Abstract
To better understand the genetic bases of postmating responses in Drosophila melanogaster females, we screened a collection of P{GS} insertion lines and identified two insertions in sarah (sra), whose misexpression in the nervous system induced high levels of ovulation in virgins. The gene sra encodes a protein similar to human Down syndrome critical region 1 (DSCR1). The ovulation phenotype was reproduced in transgenic virgins expressing UAS-sra in the nervous system. The flies also extruded the ovipositor toward courting males as seen in wild-type mated females, supporting the notion that ovulation and behavioral patterns are physiologically coupled. The sra insertions were found to be hypomorphic alleles with reduced expression levels. Females homozygous for these alleles show: (1) spontaneous ovulation in virgins, (2) sterility with impaired meiotic progression, and (3) compromised postmating responses with lower ovulation level, higher remating rate, and shorter period for restoration of receptivity. No obvious defects were observed in the homozygous males. The gene sra is predominantly expressed in oocytes, nurse cells, and the nervous system. Taken together, these results indicate that the expression level of sra is critical for ovulation and female courtship behavior, including their postmating changes.
REPRODUCTIVE behavior of Drosophila melanogaster provides an excellent model system to study genetic and molecular mechanisms underlying the higher nervous system functions (Hall 1994; Yamamoto and Nakano 1999). Mature virgin females are highly receptive to mating. Although they may initially display the rejection-behavior characteristics of virgin females, such as kicking, flicking, fending, and decamping, they become less active in locomotion and eventually allow males to mount them (Burnet et al. 1973; Fowler 1973; Tompkins et al. 1982; Spieth and Ringo 1983). In contrast, mated females are reluctant to mate; they reject courting males mainly by extrusion of the ovipositor (Connolly and Cook 1973; Spieth and Ringo 1983). Mated females also have stimulated egg production and oviposition behavior. A mature virgin female lays only a few eggs per day, whereas a mated female lays up to 60 eggs/day (Leahy and Lowe 1967).
The male accessory gland proteins (Acps) have been shown to play important roles in postmating behavior (Kubli 1992; Wolfner 1997). The sex peptide (SP or Acp70A), has been shown to be the most potent seminal peptide that induces the postmating responses in females (Chen et al. 1988; Aigaki et al. 1991). SP has been thought to be responsible for the short-term effects, since the effects of injected or ectopically expressed SP persisted for only 1 or 2 days. The “sperm effect” has been proposed to explain the long-lasting effects of mating (Manning 1962), but its mechanism has been an enigma for a long time. However, recent studies involving a knock-out and a knockdown of the SP gene unambiguously demonstrated that SP not only is responsible for the short-term effects, but also is essential for the long-term effects (Chapman et al. 2003; Liu and Kubli 2003). SP may be bound to sperm and transferred into the sperm storage organs of the female to remain active for a longer period.
Much less is known about how SP works in mated females. The importance of the nervous system has been suggested in various studies (Boulétreau-Merle 1976; Szabad and Fajszi 1982; Tompkins and Hall 1983). Nakayama et al. (1997) expressed a membrane-bound SP transgene in various patterns in the female body and found a correlation between SP responses and expression in the brain. More directly, histological studies using radiolabeled SP showed localization of putative SP-binding proteins to the sensory nerves and the genital tract (Ottiger et al. 2000; Ding et al. 2003). It has been shown that SP stimulates biosynthesis of juvenile hormone (JH) in excised corpus allatum in vitro (Moshitzky et al. 1996) and accumulation of yolk proteins into oocytes (Soller et al. 1997) as well as vitellogenic oocyte progression in sexually mature virgin females (Soller et al. 1999). However, application of a JH analog did not induce ovulation (Soller et al. 1999). Thus, it appears that oocyte progression is independent of the ovulation mechanism.
There have been a few studies on genetic bases of female receptivity and ovulation. Boulétreau-Merle (1982; Boulétreau et al. 1989) reported genetic divergence in egg-retention periods in virgin females of D. melanogaster. Chromosomal analysis of the variants revealed major components located on the third chromosome and secondary factors on the X chromosome. In addition, some alleles of the lozenge gene are known to cause high levels of ovulation and oviposition in virgin females (Fuyama 1995). Similarly, increased levels of spontaneous ovulation were also observed in some single P-element insertion lines (Ejima et al. 2001). Interestingly, virgin females of such lines frequently show extrusion of the ovipositor in response to a male's courting behavior, suggesting a common genetic basis for the regulation of ovulation level and behavioral pattern. However, no further information is available for the mechanisms involved in the postmating responses.
To dissect the postmating responses genetically, it is essential to identify mutations affecting these processes. Ovulation is an obvious phenotype and is simple and fast to score. However, there are potential limitations with the screen in a conventional method. First, looking at ovulation phenotypes of mated females may not specifically identify the candidates; the anticipated mutants would not ovulate as frequently as wild type, perhaps as a result of problems in oogenesis or development of organs involved in egg laying. In other words, a high level of noise would hamper the screen. Second, postmating phenotypes could be assessed only for females that had developed, had matured, and were also properly inseminated by males. Considering the pleiotropic effects of mutations in general, such requirements would greatly reduce the number of genes to be tested. Tissue-specific mutagenesis should overcome these problems. A systematic misexpression system for genes in the Drosophila genome has been developed using P-element vectors that contain an upstream activating sequence (UAS) for the GAL4 transcription factor (Rorth 1996, 1998; Toba et al. 1999). In addition, looking for mutant virgins that ovulate at high frequency would be feasible, instead of looking for mated ones that do not ovulate.
In the present study, we used a GAL4-dependent misexpression system to identify genes that might be involved in postmating responses. We chose GAL4 drivers that direct misexpression in the nervous system to allow restricted perturbations of neural functions, which are thought to be important for the postmating responses. We demonstrate that overexpression of sarah (sra) in the nervous system of virgin females induces high levels of ovulation and behavioral patterns characteristic of mated females. Females with a reduced level of sra function were sterile and showed compromised postmating responses. Together these results indicate that the expression level of sra is critical for ovulation and female courtship behavior, including postmating changes.
MATERIALS AND METHODS
Fly strains:
Flies were raised on a standard yeast-cornmeal-glucose-agar medium at 25°. A collection of gene search (GS) lines, each bearing a GAL4-dependent misexpression vector, P{GS}, were used for screening (Toba et al. 1999). Oregon-R (Or-R) and y w (y w67c23 Df(1)w67c23) stock, a recipient strain for the GS lines, were used as controls for the ovulation and behavioral phenotype assays. elav-GAL4 (Lin and Goodman 1994) and sca-GAL4 (Klaes et al. 1994) were used as forced expression drivers. Line EP3462 was obtained from the Szeged Drosophila melanogaster P Insertion Mutant Stock Center. GS10207, GS10900, GS12031, GS9883, and GS10522 carry a unidirectional misexpression vector, P{GSv6} (FlyBase: http://flybase.net/.bin/fbidq.html?FBtp0017434).
Ovulation assay:
Ovulation was examined in 5-day-old virgins or mated females by individually squeezing the tip of the abdomen with forceps; only ovulating females eject an egg from the ovipositor (Aigaki et al. 1991). Ovulation level was expressed as a percentage of ovulating females per total number of females tested. At least 30 individuals were used for each assay for the primary screening, and at least 100 females were used for confirmation of the phenotype.
