Abstract
We studied the activity of three multicopy insertion sequence (IS) elements in 12 populations of Lactococcus lactis IL1403 that evolved in the laboratory for 1000 generations under various environmental conditions (growth or starvation and shaken or stationary). Using RFLP analysis of single-clone representatives of each population, nine IS-mediated mutations were detected across all environmental conditions and all involving IS981. When it was assumed that these mutations were neutral, their frequency was higher under shaken than under stationary conditions, possibly due to oxygen stress. We characterized seven of the nine mutations at the molecular level and studied their population dynamics where possible. Two were simple insertions into new positions and the other five were recombinational deletions (of <1–>10 kb) among existing and new copies of IS981; in all but one case these mutations disrupted gene functions. The best candidate beneficial mutations were two deletions of which similar versions were detected in two populations each. One of these two parallel deletions, affecting a gene involved in bacteriophage resistance, showed intermediate rearrangements and may also have resulted from increased local transposition rates.
PREDICTING the long-term outcome of evolution is largely impossible due to the unpredictable nature of two key variables: the environmental conditions that will prevail and the genotypes that will arise. While uncertainties like these may seem insurmountable obstacles to the development of a predictive theory of evolution, the use of micro-organisms in laboratory evolution experiments offers a promising approach for the empirical exploration of the evolutionary role of even such stochastic variables (Elena and Lenski 2003). Their rapid generations, large populations, ease of experimental control, and wealth of genetic and physiological information are among the many benefits provided by microbes for a systematic study of both the effect of environmental conditions and the properties of mutations. Of particular interest is the identification and characterization of those rare mutations that are responsible for adaptation, as they are the ones that direct most evolution. At the phenotypic level, information on the frequency and fitness effects of beneficial mutations is needed for population-genetic models of adaptive evolution (Imhof and Schlötterer 2001; Rozen et al. 2002; Orr 2003). At the molecular level, characterization of individual beneficial mutations may lead to the identification of underlying molecular rules and constraints, as well as common adaptive pathways (Rainey et al. 2000; Travisano 2001; Otto 2002).
Insertion sequences (IS) are short DNA sequences (at most a few kilobases), consisting of a transposase gene flanked by two inverted repeats, which are able to mobilize themselves via transposition (Mahillon and Chandler 1998). IS elements in bacterial genomes can be a significant source of spontaneous mutation. By inserting into coding regions they can cause the disruption of a gene, while an increase in gene expression may occur when an element carrying an outward-reading promoter inserts directly upstream of a gene (Schneider and Lenski 2004 and references therein). Different copies of the same IS element may also form the substrate for recombination and result in chromosomal rearrangements such as deletions, duplications, or inversions (Drlica and Riley 1990; Gray 2000). The evolutionary role of IS elements is unclear; some view them as genomic parasites with mainly deleterious effects on their host (Charlesworth et al. 1994), while others have shown their involvement in host adaptation (Chao et al. 1983; Moxon et al. 1994; Riehle et al. 2001; Edwards et al. 2002; Wery et al. 2002; Bongers et al. 2003). IS elements can even be seen as adaptive mutator genes, since the mutations they mediate can occur at increased rates (Chao et al. 1983; Moxon et al. 1994; Cooper et al. 2001). This may be particularly the case during adverse conditions such as starvation or oxygen stress (Naas et al. 1994; Mahillon and Chandler 1998; Hall 1999a,b). The evolutionary role of IS elements has been studied mostly in gram-negative bacteria. These studies often involved simple homogeneous environments with conditions that either allowed regular growth (Chao et al. 1983; Papadopoulos et al. 1999; Schneider et al. 2000; Cooper et al. 2001; Edwards et al. 2002) or caused starvation (Naas et al. 1994; Kim et al. 2001); we know of only one study that compared IS-mediated mutations under growth and starvation conditions (Hall 1999a). Differences among these studies in strains and under evolutionary conditions complicate a systematic comparison of the role of IS-mediated mutations under different environmental conditions.
Here, we seek to detect and characterize IS-mediated mutations in populations of the gram-positive bacterium Lactococcus lactis IL1403 that are adapting to various environmental conditions in the laboratory. L. lactis IL1403 has an anaerobic lifestyle and the analysis of its known 2.3-Mb genome sequence predicts the presence of 43 IS elements (Bolotin et al. 2001). Variation in IS copy number and position among related L. lactis strains suggests that at least some of these elements are mobile (Romero and Klaenhammer 1993). We were particularly interested in the effect of stress due to starvation and increased oxygen levels, as some have suggested that these adverse conditions activate IS elements (Naas et al. 1994; Mahillon and Chandler 1998; Hall 1999a), thereby perhaps helping their host to adapt to these adverse conditions. Basically, stressful conditions can enhance the transposition rate of IS elements, but also the selection of the resulting IS-mediated mutations, and our approach cannot distinguish between these possibilities. However, our approach does allow a systematic comparison of these combined effects of stress on the frequency of IS-mediated mutations after evolution. Using single-clone representatives of each population, we detected nine and characterized seven IS-mediated mutations, all involving one element (IS981). The detection of these mutations by picking random clones makes it likely that they had reached a relatively high frequency, either due to the selective benefit they caused themselves or due to hitchhiking along with other beneficial mutations. We were especially interested in those mutations that were repeatedly found in independent populations, as these are thought to contribute to adaptation (Wichman et al. 1999; Wahl and Krakauer 2000; Dunham et al. 2002; Cooper et al. 2003), occur at a high rate, or both (Moxon et al. 1994; Cooper et al. 2001). While our incentive was the fundamental study of evolution, knowledge of the variables affecting genomic stability of L. lactis is relevant for the control of the industrial fermentation of dairy products, for which this species is widely used.
MATERIALS AND METHODS
Experimental evolution:
A single spontaneous streptomycin-resistant colony of plasmid-free L. lactis strain IL1403 (Bolotin et al. 2001) was used to found 12 experimental populations that were started by inoculating test tubes with 10 ml M17 broth (Difco). Six cultures were propagated under growth conditions (G) by transferring a small aliquot with 105-fold dilution (containing 3 × 104 cells) into 10 ml fresh medium three times per week. This transfer regime led to an effective population size, Ne, of 5 × 105 (Lenski et al. 1991) and 16.6 cell generations between transfers, resulting in ∼1000 generations in 5 months. The remaining six populations were maintained under starvation conditions (NG) in the original M17 culture without transfer to fresh medium during the same 5 months. (Although probably fewer generations elapsed in the NG populations than in the G populations during these 5 months, we refer to 1000 generations of evolution for both.) Three cultures each of the growing and starving populations were shaken at 100 rpm to cause oxygen stress (S), and the other three growing and three starving populations were kept without shaking (NS). All 12 populations were maintained at a constant temperature of 30° in the dark. The starved populations declined to ∼5 × 106 cells (i.e., 10−3 of the stationary-phase size of the growing populations) within 2 weeks and roughly maintained these densities after 6 and 10 weeks. [Similar densities were observed during 350 days of starvation in M17 with no additional glucose of L. lactis strain LL41-1 (Kim et al. 2001).] Because no viable cells could be isolated from two of the 6 starved populations after 5 months, the analysis was limited to the 10 surviving populations.
