Abstract
The degree to which stigmas are exserted above the stamen in flowers is a key determinant of cross-pollination (and hence allogamy) in many plant species. Most species in the genus Lycopersicon are obligate or facultative outcrossers and bear flowers with highly exserted stigmas. In contrast, the cultivated tomato (Lycopersicon esculentum) bears flowers with flush or inserted stigmas promoting self-fertilization. It has been observed that a major QTL, se2.1, on chromosome 2 is responsible for a large portion of phenotypic variation for this trait and that mutation(s) at this locus were likely involved in the evolution from allogamy to autogamy in this genus. To understand the genetic and molecular basis of stigma exsertion, we have conducted a high-resolution mapping at the chromosome region harboring the se2.1 QTL. The results indicate that this is a compound locus, comprising at least five tightly linked genes, one controlling style length, three controlling stamen length, and the other affecting anther dehiscence, a taxonomic character used to distinguish Lycopersicon species from other solanaceous species. This cluster of genes may represent the vestiges of an ancient coadapted gene complex in controlling mating behavior.
OUTCROSSING is an important and widespread strategy for maintaining genetic variability in populations of sexually reproducing organisms (Barrett 2002). Unlike animals, most plants are hermaphroditic—producing flowers with both male and female sex organs. As a result, most plants are potentially capable of self-reproduction. Genetically controlled self-incompatibility is a common mechanism in many plants, which prevents self-fertilization and hence enforces outcrossing. Two of the most common self-incompatibility mechanisms are sporophytic and gametophytic (de Nettancourt 1977). Sporophytic self-incompatibility has been thoroughly studied in the crucifers, where both the female (Stein et al. 1991) and the male (Schopfer et al. 1999) components on self-incompatibility have been cloned. Gametophytic self-incompatibility is common in the nightshade family Solanaceae, where the female components have been cloned and characterized at the molecular level (McClure et al. 1989).
While self-incompatibility is able to block self-reproduction (autogamy), it is not sufficient to ensure cross-pollination (allogamy). Conversely, lack of self-incompatibility does not guarantee self-fertilization. Changes in floral morphology are normally required to promote either autogamy or allogamy (Kalisz et al. 1999). One common floral polymorphism associated with change in mating system is the degree to which the female stigma surface is either exserted above the anthers (promoting outcrossing) or recessed below the anthers (promoting self-fertilization; Rick et al. 1977; Barrett 1992; Motten and Antonovics 1992; Karron et al. 1997; Richards 1997; Motten and Stone 2000).
The tomato genus Lycopersicon is ideal for the studies of floral variation associated with changes in mating system. It is composed of nine species that cover the full range of mating systems: allogamy (obligate cross-pollination, enforced by gametophytic self-incompatibility), facultative allogamy (self-compatible but with a wide variation in cross-pollination rate), and autogamy (obligate self-fertilization; Table 1; Rick 1988). Tomato species can be roughly classified into three categories according to the degree of stigma exsertion. The first category is largely outcrossing species bearing flowers with highly exserted stigmas and includes Lycopersicon chilense, L. peruvianum, L. pennellii, L. hirsutum, and L. chmielewskii (Rick and Lamm 1955; Rick et al. 1976; Rick 1982). It should be noticed that L. chmielewskii was classified as facultative allogamy, unlike the other tomato species in this category, because L. chmielewskii is self-compatible. L. pimpinellifolium represents the secondary category and varies greatly among accessions with respect to stigma exsertion and outcrossing rate (Rick et al. 1977). The third category, L. parviflorum, L. cheesmanii, and the cultivated tomato (L. esculentum), produces flowers in which stigma surface is only slightly exserted, flush, or recessed with respect to the anther cone, promoting self-fertilization (Rick 1963, 1995; Rick et al. 1976). On the basis of the phylogenic relationships (Palmer and Zamir 1982; Miller and Tanksley 1990), autogamous species most likely evolved from self-incompatibility ancestors that have exserted stigmas.
TABLE 1.
Mating system classification for species in the tomato genus, Lycopersicon, adapted from Rick (1988)
| Mating system | Species |
|---|---|
| Autogamy |
L. parviflorum, L. cheesmanii, L. esculentum |
| Facultative allogamy | L. chmielewskii, L. pimpinellifolium |
| Obligate allogamy |
L. chilense, L. peruvianum, L. pennellii, L. hirsutum |
While the autogamous tomato species are thought to have evolved from self-incompatible ancestors (Rick 1988), only recently have studies begun to address the genetic basis for the accompanying shift from exserted stigmas to flush or inserted stigmas. Early reports suggested that stigma exsertion is quantitatively inherited and controlled by a few genes (Rick and Dempsey 1969; Scott and George 1978; Levin et al. 1994). With the advent of molecular linkage maps, it has been possible to study the genetic basis of quantitative trait with much more rigor. Recently, several quantitative trait loci (QTL) mapping studies for stigma exsertion in tomato have been reported (Bernacchi and Tanksley 1997; Fulton et al. 1997; Georgiady et al. 2002). In the studies involving crosses between self-incompatible wild species with the self-compatible cultivated tomato, a major QTL, se2.1 or stg2.1 (hereafter called se2.1), on tomato chromosome 2 was reported (Bernacchi and Tanksley 1997; Fulton et al. 1997). In contrast, the self-incompatibility locus maps to chromosome 1 (Tanksley and Loaizafigueroa 1985). On the basis of these studies it is proposed that mutation(s) in se2.1, resulting in more recessed stigmas, accompanied the evolution from self-incompatible allogamous species to self-compatible autogamous species. If this hypothesis is correct, all self-incompatible species should contain wild-type alleles (for exserted stigmas) at the se2.1 QTL.
L. pennellii is a largely self-incompatibility tomato species that produces large showy flowers with exserted stigmas (Rick and Tanksley 1981). Near isogenic lines (ILs) have been developed in which specific chromosomal regions of one accession (LA716) of L. pennellii have been transferred into a single inbred cultivar tomato (Eshed and Zamir 1995). The ILs for chromosome 2 provide ideal genetic materials for genetic dissection of the se2.1 QTL. The goal of the current study was to produce a fine map of the region of chromosome 2 containing the L. pennellii se2.1 QTL in order to: (1) ascertain whether changes in stigma exsertion modulated by this QTL are due to a single gene or several tightly linked genes, (2) characterize the individual aspects of floral morphology controlled by the se2.1 QTL, and (3) set the stage for eventual cloning of the gene(s) underlying the se2.1 QTL.
