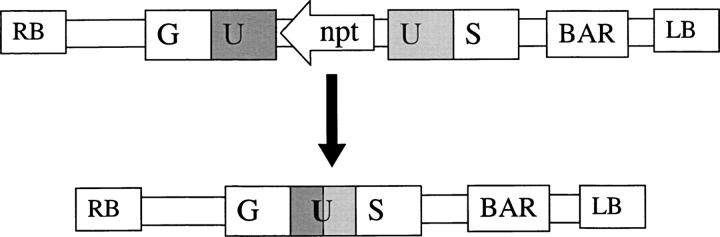
Figure 1.—
Assay for recombination between divergent repeats. A schematic of the assay construct is shown, before (top) and after (bottom) recombination. The GU-npt-US construct (top) contains two overlapping halves of the β-glucuronidase GUS gene, GU and US, with the U (618 bp) overlapping region and the NPTII neomycin phosphotransferase gene (npt) as spacer in between the direct repeats. Mutations are introduced in the U part of the GU half. Homologous recombination between the U divergent repeats gives rise to an active GUS reporter gene (bottom). Such event is recognized as a blue sector upon X-gluc histological staining in the daughters of the cell where HR occurred. RB and LB represent the right and left borders of the pMLBART binary vector, respectively. The npt arrow indicates the gene transcription direction. The BAR gene confers resistance to phosphinothricin (BASTA) and was used for transformation selection.
Site-directed mutagenesis:
The KS-GU construct was used as the template for the site-directed mutagenesis procedure. The procedure follows Stratagene (La Jolla, CA) QuickChange site-directed mutagenesis kit instruction manual using the set of primers 51329–51352 described in Table 1.
Construction of pMLBART-based constructs:
Binary vector pMLBART was kindly provided by Eshed Yuval. It contains spectinomycin resistance for selection in bacteria, BASTA (glufosinate ammonium) resistance for selection in plants, as well as a unique NotI cloning site between the RB and LB. Vector pMLBART-GUnptUS was created from KS-GUnptUS by digestion of KS- GUnptUS with NotI and cloning into the same site of pMLBART. Vectors pMLBART-GUnpt, pMLBART-nptUS and pMLBART-GUS were built in a similar manner by NotI digestion and ligation. Vectors pMLBART-GUnptUS, pMLBART-GUnpt, pMLBART-nptUS, and pMLBART-GUS were kindly provided by Dan Frumkin.
Plant transformation:
Agrobacterium tumefaciens strain Ase (containing kanamycin and chloramphenicol resistance) was kindly provided by Eshed Yuval and was transformed by electroporation using all above pMLBART constructs. For Arabidopsis transformation, cultures of 300 ml were grown. The bacteria were pelleted and resuspended in infiltration medium (10 mm MgCl2, 5% sucrose, 0.044 mm benzylaminopurine, 0.03% Silwet L77 from Lehle Seeds, and 0.112 g B5 vitamin mix from Duchefa). Twenty preanthesis Arabidopsis thaliana plants, ecotype landsberg erecta, were transformed for each construct. The plants (named T0) were dipped in the infiltration medium-Agrobacteria mixture for 5 min and then left to grow. T1 seeds were collected in a pool for each construct.
T1 seeds were sown at a density of ∼5000 seeds per punnet, germinated, and sprayed with a 1:1000 dilution of 5.78% glufosinate-ammonium. This procedure was repeated twice in the first 2 weeks following germination. Surviving plants were transferred into a fresh punnet (4 plants per punnet). Approximately 40 transgenic plants were grown for each construct and T2 seeds were harvested from each plant and mixed, creating the seed pools.
GUS staining:
T2 seedlings were grown in sterile petri dishes on 1/2 Murashige-Skoog (MS) medium plus 2% sucrose. Each petri dish contained 40–50 seedlings. Three weeks after sowing, they were stained in the following manner: First, the seedlings were completely immersed in a working solution consisting of (for every 200 ml) 5 ml 1 m Na2HPO4/NaH2PO4 buffer, pH 7.0, 5 ml 50 mm K3Fe(CN)6, 5 ml 50 mm K4Fe(CN)6, 5 ml 10% Triton, 100 ml 0.5 m EDTA, 179.9 ml H2O, and 10 ml X-gluc stock solution (25 mg/ml in N-N dimethylformamide; X-gluc from Duchefa). Immersed leaves were put in the dark and agitated at 37° for 48 hr. Then leaves were rinsed several times in 70% ethanol solution at 50°. Leaves were taken directly from the ethanol solution for viewing in bright light.
