Abstract
In Saccharomyces cerevisiae the MSH4-MSH5, MLH1-MLH3, and MUS81-MMS4 complexes act to promote crossing over during meiosis. MSH4-MSH5, but not MUS81-MMS4, promotes crossovers that display interference. A role for MLH1-MLH3 in crossover control is less clear partly because mlh1Δ mutants retain crossover interference yet display a decrease in crossing over that is only slightly less severe than that seen in msh4Δ and msh5Δ mutants. We analyzed the effects of msh5Δ, mlh1Δ, and mms4Δ single, double, and triple mutants on meiotic crossing over at four consecutive genetic intervals on chromosome XV using newly developed computer software. mlh1Δ mms4Δ double mutants displayed the largest decrease in crossing over (13- to 15-fold) of all mutant combinations, yet these strains displayed relatively high spore viability (42%). In contrast, msh5Δ mms4Δ and msh5Δ mms4Δ mlh1Δ mutants displayed smaller decreases in crossing over (4- to 6-fold); however, spore viability (18–19%) was lower in these strains than in mlh1Δ mms4Δ strains. These data suggest that meiotic crossing over can occur in yeast through three distinct crossover pathways. In one pathway, MUS81-MMS4 promotes interference-independent crossing over; in a second pathway, both MSH4-MSH5 and MLH1-MLH3 promote interference-dependent crossovers. A third pathway, which appears to be repressed by MSH4-MSH5, yields deleterious crossovers.
IN most eukaryotic organisms the correct segregation of chromosomes at the first meiotic division requires reciprocal exchange between homologs. The physical manifestations of these crossover events, chiasmata, provide the contacts between homologous chromosomes that are necessary for segregation (Jones 1987). This cohesion or “chiasma binder” function ensures the generation of a bipolar spindle in which tension is generated at the kinetochores (Maguire 1974). The subsequent “programmed release of sister connections” is thought to be critical for meiosis I segregation (Storlazzi et al. 2003). Because of their importance, crossover events are highly regulated both within and among chromosomes. This regulation is clearly seen in Saccharomyces cerevisiae where two crossovers rarely occur within the same genetic interval (positive interference) and smaller chromosomes tend to display less positive interference than larger ones (Mortimer and Fogel 1974; Kaback et al. 1999). A net result of this regulation is that every chromosome, regardless of size, receives at least one reciprocal exchange (Jones 1987).
How are crossover events generated? Genetic and physical analyses of meiosis in S. cerevisiae showed that meiotic recombination is initiated by double-strand breaks that occur at specific chromosomal positions (reviewed in Keeney 2001). The repair of these breaks, preferentially using an unbroken homolog as a template, results in both reciprocal exchanges, termed crossovers (CO), and nonreciprocal exchanges, termed noncrossovers (NCO). The classical double-strand break repair (DSBR) model proposes that these events result from alternative resolutions of a common Holliday junction intermediate (reviewed in Pâques and Haber 1999). Recent studies, however, have suggested that COs and NCOs are processed via separate pathways. In support of this idea, meiotic mutants have been identified that specifically reduce the number of COs or allow NCO formation in the absence of COs (Ross-Macdonald and Roeder 1994; Sym and Roeder 1994; Hollingsworth et al. 1995; Storlazzi et al. 1995; Hunter and Borts 1997; Chua and Roeder 1998; Nakagawa and Ogawa 1999; Agarwal and Roeder 2000; Allers and Lichten 2001a,b; Hunter and Kleckner 2001; reviewed in Bishop and Zickler 2004; Hollingsworth and Brill 2004). Furthermore, the configuration of heteroduplex DNA seen in NCOs does not fit that predicted by the DSBR model (Porter et al. 1993; Gilbertson and Stahl 1996; Merker et al. 2003). Finally, the majority of Holliday junctions detected by physical analyses of cells induced for meiosis are processed into COs (Allers and Lichten 2001a,b; Börner et al. 2004).
In the budding yeast S. cerevisiae, the MER3, EXO1, MSH4, MSH5, MLH1, MLH3, MMS4, and MUS81 genes are each required to achieve wild-type levels of meiotic crossing over (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Hunter and Borts 1997; Nakagawa and Ogawa 1999; Wang et al. 1999; Khazanehdari and Borts 2000; Borts et al. 2000; Tsubouchi and Ogawa 2000; de los Santos et al. 2001, 2003; Börner et al. 2004; Mazina et al. 2004). In each of these mutants, crossing over, as measured at specific genetic intervals, is reduced by less than threefold. The proteins encoded by these genes are thought to participate in the biochemical steps that lead to meiotic recombination. EXO1 is a 5′–3′ exonuclease that can act on duplex DNA ends (Tsubouchi and Ogawa 2000), MER3 is a meiosis-specific 3′–5′ helicase that is thought to process double-strand breaks into Holliday junction intermediates that form COs (Nakagawa and Ogawa 1999; Nakagawa and Kolodner 2002a,b; Mazina et al. 2004), and MUS81-MMS4 is an endonuclease that appears to preferentially cleave D-loops and half-Holliday junctions (Kaliraman et al. 2001; reviewed in Hollingsworth and Brill 2004). How these biochemical activities converge to regulate crossing over and interference remains a major question in the field.
Little is known about the roles of MSH4, MSH5, MLH1, and MLH3 in meiotic crossing over. Biochemical and genetic studies, however, have shown that they act in MLH1-MLH3 and MSH4-MSH5 complexes (Pochart et al. 1997; Wang et al. 1999; Wang and Kung 2002). While both MSH4 and MSH5 are homologs of the bacterial MutS mismatch repair protein, they do not appear to play a role in eukaryotic mismatch repair (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995). In S. cerevisiae, msh4Δ and msh5Δ mutants display a two- to threefold reduction in crossing over, an increase in meiosis I nondisjunction, the loss of interference, and a subsequent loss in spore viability (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Novak et al. 2001). In Caenorhabditis elegans, deletion of either the MSH4 or the MSH5 homolog results in a complete loss of crossing over that is accompanied by meiotic inviability (Zalevsky et al. 1999; Kelly et al. 2000). These observations have led to models in which MSH4-MSH5 acts to stabilize and/or resolve Holliday junction intermediates (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Pochart et al. 1997). While meiotic crossover defects in mlh1Δ and mlh3Δ mutants appear less severe than those in msh4Δ and msh5Δ mutants, these mutants still display relatively high levels of meiosis I nondisjunction (Hunter and Borts 1997; Wang et al. 1999; Argueso et al. 2003). In contrast to msh4Δ strains, interference appears intact in mlh1Δ mutants (Argueso et al. 2003). Mlh1−/− and Mlh3−/− mutant mice show severe defects in crossing over, resulting in sterility (Edelmann et al. 1996; Woods et al. 1999; Lipkin et al. 2002). These results, in conjunction with epistasis and cell biological analyses in yeast and mice, suggest that MSH4-MSH5 and MLH1-MLH3 act in a common crossover pathway, with MSH4-MSH5 functioning prior to MLH1-MLH3 (this study; Hunter and Borts 1997; Wang et al. 1999; Santucci-Darmanin et al. 2000; Moens et al. 2002; Wang and Kung 2002).
The genetic, cytological, and biochemical studies summarized above suggest that crossing over in mice and C. elegans occurs primarily through an interference-dependent (MSH4-MSH5, MLH1-MLH3) pathway. Crossing over in S. cerevisiae, however, is thought to be controlled by both interference-dependent and interference-independent (MUS81-MMS4) mechanisms (Zalevsky et al. 1999; Khazanehdari and Borts 2000; de los Santos et al. 2001, 2003). The above observations, which suggest that organisms utilize interference-dependent and -independent crossover pathways to varying degrees, are supported by the following:
Mouse and C. elegans mutants defective in MSH4-MSH5 and MLH1-MLH3 complexes display severe crossover defects relative to the equivalent S. cerevisiae mutants (Edelmann et al. 1999; Woods et al. 1999; Zalevsky et al. 1999).
Crossing over, while reduced in S. cerevisiae mus81Δ and mms4Δ strains, is still subject to interference (de los Santos et al. 2001, 2003).
Crossing over and spore viability in S. cerevisiae msh5Δ mus81Δ or msh5Δ mms4Δ double mutants is significantly lower (approximately fivefold) than that in the single mutants (de los Santos et al. 2001, 2003; this study).
Schizosaccharomyces pombe mus81Δ strains display severe defects in spore viability and crossing over that can be explained by the lack of an interference-dependent pathway in this organism (Egel 1995; reviewed in Hollingsworth and Brill 2004).
