Abstract
We study levels of X-linked vs. autosomal diversity using a model developed to analyze the hitchhiking effect. Repeated bouts of hitchhiking are thought to lower X-linked diversity for two reasons: first, because sojourn times of beneficial mutations are shorter on the X, and second, because adaptive substitutions may be more frequent on the X. We investigate whether each of these effects does, in fact, cause reduced X-linked diversity under hitchhiking. We study the strength of the hitchhiking effect on the X vs. autosomes when there is no recombination and under two different recombination schemes. When recombination occurs in both sexes, X-linked vs. autosomal diversity is reduced by hitchhiking under a broad range of conditions, but when there is no recombination in males, as in Drosophila, the required conditions are considerably more restrictive.
A long-standing debate in evolutionary biology concerns whether nearly neutral evolution (such as purifying selection against deleterious mutations) or adaptive evolution has played a larger role in shaping genome-wide patterns of genetic variation. One such pattern is the well-known positive correlation between recombination and polymorphism seen in many taxa (Begun and Aquadro 1992; Nachman 1997; Nachman et al. 1998; Stephan and Langley 1998; Cutter and Payseur 2003). Both neutral and nonneutral explanations have been offered to explain this pattern, i.e., the background selection and hitchhiking hypotheses, both of which are forms of Hill-Roberston interference (Hill and Robertson 1966). Background selection involves the constant removal of weakly deleterious mutations by purifying selection: in regions of low recombination, deleterious mutations cannot be separated from linked neutral variants, so that purifying selection tends to remove both (Charlesworth 1994, 1996). Hitchhiking due to selective sweeps also purges variation from regions of low recombination. But in this case, positively selected mutations going to fixation cannot be separated from the surrounding neutral variation, so that directional selection tends to fix both (Maynard Smith and Haigh 1974). In regions of high recombination, in contrast, only short stretches of linked neutral sites are affected by selection (either purifying or positive) at neighboring sites and neutral variation is preserved. Both models can, therefore, qualitatively explain the observed positive correlation between recombination and neutral variation.
Attempts to evaluate the relative importance of background selection and hitchhiking have naturally focused on predictions that differ between the two models (Aquadro et al. 1994; Stephan et al. 1998; Begun and Whitley 2000; Andolfatto and Przeworski 2001; Wall et al. 2002; Innan and Stephan 2003; see Table 1 in Kauer et al. 2002). One potentially powerful means of distinguishing the two models involves comparing levels of variation on X chromosomes to that on autosomes (Aquadro et al. 1994). Both types of chromosomes have presumably experienced similar (though not necessarily identical) demographic histories, but the effects of background selection and hitchhiking differ for X chromosomes and autosomes due to hemizygous selection in males (Aquadro et al. 1994). [For simplicity, we assume throughout that males are the heterogametic (XY) sex, as in Drosophila and mammals.]
Background selection is more effective on the autosomes, as the strength of background selection at a locus is proportional to the frequency of deleterious alleles under purifying selection (Charlesworth et al. 1993; Charlesworth 1994). Because deleterious alleles can reach higher frequencies on the autosomes than on the X, background selection purges more variation from the autosomes than from the X. Hitchhiking, on the other hand, may be more powerful on the X for two quite different reasons. First, the sojourn time of a beneficial mutation on its way to fixation is shorter on the X chromosome than on an autosome (Avery 1984; Aquadro et al. 1994). There are thus fewer generations in which recombination can occur during a selective sweep. Second, the adaptive substitution rate may be higher on the X than on the autosomes if the average beneficial mutation is new and partially recessive (with a heterozygote enjoying less than half of the fitness benefit enjoyed by homozygotes); under these conditions, the mean time back to the last substitution is shorter on the X than on an autosome (Charlesworth et al. 1987). Because the strength of hitchhiking increases both when sojourn times are shorter and when substitution rates are higher, both effects might reduce X-linked variation more than autosomal variation under hitchhiking.
To date, most data comparing levels of X-linked vs. autosomal variation come from Drosophila. Interestingly, the pattern observed depends on the population sampled. In African populations of Drosophila melanogaster and D. simulans, which are thought to be ancestral for these two species, X-linked diversity appears to be equal to or higher than autosomal diversity (Irvin et al. 1998; Begun and Whitley 2000; Andolfatto 2001; Kauer et al. 2002; Sheldahl et al. 2003). Outside of Africa, however, X-linked diversity may be reduced relative to autosomal diversity (Irvin et al. 1998; Begun and Whitley 2000; Andolfatto 2001; Kauer et al. 2002; Sheldahl et al. 2003; Mousset and Derome 2004). Remarkably, this contrast between African and non-African populations may be mirrored in humans, which also have an ancestral African source population (Payseur and Nachman 2002). It is tempting to suggest, as Andolfatto (2001) and Kauer et al. (2002) do, that this difference between African and non-African populations reflects rapid adaptation to temperate environments and the resulting bouts of selective sweeps.