MATERIALS AND METHODS
Genetic markers:
Molecular markers derived from cDNA and genomic DNA clones were from the tomato high-density linkage map (Tanksley et al. 1992; Fulton et al. 2002; http://www.sgn.cornell.edu). Molecular markers were also derived from the end sequences of tomato bacterial artificial chromosome (BAC) clones LE136C06 and LE27H05, which were isolated from a BAC library constructed by Budiman et al. (2000) according to published protocols (http://www.genome.clemson.edu/groups/bac/protocols/bacmanual.html) and using cLED19A24 (http://www.sgn.cornell.edu) as a probe. Positive BAC clones were obtained from the Clemson University Genomics Institute and confirmed by Southern analysis (Sambrook et al. 1989). BAC ends were isolated using the strategy of either plasmid rescue (for right end) or inverse PCR (for left end) as described by Woo et al. (1994). All the molecular markers were used as RFLP probes for mapping experiments. DNA from seedling leaf tissue was extracted as described by Fulton et al. (1995).
Plant material:
IL2-5, an L. pennellii introgression line, was provided by D. Zamir, Hebrew University of Jerusalem, Israel, and was generated as described previously in a cross between the domesticated tomato (L. esculentum) cv. M82-1-8 and the wild tomato species, L. pennellii (Eshed and Zamir 1995). The lines contain L. pennellii DNA for a segment of chromosome 2 encompassing the se2.1 QTL (Figure 2A). IL2-5 was crossed to L. esculentum cv. M82-1-8 and the F1 self-fertilized to produce F2 seeds for high-resolution mapping. Two such F2 populations were used in this study: one composed of 1535 plants and generated in Ithaca, New York, and the other containing 1169 plants provided by D. Zamir. F3 seed was generated by self-fertilization of selected recombinant F2 plants. Hereafter individuals derived from the same F2 recombinant individual are defined in the same “family” and individuals derived from the same F3 recombinant individual are defined in the same “line.”
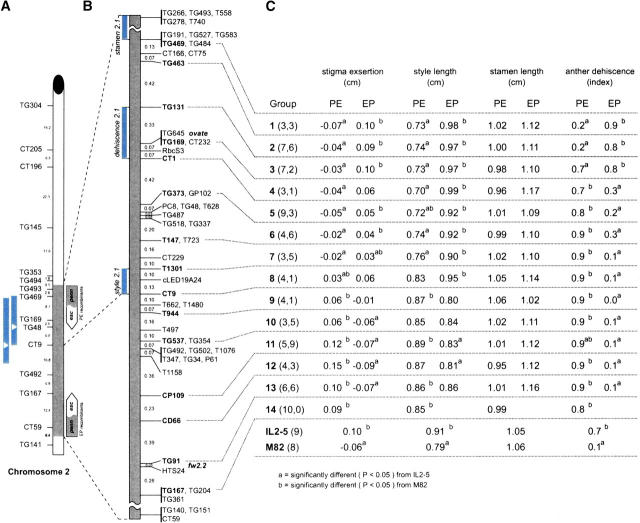
Figure 2.—
Fine mapping the se2.1 QTL on tomato chromosome 2. (A) The genetic map of chromosome 2 showing the L. pennellii introgressed region (gray) in IL2-5. The genetic distance between markers is from the tomato RFLP high-density map (Tanksley et al. 1992). Two short bars to the left show the location of se2.1 QTL from two previous mapping studies (Bernacchi and Tanksley 1997; Fulton et al. 1997). The white triangle inside a bar represents the location of the most significant marker associated with stigma exsertion in each study. Two arrow bars to the right show the portion of the L. pennellii introgression region (gray) in the PE and EP recombinant groups (esc, L. esculentum cv. M82 DNA; penn, L. pennellii DNA). (B) The high-resolution genetic map between markers TG469 and TG167 derived from this study. The order and map distance between markers is based on 123 crossovers in the interval derived from the screening of 1535 F2 plants. Markers separated by commas cosegregate. Bars on the left indicate the location of loci as deduced from this study: style length (style2.1), stamen length (stamen2.1), and anther dehiscence (dehiscence2.1). (C) The phenotypic means of each trait in different grouped recombinant families. PE groups represent the recombinants containing pennellii DNA from the proximal end of the introgression (TG493) down to the recombination point. EP groups represent the recombinants containing L. pennellii DNA from the distal end (CT59) up to the recombination point. The two numbers in parentheses represent the number of recombinant families in PE type (the first number) or in EP type (the second number). The numbers in parentheses after IL2-5 and M82 represent the number of the replicated control plants.
Recombinant screening and construction of a high-resolution genetic map:
The first set of 1535 F2 plants were screened with markers TG469 and TG167 known to flank the se2.1 region (Bernacchi and Tanksley 1997; Fulton et al. 1997; Figure 2A). 123 F2 plants with crossover in this interval were thus identified. Three F3 individuals bearing homozygous genotypes for both markers TG469 and TG167—one marker for homozygous L. pennellii genotype and the other for homozygous L. esculentum genotype—were isolated from each recombinant F2 individual via marker-assisted selection. The genotypes for all markers in the TG469-TG167 interval were determined for all F3 progeny to precisely locate the recombination points in each F2 plant and thus develop a high-resolution genetic map (Figure 2B). In addition, F4 seeds were generated from each F3 homozygous recombinant plant for later phenotypic evaluations.
On the basis of analysis of the above-described F2, F3, and F4 populations, it was possible to narrow the location of a major stigma exsertion locus between markers T1301 and CT9 (Figure 2B, also see results). Hence, a second set of 1169 F2 plants (also derived from IL2-5 × M82) were screened with markers T1301 and CT9. An additional six crossovers located between these two markers were identified (Figure 3A). Homozygous F3 and F4 recombinants were derived in a manner similar to that described for the first F2 population. All the F2 and F3 plants were grown in a greenhouse to avoid cross-pollination by insect pollinators.