Behavioral assay:
Behavioral phenotypes of females were assessed by mass or pair mating. In the mass mating assay, five virgin females were placed together with seven Or-R males in a standard vial (3 cm in diameter, 10 cm in height), and the number of copulations achieved within 30 min was scored. Mating success was expressed as a percentage of copulated females per total number of females tested. At least 50 females were tested for each line. The pair-mating assay was performed as described in Ejima et al. (2001). Briefly, a 5-day-old virgin female was placed together with an Or-R male in a mating chamber (1.3 cm in diameter, 0.5 cm in height) and allowed to acclimate to the new environment for 5 min, since newly transferred flies are hyperactive and not interested in courtship at least for the first few minutes. Courtship behavior of the male and rejection behavior of the female were then observed either for the following 10 min or until copulation occurs. Total time of courting by the male and that of extrusion by the female were recorded for each pair. The courtship index (CI)—the percentage of time within the observation period that a male displayed any element of courtship behavior—was determined as described by Tompkins et al. (1980). To express the amount of rejection behavior, we employed the extrusion index (EI) as a percentage of the time that a female showed extrusion behavior during the total time of being courted by a male (Ejima et al. 2001). At least 10 individuals were used to determine the mean CI and EI of one genotype.
Remating test:
Females were placed together with Or-R males, and mated females were separated from males and maintained in a vial with medium until the test. A mated female was placed with two Or-R males in an observation chamber (16 mm in diameter, 19 mm in height; BD FALCON Multiwell Cell Culture Plates, 24 well, BD Bioscience, San Jose, CA). The number of rematings achieved during 2 hr of observation was counted, and the remating rate was expressed as a percentage of remated females per total number of tested females.
Inverse PCR:
Genomic DNA was extracted from GS insertion lines using a Fast DNA kit (Qbiogene, Carlsbad, CA) and digested with MspI, followed by self-ligation. Polymerase chain reaction (PCR) was carried out using a KOD-PLUS kit (TOYOBO, Osaka, Japan) and a Perkin-Elmer (Norwalk, CT) Gene Amp PCR 2400 with a primer set, 5′P wing-specific primers (In 5′-CTCCGTAGACGAAGCGCCTCTATTT and Out 5′-CTGAATAGGGAATTGGGAATTCGACGACTAGTT) or 3′P wing-specific primers (In 5′-TCGAGCGCGGCCGCAAGAT and Out 5′-CCTGCAGCCAAGCTTTGGTACT). The resulting PCR products were electrophoresed on a 1% agarose [Sigma (St. Louis) type II] gel, and the amplified DNA bands were excised with a razor blade and subsequently purified using a QIAquick column (QIAGEN, Hilden, Germany). The purified DNA fragments were used as a template for sequencing reactions with the BigDye Terminator Cycle Sequencing FS Ready Reaction kit (Applied Biosystems, Foster City, CA) using either 5′P primer (5′-CACTGAATTTAAGTGTATACTTCGG) or 3′P primer (5′-CTCGCACTTATTGCAAGCATAC). Sequencing was carried out using a Applied Biosystems PRISM Genetic Analyzer 310. Sequence similarity searches were performed using the BLASTN or BLASTX program (Altschul et al. 1997).
Transgenic flies:
sra cDNA was excised from the plasmid clone (no. LD15403, Research Genetics, Huntsville, AL) with EcoRI and XhoI (TOYOBO) and subcloned into the corresponding sites of pUAST (Brand and Perrimon 1993). The resulting vector containing UAS-sra cDNA was introduced into y w stock using a P-element-mediated transformation technique (Rubin and Spradling 1982).
Whole-mount RNA in situ hybridization:
Imaginal discs, brains, and ovaries were dissected from third instar larvae and 3-day-old Or-R adult females in Ringer's solution. Dissected tissues were fixed with 4% paraformaldehyde in phosphate-buffered saline containing 0.1% Tween 20 (PBS-Tween) for 20 min at room temperature. After washing three times with PBS-Tween, the tissues were treated with 4 μg/ml proteinase K (Boehringer Mannheim, Mannheim, Germany) in PBS-Tween for 5 min at room temperature, washed three times with PBS-Tween, and refixed with 4% paraformaldehyde in PBS-Tween. Antisense RNA probe was prepared by in vitro transcription of sra cDNA cloned into pBluescript KS using digoxigenin (DIG)-11-UTP (Boehringer Mannheim) and T3 RNA polymerase (Boehringer Mannheim). Hybridization was performed essentially as described in Lehmann and Tautz (1994), and the signals were detected immunohistochemically with antidigoxigenin antibodies conjugated with alkaline phosphatase (Boehringer Mannheim) using 4-nitroblue tetrazolium chloride (Boehringer Mannheim) and 5-bromo-4-chloro-3-indolyl-phosphate (Boehringer Mannheim) as substrates.
Northern analysis:
Total RNA was isolated from adult flies using an RNeasy kit (QIAGEN) and electrophoresed on 1% agarose gel containing 20 mm MOPS (pH 7.0), 5 mm sodium acetate, 0.1 mm EDTA, and 6.5% formaldehyde. After being transferred to a nylon membrane (Hybond-N+, Amersham Biosciences, Piscataway, NJ), RNA was fixed onto the membrane by irradiation with ultraviolet light for 3 min. The membranes were hybridized with DIG-UTP-labeled RNA probes prepared using a DIG RNA labeling kit (Roche, Basel, Switzerland). Signals were visualized by a CDP-star system (Roche).
DAPI staining of eggs:
Eggs were collected within 60 min after deposition on the media, devitellinized, and fixed as described by Ashburner (1989). The eggs were treated with 1 μg/ml 4′6-diamidino-2-phenylindole (DAPI; Sigma) for 5 min at room temperature and washed with PBS three times each for 1 min and then observed under an epifluorescent microscope (BX50-FLA, Olympus, Lake Success, NY).
RESULTS
Misexpression screen for spontaneous ovulation level in virgins:
We chose GAL4 drivers that direct misexpression in the nervous system to allow perturbations of neural functions. It is anticipated that increased ovulation phenotypes could be seen only when particular genes are misexpressed in proper patterns and at proper levels in the nervous system. To increase the probability of identifying candidates, we used two distinct drivers, elav-GAL4 and sca-GAL4, both of which are expressed in the central nervous system (CNS) and peripheral nervous system (PNS), but clearly different in expression level depending on the region and the developmental stage: elav-GAL4 is expressed at a relatively high level in the CNS in both larval and adult stages, whereas sca-GAL4 is relatively high in the embryonic and larval PNS and also expressed in the proneural clusters in the embryos and the sensory organ precursors.