To gain a better understanding of the relationships between members of different crossover pathways as well as the contribution of distributive pairing to the meiosis I division, we analyzed the effect of msh5Δ, mlh1Δ, and mms4Δ single, double, and triple mutations on meiotic crossing over at four consecutive genetic intervals on chromosome XV. Data from tetrad dissection and single spores were analyzed using newly developed software. Our data suggest that meiotic crossing over in yeast can occur through three distinct crossover pathways: MUS81-MMS4 promotes interference-independent crossing over in one pathway while both MSH4-MSH5 and MLH1-MLH3 participate in a second interference-dependent pathway (Argueso et al. 2003; de los Santos et al. 2003). MSH4-MSH5 appears to repress a third pathway that yields deleterious crossovers.
MATERIALS AND METHODS
Media and strains:
Yeast strains were grown in either yeast extract-peptone-dextrose (YPD) or minimal selective media (Rose et al. 1990). Sporulation plates were prepared as described previously (Detloff et al. 1991). All incubations were performed at 30°. When required, geneticin (Invitrogen, San Diego), nourseothricin (Hans-Knoll Institute fur Naturstoff-Forschung), and hygromycin B (Calbiochem, La Jolla, CA) were included in YPD media as described (Wach et al. 1994; Goldstein and McCusker 1999).
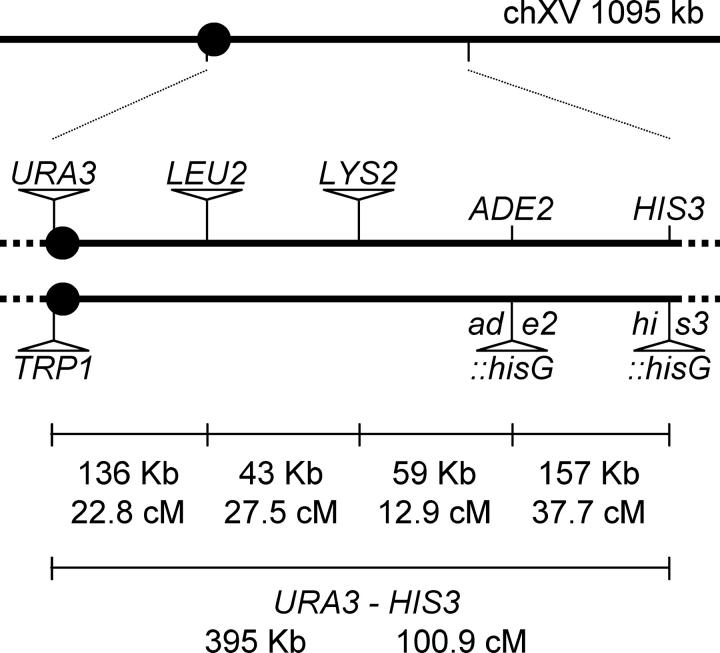
The strains used in this study were derived from the SK1-congenic strains HTY1212 and HTY1213 (Sym and Roeder 1994; Tsubouchi and Ogawa 2000). Homologous gene replacement was used to insert genetic markers near the centromere and on the right arm of chromosome XV at positions 326272 (URA3-cenXVi and TRP1-cenXVi), 462712 (LEU2-chXVi), and 504881 (LYS2-chXVi). The inserted markers are located in intergenic regions predicted to not affect the functions of neighboring genes. The resulting parental haploid strains are EAY1108 (MATa, ho::hisG, lys2, ura3, leu2::hisG, trp1::hisG, URA3-cenXVi, LEU2-chXVi, LYS2-chXVi) and EAY1112 (MATα, ho::hisG, lys2, ura3, leu2::hisG, trp1::hisG, ade2::hisG, his3::hisG, TRP1-cenXVi). These strains were mated to create the reference wild-type diploid strain (Figure 1). For the mutant analyses, at least two independent transformants for each genotype were analyzed.
Figure 1.—
Distribution of genetic markers on chromosome XV. The solid circle indicates the centromere. The distances between markers are not drawn to scale. The actual physical and genetic distances in the wild-type diploid are given numerically for each interval and for the entire region between CENXV and HIS3.
EAY1108/EAY1112 diploids homozygous for coding-region deletion mutations in MMS4, MSH5, and MLH1 were created by sequential deletion using the KanMX4, NatMX4, and HphMX4 selectable markers, respectively (Wach et al. 1994; Goldstein and McCusker 1999). Mutant derivatives of EAY1108 are: EAY1167 (mms4Δ::Kan), EAY1281 (msh5Δ::Nat), EAY1271 (mlh1Δ::Hph), EAY1288 and EAY1289 (mlh1Δ::Hph msh5Δ::Nat), EAY1273 and EAY1274 (mlh1Δ::Hph mms4Δ::Kan), EAY1284 and EAY1285 (mms4Δ::Kan msh5Δ::Nat), EAY1303 and EAY1304 (mlh1Δ::Hph mms4Δ::Kan msh5Δ::Nat), and EAY1165 (pms1Δ::Kan). Mutant derivatives of EAY1112 are: EAY1168 (mms4Δ::Kan), EAY1279 and EAY1280 (msh5Δ::Nat), EAY1276 (mlh1Δ::Hph), EAY1286 and EAY1287 (mlh1Δ::Hph msh5Δ::Nat), EAY1277 and EAY1278 (mlh1Δ::Hph mms4Δ::Kan), EAY1282 and EAY1283 (mms4Δ::Kan msh5Δ::Nat), EAY1290 and EAY1291 (mlh1Δ::Hph mms4Δ::Kan msh5Δ::Nat), and EAY1166 (pms1Δ::Kan).
Genetic analysis:
Diploids were sporulated using the zero-growth mating protocol (Argueso et al. 2003). Briefly, haploid parental strains were patched together, allowed to mate for 4 hr on complete plates, and then transferred to sporulation plates where they were incubated at 30° for 3 days. Because of our interest in comparing our data to previous studies, all strains were sporulated at 30°. Tetrads were dissected on minimal complete plates and then incubated at 30° for 3–4 days. Spore clones were replica plated onto relevant selective plates and assessed for growth after an overnight incubation.
Recently, Börner et al. (2004) examined zip1Δ, zip2Δ, zip3Δ, mer3Δ, and msh5Δ S. cerevisiae mutants for meiotic progression at 23° and 33°. Their studies suggested a coordinated formation of early meiotic recombination intermediates that is important for establishing CO and NCO products. They hypothesized that yeast meiosis can proceed through two recombination modes and that sporulation at 30° represented a mixture of the two. While we would have liked to perform tetrad analyses at 23° and 33°, the meiotic prophase arrest of msh5Δ strains at 33° (Börner et al. 2004) makes such a study untenable.
The segregation data from each replica were converted to a numeric tetrad scoring code and analyzed using the recombination analysis software (RANA, available upon request). RANA analyzes tetrad data for spore viability, genetic linkage, genetic interference, and non-Mendelian segregation. The most important feature of the system is that it allows linkage and interference analysis of data from complete tetrads (four viable spores), as well as from single spores present in incomplete tetrads (three, two, and one viable spores). This is especially useful for the analysis of meiotic recombination mutants because direct comparison of recombination frequencies between complete and incomplete tetrads provides a valuable experimental control and may uncover interesting phenotypes. Only tetrads with Mendelian segregation of all markers were used in tetrad analysis, but all spores in the data set were used in single-spore analysis. In the single-spore analysis, the program compares the marker segregation pattern for each individual spore, outside of the context of a tetrad. Spores are classified as parental or recombinant for each marker pair. The total number of recombinant spores is then counted and divided by the total number of viable spores to obtain recombination frequency (Rf) values.
Genetic map distances were determined by the formula of Perkins (1949) and the expected number of nonparental ditype tetrads (NPD) was calculated using the equation of Papazian (1952). Interference calculations from three-point intervals were conducted as described (Novak et al. 2001; de los Santos et al. 2003; Shinohara et al. 2003). Statistical analysis was done using the Stahl Laboratory Online Tools (http://groik.com/stahl/), VassarStats (http://faculty.vassar.edu/lowry/VassarStats.html), and the Categorical Statistics Packages (http://engels.genetics.wisc.edu).
RESULTS
Development of genetic and software tools to examine crossing over in S. cerevisiae:
Tetrad dissection of meiotic recombination mutants can provide information on crossover frequency, crossover interference, spore viability, and chromosome segregation efficiency. Such an analysis, however, can be difficult to perform in mutants that display poor spore viability. To overcome this, we developed a computer program (RANA) that allows us to organize, store, and share information obtained from a large set of tetrad dissections (29,000 in this study). As described in materials and methods, this allows us to analyze complete tetrads as well as single spores and to identify any inconsistencies when the data are compared.