During evolution, samples of 1 ml from each of the six G populations were stored at −80° at 100-generation intervals, after plating on selective and permissive plates to check for contaminants. To prevent exhaustion of the NG populations, samples of 0.1 ml were frozen only once, i.e., after 2 weeks of evolution (equaling 100 generations for the G populations). After 1000 generations, samples from all populations (G and NG) were plated and a single random colony was isolated, grown overnight in M17 containing 0.5% glucose, and frozen at −80°. To be able to correlate possible phenotypic and genotypic changes, all analyses were performed on these single-clone representatives of the 10 surviving populations, except where indicated differently. We assumed that, because the probability was high that these randomly picked cells belonged to the majority genotype, they should be good representatives of the evolved populations.
Fitness measurements of 1000-generation clones:
To determine the extent of adaptation, competition experiments were performed between the evolved clones and a genetically marked version of the ancestor in the same environment where the clone had evolved. A single spontaneous rifampicin-resistant colony was isolated from the ancestral strain on medium containing 50 μg/ml rifampicin; this marker was shown to be neutral under all test conditions. Competition experiments were started by growing frozen samples of both competitors overnight in M17 containing 0.5% glucose. On the next day, diluted samples were transferred separately to the medium under conditions that prevailed during experimental evolution to allow physiological acclimation for 2 days. For the G populations, both competitors were then diluted 105-fold, mixed 1:1 into fresh medium, and allowed to compete under the relevant conditions during one growth cycle of 16.6 generations (∼2 days). At the start and the end of competition, diluted samples were plated on M17 + 0.5% glucose agar with and without 50 μg/ml rifampicin to estimate the numbers of both competitors. Plates were counted after at least 2 days of incubation at 30°. For the NG populations, competitions were done similarly, except that they were started by mixing 5 ml of 2-day-old, stationary-phase acclimatized cultures of each competitor, which were then allowed to compete for 7 days under these conditions of restricted growth. In all cases, five replicate competition experiments were performed. Competitions were run in two separate blocks for G and NG populations.
Because starvation conditions do not allow net population growth (but rather cause decline of numbers), fitness could not be defined as the ratio of the Malthusian parameters of evolved over ancestral strain (Lenski et al. 1991). Rather, we used a derived measure, the selection-rate constant, which can be expressed as
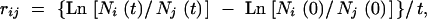 |
where Ni and Nj are the numbers of competitor i and j, respectively, measured at the start (t = 0) and end (t = t) of competition, with unit per day (Lenski et al. 1991). This measure, therefore, reflects the change in the log ratio of evolved and ancestral strain per unit time. We used this measure for all populations, G and NG, so that their adaptive improvement could be directly compared.
DNA isolation, digestion, and hybridization:
Single-clone representatives of each evolved population, as well as population samples taken at intermediate time points of three growing populations were used for DNA isolation for restriction fragment length polymorphism (RFLP) analysis. Five-milliliter overnight cultures of clones or populations in M17 broth containing 0.5% glucose were harvested, resuspended in a 6.7% sucrose 10 mm Tris-HCl buffer (pH 8), and treated with lysozyme (at 8 mg/ml), SDS (at 1%), and proteinase K (at 140 μg/ml) at 37°. Two phenol extractions were followed by a DNA precipitation, using sodium acetate (at 0.3 m) and ethanol, spooling out the DNA and resuspending in 0.05 m TBE (1 m TBE is 1 m Tris base, 1 m boric acid, and 20 mm EDTA). The DNA was then treated with RNase (at 130 μg/ml) at 37°, and a phenol/chloroform and chloroform extraction were carried out. Finally, the DNA was ethanol precipitated again, washed with 70% ethanol, and resuspended in 100 μl water. A 10-fold dilution of this DNA solution was used in PCR reactions.
For RFLP analysis, ∼3 μg DNA was digested with HindIII under conditions specified by the supplier (Biolab), and the resulting fragments (>103) were separated by electrophoresis on a 0.8% agarose gel. HindIII does not cut within the three IS elements studied. To verify conclusions drawn from changes in HindIII profiles, similar Southern hybridizations were performed using MspI and EcoRI digestions; neither enzyme cuts within IS981.
Digested DNA was blotted onto a nitrocellulose membrane using vacuum blotting. Hybridization (in 0.5 m NaHPO4, 1% BSA, 1 mm EDTA, 7% SDS, pH 7.2) was done overnight at 65° with a radioactively labeled probe (using nick translation of the PCR amplicon with [α32]ATP for each of the three IS elements (IS904, IS983, and IS981). The following primers were used to obtain the probes used for hybridization: IS904f 5′-CTTGAAGAGGGTTACTCTGT-3′; IS904r 5′-GGTCATGTCTCCCAGCCATA-3′, resulting in a probe of 641 bp; IS983f 5′-GCCAGAGCAAAGGGAAACAC-3′; IS983r 5′-CTTTGCCGCCTTATACTGC-3′, resulting in a probe of 417 bp; IS981f 5′-ACCCTTATCGCCTTCTATCA-3′; and IS981r 5′-CACTTAGTGAGTATCCAGGC-3′, resulting in a probe of 552 bp. Hybridized membranes were nonstringently washed (2 × 5 min at 20° using 5× SSC, 5 mm EDTA, pH 8.0, followed by 2 × 20 min at 20° using 2× SSC, 0.1% SDS, pH 7.2) and used to develop a phosphor screen, which was read with a STORM scanner and software (Molecular Dynamics, Sunnyvale, CA).
Molecular characterization of IS981-mediated mutations:
Two strategies were followed to identify the genetic events underlying the observed changes in IS981 hybridizations. First, fragments with changed position were isolated, and sequence analysis was performed to identify the position of IS981 in these fragments and the genes that were affected. For this, HindIII-digested DNA fragments of a size similar to the changed hybridization fragments were cut from the gel and DNA was extracted using the Concert Nucleic Acid purification system (GIBCO-BRL, Gaithersburg, MD). These fragments were ligated into vector pUC18ery and cloned into Escherichia coli strain JM109, using conditions recommended by the supplier. To detect transformants carrying fragments that contain IS981, we performed PCR using IS981 primers on pools of lysed cells of 10 transformants/reaction. On positive pools, a separate PCR on each of the 10 constituent transformants was performed to single out the transformant carrying the IS981 fragment. More than 2000 transformants were screened this way, allowing the isolation of seven of the nine novel or changed IS981 fragments. Plasmid DNA of transformants was isolated using a QIAprep Spin miniprep kit (QIAGEN, Chatsworth, CA). The DNA sequence flanking these IS981 fragments was determined using labeled primers for the left and right part of IS981 that are directed outward (IS981Lout 5′-CGGATTGACCAGAATGATAG-3′ and IS981Rout 5′-CCATCAAAGTTTAGGGTATC-3′) and a LI-COR (Lincoln, NE) DNA sequencer 4000L. Sequences were read semiautomatically and checked and corrected manually. Sequences were compared to the sequence of IL1403 (Bolotin et al. 2001) at the NCBI (http://www.ncbi.nlm.nih.gov/BLAST) and MOLOKO (http://www.spock.jouy.inra.fr/) databases using BLAST (Altschul et al. 1997).
