Abstract
We compared the pattern of nucleotide difference in 8034 genes and in their 5′ intergenic spacers between conspecific pairs of genomes from 10 species of pathogenic bacteria. Certain genes or spacers showed much greater sequence divergence between the genotypes compared to others; such divergent regions plausibly originated by recombinational events by which a gene and/or spacers was donated from a divergent genome. Different patterns of divergence in genes and spacers identified different recombinational patterns. For example, in Chlamydophila pneumoniae, there were examples of both unusually divergent spacers and unusually divergent genes, but there were no cases in which a gene and its spacer were both unusually divergent. This pattern suggests that, in C. pneumoniae, recombination events have broken up the linkage between genes and 5′ spacers. By contrast, in Streptococcus agalactiae, there were a number of cases in which both spacer and gene were unusually divergent, indicating that a number of large-scale recombination events that included both genes and 5′ spacers have occurred; there was evidence of at least two large-scale recombination events in the genomic region including the pur genes in S. agalactiae.
THE extent to which recombination occurs in free-living bacterial populations is an important factor in their population biology. In the case of pathogenic bacteria, Feil et al. (2000) argue that an understanding of the role of recombination is important for prophylaxis and treatment because recombination will have a major impact on the evolution of pathogenicity and the effectiveness of vaccines. Classical studies, using techniques such as multilocus enzyme electrophoresis, assessed recombination between protein-coding loci (Selander et al. 1986). More recently, studies focused on sequence data have tested for evidence of small-scale intralocus recombinational events (McGraw et al. 1999). However, little attention has been paid to the extent of recombination between protein-coding loci and adjacent intergenic (spacer) regions.
Intergenic regions in prokaryotes are much shorter on average than those of eukaryotes, and consequently the density of regulatory elements per intergenic region is expected to be much higher in prokaryotes than in eukaryotes (Rogozin et al. 2002). Spacers between genes in a head-to-head orientation will include promoters for both genes, while spacers between genes in a head-to-tail orientation will include the promoter of one gene. Consequently, the extent of recombination between genes and adjacent spacers may be an important factor in the evolution of intraspecific differences in gene regulation.
A number of recent reviews have focused on the need to understand the evolutionary dynamics of promoters and other genomic regions involved in the regulation of gene expression (Rodriguez-Trelles et al. 2003; Wray et al. 2003). Promoter sequences required for gene expression are expected to be subject to purifying selection (Kohn et al. 2004). Alternatively, there is evidence that natural selection may favor differentiation of promoter regions between populations within species (Rockman et al. 2003) or between closely related species (Kohn et al. 2004) when environmental differences favor changes in the pattern of gene expression. In bacteria, a number of studies have analyzed genome-wide patterns of nucleotide substitution in protein-coding genes by comparing conspecific genomic sequences (Hughes et al. 2002; Jordan et al. 2002). This approach has not been extended to intergenic spacers except in a recent study by Fuglsang (2004) analyzing the 50 nucleotides upstream of start codons and downstream of stop codons in Escherichia coli and two related species.
Here we analyze patterns of nucleotide difference in 5′ intergenic spacers and linked protein-coding genes by comparing conspecific genomic sequences from 10 species of pathogenic bacteria. This data set enables us to compare the pattern of sequence differentiation in intergenic regions with that in protein-coding genes. By comparing the sequence divergence in protein-coding genes with that in adjacent intergenic regions, we obtain evidence regarding the extent to which these two types of sequence share recent evolutionary histories. This in turn provides evidence regarding the relative prevalence of recombination between genes and their promoters in comparison to events in which the gene and promoter region are exchanged as a unit.
METHODS
Comparisons were made of orthologous pairs of genes and 5′ intergenic regions (spacers) between 10 pairs of conspecific complete genomes: Chlamydophila pneumoniae CWL029 (NC_000922) and J138 (NC_002491); E. coli K12 (NC_000913) and O157:H7 (NC_002695); Helicobacter pylori 26695 (NC_000915) and J99 (NC_000921); Mycobacterium tuberculosis CDC1551 (NC_002755) and H37Rv (NC_000962); and Neisseria meningitidis Z2491 (NC_003116) and MC58 (NC_003112); Staphylococcus aureus Mu50 (NC_002758) and N315 (NC_002745); Streptococcus agalactiae 2603V/R (NC_004116) and NEM316 (NC_004368); Streptococcus pyogenes MGAS315 (NC_004070) and SSI-1 (NC_004606); Streptococcus pneumoniae R6 (NC_003098) and TIGR4 (NC_003028); and Yersinia pestis CO92 (NC_003143) and KIM (NC_004088).
To identify orthologous pairs of sequences, we applied the BLASTCLUST program (Altschul et al. 1997) to each pair of conspecific genomes to assemble coding regions and spacers into gene families on the basis of sequence homology. Homology search for genes was done at the amino acid level, while that for spacers was done at the nucleotide level. We used the following parameters for the homology search and assembly of families: expect (E) value of 10−6 and 60% similarity across at least 80% of the alignment. We used these relatively strict search criteria to maximize the probability that orthologous rather than paralogous sequences would be identified. When sequence families were identified, we chose for further analysis only those families including a single member from each of the two conspecific genomes. This was done to avoid the problem of identifying orthologs in cases where recent gene duplication had occurred in one or both of the genomes compared. Finally, 5′ spacer sequences were linked by genomic location with adjacent coding sequences.