Firm conclusions may be premature, however, as the verbal argument given above—that hitchhiking disproportionately reduces X-linked heterozygosity—has not been systematically studied theoretically. And the theoretical work that has been performed actually suggests that hitchhiking may not explain patterns of diversity in non-African D. simulans populations (Wall et al. 2002). Here, we modify Gillespie's (2000) pseudohitchhiking model in an attempt to more thoroughly study the effect of hitchhiking on levels of X-linked vs. autosomal variation. We pay particular attention to the effect of the dominance of beneficial mutations, as this parameter determines the relative rates of adaptive substitutions, and thus the frequency of hitchhiking, on the X vs. the autosomes. Specifically, we determine the range of dominance coefficients over which hitchhiking causes a reduction in X-linked vs. autosomal diversity. We also determine whether this effect is due to shorter sojourn times on the X, to faster substitution rates on the X, or to both.
THE MODEL AND RESULTS
We consider a two-locus model, with a “selected” locus, which experiences recurrent adaptive substitutions, and a “neutral” locus, which is linked to the selected locus. Throughout we assume that adaptation involves fixation of new beneficial mutations, not segregating polymorphic alleles, for which results may differ (see Orr and Betancourt 2001). Substitutions at the selected locus reduce heterozygosity at the neutral locus via pseudohitchhiking or “genetic draft” (Gillespie 2000). In this model, the reduction of heterozygosity is caused by a series of selective sweeps, rather than by a single substitution. Each sweep is treated as instantaneous (except when calculating the increase in frequency of a “hitchhiking” neutral allele) and substitutions form a Poisson process with a rate that depends on the rate at which new mutations appear (see Gillespie 2000). The model also assumes a Wright-Fisher population, wherein genetic drift is modeled by binomial sampling of alleles from a single population. The equilibrium heterozygosity at the neutral locus is measured by the quantity ssh, the sum-of-site heterozygosities.
We consider two general cases: that in which linkage between the selected and neutral loci is complete (the no-recombination case) and that in which the linkage is partial (the recombination case). We also consider two variations on the recombination case, that in which recombination is Drosophila-like, occurring only in females, and that in which recombination occurs in both sexes.
No recombination:
With no crossing over between the selected and neutral loci, Gillespie (2000) showed that the expected sum-of-sites heterozygosity at a neutral autosomal locus is
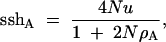 |
1 |
where ρA is the rate of adaptive substitution at the selected locus, and N and u are the population size and mutation rate at the neutral locus, respectively. As population size grows (N → ∞), genetic drift becomes negligible and recurrent hitchhiking alone acts. Equation 1 then approaches sshA = 2u/ρA.
We now find the expected sum-of-sites heterozygosity at a neutral X-linked locus that is completely linked to a selected locus experiencing a stream of adaptive substitutions. Our derivation is a trivial modification of Gillespie's (2000) derivation for an autosomal locus. The mean time back to the most recent common ancestor of two randomly chosen X-linked alleles is
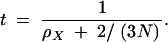 |
2 |
This reflects the fact that the two alleles will coalesce either because of a hitchhiking event at the selected locus (which occurs on average t1 = 1/ρX generations ago) or because of a coalescent event at the neutral locus (which occurs on average t2 = 3N/2 generations ago). The overall mean time to a coalescence is the minimum of these two exponentially distributed times and is itself exponentially distributed (Gillespie 1991), with a mean of t = 1/[1/t1 + 1/t2]; hence we have Equation 2. Because an average of 2ut mutations accumulates during this time,
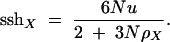 |
3 |
For large populations (N → ∞), this quantity approaches sshX = 2u/ρX.
Thus, with no recombination,
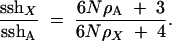 |
4 |
Two extreme cases are of interest. First, with no hitchhiking (ρA = ρX = 0), sshX/sshA = 3/4; i.e., the ratio of heterozygosities equals the ratio of effective population sizes of the X and autosome, as expected under the neutral theory. Second, when hitchhiking alone acts in a very large population (N → ∞), sshX/sshA = ρA/ρX; i.e., the ratio of heterozygosities equals the reciprocal of the ratio of rates of adaptive substitution on the two chromosomes, as one might guess intuitively.
Focusing on the large population case and using standard approximations for the rates of adaptive substitution [ρA = 4Nvhs and ρX = Nvs(1 + 2h); Charlesworth et al. 1987], where v is the mutation rate to beneficial alleles, h is the dominance coefficient, and s is the homozygous fitness advantage, we find that
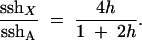 |
5 |
This is just the ratio of the X to autosomal substitution rates, first derived by Charlesworth et al. (1987). Thus, if beneficial mutations have additive effects (h = 1/2), the X and autosome will show equal heterozygosities at neutral loci given a stream of adaptive substitutions at a nearby locus (sshX/sshA = 1). But if beneficial mutations are partially recessive (h < 1/2), the X will be less variable than the autosome; conversely, if beneficial mutations are partially dominant (h > 1/2), the X will be more variable than the autosomes. In all cases, note that heterozygosities are unnormalized by differences in effective population sizes on the X vs. autosomes.
Recombination:
Recombination between the neutral and selected loci makes our problem much more difficult. Our approach is to (i) restrict attention to low rates of recombination, (ii) present analytic approximations that hopefully capture the essence of the dynamics, and (iii) check these approximations against exact computer simulations.
