Abstract
Most sexually reproducing organisms depend on the regulated formation of crossovers, and the consequent chiasmata, to accomplish successful segregation of homologous chromosomes at the meiosis I division. A robust, chromosome-wide crossover control system limits chromosome pairs to one crossover in most meioses in the nematode Caenorhabditis elegans; this system has been proposed to rely on structural integrity of meiotic chromosome axes. Here, we test this hypothesis using a mutant, him-3(me80), that assembles reduced levels of meiosis-specific axis component HIM-3 along cohesin-containing chromosome axes. Whereas pairing, synapsis, and crossing over are eliminated when HIM-3 is absent, the him-3(me80) mutant supports assembly of synaptonemal complex protein SYP-1 along some paired chromosomes, resulting in partial competence for chiasma formation. We present both genetic and cytological evidence indicating that the him-3(me80) mutation leads to an increased incidence of meiotic products with two crossovers. These results indicate that limiting the amount of a major axis component results in a reduced capacity to communicate the presence of a (nascent) crossover and/or to discourage others in response.
BIPARENTAL inheritance, a defining feature of sexual reproduction, is achieved by the fusion of two haploid gametes, one derived from each diploid parent. Haploid gametes are generated from diploid germ cells through a special type of cell division, meiosis, which accurately reduces ploidy by half by sorting chromosomes into homologous pairs and then partitioning one member of each pair to opposite poles of a bipolar spindle. In most sexually reproducing organisms, this critical reduction in chromosome number relies on crossing over between DNA molecules of a pair of homologous chromosomes: crossovers lead to formation of chiasmata that hold homologs together and allow them to orient toward opposite poles of the spindle, thereby ensuring proper disjunction (Page and Hawley 2003).
Meiotic crossing over is accomplished during prophase of meiosis I through a carefully choreographed series of chromosome interactions and DNA metabolism steps. Following completion of premeiotic DNA replication, replicated chromosomes begin to condense, and a proteinaceous structure known as the meiotic chromosome axis (or axial element), composed of cohesins and meiosis-specific axis components, assembles between sister chromatids. Homologous chromosomes pair and align along their lengths, culminating in an organization in which the coaligned axes of homologs are connected by coiled-coil proteins in a highly ordered structure known as the synaptonemal complex (SC; Zickler and Kleckner 1999). In some organisms, assembly of SC is tightly coupled to and dependent on initiation of meiotic recombination, which occurs through the enzymatic introduction of double-strand DNA breaks (DSBs; Keeney 2001); in other organisms DSB formation occurs by the same conserved mechanism but SC installation is not dependent on break formation (Dernburg et al. 1998; McKim and Hayashi-Hagihara 1998). In either case, crossing over between homologs results from a specialized recombinational repair program that proceeds in the context of assembled SC (Padmore et al. 1991; Plug et al. 1998; Hunter and Kleckner 2001; Guillon and de Massy 2002; Moens et al. 2002; Colaiacovo et al. 2003; Jang et al. 2003) and is promoted by SC central region proteins (Sym and Roeder 1994; Page and Hawley 2001; Colaiacovo et al. 2003; Börner et al. 2004). Crossovers between homologs in conjunction with sister-chromatid cohesion form the basis of chiasmata, which maintain connections between homologs following SC disassembly until selective release of cohesion distal to the chiasmata at the metaphase-anaphase transition of meiosis I (Buonomo et al. 2000; Page and Hawley 2003).
For successful chiasmate meiosis, it is necessary to ensure the formation of at least one crossover between each homolog pair. Failure to fulfill this requirement results in chromosome missegregation, leading to aneuploid gametes and consequent inviability or developmental abnormalities in resulting embryos (Page and Hawley 2003). Nature seems to have invented two distinct strategies to meet this requirement. One strategy is to make crossovers in sufficiently large numbers such that at least one crossover per homolog pair is likely to occur by apparently stochastic means; Schizosaccharomyces pombe is one of the organisms that employ this strategy (Kohli and Bahler 1994; Munz 1994). The alternative, more commonly used strategy is to make a relatively small number of crossovers (on the order of one to three per chromosome arm) and distribute them in a regulated manner (Jones 1987). Meiotic crossing over in the nematode Caenorhabditis elegans exemplifies an extreme instance of this more widespread strategy. Chiasma number per chromosome pair exhibits a very narrow, nonrandom distribution in this organism: genetic map lengths indicate an average of only one crossover per chromosome pair (WormBase 2004), yet chromosome pairs lacking chiasmata are very rare (<1%; Villeneuve 1994; Dernburg et al. 1998), implying that crossover formation must be governed by robust regulatory mechanisms (Hodgkin et al. 1979; Meneely et al. 2002; Hillers and Villeneuve 2003).
In previous work we investigated the control of meiotic crossing over in C. elegans by examining the meiotic behavior of end-to-end fusions of whole chromosomes (Hillers and Villeneuve 2003). This study revealed a remarkable capacity of the organism to modulate crossing over in response to altered karyotype. We found that in the homozygous state, fusion chromosomes composed of two or three whole chromosomes (encompassing as much as half the genome) typically enjoyed only a single crossover in most meioses, and that when double crossovers did occur, they tended to be widely spaced. The fact that only a single crossover usually occurred over a segment of the genome that would normally receive two or three crossovers implied that the fused chromosomes were being perceived as a single chromosome “unit” by the organism. These results indicated that meiotic crossovers in C. elegans are limited by a particularly robust chromosome-wide interference mechanism that operates to discourage (or interfere with) the likelihood of additional crossovers occurring “nearby” on the same chromosome. Further, our analysis of fusion chromosome heterozygotes suggested that the ability of chromosome-wide crossover control mechanisms to limit crossover formation is dependent on integrity of meiotic chromosome axis structures. We found that fusion chromosomes enjoyed more crossovers when present in a heterozygous state than in a homozygous state in hermaphrodites; we also found that in males heterozygous for a three-chromosome fusion in which two autosomal segments were separated by a partnerless X chromosome segment (which has a different axis organization and does not load SC central region proteins), the two autosomal segments behaved as two independent chromosomes, each receiving a crossover. Together these data suggested that continuity of chromosome axes, and/or structures that depend on axis continuity, plays an important role in crossover regulation in C. elegans.
Crossover interference is a long-recognized and widespread feature of meiosis (Muller 1916; Jones 1987; Carpenter 1988), and the idea that crossover regulation might depend on integrity of meiotic chromosome axes, or on structures that depend on proper axis morphogenesis, has been incorporated into interference models for decades (e.g., Egel 1978; Sym and Roeder 1994; Börner et al. 2004). An inherent impediment to testing potential involvement of meiotic axes in crossover control is the fact that these structures (or their constituent parts) are themselves required to form normal levels of crossovers in many organisms (Mason 1976; Hollingsworth et al. 1990; Rockmill and Roeder 1990, 1991; Leem and Ogawa 1992; Klein et al. 1999; Manheim and McKim 2003) and are completely required for crossing over in C. elegans (Couteau et al. 2004). Thus the usual genetic strategies of eliminating candidate components by mutation also reduce or eliminate the very crossover events whose regulation we wish to study.
Here we investigate the contribution of a conserved meiotic axis component to crossover regulation in C. elegans, taking advantage of a newly identified mutant, him-3(me80), that has allowed us to circumvent this difficulty. HIM-3 is a major component of meiotic chromosome axes in C. elegans (Zetka et al. 1999) and is one of four C. elegans members of the meiosis-specific HORMA (Hop1, Rev7, Mad2) domain protein family (Aravind and Koonin 1998) that includes meiotic axis proteins Saccharomyces cerevisiae Hop1 (Hollingsworth et al. 1990) and Arabidopsis thaliana Asy1 (Caryl et al. 2000). Whereas homolog pairing, SC formation, and interhomolog crossovers are all abolished in the absence of HIM-3 (Couteau et al. 2004), the reduced levels of HIM-3 protein that associate with meiotic chromosomes in the nonnull him-3(me80) mutant support substantial homologous synapsis and crossover formation on the X chromosomes as well as severely reduced but significant levels of synapsis and crossing over on autosomes (Couteau et al. 2004). The fact that the him-3(me80) mutant retains a substantial capacity to form crossovers despite altered chromosome axis composition allowed us to assess the integrity of crossover regulation in this mutant, revealing a role for a conserved chromosome axis component in limiting the number of crossovers per chromosome pair.
MATERIALS AND METHODS
“Green chromosome screen” for mutants with cytological defects in meiotic prophase:
Worms homozygous for integrated transgene ruIs32 expressing histone H2B::GFP under control of a germline promoter (Praitis et al. 2001) were mutagenized with ethyl methanesulfonate following standard procedures and allowed to produce F1 progeny. F1 hermaphrodites were plated individually, allowed to produce F2 progeny, and 20 F2 progeny from each F1 were mounted for microscopy. Slides containing F2 worms were screened for the presence of worms with diakinesis nuclei exhibiting more than six chromatin masses (indicating failure in bivalent formation). Candidate mutants were recovered from plates, outcrossed, reisolated, and tested using fluorescence in situ hybridization (FISH) to identify mutants defective in homolog pairing. The him-3(me80) mutation was identified in this screen and mapped between markers pkP4052 and pkP4058 by the method of Wicks et al. (2001); a complementation test with him-3(e1256) identified me80 as an allele of him-3.
Cytological analysis:
Gonad dissection, fixation for immunostaining, FISH, and imaging using the DeltaVision deconvolution microscopy system were conducted basically as in MacQueen et al. (2002) with minor modifications for immunostaining: gonads were dissected from young adult worms (24 hr post-L4 stage), fixed with 1% formaldehyde for 5 min, and then frozen in liquid nitrogen before immersion in cold (−20°) methanol for 1 min. Slides were washed several times in phosphate-buffered saline containing 0.1% Tween-20 (PBT) for 10 min each and then incubated with 1% bovine serum albumin diluted in PBT for 30 min. A hand-cut paraffin square was used to cover the tissue with 50 μl of antibody solution. Incubation was conducted in a humid chamber overnight at room temperature. Double labeling of SYP-1 (MacQueen et al. 2002) and HIM-3 (Zetka et al. 1999) or REC-8 (Pasierbek et al. 2001) was performed with both rabbit anti-HIM-3 (1:200) or REC-8 (1:100) and guinea pig anti-SYP-1 (1:200) primary antibodies present simultaneously during the first incubation step. Secondary incubations contained a mixture of appropriate secondary antibodies (Alexa-488-labeled anti-rabbit IgG and Alexa-555-labeled anti-guinea pig IgG: Molecular Probes, Eugene, OR), each at 1:400. Data were collected as a series of optical sections in increments of 0.2 μm. Probes were generated from yeast artificial chromosome (YAC) clones as in Zalevsky et al. (1999). The following YACs were used (chromosomal locations in parentheses): Y51E2 [X, extreme left (XL)], Y68A3 [X, extreme right (XR)], Y13H5 [I, left (IL)], Y48E9 [I, right (IR)], Y25B10 [II, left (IIL)], Y26G1 [II, right (IIR)], Y40H8 (IV, right), and Y44F12 (IV, left). Simultaneous FISH and immunostaining were done as in Kelly et al. (2002).