The resulting sequence data set for each species included the sequences of putatively orthologous gene pairs together with their 5′ spacers. In every case in which the data set included two genes in head-to-head orientation (thus sharing a 5′ spacer), one of the two genes was excluded at random. Preliminary analyses showed that the results were not affected by the choice of gene to exclude; similar results were obtained when the excluded gene was used. The numbers of pairs of orthologous genes and 5′ spacers for each species are shown in Table 1. Because these comparisons were between orthologous genes from pairs of closely related genomes, each nucleotide difference between the two sequences must have arisen since their most recent common ancestor. Thus, each comparison is phylogenetically and statistically independent of each other comparison (Felsenstein 1985). In statistical analyses, we assumed that each orthologous sequence pair evolves independently of other genes in the genome. However, because we used the orthologous gene pair as the unit of statistical analysis, our analyses did not require the assumption that each nucleotide site evolves independently, a biologically unrealistic assumption that is routinely made in studies of molecular evolution.
TABLE 1.
Proportions of nucleotide differences at synonymous (pS), nonsynonymous (pN), and 5′ spacer (p) sites for comparisons between conspecific genomes of 10 pathogenic bacteria
|
pS
|
pN
|
p
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | N | Mean ± SE | Median | Skew | Mean ± SE | Median | Skew | Mean ± SE | Median | Skew |
| C. pneumoniae | 494 | 0.0009 ± 0.0001 | 0.0000 | 5.13 | 0.0004 ± 0.0001 | 0.0000* | 5.93 | 0.0008 ± 0.0002 | 0.0000 | 13.03 |
| E. coli | 1453 | 0.0472 ± 0.0011 | 0.0391 | 3.59 | 0.0035 ± 0.0002 | 0.0017** | 12.18 | 0.0102 ± 0.0004 | 0.0054**,*** | 2.82 |
| H. pylori | 218 | 0.1518 ± 0.0034 | 0.1531 | 0.54 | 0.0181 ± 0.0019 | 0.0140** | 10.51 | 0.0429 ± 0.0019 | 0.0394**,*** | 0.60 |
| M. tuberculosis | 984 | 0.0074 ± 0.0017 | 0.0000 | 8.30 | 0.0087 ± 0.0020 | 0.0000 | 8.25 | 0.0003 ± 0.0001 | 0.0000*** | 10.01 |
| N. meningitidis | 436 | 0.0977 ± 0.0064 | 0.0661 | 4.73 | 0.0243 ± 0.0035 | 0.0062** | 7.90 | 0.0135 ± 0.0009 | 0.0068** | 2.31 |
| S. aureus | 1211 | 0.0010 ± 0.0006 | 0.0000 | 29.62 | 0.0007 ± 0.0005 | 0.0000 | 34.60 | 0.0002 ± 0.0000 | 0.0000 | 12.37 |
| S. agalactiae | 746 | 0.0129 ± 0.0010 | 0.0040 | 6.74 | 0.0019 ± 0.0002 | 0.0000** | 7.88 | 0.0040 ± 0.0004 | 0.0000**,*** | 3.50 |
| S. pneumoniae | 502 | 0.0213 ± 0.0022 | 0.0098 | 9.31 | 0.0042 ± 0.0007 | 0.0013** | 12.38 | 0.0183 ± 0.0020 | 0.0000** | 3.32 |
| S. pyogenes | 683 | 0.0001 ± 0.0000 | 0.0000 | 14.02 | 0.0000 ± 0.0000 | 0.0000 | 11.06 | 0.0002 ± 0.0001 | 0.0000 | 25.04 |
| Y. pestis | 1308 | 0.0002 ± 0.0001 | 0.0000 | 22.30 | 0.0001 ± 0.0001 | 0.0000 | 27.88 | 0.0002 ± 0.0001 | 0.0000 | 13.62 |
N, number of genes compared. Wilcoxon signed-rank tests of the hypothesis that the median difference between pN and pS equals zero and of the hypothesis that the median difference between p and pS equals zero: *P < 0.001; **P < 0.0001 (Bonferroni corrected). Wilcoxon signed-rank tests of the hypothesis that the median difference between pN and p equals zero: ***P < 0.0001 (Bonferroni corrected).
Both coding and spacer regions were aligned using the program ClustalW 1.83 (Thompson et al. 1994). In the case of S. agalactiae, the BLASTZ program (Schwartz et al. 2003) was used to align the complete genomic sequences. For coding regions, gaps were not allowed to occur within any codon, thus preserving the frame of translation. The proportions of synonymous differences per synonymous site (pS) and of nonsynonymous differences per nonsynonymous site (pN) were estimated between each pair of orthologous coding sequences by the Nei and Gojobori (1986) method. In spacers, the uncorrected proportion (p) of nucleotide differences was calculated. Thus, for both coding and noncoding regions no statistical correction for multiple hits was applied to the observed proportions of nucleotide difference.
Uncorrected proportions were used for a number of reasons. First, in most cases, the sequences compared were very similar; thus, correction made little if any practical difference. Second, in the present study, we were interested in comparing relative proportions of sequence difference in different regions, not in estimating the true number of substitutions accumulated over evolutionary time. By not assuming any substitution model, we can compare proportions of difference in a robust fashion that is not model dependent. Because within each species the distributions of pS, pN, and p deviated significantly from normality (P < 0.01 by Kolmogorov-Smirnov test), we used nonparametric methods for all statistical tests. In all analyses, significance levels were adjusted for multiple testing using the Bonferroni correction. We conducted nonhierarchical k-means clustering using McQueen's algorithm (Johnson and Wichern 1992). This is a method of creating clusters of observed multivariate data points such that variability within clusters is minimized and variability between clusters is maximized. All statistical analyses were conducted using the Minitab statistical package, release 13 (http://www.minitab.com/).
