Abstract
In a screen for suppressors of activated GOA-1 (Gαo) under the control of the hsp-16.2 heat-shock promoter, we identified three genetic loci that affected heat-shock-induced GOA-1 expression. The cyl-1 mutants are essentially wild type in appearance, while hsf-1 and sup-45 mutants have egg-laying defects. The hsf-1 mutation also causes a temperature-sensitive developmental arrest, and hsf-1 mutants have decreased life span. Western analysis indicated that mutations in all three loci suppressed the activated GOA-1 transgene by decreasing its expression. Heat-shock-induced expression of hsp-16.2 mRNA was reduced in cyl-1 mutants and virtually eliminated in hsf-1 and sup-45 mutants, as compared to wild-type expression. The mutations could also suppress other transgenes under heat-shock control. cyl-1 and sup-45, but not hsf-1, mutations suppressed a defect caused by a transgene not under heat-shock control, suggesting a role in general transcription or a post-transcriptional aspect of gene expression. hsf-1 encodes the C. elegans homolog of the human heat-shock factor HSF1, and cyl-1 encodes a cyclin most similar to cyclin L. We believe HSF-1 acts in heat-shock-inducible transcription and CYL-1 acts more generally in gene expression.
GENETIC screens for mutations affecting the expression or regulation of specific genes often identify genes involved in general aspects of gene expression, e.g., SNF/SW1 in yeast (reviewed by Winston and Carlson 1992) or SMG in Caenorhabditis elegans (Hodgkin et al. 1989). Our screen for suppressors of heat-shock-induced expression of the consitutively activated G protein Gαo has revealed new genes involved in signal transduction, eat-16 and dgk-1 (Hajdu-Cronin et al. 1999; Lackner et al. 1999), as well as mutations affecting gene expression, which we describe here.
RNA polymerase II (Pol II) catalyzes transcription of all protein-coding genes in eukaryotes (reviewed by Proudfoot et al. 2002). After recruitment to a promoter (“initiation”), Pol II transcribes a short RNA of ∼20–40 nucleotides and then pauses. Phosphorylation of the carboxyl terminal domain (CTD) of Pol II enables capping of the emerging RNA and resumption of transcription by Pol II (“elongation”). Phosphorylation also recruits mRNA processing factors to the CTD. During elongation, introns are removed by pre-mRNA splicing, the mRNA is packaged for export to the cytoplasm, and the CTD is dephosphorylated. While initiation, elongation, and processing were thought to occur sequentially and independently, recent research suggests that transcription and processing of mRNA function in an interdependent manner. For example, the CTD interacts with pre-mRNA processing factors and is required for efficient processing; phosphorylation of the CTD may recruit capping, splicing, and processing factors to the pre-mRNA during transcription (Hirose and Manley 2000; Orphanides and Reinberg 2002; Proudfoot et al. 2002).
One of the most-studied examples of inducible transcription in eukaryotes is heat-shock induced mRNA synthesis, a stress survival mechanism that results in elevated expression of heat-shock genes and diminished expression of other genes. In most eukaryotes, heat-shock-induced transcription requires activation of the heat-shock factor (HSF) and binding of HSF to specific sequence motifs on the promoter called heat-shock elements (HSE; Lis and Wu 1993). HSF is synthesized and sequestered at a constant level in the cells under normal conditions (Wu 1995). Saccharomyces cerevisiae and Drosophila melanogaster each have one HSF, which functions similarly to HSF1 in higher animals (Morimoto 1998). Under unstressed conditions, HSF exists as an inactive monomer in most eukaryotes. Stress causes HSF to trimerize and bind to the HSE with high affinity (Westwood et al. 1991), resulting in transcription of heat-shock proteins (hsp). In D. melanogaster, stress causes a rapid recruitment of both HSF and cyclin-dependent kinase 9 (CDK9) to heat-shock promoters, where Pol II is docked and paused after initiation of a short transcript (Rougvie and Lis 1988; Giardina et al. 1992; Rasmussen and Lis 1993). Phosphorylation of the CTD releases Pol II from the promoter to resume transcription of heat-shock proteins (Lis et al. 2000).
The cyclin-dependent kinases (CDK), which phosphorylate the CTD during transcription, are regulated by their associated cyclin partners C, H, T, and K (Rickert et al. 1996; Svejstrup et al. 1996; Price 2000). Cyclin L, a new member of the cyclin family, was identified in D. melanogaster and C. elegans (Berke et al. 2001; Boucher et al. 2001), mouse (Berke et al. 2001), and human (Dickinson et al. 2002). Cyclin L associates with CDK11, the splicing factors S-35 (mice) or SC35 (human), and phosphorylated Pol II (Berke et al. 2001; Dickinson et al. 2002). In addition to the highly conserved “cyclin box” that interacts with the CDK (Kobayashi et al. 1992), cyclin L also possesses an RS domain characteristic of splicing factors (Berke et al. 2001; Boucher et al. 2001). An antibody to cyclin L inhibits the second step of splicing in vitro (Dickinson et al. 2002). Recently, CDK11 was reported to interact with the splicing protein 9G8 in vivo and to phosphorylate 9G8 and promote splicing activity in vitro (Hu et al. 2003). Therefore, cyclin L/CDK11 might participate in a signaling pathway that links or regulates transcription and RNA processing. In mouse neurons, dopamine and glutamate induce expression of two isoforms of cyclin L (ania-6) by alternative splicing (Berke et al. 2001). C. elegans cyclin L is predicted to have two alternatively spliced isoforms similar to those of ania-6 (WS110; http://www.wormbase.org). No mutants in cyclin L have been reported in any organism.
In this study, we describe mutations of C. elegans in three loci, including those that encode the HSF and cyclin L homologs. These mutations were identified as suppressors of a heat-shock-inducible transgene. We show that the mutants have reduced expression of the transgene compared to wild type and can suppress, to different degrees, transgenes under heat-shock control while having little to no effect on non-heat-shock transgenes. The mutants also contain lower levels of hsp16.2 mRNA than wild type when heat-shocked. All three cyl-1 alleles are missense mutations in conserved residues in the cyclin box, which may disrupt binding of CYL-1 to its kinase partner. The hsf-1 mutation truncates the carboxyl terminus of HSF, eliminating the transactivation domain, and affects larval development, egg-laying behavior, and longevity, in addition to heat-shock response.
MATERIALS AND METHODS
C. elegans strains were cultured and strains mapped and constructed using standard procedures (Brenner 1974). Nematodes were cultured and characterized at 20° unless indicated. Animals were heat-shocked on petri dishes in a 33° water bath for 30 min unless otherwise indicated. Suppression of the lethality induced by syIs17 was used as the criterion to follow the suppressors during mapping and complementation experiments. In constructions involving other transgenes, double mutants were confirmed for the presence of the suppressor mutation by complementation tests, scoring heterozygous animals for ability to suppress syIs17.
The following alleles and strains were used for genetic mapping and double-mutant construction: wild type: N2 (Brenner 1974), CB4856 (Hodgkin and Doniach 1997); LGI: dpy-5(e61), goa-1(n363), unc-29(e1072), unc-75(e950), hIn1[h1040], unc-101(m1); LGII: unc-4(e120), syIs12 (Katz et al. 1995); LGIII: unc-32(e189), dpy-1(e1), daf-2(e1368ts), unc-45(e286), daf-7(e1372); LGIV: unc-31(e169), dpy-20(e1282); LGV: him-5(e1490), dpy-11(e224), rol-3(e754), unc-42(e270), lin-25(sy29), sma-1(e30), BW163 ctDf1/DnT1 [unc(n754) let(m435)] (Manser and Wood 1990), GS357 unc-42(e270)arDf1/nT1[let(m435)] (Tuck and Greenwald 1995); LGX: lin-15(n765ts), syIs1 (Hsieh et al. 1999), syIs38 (Bastiani et al. 2003); linkage unknown: syIs9; dpy-20(e1362) (Mendel et al. 1995).
Isolation and characterization of suppressors:
The genetic screen has already been described (Hajdu-Cronin et al. 1999). All suppressors were backcrossed at least three times to the parental strain syIs17 dpy-20(e1282) and outcrossed to N2 for characterization. To observe egg-laying defects, animals of each strain and wild type that had been cultured with adequate food for two generations were selected as L4 larvae. Over the course of a week, animals were observed for internal hatching of unlaid eggs. To measure the number of eggs in the uterus, animals were bleached 24 hr after being selected as L4 larvae. Bleaching dissolved the animals, leaving behind the eggs, which could then be easily counted at 100× magnification. Vulva induction and P11.p to P12.p transformations in syIs1 and syIs12 double mutants was scored using Nomarski optics (Sternberg and Horvitz 1986; Jiang and Sternberg 1998).