To evaluate pairing and synapsis in comparable regions of wild-type and him-3(me80) germ lines, we subdivided the germ lines into five zones (38-μm width for each) along the distal-proximal axis, beginning ∼10 nuclear diameters from the distal tip and ending at the late pachytene region. Occasionally the length of the germ line accommodated an additional partial zone before the region of pachytene exit; in such cases the small number of nuclei in this zone were included with zone 5, the late pachytene zone. For the quantitative analysis of pairing for chromosomes II and X shown in Figure 2, data are from zone 5. For the simultaneous analysis of FISH and SYP-1 immunostaining in Figure 3 (and for the chromosome I analysis in Figure 2), quantitation was conducted for the region in which SYP-1 stretches showed maximum development in the him-3(me80) mutant; this region corresponds to middle/late pachytene stages in wild type and straddles the boundary between zones 4 and 5 defined above. A z-series of images was collected for the appropriate germ-line zone, and for nuclei completely contained within the data stack, pairwise distances between peak intensities of FISH signals were measured using the softWoRx software package (Applied Precision). FISH signals were considered paired if the distance between their peak intensities was ≤0.7 μm. For Figure 3, three-dimensional (3-D) image stacks were also examined to determine whether FISH signals overlapped with or were immediately adjacent to SYP-1 signals.
Figure 2.—
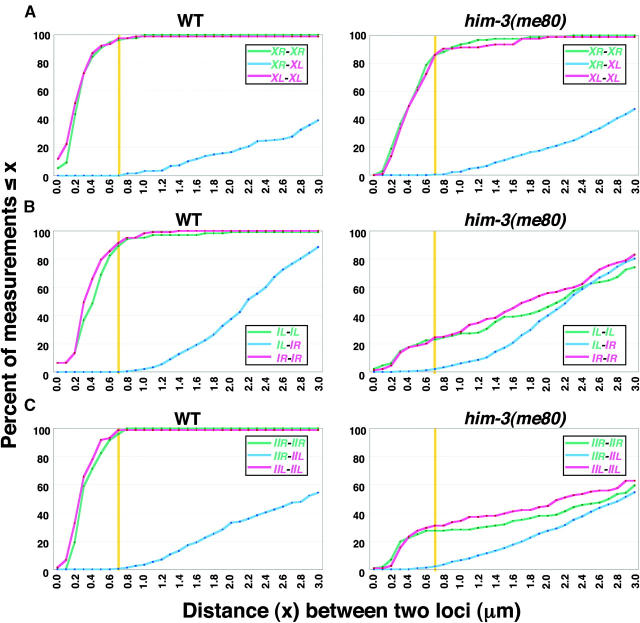
High-resolution quantitative analysis of pairing at X chromosome and autosomal loci. Cumulative distribution graphs plot data from two-probe FISH experiments monitoring pairing at opposite ends of the X chromosome (A), chromosome I (B), and chromosome II (C) in nuclei from the late (A and C) or mid/late (B) pachytene regions of whole-mount germ lines (see materials and methods). In these cumulative distribution plots, distances between each pair of homologous signals (red or green) and distances between the four possible pairwise combinations of heterologous signals (blue) were plotted against the percentage of measurements that were equal to or less than the value indicated on the x-axis. (For each chromosome, the end harboring the cis-acting “pairing center” region is indicated in red, and the opposite end is in green.) A yellow vertical line indicates the 0.7-μm threshold distance for considering loci to be paired; the blue plots show that distances between heterologous loci at opposite ends of the same chromosome rarely fell below this threshold, even when homologous pairing was severely impaired in the him-3(me80) mutant. The numbers of nuclei scored were as follows: for the X chromosome, 76 and 95 nuclei for WT and him-3(me80), respectively; for chromosome I, 103 and 143 nuclei; for chromosome II, 73 and 116 nuclei.
Figure 3.—
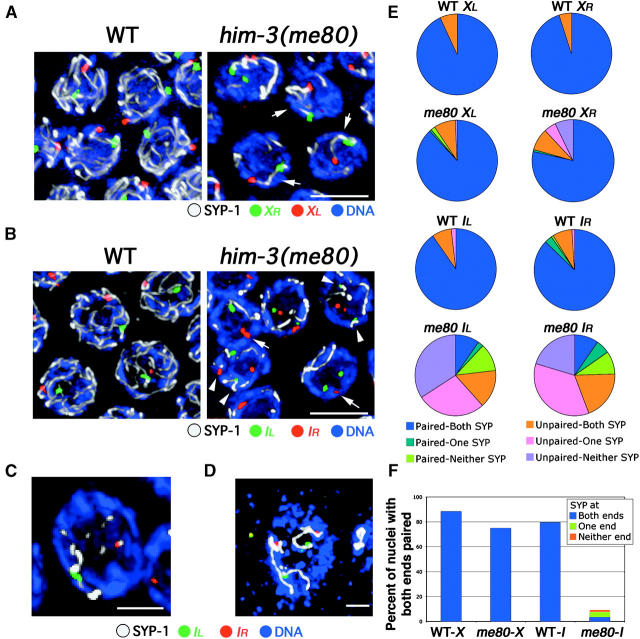
Simultaneous assessment of pairing and synapsis for chromosomes X and I. Analysis of FISH for two loci (red and green) in conjunction with SYP-1 immunostaining (white) on DAPI-stained chromosomes (blue) in mid/late-pachytene nuclei from WT and him-3(me80) hermaphrodites is shown. Images in A–C show nuclei from whole-mount germ lines, and the image in D shows a nucleus from a squash preparation; all images are projections of 3-D data stacks encompassing whole nuclei. (A) FISH for two loci at opposite ends of the X chromosome. Both XL and XR are usually paired and associated with a single continuous stretch of SYP-1 immunostaining (arrows) in the him-3(me80) mutant. Bar, 5 μm. (B–D) FISH for two loci at opposite ends of chromosome I. In B, arrows indicate examples of nuclei in the him-3(me80) mutant where IR FISH signals are paired but are not associated with a stretch of SYP-1; arrowheads indicate examples where unpaired FISH signals associate with SYP-1 stretches. (C) A him-3(me80) nucleus in which the FISH signals at both ends of chromosome I are paired, but are associated with different SYP-1 stretches. (D) A him-3(me80) nucleus in which all four chromosome I FISH signals are unpaired yet are associated with SYP-1 stretches. Bars: B and D, 5 μm; C, 2 μm. (E) Pie charts showing simultaneous quantitation of pairing of FISH signals at a given locus and association of those FISH signals with a stretch of anti-SYP-1 immunostaining. Each pie chart represents data for a single probe in either wild type or the him-3(me80) mutant; between 102 and 143 nuclei were scored in each case. Nuclei were classified with respect to whether FISH signals for the given locus were paired (i.e., the distance between their peak signal intensities was ≤0.7 μm), and nuclei were further classified according to whether both, only one, or neither of the FISH signals for that locus were associated with (i.e., touched or overlapped with) a SYP-1 stretch. One classification requires further explanation: for all probes and all genotypes, a subset of nuclei was classified as “Unpaired-Both SYP.” For all four probes in WT, and for the X probes in him-3(me80), in the vast majority of these cases the distances between the FISH signals fell in the 0.8–1.0 μm range and both signals were associated with the same SYP-1 stretch. For the chromosome I probes in him-3(me80), in contrast, most nuclei in the Unpaired-Both SYP class have the FISH signals far apart (2.3 ± 1.0 μm, n = 50) and associated with distinct SYP-1 stretches. (F) Bar graph incorporating the data for the two FISH probes targeting opposite ends of the same chromosome. Graphs indicate the percentage of nuclei showing homologous pairing at both ends of the indicated chromosome; such nuclei are further classified with respect to whether both, only one, or neither of the two sets of paired FISH signals were associated with SYP-1 staining.
Relative fluorescence intensities for HIM-3 or REC-8 signals on different chromosome segments within a given nucleus were estimated using a Zeiss LSM-510 META confocal microscope. A SYP-1-positive and a SYP-1-negative chromosome axis segment (well separated from the other chromosomes) were chosen for each of three different nuclei. For each chosen segment, the fluorescence intensity corresponding to HIM-3 (or REC-8) immunostaining was measured within a square ∼0.5 μm2 in area centered along the path of the chromosome axis; background fluorescence was measured for a comparable area lacking chromosomes. The average fluorescence intensity of three consecutive sections centered at peak intensity (minus background fluorescence) was used as the fluorescence intensity value for a given area. For each nucleus, we calculated the ratio of signal from SYP-1-positive and SYP-1-negative axis stretches; we then determined the mean ratio: 1.74 (SD 0.2) for HIM-3 and 2.13 (SD 0.35) for REC-8.
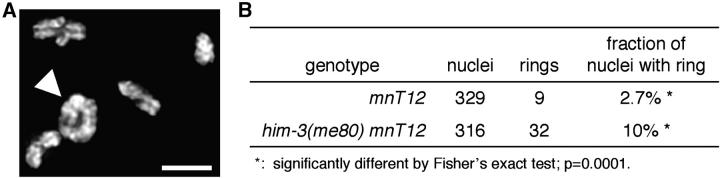
For quantitation of ring bivalents in diakinesis-stage oocytes, adult hermaphrodites (24 hr post-L4 stage) were fixed and stained with DAPI as in Villeneuve (1994), and the most uterus-proximal oocyte nucleus in each gonad arm was scored for the presence of a ring-shaped DAPI-stained body. A bivalent was scored as ring shaped if it was roughly circular in appearance, with an area of reduced staining intensity in the center; as visual detection of a ring bivalent is dependent upon its spatial orientation, frequencies reported are necessarily underestimates.
Western analysis:
A total of ≥200 worms were washed and suspended (1 worm/1 μl) in lysis buffer [10 mm Tris-Cl pH 7.5, 150 mm NaCl, 2 mm EDTA, 1.5 mm EGTA, 0.5 mm Na3VO4, 1.5 mm MgCl2, protease inhibitor cocktail: complete mini (Roche, Indianapolis)] and frozen in liquid nitrogen. Worm suspension was thawed, sonicated, boiled with SDS-PAGE sample buffer, and then separated by 10% SDS-PAGE. Western analysis was performed using standard procedures, using rabbit anti-HIM-3 antibody (1:500) or mouse anti-α-tubulin antibody: DM1A (1:2000; Sigma, St. Louis).