To analyze the above parameters in a single-strain set, two SK1 congenic strains, EAY1108 and EAY1112, were created to measure crossing over at four consecutive genetic intervals on chromosome XV (100.9 cM, 395 kB; Figure 1). Recent studies by Fung et al. (2004) indicate that chromosome XV was an appropriate choice because interference appears constant throughout its length. The diploid strain created by mating EAY1108 × EAY1112 displays high spore viability and chromosome XV genetic map distances (Table 1) that correspond well with previously published data (Saccharomyces Genome Database at http://www.yeastgenome.org/). Strains isogenic to the EAY1108/EAY1112 diploid and homozygous for the mlh1Δ, msh5Δ, and mms4Δ deletions were generated as described in materials and methods. These mutations were chosen because previous studies had shown that mutants bearing these single mutations displayed phenotypes indistinguishable from those defective in both partners (msh5Δ vs. msh5Δ msh4Δ, mlh1Δ vs. mlh1Δ mlh3Δ, and mms4Δ vs. mms4Δ mus81Δ; Hollingsworth et al. 1995; Argueso et al. 2003; de los Santos et al. 2003).
TABLE 1.
Genetic map distances (cM) in wild type,mms4Δ,msh5Δ, andmlh1Δ strains
| Relevant genotype | No. analyzed | URA3-LEU2 | LEU2-LYS2 | LYS2-ADE2 | ADE2-HIS3 | URA3-LYS2 | LYS2-HIS3 |
|---|---|---|---|---|---|---|---|
| Tetradsa | |||||||
| Wild type | 1068 | 21.8–23.8 | 26.6–28.4 | 12.1–13.7 | 36.5–38.9 | 46.5–49.9 | 46.0–49.6 |
| mms4Δ | 153 | 14.8–18.6 | 23.8–29.2 | 7.9–11.1 | 28.7–32.7 | 39.2–48.4 | 32.8–39.0 |
| mlh1Δ | 616 | 10.3–12.5 | 11.8–13.6 | 6.2–7.6 | 18.2–21.0 | 21.8–24.4 | 24.7–28.3 |
| msh5Δ | 720 | 5.0–6.4 | 11.0–13.0 | 3.7–4.7 | 17.2–20.2 | 15.5–18.1 | 20.5–23.9 |
| mlh1Δ msh5Δ | 764 | 8.7–10.9 | 14.9–17.5 | 5.2–6.6 | 19.1–21.9 | 24.1–27.9 | 24.5–27.9 |
| mlh1Δ mms4Δ | 201 | 0.1–0.9 | 1.7–3.3 | 0.7–1.7 | 1.9–3.5 | 2.2–3.8 | 2.8–4.6 |
| mms4Δ msh5Δ | 52 | 0.1–2.0 | 5.2–10.2 | 2.0–5.6 | 2.8–6.8 | 6.1–11.3 | 6.1–11.3 |
| mlh1Δ mms4Δ msh5Δ | 51 | 0.6–3.4 | 7.0–12.6 | 0.1–2.0 | 5.3–10.3 | 7.0–12.6 | 6.1–11.5 |
| Single sporesb | |||||||
| Wild type | 4644 | 20.6–23.0 | 25.8–28.4 | 11.8–13.8 | 33.3–36.1 | 38.0–40.8 | 37.7–40.5 |
| mms4Δ | 2732 | 17.1–20.0 | 22.3–25.5 | 9.3–11.7 | 27.9–31.4 | 33.6–37.3 | 32.1–35.7 |
| mlh1Δ | 3792 | 9.6–11.6 | 11.7–13.9 | 6.5–8.1 | 16.9–19.4 | 20.2–22.9 | 21.7–24.5 |
| msh5Δ | 5674 | 5.1–6.3 | 10.3–11.9 | 4.1–5.3 | 14.5–16.4 | 13.8–15.6 | 17.3–19.3 |
| mlh1Δ msh5Δ | 6509 | 8.3–9.7 | 12.1–13.7 | 5.5–6.6 | 16.2–18.1 | 18.4–20.4 | 20.3–22.3 |
| mlh1Δ mms4Δ | 2260 | 0.8–1.7 | 1.8–3.1 | 0.5–1.3 | 2.3–3.8 | 2.5–4.0 | 3.0–4.6 |
| mms4Δ msh5Δ | 1920 | 1.7–3.1 | 6.2–8.6 | 2.1–3.6 | 6.2–8.6 | 7.3–9.9 | 8.3–11.0 |
| mlh1Δ mms4Δ msh5Δ | 1790 | 2.1–3.7 | 7.1–9.7 | 1.8–3.3 | 8.6–11.4 | 8.2–11.0 | 9.9–12.9 |
All mutants are isogenic derivatives of EAY1108/EAY1112 (materials and methods).
Intervals correspond to the genetic distance calculated from tetrads ±1 standard error. Standard error was calculated using the Stahl Laboratory Online Tools website (http://groik.com/stahl/).
Data shown as 95% confidence intervals around the recombination frequency determined from single spores, calculated using the VassarStats website (http://faculty.vassar.edu/lowry/VassarStats.html). To facilitate comparisons to the tetrad data, recombination frequencies obtained from single-spore data were multiplied by 100 to yield genetic map distances (in centimorgans).
Effect of mlh1Δ, msh5Δ, and mms4Δ mutations on spore viability and chromosome segregation:
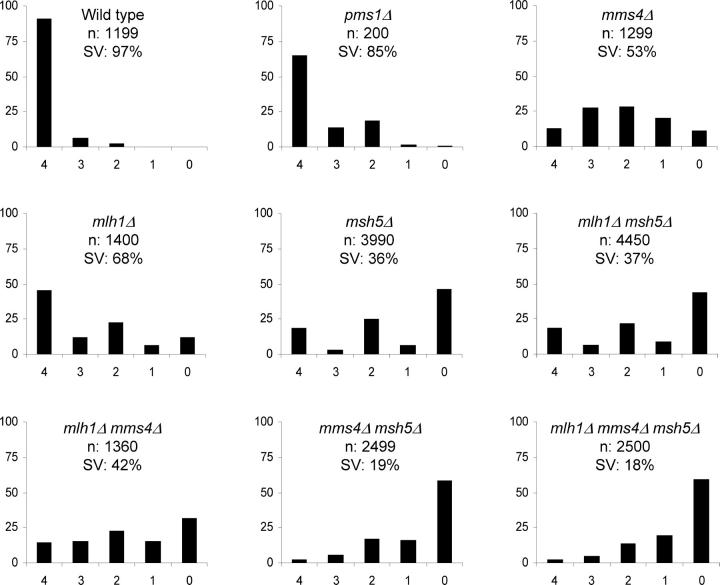
As shown in Figure 2, mlh1Δ, msh5Δ, and mlh1Δ msh5Δ strains displayed spore viability patterns (4, 2, 0 viable spores >3 and 1) consistent with high levels of meiosis I nondisjunction (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Hunter and Borts 1997; Wang et al. 1999; Argueso et al. 2003). The defect in spore viability appears more severe in msh5Δ than in mlh1Δ strains. In addition, the msh5Δ mlh1Δ strain displayed a spore viability phenotype similar to that observed in msh5Δ strains, suggesting that MSH4-MSH5 and MLH1-MLH3 act in the same pathway in meiosis (Hunter and Borts 1997; Kneitz et al. 2000; Lipkin et al. 2002; Moens et al. 2002). mlh1Δ strains displayed defects in both mismatch repair (MMR) and meiotic crossing over and showed 68% spore viability. Because PMS1, MLH1's major partner in MMR, does not appear to play a role in meiotic crossing over, we examined strains homozygous for the pms1Δ mutation with the goal of determining the contribution of defects in mismatch repair to meiotic spore viability. As shown in Figure 2, the MMR defect in pms1Δ strains contributed <10% decrease in spore viability compared to wild type, suggesting that a reduction of ∼20% in spore viability in mlh1Δ strains was due to meiotic defects.