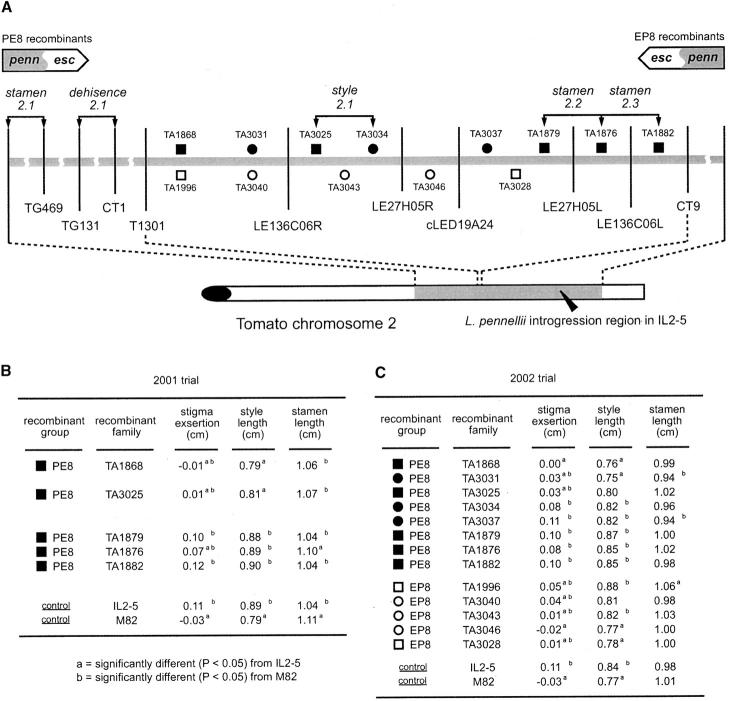
Figure 3.—
(A) Fine mapping of the style2.1, stamen2.1, -2.2, -2.3, and dehiscence2.1 on chromosome 2. Squares and circles represent the chromosomal location of crossover events in each recombinant family. Solid squares and circles represent the PE8 recombinant families. Open squares and circles represent the EP8 recombinant families. Squares represent the recombinants derived from the F2 population presented in Figure 2. Circles represent the recombinants derived from an additional F2 population. Vertical lines represent the marker locations. For those markers derived from BAC ends, an R or L was added after the BAC clone name to indicate the right or left ends of the cloned tomato chromosomal fragments, respectively, as defined by Woo et al. (1994). Two arrow bars at the top show the portion of L. pennellii introgression region (gray) in the PE8 and EP8 recombinant families (esc, L. esculentum cv. M82 DNA; penn, L. pennellii DNA). (B) Phenotypic values and comparisons for 2001 trial. (C) Phenotypic values and comparisons for 2002 trial.
Phenotypic and statistical analyses:
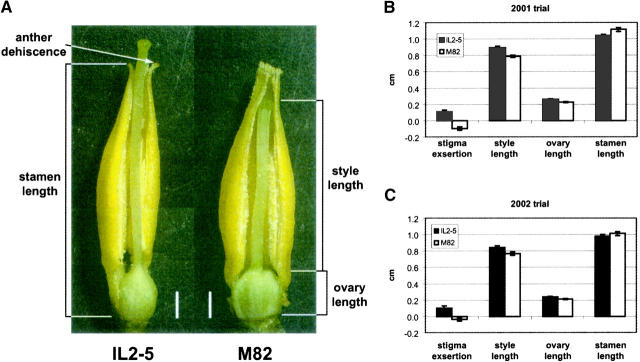
For phenotypic evaluations, homozygous F4 progeny from the first 123 recombinant plants were field grown in the summer of 2000 in Ithaca, New York. A randomized block design was used with three blocks, each block containing a single F4 homozygous plant from each recombinant line plus three plants of each parental control (IL2-5 and tomato cultivar M82-1-8). Flowers were harvested at midday on the day of anthesis and inserted, pedicel-side down, in a water-soaked sponge to keep fresh before dissecting. The pistil and stamen cone were separated from each flower and their images were scanned (Astra 600S, UMAX Data System) with 800-dpi scanning resolution and saved as JEPG files. Measurements were taken from scanned images using Adobe Photoshop v5.5 software. Flower traits were evaluated by measuring 10 flowers from each plant. Style length and ovary length measurements were taken from each flower with accuracy ±0.05 mm. The length of a single, random stamen was also measured from each flower. The stamen length subtracted from the sum of the style length plus the ovary length was used to calculate stigma exsertion. Anther dehiscence was also measured. Anther dehiscence is defined as the degree to which the tips of the stamen curve outward and can enhance the extent of stigma exsertion (Figure 1A). Anther dehiscence was scored as 0 for straight stamen tip or as 1 for curved stamen tip in each flower. L. pennellii and IL2-5 produce curved stamen tips, whereas M82 produces straight stamen (Figure 1A).
Figure 1.—
Stigma exsertion and associated floral traits in L. esculentum cv. M82-1-8 and the L. pennellii introgression line IL2-5. M82 and IL2-5 differ for stigma exsertion and style length. (A) M82 has straight fused anthers vs. the loose and tip-curved anthers of IL2-5. Bars, 1 mm. (B) The measurements of style length, ovary length, and stamen length in M82 and IL2-5 in 2001 trial. Error bars represent 95% confidence interval. Note that M82 and IL2-5 significantly differ in stamen length (P < 0.0001). (C) The same measurements as B in 2002 trial. Note that the difference in stamen length between M82 and IL2-5 was not significant.
For statistical analyses, recombinant families bearing the same marker genotypes were grouped together. The least-squares mean of each trait was calculated for each grouped recombinant family using SAS v8.2 software. Pairwise comparisons between grouped families and the two parental controls were conducted using Tukey's studentized range test to control experimentwise type I error rate at significance level <0.05 (Figure 2C). The comparisons of the stamen length between reciprocal recombinant groups were conducted using a paired t-test (Table 3). The null hypothesis was that the stamen length of a PE group is not smaller than that of its reciprocal EP group.
TABLE 3.
Pairedt-test for stamen length between PE and EP recombinant groups
| Mean
|
||||
|---|---|---|---|---|
| Group | PEa | EPa | d.f. | P-value |
| 1 | 1.02 (3) | 1.12 (3) | 4 | 0.0160 |
| 2 | 1.00 (7) | 1.11 (6) | 11 | 0.0012 |
| 3 | 0.98 (7) | 1.10 (2) | 7 | 0.0007 |
| 4 | 0.96 (3) | 1.17 (1) | — | ND |
| 5 | 1.01 (9) | 1.09 (3) | 10 | 0.0022 |
| 6 | 0.99 (4) | 1.10 (6) | 8 | 0.0002 |
| 7 | 1.02 (3) | 1.10 (5) | 6 | 0.0260 |
| 8 | 1.05 (4) | 1.14 (1) | — | ND |
| 9 | 1.06 (4) | 1.02 (1) | — | ND |
| 10 | 1.02 (3) | 1.11 (5) | 6 | 0.0059 |
| 11 | 1.01 (5) | 1.12 (9) | 12 | <0.0001 |
| 12 | 0.95 (4) | 1.12 (3) | 5 | 0.0001 |
| 13 | 1.01 (6) | 1.16 (6) | 10 | <0.0001 |
| Combined | 1.01 (13) | 1.12 (13) | 23 | <0.0001 |
ND, not determined, due to lack of multiple genotypes for either PE or EP groups.
Numbers in parentheses indicate the number of recombinants in that group.
