Abstract
The plasma membrane proton pump (H+-ATPase) found in plants and fungi is a P-type ATPase with a polypeptide sequence, structure, and in vivo function similar to the mammalian sodium pump (Na+, K+-ATPase). Despite its hypothetical importance for generating and maintaining the proton motive force that energizes the carriers and channels that underlie plant nutrition, genetic evidence for such a central function has not yet been reported. Using a reverse genetic approach for investigating each of the 11 isoforms in the Arabidopsis H+-ATPase (AHA) gene family, we found that one member, AHA3, is essential for pollen formation. A causative role for AHA3 in male gametogenesis was proven by complementation with a normal transgenic gene and rescue of the mutant phenotype back to wild type. We also investigated the requirement for phosphorylation of the penultimate threonine, which is found in most members of the AHA family and is thought to be involved in regulating catalytic activity. We demonstrated that a T948D mutant form of the AHA3 gene rescues the mutant phenotype in knockout AHA3 plants, but T948A does not, providing the first in planta evidence in support of the model in which phosphorylation of this amino acid is essential.
THE plasma membrane proton ATPase (H+-ATPase) of plants and fungi creates a proton motive force, which is used directly by most secondary transporters to mediate the movement of solutes into and out of the cell (Morsomme and Boutry 2000; Palmgren 2001). In Arabidopsis, the electrochemical potential has been recorded at very negative levels, such as −230 mV, while the corresponding protein in animal cells, the Na+,K+-ATPase, typically generates membrane potentials of only ∼−100 mV (Hirsch et al. 1998). This potential, together with a chemical gradient of protons, is thought to be essential for diverse cellular processes, including nutrient transport, cellular expansion, and osmoregulation. Thus, the proton ATPase has been hypothesized to play a crucial role in many important physiological processes.
The family of genes encoding plasma membrane H+-ATPases in Arabidopsis has 11 members (Palmgren 2001). Expression data have implicated differing roles for family members in numerous tissues, including root layers, floral organs, and vasculature (Harper et al. 1990; Houlne and Boutry 1994; DeWitt and Sussman 1995). These proteins are intriguing targets for genetic studies that test the hypothesis that they play important roles in plant physiology.
One family member, AHA3, was shown to be expressed in companion cells of the phloem and various reproductive tissues (DeWitt et al. 1991; DeWitt and Sussman 1995). Its role in the phloem is presumed to be the control of sugar loading for long-distance sucrose transport, a process that is critical for plant nutrition. While localization studies were useful in determining the expression pattern of AHA3, we wanted to more closely and directly examine the in planta role of this gene. To this purpose, we took a reverse genetic approach and characterized the effect of the absence of functional AHA3 on the plant. Through transmission studies, microscopic analysis, and functional complementation, we have identified an essential role for AHA3 in pollen development.
Knockout plants are useful for structure/function experiments, which are designed to test the roles of specific domains. Results of heterologous experiments with yeast suggested that T948, at the extreme C terminus of AHA3, is essential for activation of the pump. To test the hypothesis that this residue is important for functions of the pump in planta, we mutated T948 to either aspartate or alanine. Transgenes carrying these mutations were introduced into +/aha3-5 plants. Plants containing these transgenes in an aha3 null background were identified among the progeny and characterized. Our results indicate that the highly conserved penultimate threonine residue is not absolutely required in planta, but that complementation is dependent on the presence of a negative charge at this location since substitution of T with A did not functionally replace AHA3. However, substitution of T with D rescued aha3 plants and conferred a growth advantage to seedlings on acidic media, presumably due to hyperactivation of AHA3. These results are consistent with a predicted role for this residue in regulation of activation by an as-yet-unidentified protein kinase.
MATERIALS AND METHODS
T-DNA mutant screen and identification:
T-DNA mutagenized populations were screened for the presence of insertions in AHA3 using a PCR-based strategy (Krysan et al. 1996, 1999). The sequences of primers specific for AHA3 were 5′-CCAAATAGCCTAACGTAGTCCACCTTCAC, 5′-AATATGACTAGCACAGTAGCACCTTTACC, 5′-GAATAAGGAAGGAAGAAAAACCCCAGGAG, and 5′-TCTCGTCTTCTTGGTTTTGTTTTTGTAGC. The sequence of a primer specific for the T-DNA border was 5′-CATTTTATAATAACGCTGCGGACATCTAC (JL202). An individual plant containing the aha3-1 allele was isolated from a population of 64,860 lines mutagenized with a derivative of the T-DNA vector pD991 (Young et al. 2001). This vector carries a selectable marker gene that renders plants resistant to kanamycin. Individual plants containing the aha3-5 allele were isolated from a population of 72,960 lines mutagenized with the pSKI15 vector. This vector carries the selectable marker bar gene that renders plants resistant to glufosinate, the active ingredient in the herbicide BASTA. DNA sequencing of PCR products confirmed the locations of the junctions of genomic and T-DNA sequences. Both aha3 mutant alleles were present in the Wassilewskija (Ws) background.
Mutant transcript identification:
RNA was isolated from plants heterozygous for either the aha3-1 or the aha3-5 allele using the QIAGEN RNeasy mini kit (Valencia, CA). cDNA was synthesized using a modified oligo(dT) primer (5′-24T + A/C/G/T) and SuperScript II reverse transcriptase (Invitrogen, San Diego). Nested primers specific to AHA3 cDNA and to the T-DNA border sequence (primer JL202) were used to amplify PCR products corresponding to the mutant aha3 transcripts. The cDNA-specific primers used were 5′-ACAGACTGTACCAGAGAAAACAAAA and 5′-AGAGTCAAGCCTTCTCCAACACCAGATAG (for aha3-1) and 5′-GGTATCGTTTGTCTTTTGGTTATCA and 5′-CGAAGAAAACAATGCTGGAAATGCT (for aha3-5). DNA sequencing of PCR products confirmed the location of the junction between AHA3 cDNA and T-DNA.
Media and plant growth:
Seeds were germinated on plates containing half-strength Murashige and Skoog (MS) salts, 1% (w/v) sucrose, and 0.8% (w/v) washed agar (MS plates). Seedlings were transferred to soil after 10–14 days. Plants were housed under the following growth conditions: 21°, 24 hr light (42 μE/m2/sec ) and ∼30% humidity.
The genotype of individual plants was determined by PCR using AHA3-specific primers and primer JL202. Two AHA3-specific primers were used to detect the presence of the wild-type allele. An AHA3-specific primer and primer JL202 were used to detect the presence of the mutant allele.
To select plants containing the aha3-1 allele, seedlings were germinated on MS plates containing 50 μg/ml kanamycin. To select plants containing the aha3-5 allele, seeds were sown in soil, and seedlings were subsequently sprayed with BASTA herbicide (50 μg/ml glufosinate) at 4 and 8 days.
Determination of male gametophyte lethality:
Reciprocal crosses were performed between wild-type (Ws) and AHA3/aha3-1 or AHA3/aha3-5 plants. Progeny were germinated on MS plates containing 50 μg/ml kanamycin (for aha3-1) or 25 μg/ml glufosinate (for aha3-5).