Crossover assay:
Meiotic crossing over was assayed in control and him-3(me80) animals using single-nucleotide polymorphism (SNP) markers, as in Hillers and Villeneuve (2003). Markers and primers used are listed in Table 1. To allow detection of multiply exchanged chromosomes, we assessed recombination only during oocyte meiosis. him-3(me80)/+ males carrying chromosomes X and I derived from the Hawaiian strain CB4856 were mated to him-3(me80)/+ hermaphrodites in the Bristol (N2) background. Among the progeny of this cross were him-3(me80)/him-3(me80) hermaphrodites and control hermaphrodites [him-3(me80)/+ or +/+] that were heterozygous for Bristol- and Hawaiian-derived whole chromosomes I and X. These hermaphrodites were plated individually and mated with Bristol-derived males carrying a chromosomally integrated transgene insertion ccIs4251 expressing GFP under control of the myo-3 promoter, which drives GFP expression in body wall and vulval muscle (Fire et al. 1998); GFP expression was used to identify cross-progeny animals, each of which represents a single meiotic product of an oocyte meiosis. [him-3(me80)/him-3(me80) hermaphrodites produce dead embryos at high frequency, allowing plates containing him-3(me80)/him-3(me80) mothers to be readily distinguished from those containing control mothers.] To assess the genotypes of individual oocyte meiotic products, outcross progeny were picked individually and prepared for SNP analysis as in Wicks et al. (2001), with the following alterations: individuals were lysed in 8 μl of single-worm lysis buffer. Following lysis, samples were diluted with 8–10 μl 10 mm Tris pH 8.0. PCR was performed in a final volume of 15 μl using 0.5 μl of diluted lysate in a 1:2 dilution of TaqPCR Master Mix (QIAGEN, Valencia, CA) through 35 cycles (94°, 20 sec; 60°, 30 sec; 72°, 40 sec), followed by a 10 min extension; amplified products were digested overnight with appropriate restriction enzymes and analyzed on 2.5% agarose gels. Marker content of the chromosome X or I contributed by the oocyte was inferred from the genotype of the tested progeny. All animals received a Bristol chromosome I and nearly all (see below) received either a Bristol X or no X from the sperm of their male parent. Thus either males or hermaphrodites that were homozygous for a Bristol allele at a given chromosome I marker locus and hermaphrodites that were homozygous for a Bristol allele at a given X chromosome marker locus were inferred to have received a Bristol allele from the oocyte meiotic product contributed by their hermaphrodite parent (“mother”). Likewise, animals that were heterozygous (Bristol/Hawaiian) at a given marker locus were inferred to have received a Hawaiian allele from their mother. XO male progeny are hemizygous for all X chromosome markers and thus marker content could be scored directly. Virtually all XO male cross-progeny from control parents and the vast majority of XO males from him-3(me80) parents inherited their single X chromosome from their mothers, but ∼5–10% of XO male cross-progeny derived from him-3(me80) mothers were expected to have inherited their X chromosome from their father owing to loss of the maternal X through segregation errors during oocyte meiosis; such patroclinous XO males would contain only Bristol alleles at all X chromosome marker loci. However, the majority of XO males scored in both him-3(me80) (78%) and control (72%) data sets were derived from parents in which the Bristol-derived X chromosome also contained one or two visible markers, allowing us to recognize and exclude these patroclinous X chromosomes. Since patroclinous XO males represented 10% of the him-3(me80)-derived XO males in such experiments, we excluded a proportional number of X chromosomes with only Bristol alleles (four total) from the XO male data derived from him-3(me80) mothers that did not carry visible markers. Any animal displaying segregation of SNP markers indicative of a double-crossover chromosome was retested by repeating PCR analysis and digestion for each marker.
TABLE 1.
SNP alleles and corresponding primers
| Digested product size (bp)
|
|||||
|---|---|---|---|---|---|
| Chromosome | SNP | Primer sequence | Enzyme | Bristol | Hawaiian |
| pkP1006 | 5′-AGAAATTGAGGAGAACCTCG-3′ | DraI | 311 | 262, 50 | |
| 5′-TTTGTTCTCGGATCTCACG-3′ | |||||
| pkP1016 | 5′-GATCCGTGAAATTGTTCCG-3′ | BsrI | 440, 125 | 364, 125, 76 | |
| 5′-GACAATGACCAATAAGACG-3′ | |||||
| I | pkP1057 | 5′-CTGAACTAGTCGAACAAACCCC-3′ | NdeI | 594 | 294, 300 |
| 5′-ATCATTCTCCAGGCCACGTTAC-3′ | |||||
| pkP1069 | 5′-AGGAGCAGTAGAGTCTGAAACG-3′ | MluNI | 342, 292 | 207, 96, 63 | |
| 5′-GAGGTACAGAAAATGCTGCC-3′ | |||||
| pkP1072 | 5′-CAACAAAGGGATAGATCACGGG-3′ | HindIII | 450 | 236, 214 | |
| 5′-CACAAGTGGTTTGGAAGTACCG-3′ | |||||
| pkP6100 | 5′-TGGCAAAACACATCCCTGTG-3′ | BspHI | 208, 156 | 364 | |
| 5′-GGTATCCGATCCCTTCAACAAG-3′ | |||||
| pkP6103 | 5′-TGTCTAGTTCAAAAGCCCGG-3′ | MseI | 270, 215, 111, 74 | 381, 215, 74 | |
| 5′-TTGTAGCAGATCCTACCCTACC-3′ | |||||
| X | pkP6110 | 5′-CTCACTCTGGTCTTTTTCCG-3′ | EcoRI | 516, 259 | 776 |
| 5′-TTTCTTGACACCTCCGGTAG-3′ | |||||
| pkP6123 | 5′-TTCCTCTCACATGCACAGC-3′ | EcoRV | 469 | 303, 169 | |
| 5′-TTCTCAACGCCCCTCTATG-3′ | |||||
| pkP6093 | 5′-CCTGGGAATCCGTTTTCTCC-3′ | HincII | 330, 210 | 540 | |
| 5′-TAGATATCGTGGAACCCC-3′ | |||||
Primers and restriction enzymes for amplification and detection of SNP alleles used in this study are shown. Primer sequences are from Stephen Wicks (Wicks et al. 2001; S. Wicks, personal communication). For each allele, digestion of the amplified product using the designated restriction enzyme yields products of the specified size.
On the basis of our analysis of SNP marker content alone, we cannot formally exclude the possibility that a given apparent double crossover (DCO) might represent a noncrossover gene conversion event that occurred on an otherwise single crossover (SCO) or noncrossover (NCO) chromatid. However, there were many cases in the five-marker analysis where we could unambiguously identify one crossover event (classes 2, 3, 2 + 4, 1 + 3), and in the him-3(me80) mutant these events were accompanied by a second apparent crossover event >10% of the time; frequencies of noncrossover gene conversion at a given base pair in C. elegans are estimated to be on the order of 10−5 (Waterston et al. 1982; Bullerjahn and Riddle 1988), so we think it very unlikely that an increase in gene conversion frequency could account for the high incidence of this class of apparent DCOs. Moreover, in some experiments the parental Bristol-derived X chromosome carried a recessive visible marker between SNP markers “a” and “b” (interval 1) that could be scored in XO male progeny; scoring of this marker verified two of the 1 + 3 class as unambiguous DCO products. Finally, our cytological data provide a compelling, independent demonstration of an increased incidence of bona fide DCOs: the elevated level of ring bivalents (indicative of widely separated chiasmata on a single pair of chromosomes) observed in the him-3(me80) mutant is readily explained by an increase in DCO events but cannot be accounted for by an increase in noncrossover gene conversion events.
RESULTS
Status of meiotic chromosome structures in the him-3(me80) mutant:
HIM-3 is a major component of meiotic chromosome axes required both for establishment of homolog pairing and for assembly of the SC central region (Zetka et al. 1999; Colaiacovo et al. 2003; Couteau et al. 2004). HIM-3 is composed mainly of a HORMA domain, a motif found in several proteins involved in genome maintenance, most notably spindle checkpoint protein Mad2. In the course of a cytological screen for mutants with meiotic prophase defects (see materials and methods) we identified him-3(me80), a missense allele that results in an R to H substitution at residue 54 of HIM-3 (Couteau et al. 2004); this residue corresponds to R35 of human Mad2, which forms a salt bridge with E98 in the interior of the HORMA domain (Luo et al. 2000). Because these two residues are invariant among all HORMA domain proteins, which are predicted to share a similar tertiary structure (Aravind and Koonin 1998), the amino acid substitution in the him-3(me80) mutant is expected to weaken or disrupt a conserved salt bridge, thereby destabilizing protein structure. In this mutant, substantially reduced levels of HIM-3 were found localized to meiotic chromosome axes, accompanied by a few stretches of SC central region component SYP-1 (MacQueen et al. 2002; Couteau et al. 2004).
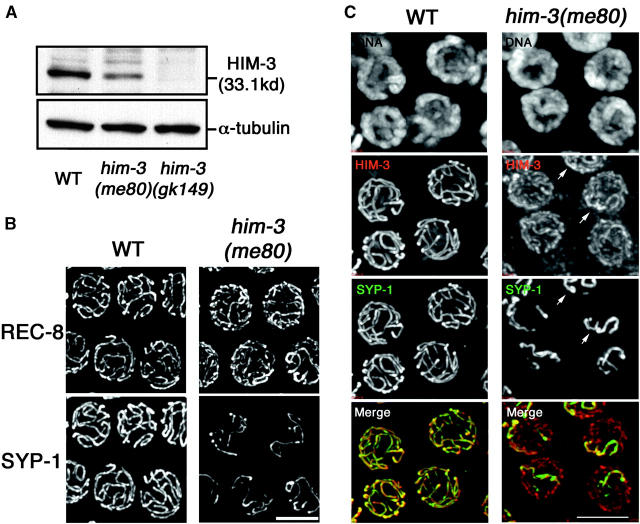
Figure 1 illustrates features of meiotic chromosome organization in the him-3(me80) mutant in greater detail. Figure 1B shows simultaneous immunolocalization of SYP-1 and REC-8, a meiosis-specific cohesin component that is concentrated at meiotic chromosome axes and is required for normal loading of HIM-3 (Pasierbek et al. 2001; Colaiacovo et al. 2003). In wild-type nuclei at the pachytene stage of meiotic prophase, mature SC extends along the full lengths of aligned homolog pairs; since the resolving power of the light microscope does not permit discrimination between chromosome axis and SC central region structures in this context, REC-8 and SYP-1 appear fully colocalized in extended contiguous linear stretches. In the him-3(me80) mutant, REC-8 also successfully localizes to meiotic chromosome axes. However, REC-8 stretches are more numerous than in wild-type nuclei, reflecting the fact that many axes (or axis segments) are not engaged in synapsis, and SYP-1 is detected only on a subset of REC-8 stretches. Figure 1C shows meiotic nuclei costained with anti-HIM-3, anti-SYP-1 antibody, and DAPI. In the wild-type pachytene nuclei, HIM-3 and SYP-1 are extensively colocalized in long contiguous linear stretches at the interface between aligned parallel tracks of DAPI staining, corresponding to fully aligned and synapsed homologs. In the him-3(me80) mutant, linear HIM-3 stretches are also observed, but signal intensities are much weaker than in wild type (Figure 1C), reflecting the reduced abundance of HIM-3 protein (Figure 1A); this reduced level of HIM-3 immunostaining is widely distributed among all chromosomes. A subset of chromosomal regions exhibits brighter-appearing HIM-3 staining; these brighter HIM-3 signals coincide with stretches of SYP-1 staining. We can frequently resolve a pair of parallel tracks of DAPI signals flanking regions of HIM-3 SYP-1 colocalization, suggesting that they represent regions where SC has assembled between a pair of closely juxtaposed axis segments. Quantitative measurements of relative fluorescence signal strengths support this interpretation; HIM-3 signal intensities in the him-3(me80) mutant were roughly twofold higher for regions with SYP-1 staining than for regions without SYP-1 staining (see materials and methods); similar results were obtained for REC-8 SYP-1 double-staining experiments.