DNA extraction from T2 plants:
Tissue (100 mg) from Basta-resistant seedlings in each T2 punnet was picked, inserted in 2-ml microcentrifuge tubes, and frozen immediately in liquid nitrogen. The DNA was then extracted using the Dneasy Plant Mini kit by QIAGEN according to the manufacturer's instruction.
Data analysis:
Two independent experiments were performed with four petri dishes per construct per experiment. In each experiment, about half of the GUS-stained seedlings were randomly counted for each construct. The distribution of number of spots per seedling is not normal but rather Poisson like. Therefore, per each experiment and each construct, seedlings were randomly grouped into samples of ∼25. The sample mean was used as the basic variable in the statistical analysis that followed.
In preliminary experiments using the zero-mismatches construct we found the measured recombination rate to be highly sensitive to different growing conditions as well as changes in the staining procedure. Indeed, the overall average number of spots measured in each of the two experiments was highly diverse. Nevertheless, we found that the ratio between the values within each experiment batch was highly reproducible. We therefore used the ratio between average values measured for the zero-mismatches construct in each experiment as the factor by which those two experiments were normalized. The ratio between those values for experiment 1 and experiment 2 is 3.88. Each value of experiment 2 was multiplied by this factor.
RESULTS
Design of an assay for recombination between divergent repeats:
The recombination assay that we developed is based on a series of constructs aimed at monitoring the effect of various aspects of sequence divergence on the frequency of somatic recombination between direct repeats. The assay construct contains two overlapping parts of the GUS gene, namely the U repeat as shown in Figure 1. The U repeat is the segment where homologous recombination can occur and where mismatches were inserted. Homologous recombination events between the two direct repeats (U sequences) leads to the formation of the intact GUS gene (Figure 1) and results in a blue sector upon incubation with the enzyme substrate X-gluc. The sector size is indicative of the timing of the recombination event, with early events generating large sectors, late events generating small ones, and germinal events giving rise to plants that are completely blue. The majority of the sectors observed in this work were small, but large sectors or whole recombinant plants were found occasionally.
The U repeat of the GU part was mutated by site-directed mutagenesis, creating sequence divergence between the recombining GU and US parts. The inserted mutations are silent; they change the DNA sequence without changing the amino acid composition of the translated protein. To address the question of how the number of mismatches affects the rate of recombination, a series of constructs was designed containing an increasing number of mutations. The number of mutations inserted was 1, 3, 6, and 10. This corresponds to a divergence level of 0.162%, 0.485%, 0.971%, and 1.618%, respectively. To assess the mutations position effect, the homologous region was conceptually divided into three parts of equal length designated sections A, B, and C (see Figures 2 and 3 for details). The four levels of divergence were independently introduced into each part. This design was aimed at testing whether mismatches in the middle part, which have the greatest reduction in maximal length of identity, also have the strongest inhibitory effect on recombination and whether a recombination tract gradient exists. In addition to these experimental constructs, the following control constructs were generated: 35S-GUS, the positive control, was constructed as described in materials and methods. T1 plants carrying this construct were grown and stained with X-gluc resulting in all-blue plants. GUnpt and nptUS, the two halves of the experimental construct, were used as negative controls. To discard any possibility that the two fragments GU and US had any β-glucuronidase activity, T1 plants carrying half (GU or US) of the assay constructs were stained and examined. None showed any GUS activity.