With no recombination between the selected and neutral loci, the sweep of a beneficial mutation through a population will drag a neutral allele from its initial frequency, x0, to a final frequency of x∞ = 1. But when recombination occurs between the selected and neutral loci, the hitchhiking neutral allele will often be separated from the beneficial mutation before reaching fixation, i.e., x∞ < 1. In Gillespie's (2000) pseudohitchhiking model, it is more useful to track the frequency of only those copies of the neutral allele that are direct descendants of the single copy that resided on the chromosome on which the beneficial mutation arose, rather than the overall frequency of the hitchhiking neutral allele. This frequency increases during a hitchhiking event from 1/(2N) (on an autosome) or 2/(3N) (on an X chromosome) to a final frequency of y when the beneficial mutation is fixed, where, usually, y < 1 because of recombination.
By a slight variation on the argument presented above for the no-recombination case, Gillespie (2000) showed that the expected sum-of-sites heterozygosity at an autosomal neutral locus with recombination is
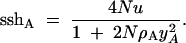 |
6 |
It is easy to show that the analogous expected sum-of-sites heterozygosity at an X-linked locus is
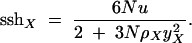 |
7 |
When yA = yX = 1, the above results collapse to those with no recombination (Equations 1 and 3), as they must.
The ratio of X-linked to autosomal heterozygosities is therefore
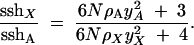 |
8 |
In the absence of hitchhiking ρA = ρX = 0, we again obtain sshX/sshA = 3/4, as expected under neutrality. But when hitchhiking alone acts in a very large population (N → ∞), we now have
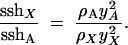 |
9 |
As we are mainly interested in the effects of hitchhiking, we focus on this large population case. Equation 9 shows that knowing the ratio of X to autosomal heterozygosities under a stream of hitchhiking events requires knowing y2A and y2X. Here, we use two approaches to calculate y2, an “exact” numerical solution and a more approximate solution that can be obtained in closed form. In fact, both of these approaches solve for y, rather than for y2, but because both approaches are deterministic, the expected value of y2 in Equation 9 simply equals the square of y.
A general solution that describes the deterministic increase of y can be written as
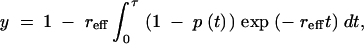 |
10 |
where p(t) is the frequency of a beneficial allele at time t such that p(0) = 1/(2N)on an autosome or 2/(3N) on an X chromosome and p(τ) = 1 (i.e., τ is the sojourn time of the beneficial mutation). The meaning of reff, the effective rate of recombination, is explained shortly. Equation 10 is easily derived from Equations 8a and 8b of Stephan et al. (1992) and is equivalent to Equations 18–20 of Maynard Smith and Haigh (1974). By modeling the deterministic increase in p(t) for arbitrary h, y can be obtained for a beneficial mutation having any dominance. This exact solution for y, and thus for y2, can be obtained numerically for both X-linked and autosomal loci (see appendix).
The above solution to y2 has the advantage of being valid over a wide range of parameter values. However, because y2 must be obtained numerically for each case, it is difficult to intuit the behavior of sshX/sshA. Therefore, we also pursue a rougher solution that, following Maynard Smith and Haigh (1974), applies only under a more restricted range of conditions, but that has the advantage of being in closed form. When the recombination rate is very small relative to the selection coefficient (r ≪ s), Maynard Smith and Haigh (1974) showed that a hitchhiking allele with an initial frequency of x0 will increase to a frequency of x∞, where, for an autosomal locus, x∞ ≈ 1 − (1 − x0)(reff,A/(hs))log(2N). Because y = (x∞ − x0)/(1 − x0) (Gillespie 2000) we get
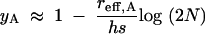 |
11 |
An analogous calculation for the X shows that
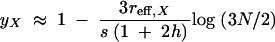 |
12 |
The calculations below use these closed-form solutions for yA and yX. Because we can write ρA, ρX, yA, and yX, we can calculate sshX/sshA by Equation 9.
Recombination in females only:
To make our solution biologically meaningful, we must demystify reff. This effective rate of recombination refers to the rate of recombination averaged over the two sexes. In Drosophila, for example, recombination between two loci might occur at a rate r per base pair per generation in females, but recombination does not occur in males. Thus in Drosophila the effective rate of recombination on the autosomes is reff,A = r/2, whereas the effective rate of recombination on the X is reff,X = 2r/3, reflecting that two-thirds of all X chromosomes reside in the recombining sex, females.
First, consider the effects of repeated hitchhiking in Drosophila when beneficial mutations have additive effects (h = 1/2) and therefore rates of X-linked and autosomal evolution are equal (ρA/ρX = 1). From Equations 9–11, we get
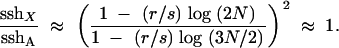 |
13 |
In words, unnormalized heterozygosities on the X and autosome are nearly equal, except for a small difference in the logarithm of population size, and sshX/sshA ≈ 1. This equality of sshX and sshA reflects the fact that when h = 1/2 (i) the rates of adaptive substitution are the same on the X and autosomes, and (ii) the ratio of r/s is the same on the X and autosomes.