Life-span analysis:
Wild-type and hsf-1 animals were selected as L4 larvae and transferred to new plates every 1–2 days until they ceased laying eggs and transferred as necessary thereafter. Animals were cultured at 20°–22° for the duration of the experiment. Animals that crawled off the plate, died from internally hatching progeny, or displayed exploded internal organs were not included in the analysis. Because most hsf-1 animals die from internally hatched progeny, an initial assay of 500 animals resulted in a sample size of 42. The graph in Figure 5 represents one of two separate trials, which gave similar results.
Figure 5.—
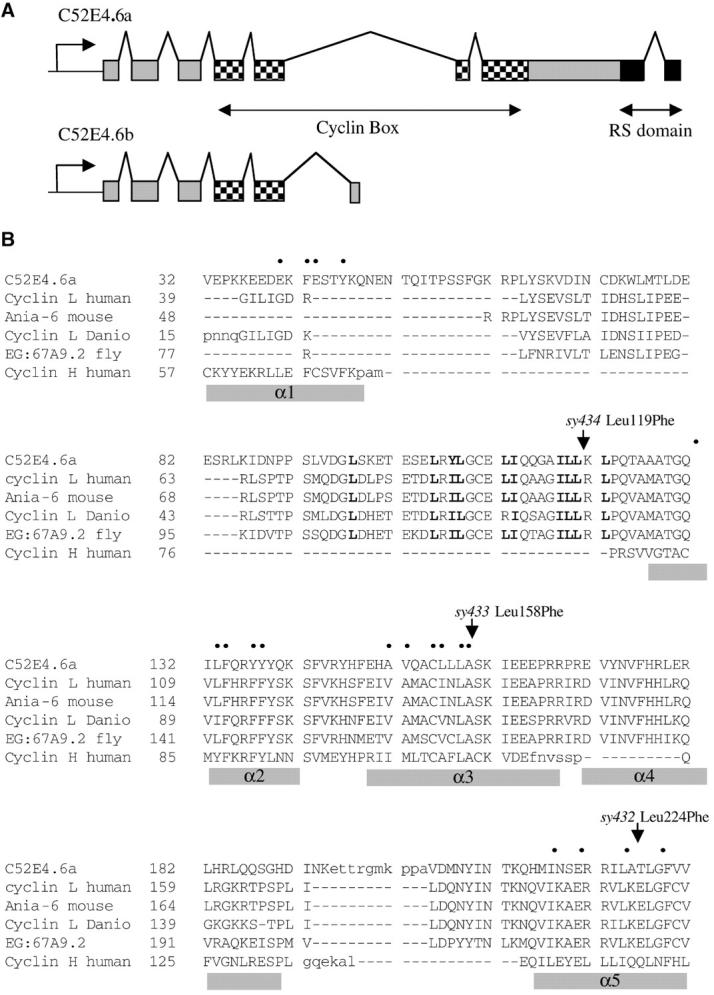
Sequence analysis of cyl-1. (A) Splicing pattern of C52E4.6a as determined by sequence analysis of cDNA. The cyclin box extends from exon 4 to exon 7. A truncated splicing variant C52E4.6b has also been identified (WS110; http://www.wormbase.org) and is depicted here for comparison. (B) Sequence alignment of the cyclin boxes of cyclin L homologs from C. elegans, human, mouse, Danio, and Drosophila, and human cyclin H. Lowercase residues are not aligned. Nineteen conserved nonpolar residues in cyclin A and H α-helices are indicated by bullets; α-helices previously described in cyclin H are marked by shaded bars (Noble et al. 1997). The α1 and α4 helices of cyclin H are not conserved on an amino acid level in cyclin L; hydrophobic residues in boldface type indicate a possible location for α1 in cyclin L. Arrows mark cyl-1 mutations.
Western blot:
Animals were collected at the L4 adult stage and dissolved in a 2× SDS sample buffer (Maniatis et al. 1982) to a concentration of 1 mg/μl, boiled for 10 min, and centrifuged for 5 min at 3000 rpm. The resulting supernatant was loaded to a 12% SDS gel (10 μl/well). The blot was performed following ECL Western blotting protocols (Amersham Biosciences). Polyclonal anti-GOA-1 (a gift from C. Bastiani and M. Simon) was produced in rabbits immunized with the C-terminal peptide ANNLRGCGLY (Cocalico Biologicals) with SulfoLink coupling gel (Pierce, Rockford, IL) according to the manufacturer's instructions. Anti-GOA-1 was used in a 1:500 dilution. Rabbit polyclonal anti-paramyosin (MH16, a gift from M. C. Hresko and R. Waterston) was used in a 1:2000 dilution. Secondary antibody (anti-rabbit IgG from donkey; Amersham Biosciences) was used in a 1:1000 dilution.
Length measurements:
We used an automated motion tracking, videotaping, and analysis software system to measure the physical lengths of the strains listed in Table 4. Each worm was videotaped for 10 sec, from which we used 5 sec of video for length measurements. “Recognizer 2.1” was used as described by C. J. Cronin, J. E. Mendel, S. Muhktar, Y.-M. Kim, R. A. Stirbl, J. Bruck and P. W. Sternberg (unpublished results) to extract 20–24 individual still images from each worm's video recording. With each still image, Recognizer identified the worm, calculated a curve representing the worm's virtual “spine,” and recorded the x-y coordinates of a set of 50 points distributed along the “spine” curve. (For these measurements we customized Recognizer 2.1 to distribute 50 points along the virtual “spine” of the worm instead of the standard 13 points to minimize length measurement error. Results with 100 points were not appreciably different from those obtained with 50 points.) We used an analysis program to sum the distances between the 50 spine points from all 20–24 still images for each worm and to calculate the average of these lengths. Mean length as recorded in Table 4 was the mean of these averages. At least 10 animals per strain were recorded.
TABLE 4.
Effect of suppressors on transgenicdpy-20 genomic DNA driven by its native promoter
| Strain | n | Mean body length (mm) | % control lengtha | P-valueb |
|---|---|---|---|---|
| N2 | 10 | 1.0 ± 0.035 | 100 | |
| hsf-1(sy441) | 10 | 1.0 ± 0.047 | 100 | 0.7394 |
| cyl-1(sy433) | 10 | 1.0 ± 0.034 | 97 | 0.0288* |
| cyl-1(sy434) | 10 | 1.1 ± 0.036 | 101 | 0.5787 |
| sup-45(sy509) | 10 | 0.94 ± 0.047 | 90 | 0.0002*** |
| dpy-20(e1282) | 12 | 0.76 ± 0.018 | 73 | <0.0001*** |
| dpy-20; syIs38 | 10 | 0.99 ± 0.027 | 100 | |
| hsf-1; dpy-20; syIs38 | 10 | 1.0 ± 0.041 | 104 | 0.0021** |
| dpy-20; cyl-1(sy433); syIs38 | 10 | 0.95 ± 0.023 | 95 | 0.0021** |
| dpy-20; cyl-1(sy434); syIs38 | 11 | 1.0 ± 0.018 | 102 | 0.0430* |
| sup-45; dpy-20; syIs38 | 11 | 0.87 ± 0.015 | 88 | <0.0001*** |
Significant;
very significant;
extremely significant.
Control for single mutants is N2; control for triple mutants is dpy-20(e1282); syIs38.
P-values were calculated using the Mann-Whitney test, comparing single mutants to N2 and triple mutants to dpy-20(e1282); syIs38.
Real-time PCR expression analysis:
Plates of synchronous populations of N2 and each mutant strain were divided at the L4 stage. Half of them were heat-shocked for 1 hr at 33°, harvested 1 hr later, separated from bacteria and debris with sucrose flotation, quick frozen in liquid N2, and stored at −80°. Animals on the other plates were harvested in the same manner with no heat-shock treatment. Total RNA was prepared from frozen worms with Trizol (Chomczynski and Sacchi 1987). First-strand cDNA was generated for each RNA sample using the TaqMan RT-PCR kit (Applied Biosystems, Foster City, CA) with random hexamers. Since hsp-16.2 is nearly identical to several other heat-shock proteins, quantitative real-time RT-PCR was run using primers specific to the unique 3′ untranslated region of hsp-16.2 (5′–3′ CGTCGAAGAGAAATCTGCTGAA and TGCAGCGAACAATACTGTAATTTATG) with SYBR green PCR kit reagents (Applied Biosystems) and analyzed using the ABI PRISM 7700 sequence detection system (Applied Biosystems). Cycle threshold (Ct) values for heat-shocked samples ranged from 26.5 to 29.5 cycles, as compared to non-heat-shocked values of 34–40+ cycles, and no template control values of 40+ cycles. All reactions were performed in triplicate and Ct values were averaged. Reactions with primers for 18S rRNA were performed on all samples in parallel as a normalization control. A standard curve was generated for each primer set by performing PCR reactions on several 10-fold serial dilutions of cDNA from heat-shocked N2. The values m and b from the best-fit line for the standard curve (equation y = −mx + b) were used to calculate the log input for each sample using the formula: log input = (average Ct − b)/−m. Input was then derived from the log input. Inputs for each sample were normalized by dividing the hsp-16.2 input by the corresponding 18S input value. The experiment was performed twice with similar results. Primer sequences for 18S rRNA (5′–3′ AAGGCGTGGAGCTTGCGGCTTAAT and TGCACCACCAACCACCAAATCGAG) were kindly provided by N. Moghal.