Figure 2.—
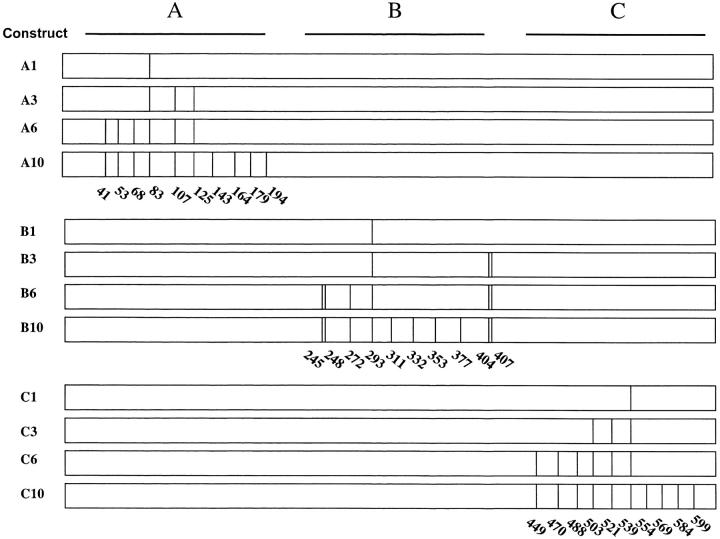
The distribution of mismatches inserted. Schematic of the 618-bp U recombination substrates that were generated by site-directed mutagenesis. Each mismatch location is indicated by a vertical line. The coordinates below the vertical lines correspond to the coordinates within the U repeat. Each construct is named on the left, according to the part of the repeat (A, B, or C) that was mutated and the number of mismatches in the repeat.
Figure 3.—
The type of mismatches inserted. The nucleotide sequence of part U of the GU-npt-US construct is shown for the wild-type sequence (upper strand) and the mutated sequence (lower strand). The clones used in this study contain one or more of the site-directed introduced mutations indicated here by their coordinates.
Rates of recombination between the divergent repeats:
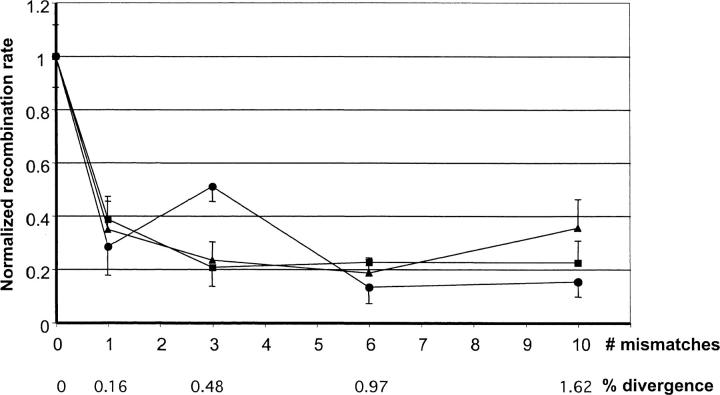
Two independent experiments were performed. The average number of blue spots per seedling was determined for each random sample of ∼25 individual seedlings (the replica unit). The number of blue spots was normalized as described in materials and methods and the average and standard deviation were calculated for each construct (Table 2). The data in Table 2 show that there is a decrease in recombination with the increase in divergence and that the region within the repeat where divergence occurs (zone A, B, or C) does not affect the recombination rate. These data are shown graphically in Figure 4. In general, an inverse relation between the level of divergence and the recombination rate was observed. The inhibitory effect of mismatches on recombination is strongest with the first mismatch and weakens thereafter. Interestingly, a single mismatch difference between the two repeats (corresponding to a 0.16% divergence) causes an approximately threefold decrease in the recombination rate. The next increase in the number of mismatches decreases the recombination rate to a smaller extent. Further increase steps exert no obvious effect on the recombination rate and the curve flattens out. t-tests showed no significant (P < 0.05) differences between the zones (at the same mismatch level), except for constructs A3 and C10 that were significantly different from B3, C3 and A10, B10, respectively (data not shown).
TABLE 2.
Frequency of recombination between divergent direct repeats
| Location within the U repeatb
|
||||
|---|---|---|---|---|
| Degree of divergencea | A zone | B zone | C zone | |
| 0 mismatches (0% divergence) | Normalized average | 1 | 1 | 1 |
| No. of seedlings | 262 | 262 | 262 | |
| SDc | 0.199 | 0.199 | 0.199 | |
| 1 mismatch (0.162% divergence) | Normalized average | 0.284 | 0.386 | 0.349 |
| No. of seedlings | 152 | 156 | 177 | |
| SDc | 0.133 | 0.109 | 0.151 | |
| 3 mismatches (0.485% divergence) | Normalized average | 0.510 | 0.206 | 0.233 |
| No. of seedlings | 185 | 145 | 173 | |
| SDc | 0.076 | 0.096 | 0.093 | |
| 6 mismatches (0.971% divergence) | Normalized average | 0.132 | 0.226 | 0.186 |
| No. of seedlings | 156 | 158 | 181 | |
| SDc | 0.070 | 0.021 | 0.074 | |
| 10 mismatches (1.618% divergence) | Normalized average | 0.153 | 0.225 | 0.355 |
| No. of seedlings | 192 | 160 | 110 | |
| SDc | 0.075 | 0.103 | 0.110 | |
The frequency of recombination events, as estimated by the number of blue spots per seedling, was normal-ized relative to the identity (0 mismatches) construct.