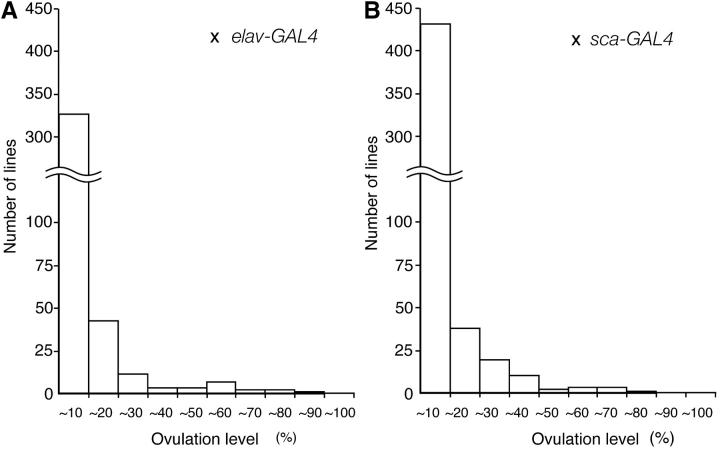
A total of 614 P{GS} insertion lines were crossed to elav-GAL4 and sca-GAL4 drivers, and the F1 virgin females carrying both GS and GAL4 transgenes were individually examined for the presence or absence of an egg in the uterus. Most of the combinations of GS insertions and GAL4 drivers had no effect on ovulation. However, some insertions induced ovulation at high levels, comparable to those observed in mated females (Figure 1). There were eight GS lines whose misexpression caused >50% of ovulation. We repeated the assay using >100 individuals for these lines and confirmed that all of them showed stimulated ovulation with at least one of the GAL4 drivers (Figure 2). There were no dominant effects of insertions on ovulation when these GS lines were crossed to Or-R or y w, a recipient stock of the GS line, indicating that the increased ovulation phenotypes were caused by the GAL4-dependent transcripts of DNA flanked by the insertions.
Figure 1.—
Classification of GS lines according to the effects of GAL4-dependent misexpression on ovulation. A total of 614 GS lines were crossed to elav-GAL4 (A) or sca-GAL4 (B), and the F1 virgin females were scored for ovulation level, which was expressed as a percentage of ovulating females per total number of tested females. Bars represent the number of lines with indicated levels of ovulation. At least 30 virgin females were examined for each GS line.
Figure 2.—
Effects of GAL4-dependent misexpression of eight GS insertions on ovulation in virgin females. GS lines, which induced ovulation in >50% of their progeny during the first screen, were reexamined using the same driver lines, elav-GAL4 (hatched) or sca-GAL4 (solid). Bars indicate the ovulation level (number of ovulated females per total). At least 100 individuals were examined for each GS line. For comparison, results of experiments with Or-R (open bar) and a UAS-sra transgene were also included. Mean ovulation levels are indicated with a 95% confidence interval. (*) Ovulation level was not determined, because of semilethality in the F1 hybrids.
Mapping of vector insertion sites:
To identify candidate genes responsible for the increased ovulation phenotypes, we determined the vector-flanking sequences of the eight insertion lines, whose ovulation levels were >50%. Vector-flanking sequences were obtained using inverse PCR and used as queries for BLAST searches against the Drosophila genome sequences. Five of them matched known genes and three matched predicted genes (Table 1). The GS1093 insertion was in the first intron of norpA encoding a 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase involved in vision and olfaction (Yoshioka et al. 1985). Line GS1116 had an insertion in the broad (br) gene, which encodes a family of BTB (broad complex, tramtrack, bric-a-brac) zinc-finger transcription factors, each containing one of four distinct zinc-finger domains, Z1, Z2, Z3, and Z4 (DiBello et al. 1991). The br gene is known to affect oogenesis and development of imaginal discs and the central nervous system (Gotwals and Fristrom 1991; Restifo and White 1991; Huang and Orr 1992). In line GS2133, the vector was inserted into the second intron of the grapes (grp) gene, which encodes a protein kinase, whose mutation causes metaphase arrest early in embryogenesis at mitotic cycle 13 (Fogarty et al. 1994). Lines GS3080 and GS3168 had insertions in the same locus, sra (A. Czank, personal communication), previously called nebula (McCormick 1999). We named the two insertions in the sra locus, sraGS3080 and sraGS3168, respectively, and characterized them in further detail.
TABLE 1.
GS insertion lines with high-ovulation phenotype
| GS lines | Genes nearby insertiona |
Insertion siteb | Gene product/protein domainc |
|---|---|---|---|
| Cloned genes | |||
| 1093 | norpA | +2835 (intron) | Phospholipase C |
| 1116 | br | +13050 (intron) | BTB-zinc-finger transcription factor |
| 2133 | grp | +4129 (intron) | Protein serine/threonine kinase |
| 3080 | sra | +2668 (intron) | DSCR1 homolog |
| 3168 | sra | +2675 (intron) | DSCR1 homolog |
| Predicted genes | |||
| 1061 | CG11700 | +1370 (downstream) | Ubiquitin-like domain |
| 2143 | CG4612 | −72 (upstream) | RNA-binding domain |
| CG30169 | −78 (upstream) | Unknown | |
| 3132 | CG3961 | −428 (upstream) | Firefly luciferase-like domain |
Nearest genes or flanking genes within 500 bp of the insertion sites were indicated.
The 5′-most end of mRNA was defined as +1.
FlyBase (http://flybase.net/).
Misexpression of sra induces high levels of ovulation:
The GS vector is a bidirectional misexpression vector; thus there are two kinds of transcripts derived from the DNA flanked by an insertion (Toba et al. 1999). Since the GS vector insertions in both sraGS3080 and sraGS3168 were in the intron, it was not clear which strand was responsible for the GAL4-dependent phenotypes. The sense-strand transcripts may encode a full-length Sra protein, whereas the antisense-strand transcripts may interfere with expression of wild-type sra. To clarify the causative transcripts, we examined several insertions in sra with a unidirectional misexpression vector, P{GSv6} or P{EP} (Table 2). Lines GS10207, GS10900, and GS12031 had P{GSv6} insertions in the intron, which would direct misexpression of the sra sense strand encoding a full-length Sra protein. Effects of the sra antisense strand were examined with two GS lines (GS9883 and GS10522) inserted in the same region of the intron and with EP3462 inserted in the 5′ UTR (Figure 3A). Ovulation was clearly stimulated in virgin females when the sra sense strand was misexpressed, whereas none of the antisense misexpression lines caused stimulated ovulation. These results strongly suggested that misexpression of the sra sense strand is responsible for the high-ovulation phenotype.
TABLE 2.
Phenotypic analysis of unidirectional misexpression vector insertions in thesra locus
| Insertion site
|
|||
|---|---|---|---|
| P insertiona | Distance from 5′ end of mRNA |
Location | Ovulation level (%) |
| Sense | |||
| GS10207 | +2621 | First intron | 58 |
| GS10900 | +2674 | First intron | 43 |
| GS12031 | +2675 | First intron | 51 |
| Antisense | |||
| GS9883 | +2743 | First intron | 6 |
| GS10522 | +2745 | First intron | 4 |
| EP3462 | +104 | 5′ UTR | 0 |
All insertions carry a single unidirectional misexpression vector, P{GSv6} or P{EP}, and were classified according to the UAS-linked strand, sense or antisense.