To confirm male gametophytic lethality, AHA3/aha3-5 plants were crossed as females to qrt1-3/qrt1-3 plants (gift from Gregory Copenhaver; lab stock CLA18). The F1's were selfed and F2 progeny heterozygous for aha3-5 were selected by spraying with BASTA herbicide (50 μg/ml glufosinate). Plants homozygous for the qrt mutation were identified by visualization of “quartets” of pollen grains (Preuss et al. 1994).
Light microscopy and staining:
To visualize mutant pollen grains, anthers from a plant heterozygous for aha3 were collected and dabbed on a microscope slide coated with a thin layer of pollen germination media (Mouline et al. 2002). Pollen grains and emerging tubes were viewed using a Nikon E400 clinical microscope. Images were recorded using a Spot Insight color digital camera (Diagnostic Instruments).
To visualize color-differentiated wild-type and mutant pollen grains, anthers were collected and squashed on a microscope slide. Alexander's stain solution, prepared according to Alexander (1969) and used at a concentration of 3 parts stain/47 parts water, was added; the sample then was covered with a cover-slip and viewed using an Olympus BX60 microscope (Olympus Optical, Tokyo). Images were captured using an Olympus DP12 digital microscope camera (Olympus Optical).
Scanning electron microscopy:
We prepared samples for scanning electron microscopy using tested methods (Fernandez et al. 2000). Briefly, flowers were collected and fixed in 4% (v/v) glutaraldehyde, rinsed in 0.05 m KPO4, dehydrated in an ethanol series, and critical point dried using CO2. Samples were mounted on steel posts covered with adhesive circles (Ted Pella, Redding, CA) and sputter coated with a thin film of gold. Samples were viewed at 10,000 accelerating voltage on a Hitachi S-570 scanning electron microscope (Hitachi, Tokyo). Images were recorded digitally using a Gatan digital image capture system and digital micrograph version 2.5 software (Gatan, Pleasanton, CA).
Flower fixation and sectioning:
We prepared flowers for sectioning using tested methods (Fernandez et al. 2000). Briefly, flower buds from all ages were collected, fixed in 4% (v/v) glutaraldehyde, rinsed in 0.05 m KPO4, dehydrated in an ethanol series, and embedded in London Resin white medium-grade resin (EMS, Fort Washington, PA). The embedded flowers were sectioned in 2μ increments using an ultramicrotome (RMC, Tucson, AZ). Sections were heat fixed on a microscope slide, stained with 0.05% (w/v) toluidine blue O, and viewed with bright-field optics using a Nikon E400 clinical microscope. Images were recorded using a Spot Insight color digital camera (Diagnostic Instruments).
To visualize β-glucuronidase (GUS) staining in sections from C-terminally modified AHA3 transgenic plants, the protocol described above was followed, with the addition of one step. Prior to fixation in glutaraldehyde, samples were incubated in modified X-Gluc buffer (0.5 mg/ml X-Gluc, 50 mm KPO4, 2.5 mm potassium ferricyanide, and 2.5 mm potassium ferrocyanide) for at least 6 hr. To view GUS color precipitates, the sections were left unstained and observed with dark-field optics using an Olympus BX60 microscope.
Molecular complementation:
A 9-kb SpeI fragment containing the entire regulatory and coding sequence of AHA3 (N. D. DeWitt, unpublished results) was ligated into the binary vector pCAMBIA3300S, a spectinomycin-resistant derivative of pCAMBIA3300. This vector contains the coding sequence of the bar gene between the T-DNA border regions. AHA3/aha3-1 plants (designated the T0 generation) were transformed with AHA3::pCAMBIA3300S using the Agrobacterium tumefaciens-based floral dip method (Clough and Bent 1998). Transformants (T1) were selected in soil by spraying with BASTA (50 μg/ml glufosinate) at 4 and 8 days postgermination. Rescued plants were identified among the T2 plants using a PCR-based strategy. Rescued plants were defined as those homozygous for the aha3-1 allele, but containing an AHA3 transgene. Primers used to identify the wild-type, native AHA3 allele were 5′-CTAACCAGTAGAGCCATCTTCCAGAGAATG and 5′-AATAACCAAGAAAGTAGCGGACCAAACAC; those used to identify the aha3-1 allele were 5′-TTATTAGTGTTGGTCGTTGGGGTATCTTG and the T-DNA-specific primer, JL202.
Molecular complementation with altered AHA3 clones:
Altered clones of AHA3 were constructed using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primers used to introduce the alterations for threonine/alanine substitution were 5′-GCTGGTCACTACGCCGTTTAATAAAGATTTAAC and its complement and for threonine/aspartate substitution, 5′-GCTGGTCACTACGACGTTTAATAAAGATTTAAC and its complement. The identities of the desired clones were confirmed by DNA sequencing of PCR products spanning the alteration. Clones were ligated into the binary vector pGreen0029, which carries the selectable marker nptII gene that confers resistance to kanamycin in transgenic plants. Plants heterozygous for aha3-5 (designated the T0 generation) were transformed with altered AHA3::pGreen0029 using the A. tumefaciens-based floral dip method (Clough and Bent 1998). T1 seedlings were selected on MS plates containing 50 μg/ml kanamycin. Rescued mutants among the T2 generation were identified using a PCR-based strategy. Rescued plants were defined as plants homozygous for the aha3-5 allele, yet alive due to the presence of the AHA3 transgene. Primers used to identify the wild-type, native AHA3 allele were 5′-TTGTCGGTATCGTTTGTCTTTTGGTTATC and 5′-AATAACCAAGAAAGTAGCGGACCAAACAC. Primers used to identify the aha3-5 allele were 5′-AATATGACTAGCACAGTAGCACCTTTACC and the T-DNA-specific primer JL202.
Assays of transgenic seedlings:
Seedlings containing altered AHA3 transgenes were germinated on growth media at pH 4.5, 5, or 5.5 according to methods described previously (Young et al. 1998). After 10 days, hypocotyls of dark-grown seedlings were measured to the nearest millimeter with a ruler.
RESULTS
Identification and characterization of aha3 alleles:
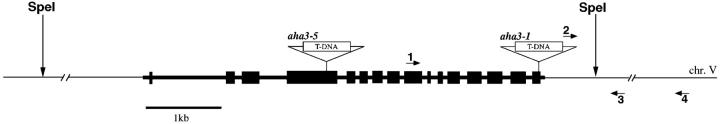
A PCR-based, reverse-genetic strategy was used to identify Arabidopsis plants containing mutant alleles of the plasma membrane transporter AHA3 (Krysan et al. 1996). In all, five aha3 alleles were identified from T-DNA-mutagenized populations of Arabidopsis. A diagram of the insertion sites for the alleles aha3-1 and aha3-5 is shown in Figure 1.