To further refine the location of se2.1 QTL from the analysis describe above, two additional trials were conducted. For the first trial, five F4 families from the original 123 recombinants corresponding to crossovers in the T1301–CT9 interval (Figure 3, A and B) were additionally field evaluated in the summer of 2001. For the second trial, the same five F4 families evaluated in 2001 plus two recombinant families from the first screen and six additional recombinant families from the second screen were field evaluated again in the summer of 2002 (Figure 3, A and C). Each recombinant family was represented by three derived F3 recombinant lines from which 6 plants were each evaluated. A randomized block design was used with six blocks, each block containing a single plant from each recombinant line plus three plants of each parental control (IL2-5 and tomato cultivar M82-1-8). Phenotypic evaluations in the summer of 2001 and 2002 were the same as those employed in the summer of 2000, but 5 flowers (instead of 10) were evaluated per plant.
The least-squares mean of each trait was calculated for each recombinant family using SAS v8.2 software. Pairwise comparisons between each family and the two parental controls were conducted using Tukey's studentized range test to control experimentwise type I error rate at significance level <0.05 (Figure 3, B and C).
Construction of subNILs and the test of gene action:
An F3 subnear isogenic line (subNIL) containing the L. pennellii introgression only for the 0.53-cM interval between T1301 and T497 (Figure 2B) was generated by screening 287 F3 plants derived from a single F2 recombinant plant containing the L. pennellii introgression region between markers T1301 and CT59. From a single plant, heterozygous for only the T1301-T497 interval, 170 F4 progeny were screened and classified according to their genotype for marker T1301: homozygous L. pennellii, homozygous L. esculentum, and heterozygous. Twenty-four plants of each genotype were evaluated in 24 blocks in the field in the summer of 2001. Each block consisted of one plant of each genotype. Flower traits were evaluated as described above by measuring five flowers from each plant. The least-squares mean of each trait was calculated for each recombinant family using SAS v8.2 software. Pairwise comparisons between different genotypes were conducted using Tukey's studentized range test to control experimentwise type I error rate at significance level <0.05 (Table 2). Dominance was also measured by the ratio d/a, where d is the difference between the phenotypic mean of the heterozygous genotype and the average of the phenotypic means of the two homozygous genotypes and a is half of the difference between the phenotypic means of the two homozygous genotypes.
TABLE 2.
Gene action (d/a) of these21 QTL and its component traits as determined by phenotypic evaluation of the chromosome 2 subNIL TA3178
| Genotype
|
||||
|---|---|---|---|---|
| TA3178 (L. pennellii subNIL)
| ||||
| Trait | Homozygous (n = 24) |
Heterozygous (n = 24) |
M82 control (L. esculentum) homozygous (n = 24) |
Gene action (d/a) |
| Stigma exsertion (cm) | −0.002 (a) | −0.034 (b) | −0.103 (c) | 0.37 |
| Style length (cm) | 0.850 (a) | 0.853 (a) | 0.765 (b) | 1.07 |
| Ovary length (cm) | 0.241 (a) | 0.240 (a) | 0.223 (b) | 0.89 |
| Stamen length (cm) | 1.093 (b) | 1.126 (a) | 1.090 (b) | 23.00 |
Means with a different letter for each trait are significantly different (P < 0.05).
RESULTS AND DISCUSSION
The segment of DNA corresponding to the stigma exsertion QTL influences several aspects of floral morphology:
The L. pennellii LA716-derived introgression line IL2-5 contains pennellii DNA for the region of chromosome 2 predicted to contain the stigma exsertion (se2.1) QTL (Figure 2A). Phenotypic evaluations of the IL2-5 and the M82 controls confirmed this prediction with the IL2-5 line producing flowers with stigmas significantly more exserted (P ≤ 0.0001) compared with the M82 control (Figure 1, B and C). Moreover, it was determined that stigma exsertion in IL2-5 is largely due to an increase in style length, and, to a lesser degree, a decrease in stamen length (Figure 1, B and C). The difference in style length between M82 and IL2-5 was highly significant in both years of evaluation. However, while the difference in stamen length was also observed in both years, it was statistically significant only in the 2001 trial and hence may be more environmentally sensitive.
M82 and IL2-5 also differ for stamen architecture, with M82 having fused, straight anthers and IL2-5 having more loosely connected dehiscent anthers with distinctive outward curvature at the tips of the anthers—a characteristic that further enhances stigma exsertion (Figure 1A). This anther dehiscence character displayed by IL2-5 is unique to L. pennellii. All other species in the genus Lycopersicon have fused anthers (Muller 1940). Anther dehiscence is a key taxonomic character used to distinguish Lycopersicon species from other solanaceous species including species in the closely related and paraphyletic genus Solanum (Muller 1940). In fact, L. pennellii was originally placed in the Solanum genus rather than in the Lycopersicon genus largely because of its unique anther dehiscence trait (Correll 1962). The fact that IL2-5 produces dehiscent anthers like L. pennellii indicates that the genetic cause of this trait maps to the segment of genome represented by the IL2-5 introgression and, as will be shown shortly, is likely attributable to a single gene.
Genetic dissection of the stigma exsertion-containing region of chromosome 2:
To fine map the se2.1 QTL, a large F2 derived from a cross between IL2-5 and M82 was screened with molecular markers (TG469 and TG167) that bracket the region thought to encompass the QTL (Bernacchi and Tanksley 1997; Fulton et al. 1997; Figure 2A). From 1535 F2 plants, 123 recombinants were confirmed. Further genotyping of the recombinant individuals with 40 markers known to be in the interval allowed ordering of those markers and construction of a high-resolution genetic map for the region (Figure 2B). The marker order in this study is consistent with the previous published map; however, due to the very large number of F2 plants examined, the map resolution is greatly enhanced in this area (Tanksley et al. 1992; Fulton et al. 2002; http://www.sgn.cornell.edu/). The major difference between the earlier published high-density map and the current study is that the genetic distance between TG469 and TG167 is 4 cM in this study and is approximately ninefold reduced compared with the earlier study (Tanksley et al. 1992). It is a well-documented phenomenon that recombination is reduced in the regions of the genome introgressed from wild species (Rick 1969; Alpert et al. 1995; Fridman et al. 2000).
A total of 123 F4 homozygous recombinant families were phenotypically evaluated for stigma exsertion, style length, ovary length, stamen length, and anther dehiscence (Figure 1A) and, for statistical analyses, were divided into 27 groups (PE1–PE14 and EP1–EP13; Figure 2C) based on their marker genotypes. Anther dehiscence was recorded at the degree to which the anthers splayed apart at the tip of the anther cone and is highly correlated with the ease with which the anthers separate from one another (Figure 1A). PE groups represent the recombinants containing L. pennellii DNA from the proximal (centromeric) end of the introgression (TG493) up to the recombination point. EP groups represent the recombinants containing L. pennellii DNA from the distal (telomeric) end of the introgression (CT59) down to the recombination point (Figure 2A). A statistical comparison of each group with the M82 and IL2-5 controls allowed fine mapping of each trait in the TG469-TG167 interval (Figure 2C).