Figure 2.—
Plots showing the distribution of viable spores in tetrads of each genotype. In all plots, the horizontal axis corresponds to the classes of tetrads with 4, 3, 2, 1, and 0 viable spores, and the vertical axis corresponds to the frequency of each class given in a percentage. The total number of tetrads dissected (n) and the overall spore viability (SV) are shown for each genotype.
mms4Δ strains did not display a spore viability pattern consistent with meiosis I misegregation despite displaying defects in meiotic crossing over (de los Santos et al. 2001, 2003). Presumably such a pattern was not observed because mms4Δ strains display defects associated with DNA metabolism that result in random spore death (Mullen et al. 2001). Double- and triple-mutant combinations involving mms4Δ, mlh1Δ, and msh5Δ yielded a spore viability pattern that appeared as a mixture of the mms4Δ and mlh1Δ/msh5Δ spore viability profiles. Consistent with this, the double- and triple-mutant analysis did not reveal an epistatic relationship between mms4Δ and mlh1Δ or msh5Δ mutations with respect to spore viability. Strikingly, spore viability was significantly lower in msh5Δ mms4Δ strains (19%) than in mlh1Δ mms4Δ strains (42%).
The presence of centromere-linked markers at chromosome XV in the EAY1108/EAY1112 diploid allows us to analyze two viable spore tetrads for a chromosome disjunction phenotype. The detection of a large percentage of sisters (Trp+/Ura−, Trp−/Ura+, or Trp+/Ura+) in this two-viable-spore class is suggestive of a meiosis I defect (e.g., Khazanehdari and Borts 2000). In contrast, a large percentage of nonsister spores (one Trp+/Ura− and the other Trp−/Ura+) suggests spore death unrelated to the meiosis I division. Only a small number of two-spore-viable tetrads, 32, were observed for wild type, with 38% displaying the sister pattern. Between 323 and 974 two-spore tetrads were observed in each mutant study. Consistent with the spore viability data (Figure 2), mlh1Δ (72%), msh5Δ (95%), and mlh1Δ msh5Δ (85%) strains displayed high percentages of two-spore-viable sister tetrads. In contrast, mms4Δ strains, which displayed a spore viability distribution consistent with random spore death, displayed a frequency of two-spore-viable sister tetrads (37%) that was similar to wild type. mlh1Δ mms4Δ (68%), mms4Δ msh5Δ (73%), and mlh1Δ msh5Δ mms4Δ (62%) strains displayed intermediate frequencies, relative to the single mutants, of two-spore-viable tetrads that were sisters.
Crossing over is reduced 13- to 15-fold in mlh1Δ mms4Δ strains:
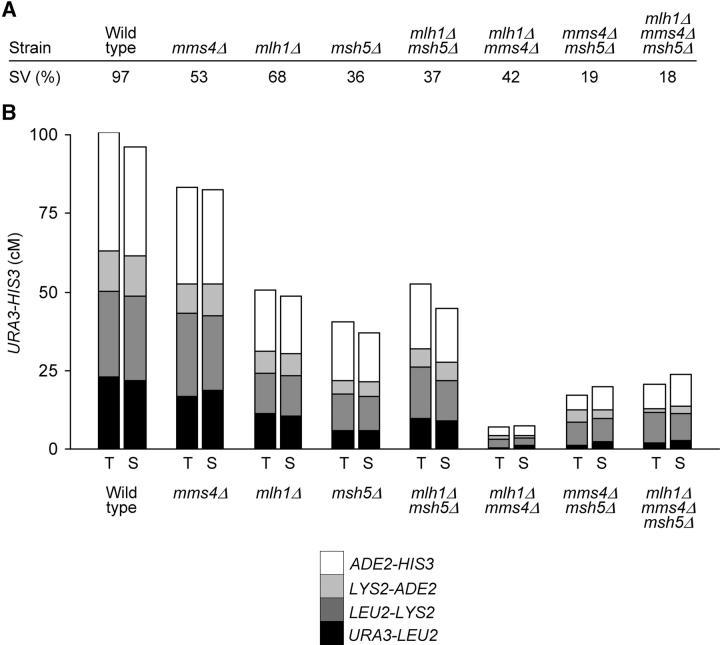
A major advantage of using the EAY1108/EAY1112 strain set is that the genetic intervals can be expanded to measure crossing over in mutants strongly defective in crossing over (Tables 1 and 2). Because each of the four genetic intervals in EAY1108/EAY1112 appeared to be similarly affected by the mms4Δ, mlh1Δ, and msh5Δ mutations, the data can be examined as composite graphs (Figure 3). It is important to note that due to high levels of spore inviability, only a small number of complete tetrads could be recovered for the double- and triple-mutant combinations containing the mms4Δ mutation. This limitation was partly overcome by the use of RANA software, which helped us recover and analyze genetic recombination data from a very large number of viable single spores (1790–2260, Table 1) from these same strains. As shown previously, the mms4Δ, msh5Δ, and mlh1Δ mutations caused small increases in the frequency of aberrant segregation events (Table 3; Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Hunter and Borts 1997; Argueso et al. 2003; de los Santos et al. 2003). However, the finding that gene conversions represented only a small proportion of events in the entire strain set allowed us to include information from single spores. As shown in Table 1 and Figure 3, the crossover frequencies obtained in the two analyses matched extremely well, suggesting that the crossover events in complete tetrads did not represent a subset of events that permitted all spores from a single tetrad to be viable.
TABLE 2.
Distribution of parental and recombinant progeny for strains presented in Table 1
| Single sporesa
|
Tetrads
|
||||||
|---|---|---|---|---|---|---|---|
| Relevant genotype | Parental | Recombinant | Rf | cM | PD | TT | NPD |
| URA3-LEU2 | |||||||
| Wild type | 3635 | 1009 | 0.217 | 22.8 | 607 | 456 | 5 |
| pms1Δ | 539 | 140 | 0.206 | 20.9 | 74 | 53 | 0 |
| mms4Δ | 2227 | 505 | 0.185 | 16.7 | 102 | 51 | 0 |
| mlh1Δ | 3393 | 399 | 0.105 | 11.4 | 486 | 128 | 2 |
| msh5Δ | 5352 | 322 | 0.057 | 5.7 | 643 | 76 | 1 |
| mlh1Δ msh5Δ | 5928.5 | 580.5 | 0.089 | 9.8 | 639 | 120 | 5 |
| mlh1Δ mms4Δ | 2234 | 26 | 0.012 | 0.5 | 199 | 2 | 0 |
| mms4Δ msh5Δ | 1876 | 44 | 0.023 | 1.0 | 51 | 1 | 0 |
| mlh1Δ mms4Δ msh5Δ | 1739.5 | 50.5 | 0.028 | 2.0 | 49 | 2 | 0 |
| LEU2-LYS2 | |||||||
| Wild type | 3388 | 1256 | 0.270 | 27.5 | 496 | 569 | 3 |
| pms1Δ | 504 | 175 | 0.258 | 26.8 | 64 | 62 | 1 |
| mms4Δ | 2081 | 651 | 0.238 | 26.5 | 77 | 75 | 1 |
| mlh1Δ | 3309 | 438 | 0.127 | 12.7 | 459 | 157 | 0 |
| msh5Δ | 5047 | 627 | 0.111 | 12.0 | 562 | 155 | 3 |
| mlh1Δ msh5Δ | 5672 | 837 | 0.129 | 16.2 | 557 | 199 | 8 |
| mlh1Δ mms4Δ | 2206 | 54 | 0.024 | 2.5 | 191 | 10 | 0 |
| mms4Δ msh5Δ | 1779 | 141 | 0.073 | 7.7 | 44 | 8 | 0 |
| mlh1Δ mms4Δ msh5Δ | 1641.5 | 148.5 | 0.083 | 9.8 | 41 | 10 | 0 |
| LYS2-ADE2 | |||||||
| Wild type | 4052 | 592 | 0.127 | 12.9 | 803 | 263 | 2 |
| pms1Δ | 591 | 88 | 0.130 | 12.2 | 96 | 31 | 0 |
| mms4Δ | 2447 | 285 | 0.104 | 9.5 | 124 | 29 | 0 |
| mlh1Δ | 3517 | 275 | 0.073 | 6.9 | 531 | 85 | 0 |
| msh5Δ | 5409 | 265 | 0.047 | 4.2 | 659 | 61 | 0 |
| mlh1Δ msh5Δ | 6118 | 391 | 0.060 | 5.9 | 679 | 84 | 1 |
| mlh1Δ mms4Δ | 2242 | 18 | 0.008 | 1.2 | 196 | 5 | 0 |
| mms4Δ msh5Δ | 1867 | 53 | 0.028 | 3.8 | 48 | 4 | 0 |
| mlh1Δ mms4Δ msh5Δ | 1745.5 | 44.5 | 0.025 | 1.0 | 50 | 1 | 0 |
| ADE2-HIS3 | |||||||