Figure 1.—
Structures of aha3 alleles and AHA3 fragment for molecular complementation. In the PCR-based strategy to identify rescued aha3-1/aha3-1 plants, primers 1 and 4 amplify a product corresponding to native AHA3; primers 2 and 3 amplify a product corresponding to aha3-1. Boxes represent exons; lines represent introns. All T-DNA borders are canonical left-border sequences. T-DNA is not drawn to scale.
aha3-1:
The AHA3 wild-type genomic sequence is ∼6 kb in length and contains 16 exons. A plant containing one copy of the mutant allele aha3-1 was isolated from a T-DNA-mutagenized population of 60,470 independently transformed lines (Young et al. 2001). DNA sequencing confirmed the location of T-DNA within the gene. In aha3-1, the T-DNA was located in the last exon, 47 bases from the stop codon. Both sides of the T-DNA insert were intact, with canonical left-border sequences facing outward (data not shown). No plants homozygous for the T-DNA insertion allele were ever found, suggesting that it was lethal for development of the embryo or one of the gametes.
To determine whether the aha3-1 allele was expressed, RNA was isolated from whole plants heterozygous for aha3-1. The sequence of the corresponding cDNA confirmed the existence of the aha3-1 transcript. The aha3-1 transcript was identical to that of wild type, minus 65 bases from the 3′ end. Starting at this site, the sequence was T-DNA encoded and extended for at least 213 bases (data not shown).
It was not determined whether an aha3-1 protein was present. On the basis of the location of the T-DNA insertion, one can predict that if aha3-1 protein were made, it would be identical to wild-type AHA3 protein, minus 21 amino acids from the C terminus. At this location, the protein sequence would contain an additional 71 T-DNA-based amino acids. Therefore, any aha3-1 protein would lack a portion of the autoinhibitory domain, which is involved in enzyme regulation (Palmgren 2001). However, it is unknown whether this loss would result in protein instability or, if stable, an alteration in enzyme catalytic activity.
aha3-5:
A plant containing a copy of the mutant allele aha3-5 was isolated from a T-DNA mutagenized population of 72,960 lines. DNA sequencing confirmed the location of the T-DNA within the fourth exon. Both sides of the T-DNA insert were intact, with canonical left-border sequences facing outward (data not shown).
To determine whether the aha3-5 allele was expressed, RNA was isolated from plants heterozygous for aha3-5. The sequence of a PCR product corresponding to the aha3-5 transcript revealed that it was just over one-third of the length of the full-length AHA3 message. It contained 17 bases of unknown origin, followed by at least 213 bases of T-DNA-encoded sequence at the 3′ end (data not shown).
Due to the location of the T-DNA in the first third of AHA3, it was unlikely that any functional AHA3 protein was made. On the basis of the sequence of the aha3-5 message, any aha3-5 protein would contain the first 353 of 949 amino acids of wild-type AHA3 protein, followed by 71 T-DNA-encoded amino acids. The first 353 amino acids include the first four transmembrane domains, but not the pore, the large cytoplasmic loop containing the ATP-binding site or regulatory C-terminal domain.
Genetic characterization of aha3 plants:
Plants heterozygous for the aha3-1 or aha3-5 allele were selfed, and progeny were tested by PCR for segregation of the mutation. Selection on media containing kanamycin or BASTA was also used to analyze segregation of aha3, since the T-DNA contained a selectable marker gene that conferred resistance to either kanamycin (aha3-1) or BASTA (aha3-5). The observed ratios of wild-type to heterozygous plants and kanamycin sensitive (KanS) to kanamycin resistant (KanR) plants were consistent with a 1:1 ratio of segregation of the aha3 mutation (Tables 1 and 2). This segregation ratio is characteristic of mutations that cause defects or are lethal during the haploid gametophytic phase of the plant life cycle. In addition, the segregation data indicated that no other T-DNA insertion was present in the background. One explanation for the discrepancy of the 1.3:1 ratio of BASTAS to BASTAR plants could be that the gene conferring resistance to BASTA was silenced in a small percentage of heterozygotes, resulting in seedling sensitivity despite the presence of the T-DNA. This silencing is a common observation (Matzke and Matzke 1998; Koprek et al. 2001). Therefore, despite a P-value of 0.05, we accepted the likelihood that the 1:1 ratio of segregation of the aha3-5 mutation was true.
TABLE 1.
PCR genotyping of progeny of seedlings heterozygous foraha3
| Progeny genotype
|
||||
|---|---|---|---|---|
| Parent genotype | AHA3/AHA3 | AHA3/aha3 | aha3/aha3 | P |
| AHA3/aha3-1 | 126 | 121 | 0 | 0.9 |
| AHA3/aha3-5 | 110 | 115 | 0 | 0.9 |
TABLE 2.
Genotyping progeny ofaha3 mutants by selection on antibiotics or herbicide
| Progeny phenotype
|
|||
|---|---|---|---|
| Parent genotype | Sensitive | Resistant | P |
| AHA3/aha3-1 | 82 | 85 | 0.9 |
| AHA3/aha3-5 | 127 | 96 | 0.05 |
Progeny of heterozygous parents were PCR genotyped and tested for the ability to grow on selective media. Kanamycin was used (50 μg/ml) to detect the presence of aha3-1; BASTA herbicide (50 μg/ml) for aha3-5.
aha3 mutant plants display male gametophytic lethality:
To determine why homozygous aha3 plants could not be found, reciprocal crosses between wild-type plants and plants heterozygous for aha3 were performed. Seeds from individual crosses were harvested, and progeny were germinated on media containing either kanamycin or BASTA (for aha3-1 and aha3-5, respectively) to test for transmission of the T-DNA. The results are shown in Table 3. Transmission of the T-DNA through the male gamete was not observed for either allele. Transmission of the T-DNA through the female gametophyte was normal for both alleles.
TABLE 3.
Determination of gametophytic lethality by reciprocal crosses
| Progeny
|
|||
|---|---|---|---|
| Female parent genotype |
Male parent genotype |
AHA3/AHA3 | AHA3/aha3 |
| AHA3/aha3-1 | AHA3/AHA3 | 23 | 25 |
| AHA3/AHA3 | AHA3/aha3-1 | 52 | 0 |
| AHA3/aha3-5 | AHA3/AHA3 | 55 | 52 |
| AHA3/AHA3 | AHA3/aha3-5 | 101 | 0 |
Cytological and morphological analysis of aha3 pollen:
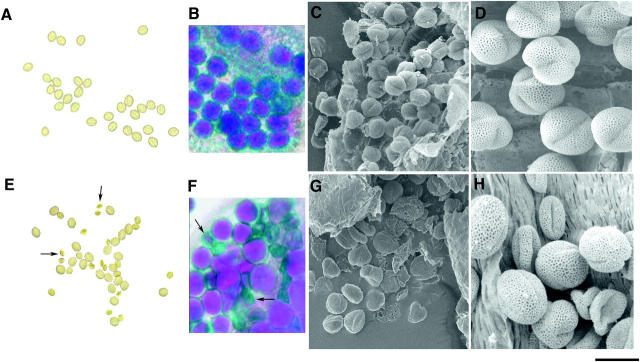
Pollen grains from wild-type and +/aha3-5 flowers were dabbed onto a slide covered with germination media and viewed using a microscope. Figure 2A shows pollen from a wild-type flower. All pollen grains were uniformly shaped and sized. Figure 2E shows pollen from a +/aha3-5 flower. A mixture of full, round pollen grains and smaller, misshapen pollen grains was observed. In some instances, pollen tubes were seen emerged from full, round pollen grains, but none from the smaller, misshapen grains. We repeatedly observed a 1:1 ratio of full, round pollen grains to smaller, misshapen pollen grains.