On the basis of analysis of these recombinants it was possible to deduce that a major genetic determinant of stigma exsertion resides in the 0.23-cM interval bounded by markers T1301 and CT9 (Figure 2). In support of this hypothesis are the following observations. Recombinants in the PE1-7 intervals all displayed stigmas significantly less exserted than those of the IL2-5 parent, but similar to those of the M82 parent (Figure 2C). Conversely, recombinants in the PE9-12 intervals had stigma exsertion values similar to those of the IL2-5 parent, but significantly greater than those of the M82 parent (Figure 2C). Examination of the reciprocal recombinants revealed that most of EP1-7 recombinants had stigmas significantly more exserted than those of M82 and similar to those of IL2-5; and conversely most of EP9-12 recombinants had stigmas significantly less exserted than those of IL2-5 and similar to those of M82 (Figure 2C). Moreover, the major cause of the stigma exsertion mapping to the TG469-TG167 interval appears to be style length, since the major determinant of style length also maps precisely to the same interval as stigma exsertion (Figure 2C). Hence, we refer to this locus as style 2.1.
Fine mapping of the style 2.1 locus:
As described above, there is strong evidence for a locus (style 2.1) in the T1301-CT9 interval controlling style length, which is a major cause of stigma exsertion. To further characterize and fine map this locus, more detailed evaluations were made for all recombinants in the T1301-CT9 interval. To have sufficient data for statistical comparisons, plants with a crossover in the T1301-CT9 interval were subjected to further replications for phenotypic measurements in the field in 2001 and 2002 (Figure 3, B and C; see materials and methods).
In total, 13 F4 lines, with independent crossovers in the T1301-CT9 interval, were evaluated over the 2 years—a subset of 5 lines in 2001 and all 13 lines in 2002. Five of these lines correspond to the 4 PE8 lines (TA1868, TA1876, TA1879, and TA1882) and 1 EP8 line (TA1996) represented in Figure 2C. The other 8 lines came from further recombinant screening (Figure 3, A and C). Two F4 lines (TA3025 and TA3028) were from the same F2 population generating recombinants for trial 2000. The other 6 F4 lines (TA3031, TA3034, TA3037, TA3040, TA3043, and TA3046) were obtained from a screen of an additional F2 population. Each recombinant F4 line, along with the two parental controls, was represented by 18 single-plant replications. Each plant was evaluated for stigma exsertion, style length, ovary length, and stamen length as described in materials and methods.
Phenotypic values for all recombinants were statistically compared with the M82 and IL2-5 controls for both 2001 and 2002 data (Figure 3). Recombinant lines TA1868, TA3031, and TA3025 contain pennellii DNA from the proximal (centromeric) end of the introgression up to the crossover point represented by TA3025 (Figure 3B). All of these lines had style lengths similar to those of M82 but significantly shorter than those of IL2-5 in one or both years (Figure 3B). These results indicate that style2.1 is located distal to marker LE136C06R and the crossover point represented by TA3025 (Figure 3B). Recombinant lines TA3034, TA3037, TA1879, TA1876, and TA1882 contain pennellii DNA extending distally (toward the telomere) from the TA3025 crossover point. All of these lines produced styles similar in length to those of IL2-5 but significantly longer than those of M82 (Figure 3B). These combined results indicate that style2.1 is located in the interval bounded by markers LE136C06R and LE27H05R and between the crossover points represented by lines TA3025 and TA3034 (Figure 3B). This conclusion is reinforced by examining the reciprocal (EP) recombinants. For example, TA3028 and TA3046 contain pennellii DNA from the distal (telomeric) end of the introgression up to the crossover point represented by TA3046 (Figure 3B). As expected, both of these lines produced styles similar to those of M82 and significantly shorter than those of IL2-5 (Figure 3). TA3043, TA3040, and TA1996 contain pennellii DNA extending proximally (toward the centromere) from the TA3046 crossover point, and all but TA3040 produced styles similar in length to those of IL2-5 and significantly shorter than those of M82. Together, these data support the placement of style2.1 in the LE136C06R-LE27H05R interval (Figure 3). The marker LE136C06R corresponds to the right end of a 120-kb BAC (LE136C06). The marker LE27H05R corresponds to the right end of a second, shorter (65 kb) BAC (LE27H05). On the basis of the sizes of the two BACs and the fact that BAC LE27H05 is encompassed entirely by BAC LE136C06 (data not shown) we estimate that the LE136C06R-LE27H05R interval containing style2.1 is <55 kb (Figure 3A). The fact that the genetic determinant of style length maps as a single locus in a very small genetic and physical interval suggests that this aspect of stigma exsertion is likely controlled by a single gene, rather than by several linked genes.
The long style allele of style 2.1 is dominant to the short style allele:
To determine the gene action of the short and long style alleles of the style2.1 locus, a subNIL line containing pennellii DNA only for the 0.56-cM interval bounded by markers TA1301 and T497 was isolated (see materials and methods for details). From F2 progeny derived from a cross of this subNIL and M82 it was possible to isolate multiple individuals for all three genotypes (homozygous L. esculentum, homozygous L. pennellii, and heterozygous). In 2001, these lines were evaluated in replicated field trials for style length, ovary length, stamen length, and stigma exsertion. Heterozygous individuals produced styles indistinguishable in length from individuals homozygous for pennellii DNA (Table 2). However, individuals homozygous for esculentum DNA produced styles significantly shorter than those of either of the aforementioned genotypes (Table 2). On the basis of these data the gene action (d/a) of the pennellii allele was calculated to be 1.07—a finding indicative of complete dominance of the long style (pennellii) allele (Table 2).
Evidence for several linked loci controlling stamen length:
Stigma exsertion is a composite trait—the interplay between the length of the styles and anthers in a flower. Hence, variation in either stamen length or style length (or both) can affect the degree to which the stigma surface extrudes above the anthers. As described earlier, a comparison of M82 and IL2-5 revealed that the stigma exsertion QTL mapping to chromosome 2 is modulated by both style length and stamen length—the former being the more significant and the latter being more environmentally sensitive (see previous section). The fact that M82 and IL2-5 differ for stamen length indicates one or more genes controlling this trait are located in the region of chromosome 2 encompassed by the IL2-5 introgression.