Figure 3.—
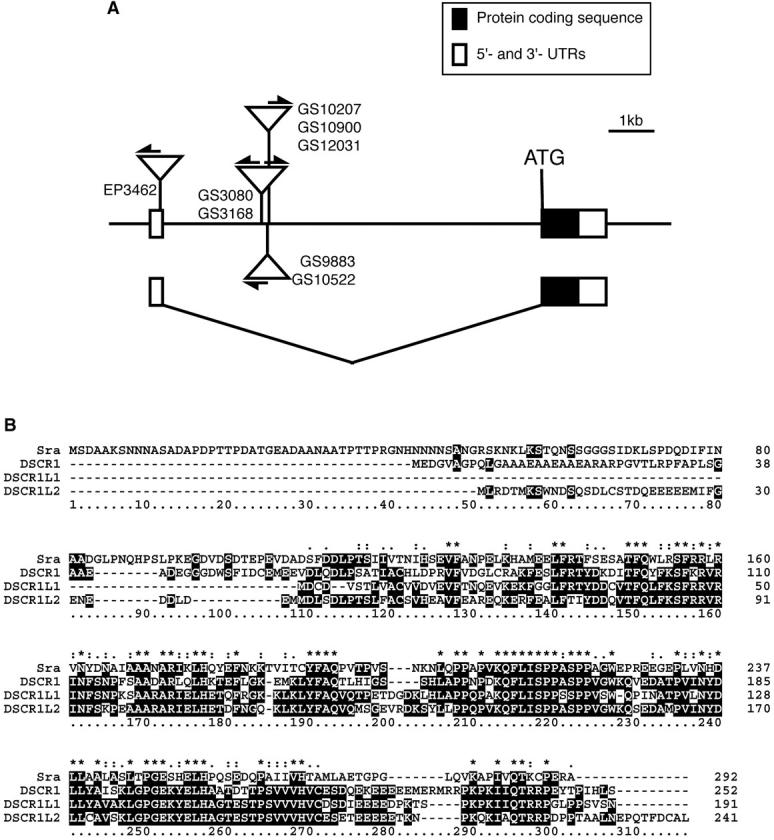
(A) Schematic of the genomic organization of the sra gene with misexpression vector insertions. Exons are indicated by boxes with protein-coding sequences (solid boxes) and untranslated regions (open boxes). Closely situated insertions are grouped and represented by open triangles with the line numbers (see Tables 1 and 2 for map positions). Arrows indicate the direction of transcription upon GAL4 activation. Lines GS3080 and GS3168 carry a bidirectional vector, P{GS}, whereas other insertion lines have the unidirectional vector P{GSv6} or P{EP}. (B) Sequence alignment of Sra and human DSCR1 family proteins. Amino acid sequences were aligned using CLUSTALX (Thompson et al. 1997). (*) A single, fully conserved residue; (: and .) the positions at which conserved residues belong to the “strong” and “weak” groups in the Gonnet Pam250 matrix, respectively (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/Top.html).
The sra gene encodes a protein consisting of 292 amino acids with sequence similarity to human genes Down syndrome critical region 1 (DSCR1), DSCR1-like1 (DSCR1L1), and DSCR1-like2 (DSCR1L2; Strippoli et al. 2000). It has a cadherin signature (amino acid residues 73–102) and an RNA recognition motif (118–188). Amino acid identities of Sra with the DSCR1 family proteins were 27% (80/297), 40% (79/197), and 36% (88/243) for DSCR1, DSCR1L1, and DSCR1L2, respectively (Figure 3B). The sra locus consists of two exons interrupted by an 8731-base-long intron, in which the GS vectors were inserted (Figure 3A).
We constructed UAS-sra transgenic flies to test whether overexpression of sra is responsible for the observed phenotype. When UAS-sra was overexpressed in the nervous system, ovulation levels were increased up to 74 and 72% for elav-GAL4 and sca-GAL4, respectively (Figure 2). These results demonstrated that misexpression of sra induced high levels of ovulation in virgin females.
Misexpression of sra affects female courtship behavior:
We determined the mating success of flies misexpressing sra to estimate whether they have any abnormality in courtship behavior. Within 30 min, 82% of wild-type virgin females mated, whereas only 20% (sraGS3080/elav-GAL4), 35% (sraGS3168/sca-GAL4), 50% (UAS-sra/elav-GAL4), and 65% (UAS-sra/sca-GAL4) of sra-misexpressing virgins succeeded in mating, implying that sra misexpression affects the behavioral phenotypes as well. To analyze the behavioral phenotype in detail, we determined the CI, which represents the attractiveness to males, and the EI, which represents the frequency of extrusion behavior toward courting males. As shown in Figure 4A, effects of misexpression on females' attractiveness were variable, depending on the line. sraGS3080/elav-GAL4, sraGS3168/sca-GAL4, UAS-sra/elav-GAL4, and UAS-sra/sca-GAL4 females elicited intermediate CI levels between wild-type virgins (CI = 83) and mated females (CI = 30) levels. CIs of sraGS3080/elav-GAL4 and UAS-sra/elav-GAL4 were significantly different from both wild-type virgins and mated females (ANOVA, P < 0.05), whereas sraGS3168/sca-GAL4 was significantly different from Or-R virgins only, and UAS-sra/sca-GAL4 was significantly different from mated ones only.
Figure 4.—
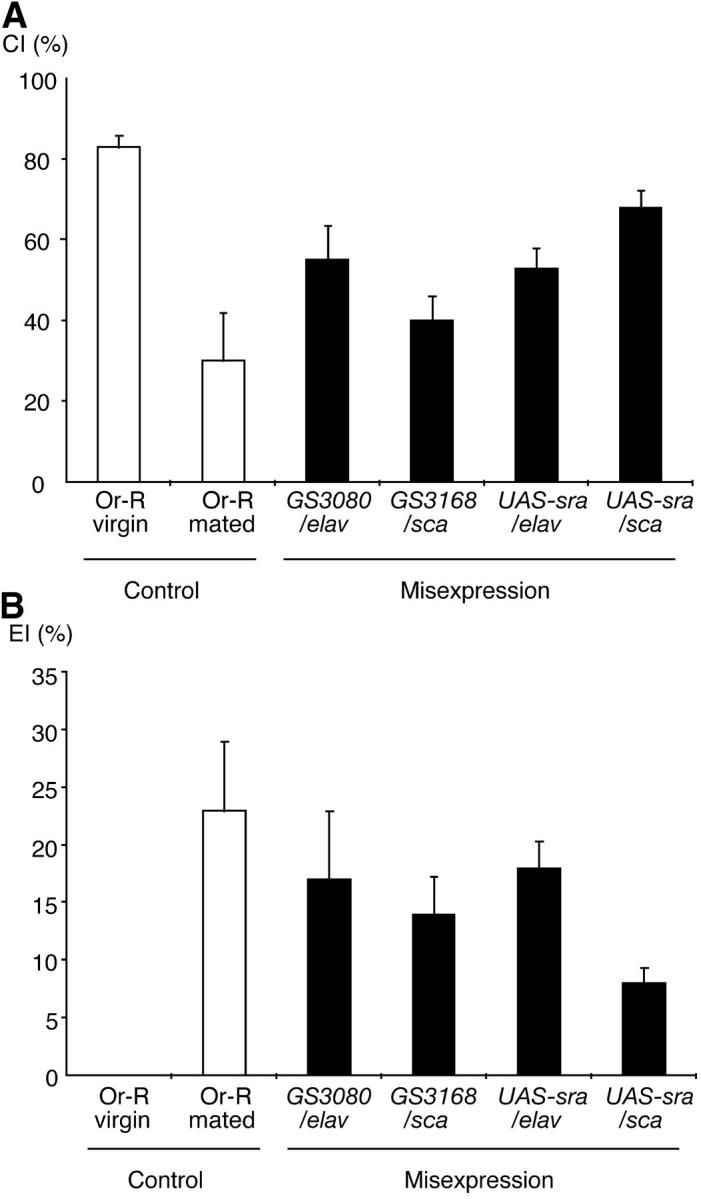
Behavioral phenotypes associated with misexpression of GS insertions in sra. (A) Courtship index. CIs of GS3080/elav-GAL4 and UAS-sra/elav-GAL4 were significantly different from both wild-type virgins and mated females (ANOVA, P < 0.05), whereas GS3168/sca-GAL4 was significantly different from Or-R virgins only, and UAS-sra/sca-GAL4 was significantly different from mated ones only. (B) Extrusion index. EIs of sra-misexpressing virgins tested were significantly higher than those of Or-R virgins. Statistic analysis showed that the level of EI for UAS-sra/sca-GAL4 was significantly lower than that of mated Or-R females. CI and EI were determined as described in materials and methods. Virgin and mated Or-R females were used as a control. Ten females were used for each assay. Mean CI or EI was indicated with a 95% confidence interval.