Figure 2.—
Microspores or pollen grains from wild-type (A–D) and +/aha3-5 (E–H) anthers. (A and E) Pollen grains applied to a glass slide. Arrows in E indicate mutant pollen grains. (B and F) Pollen grains still attached to the anther, stained with Alexander's viability stain. Arrows in F indicate mutant pollen grains. The observed frequencies of mutant pollen grains stained with Alexander's stain were 3.2% ± 0.6 in AHA3/AHA3 flowers and 50.9% ± 0.7 in AHA3/aha3-5 flowers. (C and G) Immature microspores. (D and H) Mature pollen grains. Magnification of micrographs, 1000×; bar, 15 μm.
A stain was applied to pollen grains from a +/aha3-5 plant to distinguish viable and nonviable pollen (Alexander 1969). In Alexander's stain, viable pollen grains are purple with a green outline, and aborted pollen grains are green inside and out. The basis for this differential staining may be a pH difference in the cytoplasm between live and dead pollen (Alexander 1969). Pollen grains from a wild-type plant were viewed as full, round, purple-stained grains with a green outline (Figure 2B; and, occasionally, as smaller, misshapen, green-stained grains, presumably due to a small, normal rate of wild-type pollen abortion). However, pollen grains from a +/aha3-5 plant were viewed as a mixture of purple grains with a green outline and green, misshapen grains (Figure 2F).
To examine the morphological differences between wild-type and mutant pollen grains more closely, pollen samples were prepared for scanning electron microscopy. Anthers and pollen grains from a spectrum of flower ages were analyzed. Microspores, or immature pollen, from young flowers were uniformly shaped and sized (Figure 2, C and G). All microspores appeared round with slight definitions along lobe segments. In some cases, a fraction of microspores from +/aha3-5 flowers appeared slightly smaller or collapsed. These microspores could be aha3 microspores in the early stages of abortion. Pollen from mature wild-type or +/aha3-5 anthers were also compared (Figure 2, D and H). Pollen from a mature wild-type anther was uniformly full and round, whereas pollen grains from a +/aha3-5 anther were a mixture of shapes and sizes. Roughly half of the pollen appeared full and round, and half appeared shrunken and collapsed. Overall, our microscopic studies are consistent with the hypothesis that the smaller, misshapen pollen grains probably correspond to aha3 haploid mutant grains.
To determine a more precise developmental stage at which half the microspores in +/aha3-5 plants aborted, semithin sections of fixed wild-type and +/aha3-5 inflorescences were viewed. Figure 3, A and E, shows sections through pollen mother cells of both wild-type and +/aha3-5 anthers. No differences were detected, which was expected from premeiotic cells. Figure 3, B and F, shows sections through tetrads, which correspond to the stage of pollen development immediately following meiosis. It appeared that there were no visible differences in the morphology of the tetrads. Each product of meiosis, in both wild-type and +/aha3-5 anthers, appeared identical. This indicated that the abortion associated with half of the members of each tetrad, i.e., the aha3 microspores, was not yet apparent. Figure 3, C and G, shows sections through vacuolated microspores. Again, no differences between microspores from wild-type or +/aha3-5 anthers or among microspores from +/aha3-5 anthers were detectable. Differences between microspores from wild-type or +/aha3-5 anthers or among microspores from +/aha3-5 anthers were not obvious until mature pollen sections were viewed (Figure 3, D and H). These sections very clearly revealed that roughly half of the pollen grains from +/aha3-5 flowers were misshapen and empty. The other half were full-sized and stained throughout, identical to pollen grains from wild-type anthers. Taken together, these results indicated that aha3 pollen abortion occurred between the stages corresponding to vacuolated microspores and mature pollen.
Figure 3.—
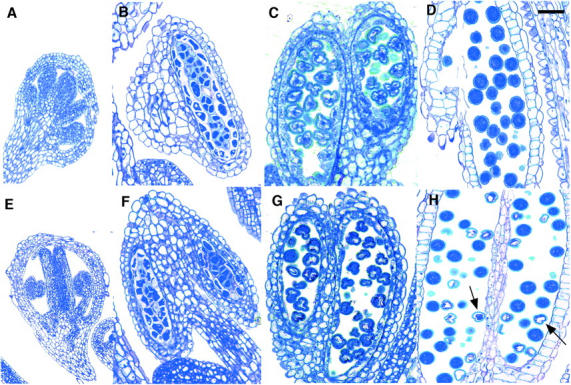
Cross sections of wild-type (A–D) and +/aha3-5 (E–H) anthers. (A and E) Pollen mother cells. (B and F) Tetrads. (C and G) Vacuolate microscores. (D and H) Mature pollen. Arrows in H point to aha3-5 pollen grains. Bar, 30 μm.
Genetic confirmation of aha3 male gametophytic lethality:
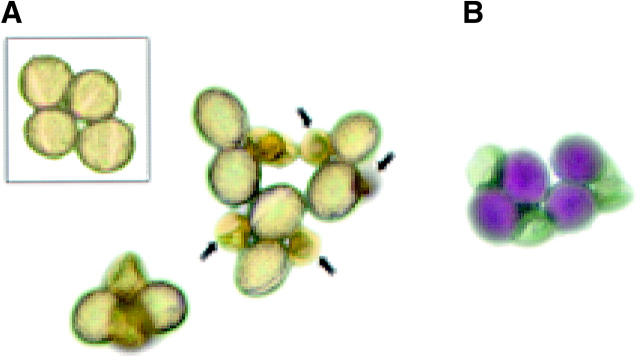
qrt mutants are pollen mutants in which the separation of the four products of meiosis is defective, resulting in “quartets” of pollen grains (Preuss et al. 1994). By expressing a pollen lethal mutation in a qrt background, it is possible to use tetrad analysis to examine the products of meiosis. A plant heterozygous for aha3-5 was crossed as a female to a qrt/qrt plant. +/aha3-5, qrt/qrt plants were identified in the F2 generation, and pollen grains were viewed microscopically. “Quartets” of mature pollen grains showed two normal and two aborted microspores (Figure 4A). For comparison, the smaller box in Figure 4A depicts a +/+, qrt/qrt pollen quartet. Figure 4B shows quartets stained with Alexander's stain.
Figure 4.—
Quartets of pollen grains from +/aha3-5, qrt/qrt plants. (A) Quartets containing two wild-type and two aha3-5 pollen grains. Arrows point to aha3-5 pollen grains (may be out of plane of focus). Inner box shows a quartet from a +/+, qrt/qrt plant. (B) Quartets containing two wild-type and two aha3-5 pollen grains, colored with Alexander's viability stain.