Figure 1.—
Cohesin-containing chromosome axes with reduced levels of HIM-3 protein support limited synapsis in the him-3(me80) mutant. (A) Western blots containing lysates from equal numbers of adult wild-type (WT) worms, him-3(me80) worms, and worms carrying the deletion allele him-3(gk149) (Couteau et al. 2004), probed with anti-HIM-3 and anti-α-tubulin antibodies. As the him-3(me80) mutation resides outside the fragment used to produce the anti-HIM-3 antibody, the reduction in the 33-kD HIM-3 band in the him-3(me80) lane reflects reduced levels of HIM-3 protein rather than absence of epitopes. (B and C) Immunostaining of REC-8 and SYP-1 (B) and HIM-3 and SYP-1 (C) in nuclei from the mid-pachytene regions of whole-mount WT and him-3(me80) germ lines. Images are projections of 3-D data stacks encompassing whole nuclei. In B, him-3(me80) shows more numerous stretches of REC-8 and fewer stretches of SYP-1 than seen in WT. In C, HIM-3 and SYP-1 immunostaining are shown in conjunction with DAPI staining of chromatin. For the him-3(me80) panels, the detection threshold for the anti-HIM-3 signal was lowered to permit imaging of the greatly reduced HIM-3 signal intensity in this mutant; consequently the background appears higher than in WT in this channel. The reduced HIM-3 signal is broadly distributed along all chromosomes in the him-3(me80) mutant; SYP-1 stretches coincide with a subset of HIM-3 stretches, typically including those with brighter HIM-3 signals (arrows). In the merged image, HIM-3 is shown in red and SYP-1 is shown in green. Bars, 5 μm.
X. chromosomes exhibit extensive, contiguous homologous synapsis in the him-3(me80) mutant:
An initial study showed that the him-3(me80) mutant is proficient for pairing at the left end of the X chromosome (Couteau et al. 2004), raising the possibility that the X chromosomes might be competent to achieve homologous synapsis despite the scarcity of HIM-3 protein on chromosome axes. Thus, we performed an in-depth analysis of X chromosome pairing and synapsis, using FISH probes targeting both ends of the chromosome and imaging FISH signals in combination with anti-SYP-1 immunostaining. This analysis demonstrated that the him-3(me80) mutant is competent to assemble SC along the full lengths of coaligned X chromosome pairs.
Figure 2A presents FISH data for a two-probe experiment in which pairing of loci at opposite ends of the X chromosome was assessed simultaneously using two probes labeled with different chromophores. The graphs report on pairing in the late pachytene region of the germ line, where maximum levels of pairing and synapsis are observed. In each nucleus, we measured distances between all pairwise combinations of the four possible FISH signals; we then generated cumulative distribution plots in which distances between (1) homologous signals from the XL locus, (2) homologous signals for the XR locus, and (3) all four heterologous distances were each plotted against the percentage of measurements that were ≤x (see materials and methods). On the basis of criteria established in previous studies (Dernburg et al. 1998), homologous loci were considered paired if the measured distance between their peak signal intensities was ≤0.7 μm. In wild-type late pachytene nuclei, >95% of distances between homologous signals were within 0.7 μm, reflecting the fact that the X chromosomes were closely juxtaposed. In the him-3(me80) mutant, the percentages of homologous distance measurements that were within 0.7 μm were only modestly lower than those seen in wild type (86% for both XL and XR), indicating a high level of success in achieving homologous alignment of the X chromosomes. The graphs in Figure 2A reveal an additional feature of the data: among measurements in the ≤0.7-μm range, the distribution of distances was significantly shifted toward larger values in the him-3(me80) mutant compared with the wild-type control (P < 0.0001 using two-tailed Mann-Whitney test). This indicates that although most X chromosome pairs are fully aligned in the him-3(me80) mutant, the associations between them may be somewhat less intimate and/or uniform than during wild-type meiosis.
Figure 3, A, E, and F, shows data from experiments in which SYP-1 immunostaining was performed in conjunction with X chromosome FISH, which revealed that most X chromosome FISH signals in the him-3(me80) mutant were associated with SYP-1 stretches. Moreover, for most X chromosomes, SYP-1 staining was continuous between FISH signals representing opposite ends, indicating that these X chromosomes were synapsed along their entire lengths. Several aspects of X chromosome synapsis in the him-3(me80) mutant differed from the wild type, however (Figure 3, E and F). In wild type, FISH signals were always associated with SYP-1 stretches, including the small fraction that were scored as “unpaired” because the distance between them exceeded the stringent 0.7-μm threshold (75% of this class had distances in the 0.8- to 1.0-μm range, suggesting that chromosomes were in fact successfully aligned in this region). In the him-3(me80) mutant, FISH signals unassociated with SYP-1 stretches were seen (in 3.2 and 12.6% of nuclei for XL and XR, respectively). Furthermore, whereas the X chromosomes could be scored unambiguously as “fully synapsed” (both ends paired and associated with SYP-1) in 88% of wild-type nuclei, X chromosomes were scored as fully synapsed in only 75% of him-3(me80) nuclei. Thus, we conclude that the him-3(me80) mutant is highly proficient in assembling extensive SC structure between aligned X chromosomes, although it does so less completely than wild type.
Limited stretches of homologous synapsis for autosomal loci:
Whereas X chromosomes are highly successful at pairing in the him-3(me80) mutant, pairing at autosomal loci is substantially impaired (Couteau et al. 2004). Figure 2, B and C, shows two-probe FISH experiments assessing pairing at opposite ends of chromosomes I and II; all four loci achieved only very modest levels of homologous pairing in the him-3(me80) mutant, with only 20–30% of homologous signal pairs located within 0.7 μm of each other [in all cases the levels of homologous association were significantly greater than the observed incidence of heterologous associations (<3%)]. These data indicate that a significant amount of pairing does occur at autosomal loci in the him-3(me80) mutant, albeit at substantially reduced levels relative to either the wild-type control or the X chromosome in him-3(me80).
To assess the extent of synapsis involving autosomal loci, we imaged FISH signals for both ends of chromosome I in conjunction with SYP-1 immunostaining. Several observations from this analysis are noteworthy. First, for each locus we found that paired FISH signals were associated with SYP-1 stretches in a significant fraction of nuclei (Figure 3, C and E; IL, 12%; IR, 15%); that segments of the chromosome were appropriately paired and synapsed suggested that such regions might be competent to undergo meiotic crossing over. Second, we found evidence for discontinuous synapsis of chromosome I: there was a class of nuclei with homologously paired FISH signals at both ends of the chromosome that were associated with two distinct SYP-1 stretches (Figure 3C), indicating discontinuity of the central region of the SC. Third, and in contrast to what was seen for the X chromosome, FISH signals for the chromosome I loci in him-3(me80) sometimes failed to associate with a SYP-1 stretch in spite of successful pairing (Figure 3B, arrows): paired FISH signals unassociated with SYP-1 staining were observed in 11 and 9% of nuclei for IL and IR, respectively, whereas they occurred in <1% and 1% of wild-type nuclei (Figure 3E). Finally, we found evidence for synapsis between nonhomologous chromosome segments (heterologous synapsis) in him-3(me80), as reported previously for the him-3(vv6) mutant (Couteau et al. 2004). Unambiguously unpaired chromosome I FISH signals (i.e., separated by >1.2 μm) were frequently associated with SYP-1 stretches in him-3(me80) (Figure 3B, arrowheads; Figure 3E), suggesting that these loci are synapsed with a heterologous chromosome segment. In squash preparations, it is obvious that such unpaired loci can be associated with several separate SYP-1 stretches (Figure 3D), supporting the idea that they are engaged in heterologous synapsis.
Competence for chiasma formation in the him-3(me80) mutant correlates with success in pairing and synapsis:
The above analyses demonstrated that the him-3(me80) mutant succeeds in forming some SC between homologously paired chromosome segments, and that it is substantially more competent in doing so for the X chromosomes than for the autosomes. We found that this X/autosome difference in the success of pairing and synapsis correlated with a difference in their proficiency for chiasma formation. We used FISH to determine the identities of individual chromosomes at diakinesis (the last stage of meiotic prophase), when the highly compact state of chromosomes and the large nuclear volume make it possible to resolve six individual bivalents (homolog pairs connected by chiasmata) during wild-type meiosis and to resolve mixtures of bivalents and achiasmate chromosomes (univalents) in a meiotic mutant. In the him-3(me80) mutant, the X chromosomes were unambiguously detected as a bivalent in 82% of nuclei (n = 56), whereas a chromosome II bivalent was detected in 17% of nuclei (n = 52) and a chromosome IV bivalent was detected in 26% of nuclei (n = 38). Further, the total number of DAPI-stained chromatin masses in diakinesis nuclei in the him-3(me80) mutant (9.6 ± 1.2, n = 148) is consistent with that expected if the remaining autosomes undergo chiasma formation at frequencies comparable to those assayed.
Increased incidence of double-crossover meiotic products in the him-3(me80) mutant:
We recently described the operation of a chromosome-wide crossover control system in C. elegans that limits the number of crossovers to one per homolog pair in most meioses (Hillers and Villeneuve 2003), and we presented evidence suggesting that this system requires continuity of axes and/or SC to function properly. The him-3(me80) mutant assembles chromosome axes that are aberrant (i.e., they have reduced levels of HIM-3 protein) and exhibits incomplete and discontinuous synapsis, yet retains a substantial capacity to form crossovers. These properties prompted us to investigate whether the him-3(me80) mutation might disrupt chromosome-wide crossover control. Specifically, we tested whether the him-3(me80) mutant exhibits an increase in the incidence of meiotic products that have enjoyed multiple crossovers. In the experiments described below, we demonstrate that the him-3(me80) mutant is indeed defective in inhibiting the formation of double crossovers.
To detect potential double-crossover events, we generated hermaphrodites that were heterozygous for whole chromosomes X and I derived from the Bristol N2 strain and the Hawaiian isolate CB4856, and we assessed crossing over in individual products of oocyte meiosis by typing SNP markers that differ in these two strain backgrounds (Figure 4; see materials and methods). In this analysis, we compared the spectrum of meiotic products produced by him-3(me80) homozygotes with that produced by a set of control animals composed of their +/+ and him-3(me80)/+ siblings.