Mapping of hsf-1 with single nucleotide polymorphisms:
CB4856 (Hodgkin and Doniach 1997), containing many single nucleotide polymorphisms (SNPs) with respect to N2 (Wicks et al. 2001), was crossed to hIn1; syIs17 dpy-20 to construct a mapping strain containing syIs17 (IV) and CB4856 DNA on the right arm of LGI, balanced by hIn1. Spontaneous males of the mapping strain were crossed to unc-75 hsf-1; syIs17 dpy-20 or hsf-1 unc-101; syIs17 dpy-20, and recombinant chromosomes were rendered homozygous. Populations of each recombinant line were incubated in lysis buffer [50 mm KCl, 10 mm Tris (pH 8.2), 2.5 mm MgCl2, 0.45% NP-40, 0.45% Tween-20, 0.01% gelatin] with Proteinase K (0.6 mg/ml) at 60° for several hours or overnight and at 95° for 15 min. The lysates were frozen at −20° and used as templates in PCR reactions.
Previously described SNPs in E03H4 and ZK1151 were tested using the published restriction enzymes (Wicks et al. 2001). SNPs in RO6C1, F58D5, and F56H6, which could be visualized by restriction digestion, were selected using the Washington University Genome Sequencing Center website (http://genome.wustl.edu/projects/celegans/?snp=1/index.html). For R06C1.21748, primers (5′–3′) CTTTGGCTTAGGCTTAGGCACAG and TCTTTTGGCGGATTTGCGTC amplified a 1.2-kb fragment in N2 and a smaller fragment, ∼1 kb, in CB4856, making the planned restriction digest unnecessary. For F58D5.22228, the primers (5′–3′) CCCGAAAATGTTGCTGCTTCTG and AAATGTTTGTCACGTCGTCCAGTG amplified a 452-nt product. A single MaeIII restriction site in the N2 fragment was eliminated in the CB4856 fragment. For F56H6.7644, the primers (5′–3′) GCTCCGATAGTTTTGATGAAAGCC and TTCTCCGCCATTTTTGGACC amplified a 423-nt fragment. Sau3A1 cut this fragment into three pieces (165, 161, and 54 bp) in N2 or two pieces (220 and 161 bp) in CB4856. These small pieces were separated on 3.5% SeaKem agarose (FMC Bioproducts, Rockland, ME) using a 100-bp ladder (New England Biolabs, Beverly, MA).
Sequencing of Y53C10.12 in hsf-1(sy441):
Four PCR products were amplified from hsf-1(sy441); syIs17 dpy-20 genomic DNA using the Expand Long Template PCR kit (Roche) in eight separate PCR reactions per product. The products were gel purified (QIAGEN, Chatsworth, CA), pooled, and sequenced. The fragment containing the single point mutation was amplified with the primers HSF-1F (5′–3′ AGATGGAAGATGGGAGAGGGGTAG) and HSF-1R (5′–3′ TGGAAAAGTGCTCATCAGTGCG) and sequenced with HSF-seq1B (5′–3′ AAGCTCCGCCCATTTATTGG). To sequence the second strand, a 600-bp fragment was amplified in six PCR reactions using Taq polymerase and primers HSF-seq1B and HSF-seq1C (5′–3′ TCCATTTTCCGGGTACTGTTGCTC). The products were gel purified as before and pooled together. HSF-seq1C was used as the sequencing primer.
Transformation rescue of cyl-1:
Ten cosmids in the interval of osm-6 and egl-10 were microinjected in pairs to syIs17 dpy-20(e1282); cyl-1(sy433); lin-15(n765ts) at concentrations of 50 ng/μl/cosmid and 50 ng/μl of pbLH98 (lin-15 rescuing plasmid; Huang et al. 1994). The total DNA concentration was normalized to 200 ng/μl with Bluescript DNA. Animals were cultured at 15° prior to injection. Injected animals were transferred to 22° to visualize the temperature-sensitive lin-15 multivulva (Muv) phenotype and select non-Muv transformants. Rescue of cyl-1(sy433) was scored by failure to suppress syIs17. At least three stable lines per cosmid were examined. Subclone pWJC1, a 6.6-kb BamHI-Asp718 fragment from C52E4 containing open reading frames C52E4.6 and C52E4.7 cloned into pBS, was injected at 43 ng/μl and gave >90% rescue (six stable lines). pWJC2, a 3.8-kb Asp718-SacI fragment containing C52E4.6 subcloned into pBS, rescued cyl-1 at both 10 ng/μl and 30 ng/μl. pWJC3 was a 4.0-kb BamHI-XhoI subclone cloned into pBS; it failed to rescue cyl-1 at 10 ng/μl (one stable line) or 30 ng/μl (three stable lines).
Identification of cyl-1 mutation sites:
A 3-kb genomic DNA fragment was amplified (Williams et al. 1992) from 8–10 L1- to L2-stage animals of cyl-1 (sy433) using Expand long-range PCR kit (Roche) with primer S4P6 (5′–3′: TACGTGACGGTGTACCGTCAAAG) and primer S4P10 (5′–3′: AACAGAACCGTGCTTGCGGAAC). The primary PCR product was gel purified and used as template for nested PCR using primers S4P10 and S4P13 (5′–3′: CGGCAACCGCTACGCAG). The secondary PCR product was gel purified and directly sequenced. The mutation was found by comparing sequence from the PCR fragment to wild-type genomic sequence from GenBank. The point mutation identified from this sample was later confirmed by directly sequencing three independent primary PCR samples using S4P6 and S4P10. The sy432 and sy434 mutants were sequenced as above except that the nested PCR primers used were S4P6 and S4P10.
cDNA sequencing:
Full-length cDNAs of hsf-1 and cyl-1 were obtained from clones yk610c7, yk609a8 (hsf-1), and yk63c9 (cyl-1), gifts from Yuji Kohara. The λZapII clones were excised in vitro and amplified in SOLR cells (Maniatis et al. 1982). Purified phagemids were then sequenced using primers for the T3 and T7 promoters and primers used for allele sequencing. The splicing patterns were inferred by comparing cDNA sequence with wild-type genomic sequence obtained from GenBank.
RESULTS
The heterotrimeric G-protein α-subunit GOA-1 (Gαo) regulates many C. elegans behaviors, including egg laying and locomotion (Mendel et al. 1995; Segalat et al. 1995). syIs17 is an integrated transgene that overexpresses full-length genomic GOA-1 containing an activating mutation (Q205L) under the control of the heat-shock promoter hsp-16.2. Under normal growth conditions, syIs17 animals appear wild type. Heat-shock treatment induces egg retention and lethargy, and after ∼4 hr the animals barely move (Mendel et al. 1995). We mutagenized 32,000 haploid genomes of syIs17 [21,000 by ethyl methanesulfonate (EMS) and 11,000 by trimethyl psoralen (TMP)]. As described previously (Hajdu-Cronin et al. 1999), we heat-shocked the grandprogeny of mutagenized syIs17 and selected moving animals 12–24 hr later. Mutants isolated in the screen were divided into three classes on the basis of phenotype. Class I included two loci, dgk-1 and eat-16, which, when mutated, resulted in a phenotype similar to that of goa-1 mutants (Hajdu-Cronin et al. 1999; Lackner et al. 1999). Class II included cyl-1 (three alleles), which appeared wild type. Class III included hsf-1 (one allele) and sup-45 (two alleles), which had defects in egg laying (Table 1). The egg-laying defect of hsf-1(sy441) was suppressed in progeny of parents that were deprived of food (data not shown). The stronger allele of sup-45, sy509, was isolated using TMP as a mutagen and might be a deletion (Yandell et al. 1994). All other alleles described here were isolated from EMS-mutagenized worms. In our unpublished abstracts describing these genes, hsf-1 was referred to as sag-3, cyl-1 as sag-4, and sup-45 as sag-5.