It is worth examining this second point further. Beneficial mutations that appear on the X chromosome enjoy an enhanced selective advantage due to hemizygous expression in males. In particular, the “effective selective advantage” for an X-linked rare allele with h = 1/2 is seff,X = (1/3)s + (2/3)(s/2) = 2s/3. An otherwise identical beneficial mutation on an autosome, however, does not enjoy the benefits of hemizygous expression and has a smaller effective advantage, with seff,A = (1/2) (s/2) + (1/2)(s/2) = s/2. Thus, all else being equal, beneficial mutations will sweep faster on the X due to their larger effective advantage. The important point, however, is that, when h = 1/2 this effect is exactly balanced by the greater effective recombination on the X chromosome (reff,X = 2r/3; reff,A = r/2). In words, the total opportunity for recombination during an adaptive sweep is about the same on an X as on an autosome since X-linked beneficial mutations sweep faster but experience more recombination per generation. Because these two tendencies trade off, the critical ratio reff/seff is the same for both the X and autosome, and (yA/yX)2 ≈ 1.
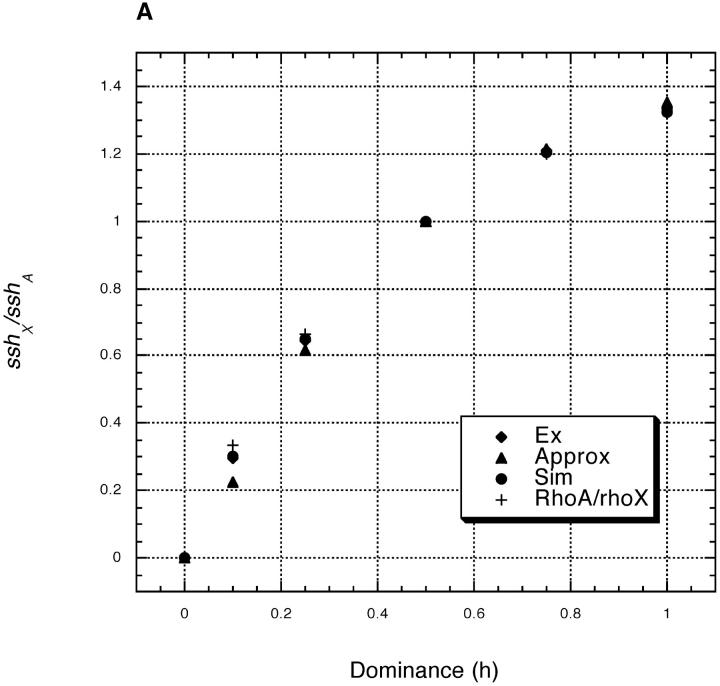
Equations 9–12 let us calculate sshX/sshA for any h among beneficial mutations. The results are shown in Figure 1A. This figure also shows the results of exact computer simulations, which agree reasonably well with theoretical predictions generated from both the exact and closed-form solutions for y. To simulate the reduction in heterozygosity at a neutral locus, we used fully stochastic simulations of sweeps in a finite, dioecious population. Starting with a single copy of a beneficial mutation, we simulated fixation or loss events at the selected locus as follows: (1) male and female parents were randomly sampled with replacement from a population in proportion to their fitness; (2) a single gamete was selected from each parent, with recombination (if appropriate), and assigned to an individual offspring; (3) when N offspring (of randomly assigned sex) were produced, we determined whether the selected allele was fixed, lost, or still segregating; and (4) if still segregating, the above process was repeated until fixation or loss. For those runs in which the beneficial mutation was fixed, we calculated y2 at a partially linked neutral locus at the time of fixation. For each value of h, the mean y2 for at least 500 sweeps was used to calculate the ratio of sshA/sshX by multiplying the y2X/y2A from the simulations by ρA/ρX for that value of h. See figure legends for more details. Our closed-form analytical results assume, however, reasonably strong selection and, not surprisingly, perform well only with appreciable selection.
Figure 1.—
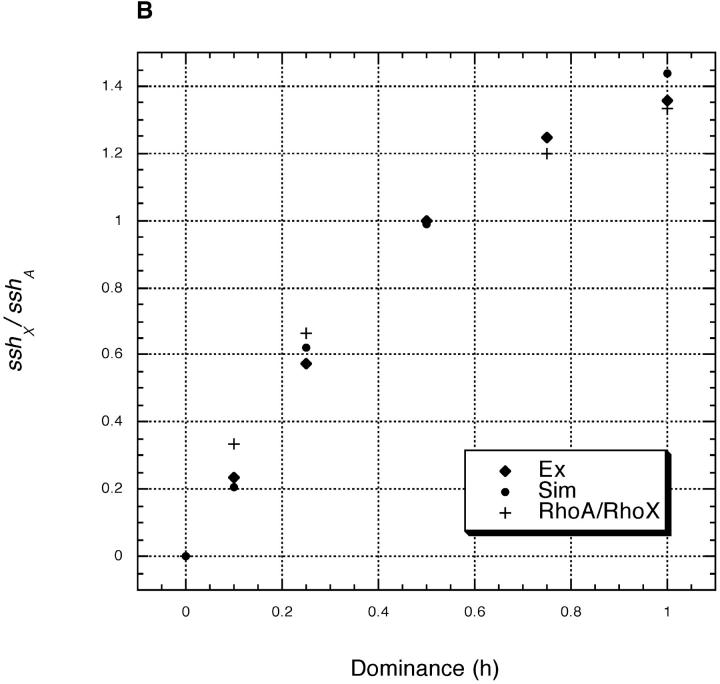
Theoretical predictions and simulations results for sshX/sshA vs. dominance (h) given Drosophila-like recombination. For both plots, sshX/sshA from Equation 9 is shown (see text), with the values of y2X and y2A determined variously from (i) the more exact numerical calculations in Equation 10 (Ex), (ii) the closed-form approximations in Equations 11 and 12 (Approx, plot A only), or (iii) two-locus forward simulations of selective sweeps in a population of N = 10,000 (with n > 500 sweeps; Sim). The recombination rate (in females) between the selected and neutral locus is r = 0.001 and the homozygous selection coefficient is either (A) s = 0.2 or (B) s = 0.02. The relative rate of evolution of autosomal and X-linked loci (ρA/ρX) is also shown.