| Wild type | 3033 | 1611 | 0.347 | 37.7 | 343 | 709 | 16 |
| pms1Δ | 477 | 202 | 0.297 | 31.5 | 57 | 68 | 2 |
| mms4Δ | 1923 | 809 | 0.296 | 30.7 | 59 | 94 | 0 |
| mlh1Δ | 3104 | 688 | 0.181 | 19.6 | 400 | 211 | 5 |
| msh5Δ | 4797 | 877 | 0.155 | 18.7 | 496 | 215 | 9 |
| mlh1Δ msh5Δ | 5394 | 1115 | 0.171 | 20.5 | 495 | 260 | 9 |
| mlh1Δ mms4Δ | 2193 | 67 | 0.030 | 2.7 | 190 | 11 | 0 |
| mms4Δ msh5Δ | 1779 | 141 | 0.073 | 4.8 | 47 | 5 | 0 |
| mlh1Δ mms4Δ msh5Δ | 1611.5 | 178.5 | 0.100 | 7.8 | 43 | 8 | 0 |
| URA3-LYS2 | |||||||
| Wild type | 2815 | 1829 | 0.394 | 48.2 | 264 | 759 | 45 |
| pms1Δ | 390 | 289 | 0.426 | 52.8 | 28 | 92 | 7 |
| mms4Δ | 1764 | 968 | 0.354 | 43.8 | 49 | 98 | 6 |
| mlh1Δ | 2976 | 816 | 0.215 | 23.1 | 351 | 261 | 4 |
| msh5Δ | 4843 | 831 | 0.146 | 16.8 | 513 | 300 | 7 |
| mlh1Δ msh5Δ | 5248.5 | 1260.5 | 0.194 | 26.0 | 481 | 260 | 23 |
| mlh1Δ mms4Δ | 2188 | 72 | 0.032 | 3.0 | 189 | 12 | 0 |
| mms4Δ msh5Δ | 1757 | 163 | 0.085 | 8.7 | 43 | 9 | 0 |
| mlh1Δ mms4Δ msh5Δ | 1619.5 | 170.5 | 0.095 | 9.8 | 41 | 10 | 0 |
| LYS2-HIS3 | |||||||
| Wild type | 2829 | 1815 | 0.391 | 47.8 | 278 | 744 | 46 |
| pms1Δ | 451 | 228 | 0.336 | 39.8 | 46 | 77 | 4 |
| mms4Δ | 1806 | 926 | 0.339 | 35.9 | 53 | 98 | 2 |
| mlh1Δ | 2917 | 875 | 0.231 | 26.5 | 344 | 261 | 11 |
| msh5Δ | 4638 | 1036 | 0.183 | 22.2 | 465 | 242 | 13 |
| mlh1Δ msh5Δ | 5127 | 1382 | 0.212 | 26.2 | 438 | 311 | 15 |
| mlh1Δ mms4Δ | 2177 | 83 | 0.037 | 3.7 | 186 | 15 | 0 |
| mms4Δ msh5Δ | 1736 | 184 | 0.096 | 8.7 | 43 | 9 | 0 |
| mlh1Δ mms4Δ msh5Δ | 1587.5 | 202.5 | 0.113 | 8.8 | 42 | 9 | 0 |
Rf refers to the recombination frequency in single spores determined by parental/(parental + recombinant) and cM indicates the genetic distance in tetrads calculated using the formula of Perkins (1949): 50 × {TT + (6 × NPD)}/(PD + TT + NPD).
In rare cases sectored spores were observed. They were assigned as half parental (0.5) and half recombinant (0.5).
Figure 3.—
Summary of the relationship between spore viability and meiotic crossing over. (A) Percentage of spore viability. (B) Cumulative genetic distances between URA3 and HIS3 measured from tetrads (T) and single spores (S). Each bar is divided into four sectors corresponding to the four genetic intervals in the region of chromosome XV analyzed. The size of the sectors is proportional to the contribution of each interval to the total URA3-HIS3 genetic distance.
TABLE 3.
Percentage of aberrant segregation events observed in tetrads from wild-type,mms4Δ,mlh1Δ, andmsh5Δ strains
| Relevant genotype | Tetrads | All markers | TRP1 | URA3 | LEU2 | LYS2 | ADE2 | HIS3 |
|---|---|---|---|---|---|---|---|---|
| Wild type | 1087 | 1.7 | 0 | 0 | 0.2 | 0.6 | 0.1 | 0.8 |
| pms1Δ | 130 | 2.3 | 0 | 0 | 0 | 0.8 | 0 | 1.5 |
| mms4Δ | 167 | 9.0 | 0 | 0 | 0.6 | 1.8 | 3.0 | 3.6 |
| mlh1Δ | 635 | 3.0 | 0 | 0.2 | 0.8 | 0.6 | 0.5 | 0.9 |
| msh5Δ | 757 | 5.0 | 0.1 | 0.1 | 1.6 | 1.2 | 0.8 | 1.2 |
| mlh1Δ msh5Δ | 815 | 7.0 | 0.4 | 0.5 | 2.1 | 1.8 | 0.9 | 1.3 |
| mlh1Δ mms4Δ | 203 | 1.0 | 0 | 0 | 0.5 | 0 | 0 | 0.5 |
| mms4Δ msh5Δ | 59 | 11.9 | 0 | 0 | 1.7 | 3.4 | 1.7 | 5.1 |
| mlh1Δ mms4Δ msh5Δ | 55 | 10.8 | 1.8 | 1.8 | 3.6 | 3.6 | 0 | 0 |
As shown in Figure 3 and Table 1, mms4Δ strains displayed an ∼20% reduction in crossing over. This value is similar to that observed by de los Santos et al. (2003) in their analysis of large chromosomes similar in size to XV. In addition, mlh1Δ (50% reduction) and msh5Δ (60% reduction) strains displayed decreases in crossing over similar to that reported previously (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995; Hunter and Borts 1997; Wang et al. 1999; Argueso et al. 2003). The mlh1Δ msh5Δ double mutant showed a decrease in crossing over that was similar to that observed in each single mutant. mms4Δ msh5Δ strains displayed a four- to sixfold decrease in crossing over that was consistent with a physical analysis of this mutant (de los Santos et al. 2003).
Strikingly, mms4Δ mlh1Δ strains displayed a 13- (single spore) to 15- (complete tetrads) fold decrease in crossing over. This and the finding that a wild-type cell experiences ∼87–95 crossovers in meiosis (Mortimer et al. 1992; Cherry et al. 1997; Winzeler et al. 1998) predicts that a mms4Δ mlh1Δ cell would experience 6–7 crossovers in meiosis. If we extrapolate the observed map for chromosome XV (100.9 cM in a 395-kb interval) over the entire yeast genome (12,300 kb), only 4.4 crossovers are predicted to occur in a mms4Δ mlh1Δ cell. It is important to note that the calculation for the total number of crossovers in mms4Δ mlh1Δ strains is based on an extrapolation of map distances obtained in a single chromosome arm. This calculation may be inaccurate if chromosomes of different size act differently with respect to crossover distribution (Kaback et al. 1999). In contrast, the mms4Δ mlh1Δ msh5Δ triple mutant displayed a decrease in crossing over (5-fold) that was similar to mms4Δ msh5Δ strains, providing further evidence that MSH5 functions upstream of MLH1.
Interference observed in mlh1Δ strains is no longer observed in msh5Δ mlh1Δ strains:
Two distinct analyses of crossover interference are shown in Table 4: (1) observed NPD/expected NPD, which represents the ratio of observed nonparental ditypes (NPDs) to NPDs predicted by the number of single crossovers detected, and (2) a coefficient of coincidence (COC), the ratio of double crossovers observed in adjacent genetic intervals to the number predicted. Because so few crossovers were observed in double-mutant combinations involving the mms4Δ mutation, statistically significant measures of interference could be obtained only in single mutants and in mlh1Δ msh5Δ strains (Table 4). The measure of interference using the COC value appeared less robust than NPD ratios because of the large genetic intervals that were examined. Such large intervals were needed to allow us to measure recombination in mutants that display a large decrease in crossing over.
TABLE 4.
