Abstract
Spontaneous mutation is the ultimate source of all genetic variation. By interacting with environmental factors, genetic variation determines the phenotype and fitness of individuals in natural populations. However, except in a few model organisms, relatively little is known about the patterns of genotype-environment interactions of spontaneous mutations. Here I examine the rates of spontaneous mutation and the patterns of genotype-environment interaction of mutations affecting vegetative growth in the human fungal pathogen Cryptococcus neoformans. Eight mutation accumulation (MA) lines were established from a single clone on the nutrient-rich medium YEPD for each of two temperatures, 25° and 37°. Cells from generations 100, 200, 400, and 600 for each of the 16 MA lines were stored and assayed for vegetative growth rates under each of four conditions: (i) 25° on SD (a synthetic dextrose minimal medium); (ii) 25° on YEPD; (iii) 37° on SD; and (iv) 37° on YEPD. Both MA conditions and assay environments for vegetative growth showed significant influence on the estimates of genomic mutation rates, average effect per mutation, and mutational heritability. Significant genotype-environment interactions were detected among the newly accumulated spontaneous mutations. Overall, clones from MA lines maintained at 37° showed less decline in vegetative fitness than those maintained at 25°. The result suggests that a high-temperature environment might be very important for the maintenance of the ability to grow at a high temperature. Results from comparisons between clinical and environmental samples of C. neoformans were consistent with laboratory experimental population analyses. This study calls into question our long-standing view that warm-blooded mammals were only occasional and accidental hosts of this human fungal pathogen.
SPONTANEOUS mutations can occur during every cell division in all living organisms. These mutations are the ultimate source of genetic variation in natural populations. Therefore, understanding the effect of spontaneous mutations on individuals and populations of living organisms is of fundamental biological importance. It has been demonstrated that most spontaneous mutations with an effect on the phenotype are typically deleterious (Crow 1992). Many important biological phenomena are thought to be at least in part the outcomes of evolutionary responses to deleterious mutations. These phenomena include: (i) the evolution and maintenance of genetic recombination and sexual reproduction (e.g., Charlesworth and Barton 1996; Kondrashov 1997); (ii) the evolution of diploidy (Kondrashov and Crow 1991); (iii) the evolution and maintenance of mating systems in fungi, plants, and animals (Charlesworth et al. 1990; Zeyl and Bell 1997; Xu 2002); (iv) aging (Partridge and Barton 1993); and (v) the viability of fragmented or captive populations (Lande 1995). Except in a few model species such as Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Escherichia coli, Saccharomyces cerevisiae, mouse, and humans, very little is known about the patterns of spontaneous mutations in the vast majority of living organisms, including human pathogens (Lynch and Walsh 1998). Even less is known about how environmental factors influence estimates of mutational parameters. The objective of this study was to investigate the patterns of spontaneous mutations and the genotype-environment interactions of these mutations in the human fungal pathogen Cryptococcus neoformans.
Genotype-environment interactions are widespread in natural populations. For example, studies in plants and animals have shown that the levels of inbreeding depression and heterosis were greater under harsher conditions than those in more benign environments (e.g., Mitton and Grant 1984; Dudash 1990). However, relatively little has been investigated on how spontaneous mutations interact with environmental factors. So far, most data have come from the model organism D. melanogaster (Fry et al. 1996; Kondrashov and Houle 1998; Mackay and Lyman 1998). Kondrashov and Houle (1998) demonstrated that spontaneous mutations accumulated in D. melanogaster showed highly significant genotype-environment interactions on development time and productivity. They found that reduced nutrition and increased population density magnified the deleterious effects of accumulated mutations (Kondrashov and Houle 1998). Similarly, Fry et al. (1996) showed significant genotype-environment interactions of spontaneous mutations for fitness. Mackay and Lyman (1998) showed that spontaneous mutations affecting bristle numbers also exhibited significant temperature-dependent effects. In each of these studies, mutations were accumulated freely in a single environment and tested over several environments. No study has examined how environments during mutation accumulation (MA) could influence the patterns of spontaneous mutations and the directions of genotype-environment interactions of these mutations.