On the other hand, virgin females misexpressing sra frequently displayed extrusion behavior as indicated by high EIs, ranging from 8 to 18, depending on the genotype (Figure 4B). The frequency of extrusion behavior observed in all cases, except UAS-sra/sca-GAL4, was nearly comparable to that of wild-type mated females (EI = 23). Statistic analysis showed that all EIs of hetero-transgenic flies were significantly higher than those of Or-R virgins (ANOVA, P < 0.05) and that the EI of UAS-sra/sca-GAL4 was significantly lower than that of mated Or-R females. The apparently associated phenotypes of ovulation and behavioral pattern suggest that sra affects a mechanism that controls both ovulation and behavioral pattern.
Reduction of sra affects female fertility:
We found that the females homozygous for sraGS3080 or sraGS3168 were sterile. Excision of the GS vector from these insertions using the Δ2-3 transposase restored fertility, suggesting that the insertion in the sra locus was the cause of the sterility (data not shown). In Northern analysis, the expression of sra mRNA was not detectable for either sraGS3080 or sraGS3168 flies (Figure 5A). However, both alleles were hypomorphic rather than null, since wild-type transcripts were detectable by RT-PCR (data not shown). The number of eggs laid by sraGS3080 or sraGS3168 females was fewer than that of wild type, and most of them were arrested at metaphase I of meiosis (Figure 5B). Some eggs showed meiotic progression, but none of them completed meiosis, suggesting that sra is required at least for female meiosis.
Figure 5.—

(A) Northern analysis of sra expression. Expression levels of sra were dramatically reduced in sraGS3080 and sraGS3168 flies. These alleles seem to be hypomorphic rather than null, since the wild-type transcript was detectable with RT-PCR (data not shown). (B) DAPI staining of eggs laid by sraGS3080 females, showing meiotic arrest. Most of the eggs were arrested at metaphase I of meiosis. Arrowhead indicates the nucleus. Eggs were collected within 1 hr after egg deposition and stained with DAPI as described in materials and methods.
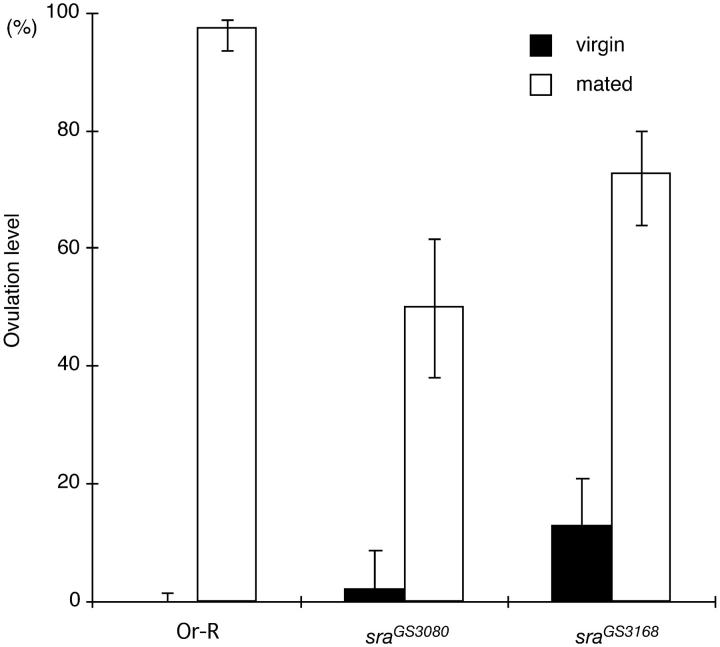
Ovulation levels in sra mutants are less strictly controlled: spontaneous ovulation occurred in 3% for sraGS3080 and in 13% for sraGS3168 virgins, whereas virtually no ovulation occurs in Or-R and y w virgins. These phenotypes suggest that sra is required for proper control of ovulation in virgins. sra females seem to accept mating normally, but their postmating changes were compromised: the level of ovulation was not as high as that of wild-type females (Figure 6); they showed reduced receptivity after mating, but their remating rate 24 hr after mating was significantly higher than that of wild type (Figure 7). Furthermore, receptivity of sra females was restored earlier than that of wild type; the remating rate 10 days after mating was significantly higher than that of wild type for both alleles, although they showed the same level of remating 15 days after mating (Figure 7). These results suggest that sra is involved in the postmating responses. On the other hand, sra males did not show any obvious defects in development or fertility. Taken together, sra appears to be involved in female-specific functions, production of eggs, ovulation, and courtship behavior.
Figure 6.—
Effects of mating on ovulation levels in the sra mutant females. Ovulation levels were determined in virgin and mated (24 hr after mating) Or-R, sraGS3080, and sraGS3168 females. Spontaneous ovulation occurred in virgin female sra mutants. Postmating stimulation of ovulation was not as dramatic as in wild-type.
Figure 7.—
Remating rate of the sra mutant females. Virgin females of Or-R (control), sraGS3080, and sraGS3168 were mated with Or-R males, and the mated females were separated and maintained in a vial with medium until testing as described in materials and methods. The remating rate was expressed as a percentage of remated females per total number of tested females. Thirty females were used for each assay, and the mean remating rates were indicated with 95% confidence intervals. Remating rate of sra mutants was higher than that of wild type, and postmating reduced receptivity was restored earlier than that of wild type. ***P < 0.001, **P < 0.01, *P < 0.02.
The gene sra is expressed in adult brain and during oogenesis:
RNA in situ hybridization showed that sra mRNA was present in the central nervous system of the third instar larvae, with a relatively intense signal in the brain and weak signals in the ventral ganglion (Figure 8, A and B). A relatively low, but ubiquitous expression level was observed in leg and wing imaginal discs, and no signal was detected in the eye-antennal discs (data not shown). A strong signal was detected in all neurons of the adult brain (Figure 8, C and D). There is no specific localization of mRNA within the brain. The gene was also expressed at high levels in the nurse cells and oocytes (Figure 8, E and F). These results support the idea that sra functions in neurons during oogenesis.