RESULTS
Patterns of nucleotide difference:
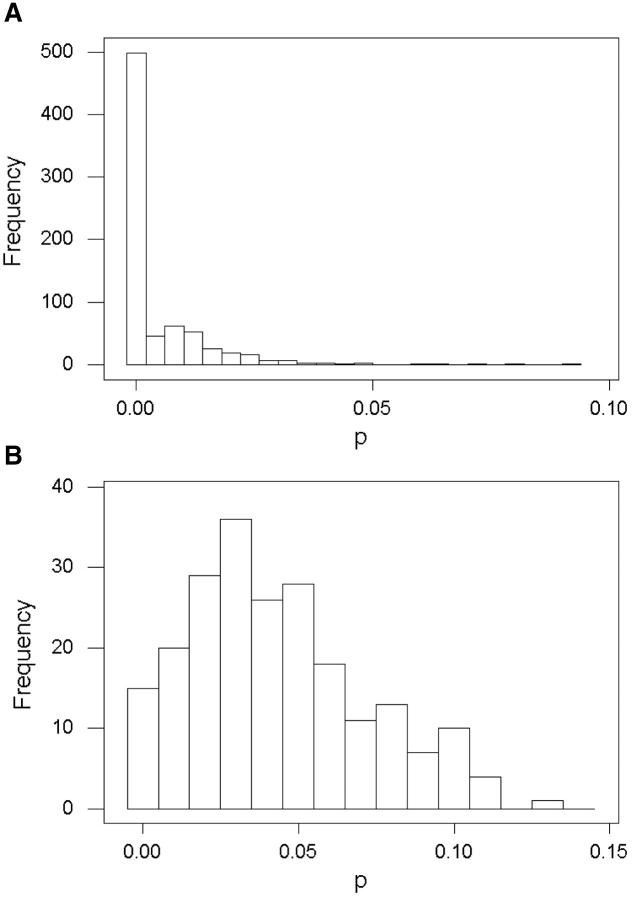
Table 1 summarizes mean and median pS and pN in coding regions and mean p in adjacent 5′ spacers for the 10 bacterial species analyzed. The distributions of all three quantities were positively skewed, in most cases extremely so; thus mean values were typically substantially higher than median values (Table 1). The distribution of p in 5′ spacers of S. agalactiae provides an example (Figure 1A). In this species, a majority of 5′ spacers compared between the two genomes showed no nucleotide differences, while a small number showed substantially higher p values, with a maximum value of 0.093 (Figure 1A). Similar distributions were seen for pS, pN, and p in most species (Table 1). The least-skewed distributions were those of pS and p in H. pylori (Table 1 and Figure 1B). The two genomes of H. pylori were much more divergent overall than were those of any other species analyzed, as indicated by the relatively high median pS and p in H. pylori (Table 1).
Figure 1.—
Frequency distributions of the proportion of nucleotide difference (p) in spacers of (A) S. agalactiae and (B) H. pylori.
In 6 of the 10 species analyzed, it was possible to reject the hypothesis that the median pairwise difference between pS and pN was equal to zero (Table 1). The pattern of pS > pN seen in these 6 species is indicative of purifying selection acting to a greater extent on nonsynonymous mutations than on synonymous mutations and is typical of most protein-coding genes (Kimura 1977; Hughes 1999). In five of these six species, median p in the 5′ spacer was also significantly less than median pS in the linked protein-coding gene (Table 1). The four species that did not show significant differences either between median pS and pN or between median pS and p were the species with the lowest mean pS and p (Table 1).
In four species, there was a significant difference between median p in the 5′ spacer and median pN in the linked gene (Table 1). In three of these species, median p was greater than median pN. This is evidence that, in these species, purifying selection on spacers was not as stringent as that on nonsynonymous sites in genes. By contrast, in M. tuberculosis, median p in the spacers was significantly lower than median pN in the linked genes (Table 1).
Nucleotide content:
Table 2 summarizes mean and median G + C content at third positions of codons in genes (GC3) and in linked 5′ spacers (GC5′). The species analyzed showed a wide range of G + C contents from very high (M. tuberculosis) to very low (S. aureus) (Table 2). In 9 of 10 species, there was a significant difference between median GC3 and median GC5′ (Table 2). In 7 species, median GC3 was significantly higher than GC5′ (Table 2). However, in 2 species, S. aureus and S. agalactiae, median GC3 was significantly lower than median GC5′ (Table 2). These two species were the two species with the lowest overall G + C levels.
TABLE 2.