Figure 4.—
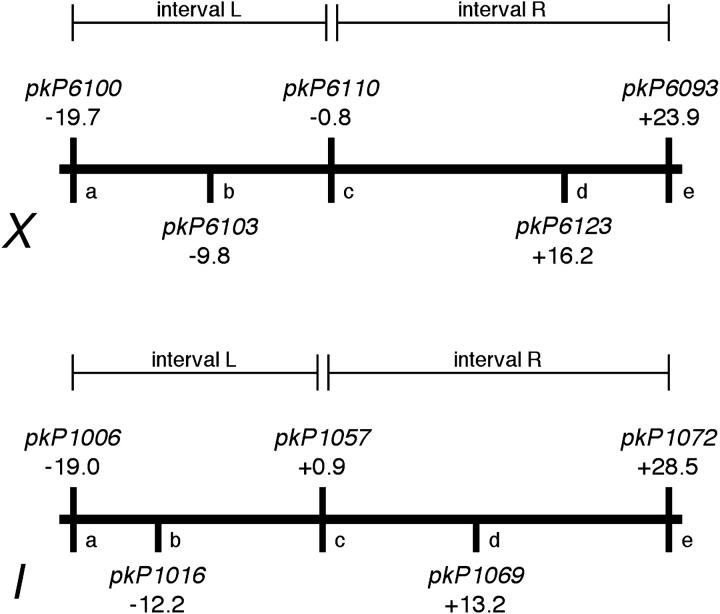
Map positions of SNP markers used in crossover assays. Schematic diagram indicates the markers used in recombination analysis and their corresponding genetic map positions. The markers used and the intervals assessed (L, R) for the three-marker analysis (Table 2) are indicated above the map; the additional markers used for the five-marker analysis and marker nicknames (a–e) used in Table 3 are indicated below the map.
In the analysis presented in Table 2, we sought to detect double crossovers by typing three markers spanning the lengths of each of the two chromosomes assayed, one near each end and one near the middle (Figure 4). In control animals, we measured total recombination frequencies of 47 and 58% for the assayed portions of the X chromosome and chromosome I, respectively. Almost all crossover products had a SCO in either interval “L” (left) or “R” (right), and DCO products were rare, representing ∼1% of all meiotic products. Moreover, both X and I exhibited robust crossover interference in control animals: the coefficients of coincidence (C; observed frequency of double crossovers/expected frequency of double crossovers) were close to 0 (0.07 and 0.08, respectively, for chromosomes X and I), indicating almost complete interference. Further, Fisher's exact test indicated strong statistical support for a lack of independence between crossovers formed in intervals L and R (P < 0.0001 for both chromosomes). These data corroborate previous findings indicating that during wild-type meiosis, C. elegans has a strong propensity to limit crossovers to one per homolog pair.
TABLE 2.
Meiotic crossing over on chromosomesX andI in control andhim-3(me80) animals
| Meiotic products
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Single CO
| ||||||||||
| Chromosome |
him-3 genotype |
Non-CO | Interval L | Interval R | Double CO | N | RF (%)a | Coefficient of coincidenceb |
P-valuec | Double CO ratio (%)d |
| X | Control | 130 | 63 | 50 | 1 | 244 | 47 | 0.07 | <0.0001 | 0.9e |
| him-3(me80) | 148 | 60 | 49 | 11 | 268 | 49 | 0.69 | 0.13 | 9.2e | |
| I | Control | 66 | 37 | 51 | 1 | 155 | 58 | 0.08 | <0.0001 | 1.1f |
| him-3(me80) | 150 | 19 | 22 | 12 | 203 | 32 | 2.31 | 0.0011 | 22.6f | |
((Total number of crossover events)/(number of meiotic products analyzed)) × 100.
((Number of observed double crossovers)/(number of expected double crossovers)), where expected double crossovers = (frequency of crossovers in interval 1) × (frequency of crossovers in interval 2) × (total meiotic products).
P-value from Fisher's exact test assessing probability of obtaining the observed set of meiotic products assuming independent behavior of intervals 1 and 2 in crossover formation.
((Number of double crossovers)/(number of single crossovers + number of double crossovers)) × 100.
,f Control and him-3(me80) differed significantly with respect to the relative incidence of double crossovers vs. single crossovers among meiotic products with crossover. P = 0.005 and P < 0.0001 for the X chromosome and chromosome I, respectively.
Strikingly, the him-3(me80) mutation leads to a significant increase in the incidence of double-crossover recombinant products, both for the X chromosome and for chromosome I. For the X chromosome, the overall recombination frequency measured was very similar to that seen in control meioses (49%). However, there was a significant excess of DCO products relative to that expected on the basis of the control: whereas DCO products represented only 0.9% of total crossover products in control meioses, they accounted for 9.2% of crossover products in him-3(me80) homozygotes (P = 0.005). Furthermore, the coefficient of coincidence calculated from the him-3(me80) homozygote data was 0.69 for the X chromosome; together these data indicate an abrogation of crossover interference.
For chromosome I, him-3(me80) homozygotes exhibited a substantially reduced overall recombination frequency (32%), which corresponds to 55% of the control value. Despite this large deficit of crossovers, however, DCOs represented a remarkable 22.6% of the total chromosome I crossover products, a level 21-fold higher than that seen for the control (P < 0.0001). An elevation in the incidence of DCOs occurring within the context of an overall reduced crossover frequency indicates a failure in a mechanism that ordinarily would prevent DCOs. The chromosome I C-value was 2.31 for the him-3(me80) data set; formally, this indicates a condition known as “negative interference,” wherein crossovers occur simultaneously in two intervals more frequently than would be predicted on the basis of the frequency of crossovers in each interval when considered separately. The observed degree of negative interference most likely reflects the fact that the meiotic products were derived from a heterogeneous population of meiotic prophase nuclei: those in which chromosome I had recruited sufficient SYP-1 protein to be competent for crossing over and those with insufficient SYP-1 protein (see discussion).
Taken together, the above data indicate that the him-3(me80) mutant is defective in inhibiting formation of double crossovers, both for the X chromosome, which is highly competent for homologous synapsis, and for chromosome I, which is strongly impaired in homologous synapsis.
For a subset of the samples analyzed in the three-marker analysis, we typed two additional SNP markers on one of the chromosomes, subdividing the chromosome into four intervals (Figure 4 and Table 3). This analysis allowed us to detect several classes of double crossovers in the him-3(me80) mutant that were invisible in the initial analysis. In addition, this analysis revealed an additional difference between the behavior of the X chromosome and the autosomes in the him-3(me80) mutant. Specifically, we observed that double crossovers on chromosome I occurred mainly in adjacent intervals (classes 1 + 2, 2 + 3, and 3 + 4 in Table 3) whereas they were typically separated by one or two interval(s) on the X (e.g., classes 1 + 3 and 1 + 4 in Table 3).
TABLE 3.
Five-marker recombination analysis
|
X chromsome
|
Chromosome I
|
||||
|---|---|---|---|---|---|
| Recombinant interval |
Marker configuration (a-b-c-d-e) |
Control | him-3(me80) | Control | him-3(me80) |
| No crossover | B-B-B-B-B | 54 | 62 | 33 | 95 |
| H-H-H-H-H | |||||
| 1 | B-H-H-H-H | 17 | 7 | 10 | 2 |
| H-B-B-B-B | |||||
| 2 | B-B-H-H-H | 13 | 14 | 14 | 4 |
| H-H-B-B-B | |||||
| 3 | B-B-B-H-H | 8 | 12 | 13 | 3 |
| H-H-H-B-B | |||||
| 4 | B-B-B-B-H | 7 | 5 | 19 | 10 |
| H-H-H-H-B | |||||
| 1 + 2 | B-H-B-B-B | 0 | 0 | 0 | 1 |
| H-B-H-H-H | |||||
| 1 + 3 | B-H-H-B-B | 0 | 4 | 0 | 0 |
| H-B-B-H-H | |||||
| 1 + 4 | B-H-H-H-B | 0 | 2 | 0 | 0 |
| H-B-B-B-H | |||||
| 2 + 3 | B-B-H-B-B | 1 | 2 | 0 | 1 |
| H-H-B-H-H | |||||
| 2 + 4 | B-B-H-H-B | 0 | 0 | 0 | 1 |
| H-H-B-B-H | |||||
| 3 + 4 | B-B-B-H-B | 0 | 0 | 0 | 1 |
| H-H-H-B-H | |||||
| Total: | 100 | 108 | 89 | 118 | |
Meiotic products were genotyped and chromatids were classified as having no crossover; one crossover in intervals 1, 2, 3, or 4; or two crossovers in the indicated pairs of intervals (1 + 2, 1 + 3, 1 + 4, 2 + 3, 2 + 4, and 3 + 4). For each class, the two possible alternative configurations of Bristol (B) and Hawaiian (H) marker alleles are indicated; markers a–e are as defined in Figure 4. The number of assayed chromatids falling into each class is shown for control and him-3(me80) animals, for both the X chromosome and chromosome I.
Increased incidence of ring bivalents in diakinesis-stage nuclei in the him-3(me80) mutant:
Crossing over at the DNA level is coupled to the formation of chiasmata connecting homologous chromosomes. Thus at least a subset of DCO events is expected to result in the presence of diakinesis-stage bivalents with two chiasmata. If two chiasmata on a bivalent are well separated and the chromosome is long enough, the bivalent may assume a ring shape at diakinesis (Figure 5). In otherwise wild-type animals homozygous for an end-to-end fusion of chromosomes X and IV (mnT12), we detected ring-shaped bivalents at diakinesis in 2.7% (9/329) of nuclei; this cytologically observed frequency of ring bivalents corresponds well with the 55-cM genetic map of mnT12 (Hillers and Villeneuve 2003) (Figure 5, A and B). Given the above evidence for an increased incidence of DCO chromosomes in the him-3(me80) mutant, we tested whether the him-3(me80) mutation would result in an increased frequency of ring-shaped bivalents at diakinesis in worms carrying the mnT12 fusion chromosome. In him-3(me80) mnT12 animals, we detected ring-shaped bivalents in 10% (32/316) of diakinesis nuclei examined, a frequency almost fourfold higher than that seen in mnT12 alone (Figure 5B). This extremely significant elevation in the incidence of ring-shaped bivalents (P = 0.0001) provides independent support for the conclusion that the him-3(me80) mutation impairs the capacity to inhibit formation of DCOs.
Figure 5.—
Cytological evidence of chromosomes with double crossovers. (A) DAPI-stained chromosomes in a diakinesis-stage oocyte nucleus from a hermaphrodite homozygous for the mnT12(IV; X) two-chromosome fusion; image is a projection of a 3-D data stack encompassing all chromosomes. The fusion chromosome bivalent is indicated by the arrowhead; the ring-like configuration of this bivalent is most easily explained by the presence of two chiasmata. Bar, 3 μm. (B) Table indicating the incidence of diakinesis nuclei containing a ring-shaped bivalent in mnT12 and him-3(me80) mnT12 animals.