Microbes are excellent organisms for the analysis of the patterns and consequences of spontaneous mutations. Large population sizes can be grown and maintained relatively easily in the laboratory, and cells at various stages of an experiment can be stored permanently in a freezer. In addition, because individual genotypes can be clonally replicated, the same genotype can be tested in a large number of environments, thus allowing robust estimates of environmental influences on examined traits. These features make microbes ideal organisms on which to perform experimental evolutionary studies to test potential genotype-environment interactions of spontaneous mutations. Surprisingly, little information on microorganisms is available in this area.
Spontaneous mutations can be obtained using mutation accumulation conditions (e.g., Mukai 1964; Kibota and Lynch 1996; Zeyl and de Visser 2001; Xu 2002). In these experiments, spontaneous mutations are allowed to accumulate in replicate lines with severe population bottlenecks to minimize selection. Estimates of mutational parameters for fitness traits rely on two typical assumptions. The first is that, in the absence of selection, deleterious mutations will continuously accumulate, leading to declined fitness for a given trait, and the second is that divergence among replicate lines in trait values should increase over time. The relative rates of decrease in fitness and among-line divergence in trait values allow estimations of the genomic mutation rate, the average mutation effect, and the mutational heritability of individual traits (Lynch and Walsh 1998).
C. neoformans is an encapsulated basidiomycetous yeast and an emerging model organism for studying molecular and evolutionary biology. Its natural reservoir is assumed to be in soil and other habitats such as bird droppings and certain tree species (Casadevall and Perfect 1998). Two factors have contributed to our increased interest in C. neoformans. The first is that it has a highly tractable genetic system for molecular and evolutionary studies. Unlike many other basidiomycetes, C. neoformans has a very well-characterized mating system and gene delivery system. The second factor is that C. neoformans is a significant human pathogen. It is one of the leading causes of human fungal infection: up to 15% of immunocompromised patients die from cryptococcal infection, the most common form being cryptococcal meningitis (Casadevall and Perfect 1998). As a result, it has attracted significant medical attention.
Despite the significant health burden on humans and other mammals, it is generally assumed that humans and other mammalian hosts are not an essential part of the life cycle of this pathogenic fungus (Casadevall and Perfect 1998). Its pathogenicity to mammals is thought to have evolved as a by-product of interactions with soil protozoa and/or other soil organisms (Steenbergen et al. 2001). One pathogenicity trait found in all human pathogens is their ability to grow vigorously at the mammalian body temperature of 37°. Because soil environments typically have temperatures below that of human hosts, how could human pathogens with soil or nonmammalian hosts as the primary reservoir maintain their ability to grow vigorously at 37° in humans? Using experimental approaches, this study investigated the influence of temperature during mutation accumulation on mutational parameters and on genotype-environment interactions.
This study was designed to address the following questions. First, what are the estimates of genomic mutation rate per mitotic generation (U), the effects per mutation (a), and the mutational heritability h2m affecting vegetative growth in C. neoformans? Second, do test conditions affect estimates of mutational parameters in C. neoformans, similar to the observations for D. melanogaster? If so, what are the patterns of genotype-environment interactions for spontaneous mutations in C. neoformans? Third, under different mutation accumulation conditions (25° and 37°, both on the same rich medium), do estimated mutational parameters and the patterns of genotype-environment interactions for spontaneous mutations differ? If differences are found, how do they correspond to vegetative growth patterns of environmental and clinical populations of C. neoformans? And finally, how can the analysis shed light on the life history of C. neoformans and on the evolution and maintenance of pathogenicity traits in human pathogens?
MATERIALS AND METHODS
Strains:
Strain JEC21 was used to set up the MA lines. JEC21 is serotype D and mating-type α (MATα). It was created in 1992 through genetic crossing (Kwon-Chung et al. 1992). The parental strains for JEC21 were NIH433, a serotype D, MATa environmental isolate from pigeon droppings in Denmark, and NIH12, a serotype D, MATα clinical isolate from a patient in the United States. Therefore, strain JEC21contains genetic materials from both environmental (NIH433) and clinical (NIH12) sources. However, which sections of the JEC21 genome came from which parental strains is unknown. JEC21 was chosen here because it was one of the most widely used laboratory strains for genetic and molecular studies and its genome has been targeted for complete sequencing by the Stanford DNA Sequencing and Technology Center and by The Institute of Genome Research. These features would make potential follow-up studies (e.g., using microarrays to trace gene expression changes) more effective and manageable. Forty-five environmental and 35 clinical strains of C. neoformans from the McMaster University Fungal Collection were also included for comparison. These 80 strains were isolated from a variety of geographical locations in Europe, Asia, and North America over the last 10 years and were stored in freezers with minimal laboratory propagation and no genetic manipulation.