We also simulated selective sweeps under weaker selection, where our closed-form approximation is inappropriate. As Figure 1B shows, the simulations agree well with our exact numerical solution.
From Figure 1, A and B, it is clear that when beneficial mutations are partially recessive (h < 1/2), sshX/sshA < 1 and when beneficial mutations are partially dominant (h > 1/2), sshX/sshA > 1. Figure 1 also plots ρA/ρX = 4h/(1 + 2h). The values of sshX/sshA closely track ρA/ρX, showing that relative heterozygosities on the X vs. autosome are largely determined by the relative rates of adaptive evolution on the two types of chromosomes, not by (yA/yX)2. The reason, once again, is that (yA/yX)2 ≈ 1, since the increased effectiveness of selection on the X is roughly balanced by the increased opportunity for recombination on the X. (This trade-off is essentially exact when h = 1/2 but holds roughly for most h; see Figure 1.) Thus, roughly at least, sshX/sshA ≈ ρA/ρX = 4h/(1 + 2h).
Recombination in both sexes:
We now turn to species that have recombination in both sexes. Assuming that rates of recombination per base pair are the same in males and females, reff,A = r and reff,X = 2r/3 (as the X still cannot recombine in the XY sex). If beneficial mutations have additive effects (h = 1/2),
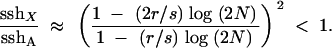 |
14 |
Equation 14 shows that, with recombination in both sexes, the ratio of effective recombination to effective selection is not the same on the X and autosomes. As a result, sweep times and recombination rates do not trade off between the X and autosomes when recombination occurs in both sexes. Consequently, in contrast to Drosophila, sshX/sshA < 1 even when h = 1/2.
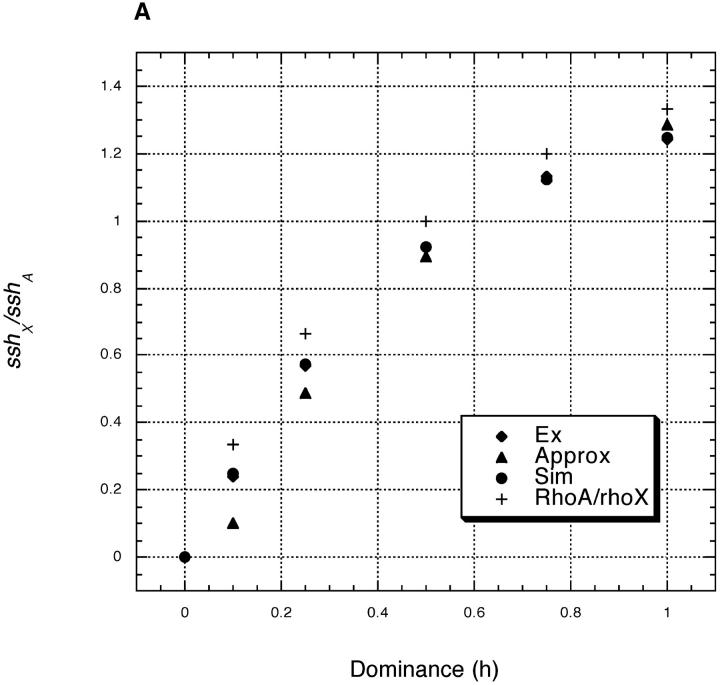
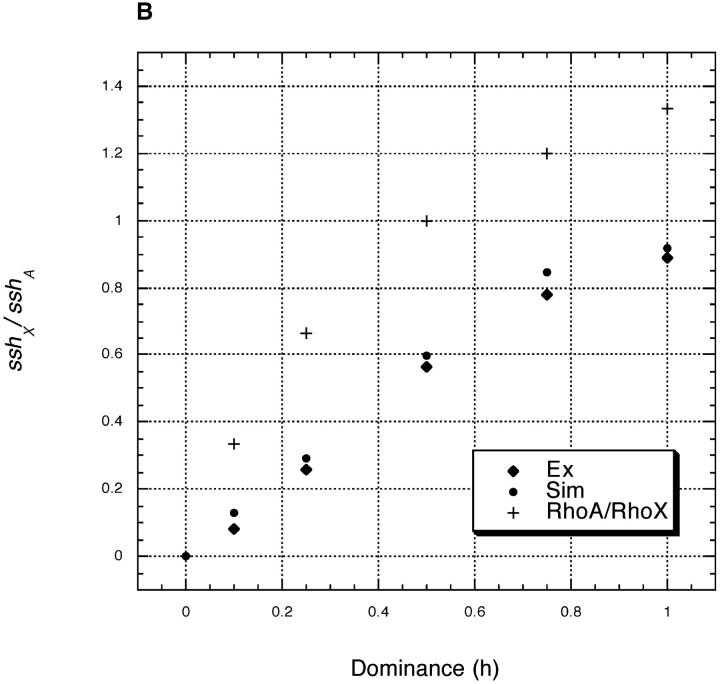
Equations 9–12 again allow us to find sshX/sshA for arbitrary h among beneficial mutations. Figure 2 shows the results, with exact simulation results and results from our analytical solutions plotted as before. The theory again performs well. In general, sshX/sshA is smaller with recombination in both sexes than with Drosophila-like recombination (compare Figures 1 and 2). Figure 2 also shows a plot of ρA/ρX = 4h/(1 + 2h). With recombination in both sexes, ρA/ρX no longer predicts sshX/sshA.