Mean and median percentage of G + C at GC3 and in GC5′
| GC3
|
GC5′
|
|||
|---|---|---|---|---|
| Species | Mean ± SE | Median | Mean ± SE | Median |
| C. pneumoniae | 34.5 ± 0.2 | 34.8 | 32.2 ± 0.3 | 32.4a |
| E. coli | 54.5 ± 0.2 | 55.4 | 42.5 ± 0.2 | 42.3a |
| H. pylori | 43.0 ± 0.3 | 43.2 | 27.9 ± 0.3 | 27.3a |
| M. tuberculosis | 79.2 ± 0.2 | 79.8 | 62.1 ± 0.2 | 62.6a |
| N. meningitides | 61.7 ± 0.5 | 63.9 | 44.6 ± 0.3 | 45.1a |
| S. aureus | 22.7 ± 0.1 | 22.2 | 27.1 ± 0.1 | 26.5a |
| S. agalactiae | 25.0 ± 0.1 | 24.8 | 28.1 ± 0.2 | 27.6a |
| S. pneumoniae | 35.2 ± 0.3 | 35.8 | 31.4 ± 0.3 | 31.1a |
| S. pyogenes | 31.5 ± 0.2 | 31.4 | 32.0 ± 0.2 | 31.6 |
| Y. pestis | 48.0 ± 0.2 | 48.8 | 40.5 ± 0.2 | 40.8a |
Wilcoxon signed-rank tests of the hypothesis that the median GC3 equals the median GC5′ (P < 0.001, Bonferroni corrected).
Recombinational patterns:
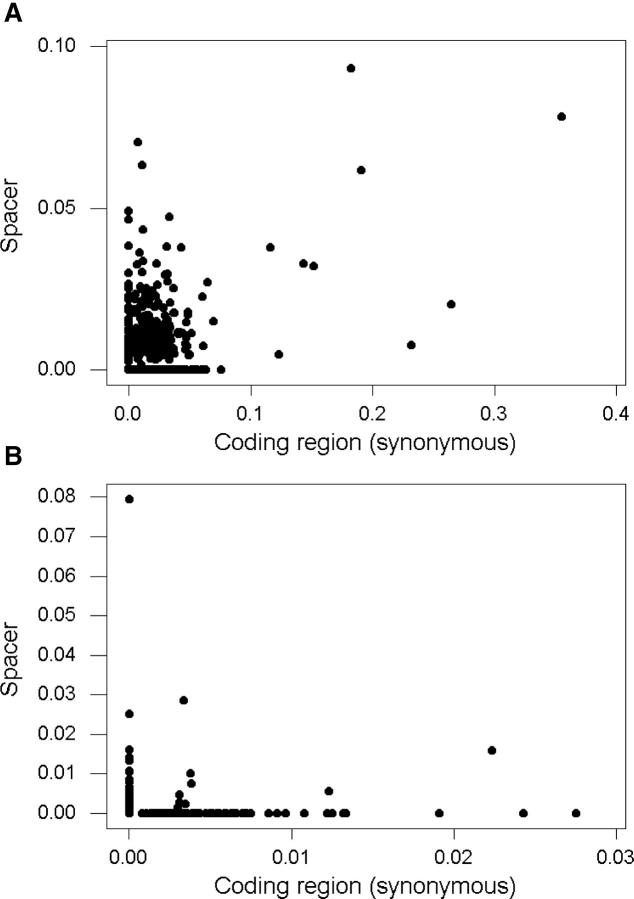
The relationship between p in 5′ spacers and pS in the linked genes differed strikingly among species. In S. agalactiae, there were numerous genes at which the two genomes showed few nucleotide differences either at synonymous sites or in the 5′ spacer. In fact, in 297 of 746 genes (39.8%), the two genomes were identical in both synonymous sites and in the 5′ spacer. On the other hand, there were a number of other cases in S. agalactiae in which both a gene and its linked 5′ spacer were relatively highly divergent between the two genomes (Figure 2A). As a consequence, in S. agalactiae, there was a strong positive correlation between p and pS (Figure 2A).
Figure 2.—
Plots of p in 5′ spacers vs. pS in the linked coding region for (A) S. agalactiae (Spearman rank correlation coefficient rS = 0.397; P < 0.001) and for (B) C. pneumoniae (rS = 0.010; n.s.).
C. pneumoniae provided an example of a very different pattern, in which genes with high pS generally had low p in the 5′ spacer and vice versa (Figure 2B). In C. pneumoniae, the two genomes compared were identical in both synonymous sites and in the 5′ spacer in 366 of 494 genes (74.1%). There were 87 genes with nonzero pS and 50 genes with nonzero p in the 5′ spacer, but only 9 genes had both nonzero pS and nonzero p (Figure 2B). As a consequence, there was not a significant correlation between p and pS in C. pneumoniae (Figure 2B). M. tuberculosis and S. pyogenes (data not shown) provided additional examples of a pattern similar to that seen in C. pneumoniae (Figure 2B).
These different patterns can be interpreted as resulting from different patterns of past recombination. If a certain sequence region shows substantially greater divergence between two conspecific genomes than do most other regions, this pattern might be explained by a recombinational event whereby one of the genomes obtained the divergent region from a distantly related donor genome (Hughes et al. 2002). The pattern seen in S. agalactiae (Figure 2A) suggests that in a number of cases both genes and their 5′ spacers have been derived by recombination with divergent genomes. In C. pneumoniae, by contrast, certain genes and certain spacers appear to be derived from divergent genotypes, but the plot of p vs. pS (Figure 2B) suggests that there were few if any cases in which a divergent gene and 5′ spacer were acquired as a unit.