Analysis of crossover interference in wild-type (EAY1108/EAY1112) andmms4Δ,mlh1Δ, andmsh5Δ derivatives
| Coefficients of coincidence (COC observed/COC expected)
|
|||||||
|---|---|---|---|---|---|---|---|
| NPD ratios (NPD observed/NPD expected)
|
URA3-LEU2-LYS2
|
LEU2-LYS2-ADE2
|
|||||
| Relevant genotype | ADE2-HIS3 | URA3-LYS2 | LYS2-HIS3 | Tetrads | Spores | Tetrads | Spores |
| Wild type | 0.097** (16/165.6) |
<0.253** (45/>178) |
<0.258** (46/>178) |
0.717** (177/246.9) |
0.799** (218/272.9) |
0.458** (65/141.9) |
0.550** (88/160.1) |
| mms4Δ | 0** (0/15.5) |
0.321** (6/18.7) |
0.107** (2/18.7) |
0.789 (20/25.3) |
0.781** (94/120.3) |
0.555 (8/14.4) |
0.633** (43/67.9) |
| mlh1Δ | 0.415* (5/12.1) |
0.197** (4/20.4) |
0.540* (11/20.4) |
0.573** (19/33.1) |
0.649** (33/50.8) |
0.646 (14/21.7) |
0.628* (22/35.0) |
| msh5Δ | 0.880 (9/10.2) |
0.808 (7/8.7) |
0.965 (13/13.5) |
1.184 (20/16.9) |
1.658a (59/35.6) |
0.747 (10/13.4) |
0.990 (29/29.3) |
| mlh1Δ msh5Δ | 0.611 (9/14.7) |
1.562a (23/14.7) |
0.658 (15/22.8) |
1.092 (37/33.9) |
1.052 (78.5/74.6) |
0.651 (15/23.0) |
1.352a (68/50.3) |
Interference was calculated from data presented in Tables 1 and 2. Asterisks indicate that the observed number of NPDs or COCs deviated significantly from the expected number based on a two-tail binomial test (Categorical Statistics Package, http://engels.genetics.wisc.edu), suggesting that interference is present in the interval (*P < 0.05; **P < 0.01).
Although double crossovers deviated significantly from the expected number in this interval, the COC (NPD ratio) is >1, indicating negative interference.
In wild type, interference was significant at all intervals analyzed in chromosome XV, with NPD ratios (<0.258) and COC values (0.458–0.799) significantly <1.0. These values did not significantly change in mlh1Δ and mms4Δ strains, which were shown previously to maintain interference. msh5Δ strains, however, displayed NPD ratios and COC values that were not significantly <1.0, indicating that interference could not be detected. A similar situation was observed in mlh1Δ msh5Δ strains. It is important to note that for the URA3-LYS2-HIS3 interval the wild type, mms4Δ, mlh1Δ, msh5Δ, and mlh1Δ msh5Δ strains all displayed 1:2:1 ratios for single crossovers involving two, three, and four chromatids, respectively (Table 5). This indicates an absence of chromatid interference. Together, the NPD ratios and COC values for all the intervals analyzed provide further evidence that MSH5 functions upstream of MLH1.
TABLE 5.
Analysis of chromatid interference at theURA3-LYS2-HIS3 interval
| No. of double-crossover tetrads involving
|
||||
|---|---|---|---|---|
| Relevant genotype | Two strands | Three strands | Four strands | P-value |
| Wild type | 138 | 287 | 114 | 0.11 |
| mms4Δ | 16 | 25 | 19 | 0.37 |
| mlh1Δ | 30 | 55 | 26 | 0.86 |
| msh5Δ | 17 | 24 | 16 | 0.48 |
| mlh1Δ msh5Δ | 24 | 48 | 27 | 0.87 |
Tetrads displaying the tetratype class at both the URA3-LYS2 and the LYS2-HIS3 intervals were examined for chromatid interference. P-values derived from χ2 analysis indicate the probability that the number of tetrads with exchanges involving two, three, and four chromatids follows a 1:2:1 neutral distribution of double crossovers. P-values <0.05 indicate a deviation from neutrality.
DISCUSSION
This study was initiated to understand how MLH1 acts in meiotic crossover control. In particular, we were interested in understanding why the mlh1 meiotic crossover defect is less severe in S. cerevisiae compared to mice (Hunter and Borts 1997; Woods et al. 1999). To determine this, we examined the effect of mlh1Δ, msh5Δ, and mms4Δ single, double, and triple mutations on meiotic crossing over at four consecutive genetic intervals on chromosome XV. As shown in Table 1 and Figure 3, mlh1Δ mms4Δ double mutants displayed a decrease (13- to 15-fold) in crossing over that was similar to that observed in mouse Mlh1−/− female meiosis (Woods et al. 1999). In contrast, msh5Δ mms4Δ and msh5Δ mms4Δ mlh1Δ mutants displayed smaller decreases in crossing over, 4- to 6-fold, yet were less viable than mlh1Δ mms4Δ strains (18–19% vs. 42%). We hypothesize that competing and overlapping crossover pathways exist in yeast, some of which are deleterious to meiosis.
Recently de los Santos et al. (2003) showed in physical and genetic analyses that the MUS81-MMS4 complex acts in an interference-independent crossover pathway during S. cerevisiae meiosis. Their physical analysis of crossover products in mms4Δ msh5Δ double mutants showed that crossing over was reduced ∼5-fold compared to wild type. Genetic analysis of mms4Δ msh5Δ strains, which revealed a 5-fold decrease in crossing over compared to wild type, is consistent with their physical studies (Table 1, Figure 3). In addition, our study of msh5Δ mlh1Δ mutants suggested that MSH5 and MLH1 act in the same crossover pathway, with MSH5 acting in an upstream step that enforces the crossover interference decision and MLH1 acting in a step after which crossover interference has been established (Börner et al. 2004; Fung et al. 2004). Surprisingly, we found that mlh1Δ mms4Δ strains displayed a much more severe defect in crossing over (13- to 15-fold decrease) than msh5Δ mms4Δ strains did, but showed significantly higher spore viability. The introduction of the msh5Δ mutation to mlh1Δ mms4Δ strains resulted in an increase in crossing over and a decrease in spore viability that was indistinguishable from that seen in msh5Δ mms4Δ strains. These data provide additional support for the idea that MSH4-MSH5 acts upstream of MLH1-MLH3 (Hunter and Borts 1997; Santucci-Darmanin et al. 2000; Moens et al. 2002); more significantly, they support the idea that compensating and competing crossover pathways function during yeast meiosis (Zalevsky et al. 1999; de los Santos 2003; reviewed in Hollingsworth and Brill 2004).
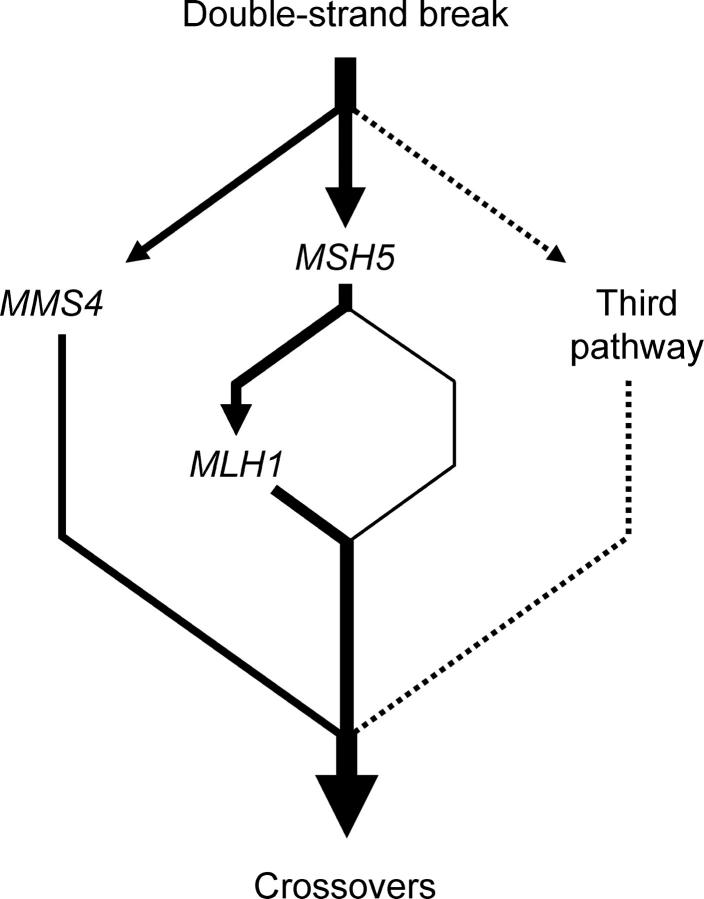
In Figure 4 we present a model consistent with the presented data. In this model, crossing over in wild-type yeast occurs primarily by MUS81-MMS4- and MSH4-MSH5-dependent pathways with MLH1-MLH3 acting in a downstream step in the MSH4-MSH5 pathway. In the absence of MUS81-MMS4, only the interference-independent pathway is compromised. The net result is a mild defect in crossing over and a spore inviability phenotype that is difficult to distinguish from inviability due to defects in DNA metabolism previously seen in mms4 and mus81 mutants (Mullen et al. 2001; de los Santos et al. 2001, 2003). In the absence of MSH4-MSH5, a significant defect in a crossover pathway subject to interference is observed, but this defect is partly compensated for by the MUS81-MMS4 pathway. In mms4Δ msh5Δ mutants, however, the two critical pathways for crossing over are absent, resulting in a modest 4- to 6-fold decrease in crossing over. The fact that a significantly higher (13- to 15-fold) decrease in crossing over was observed in mms4Δ mlh1Δ mutants suggests that recombination intermediates destined to become crossovers are shunted in mms4Δ msh5Δ mutants to a deleterious crossover pathway that results in increased spore death. According to this idea, deleterious crossovers do not arise in mms4Δ mlh1Δ but do so in mms4Δ msh5Δ mlh1Δ mutants because commitment to a MSH4-MSH5-dependent crossover pathway prevents the activation of the deleterious pathway. Under this model, crossing over, but not spore viability, was dramatically decreased in mms4Δ mlh1Δ strains because a deleterious crossover pathway was not activated. At present we do not have a sense of what genes or mechanisms could function in such a deleterious pathway. We cannot exclude the possibility that the high level of spore inviability in msh5Δ mms4Δ strains was due to a general defect in DNA metabolism unrelated to meiotic crossing over. However, the facts that MSH4 and MSH5 are specifically expressed in meiosis and msh4Δ and msh5Δ strains do not display a vegetative growth defect suggest that this was not the case (Ross-Macdonald and Roeder 1994; Hollingsworth et al. 1995).