In an effort to determine the genetic basis of the stamen length difference between IL2-5 and M82, we first examined the phenotypes of recombinants throughout the IL2-5 region (Figure 2). The most striking observation from this data is that recombinants containing pennellii DNA from the proximal (centromeric) end of the introgression (PE) consistently produced stamens shorter than those of their reciprocal counterparts (EP), which had pennellii DNA extending from the distal (telomeric) end of the introgression (Figure 2). A paired t-test (using reciprocal recombinants as pairs—e.g., PE1 vs. EP1, PE2 vs. EP2, etc.) revealed that these differences are highly significant (P < 0.05; Table 3). This result can be explained by two equally likely hypotheses—the existence of a stamen length gene either on the proximal end (beyond marker TG469 and toward the centromere) or on the distal end (beyond marker TG167 and toward the telomere) of the IL2-5 introgression. In the first case (gene on the proximal end) the pennellii allele would need to specify stamens shorter than those of the esculentum allele. In the second case (gene on the distal end), the pennellii allele would need to specify stamens longer than those of the esculentum allele. The fact that IL2-5 has shorter stamens than M82 supports the first hypothesis—a gene on the proximal end for which the pennellii allele specifies shorter stamens. This hypothesis predicts that stocks containing the pennellii allele at the style2.1 locus, but esculentum DNA at the putative stamen length locus proximal to TG469, should have less-exserted stigmas than stocks containing the pennellii allele at both loci. The short introgression stock (TA3178), generated for the evaluation of style2.1 gene action contains pennellii DNA at style2.1, but M82 DNA for TG469 and beyond (see previous section). As predicted, this stock has less-exserted stigmas (−0.002 cm; Table 2) than IL2-5 (0.11 cm; Figure 3B). On the basis of these combined lines of evidence, we hypothesize the existence of a stamen length-controlling locus (hereafter referred to as stamen2.1), located at the proximal end of the introgressed segment and between markers TG469 and TG266 (Figure 2B).
A more detailed analysis of stamen phenotypes for recombinants in the PE8 interval also supports the existence of the stamen2.1 locus and also suggests the existence of two additional stamen length genes closely linked to style 2.1. In the 2001 trial of PE8 recombinants, IL2-5 produced stamens significantly shorter than those of M82 (Figure 3B). In this same year, the PE8 recombinants (with pennellii DNA from the proximal end of the introgression containing the stamen2.1 locus) all produced stamens with lengths similar to those of IL2-5 but much shorter than those of M82. This would be expected if the pennellii allele for stamen2.1 specified short anthers (compared with the esculentum allele). The only exception to this observation was TA1876, which also contains pennellii DNA from the proximal end of the introgression, but has a point of recombination between markers LE27H05L and LE136C06L. This line was unusual in that it produced stamens significantly longer than those of IL2-5 and similar to those of M82, despite the fact that it contains the pennellii allele for stamen2.1 (Figure 3B). We can imagine two genetic models to explain these data—both require the existence one or more stamen length loci in the LE27H05L-LE136C06L interval. The first model hypothesizes the existence of two tightly linked stamen length loci in the interval. We designate these hypothetical loci as stamen2.2 (more proximal) and stamen2.3 (more distal). For stamen2.2, the pennellii allele would need to specify longer stamens than those of the esculentum allele and the reverse would need to be true for the stamen2.3 locus. When these alleles are in cis they would largely balance out each other's effects. However, a recombination between these loci, which would put the pennellii and esculentum alleles in cis could potentially generate longer or shorter stamens. TA1876 is the only stock containing a crossover in the LE27H05L-LE136C06L interval. If the crossover in TA1876 occurred between the hypothetical stamen2.2 and stamen2.3 loci, it would contain a pennellii allele at stamen2.2 in cis with an esculentum allele at stamen2.3—both of which would specify longer stamens, explaining the long stamen phenotype of TA1876.
An alternative explanation for these observations would be for the existence of a single stamen length gene in the LE27H05L-LE136C06L interval. In this model, the crossover in TA1876 would have to result in an inactivation or modified gene that must then cause longer stamens. While this explanation is plausible, it is contingent upon a very low probability event—a crossover in a gene that results in a dysfunctional chimeric gene. An additional piece of information from the gene action study may also support the two-gene (stamen2.2 and stamen2.3) hypothesis. The short introgression stock (TA3178) used for the evaluation of style2.1 gene action contains pennellii DNA at not only style2.1, but also the hypothetical stamen2.2 and stamen2.3 loci. While the gene action for style2.1 was dominant (which is consistent with a single gene), stamen length showed a highly significant overdominant (heterotic) gene action (d/a = 23) that could be explained by two loci (e.g., stamen2.2 and stamen2.3) each with dominant (or partially dominant) alleles in cis with recessive (or partially recessive) alleles (Table 2). This interpretation is consistent with the dominance model for heterosis, which is now supported by a larger body of empirical data (Xiao et al. 1995; Stuber 1997; Monforte and Tanksley 2000). The overdominance effect is not readily explained by the single-gene model. While the issue of one stamen length gene vs. two tightly linked genes in the LE27H05L-LE136C06L interval requires further study, the parsimony principle favors the two-gene model.
Anther dehiscence is controlled by a locus tightly linked to, but separated from style2.1:
IL2-5 produces dehiscent anthers similar to L. pennellii whereas M82 has fused, straight anthers characteristic of the cultivated tomato and other Lycopersicon species (see previous section). On the basis of the analysis of recombinants in the IL2-5 region, it is possible to conclude that the genetic cause of anther dehiscence maps to the TG131-CT1 interval, which is 1.22 cM from the style2.1 locus (Figure 2B). This conclusion is based on the following observations. Recombinants in the PE1-3 intervals produce fused, straight anthers similar to the M82 parent but significantly different from IL2-5, which produces dehiscent, curved anthers (Figure 2C). Conversely, recombinants in the PE4-14 intervals displayed fused, straight anthers, similar to IL2-5 but significantly different from M82 (Figure 2C). Examinination of the reciprocal recombinants revealed that all recombinants in EP1-3 intervals had dehiscent, curved anthers similar to IL2-5 but different from M82. Conversely, all recombinants in EP4-14 intervals had fused, straight anthers similar to the M82 parent but significantly different from IL2-5. We hereafter refer to this stamen architecture locus designated to the TG131-CT1 interval as dehiscence2.1.
The stigma exsertion region of chromosome 2—remnants of a coadapted gene complex?