The degree of divergence is expressed as the number of nucleotides that did not match (mismatches) or as the percentage of divergence [(number of mismatches) × 100/618] between the two 618-bp U repeats.
The location within the U repeat is shown in detail in Figure 2.
The standard deviation (SD) was calculated from the variation among the eight samples used in each treatment (mismatch-region combination). Recombination values, in each sample, were based on the average number of spots per seedling in ∼25 seedlings.
Figure 4.—
The effect of sequence divergence on homologous recombination between direct repeats. The normalized recombination rate (y-axis) is calculated from the normalized number of spots per seedling (relative to the zero-mismatch construct) as a function of divergence between the two repeats (x-axis). Sequence divergence is expressed as the number of mismatches in the 618-bp repeat (# mismatches) or as the percentage of mismatches (% divergence). The different segments within the repeats are shown as circles, squares, and triangles for segments A, B, and C, respectively (see Figure 2). Bars represent 1.96 SE.
DISCUSSION
We designed an assay that enabled us to quantify the rate of intrachromosomal recombination between two divergent direct repeats through formation of a recombinant active GUS reporter gene. In principle, GUS reactivation might also be achieved through unequal crossover between sister chromatids or homologous chromosomes or between ectopic sequences in the case of multiple T-DNA insertions. These types of recombination, however, are much less frequent than intrachromosomal recombination (Shalev and Levy 1997; Puchta 1999; Molinier et al. 2004). It is therefore probable that the assay used here measures mainly somatic recombination between divergent repeats in cis.
Sequence divergence and recombination rates—the yeast vs. plant comparison:
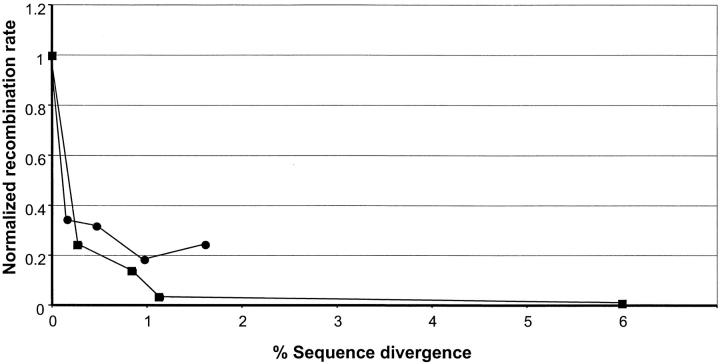
An increase in sequence divergence correlates with a decrease in homologous recombination rates in Arabidopsis. This is similar to the general trends previously reported in other species (see Introduction). However, in a detailed comparison of normalized recombination rates between our Arabidopsis results and similar data from yeast (Datta et al. 1997), one can appreciate the extent of similarity despite the interkingdom distance (Figure 5). We identified the existence of the rapid drop-off in recombination rates as previously reported in yeast (Datta et al. 1997; Chen and Jinks-Robertson 1999). In our experiment, the introduction of a single mismatch lowered the recombination rate by threefold, while additional mismatches reduced the recombination rate to a lesser extent. This threefold drop is similar in magnitude but slightly lower than the approximately fourfold drop reported in yeast (Datta et al. 1997; Chen and Jinks-Robertson 1999). In both species, as divergence increases further beyond the drop-off, the recombination rate levels off (Figure 5). A notable difference between yeast and Arabidopsis is that above 1% divergence, the yeast normalized recombination values level off to almost zero while the plant values also seem to level off, but at a higher level, namely ∼20% from the rate of the identical substrates (Figure 5). This is surprising as plant genomes are laden with repeats so one might expect a stronger inhibitory effect resulting from sequence divergence. It was shown recently that somatic mutation rates are higher in plants than in other organisms, including yeast (Kovalchuk et al. 2000). It is possible that both phenomena, namely the higher normalized recombination rates under any sequence divergence level and the higher somatic mutation rates in plants, are caused by a low efficiency of the plant MMR machinery.