Figure 4.—
Proposed organization of the meiotic crossover control pathway. The thickness of each line corresponds roughly to the relative contribution of each branch to the overall generation of crossovers. See text for details.
It is important to note that the model in Figure 4 proposes the presence of a MSH4-MSH5-dependent, MLH1-independent recombination pathway (represented by the thin line). This is based on the low level of crossing over observed in mms4Δ mlh1Δ strains and the observation that crossing over in this mutant is roughly equivalent to the difference in crossing over between mlh1Δ and msh5Δ strains. We hypothesize that this MSH5-dependent, MLH1-independent branch is the only path that is available in the mlh1Δ mms4Δ mutant; the crossovers that occur in this branch are capable of promoting meiosis I disjunction.
We were initially surprised by the high spore viability observed in mms4Δ mlh1Δ strains, which are predicted to experience only a small number of total crossovers (four to seven) in a single meiosis. Genetic studies performed in Drosophila females and S. cerevisiae, however, have shown that unrecombined chromosomes can properly segregate with varying levels of efficiency. In female Drosophila, a distributive segregation system allows chromosome IV to segregate with high fidelity even though this chromosome never undergoes reciprocal exchange (reviewed in Hawley and Theurkauf 1993; Harris et al. 2003). This distributive segregation system is disrupted in nod and mtrm mutants (Carpenter 1973; Rasooly et al. 1991; Harris et al. 2003). In S. cerevisiae, studies performed with both artificial and homeologous chromosome pairs suggest the presence of a distributive pairing system that allows for a relatively high level of disjunction at meiosis I, estimated at 89–93%, for nonexchanged chromosomes (Dawson et al. 1986; Mann and Davis 1986; Guacci and Kaback 1991; Sears et al. 1992; Ross et al. 1996; Maxfield Boumil et al. 2003).
Can the high spore viability observed in mms4Δ mlh1Δ strains be reconciled by an efficient distributive segregation system? If we assume that S. cerevisiae strains display a distributive segregation system in which each of the 16 chromosomes has an 89–93% probability of undergoing meiosis I disjunction in the absence of exchange, then 15–28% (0.8916–0.9316) of yeast cells undergoing a crossover-deficient meiosis would yield four-spore-viable tetrads in which all 16 chromosome pairs would disjoin correctly. While this calculation is simplistic, it is interesting to note that the calculated spore viability is not significantly different from that observed in mms4Δ mlh1Δ strains (14.9%). However, this correlation is complicated by the fact that residual crossing over, defects in MMR, and increased chromosome instability influence spore viability in mms4Δ mlh1Δ strains.
In mutants such as spo11Δ, which are completely defective in initiating both meiotic gene conversion and crossing over, spore viability is significantly lower than in mlh1Δ mms4Δ strains (e.g., Keeney et al. 1997). What accounts for this difference in viability? Unlike spo11Δ, mlh1Δ and msh4Δ mutants display gene conversion frequencies that are not dramatically different from wild type, and msh5Δ and mms4Δ mutants display wild-type levels of meiotically induced double-strand breaks (DSBs; Ross-Macdonald and Roeder 1994; Hunter and Borts 1997; de los Santos et al. 2001; Argueso et al. 2003; Börner et al. 2004). These observations suggest that, despite showing defects in promoting crossing over, msh4Δ, msh5Δ, and mms4Δ strains are functional in the formation of interstitial connections that appear between homologs in early meiotic prophase (Giroux et al. 1989; Ross-Macdonald and Roeder 1994; Weiner and Kleckner 1994; Hollingsworth et al. 1995; de los Santos et al. 2001; Börner et al. 2004). In spo11Δ strains, however, recombination initiation is disrupted and the interstitial connections are absent (Giroux et al. 1989; Weiner and Kleckner 1994). An attractive possibility is that these connections are important for a DNA homology search in early meiotic prophase that is essential for distributive meiosis I segregation (Weiner and Kleckner 1994; Keeney et al. 1997).
In mms4Δ msh5Δ mutants, crossing over is approximately three times higher, but spore viability is twofold lower, than that in mms4Δ mlh1Δ strains. Studies in a variety of organisms have indicated that crossing over alone does not guarantee the proper disjunction of paired homologs in meiosis I (see Ross et al. 1996 and references therein). This work also suggests that the location of a crossover in a chromosome pair can affect the efficiency of disjunction. For a crossover to mediate meiosis I segregation, it should be present within the context of sister chromatids that are held together along their lengths or at least at the site of exchange. On the basis of this information, we hypothesize that crossing over in mms4Δ msh5Δ strains (“the third pathway”) interferes with the distributive pairing system. This could occur if crossovers in this strain are not resolved, are resolved after the programmed release of sister connections, or if resolution does not occur through the generation of a chiasma binder at the site of exchange. In such a model the MSH4-MSH5 pathway ensures both the formation and the dissolution of a “chiasma binder.” Alternatively, excessive crossing over takes place in mms4Δ msh5Δ mutants (negative interference) that results in inviability due to a difficulty in separating homologs at anaphase I (Carpenter 1987). A physical analysis of mms4Δ mlh1Δ and mms4Δ msh5Δ strains in meiosis that allows for the measure of DSB formation, single- and double-ended invasion intermediates, as well as physical crossovers, would allow one to test these ideas.
Acknowledgments
We thank Nancy Hollingsworth and Neil Hunter for helpful discussions during the early stages of this project, Todd Schlenke for advice on analyzing the interference data, and Jennifer Surtees and Julie Heck for comments on the manuscript. E.A. and J.W. were supported by National Institutes of Health grant GM-53085. J.L.A. was supported in part by a Coordenacao de Aperfeicoamento de Pessoal de Nivel Superior fellowship awarded by the Brazilian government. Z.G. was supported by a Cornell-Howard Hughes Undergraduate Fellowship and a Cornell Presidential Research Scholars Fellowship.