Figure 2.—
Plots and parameter values are as in Figure 1, but with recombination occurring in both sexes (r = 0.001 in both sexes). (A) s = 0.2 and (B) s = 0.02.
CONCLUSIONS
Our results let us assess the validity of the verbal claim that X-linked diversity is reduced relative to autosomal diversity given repeated sweeps of positively selected mutations. The two reasons commonly given for this reduction—that X-linked substitution rates may be higher than autosomal rates and that X-linked sojourn times are shorter than autosomal times—hold under different conditions. X-linked substitution rates are higher than autosomal ones only when beneficial mutations are partially recessive (h < 1/2); X-linked sojourn times, on the other hand, are always shorter than autosomal ones, regardless of dominance (confirmed in our simulations, data not shown).
Our analysis incorporates both of these effects and shows that—when recombination occurs only in females, as in Drosophila—X-linked diversity is lower than autosomal diversity only when beneficial mutations are partially recessive (h < 1/2). Roughly speaking, then, sojourn time has little effect in Drosophila. The reason is that there is an approximate trade-off between sojourn time and recombination rate in Drosophila: although sojourn times are shorter on the X, per-generation recombination rates are higher on the X (since two-thirds of all X chromosomes reside in the recombining sex). The total opportunity for recombination during a selective sweep is thus nearly the same for most beneficial mutations whether they appear on the X or on an autosome, at least for the reasonably strong selection examined here. (The trade-off depends somewhat on h, being essentially exact when h = 1/2.) Thus in Drosophila, repeated hitchhiking depresses X-linked diversity only when beneficial mutations are partially recessive. The fact that, in the Drosophila-like recombination case, the approximation sshX/sshA ≈ 4h/(1 + 2h) predicts simulation results reasonably well suggests that we might be able to infer the mean dominance of new beneficial mutations from the observed sshX/sshA in natural populations of Drosophila (or any other species with a Drosophila-like recombination scheme). Recall that non-African Drosophila populations show depressed variation on the X chromosome, suggesting that hitchhiking may be the predominant force in these populations. If true, the implication is that new beneficial mutations are somewhat recessive. Indeed published estimates of ratios of X-autosome heterozygosities in non-African Drosophila yield estimates of h that vary between 0.16 and 0.38 (from data reviewed in Mousset and Derome 2004). Although these estimates of dominance may seem surprisingly low, it should be noted that these estimates refer to dominance among new beneficial mutations, i.e., before mutations are acted on by selection and subjected to a dominance sieve (Haldane 1927). In any case, these low estimates are in at least qualitative agreement with other evidence suggesting the recessivity of beneficial mutations (Charlesworth 1992; Thornton and Long 2002; Zeyl et al. 2003; Counterman et al. 2004; but see Betancourt et al. 2002).
However, even if hitchhiking is the sole force differentially affecting X-linked vs. autosomal variation in non-African Drosophila populations, such estimates of h may be inaccurate as we have ignored several complicating factors. We have assumed, for example, that both recombination rates per base pair (in females) and the density of selective targets are equivalent between X chromosomes and autosomes. Recombination rates are somewhat higher on the X in D. melanogaster (2.92 cM/Mb for the X, 2.17 cM/Mb for the autosomes excluding the tiny nonrecombining fourth; estimated from data in http://flybase.bio.indiana.edu:82/maps/lk/genome-cyto-seq-map/ and http://flybase.bio.indiana.edu/maps/lk/cytotable.txt). [Recombination data are sparser for D. simulans, where non-African X-autosome differences are more pronounced, but recombination is probably more similar between X's and autosomes than in D. melanogaster (True et al. 1996).] The density of selective targets may be somewhat lower on the X (Noor et al. 2001), particularly for male-expressed genes (Swanson et al. 2001; Parisi et al. 2003), which may be especially important as they are unusually rapidly evolving (Civetta and Singh 1995; Swanson et al. 2001). Thus, for hitchhiking to result in the observed reduction in X-linked variation in non-African Drosophila, the actual value of h may have to be lower than the above estimate of 0.16–0.38.