Mutation accumulation experiment:
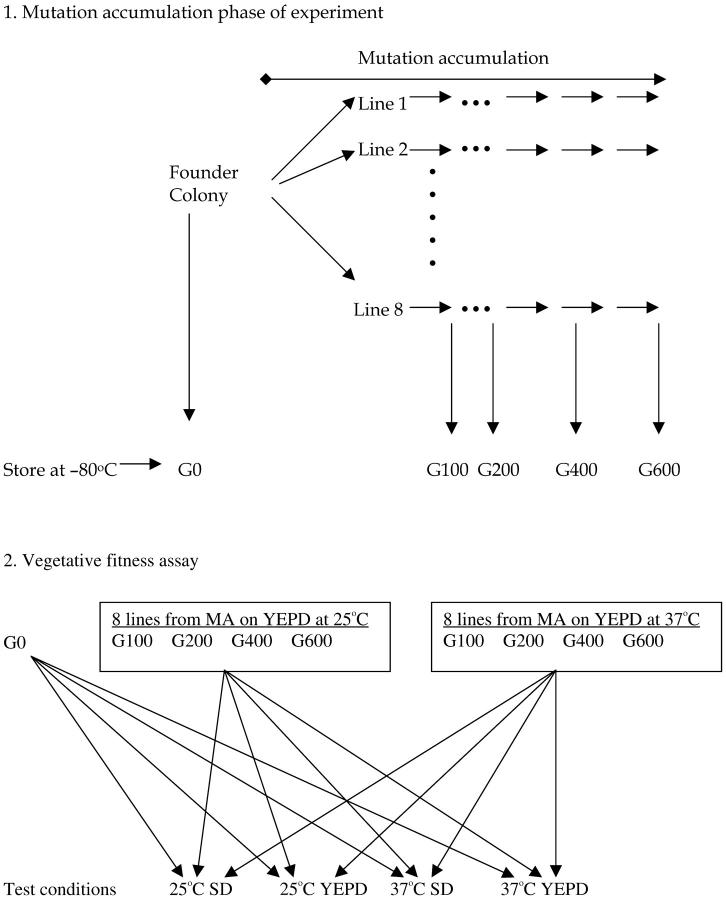
The overall experimental design is shown in Figure 1. Briefly, a single colony (the founder colony) of strain JEC21 was picked and streaked onto YEPD agar plates (1% yeast extract, 2% Bacto-peptone, 2% dextrose, and 2% Bacto-agar in distilled water) to establish 16 independent MA lines. Eight lines were incubated at 25° and the other 8 at 37°. Each of the 16 MA lines was maintained by repeating the picking-streaking-incubating procedure (called a growth cycle) of a single colony, approximately every 72 hr for lines maintained at 25° and every 48 hr for lines maintained at 37°. Within each growth cycle, each colony increased from 1 cell to ∼106 cells. This cell count was obtained by cutting agar pieces containing entire colonies and resuspending them individually in 1 ml sterile water through vigorous vortexing. The cell suspensions were then diluted and plated on YEPD plates for viable cell counts. The growth from 1 cell to ∼1 million represented ∼20 mitotic divisions (±1 cell division). These 16 MA lines were maintained for 30 growth cycles, equivalent to ∼600 mitotic divisions.
Figure 1.—
Schematic of experimental design. A single colony of the standard laboratory strain JEC21 was used to establish eight MA lines under each of two incubation conditions (25° on YEPD and 37° on YEPD). For each of the 16 MA lines, subcultures were stored after 5, 10, 20, and 30 transfers, roughly corresponding to 100, 200, 400, and 600 mitotic generations, respectively. The founder clone and the 64 derived MA clones (16 lines × 4 time points) were then tested for vegetative fitness on two solid media (SD and YEPD) incubated under two different temperatures, 25° and 37°.