References
- Agarwal, S., and G. S. Roeder, 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. [DOI] [PubMed] [Google Scholar]
- Allers, T., and M. Lichten, 2001. a Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Allers, T., and M. Lichten, 2001. b Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8: 225–231. [DOI] [PubMed] [Google Scholar]
- Argueso, J. L., A. W. Kijas, S. Sarin, J. Heck, M. Waase et al., 2003. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol. Cell. Biol. 23: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D. K., and D. Zickler, 2004. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. [DOI] [PubMed] [Google Scholar]
- Börner, G. V., N. Kleckner and N. Hunter, 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Borts, R. H., S. R. Chambers and M. F. Abdullah, 2000. The many faces of mismatch repair in meiosis. Mutat. Res. 451: 129–150. [DOI] [PubMed] [Google Scholar]
- Carpenter, A. T., 1973. A meiotic mutant defective in distributive disjunction in Drosophila melanogaster. Genetics 73: 393–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. T., 1987. Gene conversion, recombination nodules and the initiation of meiotic synapsis. BioEssays 6: 232–236. [DOI] [PubMed] [Google Scholar]
- Cherry, J. M., C. Ball, S. Weng, G. Juvik, R. Schmidt et al., 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67–73. [PMC free article] [PubMed] [Google Scholar]
- Chua, P. R., and G. S. Roeder, 1998. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359. [DOI] [PubMed] [Google Scholar]
- Dawson, D. S., A. W. Murray and J. W. Szostak, 1986. An alternative pathway for meiotic chromosome segregation in yeast. Science 234: 713–717. [DOI] [PubMed] [Google Scholar]
- de los Santos, T., J. Loidl, B. Larkin and N. M. Hollingsworth, 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159: 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos, T., N. Hunter, C. Lee, B. Larkin, J. Loidl et al., 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff, P., J. Sieber and T. D. Petes, 1991. Repair of specific base pair mismatches formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, S. S., E. B. Baart, M. Dekker, S. Siezen, D. G. de Rooij et al., 1999. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann, W., P. E. Cohen, M. Kane, K. Lau, B. Morrow et al., 1996. Meiotic pachytene arrest in MLH1-deficient mice. Cell 85: 1125–1134. [DOI] [PubMed] [Google Scholar]
- Edelmann, W., P. E. Cohen, B. Kneitz, N. Winand, M. Lia et al., 1999. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21: 123–127. [DOI] [PubMed] [Google Scholar]
- Egel, R., 1995. The synaptonemal complex and the distribution of meiotic recombination events. Trends Genet. 11: 206–208. [DOI] [PubMed] [Google Scholar]
- Fung, J. C., B. Rockmill, M. Odell and G. S. Roeder, 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Gilbertson, L. A., and F. W. Stahl, 1996. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics 144: 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux, C. N., M. E. Dresser and H. F. Tiano, 1989. Genetic control of chromosome synapsis in yeast meiosis. Genome 31: 88–94. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Guacci, V., and D. B. Kaback, 1991. Distributive disjunction of authentic chromosomes in Saccharomyces cerevisiae. Genetics 127: 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, D., C. Orme, J. Kramer, L. Namba, M. Champion et al., 2003. A deficiency screen of the major autosomes identifies a gene (matrimony) that is haplo-insufficient for achiasmate segregation in Drosophila oocytes. Genetics 165: 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, R. S., and W. E. Theurkauf, 1993. Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet. 9: 310–317. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, N. M., and S. J. Brill, 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N. M., L. Ponte and C. Halsey, 1995. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9: 1728–1739. [DOI] [PubMed] [Google Scholar]
- Hunter, N., and R. H. Borts, 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11: 1573–1582. [DOI] [PubMed] [Google Scholar]
- Hunter, N., and N. Kleckner, 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Jones, G. H., 1987 Chiasmata, pp. 213–238 in Meiosis, edited by P. Moens. Academic Press, New York/London.
- Kaback, D. B., D. Barber, J. Mahon, J. Lamb and J. You, 1999. Chromosome size-dependent control of meiotic reciprocal recombination in Saccharomyces cerevisiae: the role of crossover interference. Genetics 152: 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower and S. J. Brill, 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15: 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kelly, K. O., A. F. Dernburg, G. M. Stanfield and A. M. Villeneuve, 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazanehdari, K. A., and R. H. Borts, 2000. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma 109: 94–102. [DOI] [PubMed] [Google Scholar]
- Kneitz, B., P. E. Cohen, E. Avdievich, L. Zhu, M. F. Kane et al., 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14: 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Lipkin, S. M., P. B. Moens, V. Wang, M. Lenzi, D. Shanmugarajah et al., 2002. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31: 385–390. [DOI] [PubMed] [Google Scholar]
- Maguire, M. P., 1974. The need for a chiasma binder. J. Theor. Biol. 48: 485–487. [DOI] [PubMed] [Google Scholar]
- Mann, C., and R. W. Davis, 1986. Meiotic disjunction of circular minichromosomes in yeast does not require DNA homology. Proc. Natl. Acad. Sci. USA 83: 6017–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield Boumil, R., B. Kemp, M. Angelichio, T. Nilsson-Tillgren and D. S. Dawson, 2003. Meiotic segregation of a homeologous chromosome pair. Mol. Genet. Genomics 268: 750–760. [DOI] [PubMed] [Google Scholar]
- Mazina, O. M., A. V. Mazin, T. Nakagawa, R. D. Kolodner and S. C. Kowalczykowski, 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ extension by Rad51: implications for crossover control in meiotic recombination. Cell 117: 47–56. [DOI] [PubMed] [Google Scholar]
- Merker, J. D., M. Dominska and T. D. Petes, 2003. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 165: 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, P. B., N. K. Kolas, M. Tarsounas, E. Marcon, P. E. Cohen et al., 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115: 1611–1622. [DOI] [PubMed] [Google Scholar]
- Mortimer, R., and S. Fogel, 1974 Genetical interference and gene conversion, pp. 263–275 in Mechanisms in Recombination, edited by R. Grell. Plenum Press, New York.
- Mortimer, R. K., C. R. Contopoulou and J. S. King, 1992. Genetic and physical maps of Saccharomyces cerevisiae, ed. 11. Yeast 8: 817–902. [DOI] [PubMed] [Google Scholar]
- Mullen, J. R., V. Kaliraman, S. S. Ibrahim and S. J. Brill, 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., and R. D. Kolodner, 2002. a The MER3 DNA helicase catalyzes the unwinding of Holliday junctions. J. Biol. Chem. 277: 28019–28024. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., and R. D. Kolodner, 2002. b Saccharomyces cerevisiae Mer3 is a DNA helicase involved in meiotic crossing over. Mol. Cell. Biol. 22: 3281–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., and H. Ogawa, 1999. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 18: 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak, J. E., P. B. Ross-Macdonald and G. S. Roeder, 2001. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian, H. P., 1952. The analysis of tetrad data. Genetics 37: 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques, F. and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D., 1949. Biochemical mutants in the smut fungus Ustilago maydis. Genetics 34: 607–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochart, P., D. Woltering and J. M. Hollingsworth, 1997. Conserved properties between functionally distinct MutS homologs in yeast. J. Biol. Chem. 272: 30345–30349. [DOI] [PubMed] [Google Scholar]
- Porter, S. E., M. A. White and T. D. Petes, 1993. Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics 134: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasooly, R. S., C. M. New, P. Zhang, R. S. Hawley and B. S. Baker, 1991. The lethal(1)TW-6cs mutation of Drosophila melanogaster is a dominant antimorphic allele of nod and is associated with a single base change in the putative ATP-binding domain. Genetics 129: 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Hieter, 1990 Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ross, L. O., R. Maxfield and D. Dawson, 1996. Exchanges are not equally able to enhance meiotic chromosome segregation in yeast. Proc. Natl. Acad. Sci. USA 93: 4979–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald, P., and G. S. Roeder, 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin, S., D. Walpita, F. Lespinasse, C. Desnuelle, T. Ashley et al., 2000. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 14: 1539–1547. [DOI] [PubMed] [Google Scholar]
- Sears, D. D., J. H. Hegemann and P. Hieter, 1992. Meiotic recombination and segregation of human-derived artificial chromosomes in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89: 5296–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, M., K. Sakai, A. Shinohara and D. K. Bishop, 2003. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics 163: 1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi, A., L. Xu, L. Cao and N. Kleckner, 1995. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc. Natl. Acad. Sci. USA 92: 8512–8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi, A., S. Tesse, S. Gargano, F. James, N. Kleckner et al., 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym, M., and G. S. Roeder, 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292. [DOI] [PubMed] [Google Scholar]
- Tsubouchi, H., and H. Ogawa, 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 2221–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang, T. F., and W. M. Kung, 2002. Supercomplex formation between Mlh1-Mlh3 and Sgs1-Top3 heterocomplexes in meiotic yeast cells. Biochem. Biophys. Res. Commun. 296: 949–953. [DOI] [PubMed] [Google Scholar]
- Wang, T. F., N. Kleckner and N. Hunter, 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96: 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, B. M., and N. Kleckner, 1994. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell 77: 977–991. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., D. R. Richards, A. R. Conway, A. L. Goldstein, S. Kalman et al., 1998. Direct allelic variation scanning of the yeast genome. Science 281: 1194–1197. [DOI] [PubMed] [Google Scholar]
- Woods, L. M., C. A. Hodges, E. Baart, S. M. Baker, M. Liskay et al., 1999. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol. 145: 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky, J., A. J. MacQueen, J. B. Duffy, K. J. Kemphues and A. M. Villeneuve, 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]