Figure 5.—
Comparison of recombination rates as a function of nucleotide divergence in Arabidopsis and yeast. The two data sets were normalized for comparison relative to the recombination rates obtained in the wild type in absence of mismatches. For the yeast curve (squares), the HR rates for the cβ2a substrate were used on the basis of data from two articles from the Jinks-Robertson group (Datta et al. 1997; Chen and Jinks-Robertson 1999). In cases where the same substrate was used in both yeast articles, the average was calculated. For Arabidopsis (circles) the mean value of the three zones (A, B, and C in Figure 4) is given for each divergence level.
The number of spots in constructs A3 and C10 was somewhat anomalous (Figure 4). The batches of seeds used in our experiments were collected from ∼40 T1 plants having independent integration sites. Therefore, the higher-than-expected recombination rate in these constructs is most probably not caused by a position effect of the construct integration into a recombination hotspot. Moreover, the average number of spots in A3 and C10 was reproducible in different experiments and was not biased by some exceptional plants having extremely high spot counts (data not shown). Another plausible reason for the unique recombination rate for A3 and C10 would be the type of mismatches, but the A3 mismatches are identical to those of B3. Finally, clones A3 and C10 were resequenced from the transgenic plants that contained them to confirm that there was no mutation or cross-contamination. The odd behavior of these clones seems, therefore, to have a real biological basis that is not understood and would be worth investigating. There have been similar reports in mammalian cells showing that the combination of certain mismatches might have an unexpected effect on gene targeting rates (Lukacsovich and Waldman 1999).
Mismatch position had no effect—implications on the length of the recombination intermediate tracts:
The direct repeat sequence was divided into three equal-length segments and the effect of the segment type (A, B, or C) on the rate of recombination was measured for every level of heterogeneity (Figure 4). Overall, the location of the same number of mismatches in different zones did not change the recombination rate. This positional neutrality is most likely the result of a heteroduplex intermediate tract that covers most (if not all) of the 618-bp repeat. The cases of A1, B1, and C1 with a single mismatch between repeats are particularly instructive (see examples in Figure 6). Had the intermediate tract been shorter than the repeat length, e.g., half of it, and dispersed around the center, we would expect the mismatch in B1 (the central part) to be included in most recombination intermediates while A1 and C1 (the distal parts) would be less frequently included and thus B1 would have the strongest antirecombination effect. The distance between the A1 mismatch and the C1 mismatch is 456 bp that covers ∼75% of the repeat length. Had the intermediate tract been very short, e.g., tens of base pairs, then only a small fraction of all the recombination events would contain mismatches and a single mismatch would not have had such a profound effect (the threefold rapid drop-off) on the rate of recombination. Hence, the most plausible explanation for the similar recombination drop in A1, B1, and C1 is that the length of the intermediate tract is close to the total repeat length. Therefore, no matter what is the position of the mismatches along the repeat, they will be included in the heteroduplex and affect the recombination rate to the same extent. There are two scenarios for the inclusion of the mismatch in the intermediate (Figure 6). The mismatched base pair can be formed as a result of strand invasion (Figure 6C). Alternatively, it can be formed after strand invasion and Holliday junction formation, as a result of branch migration (e.g., Figure 6B). The latter, in Neurospora asci would give the 4:4 aberrant ratio (in the absence of mismatch repair) while the former would give the 5:3 ratio (in the absence of mismatch repair). In plants it is still not possible to distinguish between these possibilities. Several alternative models of recombination between direct repeats are not discussed here (Prado et al. 2003). Interestingly, it was shown recently for one of these mechanisms, the single-strand annealing pathway, that the MMR machinery is also involved in rejection of heteroduplex DNA (Sugawara et al. 2004).