Figure 8.—

Expression of sra in the third instar larva and the adult fly. Whole-mount in situ hybridization was performed as described in materials and methods. Larval CNS (A and B); adult brain (C and D); ovariole (E and F). Digoxigenin-labeled antisense-strand RNA was used as a probe (B, D, and F), and sense-strand RNA was used as control (A, C, and E). Relatively intense sra mRNA signals were detected in the central nervous system and weak signals in the ventral ganglion in the central nervous system of third instar larvae (A and B). Intense expression was observed in the adult brain (C and D) and in nurse cells and oocytes (E and F). Relatively low, but ubiquitous levels of expression were observed in leg and wing imaginal discs (data not shown).
DISCUSSION
Little has been known about the genes involved in postmating changes in Drosophila females. In the present study, we used a tissue-specific misexpression system to screen for genes that, when expressed in the nervous system, stimulate ovulation in virgin females. Among eight candidate genes identified, we focused on sra, since two independent insertions in this locus showed the same phenotype. Using insertions of the unidirectional misexpression vector in the same locus, we demonstrated that misexpression of the sra sense strand is responsible for the phenotype and then, using transgenic virgins expressing a UAS-sra cDNA transgene, confirmed that overexpression of the sra ORF is indeed the cause of stimulated ovulation.
Some GS lines showed increased levels of ovulation with only one of the GAL4 lines, suggesting that the expression level and/or patterns were critical for the phenotypes. However, the situation is rather complex: both GAL4 transgenes are expressed in a broad region of the nervous system and which GAL4 is effective is variable, depending on the GS line. Therefore, it is hard to clarify the relationship between the expression patterns and phenotypic effects. To analyze such a relationship, it would be feasible to choose GAL4 lines whose expression is more restricted.
The high-ovulation phenotype of sra-misexpressing virgin flies was associated with reduced receptivity; these flies rejected courting males frequently with extrusion behavior characteristics of mated females, while their attractiveness was close to virgins. These features are similar to the females mated with males lacking the accessory glands (Tram and Wolfner 1998) and those of females injected with SP (Chen et al. 1988; Schmidt et al. 1993) or ectopically expressing SP transgenes (Aigaki et al. 1991). Nakayama et al. (1997) expressed a membrane-bound form of SP in various patterns using many distinct P[GAL4] enhancer-trap lines and showed that the two postmating responses were tightly coupled. More recently, Ejima et al. (2001) have searched for genetic variations in spontaneous ovulation levels in virgins using P-element insertion lines, demonstrating a phenotypic association between ovulation level and frequency of extrusion behavior. Therefore, coupling of ovulation and the changes in courtship behavior appears to be rather general. There must be a common pathway involved in ovulation and receptivity of virgin females. Although the target of SP has yet to be identified, the most likely candidates are the nervous system and the reproductive system; membrane-tethered SP induces the postmating response most efficiently when expressed in the central nervous system (Nakayama et al. 1997) and radiolabeled SP binds to the sensory nerves and the genital tracts (Ottiger et al. 2000). Overexpression phenotype of sra might be closely associated with the SP-induced pathway.
The phenotypes of females homozygous for the hypomorphic alleles sra3080 and sra3168 were significantly different from those of wild type. They showed: (1) spontaneous ovulation in virgins, (2) sterility with impaired meiotic progression, and (3) compromised postmating responses with lower ovulation level, higher remating rate, and shorter period for restoration of receptivity. These phenotypes suggest that sra is required for female-specific functions: production of eggs and proper control of both receptivity and ovulation, depending on the status of mating (“virgin vs. mated”). sra mutants used in this study still responded to mating stimuli, laying more eggs and reducing their receptivity. These moderate phenotypes suggest that the insertions did not completely abolish the function of sra or that there may be other genes, which also contribute to the mechanism of the postmating response. It is also possible that the inserted P{GS} elements per se might have some contributions to these complex phenotypes. A null mutation of sra is essential to assess the roles of sra in the female-specific functions.
Both hypomorphy for and neural overexpression of sra increased ovulation in virgins. The apparent phenotypic consistency between the two distinct mechanisms supports the idea that ovulation is negatively regulated in virgins as the default. Disruption of this inhibitory mechanism by either reduction or overexpression of sra could result in deregulated ovulation. Since the ovulation level was higher in sra-overexpressing females compared to hypomorphic sra mutants, it is likely that sra-overexpressing females affected the ovulation mechanism only, whereas hypomorphic sra mutants had defects in both ovulation and oogenesis.
sra might be essential for resumption and progression of female meiosis. High levels of sra expression in both nurse cells and oocytes support this possibility. Meiotic arrest of oocytes and resumption at the time of ovulation or fertilization is rather common in both vertebrates and invertebrates (Gebauer and Richter 1997; Colas and Dube 1998; Russo et al. 1998; Yamashita 1998; Kishimoto 1999). So far, much of the knowledge on this topic has come from studies of vertebrates, which have revealed several important molecules, such as maturation-promoting factor, c-mos proto-oncogene product, and MAP kinase (Yamashita 1998; Kajiura-Kobayashi et al. 2000; Kotani and Yamashita 2002). In Drosophila, mature oocytes in ovaries are arrested at metaphase I of meiosis, which resumes after activation involving changes in eggshell permeability (Dävring and Sunner 1973; Foe et al. 1993). Heifetz et al. (2001) observed that an egg becomes impermeable as it proceeds down the oviducts, and the process is complete by the time the egg is in the uterus. Activation also triggers meiosis to resume before the egg reaches the uterus. It seems that these events are sequentially induced within a short time period. sra might be involved in multiple steps during these processes.
Sra is a cytoplasmic protein with sequence similarity to human DSCR1 and related proteins (Fuentes et al. 1995; Strippoli et al. 2000). A high level of DSCR1 expression is associated with Down syndrome (Fuentes et al. 2000) and Alzheimer's disease (Ermak et al. 2001). DSCR1 has been thought to function as an endogenous inhibitor of calcineurin, a calcium/calmodulin-dependent phosphatase, and protects cells from oxidative and calcium stress (Miyazaki et al. 1996; Crawford et al. 1997; Rothermel et al. 2000; Ermak et al. 2001, 2002; Lin et al. 2003). Recently, Chang et al. (2003) showed that both loss-of-function and overexpression mutants of sra exhibit severe learning defects. Overexpression of sra could have indirect effects on calcium signaling in the neurons that control the ovulation. With respect to the oocyte activation process, it has been shown in mice that Ca2+ calmodulin-dependent protein kinase II is activated in fertilized oocytes, which appeared to link the fertilization signal to resumption of meiosis (Tatone et al. 2002). Sra might be involved in the progression of meiosis through regulation of the calcium-dependent process. In addition, overexpression of sraGS3168 has been shown to extend longevity and increase oxidative stress resistance (Seong et al. 2001), implying a protective role in oxidative stress. Further investigation of sra, including isolation of a null mutation, should facilitate the understanding of its specific roles in the reproductive and the nervous system functions.