Numerous factors, in addition to recombination, can potentially influence the pattern of sequence divergence in genomic regions. These include the relative strength of purifying selection and bias in nucleotide composition. Because these factors are likely to interact in complex ways, we computed rank partial correlation coefficient between p in 5′ spacers and a set of other variables, simultaneously controlling for other variables in the set (Table 3). In these analyses, a significant positive partial correlation between p in 5′ spacers and pS in genes was observed in three species: E. coli, S. aureus, and S. agalactiae (Table 3). In E. coli and S. agalactiae, a significant positive partial correlation was also observed between p in 5′ spacers and pN in genes (Table 3).
TABLE 3.
Fourth-order rank partial correlation coefficients between the proportion of nucleotide difference (p) in 5′ spacers and five variables describing the spacer and the adjacent protein-coding gene (simultaneously controlling for other variables)
| Species | pS | pN | GC5′ | GC3 | Spacer length (bp) |
|---|---|---|---|---|---|
| C. pneumoniae | 0.0234 | −0.0089 | 0.0016 | −0.0721 | 0.2024** |
| E. coli | 0.1011* | 0.1206** | −0.0750* | −0.0832 | 0.3546** |
| H. pylori | 0.0780 | 0.0296 | 0.1343 | 0.0801 | 0.3951** |
| M. tuberculosis | 0.0469 | −0.0564 | −0.0202 | −0.0085 | 0.1664** |
| N. meningitidis | 0.0925 | 0.0472 | 0.1316 | −0.0219 | 0.2866** |
| S. aureus | 0.1316** | 0.0721 | 0.0001 | −0.0159 | 0.1415** |
| S. agalactiae | 0.2850** | 0.1650** | −0.0055 | 0.0087 | 0.2551** |
| S. pneumoniae | 0.0930 | 0.1030 | −0.0734 | −0.0378 | 0.3620** |
| S. pyogenes | −0.0413 | 0.0982 | 0.0275 | 0.0464 | 0.0939 |
| Y. pestis | −0.0249 | −0.0321 | −0.0210 | −0.0332 | 0.1059* |
P < 0.01;
P < 0.0001 (Bonferroni corrected).
The G + C content in 5′ spacers (GC5′) and at third positions of codons in coding regions (GC3) was not significantly correlated with p in all cases except that there was a negative partial correlation between p and GC5′ in E. coli (Table 3). In all but one species, there was a significant positive partial correlation between p in the spacer and length of the spacer (Table 3). When a similar partial correlation analysis was used to examine the relationship between pS and GC3 (simultaneously controlling for p, pN, GC5′, and spacer length), no significant relationships were found except for a highly significant positive correlation between pS and GC3 (0.2454, P < 0.001) in E. coli.
These analyses show that the positive correlation between p in 5′ spacers and pS in genes in S. agalactiae and two other species persists even when other variables are controlled for statistically. Thus, they are consistent with the hypothesis that recombination events involving both a gene and its 5′ spacer have occurred in the evolutionary history of the S. agalactiae genomes compared here.
Cluster analysis:
In addition to patterns of recombination, chance variation in the pattern of nucleotide substitution can potentially cause different levels of sequence divergence to occur in different regions of two related genomes. However, it is expected that sequence differences due to chance variation will be relatively small. Hughes et al. (2002) tested the hypothesis that certain values of pS observed between two genomes of M. tuberculosis were too high to be accounted for by chance, assuming that nucleotide differences follow a binomial distribution. Here we take a different approach, which does not make any assumptions about the underlying model of sequence evolution.
Individual genes were clustered using a k-means clustering algorithm (with k = 3) on the basis of p in the 5′ spacer and pS in the coding region. Then the equality of median p and pS values across clusters was tested by Kruskal-Wallis test.
Recombination that does not break up the linkage between genes and 5′ spacers is expected to yield the presence of one or more clusters of genes in which both p and pS values are relatively high. This pattern was seen in S. agalactiae (Table 4), as expected on the basis of the plot of p vs. pS (Figure 2A). In S. agalactiae, there were nine genes in a cluster with relatively high values of both p and pS (Table 5). A second, larger cluster (232 genes) had intermediate values of p and pS, while the largest cluster (505 genes) had median values of zero for both p and pS (Table 4). Median p and pS were significantly different among these clusters (P < 0.01; Bonferroni corrected). In E. coli, there were also three clusters with significantly different medians for both p and pS (Table 5). As in S. agalactiae, the clusters in E. coli were characterized by increasing numbers of genes and decreasing median values of p vs. pS (Table 5).
TABLE 4.
Medianp in the 5′ spacer andpS in the coding region for clusters of genes identified byk-means clustering
| Species | Cluster no. | N | p | pS |
|---|---|---|---|---|
| C. pneumoniae | 1 | 3 | 0.0286 | 0.0000 |
| 2 | 458 | 0.0000 | 0.0000 | |
| 3 | 33 | 0.0000 | 0.0070 | |
| E. coli | 1 | 78 | 0.0091 | 0.1396 |
| 2 | 525 | 0.0065 | 0.0647 | |
| 3 | 850 | 0.0041 | 0.0245 | |
| S. agalactiae | 1 | 9 | 0.0328 | 0.1820 |
| 2 | 232 | 0.0063 | 0.0256 | |
| 3 | 505 | 0.0000 | 0.0000 | |
| S. pyogenes | 1 | 1 | 0.0844 | 0.0000 |
| 2 | 679 | 0.0000 | 0.0000 | |
| 3 | 3 | 0.0000 | 0.0133 | |
| Y. pestis | 1 | 7 | 0.0208 | 0.0000 |
| 2 | 1298 | 0.0000 | 0.0000 | |
| 3 | 3 | 0.0000 | 0.0387 |
Kruskal-Wallis tests of the equality of medians across clusters: P < 0.01 in all cases (Bonferroni corrected).