Second, we performed PCR analysis on clones with changed IS981 hybridization profiles with primers that amplify the junction between each of the 10 ancestral IS981 copies and their flanking sequence on both sides. This was done to confirm the information obtained from sequence analysis, to distinguish between conservative and replicative transposition, and to reveal possible involvement of the ancestral IS981 copies in chromosomal rearrangements. PCR analysis was performed with one primer directed outward from IS981 (IS981 Right 5′-GTTTGTATGGTTTGAGCC-3′, and IS981 Left 5′-CTTGAATCTTGGCATGGAC-3′) and another primer corresponding to a neighboring gene on both sides of each ancestral IS981 copy (see Table 1). Absence of flanking sequences would suggest the involvement of particular IS981 copies in rearrangements such as deletions. To determine the extent of a deletion, PCR analysis using primers at ∼2, 4, 6, 8, and 10 kb from the IS981 copy, involved together with the appropriate outward-directed IS981 primer, was performed.
TABLE 1.
Primers used to amplify both flanking sequences of the 10 ancestral IS981 copies in the evolved populations
| Left
|
Right
|
|||
|---|---|---|---|---|
| IS981 copy | Primer | Gene | Primer | Gene |
| A | 5′-GGCTTTTGCTTACCCTTG-3′ | yajA | 5′-GAGTTTGGCGGTAACGTT-3′ | yajF |
| B | 5′-GGAACTGTATAAACTGGC-3′ | yajF | 5′-AGTTCATTGGCTTTGTCG-3′ | yajB |
| C | 5′-GGATTGGCTTCGTAAGCA-3′ | ygeD | 5′-GCAACAATCATGGTCACT-3′ | ygfA |
| D | 5′-GAGTCCAAGTAGAATCGG-3′ | yhcI | 5′-GTTGATGTCTAAGTTGCAG-3′ | yhcK |
| E | 5′-CTGCGATGTTTGCACCTC-3′ | ymbH | 5′-GTTTTACTTTAACGCCTCC-3′ | ymbJ |
| F | 5′-AGCGAAGCTAACATGTAC-3′ | ymcC | 5′-GCAACATCCAGGTTTGCG-3′ | ymcE |
| G | 5′-GATTCACTGAGTAGCAAG-3′ | ymhA | 5′-GAACTCAACTTGCACATG-3′ | ymhC |
| H | 5′-CCATTACCATTTTCTCCG-3′ | ypiH | 5′-CCTCGGCCATATTATTTG-3′ | ypiJ |
| I | 5′-CATCAAAGCTATCCCACTG-3′ | yreE | 5′-GTAGTGACGAGATAAGGG-3′ | yrdB |
| J | 5′-CTGGAGAACCACCAATAG-3′ | dpsA | 5′-GGAGCCAAAAATACTTCC-3′ | comC |
RESULTS
Adaptation to laboratory environments:
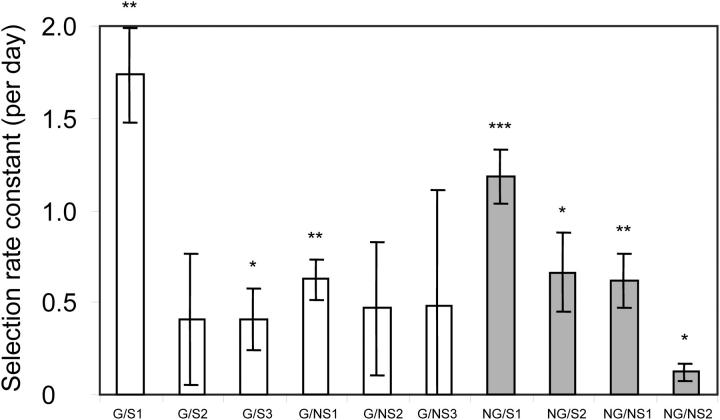
Three replicate populations of L. lactis strain IL1403 were allowed to adapt to each of four conditions during 1000 generations of evolution in an undefined rich medium (M17): growth (G) under shaken (S) and stationary (NS) conditions and starvation (NG) under S and NS conditions. After evolution, a single random clone was isolated from each of the 10 surviving populations for phenotypic and genetic analyses. To determine the fitness of these clones, we estimated the selection-rate constant from direct competition experiments against a marked version of the ancestor (Lenski et al. 1991). (When fitness of the evolved G clones is expressed as their Malthusian parameter relative to that of the ancestor, fitness improvement is on average 22%.) Although all 10 clones show improved fitness (Figure 1), for only 4 of the 10 clones is the improvement relative to the ancestor significant [using a one-tailed t-test of the five replicate estimates per clone and correcting for multiple testing (Rice 1989)]. Despite large variation of fitness among replicate assays and among populations (F9,39 = 2.98, P = 0.0086), the average fitness improvement of the 10 clones is highly significant (t = 4.61, d.f. = 9, two-tailed P = 0.0013). The reason for the large fitness variance is unknown, but might be related to their high death rate in stationary phase in rich media (see Korona 1996). Indirect support based on the number of mutations observed (see discussion) also indicates that these populations have adapted to their environments through the selection of spontaneous beneficial mutations. Using two-way analysis of variance of the mean relative fitness per population, no significant difference could be detected between growth and starvation conditions or between shaken and nonshaken conditions (Table 2).
Figure 1.—
Fitness improvement of random clones, isolated from the 10 viable populations after 5 months of laboratory evolution, relative to their common ancestor. Shown are the mean and standard error of five replicate estimates of the selection-rate constant. Open bars, populations adapted to growth conditions; shaded bars, populations adapted to starvation conditions. Asterisks above the bars reflect the P-value of a one-tailed t-test of the five replicate estimates: *P < 0.05, **P < 0.01, and ***P < 0.001.
TABLE 2.
Analysis of variance of the mean of five fitness estimates of the evolved populations relative to their common ancestor
| Source | d.f. | Mean square |
F | P |
|---|---|---|---|---|
| Growth | 1 | 0.0040 | 0.0167 | 0.901 |
| Shaken | 1 | 0.4586 | 1.9033 | 0.217 |
| Interaction | 1 | 0.0303 | 0.1258 | 0.735 |
| Residual | 6 | 0.2410 |
RFLP analysis with IS elements as probes:
The ancestral genome contains a total of 43 copies of the IS elements IS1077 (7 copies), IS904 (9 copies), IS905 (1 copy), IS981 (10 copies), IS982 (1 copy), and IS983 (15 copies; Bolotin et al. 2001). We performed RFLP analysis of the genome from the ancestor and the single-clone representatives of each evolved population, digested with HindIII, using probes for the three elements with most copies, IS904, IS981, and IS983. Hybridizations with probes for IS904 and IS983 did not reveal any variation in the banding pattern (data not shown). However, Southern hybridization with IS981 showed at least nine changes of band positions, including two apparent losses, three shifts, and four gains of fragment positions (Table 3).
TABLE 3.