Figure 6.—
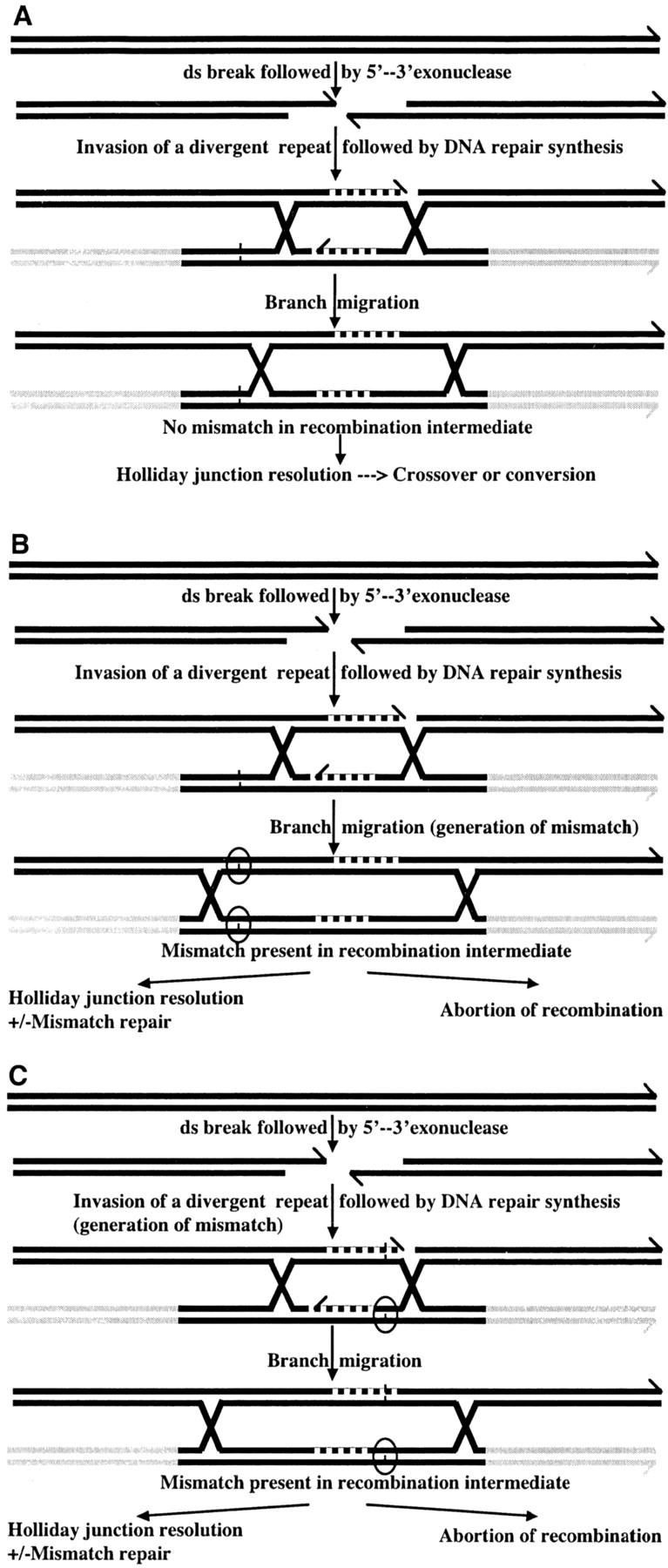
Examples of recombination intermediates formed upon recombination between divergent repeats. Recombination is shown here following the yeast DNA double-strand-break repair model (Szostak et al. 1983). It is initiated by a double-strand (ds) break, followed by 5′–3′ exonuclease activity and formation of 3′ overhangs. The 3′ ends (shown by a diagonal line) can invade a homologous template and extend through DNA synthesis. In the examples shown here the repeat is identical to the invading strand in the region shown by the black line, except for a single nucleotide polymorphism shown as a small vertical line. Beyond the repeat, the regions shown in gray are not homologous. Following strand invasion, a Holliday junction is formed and extends via branch migration to form the final recombination intermediate. In A, the polymorphic nucleotide is not included in the recombination intermediate and no mismatch nucleotide pairs are formed. In B and C, mismatch nucleotide pairs are formed (indicated by the circles), either as a result of branch migration (B) or directly through strand invasion (C). Mismatches can cause abortion of the recombination process or recombination might proceed and the recombination intermediate might be resolved, with or without mismatch repair, giving rise to gene conversion or crossover products.