AHA3 expression in male gametophyte tissues:
Previous work using immunolocalization of epitope-tagged AHA3 in transgenic plants or histochemical analysis of plants containing AHA3 reporter constructs depicted AHA3 localization in a variety of plant tissues, including vasculature and reproductive tissues (DeWitt et al. 1991; DeWitt and Sussman 1995). However, depictions of AHA3 expression in male gametophyte tissues were limited to mature pollen. No reports of AHA3 expression in younger male tissues were made. Therefore, to explain the appearance of an aha3-related phenotype during early microspore development, it was necessary to determine whether AHA3 was expressed in younger, male tissues.
Flowers from plants containing a modified AHA3 transgene, in which the reporter protein GUS was fused to the C terminus of AHA3, were examined. The flowers were incubated with GUS substrate, fixed, and sectioned using an ultramicrotome. Sections were viewed with dark-field optics to visualize GUS color precipitates. GUS expression was seen in several stages of microspore development (Figure 5). Figure 5, A–C, shows the lack of expression of AHA3 in pollen mother cells, tetrads, and prevacuolate microspores, respectively. Figure 5, D–F, shows AHA3 expression in the early developmental stages corresponding to early vacuolate microspores, vacuolate microspores, and microspores undergoing the first mitotic division (Regan and Moffatt 1990). The level of expression was strongest in the early vacuolate microspore stage and diminished with each stage. In mitotically active pollen, staining was nearly absent (Figure 5G). In mature pollen, expression was completely absent (Figure 5H). The pattern of expression was significant because it confirmed that AHA3 is indeed expressed during the early stage of microspore development at which lethality was incurred in AHA3 knockout plants. It should be noted that our findings contradict our own earlier reports of AHA3 expression in mature pollen.
Figure 5.—
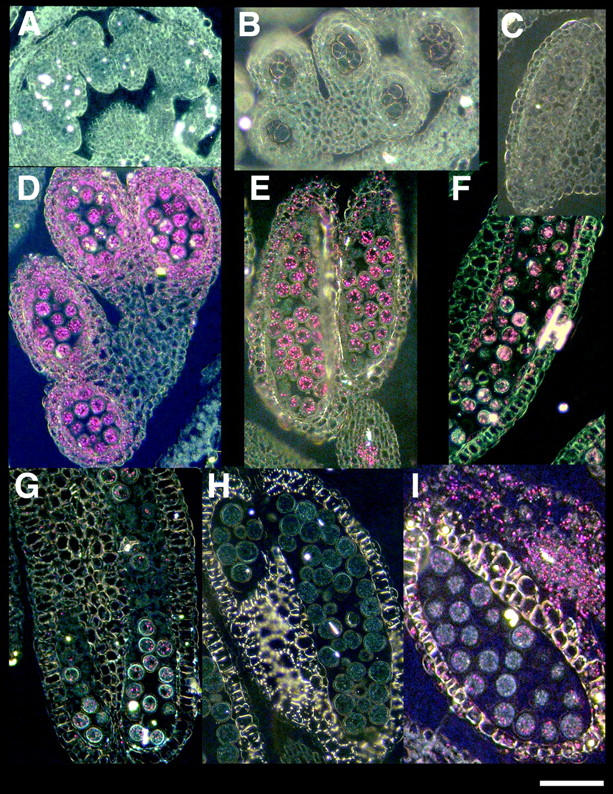
Cross sections of an anther from a plant expressing AHA3::GUS, viewed through dark-field optics. (A) Pollen mother cells. (B) Tetrads. (C) Prevacuolate microspores. (D) Early vacuolated microspores. (E) Vacuolate microspores. (F) Late vacuolated microspores. (G) Mitotic microspores. (H) Mature pollen. (I) Cross section of an anther from a plant expressing CPK28::GUS. Bar, 30 μm.
It was possible that the GUS substrate was unable to penetrate the mature pollen grains, resulting in a perceived absence of AHA3 expression in these tissues. To confirm that AHA3 expression was truly absent in mature pollen, flowers containing a CPK28::GUS construct were sectioned and analyzed. CPK28, a gene encoding a member of the calcium-dependent protein kinase family, is expressed in various floral tissues, including mature pollen (C. W. Chan, personal communication). Sections of CPK28::GUS flowers revealed GUS activity in mature pollen, seen as bright-pink crystals when viewed using dark-field optics (Figure 5I). This confirmed that GUS substrate was able to reach mature pollen and confirmed the absence of AHA3 expression in these tissues.
Molecular complementation of aha3-1:
Plants containing two copies of the aha3-1 allele were not recoverable, presumably due to the lethal effect of the aha3-1 mutation. To demonstrate causality, a full-length copy of AHA3 was reintroduced, and aha3-1/aha3-1 plants containing the transgene were identified among the transformants.
A full-length copy of AHA3, including 3 kb of upstream regulatory sequence and 500 bp of downstream regulatory sequence, was introduced into plants heterozygous for the aha3-1 allele by A. tumefaciens-mediated transformation (Figure 1). We used a PCR-based strategy to identify rescued plants. This strategy utilized two pairs of primers—one pair specific to AHA3 in its native, chromosomal location and a second pair specific to aha3-1. A rescued plant was defined as a plant from which PCR products specific to aha3-1 could be amplified, but from which PCR products specific to native, wild-type AHA3 could not be obtained.
The results of the complementation analysis are presented in Table 4. Analysis of KanR T2 plants from eight independent lines revealed that the transmission of aha3-1 increased following complementation with a wild-type copy of AHA3. Whereas transmission of aha3-1 was originally 50% (through the female only), following complementation it was between 55 and 65%. This increase reflected transmission of aha3-1 through both the female gamete, and, to some degree, the male gamete. Three lines were selected for further analysis. In each case, multiple plants homozygous for the aha3-1 allele, yet alive due to the presence of the AHA3 transgene, were identified.
TABLE 4.
Molecular complementation ofaha3-1
| T0 genotype | KanS:KanR | % T-DNA transmission |
Rescued mutants |
|---|---|---|---|
| AHA3/aha3-1 | 93:151 | 61.9 | |
| AHA3/aha3-1 | 98:184 | 65.2 | |
| AHA3/aha3-1 | 60:80 | 57.1 | |
| AHA3/aha3-1 | 91:122 | 57.3 | 5 (16) |
| AHA3/aha3-1 | 80:111 | 58.1 | |
| AHA3/aha3-1 | 161:204 | 55.9 | 3 (16) |
| AHA3/aha3-1 | 91:115 | 55.8 | |
| AHA3/aha3-1 | 70:95 | 57.6 | 6 (16) |
T0 refers to plants whose progeny were transformed with pCAMBIA3300S::AHA3. KanS:KanR refers to the ratio of sensitivity:resistance to 50 μg/ml kanamycin observed among the progeny (T2) of KanR transformants. KanR implies the presence of the aha3-1 allele. % T-DNA transmission is the percentage of KanR of the total sampled. Rescued mutants were identified among the T2 plants (number sampled).