IS981-mediated mutations detected in single-clone representatives of the evolvedL. lactis populations
| First detected using
|
|||||
|---|---|---|---|---|---|
| Population | Band changea |
PCRb | Southern hybridizationc |
Positiond (kb) |
Mutational event |
| Growth | |||||
| G/S1 | Loss 1.8 kb | ND | 800 | 1216 | Deletion of >11.3 kb at left flank of IS981 E, affecting at least ymbCDG and citRCDEF (Figure 2) |
| G/S3 | Gain 2.5 kb | 300 | 800 | 1969 | Insertion of IS981 107 bp upstream of ytgF (Figure 3A) |
| Shift 6.6 kb | 500e | 400 | 1277 | Deletion of 5.1 kb at right flank of IS981 G, affecting ymhC, amyL, lctO, and aroH (Figure 3B) |
|
| G/NS1 | Shift 8.7 kb | 300f | Never | 92 | Deletion of 1002 bp between IS981A and IS981B, affecting yajF (Figure 4) |
| Starvation | |||||
| NG/S1 | Shift 8.7 kb | >14 days | ND | 92 | Deletion of ∼800 bp between IS981 A and IS981 B, affecting yajF (Figure 4) |
| NG/S2 | Gain 3.2 kb | >14 days | ND | 896 | Insertion of IS981 into mleR (Figure 5) |
| NG/NS1 | Loss 6.6 kb | ND | ND | 1277 | Deletion of >10 kb at right flank of IS981 G (Figure 3B) |
G, growth; NG, starvation; S, shaken; NS, nonshaken; ND, not determined.
Detected on genomic HindIII digestions hybridized with a probe against IS981 (see materials and methods).
Time in generations (growing populations) or days (starving populations), based on PCR analysis of population samples isolated at 100-generation intervals; for starving populations, only a single sample taken after 14 days of evolution was available for the timing of the mutational events.
Time in generations, using Southern hybridizations of population samples isolated at 200-generation intervals.
Chromosomal coordinates of (one of the) IS981 element(s) involved in the mutation.
The 5.1-kb deletion in G/S3 probably happened via at least one intermediate step, as a (probably smaller) deletion was visible on Southern hybridizations from generation 400, while PCR amplification of the final 5.1-kb deletion end point was possible only with samples from generation 500 and later.
The deletion between existing elements IS981A and IS981B happened via at least one intermediate step (a deletion of 878 bp) in this population, which was detectable from generation 300 to 1000 (see Figure 4B).
The nine observed changes in IS981 hybridization profiles occurred in 6 of the 10 populations, with one population showing two and another three changes, while the other four changes each happened in a different population. Using Fisher's exact probability test, we found no significant effects on the number of IS981-mediated mutations that were due to starvation (three of four NG populations vs. three of six G populations; P = 0.381) or to shaking (four of five S populations vs. two of five NS populations; P = 0.238). However, this test lacks power, because it ignores multiple mutations in the same population. Assuming that these mutations were neutral, we then calculated the Poisson expectation for the observed frequency under one treatment on the basis of the observed frequency for the other treatment. Then, the frequency of mutations in NG and G populations remained nonsignificantly different (P = 0.133 to find at least five mutations in the NG populations based on the Poisson expectation for the G populations), but the frequency in S populations was significantly higher than that in NS populations (P = 0.0011 to observe at least seven mutations in the S populations based on the Poisson expectation for the NS populations). No significant relationship could be shown between the number of fragment changes of a population and its level of fitness improvement (Spearman-rank correlation coefficient = 0.466, n = 10, P > 0.05).
Molecular characterization and timing of IS981-mediated mutations:
To understand the molecular mechanisms responsible for the observed IS981-mediated mutations, seven of the nine fragment changes in the IS981 hybridization profiles were further characterized. Screening >2000 transformants containing HindIII-digested genomic fragments of approximately the predicted size with PCR analysis was insufficient to isolate the remaining two changes (both novel IS981 fragments in the clone from population NG/S2). It is unlikely, however, that these two changes were the result of recombinational deletions involving IS981, because PCR analysis showed that the flanking sequences of the 10 ancestral IS981 copies were all still present. The loss of HindIII restriction sites can also be ruled out as the cause of these two changes, because all HindIII fragments carrying the 10 ancestral IS981 copies were still present. Table 3 summarizes the results of the molecular analyses of the seven IS981-mediated mutations, which are described in more detail below.
Deletion of >11.3 kb of the left flank of IS981E in population G/S1:
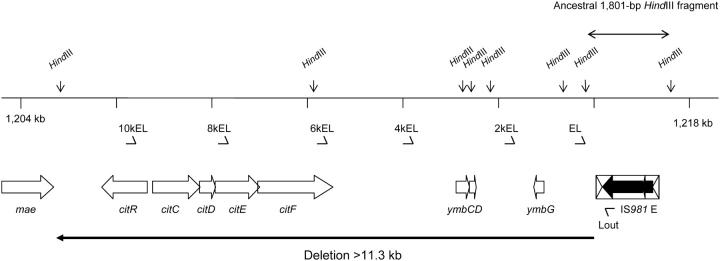
This rearrangement is illustrated in Figure 2. It was detected as the apparent loss of the 1.8-kb HindIII fragment containing IS981E (the 10 ancestral copies of IS981 are designated A–J). PCR amplification revealed the absence of the immediate left flank of IS981E only. However, attempts to amplify the region 10 kb to the left of IS981E (with primers at 2-kb intervals) were not successful. As no new HindIII fragment containing IS981 <4.2 kb was found, the deletion must include the HindIII site at 11.3 kb to the left of IS981E and thus be >11.3 kb. The rearrangement probably resulted from the replicative transposition of IS981 to a position >11.3 kb to the left of IS981E, followed by recombination between the new copy and IS981E and deletion of the intervening sequence. The genes that are affected by this deletion include at least three genes with unknown function: ymbCDG, the citrate lyase operon citCDEF (converts citrate into pyruvate) and its regulator citR, and possibly mae encoding malate oxidoreductase. As we do not know the deletion end point, PCR amplification could not be used to find when the mutation arose. The 1.8-kb HindIII fragment became visible on Southern hybridizations after 800 generations, suggesting that the mutation had reached a relatively high frequency by then.
Figure 2.—
Deletion of >11.3 kb directly left of ancestral copy IS981E, causing the apparent loss of a 1.8-kb HindIII fragment (indicated by the double-headed arrow) in population G/S1. The deletion (indicated by the bold arrow below) affected at least three genes coding for unknown proteins (ymbCDG) and the citrate lyase operon (citRCDEF). The next gene upstream, mae encoding malate oxidoreductase, may also be affected. Block arrows indicate the direction of transcription in this and all other figures. The mutation had reached a sufficiently high frequency to cause the visible loss of the ancestral 1.8-kb HindIII fragment on Southern hybridization after 800 generations. Primers Lout and EL (arrowheads) were used to amplify the junction between IS981E and its left flank; primer EL was used in combination with 2kEL, 4kEL, 6kEL, 8kEL, and 10kEL (all arrowheads) to try to determine the size of the deletion, but did not yield a product.