TABLE 5.
Genes ofS. agalactiae with highpS in coding region and highp in adjacent 5′ spacer
| Protein function | 5′ spacera | p | Coding regiona | pS | pN |
|---|---|---|---|---|---|
| Phosphoribosylformylglycinamidine synthase | 36461–36582 | 0.0328 | 36583–40408 | 0.1433 | 0.0155 |
| Phosphoribosylamine-glycine ligase | 57110–57990 | 0.0320 | 57991–59256 | 0.1520 | 0.0073 |
| Phosphoribosylaminoimidazole carboxylase | 59257–59536 | 0.0618 | 59537–60025 | 0.1909 | 0.0219 |
| Integrase | 232951–233014 | 0.0781 | 231796–232950 | 0.3554 | 0.0257 |
| Unknown | 240364–240869 | 0.0378 | 240076–240363 | 0.1163 | 0.0133 |
| Acetyl transferase | 262238–262336 | 0.0202 | 262337–262915 | 0.2649 | 0.0408 |
| Unknown | 901703–902089 | 0.0078 | 902090–906202 | 0.2320 | 0.0743 |
| dTDP-glucose-4,6-dehydratase | 12058814–1206019 | 0.0049 | 1204767–1205813 | 0.1230 | 0.0143 |
| Hypothetical transcriptional regulator | 1340567–1340839 | 0.0933 | 1340126–1340566 | 0.1823 | 0.0203 |
Nucleotide positions in the 2603V/R genome (NC_004116).
We predicted that recombination breaking up the linkage between genes and 5′ spacers would lead to the presence of a cluster with relatively high p but low pS and another cluster with relatively low p but high pS. This pattern was seen in C. pneumoniae (Table 4), as expected on the basis of the plot of p vs. pS (Figure 2B). A similar pattern was seen in S. pyogenes and Y. pestis (Table 4). In these species, although the numbers of genes with unusually high p and pS were relatively few, the clusters were nonetheless significantly different with respect to median p and pS values (Table 4).
Recombination regions in S. agalactiae:
To examine further the recombinational history of S. agalactiae, we examined the chromosomal locations of the nine genes from the cluster (Table 4) most divergent between the two genomes in the 5′ spacers and at synonymous sites in the coding regions (Table 5). Three of these genes (encoding phosphoribosylformylglycanimide synthase, phosophoribosylamine-glycine ligase, and the phosphoribosylaminoimidazole carboxylase catalytic subunit) are involved in purine metabolism (Table 5). Homologs of these and other genes involved in purine synthesis are linked and regulated as an operon (the pur operon) in certain bacteria (Ebbole and Zalkin 1987). In S. agalactiae, the corresponding genes are scattered over a region that occupies ∼25 kb and includes about 23 genes, apparently not all of them involved in purine synthesis. We refer to this region as the pur region, recognizing that it is probably not regulated as an operon in S. agalactiae, given the presence of genes not involved in purine synthesis.
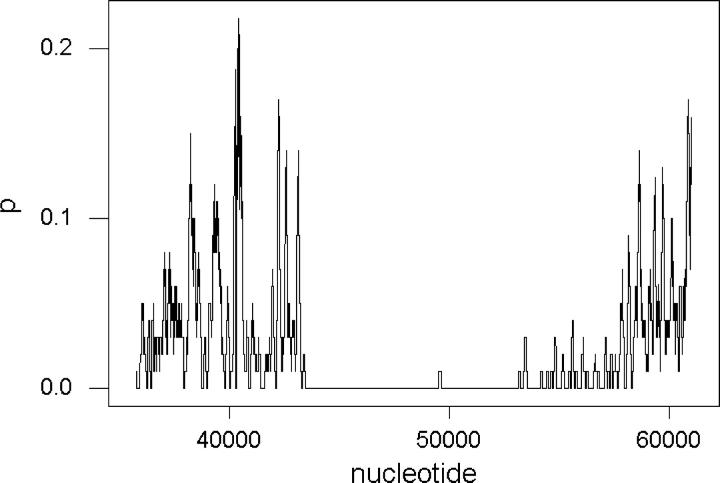
Figure 3 shows a sliding window plot of p across the pur region in comparison with the S. agalactiae genomes. There were two portions of the pur region, approximately positions 35929–43467 and 53145–61023, in which the two genomes showed substantial sequence divergence (Figure 3). By contrast, in the center portion of the pur region, the two genomes were virtually identical (Figure 3). The first divergent region (35929–43467) spanned six predicted protein-coding genes, including one of the genes in our data set with high p in 5′ spacers and high pS in coding regions (Table 5). The second divergent region (53145–61023) included eight predicted genes, including two of the genes in our data set with high p in 5′ spacers and high pS in coding regions (Table 5). Because these two regions showed higher sequence divergence than is typical for these two genomes (Tables 1 and 4), it seems likely that these two portions of the pur region have been involved in recombination events that introduced divergent sequences into one of the two genomes. Because there are two distinct divergent regions, it seems plausible that there were at least two such recombination events, although there may have been more than two.