Acknowledgments
We thank Y. Fuyama and M. Hatsumi for helpful discussions and the Szeged, Bloomington, and Kyoto Stock Centers for fly stocks. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas in “Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (no. 12202002), and a grant from The Naito Foundation to T.A.
References
- Aigaki, T., I. Fleischmann, P. S. Chen and E. Kubli, 1991. Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron 7: 557–563. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids. Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., 1989 Drosophila: A Laboratory Manual, pp. 201–202, protocol 86. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Boulétreau-Merle, J., 1976. Destruction of the pars intercerebralis in Drosophila melanogaster: effect on the fecundity and the stimulation through copulation. J. Insect Physiol. 22: 933–940 (in French). [DOI] [PubMed] [Google Scholar]
- Boulétreau-Merle, J., 1982. Variability of initial retention in natural populations of D. melanogaster. Dros. Inf. Serv. 56: 28–29. [Google Scholar]
- Boulétreau-Merle, J., O. Terrier and P. Fouillet, 1989. Chromosomal analysis of initial retention capacity in virgin Drosophila melanogaster females. Heredity 62: 145–151. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Burnet, B., K. Connolly, M. Kearney and R. Cook, 1973. Effects of male paragonial gland secretion on sexual receptivity and courtship behaviour of female Drosophila melanogaster. J. Insect Physiol. 19: 2421–2431. [DOI] [PubMed] [Google Scholar]
- Chang, K. T., Y. J. Shi and K. T. Min, 2003. The Drosophila homolog of Down's syndrome critical region 1 gene regulates learning: implications for mental retardation. Proc. Natl. Acad. Sci. USA 100: 15794–15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T., J. Bangham, G. Vinti, B. Seifried, O. Lung et al., 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100: 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P. S., E. Stumm-Zollinger, T. Aigaki, J. Balmer, M. Bienz et al., 1988. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell 54: 291–298. [DOI] [PubMed] [Google Scholar]
- Colas, P., and F. Dube, 1998. Meiotic maturation in mollusc oocytes. Semin. Cell Dev. Biol. 9: 539–548. [DOI] [PubMed] [Google Scholar]
- Connolly, K., and R. Cook, 1973. Rejection responses by female Drosophila melanogaster: their ontogeny, causality and effects upon the behavior of the courting male. Behaviour 44: 142–167. [Google Scholar]
- Crawford, D. R., K. P. Leahy, N. Abramova, L. Lan, Y. Wang et al., 1997. Hamster adapt78 mRNA is a Down syndrome critical region homologue that is inducible by oxidative stress. Arch. Biochem. Biophys. 342: 6–12. [DOI] [PubMed] [Google Scholar]
- Dävring, L., and M. Sunner, 1973. Female meiosis and embryonic mitosis in Drosophila melanogaster. I. Meiosis and fertilization. Hereditas 73: 51–64. [DOI] [PubMed] [Google Scholar]
- DiBello, P. R., D. A. Withers, C. A. Bayer, J. W. Fristrom and G. M. Guild, 1991. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics 129: 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Z., I. Haussmann, M. Ottiger and E. Kubli, 2003. Sex-peptides bind to two molecularly different targets in Drosophila melanogaster females. J. Neurobiol. 55: 372–384. [DOI] [PubMed] [Google Scholar]
- Ejima, A., S. Nakayama and T. Aigaki, 2001. Phenotypic association of spontaneous ovulation and sexual receptivity in virgin females of Drosophila melanogaster mutants. Behav. Genet. 31: 437–444. [DOI] [PubMed] [Google Scholar]
- Ermak, G., T. E. Morgan and K. J. Davies, 2001. Chronic overexpression of the calcineurin inhibitory gene DSCR1 (Adapt78) is associated with Alzheimer's disease. J. Biol. Chem. 276: 38787–38794. [DOI] [PubMed] [Google Scholar]
- Ermak, G., C. D. Harris and K. J. Davies, 2002. The DSCR1 (Adapt78) isoform 1 protein calcipressin 1 inhibits calcineurin and protects against acute calcium-mediated stress damage, including transient oxidative stress. FASEB J. 16: 814–824. [DOI] [PubMed] [Google Scholar]
- Foe, V. E., G. M. Odell and B. A. Edgar, 1993 Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint, pp. 149–300 in The Development of Drosophila melanogaster, Vol. 1, edited by M. Bate and A. M. Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Fogarty, P., R. F. Kalpin and W. Sullivan, 1994. The Drosophila maternal-effect mutation grapes causes a metaphase arrest at nuclear cycle 13. Development 120: 2131–2142. [DOI] [PubMed] [Google Scholar]
- Fowler, G., 1973. Some aspects of the reproductive biology of Drosophila: sperm transfer, sperm storage and sperm utilization. Adv. Genet. 17: 293–360. [Google Scholar]
- Fuentes, J. J., M. A. Pritchard, A. M. Planas, A. Bosch, I. Ferrer et al., 1995. A new human gene from the Down syndrome critical region encodes a proline-rich protein highly expressed in fetal brain and heart. Hum. Mol. Genet. 4: 1935–1944. [DOI] [PubMed] [Google Scholar]
- Fuentes, J. J., L. Genesca, T. J. Kingsbury, K. W. Cunningham, M. Perez-Riba et al., 2000. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum. Mol. Genet. 9: 1681–1690. [DOI] [PubMed] [Google Scholar]
- Fuyama, Y., 1995. Genetic evidence that ovulation reduces sexual receptivity in Drosophila melanogaster females. Behav. Genet. 25: 581–587. [DOI] [PubMed] [Google Scholar]
- Gebauer, F., and J. D. Richter, 1997. Synthesis and function of Mos: the control switch of vertebrate oocyte meiosis. BioEssays 19: 23–28. [DOI] [PubMed] [Google Scholar]
- Gotwals, P. J., and J. W. Fristrom, 1991. Three neighboring genes interact with the Broad-Complex and the Stubble-stubbloid locus to affect imaginal disc morphogenesis in Drosophila. Genetics 127: 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J. C., 1994. The mating of a fly. Science 264: 1702–1714. [DOI] [PubMed] [Google Scholar]
- Heifetz, Y., J. Yu and M. F. Wolfner, 2001. Ovulation triggers activation of Drosophila oocytes. Dev. Biol. 234: 416–424. [DOI] [PubMed] [Google Scholar]
- Huang, R. Y., and W. C. Orr, 1992. Broad-complex function during oogenesis in Drosophila melanogaster. Dev. Genet. 13: 277–288. [DOI] [PubMed] [Google Scholar]
- Kajiura-Kobayashi, H., N. Yoshida, N. Sagata, M. Yamashita and Y. Nagahama, 2000. The Mos/MAPK pathway is involved in metaphase II arrest as a cytostatic factor but is neither necessary nor sufficient for initiating oocyte maturation in goldfish. Dev. Genes Evol. 210: 416–425. [DOI] [PubMed] [Google Scholar]
- Kishimoto, T., 1999. Activation of MPF at meiosis reinitiation in starfish oocytes. Dev. Biol. 214: 1–8. [DOI] [PubMed] [Google Scholar]
- Klaes, A., T. Menne, A. Stollewerk, H. Scholz and C. Klambt, 1994. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell 78: 149–160. [DOI] [PubMed] [Google Scholar]
- Kotani, T., and M. Yamashita, 2002. Discrimination of the roles of MPF and MAP kinase in morphological changes that occur during oocyte maturation. Dev. Biol. 252: 271–286. [DOI] [PubMed] [Google Scholar]
- Kubli, E., 1992. The sex-peptide. BioEssays 14: 779–784. [DOI] [PubMed] [Google Scholar]
- Leahy, M. G., and M. L. Lowe, 1967. Purification of the male factor increasing egg deposition in D. melanogaster. Life Sci. 6: 151–156. [PubMed] [Google Scholar]
- Lehmann, R., and D. Tautz, 1994. In situ hybridization to RNA. Methods Cell Biol. 44: 575–598. [DOI] [PubMed] [Google Scholar]
- Lin, D. M., and C. S. Goodman, 1994. Ectopic and increased expression of fasciclin II alters motoneuron growth cone guidance. Neuron 13: 507–523. [DOI] [PubMed] [Google Scholar]
- Lin, H. Y., H. J. Michtalik, S. Zhang, T. T. Andersen, D. A. Van Riper et al., 2003. Oxidative and calcium stress regulate DSCR1 (Adapt78/MCIP1) protein. Free Radic. Biol. Med. 35: 528–539. [DOI] [PubMed] [Google Scholar]
- Liu, H., and E. Kubli, 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, A., 1962. A sperm factor affecting the receptivity of Drosophila melanogaster females. Nature 194: 252–253. [Google Scholar]
- McCormick, A. V., 1999 Drosophila melanogaster sarah (sra) mRNA, complete cds., GenBank/EMBL/DDBJ no. AF147700.