In the current study, the phenotype of the stigma exsertion QTL originally reported on chromosome 2 may be the result of the action of up to five linked genes, style2.1, stamen2.1, stamen2.2, stamen2.3, and dehiscence2.1—all with potential to affect stigma exsertion and hence mating behavior. These genes affect three different characters/components of stigma exsertion. style 2.1, with by far the largest effect, controls the length of styles. stamen2.1, stamen2.2, and stamen2.3 all affect stamen length; however, from the data presented, it is not possible to determine whether or not these genes exercise their control on stamen growth through common developmental mechanisms. Finally, dehiscence2.1 controls the degree to which anthers adhere to each other and thus form either a straight anther cone (as seen in L. esculentum and most other tomato species) or loosely connected, curved anthers, which might exaggerate stigma exsertion.
The cluster of genes controlling various aspects of stigma exsertion reported herein is reminiscent of “coadapted gene complexes” or “supergenes” conjectured since the early days of genetics. Darlington and Mather (1949) proposed that such groups of genes might cooperatively produce some adapted characteristics through coadaptation if they mechanically held together on a chromosome usually inherited as a unit. Despite the early prediction of coadapted gene complexes, there are still few cases in which the existence of such clusters has been proven in higher eukaryotes. Perhaps the best-known example is that of sex chromosomes in animals (for review see Lahn et al. 2001). Sexual dimorphic traits in gonadal tissue and brain are the adapted characteristics, and the genes on the nonrecombining region between Y and X chromosomes are the coadapted gene complex, which differentially express in male and female in those specific tissues (Lahn et al. 2001; Xu et al. 2002). Another well-known example is the S-locus in Brassica plants (Kachroo et al. 2001; Sato et al. 2002). The sporophytic self-incompatibility is the adapted characteristic and the genes in the S-locus formed a coadapted gene complex in which male components and female components showed the specificity of ligand (male)-receptor (female) interaction (Kachroo et al. 2001).
One of the earliest hypothesized and widely known cases of a coadapted gene complex in plants is that of heterostyly. Heterostyly is similar to morphological sex differentiation in animals. Only in this case, the preference for mating between individuals is governed by discrete polymorphisms for floral morphology [for review see Barrett (1992) and Richards (1997)]. Distyly is the simplest and more widely occurring form of heterostyly and was first described and studied in depth by Darwin (1877) in the genus Primula. With distyly, two types of plants occur in a population, those with long stamens and short styles, and those with short stamens and long styles. Because of the different juxtaposition of stigma and anther, mating via insect pollinators is favored between the two different flower morphs rather than between individuals of the same morph. Genetic and developmental studies have shown that distyly is likely due to tightly linked genes affecting both stamen and style development (Stirling 1932; Ernst 1955; Dorwrick 1956). However, despite the fact that heterostyly is one of the longest known purported examples of a coadapted gene complex in eukaryotes, the genes underlying this phenomenon have not yet been identified.
While the current study was directed toward understanding the genetic and molecular events that facilitate the transition from outcrossing to self-pollinating species, we cannot help noting some striking similarities between the genetic control of this phenomenon and that of heterostyly. We show herein that that modulation of stigma exsertion in tomato is due to a major gene for style length (style2.1) as well as closely linked genes controlling stamen length (stamen2.1, stamen2.2, and stamen2.3) and stamen architecture (dehiscence2.1). The tight linkage of genes controlling style and stamen length is hypothesized to form the genetic basis of the heterostyly coadapted gene complex. Whether the cluster of genes controlling stigma exsertion in tomato shares a common origin with the hypothetical clusters of genes controlling heterostyly remains to be determined. However, we cannot help speculating that this is a possibility and that the heterostyly gene cluster arose early in the evolution of flowering plants and has been maintained in at least some lineages with impact on mating behavior within species (as the case with heterostyly) as well as on evolution of mating system and reproductive isolation between species (as the case with stigma exsertion in the tomato genus).
Acknowledgments
We thank Dani Zamir for generously providing plant materials to conduct this experiment, Nick van Eck for great assistance with the greenhouse and fieldwork, and Amy Frary for helpful comments on the manuscript. This work was supported by grants from the National Science Foundation (DBI-9872617), the U.S. Department of Agriculture Plant Genome Program (97-35300-4384), and the U.S.-Israel Binational Agriculture Research and Development Fund (IS-3009-98C) to S.D.T. and K.-Y.C. and a scholarship from the Taiwan Ministry of Education to K.-Y.C. This work represents partial fulfillment of requirements for the Ph.D. degree for K.-Y.C.
References
- Alpert, K. B., S. Grandillo and S. D. Tanksley, 1995. fw2.2: a major QTL controlling fruit weight is common to both red- and green-fruited tomato species. Theor. Appl. Genet. 91: 994–1000. [DOI] [PubMed] [Google Scholar]
- Barrett, S. C. H., 1992 Heterostylous genetic polymorphisms: model systems for evolutionary analysis, pp.1–29 in Evolution and Function of Heterostyly, edited by S. C. H. Barrett. Springer-Verlag, Berlin.
- Barrett, S. C. H., 2002. The evolution of plant sexual diversity. Nat. Rev. Genet. 3: 274–284. [DOI] [PubMed] [Google Scholar]
- Bernacchi, D., and S. D. Tanksley, 1997. An interspecific backcross of Lycopersicon esculentum × L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147: 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budiman, M. A., L. Mao, T. C. Wood and R. A. Wing, 2000. A deep-coverage tomato BAC library and prospects toward an STC framework for genome sequencing. Genome Res. 10: 129–136. [PMC free article] [PubMed] [Google Scholar]
- Correll, D. S., 1962 The Potato and Its Wild Relatives. Texas Research Foundation, Renner, TX.
- Darlington, C. D., and K. Mather, 1949 The Elements of Genetics. Allen & Unwin, London.
- Darwin, C., 1877 The Different Forms of Flowers on Plants of the Same Species. John Murray, London.
- de Nettancourt, N., 1977 Monographs on Theoretical and Applied Genetics 3: Incompatibility in Angiosperms. Springer-Verlag, Berlin.