Figure 3.—
Plot of the proportion of nucleotide difference (p) between two genomes of S. agalactiae in a sliding window of 100 nucleotides across the genomic region including the pur genes. Nucleotide position of the starting point of each window on the x-axis is numbered as in the 2603V/R genome (NC_004116; positions 35756–61023).
When a similar sliding window analysis was applied to the entire genome of S. agalactiae, it likewise showed regions of sequence identity interspersed with divergent regions (not shown). Overall, regions of sequence identity predominated; in 1,392,801 of 1,955,174 (71.2%) genomic windows, the two genomes were identical. On the other hand, there were interspersed regions of greater divergence, in which the genes with high values of both p in the 5′ spacer and pS in the coding region (Table 5) were found.
DISCUSSION
We compared the patterns of nucleotide difference in genes and in the spacer regions located 5′ to genes between 10 pairs of conspecific bacterial genomes to obtain evidence regarding the comparative evolutionary dynamics of genes and spacers. In five of the species analyzed, the median proportion of nucleotide difference (p) in the 5′ spacer was significantly lower than the median proportion of synonymous difference (pS) in the coding region (Table 1). These were the five species for which the genome pairs showed the greatest overall sequence divergence (Table 1), suggesting that a certain level of sequence divergence is required for the difference between genes and spacers to be detected.
Evidence for a reduced level of nucleotide divergence in spacers compared to synonymous sites in genes suggests that intergenic spacers in general are subject to purifying selection. Furthermore, the selection on spacers at least in these five species is stronger than any purifying selection (due, for example, to selection on codon usage) that may act on synonymous sites in genes. The five species in which there was no significant difference between median p in 5′ spacers and median pS in genes were also the species with the lowest overall sequence divergence (in both spacers and genes) between the two genomes compared (Table 1). Because of this low level of sequence divergence, it may not be possible to detect differences between spacers and genes in these species even though purifying selection is present on the former.
A major factor responsible for purifying selection on spacers is presumably the presence of regulatory elements. Consistent with this hypothesis, in 9 of 10 species a significant positive partial correlation was observed between p in spacers and the length of the spacer (Table 3). This correlation may be explained by the fact that the proportion of sites that constitute conserved promoter sequences is expected to be lower in longer spacers.
Additional evidence for functional constraint on spacers was provided by data on nucleotide content. In a majority of the species examined, the median proportion G + C in 5′ spacers was significantly lower than that at synonymous sites in genes (Table 2). It is well known that bacterial promoters include certain conserved AT-rich regulatory elements, including not only the −10 and −35 hexamers but also the UP elements (Gourse et al. 2000). The reduction of G + C content in spacers relative to synonymous sites in genes may at least reflect in part the conservation of such regulatory elements and surrounding A- and T-rich sequences. In two species (S. aureus and S. agalactiae) with very low overall G + C content, the median proportion G + C in 5′ spacers was actually significantly greater than that at synonymous sites in genes (Table 2). In these cases, the conservation of regulatory elements may require a G + C content greater than that of synonymous sites in genes, since these elements include G and C at certain positions (Gourse et al. 2000).
Certain genes showed unusually high pS, unusually high p in the 5′ spacer, or both, in comparison to other genes in the same pair of conspecific genomes. The most plausible explanation for such cases is recombination with divergent genomes (Hughes et al. 2002). One limitation of using the nonuniformity of nucleotide differences as evidence for past recombination is that such evidence will not be available if recombination has involved very similar genomes; but of course this is true of all kinds of evidence of recombination, including that obtained from classical genetics, which requires differentiation of markers. Nonetheless, the presence of certain unusually high values of pS and p in the comparison of two otherwise closely related genomes (such as the two genomes of C. pneumoniae compared here) is suggestive of divergent origins for these divergent genes. Alternative hypotheses to explain such a pattern include (1) chance variation among genes with respect to the number of nucleotide differences and (2) the effects of changes in natural selection. However, there are reasons for believing that neither of these hypotheses is likely to explain the majority of cases observed in the present data set.
Assuming a probability model, it is possible to compute the probability of a given degree of sequence divergence. For example, Hughes et al. (2002) assumed a binomial model and showed that, under this model, the pS values obtained for certain genes in the comparison of the two available complete genomes of M. tuberculosis were extremely unlikely to occur as a result of random variations in the pattern of nucleotide substitution. In the present study, we used an alternative approach based on k-means clustering, which has the advantage that it does not depend on any assumptions about the underlying model of sequence evolution. In the case of S. agalactiae, this approach isolated a cluster of nine genes with relatively high values of p in the 5′ spacer and pS in the coding region (Tables 4 and 5). In C. pneumoniae and three other species, this approach yielded both clusters of genes with high p in the 5′ spacer but low pS and clusters of genes with low p in the 5′ spacer and high pS (Table 4). The fact that the median p and pS values differed significantly among these clusters is strong evidence that they are not simply the result of random variations in the pattern of nucleotide substitution.
It is uncertain how natural selection might cause an increase in pS, since natural selection acts primarily at the level of protein sequence and structure (Hughes 1999). The only plausible such effect might be the relaxation in certain genes of constraints on codon usage present in most genes in a genome. This seemed unlikely to be a major factor in the present analyses for a number of reasons. First, in 9 of 10 species there was not a significant partial correlation between pS and G + C content at third-codon positions. In E. coli, there was a significant positive partial correlation between pS and G + C content at third-codon positions. However, since the preferred codons in E. coli are usually those ending in G or C (Sharp and Li 1986), this positive correlation is not consistent with a relaxation of selection. Similarly, an increase in p might be explained by a relaxation of purifying selection on a given spacer. However, the fact that median values of p in spacers remained substantially lower than median pS in linked genes—even in the S. agalactiae genes in which both values were unusually high (Tables 4 and 5)—was evidence against the hypothesis that relaxation of selection can account for most cases of unusually high p in spacers.