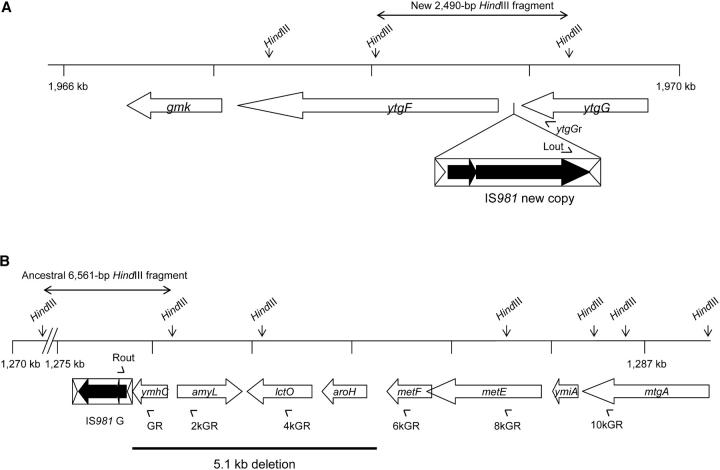
Insertion of IS981 upstream of ytgF in population G/S3:
This rearrangement is shown in Figure 3A. We observed a new 2490-bp HindIII fragment containing IS981 in this clone. Sequence analysis showed that IS981 had inserted between two genes with unknown function, ytgF and ytgG, 107 bp upstream of ytgF. BLAST analysis suggested that ytgF is a member of the HD superfamily of hydrolases that include well-studied enzymes such as the stringent response genes spoT and relA of E. coli (Aravind and Koonin 1998). ytgG shows homology to bacterial glucosamine-6-phosphate isomerases. Sequence analysis did not reveal a new hybrid promoter caused by the insertion of IS981, as was recently reported for this element in another strain of L. lactis, where it caused the transcriptional activation of lactate dehydrogenase gene ldhB (Bongers et al. 2003). RT-PCR analysis showed that the insertion had also not disrupted transcription of ytgF. The insertion resulted from replicative transposition of IS981, because PCR amplification of the flanking regions verified that all ancestral copies were still present (except the right flank of IS981G; see below). PCR amplification showed that the insertion was first detected in a sample from 300 generations. Southern hybridizations indicated that the insertion had become relatively common by generation 800.
Figure 3.—
Two rearrangements detected in population G/S3. (A) Insertion of IS981 107 bp upstream of ytgF, causing a new 2490-bp HindIII fragment (indicated by the double-headed arrow). ytgF and ytgG are genes with unknown function, but ytgF resembles members of the HD superfamily of hydrolases and ytgG shows homology to glucosamine-6-phosphate isomerases (pentose phosphate pathway). gmk encodes a guanylate kinase (nucleotide transport and metabolism). The mutation was detectable with PCR analysis using primers Lout and ytgGr (arrowheads) from generation 300 and was common by 800 generations. (B) Deletion of 5.1 kb of the right flank of ancestral copy IS981G, causing the shift of a 6.6-kb fragment to an 8.1-kb HindIII fragment. The deletion affected four genes: ymhC with homology to a rhamnosyltransferase (synthesis of lipopolysaccharides), amyL encoding an α-amylase (degradation of polysaccharides), lctO encoding l-lactate oxidase (lactate utilization), and aroH encoding a trp-sensitive aldolase (biosynthesis of aromatic amino acids). metE and metF encode enzymes involved in methionine biosynthesis, ymiA encodes a metalloregulator, and mtgA a cation-transporting P-ATPase. An unknown intermediate deletion was detectable on Southern hybridizations from generation 400, because amplification of the deletion end point with primers Rout and 6kGR (arrowheads) showed that the final rearrangement was present only from generation 500. A similar, but larger, deletion of >10 kb involving the right flank of IS981G was detected in population NG/NS1.
Deletion of 5.1 kb of the right flank of IS981G in population G/S3:
This IS981-mediated deletion is shown in Figure 3B. The deletion involves 5.1 kb of the right flank of IS981G and caused the shift of the ancestral 6.6-kb to an 8.1-kb HindIII fragment. The affected four genes are ymhC with homology to the rhamnosyl transferase gene eps9K of Streptococcus thermophilus (synthesis of lipopolysaccharides); amyL encoding an α-amylase (degradation of polysaccharides); lctO encoding l-lactate oxidase (lactate utilization); and aroH encoding a tryptophane-sensitive aldolase (biosynthesis of aromatic amino acids). PCR analysis showed that the mutation is the result of a replicative transposition of IS981 to a position 5.1 kb to the right of IS981G, followed by recombination between these two elements and deletion of the intervening sequence. The mutation was first detected by the shift of the fragment containing IS981G (from 6.6 to 8.1 kb) on Southern hybridizations in a population sample from generation 400, suggesting that it was common by then. However, PCR analysis of the deletion end point showed the presence of the final rearrangement only in population samples from generation 500 onward. The deletion thus probably happened via one or more unknown intermediate rearrangements.
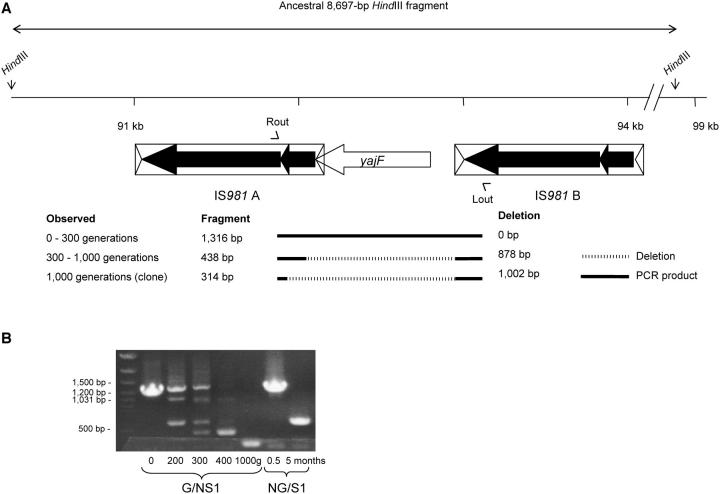
Deletion of 1.0 kb between IS981A and IS981B in population G/NS1:
The details of this rearrangement are given in Figure 4. The rearrangement was detected on Southern hybridizations, where it appeared as a shift of the ancestral 8.7-kb HindIII fragment to a 7.7-kb fragment. Sequence analysis revealed that the shift was due to a deletion of 1002 bp between (and partly of) the ancestral elements IS981A and IS981B, affecting yajF with 28% homology to orfX of L. lactis cremoris associated with an abortive infection mechanism against bacteriophages (O'Connor et al. 1996).
Figure 4.—
Deletion of 1.0 kb between IS981A and IS981B, causing the shift of the ancestral 8.7-kb HindIII fragment containing both IS981 copies to a 7.7-kb fragment, in population G/NS1. (A) The deletion affected gene yajF with unknown function, but associated with genes involved in phage resistance through abortive infection. (B) PCR analysis using primers Lout and Rout showed that the final deletion of 1002 bp occurred only at generation 1000 after at least one intermediate deletion of 878 bp in this population. (Two products of ∼1 and 0.5 kb appeared from generation 200 and disappeared after 400, but their molecular bases were not characterized.) The deletion of 878 bp was first detected at generation 300 and was dominant from generation 400 onward. A PCR product of ∼0.5 kb consistent with a deletion of ∼0.8 kb was observed in the clone from population NG/S1. This deletion was not yet detectable after 2 weeks.
PCR amplification of the region between IS981A and IS981B using population samples at 100-generation intervals revealed five products over the course of evolution (Figure 4B). PCR analysis of 20 randomly picked clones from generation 300 showed that the different products were the result of a polymorphism with different clones each expressing a single product. The ancestral 1316-bp fragment was visible through generation 300. From generation 200 through 400, two additional PCR products of ∼1 and 0.5 kb were visible at similar intensity; these products were not further characterized. Consistent with a shift on Southern hybridizations (data not shown), PCR analysis showed that from generation 400 onward the clone with the 878-bp deletion dominated the population. Hence, the clone from generation 1000 with the 1002-bp deletion was a minority genotype. Both the final 1002-bp and intermediate 878-bp deletion probably resulted from the replicative transposition of IS981 into (different positions within) the right end of IS981A, followed by recombination between the new copy and IS981B. Comparison of the deletion end point and the ancestral sequence did not reveal other repetitive sequences that could have served as substrate for recombination.