Our results for mammals—in which recombination occurs in both sexes—are more liberal than those for Drosophila: unnormalized heterozygosities are lower on the X than on autosomes even when h = 1/2 (see Figure 2). This reflects the fact that the above trade-off between sweep time and per-generation recombination rate does not occur when recombination is mammal-like. It may, therefore, be more fruitful to look in mammals for data to distinguish between hitchhiking and background selection. There are two issues to keep in mind, however, when applying this model to mammalian data. First, although we have assumed that recombination rates are equal in both sexes, this may not be true. In humans, for example, although recombination occurs in both sexes, rates are two times higher in females (Kong et al. 2002). The contrast between Drosophila and mammals may thus be less extreme than that presented here. Second, because mammals have small population sizes, our large-population, hitchhiking-only solutions may be inappropriate. A more conservative approach would be to use normalized X-linked heterozygosities (multiplied by four-thirds) to compensate for the expected effects of genetic drift.
The best relevant mammalian data come from humans. Unfortunately, the evidence that hitchhiking and/or background selection affect levels of diversity in humans is weak (Hellmann et al. 2003). However, because recombination in humans is both complex (occurring in a “block-like” fashion; see McVean et al. 2004) and apparently mutagenic (Hellmann et al. 2003), the linked selection that may give rise to a correlation between recombination rate and diversity in other organisms (Begun and Aquadro 1992; Nachman 1997; Stephan and Langley 1998; Cutter and Payseur 2003) might be obscured in humans. Nevertheless, there is some reason to believe that linked selection still affects levels of X vs. autosomal diversity in humans: in a worldwide sample of humans, diversity on the X is reduced, even when conservative corrections for differences in effective population size and mutation rate are used (Sachidanandam et al. 2001). Only hitchhiking—and not background selection—can easily explain this reduced X-linked diversity (as background selection acts to increase relative X-linked diversity; Aquadro et al. 1994).
It is entirely possible, of course, that other forces—including demography (Kimmel et al. 1998; Wall et al. 2002), male-biased mutation (Miyata et al. 1987), background selection (Charlesworth 1994), inversion frequencies (Andolfatto 2001), and sexual selection (Charlesworth 2001)—also contribute to different diversity levels on the X vs. autosomes. In particular, demography (in both humans and Drosophila; Kimmel et al. 1998; Fay and Wu 1999; Wall et al. 2002), inversion frequencies (Andolfatto 2001), and sexual selection (in Drosophila; Charlesworth 2001) may explain differences between African and non-African populations. Weighing the relative roles of these forces will require both more data and more explicit—and biologically realistic—theory.
Acknowledgments
We thank P. Andolfatto, C. Aquadro, D. Begun, K. Dyer, J. P. Masly, M. Noor, D. Presgraves, and two anonymous reviewers for helpful comments and discussion. This work was supported by National Institutes of Health grant GM-51932.
APPENDIX
To calculate Equation 10, an approximation to the trajectory of the beneficial mutation, p(t), is required. Under strong selection, this trajectory can be approximated by assuming a deterministic increase in allele frequency. However, the deterministic approach underestimates the increase in frequency for those alleles that are ultimately fixed, as these alleles are disproportionately sampled from ones that experienced an especially rapid early rise in frequency due to genetic drift (Maynard Smith and Haigh 1974; Barton 1998). We use a standard correction for this underestimation, as follows. After one copy of the beneficial allele is introduced into the population, the expected number of its descendants t generations later is given by p*(t) ≈ (1/2N)(1 + s)t, as predicted by deterministic theory. With sufficiently large t, the beneficial mutation either goes extinct or reaches a frequency that is high enough to ensure its eventual fixation; i.e., the allele enters the “deterministic phase.” At this point, p*(t) = φpf + (1 − φ)pe(t) = φpf(t), where φ is the fixation probability of the beneficial mutation and pe(t) and pf(t) are the frequencies at time t of the allele given its extinction or eventual fixation, respectively. Therefore, the early increase in frequency of the beneficial mutation destined for fixation is elevated by a factor 1/φ relative to the deterministic increase (Maynard Smith 1971; Barton 1998). This suggests a simple way to accommodate the early drift of the beneficial allele: we may model the trajectory using the deterministic solution, but with an initial frequency of 1/(2Nφ) instead of 1/(2N) for an autosomal locus or 2/(3Nφ) instead of 2/(3N) for an X-linked locus.
For autosomal loci, the trajectory is approximately
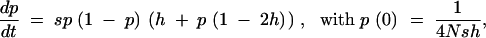 |
assuming s ≪ 1 and φ ≈ 2sh. Similarly, for X-linked loci,
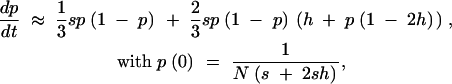 |
since φ ≈ (1/3)(2s) + (2/3)(2sh). To calculate y, we numerically solve these differential equations using the NDSolve function of Mathematica (Wolfram Research 2003) and then numerically calculate Equation 10.