Prediction of the mismatch-mediated antirecombination efficiency:
On the basis of the above conclusion that the recombination intermediate tract covers most if not all the repeat length, we can predict the efficiency of the mismatch-mediated antirecombination machinery. We refer here to the term “antirecombination efficiency” as the probability of recombination rejection given that one mismatch exists in the heteroduplex. Therefore, one may express the probability of recombination rejection P(rec-reject), in our case ∼0.66 with one mismatch, as the multiplication of the probability of mismatch formation [P(mmf)] by the antirecombination efficiency (E): P(rec-reject) = P(mmf) × E. Given the above deduction that the recombination intermediate tract covers almost all the length of the repeat, this means that P(mmf) = ∼1 and thus implies that the efficiency of the antirecombination system (E) is ∼0.66.
Mismatch composition does not affect the antirecombination effect:
Our work clearly shows that differences in mismatch composition do not alter the recombination rate, at least for the mismatch compositions examined. For example, the C1 construct C/G mismatch differs from the A1 and B1 constructs T/C mismatch, yet all showed the same decrease in HR frequency. Therefore, a C/G mismatch is recognized by the antirecombination machinery just as efficiently as a T/C mismatch. Similarly, the comparison of B3 and C3 shows that an A/G mismatch is recognized as efficiently as an A/C mismatch. The three constructs A6, B6, and C6 differ in only one mismatch type: C/T in A6, G/A in B6, and T/G in C6; but the recombination rate is again the same.
Models and mechanisms:
It is interesting to assess our findings in light of existing models that address the relation between divergence and recombination rate. One such model is the minimal efficient processing segment (MEPS) concept, originally suggested for Escherichia coli recombination and later adopted for other organisms (Shen and Huang 1986).
The MEPS model predicts an exponential decrease in recombination rate in relation to the level of divergence. Datta et al. (1997) extended the original MEPS theory to explain the rapid drop-off by assuming that if the heteroduplex has elongated less than β base pairs before encountering the first mismatch then the MMR machinery is always triggered and otherwise it is triggered with a probability R0. Another parameter, ℱ, was introduced to denote the probability of the recombination event being rejected following the MMR trigger. As discussed above, our results are similar to those for yeast (Figure 5) and thus fit this extended MEPS model just as well. An alternative model (Fujitani and Kobayashi 1999) used a “random walk” to explain the rapid drop-off. We find this model to disagree with the rates measured in our experiments. For example, the first introduced mismatch is predicted to reduce the recombination rate by eightfold, which is significantly different from the approximately threefold decrease reported here and in yeast.
Although we do not have any evidence for the antirecombination mechanism triggered by sequence divergence in Arabidopsis, by analogy to yeast we would assume that such a mechanism is mediated by the MMR machinery. The antirecombination mechanism remains one of the intriguing enigmas in the recombination field. The MMR machinery might be necessary for mismatch recognition but we do not know what succeeding steps are required for recombination rejection.
Another interesting feature of the antirecombination machinery can be deduced from the findings that the curve we measured is not decreasing exponentially but rather flattens out rapidly. The effect of multiple mismatches is not additive but rather reaches saturation rapidly with increase in divergence. It seems that the effect of the first mismatch, presumably through recognition by the MMR proteins, reduces the effect of additional mismatches. This may result from an inhibitory effect of one MMR unit, already bound to the heteroduplex DNA, on the binding efficiency of additional mismatches by other MMR units.
In summary, our work shows a strong similarity to the yeast data, namely the rapid drop-off caused by a single mismatch (approximately threefold reduction in recombination rate) followed by a rapid leveling off. An apparent difference might be the plant higher recombination rate at which the leveling off occurs. The best current model that can explain our results is therefore the extended MEPS model proposed by Datta et al. (1997). We extended the yeast data by showing, in a systematic manner, the lack of sensitivity to mismatch position. This insensitivity across the repeat suggested that the recombination intermediate length is as long as the repeat and enabled us to deduce an antirecombination efficiency of ∼66% for the first mismatch in the repeat. Moreover, we showed that the type of mismatch had no effect on the efficiency of the antirecombination machinery. Future studies in plants should address the role of MMR genes in the antirecombination effect, using Arabidopsis MMR mutants. New assays should be designed, using longer repeats, increasing divergence between repeats, and enabling rescue and molecular analysis of the recombination products. These assays should enable an in-depth comparison of the role(s) of the MMR machinery in the recombination process in plants vs. other kingdoms and shed light on the antirecombination mechanism.