TABLE 1.
Phenotypic characterization of suppressors
| % whose eggs hatch internally
|
|||||
|---|---|---|---|---|---|
| Strain | Egg retention (eggs in uterus)a |
n | Within 3 daysb | Within 6 days | n |
| N2 | 9.60 ± 3.1 | 15 | 0 | 2 | 50 |
| hsf-1(sy441) I | 16.5 ± 4.1*** | 23 | 85 | 90 | 484 |
| cyl-1(sy433) V | 11.5 ± 2.7 | 12 | 0 | 16 | 50 |
| cyl-1(sy434) V | 9.67 ± 1.8 | 15 | 0 | 2 | 50 |
| sup-45(sy509) III | 12.6 ± 6.8* | 39 | 69 | 94 | 196 |
Significant compared to N2 (P = 0.0175, Mann-Whitney test);
extremely significant compared to N2 (P < 0.0001, Mann-Whitney test).
Animals were examined 24 hr after selecting as L4 larvae.
Animals whose retained eggs hatched internally, killing the mother, within 3 or 6 days of adulthood.
In addition to the egg-laying phenotype, hsf-1(sy441) had a temperature-sensitive developmental defect. Skinny, opaque larvae reminiscent of dauer larvae were observed on plates of hsf-1 cultured at 20°, suggesting a possible role for HSF-1 in dauer formation. When viewed with Nomarski optics, these larvae did not have alae or compressed pharynges like true dauers (Cassada and Russell 1975; Vowels and Thomas 1992; data not shown), but their presence prompted us to ask if the mutant was dauer constitutive at higher temperatures. A cohort of eggs laid within 1 hr was cultured at 25° for 3 days. All the hsf-1 larvae arrested at the L2–L3 stage (n = 319), whereas no N2 larvae cultured in parallel arrested or formed dauers (n = 283). The arrested larvae were pale and vacuolated, not dauer-like. Culturing eggs at 27° yielded a similar result: cyl-1 (sy433) and sup-45(sy509) did not arrest or form dauers at 27° (n = 292 and 70, respectively), nor did N2 (n = 195), but all the hsf-1 larvae arrested at the L1–L2 stage (n = 44).
Class II and III mutants suppress syIs17 by a mechanism different from that of class I:
We performed Western analysis to measure the induction of activated GOA-1 in our suppressor strains and found that class II and III mutants had reduced expression of activated GOA-1 compared to the parental strain (Figure 1, A and B). Class I suppressors, on the other hand, had uniform levels of GOA-1 expression (data not shown), consistent with their molecular identities as members of the Gαo/Gαq signaling pathway (Hajdu-Cronin et al. 1999; Lackner et al. 1999). Therefore, hsf-1, cyl-1, and sup-45 suppressed syIs17 by reducing heat-shock-induced GOA-1 expression. We also performed Western analysis to determine if endogenous expression of GOA-1 is affected by a mutation in cyl-1. We found no significant change in the levels of endogenous GOA-1 expression in syIs17; cyl-1(sy433) without heat shock (Figure 1C). Thus, the sy433 mutation has a much greater effect on transgenic GOA-1 expression driven by the hsp-16.2 promoter/enhancer compared to endogenous expression. Supporting this result, we found that cyl-1 failed to suppress the phenotype of syIs9, an integrated transgene of activated Gαo similar to syIs17 but with the native promoter (Mendel et al. 1995). Strains carrying both syIs9 and cyl-1(sy433) looked as lethargic as syIs9 by itself (data not shown).
Figure 1.—
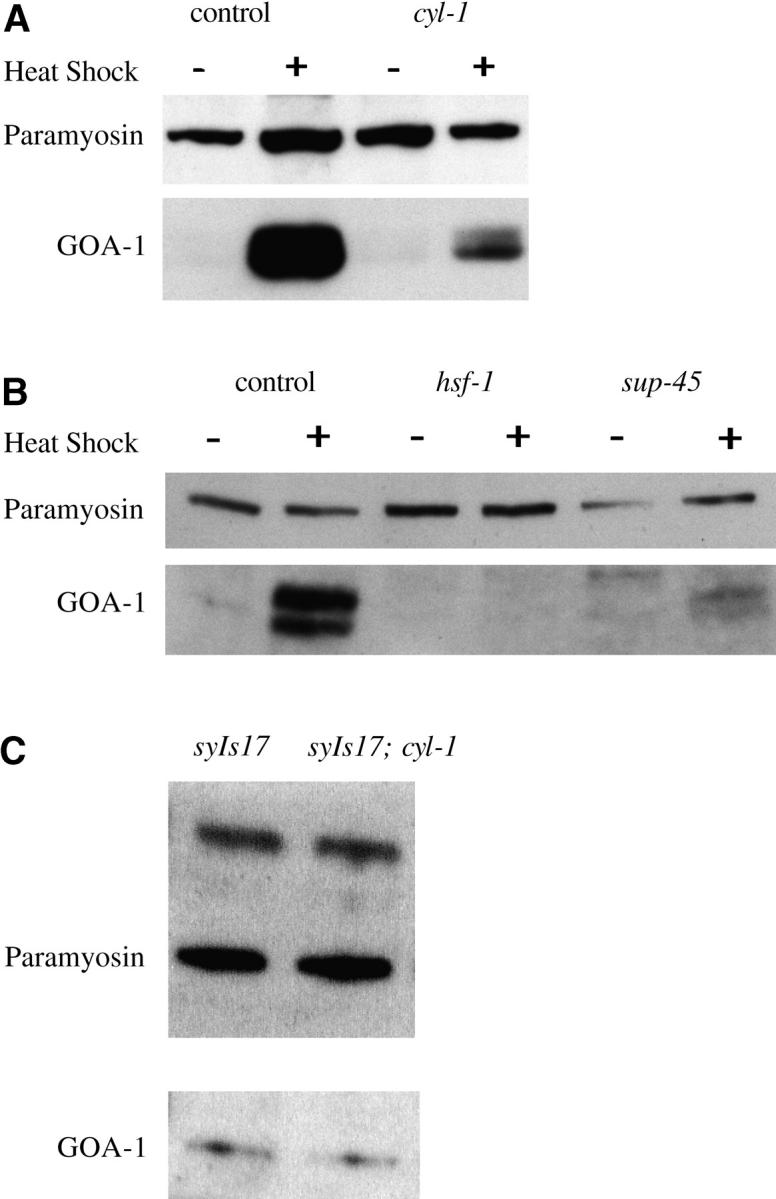
Western blots showing reduced expression of activated GOA-1 in class II and class III suppressor mutants. (A) All strains were syIs17 background. (B) These strains contained n363, a deletion of goa-1, to eliminate endogenous goa-1 expression, in addition to syIs17. (C) Endogenous wild-type GOA-1 expression in unstressed syIs17 animals is unaffected by cyl-1(sy433).
Specificity of suppression:
Since Western analysis suggested that hsf-1, cyl-1, and sup-45 might be heat shock specific, we tested this hypothesis by constructing double mutants with other transgenic lines expressing reporter genes under heat-shock and non-heat-shock control.
syIs38 contains a cDNA encoding activated Gαq (Q205L) driven by the hsp-16.2 promoter/enhancer. Two hours after heat shock, syIs38 animals become very hyperactive, moving with such extremely deep flexions that they curl up on themselves. Eventually the worms become hypercontracted and arrest (Bastiani et al. 2003). The movement defect of hsf-1; syIs38, cyl-1; syIs38 and sup-45; syIs38 was much less severe than that of syIs38 2 hr after heat-shock treatment. However, the effect of cyl-1(sy434) was only temporary: 1 hr later most of the cyl-1; syIs38 animals moved like syIs38, and after 16 hr, there was no visible difference between the two strains. In contrast, the hsf-1; syIs38 and sup-45; syIs38 double mutants did not hypercontract (Table 2). The sup-45; syIs38 animals were sterile, but the hsf-1; syIs38 animals looked like hsf-1 single mutants. Thus, hsf-1, cyl-1, and sup-45 mutations suppress hsp-16.2-driven Gαq to some extent, but cyl-1 had a minor effect. Since Gαo and Gαq have antagonistic effects (Hajdu-Cronin et al. 1999; Lackner et al. 1999; Miller et al. 1999), it is unlikely that HSF-1, CYL-1, and SUP-45 act in G-protein signaling.
TABLE 2.