- Dorwrick, V. P. J., 1956. Heterostyly and homostyly in Primula obconica. Heredity 10: 219–236. [Google Scholar]
- Ernst, A., 1955. Self-fertility in monomorphic Primulas. Genetica 27: 391–448. [Google Scholar]
- Eshed, Y., and D. Zamir, 1995. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, E., T. Pleban and D. Zamir, 2000. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. USA 97: 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton, T. M., J. Chunwongse and S. D. Tanksley, 1995. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 13: 207–209. [Google Scholar]
- Fulton, T. M., T. Beck-Bunn, D. Emmatty, Y. Eshed, J. Lopez et al., 1997. QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTLs found in other wild species. Theor. Appl. Genet. 95: 881–894. [Google Scholar]
- Fulton, T. M., R. Van der Hoeven, N. T. Eannetta and S. D. Tanksley, 2002. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14: 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiady, M. S., R. W. Whitkus and E. M. Lord, 2002. Genetic analysis of traits distinguishing outcrossing and self-pollinating forms of currant tomato, Lycopersicon pimpinellifolium (Jusl.) Mill. Genetics 161: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A., C. R. Schopfer, M. E. Nasrallah and J. B. Nasrallah, 2001. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293: 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kalisz, S., D. Vogler, B. Fails, M. Finer, E. Shepard et al., 1999. The mechanism of delayed selfing in Collinsia verna (Scrophulariaceae). Am. J. Bot. 86: 1239–1247. [PubMed] [Google Scholar]
- Karron, J. D., R. T. Jackson, N. N. Thumser and S. L. Schlicht, 1997. Outcrossing rates of individual Mimulus ringens genets are correlated with anther-stigma separation. Heredity 79: 365–370. [Google Scholar]
- Lahn, B. T., N. M. Pearson and K. Jegalian, 2001. The human Y chromosome, in the light of evolution. Nat. Rev. Genet. 2: 207–216. [DOI] [PubMed] [Google Scholar]
- Levin, I., A. Cahaner, H. D. Rabinowitch and Y. Elkind, 1994. Effects of the ms10 gene, polygenes and their interaction on pistil and anther-cone lengths in tomato flowers. Heredity 73: 72–77. [Google Scholar]
- McClure, B. A., V. Haring, P. R. Ebert, M. A. Anderson, R. J. Simpson et al., 1989. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342: 955–957. [DOI] [PubMed] [Google Scholar]
- Miller, J. C., and S. D. Tanksley, 1990. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 80: 437–448. [DOI] [PubMed] [Google Scholar]
- Monforte, A. J., and S. D. Tanksley, 2000. Fine mapping of a quantitative trait locus (QTL) from Lycopersicon hirsutum chromosome 1 affecting fruit characteristics and agronomic traits: breaking linkage among QTLs affecting different traits and dissection of heterosis for yield. Theor. Appl. Genet. 100: 471–479. [Google Scholar]
- Motten, A. F., and J. Antonovics, 1992. Determinants of outcrossing rate in a predominantly self-fertilizing weed, Dautura stramonium (Solanaceae). Am. J. Bot. 79: 419–427. [PubMed] [Google Scholar]
- Motten, A. F., and J. L. Stone, 2000. Heritability of stigma position and the effect of stigma-anther separation on outcrossing in a predominantly self-fertilizing weed, Dautura stramonium (Solanaceae). Am. J. Bot. 87: 339–347. [PubMed] [Google Scholar]
- Muller, C. H., 1940 A Revision of the Genus Lycopersicon. Misc. Pub. 382, U.S. Department of Agriculture, Washington, DC.
- Palmer, J. D., and D. Zamir, 1982. Chloroplast DNA evolution and phylogenetic relationships in Lycopersicon. Proc. Natl. Acad. Sci. USA 79: 5006–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, A. J., 1997 Plant Breeding Systems, Ed. 2, pp. 242–296. Chapman & Hall, London.
- Rick, C. M., 1963. Biosystematic studies on Galápagos tomatoes. Occas. Pap. Calif. Acad. Sci. 44: 59–77. [Google Scholar]
- Rick, C. M., 1969. Controlled introgression of chromosomes of Solanum pennellii into Lycopersicon esculentum: segregation and recombination. Genetics 62: 753–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, C. M., 1982. Genetic relationship between self-incompatibility and floral traits in tomato species. Biol. Zent. Bl. 101: 185–198. [Google Scholar]
- Rick, C. M., 1988 Evolution of mating systems in cultivated plants, pp. 133–147 in Plant Evolutionary Biology, edited by L. D. Gottlieb and S. K. Jain. Chapman & Hall, London.
- Rick, C. M., 1995 Tomato, pp. 452–457 in Evolution of Crop Plants, edited by J. Smartt and N. W. Simmonds. Longman Scientific & Technical, New York.
- Rick, C. M., and W. H. Dempsey, 1969. Position of the stigma in relation to fruit setting of tomato. Bot. Gaz. 130: 180–186. [Google Scholar]
- Rick, C. M., and R. Lamm, 1955. Biosystematic studies on the status of Lycopersicon chilense. Am. J. Bot. 47: 663–675. [Google Scholar]
- Rick, C. M., and S. D. Tanksley, 1981. Genetic variation in Solanum pennellii: Comparisons with two other sympatric tomato species. Plant Syst. Evol. 139: 11–45. [Google Scholar]
- Rick, C. M., E. Kesicki, J. F. Fobes and M. Holle, 1976. Genetic and biosystematic studies on two new sibling species of Lycopersicon from Interandean Perú. Theor. Appl. Genet. 47: 55–68. [DOI] [PubMed] [Google Scholar]
- Rick, C. M., J. F. Fobes and M. Holle, 1977. Genetic variation in Lycopersicon pimpinellifolium: evidence of evolutionary change in mating systems. Plant Syst. Evol. 127: 139–170. [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sato, K., T. Nishio, R. Kimura, M. Kusaba, T. Suzuki et al., 2002. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics 162: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer, C. R., M. E. Nasrallah and J. B. Nasrallah, 1999. The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Scott, J. W., and W. L. George, 1978. Breeding and combining ability of heterostylous genotypes for hybrid seed production in Lycopersicon esculentum Mill. Euphytica 29: 135–144. [Google Scholar]
- Stein, J. C., B. Howlett, D. C. Boyes, M. E. Nasrallah and J. B. Nasrallah, 1991. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88: 8816–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling, J., 1932. Studies of flowering in heterostyled and allied species. Part I. The Primulaceae. Publ. Hartley Bot. Lab. 8: 3–42. [Google Scholar]
- Stuber, C. W., 1997 Case history in crop improvement: yield heterosis in maize, pp. 197–205 in Molecular Dissection of Complex Traits, edited by A. H. Paterson. CRC Press, Boca Raton, FL.
- Tanksley, S. D., and F. Loaizafigueroa, 1985. Gametophytic self-incompatibility is controlled by a single major locus on chromosome 1 in Lycopersicon peruvianum. Proc. Natl. Acad. Sci. USA 82: 5093–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S. D., M. W. Ganal, J. P. Prince, M. C. de Vicente, M. W. Bonierbale et al., 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, S. S., J. Jiang, B. S. Gill, A. H. Paterson and R. A. Wing, 1994. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 22: 4922–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J. H., J. M. Li, L. P. Yuan and S. D. Tanksley, 1995. Dominance is the major genetic basis of the heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., P. S. Burgoyne and A. P. Arnold, 2002. Sex differences in sex chromosome gene expression in mouse brain. Human Mol. Genet. 11: 1409–1419. [DOI] [PubMed] [Google Scholar]