Acknowledgments
We thank Dan Frumkin for help in parts of the cloning step and for fruitful discussions and Naomi Avivi-Ragolsky for help with the tissue culture. We also thank Dean Goodman and Giel Van Dooren for critically reading the manuscript. This work was supported by a grant from the Israeli Science Foundation to A.A.L. and by fellowships from the Feinberg Graduate School of the Weizmann Institute of Science to R.O. and E.E.
References
- Ahn, B. Y., K. J. Dornfeld, T. J. Fagrelius and D. M. Livingston, 1988. Effect of limited homology on gene conversion in a Saccharomyces cerevisiae plasmid recombination system. Mol. Cell. Biol. 8: 2442–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, J. S., and R. McCulloch, 2003. Mismatch repair regulates homologous recombination, but has little influence on antigenic variation, in Trypanosoma brucei. J. Biol. Chem. 278: 45182–45188. [DOI] [PubMed] [Google Scholar]
- Chen, W., and S. Jinks-Robertson, 1999. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics 151: 1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat, R. T., V. Meglic and P. Cisneros, 2000. A genetic map of tomato based on BC(1) Lycopersicon esculentum × Solanum lycopersicoides reveals overall synteny but suppressed recombination between these homeologous genomes. Genetics 154: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys, J. P., and S. A. Lacks, 1986. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol. Rev. 50: 133–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, A., M. Hendrix, M. Lipsitch and S. Jinks-Robertson, 1997. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl. Acad. Sci. USA 94: 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, C., and M. R. Capecchi, 1992. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol. 12: 3365–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani, Y., and I. Kobayashi, 1999. Effect of DNA sequence divergence on homologous recombination as analyzed by a random-walk model. Genetics 153: 1973–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R. P., E. A. Edwards, N. R. Leyland, S. Bean and P. M. Mullineaux, 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson, S., M. Michelitch and S. Ramcharan, 1993. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 3937–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk, I., O. Kovalchuk and B. Hohn, 2000. Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J. 19: 4431–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, J. M., S. R. Bollmann and J. B. Hays, 2003. Reduction of stability of Arabidopsis genomic and transgenic DNA-repeat sequences (microsatellites) by inactivation of AtMSH2 mismatch-repair function. Plant Physiol. 133: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay, R. M., A. Letsou and J. L. Stachelek, 1987. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics 115: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacsovich, T., and A. S. Waldman, 1999. Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics 151: 1559–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich, P., and R. Lahue, 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101–133. [DOI] [PubMed] [Google Scholar]
- Molinier, J., G. Ries, S. Bonhoeffer and B. Hohn, 2004. Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado, F., F. Cortes-Ledesma, P. Huertas and A. Aguilera, 2003. Mitotic recombination in Saccharomyces cerevisiae. Curr. Genet. 42: 185–198. [DOI] [PubMed] [Google Scholar]
- Puchta, H., 1999. Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152: 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta, H., and B. Hohn, 1991. A transient assay in plant cells reveals a positive correlation between extrachromosomal recombination rates and length of homologous overlap. Nucleic Acids Res. 19: 2693–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayssiguier, C., D. S. Thaler and M. Radman, 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342: 396–401. [DOI] [PubMed] [Google Scholar]
- Rick, C. M., 1969. Controlled introgression of chromosomes of Solanum pennellii into Lycopersicon esculentum: segregation and recombination. Genetics 62: 753–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz, J., and S. Subramani, 1984. The minimum amount of homology required for homologous recombination in mammalian cells. Mol. Cell. Biol. 4: 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, E. R., 1976. Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10: 31–51. [DOI] [PubMed] [Google Scholar]
- Shalev, G., and A. A. Levy, 1997. The maize transposable element Ac induces recombination between the donor site and an homologous ectopic sequence. Genetics 146: 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, P., and H. V. Huang, 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112: 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., T. Goldfarb, B. Studamire, E. Alani and J. E. Haber, 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101: 9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda, P., S. Gal, B. Hohn and H. Puchta, 1994. Intrachromosomal homologous recombination in whole plants. EMBO J. 13: 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl, 1983. The double-strand break repair model of recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Vulic, M., F. Dionisio, F. Taddei and M. Radman, 1997. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl. Acad. Sci. USA 94: 9763–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki, P., M. S. Roberts and F. M. Cohan, 1995. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics 140: 917–932. [DOI] [PMC free article] [PubMed] [Google Scholar]