Simultaneous visualization of SYP-1 and RAD-51 in the him-3(me80) mutant:
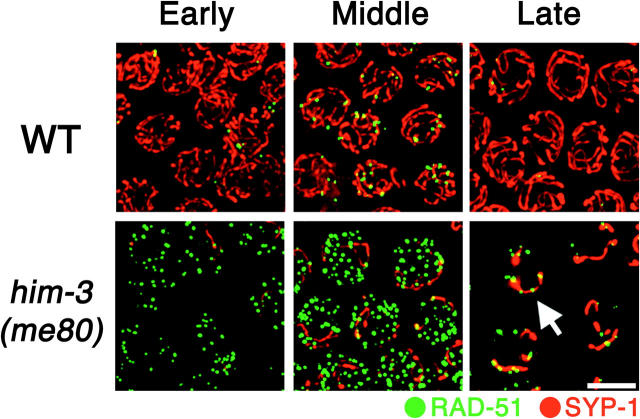
The above two assays monitor outcomes of the recombination process, i.e., formation of crossover recombination products and consequent chiasmata. We also wished to assess the relationship of the limited synapsis that occurs in the him-3(me80) mutant to the appearance and disappearance of nascent recombination intermediates, so we performed double-labeling experiments to allow simultaneous visualization of SYP-1 and DNA strand exchange protein RAD-51. RAD-51 is a highly conserved member of the RecA protein family required for repair of meiotic DSBs, the initiating events of meiotic recombination (Ogawa et al. 1993a); Rad51 assembles a filament (Ogawa et al. 1993b) along 3′ ssDNA overhangs formed by resection of DSB ends and promotes invasion of a homologous DNA duplex (Sung 1994). In wild-type C. elegans, RAD-51 foci representing nascent meiotic recombination events arise during late zygotene and early pachytene stages, peak in abundance in early/mid pachytene, and diminish in number during mid/late pachytene as meiotic DSB repair progresses (Alpi et al. 2003; Colaiacovo et al. 2003). In nuclei from the early pachytene region of him-3(me80) mutant germ lines, which contain only very limited stretches of SYP-1 staining, RAD-51 foci are already abundant (Figure 6). Further, by the time RAD-51 foci peak in abundance [later and at levels three- to fourfold higher than the wild-type peak (Couteau et al. 2004)], several additional features are evident. First, the RAD-51 foci detected in the mid/late pachytene region in the him-3(me80) mutant appear bigger and brighter than those in the early pachytene region; such a distinction is not evident in wild-type germ lines. This increase in intensity of RAD-51 signals during prophase progression is consistent with impeded progression of recombination: such a change could reflect a delay prior to the strand exchange step leading to persistence of a normally transient intermediate and/or continued resection of DSB ends to yield elongating ssDNA segments that load larger amounts of RAD-51 protein. Second, although several extensive SYP-1 stretches are present in nuclei in the mid-pachytene region in the him-3(me80) mutant, most RAD-51 foci are not associated with SYP-1 stretches; this indicates that DSBs are neither limited to nor concentrated at synapsed regions of chromosomes. In contrast, at the very end of the pachytene stage, when most RAD-51 foci abruptly disappear, the remaining RAD-51 foci preferentially colocalize with or are adjacent to SYP-1 stretches.
Figure 6.—
Simultaneous visualization of RAD-51 foci and SC central region protein SYP-1. Immunostaining of RAD-51 (green) and SYP-1 (red) in pachytene region nuclei from whole-mount germ lines of wild-type and him-3(me80) hermaphrodites is shown. Images are projections of 3-D data stacks encompassing whole nuclei; for each genotype, the three consecutive panels show different regions of the same germ line. Left, the early pachytene region, where very few SYP-1 stretches are seen in the him-3(me80) mutant and RAD-51 foci are already more abundant than in wild type. Middle, the mid-pachytene region, where RAD-51 foci in the him-3(me80) mutant appear larger than those in the left panel; some RAD-51 foci show colocalization with SYP-1 stretches, but foci are abundant on regions that lack SYP-1. Right, late pachytene region, where RAD-51 foci are greatly diminished in number compared to earlier stages but are preferentially retained at SYP-1 stretches (arrows) in the him-3(me80) mutant. Bar, 5 μm.
Figure 6 emphasizes an additional point regarding the limited SC installation that occurs in the him-3(me80) mutant. That is, synapsis not only is reduced in extent, but also reaches its maximal levels with substantially delayed kinetics. Whereas synapsis is by definition complete in the early pachytene region of wild-type germ lines, SC polymerization has begun in the corresponding region in him-3(me80) mutant germ lines but does not reach maximal levels until the region corresponding to mid/late pachytene in wild type.
DISCUSSION
We have used the nonnull him-3(me80) mutant to investigate the contribution of meiotic chromosome axes, or structures that depend on axes (e.g., SC), to regulatory mechanisms that govern the formation of meiotic crossovers. As discussed below, our analysis of the him-3(me80) mutant provides strong evidence that integrity of meiotic axis structures plays an important role in limiting the number of crossovers per homologous chromosome pair. In addition, our analysis has revealed insights into the process of SC assembly itself and its relationship to other aspects of meiotic prophase chromosome organization and metabolism.
Synapsis in the presence of limiting HIM-3:
The synapsis phenotype of the him-3(me80) mutant strongly suggests that assembly of the SC central region is a highly cooperative process. SC assembly appears to occur very rapidly during wild-type C. elegans meiosis. Loading of central region proteins SYP-1 and SYP-2 depends on prior loading of axis components such as HIM-3 (MacQueen et al. 2002; Colaiacovo et al. 2003), but SYP-1 and HIM-3 exhibit virtually complete colocalization (at the light microscope level) almost immediately after SYP-1 is first detected in association with chromosome axes (MacQueen et al. 2002). In the him-3(me80) mutant, the reduced amounts of HIM-3 present are widely dispersed along all chromosomes and there is a delay in loading of SYP-1. The SYP-1 stretches that do form are neither scattered nor tenuous, however, but rather are robust, continuous stretches comparable in intensity to those seen in wild-type controls. These observations strongly suggest that assembly of the SC central region is a highly cooperative process: it may be difficult to nucleate assembly under conditions of limiting amounts of a mutant HIM-3 protein, but once SC assembly is nucleated, extensive polymerization ensues. This property is consistent with EM observations that SC components are prone to self-assembly into highly ordered arrays known as polycomplexes under conditions where the protein is present but not assembled onto chromosomes (Goldstein 1987).
Given this cooperative nature of SYP-1 assembly, why then are only a few stretches formed per nucleus in the him-3(me80) mutant? This property suggests that the ability to nucleate SC central region assembly is limited by another event, presumably the close juxtaposition of chromosome axes. Because initial establishment of homolog pairing for all chromosomes is dependent on HIM-3, and overall proficiency in pairing is severely reduced in mutants with partial loss of HIM-3 function (Couteau et al. 2004), we suggest that the number of opportunities for SC nucleation events may be initially limited in him-3(me80) by a low incidence of closely juxtaposed axes, particularly for the autosomes. This notion is supported by the differential pairing and synapsis behavior of different chromosome pairs in the him-3(me80) mutant. The X chromosome is highly proficient for pairing in the presence of reduced HIM-3 function, while autosomal loci show significantly reduced levels of pairing relative to wild type (Couteau et al. 2004). Here, we demonstrate that the pairing behavior of chromosomes in him-3(me80) anticipates their synapsis phenotype; the X chromosomes synapse with near wild-type efficiency, while the autosomes engage in low levels of synapsis. Although we do not know the reason why the X chromosome is less reliant on HIM-3 to establish pairing, we suggest that proficiency in both initial juxtaposition and local stabilization of pairing, coupled with the cooperative nature of central region assembly, results in the high degree of success in accomplishing full homologous synapsis along the entire length of the X chromosome.
Interestingly, whereas the pairing kinetics at the left end of the X chromosome are essentially identical for the him-3(me80) mutant and wild-type controls (Couteau et al. 2004), the right end of the X chromosome tends to achieve maximal pairing with slightly lagging kinetics in the him-3(me80) mutant (K. Nabeshima, unpublished data). The left end of the X harbors a cis-acting chromosomal domain termed a meiotic pairing center, which has been proposed to play dual roles in promoting homolog synapsis: one in mediating local, SC-independent stabilization of pairing, and a second in nucleating SC assembly (McKim et al. 1988, 1993; Herman and Kari 1989; Villeneuve 1994; MacQueen et al. 2002). The delay in pairing at the right end of the X suggests a lag in stabilization of pairing through synapsis; this observation fulfills expectations of models proposing directional assembly of the SC initiating at the pairing center end. Whereas directional assembly of SC nucleated at pairing centers may predominate during wild-type meiosis, however, our data suggest that assembly initiated at other sites is also possible, as evidenced by nuclei in which two paired regions of the same chromosome are associated with separate SC stretches, clearly indicating lack of continuity of the central region.
Even though extensive SYP-1 assembly occurs along the length of nearly all X chromosome pairs in the him-3(me80) mutant, there are several indications of abnormalities in organization even for this chromosome pair, presumably reflecting the underlying defect in axis organization. First, SYP-1 stretches associated with both the X and the autosomes appear thicker than those in wild type. Second, the average distance between paired, synapsed homologous loci on the X chromosome is significantly larger in the him-3(me80) mutant than in wild type (Figure 3, A and B). These phenotypes may reflect reduced constraints on the dimensions of the central region resulting from a loss of rigidity or continuity of underlying axes, and/or a change in organization or size of chromatin loops, again conferred by impaired axis organization.
We note that synapsed and unsynapsed chromosome segments in the him-3(me80) mutant differ with respect to their association with RAD-51 foci. During early pachytene, foci are broadly distributed among chromosomes, indicating that DSB formation early in prophase does not correlate with regions of synapsis. This result was expected, since previous work had shown that DSB formation does not depend on either pairing or synapsis (Colaiacovo et al. 2003; Couteau et al. 2004). During late pachytene, however, the few remaining RAD-51 foci are found preferentially on SYP-1 stretches. It seems likely that disappearance of foci from the unsynapsed regions reflects repair using sister chromatids, which may be permitted prematurely under conditions of limiting HIM-3 since this protein appears to constitute part of a barrier that normally prevents the use of sister chromatids as recombination partners during most of meiotic prophase (Colaiacovo et al. 2003; Couteau et al. 2004). (Absence of SC central region proteins in the presence of intact axes results in prolonged persistence of RAD-51 foci, implying the presence of a barrier that impedes progression of recombinational repair using the sister chromatid; absence of HIM-3 allows removal of RAD-51 foci, suggesting loss of this barrier.) One possible interpretation for the preferential retention of RAD-51 foci on synapsed regions in the him-3(me80) mutant is that repair is actually impeded by the presence of SYP proteins in regions engaged in nonhomologous synapsis. Alternatively, this observation could reflect continued DSB formation preferentially in synapsed regions. Interestingly, we find that accumulation of the dimethyl-K9 modification of Histone H3 occurs first on chromosomal regions lacking SYP-1 in the him-3(me80) mutant (K. Nabeshima, unpublished results), a result anticipated by the observations of Bean et al. (2004). Accumulation of cytologically detectable levels of this histone modification is temporally correlated with a decline in the number of RAD-51 foci during wild-type meiosis (Kelly et al. 2002; Colaiacovo et al. 2003) and was recently proposed to correlate with loss of DSB competence (Reddy and Villeneuve 2004).