Visualization of pollen grains stained with Alexander's stain was used to further confirm that the rescued plants were no longer affected by male gametophyte lethality due to the absence of a functional copy of AHA3. Pollen grains from a rescued aha3-1/aha3-1 plant were stained and viewed as a uniform assortment of full, round, purple grains with green outlines (data not shown).
Characterization of plants containing site-directed AHA3 mutants:
To test the requirement for the penultimate threonine residue (T948), we replaced it with alanine (T948A) or aspartate (T948D) via site-directed mutagenesis. Constructs containing these mutations (AHA3T948A, AHA3T948D) were introduced into AHA3/aha3-5 plants using the Agrobacterium-mediated floral dip method. Transformants (T1) were selected and PCR genotyping was performed to determine the identity of each transformant. As expected, an approximately equal ratio of plants homozygous for the wild-type AHA3 allele or heterozygous for aha3-5 was recovered from each parent plant. However, the morphology of these plants differed depending on the genetic background and the specific transgene. While all +/+; AHA3T948A plants appeared normal, three of five +/aha3-5; AHA3T948A plants were completely male sterile (Figure 6). All siliques were empty, and no pollen could be detected. Pistils fertilized with wild-type pollen filled out, indicating that the female organs were unaffected in these plants (data not shown). Due to the sterility of these plants, no progeny were obtained, and thus it was not possible to identify rescued mutants containing the AHA3T948A transgene (aha3-5/aha3-5; AHA3T948A). The remaining two of the five +/aha3-5; AHA3T948A plants were normal in appearance, with fully developed siliques. Progeny from both of these lines were PCR analyzed, but no plants homozygous for aha3-5 were identified.
Figure 6.—

Aerial tissues of a +/aha3-5; AHA3T948A plant (left) and a +/+; AHA3T948A plant. The +/aha3-5; AHA2T498A plant is male sterile. Arrow points to an empty silique. Anthers from this plant lack pollen. The +/+; AHA3T948A plant looks normal and is fertile.
The results were different for plants transformed with the AHA3T948D construct. In the T1 generation, both +/+; AHA3T948D plants and +/aha3-5; AHA3T948D plants appeared normal and fertile. Progeny from +/aha3-5; AHA3T948D plants were PCR analyzed, and plants homozygous for aha3-5 were identified.
The growth of seedlings from one aha3-5/aha3-5; AHA3T948D line was tested on media at pH 4.5, 5, and 5.5. An earlier study indicated that removal of a C-terminal fragment of AHA3, including T948, conferred a growth advantage to seedlings on media at acidic pH (Young et al. 1998). We analyzed aha3-5/aha3-5; AHA3T948D seedlings for similar growth characteristics. Hypocotyl length was measured as an indicator of growth. The results are presented in Table 5. The average hypocotyl length of both transgenic and wild-type seedlings increased with greater pH, but at each pH value, the average hypocotyl length of AHA3T948D seedlings was significantly greater than that of wild-type seedlings.
TABLE 5.
Growth of transgenic and wild-type seedlings on media at different pH
| Average hypocotyl length (cm)a at pH:
|
|||
|---|---|---|---|
| Seedling genotype | 4.5 | 5 | 5.5 |
| aha3-5/aha3-5; AHA3T948D | 0.95 ± 0.17 | 1.22 ± 0.22 | 1.37 ± 0.25 |
| AHA3/AHA3 | 0.63 ± 0.11 | 0.74 ± 0.13 | 0.89 ± 0.16 |
30 seedlings were tested per treatment.
DISCUSSION
In this report, we used a reverse genetic strategy to explore the in planta requirement of a plasma membrane H+-ATPase. We also tested the requirement for a highly conserved threonine residue for pump activation. Our results suggest that expression of the H+-ATPase and a negative charge at the site of this threonine residue are essential.
Using different insertional mutant alleles, we demonstrated that eliminating the expression of a specific member of the Arabidopsis family of plasma membrane H+-ATPases was lethal. The presence of a T-DNA insertion in AHA3 resulted in a 1:1 segregation of wild-type plants to heterozygotes from a heterozygous parent. This ratio is symptomatic of gametophytic lethality. Reciprocal crosses revealed that the source of the lethality was the male gamete. Null aha3 alleles resulted in pollen abortion during the early vacuolated microspore stage. Because transmission of the mutant allele through the male gamete was impossible, no homozygous aha3 mutant plants were recovered.
The timing of pollen abortion in the aha3 plant most likely coincided with the onset of AHA3 expression during the early vacuolated microspore stage of microgametogenesis. Our observation of AHA3 expression in early stages of microgametogenesis is supported by microarray analysis of transcripts from unicellular, bicellular, tricellular, and mature pollen (Honys and Twell 2003). The most likely role of AHA3 in growing microspores is the control of nutrient uptake. As their dependence on the surrounding sporophytic tissues (tapetum) for nourishment decreases, early vauolated microspores rely increasingly on the activity of the plasma membrane H+-ATPase for the transport of nutrients. If this protein is nonfunctional, the microspores starve and die. In agreement with this, the morphological contrast between the aborted mutant microspores and the full-sized, wild-type microspores is most obvious at the mature pollen stage.
The essentiality of a plasma membrane H+-ATPase was originally demonstrated in yeast. Yeast contain two genes encoding plasma membrane H+-ATPases, and knockouts of one of these genes are lethal (Serrano et al. 1986). Of the 11 members of the Arabidopsis plasma membrane H+-ATPase family, T-DNA mutants have been reported previously for only one, AHA4. AHA4 was shown to be localized to root endodermis and flowers, and growth of an aha4 knockout was significantly reduced only when plants were grown in high salt (Vitart et al. 2001). This suggested a role for this gene in resistance to salt stress. However, the phenotype was subtle and the mutants were not fully rescuable, indicating that the mutation was leaky and/or conferred semidominance. The aha3 mutation described herein is the first among the AHA family members to have a clear and essential phenotype, and similar observations with other single AHA knockouts have not been seen (Young et al. 2001).
On the basis of previous work describing AHA3 localization in companion cells, we predicted a role for this gene in phloem loading, and consequently that an AHA3 knockout plant would be deficient in this process. Along these lines, transgenic tobacco plants, in which the phloem-localized plasma membrane H+-ATPase is cosuppressed, are stunted and contain excess sugar in their leaves, among other phenotypes (Zhao et al. 2000). Our results, which revealed male gametophyte lethality, were unexpected. The essentiality of AHA3 in pollen precluded studies in which the role of AHA3 in phloem function could be examined in a null background. For these experiments, it will be necessary to express wild-type copies of AHA3 in mutant plants using a pollen-specific promoter or to identify sublethal alleles of AHA3.