Suppression of heat-shock-activated Gαq cDNA (syIs38)
| Time after heat-shock treatment
|
|||||
|---|---|---|---|---|---|
| Straina | n | 2 hr | 3 hr | 5 hr | Next day |
| syIs38 | 12 | Uncb (11) | Unc (12) | Unc (12) | Dead (12) |
| hsf-1(sy441); syIs38 | 13 | Unc (1) | Normal (13) | Normal (13) | Normal movement, fertile (13) |
| cyl-1(sy434); syIs38 | 16 | Unc (1) | Unc (7) | Unc (12) | Dead (16) |
| sup-45(sy509); syIs38 | 10 | Unc (3) | Unc (4) | Transient Unc (7) | Normal movement, sterile (10) |
The number in parentheses indicates the number of animals observed with the phenotype listed.
All strains contained dpy-20(e1282).
syIs38 heat-shocked animals move with abnormally deep flexions and a curly posture, abbreviated as Unc (uncoordinated).
The hsp-16.2 and hsp-16.41 genes are divergently transcribed, sharing a 346-bp intergenic region that contains their regulatory elements (Jones et al. 1986). The hsp-16.2 promoter/enhancer expresses most strongly in neural and hypodermal cells, while the hsp-16.41 promoter/enhancer drives expression primarily in the gut (Stringham et al. 1992). To test whether our mutations act on hsp-16.41, we built strains carrying syIs12, an integrated transgene that overexpresses the epidermal growth factor (EGF) domain of lin-3 driven by the hsp-16.41 promoter/enhancer (Katz et al. 1995). lin-3 is an EGF homolog that induces three of six vulval precursor cells to initiate vulva development in C. elegans (Hill and Sternberg 1992). Vulva induction is sensitive to the expression level of lin-3: overexpression of lin-3 results in a Muv phenotype. We found that hsf-1(sy441) and sup-45(sy509) suppressed syIs12 from Muv to wild type under both mild (29°, 30 min) and strong (33°, 30 min) heat-shock conditions, while cyl-1(sy434) did not significantly suppress syIs12 under either condition (Table 3). Thus, hsf-1 and sup-45 mutations reduce expression from the hsp-16.41 promoter/enhancer.
TABLE 3.
Suppression of the heat-shock-driven LIN-3 EGF domainsyIs12
| Strain | Heat shocka | Vulva cells inducedb | n | P-valuec |
|---|---|---|---|---|
| syIs12 | 29° | 4.97 ± 0.72 | 16 | |
| hsf-1(sy441); syIs12 | 29° | 3.0 ± 0 | 16 | NA |
| syIs12; cyl-1(sy434) | 29° | 4.68 ± 0.82 | 11 | 0.3575 |
| syIs12; sup-45(sy509) | 29° | 3.0 ± 0 | 20 | NA |
| syIs12 | 33° | 5.42 ± 0.63 | 19 | |
| hsf-1(sy441); syIs12 | 33° | 3.0 ± 0 | 7 | NA |
| syIs12; cyl-1(sy434) | 33° | 4.61 ± 0.86 | 19 | 0.0982* |
| syIs12; sup-45(sy509) | 33° | 4.19 ± 0.75 | 19 | <0.0001*** |
Not quite significant;
extremely significant.
L2 Animals were heat-shocked for 30 min in a water bath at the indicated temperature.
In wild type, three cells are induced.
P-values were calculated using the Mann-Whitney test, comparing the number of cells induced in each double-mutant animal to the number of cells induced in each syIs12 animal.
To test if the suppressors nonspecifically affect the expression of reporter genes, we examined two integrated transgenes: syIs38, which contains genomic dpy-20 as a transformation marker, and syIs1, which overexpresses genomic lin-3 under the control of its native promoter (Hsieh et al. 1999). syIs38 rescues the phenotype of dpy-20(e1282) from 73 to 95% of wild-type length (see Table 4). We measured the lengths of hsf-1; dpy-20; syIs38, cyl-1; dpy-20; syIs38 and sup-45; dpy-20; syIs38 and compared them to the length of the dpy-20; syIs38 control strain. While the cyl-1(sy433) and sup-45(sy509) strains were somewhat shorter than dpy-20; syIs38, the difference in length was proportional to the difference between the mutants and N2; i.e., cyl-1(sy433) was 97% as long as wild type, and dpy-20(e1282); cyl-1(sy433); syIs38 was 95% as long as dpy-20(e1282); syIs38. Likewise, sup-45(sy509) was 90% of wild-type length, and sup-45(sy509); dpy-20(e1282); syIs38 was 88% the length of dpy-20(e1282); syIs38 (Table 4). Therefore, we found no significant suppression of the rescue of dpy-20(e1282) by the dpy-20(+) genomic clone by any of our mutants.
syIs1 has a Muv phenotype, with all six vulval precursor cells induced. Double mutants with hsf-1(sy441), cyl-1(sy434), sup-45(sy439), and sup-45(sy509) also exhibited six-cell induction (data not shown). In addition to vulval induction, we examined P11.p, a cell whose fate is often transformed to that of its neighbor, P12.p, when LIN-3 levels are increased (Jiang and Sternberg 1998). About one-third of syIs1 animals exhibited this transformation, which was unaffected by hsf-1(sy441) and the weaker allele of sup-45, sy439. However, cyl-1(sy434) and sup-45(sy509) suppressed the fate transformation (Table 5). Therefore, cyl-1 and sup-45 had a small but significant effect on phenotype, which suggests an effect on gene expression driven by a non-heat-shock promoter/enhancer, although it is possible that the observed difference in phenotype might be due to effects on other genes. In contrast, mutations involved in context-dependent gene silencing (e.g., tam-1) can suppress syIs1 to wild type (Hsieh et al. 1999).
TABLE 5.
Effects of suppressors on the non-heat-shock transgenesyIs1(genomiclin-3)
| Strain | n | P11.p → P12.p fate transformation | P-value | Vulva |
|---|---|---|---|---|
| syIs1 | 63 | 14 | Muv | |
| hsf-1(sy441); syIs1 | 74 | 34 | 0.0042** | Muv |
| cyl-1(sy434); syIs1 | 62 | 1 | 0.0005*** | Muv |
| sup-45(sy509); syIs1 | 65 | 6 | 0.0303* | Muv |
P-values were calculated using Fisher's exact test, comparing double mutants to syIs1.
Significant;
very significant;
extremely significant.
Reduction of heat-shock-induced hsp-16.2 mRNA expression:
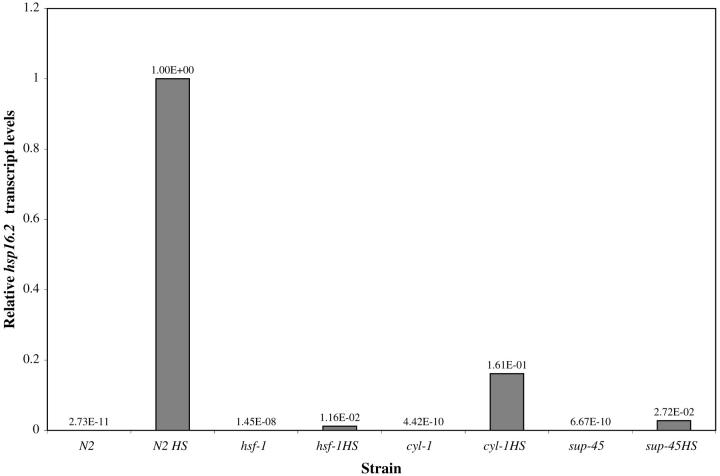
We prepared total RNA from heat-shocked and unstressed N2, cyl-1, and sup-45 animals, which we then used to generate first-strand cDNA using reverse transcriptase PCR with random hexamers as primers. Quantitative real-time PCR (Q-PCR) experiments detected hsp-16.2 mRNA in heat-shocked samples, but not from unstressed samples. The expression of hsp-16.2 mRNA detected in each mutant sample was significantly reduced as compared to N2 (Figure 2). Two trials gave results that fell into the same overall pattern. In one experiment, induction levels in cyl-1(sy433) were sixfold lower than those in N2, and in a second experiment they were 10-fold lower. Induction levels in sup-45(sy509) were reduced 37- and 200-fold in the two experiments. In hsf-1(sy441) induction levels were reduced 86- and 300-fold.