Deletion of 0.8 kb between IS981A and IS981B in population NG/S1:
A deletion involving IS981A and IS981B similar to that in population G/NS1 was observed in a clone isolated from population G/NS1. Both Southern hybridizations and PCR analysis (Figure 4B) of the region between the two IS981 copies are consistent with a deletion of ∼800 bp. Although it was not verified with sequence analysis, the deletion probably resulted from replicative transposition of IS981 and subsequent deletion caused by homologous recombination. It is unclear whether this rearrangement was preceded by intermediate rearrangements such as in population G/NS1; at the time of the only sample, after 2 weeks of evolution, the ancestral arrangement still dominated the population (Figure 4B).
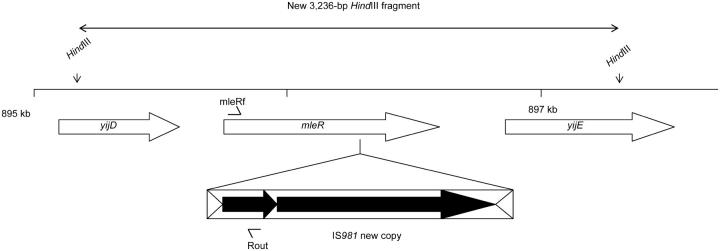
Insertion of IS981 into mleR in population NG/S2:
This rearrangement is shown in Figure 5. It was detected as a new 3236-bp HindIII fragment on Southern hybridizations and characterized by sequence analysis of the regions flanking IS981 in this fragment. PCR analysis showed that none of the ancestral IS981 copies had disappeared, indicating that the insertion resulted again from replicative transposition. The function of the mleR gene, encoding the activator of the malolactic fermentation and a member of the lysR family of regulators (Renault et al. 2000), was most likely disrupted by the insertion. PCR analysis indicated that mleR in this population did not yet contain the insertion after 14 days of starvation.
Figure 5.—
Insertion of IS981 into mleR, the regulator of the malolactic fermentation, resulting in a new 3236-bp HindIII fragment in starved population NG/S2. yijD encodes an ABC transporter ATP-binding protein; yijE is a hypothetical protein (with similarity to a conserved domain in spoT of Clostridium acetobutylicum). PCR analysis using primers Rout and mleRf (arrowheads) showed that this insertion was not yet present in this population after 2 weeks.
Deletion of >10 kb of the right flank of IS981G in population NG/NS1:
This rearrangement was detected by the apparent loss of the 6.6-kb HindIII fragment containing IS981G. PCR analysis showed that only the right flanking region of IS981G had disappeared, but we were unable to verify the size of the deletion using PCR amplification with primers up to 10 kb to the right of IS981G. The most likely scenario is a deletion caused by the replicative transposition of IS981 to a position >10 kb to the right of IS981G, followed by recombination between IS981G and the new copy and the deletion of the intervening sequence. The rearrangement thus resembles that involving IS981G observed in population G/S3 (Figure 3B), except that the deletion was larger and affected at least four additional genes: metE and metF involved in methionine biosynthesis, the metalloregulator ymiA, and the cation-transporting P-ATPase mgtA. PCR analysis on a population sample taken after 14 days of starvation showed that the right flank of IS981G was still present then.
DISCUSSION
We studied the activity of three multicopy IS elements in L. lactis IL1403 during 5 months and 1000 generations of adaptation to various conditions in the laboratory. Our interests were twofold: (i) to study the effect of starvation and oxygen stress on the frequency of IS-mediated mutations during adaptation to these stresses and (ii) to find similar IS-mediated mutations in independent populations. Once discovered, these IS-mediated mutations, especially if they occurred in multiple populations, are then candidates for the further study of their involvement in adaptation (Schneider and Lenski 2004). Using RFLP analysis, we found that only IS981 showed activity, with nine fragment changes in 6 of the 10 populations. We characterized seven of these fragment changes at the molecular level and found that all seven resulted from the mobility of IS981 rather than from the loss of restriction sites. Because the two uncharacterized changes also were not caused by the loss of restriction sites as all ancestral IS981 fragments were still present, these, too, were the likely result of IS981 activity. PCR analysis of the flanking sequences of the 10 ancestral IS981 copies showed that all nine IS981-mediated mutations started with the replicative transposition of IS981, suggesting that this is the preferred mechanism of mobility of this element.
IS-mediated mutations are of particular interest for the study of bacterial adaptation for two reasons. First, IS elements provide a molecular handle that allows the detection and manipulation of IS-mediated mutations in experiments. Second, IS-mediated mutations are a significant source of mutations in bacteria (see Schneider and Lenski 2004), and their involvement in adaptation has frequently been observed. In this light, the great variation in the frequency of IS elements in bacterial genomes, with some species harboring tens to hundreds of copies and others none, is intriguing. Correlating the presence of IS elements with the ecology of the species involved may shed light on the evolutionary role of IS elements in the future, but is at present impossible due to a general lack of knowledge of the ecology of most bacterial species.
Frequency of IS-mediated mutations:
Our results suggest that IS981 is a more active element than IS904 and IS983 under a variety of environmental conditions or is more often involved in adaptive mutations than the other two elements, or both. The frequency of IS-mediated mutations that we observed for L. lactis is in general agreement with observations in E. coli under conditions of regular growth. Papadopoulos et al. (1999) found on average 0.03076 changes/1000 generations/copy for 38 copies of seven IS elements in two evolving E. coli populations. Assuming that these mutations are neutral (and thus hitchhiked to fixation with other beneficial mutations), the Poisson probability that we observe at least four changes during 1000 generations in six growing L. lactis populations for 34 copies of three IS elements is 0.8736. Moreover, like in E. coli, we observed substantial variation in the activity among genetically different IS elements, as well as in the number of IS-mediated mutations per population.
Although based on low numbers, we found roughly similar frequencies of mutations for conditions of growth and starvation. Our results corroborate earlier findings that starving bacterial populations are dynamic due to natural selection of spontaneous mutants with improved adaptation to these adverse conditions (Zambrano et al. 1993; Naas et al. 1994; Finkel and Kolter 1999; Kim et al. 2001). The fact that two of the six starved populations went extinct is consistent with this notion and emphasizes that long-term survival under these conditions relies on the chance occurrence of relatively rare beneficial mutations. However, our results do not support the suggestion that IS-mediated mutations occur more frequently or are more often associated with beneficial effects during starvation than during growth (Naas et al. 1994; Hall 1999a). Papadopoulos et al. (1999) observed a similar frequency of IS-mediated mutations after 4 years of adaptation of E. coli B to conditions of regular growth as did an earlier study for E. coli K12 after 30 years of starvation (Naas et al. 1994). Neglecting differences in evolutionary conditions, the different time scales for these two studies suggest a lower frequency under starvation. Analysis of the four mutations with an advantage in stationary phase in E. coli identified so far also indicates that they are of diverse nature, with one of the four mutations involving IS activity (Zinser and Kolter 2004). Note, however, that these studies as well as ours observed the combined effects of mutation and natural selection on the frequency of IS-mediated mutations.