Impaired crossover regulation in the him-3(me80) mutant:
The him-3(me80) mutation creates a situation wherein functional HIM-3 protein is limiting for assembly of meiotic chromosome structures, yet is sufficient to support crossing over and chiasma formation for a subset of chromosome pairs. These conditions made it possible to investigate the consequences of reducing the levels of this major axis component on meiotic crossover control. We found that chromosome-wide crossover regulation is indeed impaired in the him-3(me80) mutant, both for the X chromosome, which is highly proficient for pairing and synapsis, and for chromosome I, which is substantially defective.
A classical metric for evaluating crossover regulation is the coefficient of coincidence (C), calculated as the ratio of the observed number of coincident crossovers in two intervals to the expected number; “expected” is calculated on the basis of the frequencies of crossovers in each interval when considered independently (Muller 1916). If crossing over in one interval is never associated with the formation of a crossover in the other, no double crossovers are observed and C = 0; under such circumstances, the intervals are said to display complete crossover interference. If crossovers occur independently in two intervals, C = 1 and the intervals are said to display no interference. Analysis of our control X chromosome and chromosome I data using this traditional metric yielded C-values near zero, indicative of robust crossover interference; this conclusion is reinforced by strong statistical support for a lack of independence between crossovers formed in the two intervals.
The X chromosomes are highly competent for pairing and synapsis in the him-3(me80) mutant, and most X chromosomes succeed in acquiring crossovers and chiasmata. This allowed us to assess the effects of altered axis composition on crossover regulation in a context where the SC central region appeared largely contiguous and crossover levels were roughly normal. Impaired crossover regulation in this context was evidenced by an elevated incidence of DCO meiotic products, which are rare during wild-type meiosis. We calculated a C-value close to 1 and statistical analysis indicated no evidence for departure from independent behavior of the two intervals, indicating a severe abrogation of crossover interference.
For chromosome I, we also saw an elevated incidence of DCOs in the him-3(me80) mutant despite the fact that the overall crossover levels were substantially lower than control levels. The data yield a C-value significantly >1, formally indicative of negative interference. The high negative interference in this case likely reflects the heterogeneous nature of the meiocyte population: only the subset of chromosome pairs that succeed in engaging in some extent of homologous synapsis are competent to receive a crossover, while those that fail to synapse are ineligible to participate in crossing over. The presence of a class of meiocytes that are incompetent for crossing over (and thus produce only noncrossover meiotic products) serves to depress the calculated recombination frequencies, lowering the expected frequency of doubles and inflating calculated C-values (as discussed in Sall and Bengtsson 1989). [The coefficient of coincidence (C) is the ratio of the observed number of double crossovers in two intervals (DCOobs) to the expected number (DCOexp) (C = DCOobs/DCOexp). DCOexp is calculated from the observed number of crossovers in each interval when considered separately; DCOexp = (COI/n)(COII/n)(n) = (COI)(COII)/n, where COI and COII are the observed number of crossovers in intervals I and II, respectively, and n is total number of meiotic products assayed. After simplification, then, C = ((DCOobs)(n))/((COI)(COII)) and can be seen to vary directly with n. The presence of a class of meiocytes incompetent for recombination will increase n without altering DCOobs, COI, or COII. This inflates C and can produce apparent negative interference.] If the fraction of “incompetent” meiocytes were known, it would be possible to correct for this in calculating the C-value; however, pairing and synapsis data for two loci on a chromosome do not permit a reliable estimate of the percentage of chromosomes harboring a stretch of homologous synapsis. This points to the inadequacy of traditional interference metrics for evaluating potential defects in crossover regulation: such measurements are inherently self-referential, with expected values derived from the empirical data on residual crossovers, and they have no means to account for or mitigate against the effects of population heterogeneity. Fortunately, we are not limited to using to these traditional metrics. Because DCOs are so infrequent during normal oocyte meiosis in C. elegans, the fraction of DCO products/total CO products can serve as a sensitive indicator of defects in crossover regulation that is robust against population heterogeneity effects. Whereas the measured recombination frequencies per se will be influenced by population heterogeneity, and possibly by other potential biases (such as selective recovery of chromatids derived from chiasmate bivalents or selective removal of a subset of meiocytes through apoptosis), the greatly elevated DCO/total CO ratio obtained for chromosome I in the him-3(me80) mutant can be explained only by disruption of a mechanism that normally prevents double crossovers.
Disruption of crossover regulation in the him-3(me80) mutant was also demonstrated by a very different approach, using a cytological assay measuring the frequency of “ring bivalents,” or diakinesis chromosome pairs connected by two widely spaced chiasmata. The three- to fourfold increase in the incidence of ring bivalents detected in him-3(me80) animals with this assay is especially dramatic given that it was detected against a backdrop of chromosome pairs lacking chiasmata. Moreover, this excess of ring bivalents almost certainly represents an underestimate of the frequency of double crossovers on the him-3(me80) fusion chromosome pair, since closely spaced double crossovers would not be detected.
We note that our results cannot be explained trivially as a consequence of crossover frequencies increasing proportionally to an increase in the number of DSBs, since the overall frequency of crossovers is not significantly elevated in the him-3(me80) mutant. Rather, the relationship between crossovers is altered, in a manner that reduces or eliminates interference between them. If this disruption of interference is in fact a consequence of an increase in DSB formation per se, the necessary implication is that part or all of crossover interference in this organism must involve regulation occurring at the DSB step, with nascent crossover events inhibiting the formation of additional DSBs on the same chromosome pair.
Implications for mechanisms of crossover regulation:
Clearly, crossover regulation is impaired in the him-3(me80) mutant. Which aspects of altered chromosome structure in the mutant might be responsible for loss of crossover control, and what does this tell us about how crossover control is exerted during wild-type meiosis?
Although mature SC had long been a favored suspect in mediating crossover interference, a growing body of evidence now suggests that at least some aspects of crossover regulation are exerted prior to and independently of SC assembly (reviewed in Bishop and Zickler 2004). Two recent reports found evidence that crossover/noncrossover differentiation in budding yeast actually precedes SC assembly. Fung et al. (2004) showed that synapsis initiation complexes (SICs), detected as chromosomal foci of the Zip2 protein and thought to correspond to sites of both crossovers and SC nucleation (Rockmill et al. 2003), exhibit a nonrandom, interference distribution that precedes and is independent of SC assembly. Further, Börner et al. (2004) demonstrated that commitment of early meiotic recombination intermediates to enter the “regulated crossover” pathway also occurs prior to and independently of SC formation. In addition, the finding that interference still occurs in Drosophila females expressing a mutant form of SC central region protein C(3)G suggests that full-length SC is not required to mediate interference during Drosophila oocyte meiosis (Page and Hawley 2001). These results are consistent with models proposing that the chromosome axis, rather than the fully elaborated SC, is the relevant conduit of communication regarding the status of nascent recombination events along a chromosome (Börner et al. 2004; Kleckner et al. 2004).
The idea that the chromosome axis is an important structure for crossover regulation is appealing, but is difficult to test in the budding yeast system since impairment of known axis components results in a severe reduction in recombination (Hollingsworth et al. 1990; Rockmill and Roeder 1990, 1991; Leem and Ogawa 1992). Our previous work analyzing the meiotic behavior of fusion chromosomes in C. elegans provided evidence suggesting that chromosome axes, or structures that depend on axial continuity, are a functional unit for crossover regulation during nematode meiosis (Hillers and Villeneuve 2003, see Introduction). The him-3(me80) mutant provided an opportunity to partially cripple axis organization while still retaining competence for DSB formation and recruitment of SC central region components essential for crossover formation.
Our results provide strong support for the conclusion that a defect in axis organization results in a breakdown in communication of recombination status along chromosomes. Further, several observations lead us to favor the view that this communication defect is due to a defect in the integrity of the axes per se, rather than to consequent defects in SC structure in the him-3(me80) mutant. SYP-1 stretches appear robust and contiguous along the entire length of the X chromosomes in the him-3(me80) mutant, whereas underlying HIM-3 levels are greatly reduced. Moreover, the negative interference and tendency of DCOs to occur in adjacent intervals on chromosome I, together with our evidence for cooperativity of SC central region assembly, suggest that chromosome I DCOs may often occur in the context of a single contiguous stretch of assembled SC central region. According to this view, the underlying defect in axis organization would be responsible for the apparent loss of communication between adjacent intervals. The SYP-1 stretches formed in the mutant may be structurally abnormal in spite of their cooperative assembly, so we cannot exclude the possibility that the crossover regulation defects could be a secondary consequence of effects on mature SC structure. However, our findings are fully consistent with the emerging view that the SC central region is dispensable for crossover interference and that the chromosome axis is the pertinent conduit of information regarding the recombination status of a chromosome pair.
Crossover regulation has at least two concrete manifestations: obligate chiasma, referring to the fact that every chromosome pair gets at least one crossover (Jones 1987), and crossover/chiasma interference, the ability of meiotic crossovers to discourage additional nearby crossovers (Muller 1916). Conceptually, disruption of crossover control in the him-3(me80) mutant could occur in a number of ways. One scenario is that discontinuities in meiotic chromosome structures cause the cell to recognize a single chromosome as being composed of multiple chromosome units, each of which warrants an obligate chiasma. Alternatively, the him-3(me80) mutation may alter the physical properties of the unit such that the formation/designation of one crossover no longer has the capacity to generate and/or communicate an inhibitory influence to interfere with the formation of additional crossovers on the same unit. Cosmetically, the outcome is the same: formation of multiple crossovers along a given pair of homologous chromosomes.
Whereas crossover regulation is generally thought of in terms of a crossover/noncrossover decision regarding the choice of repair pathway and outcome for extant DSBs, our data raise the intriguing possibility that DSB formation may also be a step subject to regulation by chromosome-wide regulatory mechanisms. We argue above that prolonged persistence of DSBs accounts for at least part of the increase in RAD-51 foci in the him-3(me80) mutant, but we also suggest that concentration of foci on synapsed regions in late pachytene might reflect late-appearing DSBs formed preferentially in synapsed regions. Under the scenario that discontinuity in axial structures results in a cell perceiving a given chromosome pair to be composed of more than one crossover-competent unit deserving of a crossover, continued formation of DSBs could reflect a capacity to recognize whether a crossover-eligible chromosome pair has suffered a DSB and to modulate DSB-forming machinery accordingly. Such an ability to regulate DSB formation could also help explain an earlier observation regarding the meiotic behavior of C. elegans chromosome rearrangements: in rearrangement heterozygotes in which homologous synapsis (and thus the opportunity for crossing over) is predicted to be limited to only 10% of the length of a chromosome pair, that pair nevertheless apparently succeeds in forming a crossover in most meioses (Zetka and Rose 1992). We speculate that regulation of DSB formation to ensure that each chromosome unit receives at least one DSB could be a means to guarantee formation of the obligate chiasma in other organisms as well. This possibility is suggested by circumstantial evidence from S. cerevisiae, where minichromosomes constituting as little as 0.4% of the genome (60 kb) were shown to average more than one crossover per meiosis, with few pairs failing to enjoy at least one crossover (and thus at least one DSB; Kaback et al. 1999).