Figure 2.—
Decreased levels of hsp-16.2 mRNA in suppressor mutants, as measured by quantitative real-time PCR. Input levels for each sample were normalized with control PCR reactions using 18S rRNA. This figure shows one of two trials, each of which gave similar results.
hsf-1 encodes C. elegans heat-shock factor:
We mapped hsf-1 to the right arm of chromosome I, to the right of unc-75 and to the left of unc-101, by three-factor mapping with genetic markers, and between the cosmids R06C1 and F58D5 by three-factor mapping with SNPs (data submitted to http://www.wormbase.org). There is one clone between R06C1 and F58D5 in the contig, the YAC subclone Y53C10A, which has 12 predicted open reading frames (ORFs; C. elegans Sequencing Consortium 1998; http://www.wormbase.org). One of these ORFs, Y53C10A.12, encodes a putative heat-shock transcription factor that is 27% identical in its predicted amino acid sequence to human HSF1. We sequenced the exons of Y53C10A.12 in hsf-1(sy441) animals and found a single G-to-A mutation in the seventh exon (AGT AAT CAT AAT TGG GAT GAT TTT GGG AAT), which would convert residue 585 (tryptophan) to a stop codon, truncating the last 86 amino acids (Figure 3A). Sequencing of full-length cDNA clones (gifts from Y. Kohara) confirmed the exon structure predicted by Genefinder (WS110; http://www.wormbase.org).
Figure 3.—
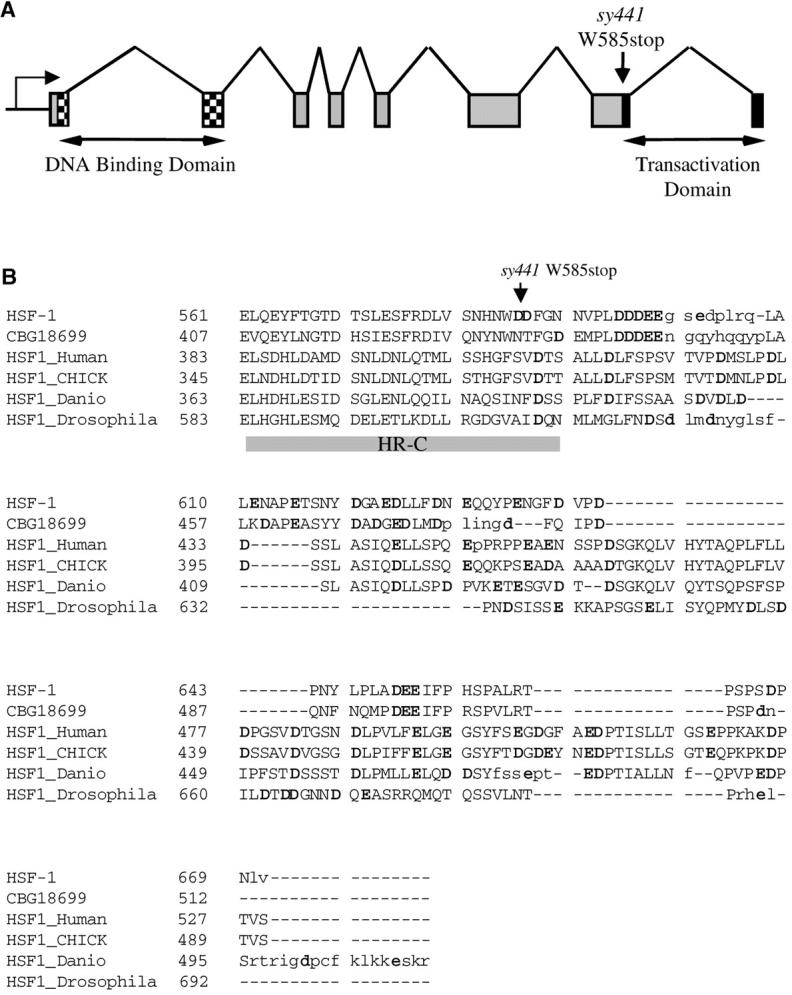
Sequence analysis of hsf-1. (A) Intron-exon structure of hsf-1. (B) Alignment of hsf-1 with other HSFs. CBG18699 is the C. briggsae ortholog of hsf-1 (Stein et al. 2003). Lowercase residues are not aligned. The heptad repeat HR-C is denoted by a shaded bar. Acidic residues are shown in boldface type.
Heat-shock transcription factors contain a number of conserved domains (reviewed in Wu 1995). While the DNA-binding domain is responsible for the binding of HSF to HSE, transcription at the heat-shock promoter also requires two domains at the C terminus of HSF1: HR-C, a hydrophobic heptad repeat, and the adjacent transcriptional activation domain (Green et al. 1995; Shi et al. 1995; Zuo et al. 1995). An alignment between HSF-1 and C. briggsae, human, chick, Danio, and Drosophila HSFs using Dialign 2.2.1 (Morgenstern 1999) confirmed that the sy441 truncation of HSF-1 occurs at the 3′ end of HR-C (Figure 3B). Two of the leucines in the HR-C repeat conserved in the other animal species are replaced by phenylalanines in HSF-1 and its homolog in C. briggsae (Figure 3B), as well as in the HSF homologs in Schizosaccharomyces pombe, Kluyveromyces lactis and Arabidopsis thaliana (not shown). Beyond HR-C, the C-terminal end of the protein is a hydrophobic, acid-rich transcriptional activation domain (Shi et al. 1995) that is conserved functionally but not at the primary sequence level (Green et al. 1995). This domain is eliminated in the sy441 mutant (Figure 3B).
hsf-1 mutants have shortened life span:
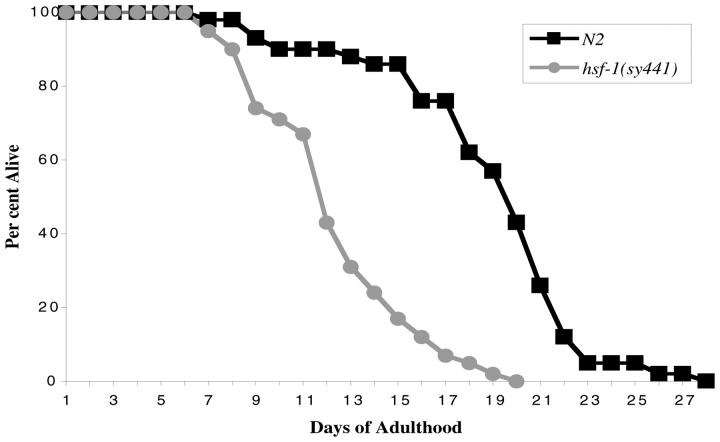
Previous work has shown that wild-type animals cultured on bacteria containing hsf-1 double-stranded RNA interference (RNAi) had a significantly shorter life span than animals grown on control bacteria (Garigan et al. 2002; Morley and Morimoto 2004), whereas animals overexpressing hsf-1 live longer than wild type (Hsu et al. 2003; Morley and Morimoto 2004). We examined the life span of hsf-1(sy441) and N2 and found that the mutants lived an average of 7 fewer days than wild type (Figure 4). Mean life span for the first trial was 12.4 ± 3.20 days of adulthood for hsf-1 vs. 19.0 ± 4.39 days for N2 (35% shorter; P < 0.0001, Mann-Whitney test). The second trial gave a similar result: 15.2 ± 3.45 days for hsf-1 vs. 22.3 ± 3.80 days for N2 (32% shorter; P < 0.0001, Mann-Whitney test). Fifty percent mortality occurred 8 days sooner in hsf-1 than in wild type. These results are very similar to those obtained using hsf-1 RNAi (Garigan et al. 2002).
Figure 4.—
Life spans of N2 and hsf-1(sy441) animals cultured at 20° (n = 42 for both; P < 0.0001, Mann-Whitney test). This figure shows one of two trials, each of which gave similar results.
cyl-1 encodes a homolog of cyclin L:
We mapped cyl-1 to the interval between sma-1 and egl-10 on chromosome V by a combination of three-factor and deficiency mapping (data submitted to http://www.wormbase.org). Of the cosmids in this interval, only C52E4 could rescue cyl-1: four stable lines had 40–70% rescued animals. C52E4 contains seven predicted ORFs (C. elegans Sequencing Consortium 1998; http://www.wormbase.org). A 6.6-kb BamHI-Asp718 subclone from C52E4 containing the last two open reading frames (pWJC1, 43 ng/μl) gave >90% rescue in six stable lines. pWJC2, a 3.8-kb Asp718-SacI subclone from pWJC1 containing C52E4.6, a cyclin L homolog, also rescued cyl-1(sy433), whereas pWJC3, a 4.0-kb BamHI-XhoI subclone containing C52E4.7 and truncated C52E4.6, failed to rescue cyl-1. sup-45 was mapped to a 1-MU interval on chromosome III that contained no predicted cyclins or CDKs (data submitted to http://www.wormbase.org).