When the IS981-mediated mutations were assumed to be neutral, their observed frequency was higher in shaken than in stationary cultures. Two different mechanisms can explain a higher frequency. First, shaken conditions could allow for greater adaptive improvement than stationary conditions. If so, shaken conditions would provide more opportunities for IS-mediated mutations to hitchhike with other beneficial mutations. Our data do not support this explanation, because fitness improvement was not higher under shaken than under stationary conditions (Table 2). Second, shaken conditions could lead to a higher rate of IS981-mediated mutations, possibly stimulated by increased oxygen stress. L. lactis is an obligatory anaerobic species, whose low tolerance of oxygen may imply high intracellular concentrations of oxygen radicals under the aerobic conditions of the evolution experiment, possibly enhanced by a slightly increased growth rate (see Bolotin et al. 2001). Environmental stimuli for transposition of IS981 are unknown, but the involvement of the SOS system in the activation of other transposable elements (Mahillon and Chandler 1998) suggests that cellular mechanisms by which oxygen radicals can activate transposition do exist. Alternatively, oxygen radicals could enhance the mutation rate of the transposase or its regulatory elements, which may sometimes lead to mutants with increased IS activity.
Role of IS981-mediated mutations in adaptation:
Can we be certain that the clones that we studied improved their fitness, despite the lack of significant competition results for some clones? Had adaptation not occurred, neutral theory (Kimura 1983) predicts that it would take each mutation in the order of Ne (the effective population size) generations to increase to fixation. Since the populations were maintained at an Ne of at least 5 × 105, it would be extremely unlikely to find any fixed mutation after 1000 generations by picking random clones from these populations. That most mutations we observed had reached a relatively high frequency was suggested by their detection with Southern hybridizations using whole-population DNA. Moreover, three of the four mutations in the G populations were shown to persist at a detectable frequency on Southern hybridizations after their first discovery, suggesting again the systematic effect of selection rather than the erratic effect of drift. Hence, genetic drift alone is not a sufficient explanation, and we need to invoke natural selection to explain the detection of nine IS981-mediated mutations in the evolved populations. Two possibilities remain: (i) these mutations themselves caused a fitness benefit or (ii) the IS-mediated mutations were without fitness benefit, and hitchhiked to high frequency along with beneficial mutations elsewhere on the chromosome.
To distinguish between these two possibilities, we would need to move the mutations into the ancestral strain and perform competition experiments in the environment where the mutations arose (Schneider et al. 2000; Cooper et al. 2003; Schneider and Lenski 2004). While we may do so in the future, at present we can only discuss the possible adaptive nature of these mutations on the basis of their detection in multiple populations and on the genes that are affected.
Insertions:
Two of the seven mutations were simple insertions after replicative transposition of IS981 into new positions. In population G/S3, IS981 inserted 107 bp upstream of ytgF, a gene with unknown function that contains a domain typical for the HD superfamily of hydrolases that include the stringent response regulators spoT and relA of E. coli (Aravind and Koonin 1998). The stringent response of L. lactis is involved in stress tolerance (Rallu et al. 2000) and may thus affect fitness through a general change of cell physiology. However, there were no indications that the insertion affected transcription of ytgF: it did not establish a new hybrid promoter resulting in increased expression (cf. Bongers et al. 2003), nor was expression disrupted. The insertion was thus probably without fitness effects.
The other simple insertion was observed in the clone isolated from population NG/S2, where IS981 inserted into the coding region of mleR, the positive regulator of the malolactic fermentation and member of the lysR family of positive regulators (Renault et al. 2000). Since no malate was present in the medium, this function was unnecessary. However, it is unlikely that disruption of this function due to IS981 insertion in its regulator liberated energy, because in the absence of malate the genes involved are not transcribed. Therefore, unless mleR had other physiological effects, it is most likely that this insertion was also without fitness consequences.
Deletions:
Five of the seven mutations were replicative transpositions of IS981 followed by recombinational deletions between the new copy and an existing one. Of these five mutations, four consisted of two different rearrangements of which similar variants were observed in two independent populations each. The excess of genomic deletions that we found is consistent with a pervasive bias toward higher frequencies of deletions rather than insertions shaping the evolution of bacterial genomes (Mira et al. 2001).
The only unique event was a deletion of >11.3 kb left of IS981E in the clone from population G/S1. The affected genes included three genes with unknown function: the citrate lyase operon citCDEF with its regulator citR involved in the metabolization of citrate and the malate oxidoreductase gene mae that catalyzes the conversion between malate and oxaloacetate. There is no simple physiological reason why the deletion of these functions should improve fitness in the evolutionary environment. Although both the citrate lyase operon and the malate oxidoreductase were dispensable due to the lack of citrate and malate in the medium, it is uncertain that their deletion liberated energy for other functions. This could be so for the citrate lyase operon, since it is constitutively expressed in another L. lactis strain (Magni et al. 1999); the regulation of transcription of mae is unknown. Due to additional uncertainties about the extent of the deletion and the function of three of the deleted genes, the fitness consequences of this deletion are unpredictable.
The first deletion of which similar variants were found in two different populations involved the right flanking sequence of IS981G: deleting 5.1 kb in population G/S3 and >10 kb in population NG/NS1. The genes that were affected in both populations were ymhC with homology to a rhamnosyl transferase gene, amyL encoding an α-amylase, lctO encoding l-lactate oxidase, and aroH encoding a trytophane-sensitive aldolase. Although the repeated deletion of genes in independent populations may be indicative of their adaptive nature (Wichman et al. 1999; Wahl and Krakauer 2000; Dunham et al. 2002; Cooper et al. 2003), here repeatability was limited. The deletion was found in only two populations, which furthermore adapted to rather different selective conditions (one was a shaken culture undergoing regular growth and the other a starving stationary culture). Despite the need for different cell physiologies under these contrasting conditions, it is conceivable that the deletion of each of these four genes conferred a fitness advantage under both conditions. ymhC is likely involved in the synthesis of lipopolysaccharides, and the deletion of genes involved in lipopolysaccharides in E. coli populations that were adapting to conditions of regular growth at increased temperature was associated with a 25% fitness benefit (Riehle et al. 2001). Genes involved in the degradation of polysaccharides (amyL) and in the biosynthesis of amino acids (aroH) seem, at first glance, obsolete, because polysaccharides are not and amino acids are provided in the medium at nonlimiting concentrations. However, lactate is an important fermentation end product and thus a likely component in the medium, and the deletion of lctO involved in the utilization of lactate may have been deleterious. The fitness effect of deleting all four genes together is unpredictable and can be addressed only in further experiments.
The other repeated deletion involved a sequence between the closely residing IS981 copies A and B, affecting yajF with homology to abortive infection genes, observed in clones from populations G/NS1 and NG/S1. Both deletions were similarly small (1.0 kb in population G/NS1 and 0.8 kb in population NG/S1), while the variant observed in growing population G/NS1 resulted in at least one intermediate rearrangement that reached high frequency within 400 generations (see Figure 4). The repeatability of this deletion was again limited to two populations adapting to rather different conditions, and hence it is not immediately likely that the deletion was adaptive. However, the fact that IS981 was first discovered in association with genes involved in abortive infection resistance against prolate-headed phage (Romero and Klaenhammer 1993) suggests a long-term adaptive role of these genes and thus perhaps of their deletion in these two populations. The close presence of the two IS981 copies involved and the detection of intermediate rearrangements is also consistent with an increased transposition rate, possibly due to the presence of a second transposase promoter nearby (Mahillon and Chandler 1998). The repeated detection of this deletion may thus have resulted from a combination of a (locally) increased rate of transposition and an adaptive benefit.