- Miyazaki, T., Y. Kanou, Y. Murata, S. Ohmori, T. Niwa et al., 1996. Molecular cloning of a novel thyroid hormone-responsive gene, ZAKI-4, in human skin fibroblasts. J. Biol. Chem. 271: 14567–14571. [DOI] [PubMed] [Google Scholar]
- Moshitzky, P., I. Fleischmann, N. Chaimov, P. Saudan, S. Klauser et al., 1996. Sex-peptide activates juvenile hormone biosynthesis in the Drosophila melanogaster corpus allatum. Arch. Insect Biochem. Physiol. 32: 363–374. [DOI] [PubMed] [Google Scholar]
- Nakayama, S., K. Kaiser and T. Aigaki, 1997. Ectopic expression of sex-peptide in a variety of tissues in Drosophila females using the P[GAL4] enhancer-trap system. Mol. Gen. Genet. 254: 449–455. [DOI] [PubMed] [Google Scholar]
- Ottiger, M., M. Soller, R. F. Stocker and E. Kubli, 2000. Binding sites of Drosophila melanogaster sex peptide pheromones. J. Neurobiol. 44: 57–71. [PubMed] [Google Scholar]
- Restifo, L. L., and K. White, 1991. Mutations in a steroid hormone-regulated gene disrupt the metamorphosis of the central nervous system in Drosophila. Dev. Biol. 148: 174–194. [DOI] [PubMed] [Google Scholar]
- Rorth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth, P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78: 113–118. [DOI] [PubMed] [Google Scholar]
- Rothermel, B., R. B. Vega, J. Yang, H. Wu, R. Bassel-Duby et al., 2000. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem. 275: 8719–8725. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Russo, G. L., M. Wilding, M. Marino and B. Dale, 1998. Ins and outs of meiosis in ascidians. Semin. Cell Dev. Biol. 9: 559–567. [DOI] [PubMed] [Google Scholar]
- Schmidt, T., Y. Choffat, S. Klauser and E. Kubli, 1993. The Drosophila melanogaster sex-peptide: a molecular analysis of structure-function relationships. J. Insect Physiol. 39: 361–368. [Google Scholar]
- Seong, K. H., T. Ogashiwa, T. Matsuo and T. Aigaki, 2001. Application of the gene search system to screen for longevity genes in Drosophila. Biogerontology 2: 209–217. [DOI] [PubMed] [Google Scholar]
- Soller, M., M. Bownes and E. Kubli, 1997. Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster. Eur. J. Biochem. 243: 732–738. [DOI] [PubMed] [Google Scholar]
- Soller, M., M. Bownes and E. Kubli, 1999. Control of oocyte maturation in sexually mature Drosophila females. Dev. Biol. 208: 337–351. [DOI] [PubMed] [Google Scholar]
- Spieth, H. T., and J. M. Ringo, 1983 Mating behavior and sexual isolation in Drosophila, pp. 223–284 in The Genetics and Biology of Drosophila, Vol. 3c, edited by M. Ashburner, H. L. Carson and J. N. Thompson. Academic Press, New York.
- Strippoli, P., M. Petrini, L. Lenzi, P. Carinci and M. Zannotti, 2000. The murine DSCR1-like (Down syndrome candidate region 1) gene family: conserved synteny with the human orthologous genes. Gene 257: 223–232. [DOI] [PubMed] [Google Scholar]
- Szabad, J., and C. Fajszi, 1982. Control of female reproduction in Drosophila: genetic dissection using gynandromorphs. Genetics 100: 61–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatone, C., S. Delle Monache, R. Iorio, D. Caserta, M. Di Cola et al., 2002. Possible role for Ca(2+) calmodulin-dependent protein kinase II as an effector of the fertilization Ca(2+) signal in mouse oocyte activation. Mol. Hum. Reprod. 8: 750–757. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba, G., T. Ohsako, N. Miyata, T. Ohtsuka, K. H. Seong et al., 1999. The gene search system: a method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins, J., J. C. Hall and L. M. Hall, 1980. Courtship-stimulating volatile compounds from normal and mutant Drosophila. J. Insect Physiol. 26: 689–697. [Google Scholar]
- Tompkins, L., and J. C. Hall, 1983. Identification of brain sites controlling female receptivity in mosaics of Drosophila melanogaster. Genetics 103: 179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins, L., A. C. Gross, J. C. Hall, D. A. Gailey and R. W. Siegel, 1982. The role of female movement in the sexual behavior of Drosophila melanogaster. Behav. Genet. 12: 295–307. [DOI] [PubMed] [Google Scholar]
- Tram, U., and M. F. Wolfner, 1998. Seminal fluid regulation of female sexual attractiveness in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 95: 4051–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner, M. F., 1997. Tokens of love: functions and regulation of Drosophila male accessory gland products. Insect Biochem. Mol. Biol. 27: 179–192. [DOI] [PubMed] [Google Scholar]
- Yamamoto, D, and Y. Nakano, 1999. Sexual behavior mutants revisited: molecular and cellular basis of Drosophila mating. Cell. Mol. Life Sci. 56: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, M., 1998. Molecular mechanisms of meiotic maturation and arrest in fish and amphibian oocytes. Semin. Cell Dev. Biol. 9: 569–579. [DOI] [PubMed] [Google Scholar]
- Yoshioka, T., H. Inoue and Y. Hotta, 1985. Absence of phosphatidylinositol phosphodiesterase in the head of a Drosophila visual mutant, norpA (no receptor potential A). J. Biochem. 97: 1251–1254. [DOI] [PubMed] [Google Scholar]