Male gametophyte lethality in Arabidopsis can be manifested a number of times during pollen development and function. For example, the male gametophyte mutants mad2 and mad3 are defective during pollen mitosis I, which occurs after reabsorption of the vacuoles in vacuolated microspores (Grini et al. 1999); mad1 is defective during pollen mitosis II (Grini et al. 1999), as is ttd38 (Procissi et al. 2001); mad4 is defective in pollen tube elongation (Grini et al. 1999); and raring-to-go causes precocious germination in the anther (Johnson and McCormick 2001). An estimate of the frequency of male gametophytic lethality in a T-DNA-mutagenized population is 0.2% (Procissi et al. 2001), but in most cases the effect is not completely penetrant. In addition, female gametophyte lethality is also often observed. These cases therefore do not represent fully penetrant, male-specific gametophyte lethality. Only seven cases cite mutations that cause fully penetrant, male-specific gametophyte lethality; for only one of these mutations was the gene identified (ADL1C, a dynamin-like gene; Kang et al. 2003). AHA3 is therefore only the second known gene for which fully penetrant, male gametophyte lethality is observed in the knockout.
We tested the requirement for a residue thought to play a critical role in H+-ATPase activation. According to the current model H+-ATPase activity under resting conditions is reduced because of an autoinhibitory effect applied by the C terminus (Morsomme and Boutry 2000; Palmgren 2001). The role of this domain was confirmed by several studies, which showed that truncation or displacement of the C terminus of plant H+-ATPases yielded a constitutively activated enzyme when expressed in yeast (Portillo and Serrano 1989; Palmgren et al. 1990; Portillo et al. 1991; Palmgren and Christensen 1993; Regenberg et al. 1995). Activation in vivo is initiated by phosphorylation of the highly conserved penultimate threonine residue (T948 in AHA3). The phosphorylated threonine is recognized and bound by a 14-3-3 protein dimer (Fuglsang et al. 1999; Svennelid et al. 1999; Maudoux et al. 2000). This binding is thought to alter the orientation of the autoinhibitory C-terminal domain so that it is displaced from the active site of the ATPase, which relieves inhibition and activates the pump (Olsson et al. 1998; Fuglsang et al. 1999; Camoni et al. 2000).
To test the role of T948 in AHA3 regulation in planta, we replaced it with alanine or aspartate and characterized the effects of these mutant transgenes in plants with a null aha3 background. We predicted that substitution with alanine would produce a constitutively inactive enzyme, as it is not possible to phosphorylate alanine residues. Conversely, we predicted that the negatively charged aspartate would mimic a constitutively phosphorylated threonine residue, producing a constitutively activated, or hyperactive, enzyme.
Plants with a null aha3 background but that contained the AHA3T948A transgene were never identified among the progeny of +/aha3-5; AHA3T948A plants. We proposed that substitution of threonine with alanine at the penultimate location in AHA3 prevents phosphorylation of the 14-3-3 binding target, binding of the 14-3-3 protein dimer, and activation of the H+-ATPase. We argue that the constitutive inactivity of the altered H+-ATPase produces a perceived functional absence of this enzyme. However, while two of the five +/aha3-5; AHA3T948A plants identified were normal in appearance, the remaining three were male sterile, suggesting that the AHA3T948A transgene behaved in a dominant fashion. One possible explanation for the observed male sterility could be the presence of this transgene in the tapetal cells, which are known to be extremely sensitive to alterations in enzyme activity (Wu and Cheung 2000). Metabolic perturbations in these cells, caused by unusual AHA3T948A functions, could inhibit nourishment of developing microspores, resulting in male sterility. In nonsterile +/aha3-5; AHA3T948A plants, expression of the transgene may have been insufficient to cause sterility due to copy number or position effect. The use of a pollen-specific promoter to drive AHA3T948A expression in aha3 plants would directly test the effect of the amino acid substitution and remove the effects of such confounding factors.
Plants with an aha3 null background but containing AHA3T948D were identified by consecutive selection on kanamycin and BASTA, and their identities were confirmed by PCR genotyping. This result indicates that the conservation of negative charge at the penultimate threonine residue yielded a functional enzyme that could complement the lethal effect of aha3. However, identification of these plants at less than the expected Mendelian frequency of 25% from a heterozygous parent suggests that this transgene did not rescue with full penetrance, perhaps because our AHA3 transgene requires additional flanking sequence and/or its native chromosomal locus for optimal expression.
Seedlings containing the AHA3T948D transgene appeared to have a significant growth advantage over wild-type seedlings on media at low pH. This result suggests that the substitution at T948 produced a hyperactive H+-ATPase that may drive increased removal of toxic protons from the cytoplasm of vascular cells of seedlings, “insensitivity” to the acidic media, and superior growth (Young et al. 1998).
The role of the penultimate threonine residue of plant H+-ATPases was previously tested by expression of mutants in yeast lacking their own pump or in cell-free extracts. Replacement of threonine with alanine in the tobacco plasma membrane H+-ATPase, PMA2, prevented complementation of yeast H+-ATPase mutants (Svennelid et al. 1999; Maudoux et al. 2000). Surprisingly, replacement of threonine with aspartate also prevented complementation, but a negative result is difficult to interpret since effects of this mutation on biogenesis or turnover in this heterologous organism were not determined. In another study, replacement of threonine with alanine in AHA2 prevented complementation of mutant yeast unless additional mutations in the C terminus were also present (Jahn et al. 2002). When the 14-3-3 protein-binding target was destroyed, but mutations in autoinhibitory regions in the C terminus were present, the mutant yeast was rescued. This demonstrated that activation of the H+-ATPase is not dependent on 14-3-3 protein binding per se, but on displacement or neutralization of the C terminus. Finally, substitution of alanine for the penultimate threonine of AHA2 prevented phosphorylation at this site, 14-3-3 binding, and ATP hydrolysis in vitro, further illustrating the requirement for threonine in pump function (Fuglsang et al. 1999).
Our results help define the role of the penultimate threonine residue in plasma membrane H+-ATPase activity in planta. First, we showed that substitution of the penultimate threonine residue with alanine prevents complementation of the mutant plant. This finding is in agreement with results from the heterologous experiments described above. Second, we showed that replacement of the penultimate threonine with aspartate produces an active H+-ATPase that is capable of rescuing transmission of aha3 through the male gamete.
Through segregation analyses, microscopic analysis of mutant reproductive tissues, confirmation of AHA3 expression in the appropriate microspore stages, complementation of the male gametophyte lethality with a wild-type copy of AHA3, and confirmation of gametophytic lethality by crossing to qrt, we have identified an essential function for AHA3 in microspore development. We suggest that this role involves control of nutrient acquisition, although other functions, such as in cell-wall deposition or intercellular signaling, cannot be ruled out. We have also confirmed the requirement for a negatively charged amino acid at the penultimate location in the regulatory C terminus.
Acknowledgments
We gratefully acknowledge Melissa Lehti-Shiu and Sara Patterson for technical assistance. This work was supported by grants from the National Institutes of Health/National Institute of General Medical Sciences Predoctoral Training Grant in Molecular Biosciences T32 GM007125 (to W.R.R), the United States Department of Energy (DE-FG02-88ER13938), and the University of Wisconsin College of Agricultural and Life Sciences Hatch Funds (WIS04791).