We sequenced cyl-1 cDNA clones (gifts from Y. Kohara) to determine the intron-exon boundaries. The longest cyl-1 cDNA, clone yk63c9, is 1.6 kb in length with eight exons and a 140-bp 3′ untranslated region, corresponding to the predicted full-length isoform C52E4.6a (WS110; http://www.wormbase.org). The 180-amino-acid cyclin box starts from exon 4 and ends in exon 7 (Figure 5A). Berke et al. (2001) previously described an RS domain characteristic of splicing factors at the carboxy terminus of CYL-1/cyclin L.
cyl-1 alleles are missense mutations:
To test if the recessive cyl-1 alleles reduced its function, we examined their phenotypes in trans to a deficiency of the cyl-1 region. All cyl-1 alleles and cyl-1 (sy432, sy433, or sy434)/ctDf1 looked wild type in a syIs17 background without heat shock. After heat shock, syIs17/+; cyl-1(sy433, sy434 or sy432)/ctDf1 behaved similarly to syIs17; cyl-1(sy433) (n > 12 animals for each strain), consistent with all three alleles of cyl-1 being severe hypomorphs. Multi-copy transgenic expression of cyl-1 did not cause any phenotype in appearance or behavior (data not shown).
All three cyl-1 alleles contained C-to-T mutations in the cyclin box. sy433 had the mutation Leu158Phe (CAA GCT TGT CTA CTT CTT GCA TCC AAA ATC); sy432 was Leu224Phe (TCC GAA AGA AGA ATA CTT GCA ACT CTG GGA); and sy434 was Leu119Phe (CAA CAA GGA GCA ATC CTT TTA AAA CTT CCA). These leucines are conserved in all cyclin L homologs that we examined (Figure 5B). The residues mutated in sy432 and sy433 are also conserved in T-, K-, and C-type cyclins (data not shown).
Sequence analysis of the cyclin box domain (Bazan 1996) and crystal structures of cyclins A and H (Brown et al. 1995; Andersen et al. 1996) revealed two α-helical repeats (α1–α5, and α1′–α5′), which form a pocket or fold where the cyclin interacts with its kinase partner. Sequence comparisons between the cyclin box folds of cyclins A and H, along with similar pockets in the retinoblastoma tumor suppressor protein and TFIIB, revealed that in all these proteins the α-helices contain 19 positions with structurally conserved nonpolar residues (Noble et al. 1997). An alignment (Dialign 2.2) among CYL-1, human cyclin H, and cyclin L homologs from human, Danio, mouse, and Drosophila showed that the cyclin L homologs all contain a conserved pattern of nonpolar repeats similar to those in cyclin H. The leucine residues mutated in sy432 and sy433 are in 2 of the 19 nonpolar residues mentioned above. The mutation in sy434 is in a region where cyclin L and cyclin H are more divergent, but is part of a similar nonpolar repeat that might correspond to the α1 helix for cyclin L (Figure 5B). It is possible that mutating these leucines causes conformational changes in the helical repeats that would disrupt binding of CYL-1 to its kinase partner. The predicted C52E4.6b isoform eliminates α1′–α5′ and the RS domain (WS110; http://www.wormbase.org) and is probably nonfunctional. The “b” isoform of ania-6 also truncates within the cyclin box (Berke et al. 2001) and therefore is similar to C52E4.6b.
DISCUSSION
In this study we used a simple but powerful screen to isolate mutants in genes encoding two proteins that previously had been studied primarily biochemically. From 32,000 mutagenized haploid genomes screened for suppression of a heat-inducible transgene, we identified several loci including a C-terminal truncation of the heat-shock factor hsf-1 and three missense mutations in the cyclin L homolog cyl-1. While one allele of sup-45 might be a deletion (Yandell et al. 1994), all alleles of hsf-1 and cyl-1 were point mutations. No alleles of hsf-1 or cyl-1 were isolated using TMP. Animals fed cyl-1 RNAi display phenotypes including slow growth, larval lethality, and adult lethality (Kamath and Ahringer 2003). In contrast, our cyl-1 mutants are almost wild type in appearance and behavior. The screen as it was designed and executed would favor the isolation of hypomorphs of essential genes involved in transcription and related processes. It is therefore likely that we isolated only one allele in hsf-1 because a stronger mutation affecting the DNA-binding domain would have been lethal or had so many defects that it could not be easily maintained. Indeed, animals fed hsf-1 RNAi display several phenotypes, including larval arrest or lethality, slow growth, protruding vulva, sterility, and adult lethality (Maeda et al. 2001; Simmer et al. 2003). Elimination of the C-terminal transcriptional activation domain of HSF in hsf-1(sy441) resulted in animals that are fairly healthy but with defects in egg laying, longevity, growth, and development. The temperature-sensitive larval arrest that we observed in hsf-1 mutants (and in the dauer-like larvae seen at the permissive temperature) suggests that HSF plays an important role in growth and development, and the transcriptional activation domain missing in sy441 is required for viability at high temperatures. Yeast cells bearing a C-terminal truncated form of yeast HSF are also temperature sensitive for growth, indicating that the requirement of the transcriptional activation domain for growth under stress is conserved across species (Morano et al. 1999).
The three loci that we isolated appear to function in different aspects of gene expression. Heat-induced GOA-1 expression was reduced in cyl-1 and sup-45 mutants and virtually eliminated in hsf-1 mutants. Not surprisingly, the HSF mutation completely suppressed every heat-inducible transgene that we tested, suggesting that the suppression we see in hsf-1(sy441) is heat shock specific. The demonstrated role of cyclin L as a splicing factor in other systems raises the question of whether CYL-1 acts in a general fashion in C. elegans. Suppression of the P11.p fate caused by the lin-3 genomic transgene syIs1 by cyl-1 and sup-45 suggests that these two genes function in processes common to both general and inducible mRNA accumulation, possibly at a post-transcriptional level. Unlike sup-45(sy509), the cyl-1 mutations did not consistently suppress every heat-shock transgene that we tested. Suppression of full-length genomic goa-1 was strong, of Gαq cDNA, transient, and of the lin-3 gene fragment in syIs12, insignificant. These observations suggest that a long message and/or many introns might be necessary to observe a phenotype in our hypomorphs, a model consistent with the reported role of cyclin L in promoting pre-mRNA splicing in mammals (Berke et al. 2001; Dickinson et al. 2002). However, there was little to no effect on the genomic non-heat-shock transgenes dpy-20, goa-1, or lin-3 in cyl-1 mutants. One explanation for this difference is that an observable effect of reduced CYL-1 function depends on message stability, and the heat-shock-driven messages are more unstable. It is also possible that stresses such as heat shock might result in alternative splicing of cyl-1. The similarity between the ania-6b and C52E4.6b isoforms supports a hypothesis in which stresses such as heat shock drive preferential expression of the truncated “b” isoform, which lacks the activity of full-length CYL-1.
The mutations described here are examples of informational suppressors that have transgene-specific effects. Like other informational suppressors, e.g., mutations that disrupt nonsense-mediated decay, they can inform us as to the mechanisms of gene expression. For example, sup-45 might encode another protein involved in post-transcriptional regulation of gene expression. Previously an informational suppressor, TAM-1, was identified in a screen for suppressors of a transgenic phenotype (Hsieh et al. 1999). Similarly, our screen yielded these informational suppressors in addition to genes involved in G-protein signaling. This screen was easy to perform, and it or a similar screen could be used to identify other genes involved in the heat-shock response. On the other hand, knowledge of the existence of these informational suppressors can allow for the design of screens to avoid them if one is more interested in pathway-specific suppressors.
Acknowledgments
We thank C. Bastiani for C. elegans GOA-1 antibody, M. C. Hresko and R. Waterston for MH16 anti-paramyosin antibody, the C. elegans Genetics Center for strains, A. Coulson for cosmids used in positional cloning, and Y. Kohara for cDNA clones. We thank H. Yu, G. Schindelman, E. Schwarz, N. Moghal, L. R. Garcia, C. van Buskirk, C. Bastiani, R. Lee, B. Gupta, and T. Inoue for advice and helpful discussions. We thank E.-S. David, R. Pant, and C. Dionne for guidance and E. Rothenberg for use of her facility for Q-PCR. C. Cronin provided the software for analyzing the length of worms. Most mapping information was obtained from WormBase (http://www.wormbase.org). Research was supported by the Howard Hughes Medical Institute, with which P.W.S. is an investigator. W.J.C. was an Amgen graduate Fellow.