The pattern of nucleotide divergence in spacers and at synonymous sites in linked genes provided evidence regarding the extent to which genes and 5′ spacers are transferred together by recombination events. Different patterns were seen in different species, indicative of distinct recombinational histories. In C. pneumoniae, S. pyogenes, and Y. pestis there were genes in which the 5′ spacer was substantially more divergent between the two genomes than were synonymous sites in the linked gene, as well as cases in which synonymous sites in the gene were substantially more divergent than the 5′ spacer, but there were no cases in which both gene and spacer were unusually divergent (Table 4). By contrast, in S. agalactiae and E. coli, there were a number of cases in which both spacer and gene were unusually divergent (Table 4). Partial correlation analyses indicated that factors such as nucleotide content could not account for this pattern (Table 3). Therefore, the most reasonable interpretation seems to be that, in C. pneumoniae, recombination events have broken up the linkage between genes and 5′ spacers, whereas in the S. agalactiae genomes a number of large-scale recombination events that included both genes and 5′ spacers have occurred.
The latter hypothesis was supported by a sliding window comparison of the two S. agalactiae genomes. The portion of the genome including the pur genes showed evidence of two extensive recombination regions, including multiple genes and spacers (Figure 3). Although these genes evidently do not constitute an operon in S. agalactiae, their expression must be coordinated in some fashion. In such cases, recombination events that include several genes along with their 5′ spacers may have more chance of success than the exchange of shorter regions because genes and regulatory regions will be transferred together. Furthermore, such exchanges have the potential to introduce new regulatory pathways to a genotype and thus contribute to adaptive evolution of bacterial phenotypes.
As more genomes of S. agalactiae are sequenced, it should be possible to identify the source of divergent genomic regions, thereby providing definitive evidence in support of the hypothesis of recombination.
Acknowledgments
This research was supported by grant GM43940 from the National Institutes of Health.
References
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole, D. J., and H. Zalkin, 1987. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 262: 8274–8287. [PubMed] [Google Scholar]
- Feil, E. J., M. C. Enright and B. G. Spratt, 2000. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitides and Streptococcus pneumoniae. Res. Microbiol. 151: 465–469. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1985. Phylogenies and the comparative method. Am. Nat. 125: 1–15. [Google Scholar]
- Fuglsang, A., 2004. Evolution of prokaryotic DNA: intragenic and extragenic divergences observed with orthologs from three related species. Mol. Biol. Evol. 21: 1152–1159. [DOI] [PubMed] [Google Scholar]
- Gourse, R. L., W. Ross and T. Gaal, 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37: 687–695. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., 1999 Adaptive Evolution of Genes and Genomes. Oxford University Press, New York.
- Hughes, A. L., R. Friedman and M. Murray, 2002. Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerg. Infect. Dis. 8: 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R., and D. Wichern, 1992 Applied Multivariate Statistical Methods, Ed 3. Prentice-Hall, Englewood Cliffs, NJ.
- Jordan, I. K., I. B. Rogozin, Y. I. Wolf and E. V. Koonin, 2002. Microevolutionary genomics of bacteria. Theor. Popul. Biol. 61: 435–444. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1977. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267: 275–276. [DOI] [PubMed] [Google Scholar]
- Kohn, M. H., S. Fang and C.-I Wu, 2004. Inference of positive and negative selection on the 5′ regulatory regions of Drosophila genes. Mol. Biol. Evol. 21: 374–383. [DOI] [PubMed] [Google Scholar]
- McGraw, E. A., J. Li, R. K. Selander and T. S. Whittam, 1999. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16: 12–22. [DOI] [PubMed] [Google Scholar]
- Nei, M., and T Gojobori, 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Rockman, M. V., M. W. Hahn, N. Sorzano, D. B. Goldstein and G. A. Wray, 2003. Positive selection on a human-specific transcription factor binding site regulating IL4 expression. Curr. Biol. 13: 2116–2123. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles, F., R. Tarrio and F. J. Ayala, 2003. Evolution of cis-regulatory elements versus codifying regions. Int. J. Dev. Biol. 47: 665–673. [PubMed] [Google Scholar]
- Rogozin, I. B., K. S. Makarova, D. A. Natale, A. N. Spiridonov, R. L. Tatusov et al., 2002. Congruent evolution of different classes of non-coding DNA in prokaryotic genomes. Nucleic Acids Res. 30: 4264–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., W. J. Kent, A. Smit, Z. Zhang, R. Baertsch et al., 2003. Human-mouse alignments with BLASTZ. Genome Res. 13: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour et al., 1986. Methods of multilocus enzyme electrophoresis for bacterial genetics and systematics. Appl. Environ. Microbiol. S1: 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P. M., and W.-H. Li, 1986. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 24: 28–38. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. Gibson, 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G. A., M. W. Hahn, A. Abouheif, J. P. Balhoff, M. Pizer et al., 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377–1419. [DOI] [PubMed] [Google Scholar]