To conclude, of the seven IS981-mediated mutations that we characterized, two similar deletions that were found in two independent populations each are the best candidates of beneficial mutations (Wichman et al. 1999; Wahl and Krakauer 2000; Dunham et al. 2002; Cooper et al. 2003). Competition experiments between ancestral strains with and without these mutations should be performed to be able to distinguish between the possibility that these mutations conferred a selective advantage themselves or hitchhiked along with other beneficial mutations. Only then can we learn whether IS-mediated mutations are a common cause or a consequence of bacterial adaptation.
Acknowledgments
We thank N. Fitzsimons, K. Fritsche, H. Heilig, and B. Koopmanschap for help in the laboratory and J. van der Oost, M. Maas, and D. Rozen for discussion and comments. J.A.G.M.V. was supported by a grant from the Netherlands Organization for Scientific Research.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., and E. V. Koonin, 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23: 469–472. [DOI] [PubMed] [Google Scholar]
- Bolotin, A., P. Winkler, S. Mauger, O. Jaillon, K. Malarme et al., 2001. The complete genome sequence of the lactic acid bacterium Lactococcu lactis ssp. lactis IL1403. Genome Res. 11: 731–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers, R. S., M. H. N. Hoefnagel, M. J. C. Starrenburg, M. A. J. Siemerink, J. G. A. Arends et al., 2003. IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis. J. Bacteriol. 185: 4499–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, L., C. Vargas, B. B. Spear and E. C. Cox, 1983. Transposable elements as mutator genes in evolution. Nature 303: 633–635. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., P. Sniegowski and W. Stephan, 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Cooper, T. F., D. E. Rozen and R. E. Lenski, 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 100: 1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, V. S., D. Schneider, M. Blot and R. E. Lenski, 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183: 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica, K., and M. Riley, 1990 The Bacterial Chromosome. American Society for Microbiology, Washington, DC.
- Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown et al., 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R. J., R. E. Sockett and J. F. Y. Brookfield, 2002. A simple method for genome-wide screening for advantageous insertions of mobile DNAs in Escherichia coli. Curr. Biol. 12: 863–867. [DOI] [PubMed] [Google Scholar]
- Elena, S. F., and R. E. Lenski, 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4: 457–469. [DOI] [PubMed] [Google Scholar]
- Finkel, S. E., and R. Kolter, 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96: 4023–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, Y. H. M., 2000. It takes two transposons to tango. Trends Genet. 16: 461–468. [DOI] [PubMed] [Google Scholar]
- Hall, B. G., 1999. a Spectra of spontaneous growth-dependent and adaptive mutations at ebgR. J. Bacteriol. 181: 1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, B. G., 1999. b Transposable elements as activators of cryptic genes in E. coli. Genetica 107: 181–187. [PubMed] [Google Scholar]
- Imhof, M., and C. Schlötterer, 2001. Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc. Natl. Acad. Sci. USA 98: 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W. S., J. H. Park, J. Ren, P. Su and N. W. Dunn, 2001. Survival response and rearrangement of plasmid DNA of Lactococcus lactis during long-term starvation. Appl. Environ. Microbiol. 67: 4594–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1983 The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- Korona, R., 1996. Adaptation to structurally different environments. Proc. R. Soc. London Ser. B 263: 1665–1669. [Google Scholar]
- Lenski, R. E., M. R. Rose, S. C. Simpson and S. C. Tadler, 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- Magni, C., D. de Mendoza, W. N. Konings and J. S. Lolkema, 1999. Mechanism of citrate metabolism in Lactococcus lactis: resistance against lactate toxicity at low pH. J. Bacteriol. 181: 1451–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon, J., and M. Chandler, 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62: 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, A., H. Ochman and N. A. Moran, 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17: 589–596. [DOI] [PubMed] [Google Scholar]
- Moxon, E. R., P. B. Rainey, M. A. Nowak and R. E. Lenski, 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4: 24–33. [DOI] [PubMed] [Google Scholar]
- Naas, T., M. Blot, W. M. Fitch and W. Arber, 1994. Insertion sequence-related genetic rearrangements in resting Escherichia coli K-12. Genetics 136: 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, L., A. Coffey, C. Daly and G. F. Fitzgerald, 1996. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl. Environ. Microbiol. 62: 3075–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., 2002. Evolving beyond point mutations. Trends Ecol. Evol. 17: 110. [Google Scholar]
- Papadopoulos, D., D. Schneider, J. Meier-Eiss, W. Arber, R. E. Lenski et al., 1999. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl. Acad. Sci. USA 96: 3807–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey, P. B., A. Buckling, R. Kassen and M. Travisano, 2000. The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol. Evol. 15: 243–247. [DOI] [PubMed] [Google Scholar]
- Rallu, F., A. Gruss, S. D. Ehrlich and E. Maguin, 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35: 517–528. [DOI] [PubMed] [Google Scholar]
- Renault, P., C. Gaillardin and H. Heslot, 2000. Product of the Lactococcus lactis gene required for malolactic fermentation is homologous to a family of positive regulators. J. Bacteriol. 171: 3108–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W. R., 1989. Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Riehle, M. M., A. F. Bennett and A. D. Long, 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. USA 98: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, D. A., and T. R. Klaenhammer, 1993. Transposable elements in Lactococci: a review. J. Dairy Sci. 76: 1–19. [DOI] [PubMed] [Google Scholar]
- Rozen, D. E., J. A. G. M. de Visser and P. J. Gerrish, 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Schneider, D., and R. E. Lenski, 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 155: 319–327. [DOI] [PubMed] [Google Scholar]
- Schneider, D., E. Duperchy, E. Coursange, R. E. Lenski and M. Blot, 2000. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano, M., 2001. Evolution: towards a genetical theory of adaptation. Curr. Biol. 11: R440–R442. [DOI] [PubMed] [Google Scholar]
- Wahl, L. M., and D. C. Krakauer, 2000. Models of experimental evolution: the role of genetic chance and selective necessity. Genetics 156: 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wery, J., B. Hidayat, J. Kieboom and J. A. M. de Bont, 2002. An insertion sequence prepares Pseudomonas putida S12 for severe solvent stress. J. Biol. Chem. 276: 5700–5706. [DOI] [PubMed] [Google Scholar]
- Wichman, H. A., M. R. Badgett, L. A. Scott, C. M. Boulianne and J. J. Bull, 1999. Different trajectories of parallel evolution during viral adaptation. Science 285: 422–424. [DOI] [PubMed] [Google Scholar]
- Zambrano, M. M., D. A. Siegele, M. Almirón, A. Tormo and R. Kolter, 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259: 1757–1760. [DOI] [PubMed] [Google Scholar]
- Zinser, E. R., and R. Kolter, 2004. Escherichia coli evolution during stationary phase. Res. Microbiol. 155: 328–336. [DOI] [PubMed] [Google Scholar]