References
- Alexander, M. P., 1969. Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Camoni, L., V. Iori, M. Marra and P. Aducci, 2000. Phosphorylation-dependent interaction between plant plasma membrane H+-ATPase and 14-3-3 proteins. J. Biol. Chem. 275: 9919–9923. [DOI] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- DeWitt, N. D., and M. R. Sussman, 1995. Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell 7: 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt, N. D., J. F. Harper and M. R. Sussman, 1991. Evidence for a plasma membrane proton pump in phloem cells of higher plants. Plant J. 1: 121–128. [DOI] [PubMed] [Google Scholar]
- Fernandez, D. E., G. R. Heck, S. E. Perry, S. E. Patterson, A. B. Bleecker et al., 2000. The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12: 183–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang, A. T., S. Visconti, K. Drumm, T. Jahn, A. Stensballe et al., 1999. Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947). J. Biol. Chem. 274: 36774–36780. [DOI] [PubMed] [Google Scholar]
- Grini, P. E., A. Schnittger, H. Schwarz, I. Zimmermann, B. Schwab et al., 1999. Isolation of ethyl methanesulfonate-induced gametophytic mutants in Arabidopsis thaliana by a segregation distortion assay using the multimarker chromosome 1. Genetics 151: 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J. F., L. Manney, N. D. DeWitt, M. H. Yoo and M. R. Sussman, 1990. The Arabidopsis thaliana plasma membrane H+-ATPase multigene family. Genomic sequence and expression of a third isoform. J. Biol. Chem. 265: 13601–13608. [PubMed] [Google Scholar]
- Hirsch, R. E., B. D. Lewis, E. P. Spalding and M. R. Sussman, 1998. A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921. [DOI] [PubMed] [Google Scholar]
- Honys, D., and D. Twell, 2003. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132: 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlne, G., and M. Boutry, 1994. Identification of an Arabidopsis thaliana gene encoding a plasma membrane H+-ATPase whose expression is restricted to anther tissue. Plant J. 5: 311–317. [DOI] [PubMed] [Google Scholar]
- Jahn, T. P., A. Schulz, J. Taipalensuu and M. G. Palmgren, 2002. Post-translational modification of plant plasma membrane H+-ATPase as a requirement for functional complementation of a yeast transport mutant. J. Biol. Chem. 277: 6353–6358. [DOI] [PubMed] [Google Scholar]
- Johnson, S. A., and S. McCormick, 2001. Pollen germinates precociously in the anthers of raring-to-go, an Arabidopsis gametophytic mutant. Plant Physiol. 126: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B. H., D. M. Rancour and S. Y. Bednarek, 2003. The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J. 35: 1–15. [DOI] [PubMed] [Google Scholar]
- Koprek, T., S. Rangel, D. McElroy, J. D. Louwerse, R. E. Williams-Carrier et al., 2001. Transposon-mediated single-copy gene delivery leads to increased transgene expression stability in barley. Plant Physiol. 125: 1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P. J., J. C. Young, F. Tax and M. R. Sussman, 1996. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 93: 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P. J., J. C. Young and M. R. Sussman, 1999. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, A. J., and M. A. Matzke, 1998. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1: 142–148. [DOI] [PubMed] [Google Scholar]
- Maudoux, O., H. Batoko, C. Oecking, K. Gevaert, J. Vandekerckhove et al., 2000. A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275: 17762–17770. [DOI] [PubMed] [Google Scholar]
- Morsomme, P., and M. Boutry, 2000. The plant plasma membrane H+-ATPase: structure, function and regulation. Biochim. Biophys. Acta 1465: 1–16. [DOI] [PubMed] [Google Scholar]
- Mouline, K., A. A. Very, F. Gaymard, J. Boucherez, G. Pilot et al., 2002. Pollen tube development and competitive ability are impaired by disruption of a Shaker K+ channel in Arabidopsis. Genes Dev. 16: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, A., F. Svennelid, B. Ek, M. Sommarin and C. Larsson, 1998. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 118: 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren, M. G., 2001. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 817–845. [DOI] [PubMed] [Google Scholar]
- Palmgren, M. G., and G. Christensen, 1993. Complementation in situ of the yeast plasma membrane H+-ATPase gene PMA1 by an H+-ATPase gene from a heterologous species. FEBS Lett. 317: 216–222. [DOI] [PubMed] [Google Scholar]
- Palmgren, M. G., C. Larsson and M. Sommarin, 1990. Proteolytic activation of the plant plasma membrane H+-ATPase by removal of a terminal segment. J. Biol. Chem. 265: 13423–13426. [PubMed] [Google Scholar]
- Portillo, F., and R. Serrano, 1989. Growth control strength and active site of yeast plasma membrane ATPase studied by site-directed mutagenesis. Eur. J. Biochem. 186: 501–507. [DOI] [PubMed] [Google Scholar]
- Portillo, F., P. Eraso and R. Serrano, 1991. Analysis of the regulatory domain of yeast plasma membrane H+-ATPase by directed mutagenesis and intragenic suppression. FEBS Lett. 287: 71–74. [DOI] [PubMed] [Google Scholar]
- Preuss, D., S. Y. Rhee and R. W. Davis, 1994. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460. [DOI] [PubMed] [Google Scholar]
- Procissi, A., S. de Laissardiere, M. Ferault, D. Vezon, G. Pelletier et al., 2001. Five gametophytic mutations affecting pollen development and pollen tube growth in Arabidopsis thaliana. Genetics 158: 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan, S. M., and B. A. Moffatt, 1990. Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg, B., J. M. Villalba, F. C. Lanfermeijer and M. G. Palmgren, 1995. C-terminal deletion analysis of plant plasma membrane H+-ATPase: yeast as a model system for solute transport across the plant plasma membrane. Plant Cell 7: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, R., M. C. Kielland-Brandt and G. R. Fink, 1986. Yeast plasma membrane ATPase is essential for growth and has homology with Na+/K+, K+- and Ca2+-ATPases. Nature 319: 689–693. [DOI] [PubMed] [Google Scholar]
- Svennelid, F., A. Olsson, M. Piotrowski, M. Rosenquist, C. Ottman et al., 1999. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11: 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitart, V., I. Baxter, P. Doerner and J. F. Harper, 2001. Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis. Plant J. 27: 191–201. [DOI] [PubMed] [Google Scholar]
- Wu, H., and A. Y. Cheung, 2000. Programmed cell death in plant reproduction. Plant Mol. Biol. 44: 267–281. [DOI] [PubMed] [Google Scholar]
- Young, J. C., N. D. DeWitt and M. R. Sussman, 1998. A transgene encoding a plasma membrane H+-ATPase that confers acid resistance in Arabidopsis thaliana seedlings. Genetics 149: 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J. C., P. J. Krysan and M. R. Sussman, 2001. Efficient screening of Arabidopsis T-DNA insertion lines using degenerate primers. Plant Physiol. 125: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R., V. Dielen, J. M. Kinet and M. Boutry, 2000. Cosuppression of a plasma membrane H+-ATPase isoform impairs sucrose translocation, stomatal opening, plant growth, and male fertility. Plant Cell 12: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]