References
- Andolfatto, P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., and M. Przeworski, 2001. Regions of lower crossing over harbor more rare variants in African populations of Drosophila melanogaster. Genetics 158: 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro, C. F., D. J. Begun and E. C. Kindahl, 1994 Selection, recombination, and DNA polymorphism in Drosophila, pp. 46–56 in Non-Neutral Evolution: Theories and Molecular Data, edited by B. Golding. Chapman & Hall, New York.
- Avery, P. J., 1984. The population genetics of haplo-diploids and X-linked genes. Genet. Res. 44: 321–341. [Google Scholar]
- Betancourt, A. J., D. C. Presgraves and W. J. Swanson, 2002. A test for faster X evolution in Drosophila. Mol. Biol. Evol. 19: 1816–1819. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and P. Whitley, 2000. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc. Natl. Acad. Sci. USA 97: 5960–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1992. Evolutionary rates in partially self-fertilizing species. Am. Nat. 140: 126–148. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63: 213–227. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1996. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. 68: 131–150. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 2001. The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res. 77: 153–166. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., J. A. Coyne and N. Barton, 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130: 113–146. [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1995. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis group species. J. Mol. Evol. 41: 1085–1095. [DOI] [PubMed] [Google Scholar]
- Counterman, B. A., D. Ortiz-Barrientos and M. A. Noor, 2004. Using comparative genomic data to test for fast-X evolution. Evol. Int. J. Org. Evol. 58: 656–660. [PubMed] [Google Scholar]
- Cutter, A. D., and B. A. Payseur, 2003. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol. Biol. Evol. 20: 665–673. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C.-I Wu, 1999. A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol. Biol. Evol. 16: 1003–1005. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 1991 The Causes of Molecular Evolution. Oxford University Press, Oxford.
- Gillespie, J. H., 2000. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics 155: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1927. A mathematical theory of natural and artificial selection, part V: selection and mutation. Proc. Camb. Philos. Soc. 28: 838–844. [Google Scholar]
- Hellmann, I., I. Ebersberger, S. E. Ptak, S. Paabo and M. Przeworski, 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1966. The effect of linkage on the limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Innan, H., and W. Stephan, 2003. Distinguishing the hitchhiking and background selection models. Genetics 165: 2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin, S. D., K. A. Wetterstrand, C. M. Hutter and C. F. Aquadro, 1998. Genetic variation and differentiation at microsatellite loci in Drosophila simulans: evidence for founder effects in new world populations. Genetics 150: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M., B. Zangerl, D. Dieringer and C. Schlotterer, 2002. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics 160: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., 1971. What use is sex? J. Theor. Biol. 30: 319–335. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McVean, G. A., S. R. Myers, S. Hunt, P. Deloukas, D. R. Bentley et al., 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Miyata, T., H. Hayashida, K. Kuma, K. Mitsuyasu and T. Yasunaga, 1987. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harbor Symp. Quant. Biol. 52: 863–867. [DOI] [PubMed] [Google Scholar]
- Mousset, S., and N. Derome, 2004. Molecular polymorphism in Drosophila Melanogaster and D. simulans: What have we learned from recent studies? Genetica 120: 79–86. [DOI] [PubMed] [Google Scholar]
- Nachman, M., 1997. Patterns of DNA variablility at X-linked loci in Mus domesticus. Genetics 147: 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M. W., V. L. Bauer, S. L. Crowell and C. F. Aquadro, 1998. DNA variability and recombination rates at X-linked loci in humans. Genetics 150: 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor, M. A., A. L. Cunningham and J. C. Larkin, 2001. Consequences of recombination rate variation on quantitative trait locus mapping studies: simulations based on the Drosophila melanogaster genome. Genetics 159: 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and A. J. Betancourt, 2001. Haldane's sieve and adaptation from the standing genetic variation. Genetics 157: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur, B. A., and M. W. Nachman, 2002. Natural selection at linked sites in humans. Gene 300: 31–42. [DOI] [PubMed] [Google Scholar]
- Sachidanandam, R., D. Weissman, S. C. Schmidt, J. M. Kakol, L. D. Stein et al., 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928–933. [DOI] [PubMed] [Google Scholar]
- Sheldahl, L. A., D. M. Weinreich and D. M. Rand, 2003. Recombination, dominance and selection on amino acid polymorphism in the Drosophila genome: contrasting patterns on the X and fourth chromosomes. Genetics 165: 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W., and C. H. Langley, 1998. DNA polymorphism in Lycopersicon and crossing-over per physical length. Genetics 150: 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W., T. H. E. Wiehe and M. W. Lenz, 1992. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theor. Popul. Biol. 41: 237–254. [Google Scholar]
- Stephan, W., L. Xing, D. A. Kirby and J. M. Braverman, 1998. A test of the background selection hypothesis based on nucleotide data from Drosophila ananassae. Proc. Natl. Acad. Sci. USA 95: 5649–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K., and M. Long, 2002. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol. Biol. Evol. 19: 918–925. [DOI] [PubMed] [Google Scholar]
- True, J. R., J. M. Mercer and C. C. Laurie, 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram Research, 2003 Mathematica. Wolfram Research, Champaign, IL.
- Zeyl, C., T. Vanderford and M. Carter, 2003. An evolutionary advantage of haploidy in large yeast populations. Science 299: 555–558. [DOI] [PubMed] [Google Scholar]