References
- Andersen, G., A. Poterszman, J. M. Egly, D. Moras and J.-C. Thierry, 1996. The crystal structure of human cyclin H. FEBS Lett. 397: 65–69. [DOI] [PubMed] [Google Scholar]
- Bastiani, C. A., S. Gharib, M. I. Simon and P. W. Sternberg, 2003. C. elegans Gαq regulates egg-laying behavior via a PLCβ-independent and serotonin-dependent signaling pathway, and likely functions in both the nervous system and in muscle. Genetics 165: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan, J. F., 1996. Helical fold prediction for the cyclin box. Proteins 24: 1–17. [DOI] [PubMed] [Google Scholar]
- Berke, J. D., V. Sgambato, P. P. Zhu, B. Lavoie, M. Vincent et al., 2001. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron 32: 277–287. [DOI] [PubMed] [Google Scholar]
- Boucher, L., C. A. Ouzounis, A. J. Enright and B. J. Blencowe, 2001. A genome-wide survey of RS domain proteins. RNA 7: 1693–1701. [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, N. R., M. E. Noble, J. A. Endicott, E. F. Garman, S. Wakatsuki et al., 1995. The crystal structure of cyclin A. Structure 3: 1235–1247. [DOI] [PubMed] [Google Scholar]
- Cassada, R. C., and R. L. Russell, 1975. The dauer larva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46: 326–342. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P., and N. Sacchi, 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Dickinson, L. A., A. J. Edgar, J. Ehley and J. M. Gottesfeld, 2002. Cyclin L is an RS domain protein involved in pre-mRNA splicing. J. Biol. Chem. 277: 25465–25473. [DOI] [PubMed] [Google Scholar]
- Garigan, D., A.-L. Hsu, A. G. Fraser, R. S. Kamath, J. Ahringer et al., 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161: 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina, C., M. Perez-Riba and J. T. Lis, 1992. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 6: 2190–2200. [DOI] [PubMed] [Google Scholar]
- Green, M., T. J. Schuetz, E. K. Sullivan and R. E. Kingston, 1995. A heat shock-responsive domain of human HSF1 that regulates transcription activation domain function. Mol. Cell. Biol. 15: 3354–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin, Y. M., W. J. Chen, G. Patikoglou, M. R. Koelle and P. W. Sternberg, 1999. Antagonism between Goα and Gqα in Caenorhabditis elegans: the RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes Dev. 13: 1780–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, R. J., and P. W. Sternberg, 1992. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature 358: 470–476. [DOI] [PubMed] [Google Scholar]
- Hirose, Y., and J. L. Manley, 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Hodgkin, J., and T. Doniach, 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146: 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J. A., A. Papp, R. A. Pulak, V. R. Ambros and P. Anderson, 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, J., J. Liu, S. A. Kostas, C. Chang, P. W. Sternberg et al., 1999. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13: 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, A.-L., C. T. Murphy and C. Kenyon, 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300: 1142–1145. [DOI] [PubMed] [Google Scholar]
- Hu, D., A. Mayeda, J. H. Trembley, J. M. Lahti and V. J. Kidd, 2003. CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem. 278: 8623–8629. [DOI] [PubMed] [Google Scholar]
- Huang, L. S., P. Tzou and P. W. Sternberg, 1994. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. I., and P. W. Sternberg, 1998. Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development 125: 2337–2347. [DOI] [PubMed] [Google Scholar]
- Jones, D., R. H. Russnak, R. J. Kay and E. P. Candido, 1986. Structure, expression, and evolution of a heat shock gene locus in Caenorhabditis elegans that is flanked by repetitive elements. J. Biol. Chem. 261: 12006–12015. [PubMed] [Google Scholar]
- Kamath, R. S., and J. A. Ahringer, 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Katz, W. S., R. J. Hill, T. R. Clandinin and P. W. Sternberg, 1995. Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell 82: 297–307. [DOI] [PubMed] [Google Scholar]
- Kobayashi, H., E. Stewart, R. Poon, J. P. Adamczewski, J. Gannon et al., 1992. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol. Biol. Cell 3(11): 1279–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner, M. R., S. J. Nurrish and J. M. Kaplan, 1999. Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron 24: 335–346. [DOI] [PubMed] [Google Scholar]
- Lis, J., and C. Wu, 1993. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell 74: 1–4. [DOI] [PubMed] [Google Scholar]
- Lis, J. T., P. Mason, J. Peng, D. H. Price and J. Werner, 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14: 792–803. [PMC free article] [PubMed] [Google Scholar]
- Maeda, I., Y. Kohara, M. Yamamoto and A. Sugimoto, 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11(3): 171–176. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E. F. Fritsch and J. Sambrook, 1982 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Manser, J., and W. B. Wood, 1990. Mutations affecting embryonic cell migrations in Caenorhabditis elegans. Dev. Genet. 11: 49–64. [DOI] [PubMed] [Google Scholar]
- Mendel, J. E., H. C. Korswagen, K. S. Liu, Y. M. Hajdu-Cronin, M. I. Simon et al., 1995. Participation of the protein Go in multiple aspects of behavior in C. elegans. Science 267: 1652–1655. [DOI] [PubMed] [Google Scholar]
- Miller, K. G., M. B. Emerson and J. B. Rand, 1999. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron 24: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano, K. A., N. Santoro, K. A. Koch and D. J. Thiele, 1999. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 19: 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, B., 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15: 211–218. [DOI] [PubMed] [Google Scholar]
- Morimoto, R. I., 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12: 3788–3796. [DOI] [PubMed] [Google Scholar]
- Morley, J., and R. I. Morimoto, 2004. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell 15: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, M. E. M., J. A. Endicott, N. R. Brown and L. N. Johnson, 1997. The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biochem. Sci. 22: 482–487. [DOI] [PubMed] [Google Scholar]
- Orphanides, G., and D. Reinberg, 2002. A unified theory of gene expression. Cell 108: 439–451. [DOI] [PubMed] [Google Scholar]
- Price, D. H., 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20: 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N. J., A. Furger and M. J. Dye, 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Rasmussen, E. B., and J. T. Lis, 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. USA 90: 7923–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert, P., W. Seghezzi, F. Shanahan, H. Cho and E. Lees, 1996. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene 12: 2631–2640. [PubMed] [Google Scholar]
- Rougvie, A. E., and J. T. Lis, 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804. [DOI] [PubMed] [Google Scholar]
- Segalat, L., D. A. Elkes and J. M. Kaplan, 1995. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Science 267: 1648–1651. [DOI] [PubMed] [Google Scholar]
- Shi, Y., P. E. Kroeger and R. I. Morimoto, 1995. The carboxyl-terminal transactivation domain of heat shock factor 1 is negatively regulated and stress responsive. Mol. Cell. Biol. 15: 4309–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., C. Moorman, A. M. Van Der Linden, E. Kuijk, P. V. Van Den Berghe et al., 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1(1): E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L. D., Z. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1(2): E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, P. W., and H. R. Horvitz, 1986. Pattern formation during vulval development in C. elegans. Cell 44: 761–772. [DOI] [PubMed] [Google Scholar]
- Stringham, E. G., D. K. Dixon, D. Jones and E. P. Candido, 1992. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell 3: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup, J. Q., P. Vichi and J. M. Egly, 1996. The multiple roles of transcription/repair factor TFIIH. Trends Biochem. Sci. 21: 346–350. [PubMed] [Google Scholar]
- Tuck, S., and I. Greenwald, 1995. lin-25, a gene required for vulval induction in Caenorhabditis elegans. Genes. Dev. 9: 341–357. [DOI] [PubMed] [Google Scholar]
- Vowels, J. J., and J. H. Thomas, 1992. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130: 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood, J. T., J. Clos and C. Wu, 1991. Stress-induced oligomerization and chromosomal relocalization of heat shock factor. Nature 353: 822–827. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. A. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Williams, W. V., A. Sato, M. Rossman, Q. Fang and D. B. Weiner, 1992. Specific DNA amplification utilizing the polymerase chain reaction and random oligonucleotide primers: application to the analysis of antigen receptor variable regions. DNA Cell Biol. 11: 707–720. [DOI] [PubMed] [Google Scholar]
- Winston, F., and M. Carlson, 1992. Yeast SNF/SW1 transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8: 387–391. [DOI] [PubMed] [Google Scholar]
- Wu, C., 1995. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11: 441–469. [DOI] [PubMed] [Google Scholar]
- Yandell, M. D., L. G. Edgar and W. B. Wood, 1994. Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 91: 1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., D. Rungger and R. Voellmy, 1995. Multiple layers of regulation of human heat shock transcription factor 1. Mol. Cell. Biol. 15: 4319–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]