Concluding remarks:
The structural organization of meiotic chromosome axes has been demonstrated to play multiple roles crucial for successful homolog segregation at the meiosis I division, including roles in: promoting normal levels of DSB formation (Mao-Draayer et al. 1996; Schwacha and Kleckner 1997); counteracting destabilizing influences of DSBs (Storlazzi et al. 2003); promoting both initial establishment of homolog alignment and installation of SC central region components (Colaiacovo et al. 2003; Couteau et al. 2004); and promoting use of the homolog while inhibiting use of a sister chromatid as a recombination partner (Schwacha and Kleckner 1997; Bishop et al. 1999; Thompson and Stahl 1999; Couteau et al. 2004; Webber et al. 2004). Models for crossover regulation have also speculated that structural features of meiotic chromosome axes form the underlying basis of crossover interference mechanisms, but while there are hints that crossover regulation may be impaired in female mice lacking axis component Scp3 (Yuan et al. 2002), there has been no definitive evidence to date linking axis structural components to crossover regulation. Here we have shown that reducing the levels of a major conserved meiotic chromosome axis component results in a substantial increase in the production of double-crossover recombination products during meiosis in C. elegans, an organism that is normally inclined to limit crossovers to one per homolog pair. These results expand the repertoire of functions conferred by structural differentiation of meiotic chromosome axes by demonstrating that a meiosis-specific chromosome axis protein does indeed play a role in chromosome-wide mechanisms of crossover regulation.
Acknowledgments
We thank F. Couteau, M. Zetka, and the Caenorhabditis Genetics Center for strains; and M. Zetka, J. Loidl, and A. La Volpe for HIM-3, REC-8, and RAD-51 antibodies, respectively. We thank A. MacQueen, E. Martinez-Perez, M. Colaiacovo, K. Lee, and J. Mulholland for technical advice and suggestions; members of the Villeneuve lab and J. Engebrecht for many helpful discussions; and G. Stanfield for comments on the manuscript. This work was supported by National Institutes of Health grants R01GM53804 and R01GM67268 and the Esther Ehrman Lazard Faculty Scholar Award to A.M.V. and a Uehara Memorial Foundation Research Fellowship and postdoctral training support from Osaka University, Japan to K.N.
References
- Alpi, A., P. Pasierbek, A. Gartner and J. Loidl, 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and E. V. Koonin, 1998. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 23: 284–286. [DOI] [PubMed] [Google Scholar]
- Bean, C. J., C. E. Schaner and W. G. Kelly, 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D. K., and D. Zickler, 2004. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. [DOI] [PubMed] [Google Scholar]
- Bishop, D. K., Y. Nikolski, J. Oshiro, J. Chon, M. Shinohara et al., 1999. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells 4: 425–444. [DOI] [PubMed] [Google Scholar]
- Bullerjahn, A. M., and D. L. Riddle, 1988. Fine-structure genetics of ama-1, an essential gene encoding the amanitin-binding subunit of RNA polymerase II in Caenorhabditis elegans. Genetics 120: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo, S. B., R. K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann et al., 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103: 387–398. [DOI] [PubMed] [Google Scholar]
- Börner, G. V., N. Kleckner and N. Hunter, 2004. Crossover/noncrossover differentiation, synaptonemal complex formation and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Carpenter, A. T. C., 1988 Thoughts on recombination nodules, meiotic recombination, and chiasmata, pp. 529–548 in Genetic Recombination, edited by R. Kucherlapati and G. R. Smith. American Society for Microbiology, Washington, DC.
- Caryl, A. P., S. J. Armstrong, G. H. Jones and F. C. Franklin, 2000. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71. [DOI] [PubMed] [Google Scholar]
- Colaiacovo, M. P., A. J. MacQueen, E. Martinez-Perez, K. McDonald, A. Adamo et al., 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Couteau, F., K. Nabeshima, A. Villeneuve and M. Zetka, 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14: 585–592. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser et al., 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Egel, R., 1978. Synaptonemal complex and crossing-over: Structural support or interference? Heredity 41: 233–237. [DOI] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fung, C. J., B. Rockmill, M. Odell and G. S. Roeder, 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Goldstein, P., 1987. Multiple synaptonemal complexes (polycomplexes): origin, structure and function. Cell Biol. Int. Rep. 11: 759–796. [DOI] [PubMed] [Google Scholar]
- Guillon, H., and B. de Massy, 2002. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat. Genet. 32: 296–299. [DOI] [PubMed] [Google Scholar]
- Herman, R. K., and C. K. Kari, 1989. Recombination between small X chromosome duplications and the X chromosome in Caenorhabditis elegans. Genetics 121: 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers, K. J., and A. M. Villeneuve, 2003. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 13: 1641–1647. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J. H., R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N. M., L. Goetsch and B. Byers, 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61: 73–84. [DOI] [PubMed] [Google Scholar]
- Hunter, N., and N. Kleckner, 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Jang, J. K., D. E. Sherizen, R. Bhagat, E. A. Manheim and K. S. McKim, 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069–3077. [DOI] [PubMed] [Google Scholar]
- Jones, G. H., 1987 Chiasmata, pp. 213–244 in Meiosis, edited by P. B. Moens. Academic Press, Orlando, FL.
- Kaback, D. B., D. Barber, J. Mahon, J. Lamb and J. You, 1999. Chromosome size-dependent control of meiotic reciprocal recombination in Saccharomyces cerevisiae: the role of crossover interference. Genetics 152: 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Kelly, W. G., C. E. Schaner, A. F. Dernburg, M. H. Lee, S. K. Kim et al., 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner, N., D. Zickler, G. H. Jones, J. Henle, J. Dekker et al., 2004. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA 101: 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S. B. Buonomo, C. Michaelis et al., 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Kohli, J., and J. Bahler, 1994. Homologous recombination in fission yeast: absence of crossover interference and synaptonemal complex. Experientia 50: 295–306. [DOI] [PubMed] [Google Scholar]
- Leem, S. H., and H. Ogawa, 1992. The MRE4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 20: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X., G. Fang, M. Coldiron, Y. Lin, H. Yu et al., 2000. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 7: 224–229. [DOI] [PubMed] [Google Scholar]
- MacQueen, A. J., M. P. Colaiacovo, K. McDonald and A. M. Villeneuve, 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manheim, E. A., and K. S. McKim, 2003. The synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr. Biol. 13: 276–285. [DOI] [PubMed] [Google Scholar]
- Mao-Draayer, Y., A. M. Galbraith, D. L. Pittman, M. Cool and R. E. Malone, 1996. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics 144: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, J. M., 1976. Orientation disruptor (ord): a recombination-defective and disjunction-defective meiotic mutant in Drosophila melanogaster. Genetics 84: 545–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., and A. Hayashi-Hagihara, 1998. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12: 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., A. M. Howell and A. M. Rose, 1988. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., K. Peters and A. M. Rose, 1993. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics 134: 749–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely, P. M., A. F. Farago and T. M. Kauffman, 2002. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics 162: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, P. B., N. K. Kolas, M. Tarsounas, E. Marcon, P. E. Cohen et al., 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115: 1611–1622. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1916. The mechanism of crossing over. Am. Nat. 50: 193–221, 284–305, 350–366, 421–434. [Google Scholar]
- Munz, P., 1994. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137: 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, T., A. Shinohara, A. Nabetani, T. Ikeya, X. Yu et al., 1993. a RecA-like recombination proteins in eukaryotes: functions and structures of RAD51 genes. Cold Spring Harbor Symp. Quant. Biol. 58: 567–576. [DOI] [PubMed] [Google Scholar]
- Ogawa, T., X. Yu, A. Shinohara and E. H. Egelman, 1993. b Similarity of the yeast Rad51 filament to the bacterial RecA filament. Science 259: 1896–1899. [DOI] [PubMed] [Google Scholar]
- Padmore, R., L. Cao and N. Kleckner, 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2003. Chromosome choreography: the meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer et al., 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plug, A. W., A. H. Peters, K. S. Keegan, M. F. Hoekstra, P. de Boer et al., 1998. Changes in protein composition of meiotic nodules during mammalian meiosis. J. Cell Sci. 111: 413–423. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar and J. Austin, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, K. C., and A. M. Villeneuve, 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., and G. S. Roeder, 1990. Meiosis in asynaptic yeast. Genetics 126: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., and G. S. Roeder, 1991. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev. 5: 2392–2404. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., J. C. Fung, S. S. Branda and G. S. Roeder, 2003. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 13: 1954–1962. [DOI] [PubMed] [Google Scholar]
- Sall, T., and B. O. Bengtsson, 1989. Apparent negative interference due to variation in recombination frequencies. Genetics 122: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha, A., and N. Kleckner, 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- Storlazzi, A., S. Tesse, S. Gargano, F. James, N. Kleckner et al., 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, P., 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast Rad51 protein. Science 265: 1241–1243. [DOI] [PubMed] [Google Scholar]
- Sym, M., and G. S. Roeder, 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292. [DOI] [PubMed] [Google Scholar]
- Thompson, D. A., and F. W. Stahl, 1999. Genetic control of recombination partner preference in yeast meiosis: isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics 153: 621–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve, A. M., 1994. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136: 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R. H., K. C. Smith and D. G. Moerman, 1982. Genetic fine structure analysis of the myosin heavy chain gene unc-54 of Caenorhabditis elegans. J. Mol. Biol. 158: 1–15. [DOI] [PubMed] [Google Scholar]
- Webber, H. A., L. Howard and S. E. Bickel, 2004. The cohesion protein ORD is required for homologue bias during meiotic recombination. J. Cell Biol. 164: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- WormBase, 2004 Release WS121, March 12, 2004 (http://www.wormbase.org).
- Yuan, L., J. G. Liu, M. R. Hoja, J. Wilbertz, K. Nordqvist et al., 2002. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296: 1115–1118. [DOI] [PubMed] [Google Scholar]
- Zalevsky, J., A. J. MacQueen, J. B. Duffy, K. J. Kemphues and A. M. Villeneuve, 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka, M. C., and A. M. Rose, 1992. The meiotic behavior of an inversion in Caenorhabditis elegans. Genetics 131: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka, M. C., I. Kawasaki, S. Strome and F. Muller, 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13: 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner, 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]