To ensure that picked colonies descended from a single cell in each growth cycle, a small cluster of cells (∼104–105) from a colony was resuspended in 1 ml sterile water by vigorous vortexing, and 1 μl of the suspension was streaked onto agar medium. Streaked single cells were confirmed by microscopy and one cell from each MA line was randomly marked for future transfers. Cells from generations 0, 100, 200, 400, and 600 (abbreviated G0, G100, G200, G400, and G600, respectively) were stored in 18% glycerol in a −80° freezer. This protocol intensified genetic drift by forcing each line through one random cell in each growth cycle. Therefore, barring selection and competition within each colony during the clonal asexual growth, mutations were expected to accumulate freely in each line. Because different lines were maintained independently of each other, different numbers and types of mutations were expected to accumulate in different lines.
Vegetative fitness:
The vegetative fitness of the evolved clones relative to the original was determined in each of four environments: (i) 25° on YEPD, the nutrient-rich medium originally used for MA; (ii) 37° on YEPD; (iii) 25° on SD without any organic nitrogen source (i.e., no amino acids and no nucleotide bases); and (iv) 37° on SD. Here, colony size was used as the indicator of vegetative fitness. The use of colony size to measure vegetative fitness in fungi has a long tradition dating back several decades (e.g., Simchen and Jinks 1964) and has been extensively used in recent and current work (e.g., Xu 1995, 2002; Zeyl and Bell 1997; Xu et al. 1998; Larraya et al. 2002; Pringle and Taylor 2002). In this method, cell suspension and streaking were performed as described above for colony transfers during MA. Immediately after streaking, 10 well-separated cells were randomly marked for later measurement. Cells were incubated for 72 hr for the two 25° environments and 48 hr for the two 37° environments. The diameters of individual colonies were measured under the microscope using an ocular scale. Ten random colonies were measured for each clone in each of the four environments. Because of the large number of colonies to be measured, the evolved clones were divided into 16 sets with eight MA clones in each set. The original clone was included in each set for standardization. No difference was observed between different sets for the original clone grown in each of the four environments.
To determine the vegetative growth for the 35 clinical and 45 environmental strains, cells were first retrieved from −80° freezer stocks and incubated on YEPD plates at 25° for 11/2 days (∼8–10 cell divisions) until visible colonies appeared on the agar surface. One random colony from each of the 80 strains was then resuspended in 50 μl sterile water and streaked onto solid media for vegetative fitness testing. The remaining steps were identical to those described above for vegetative fitness determination of evolved clones. However, because there was no significant interaction between temperature and medium independent of genotype on vegetative fitness (see below and Table 3), only the rich medium YEPD was used for these 80 strains. In addition, to make measurements comparable for direct testing of the effects of strain source (environmental vs. clinical) and temperature (25° vs. 37°) on vegetative fitness, all vegetative fitness measurements were made after 48 hr of incubation.
TABLE 3.
Three-way ANOVA for genotype-environment interactions of spontaneous mutations in experimental populations ofC. neoformans
| MA condition
|
|||||
|---|---|---|---|---|---|
| T25/YEPD
|
T37/YEPD
|
||||
| Source of variation | d.f. | Mean square | F | Mean square | F |
| MA lines (L) | 7 | 0.1241 | 29.188*** | 0.0618 | 15.096*** |
| Temperature (T) | 1 | 0.6362 | 149.70*** | 0.0032 | 0.7805NS |
| Medium (M) | 1 | 0.4084 | 96.104*** | 0.0293 | 7.1543** |
| L × T | 7 | 0.0396 | 9.3194*** | 0.0139 | 3.4151** |
| L × M | 7 | 0.0488 | 11.483*** | 0.0274 | 6.6967*** |
| T × M | 1 | 0.0012 | 0.2858NS | 0.0090 | 2.2015NS |
| L × T × M | 7 | 0.0451 | 10.615*** | 0.0197 | 4.8172*** |
| Within treatment | 288 | 0.0043 | 0.0041 | ||
Only data from G600 were used in these analyses. **P < 0.01; ***P < 0.001; NS, not significant.
Estimates of mutational parameters:
To standardize our calculations and estimations, the vegetative fitness of the starting clone in each of the four testing environments was defined as one and the relative fitness of each evolved clone was expressed accordingly as a simple ratio of colony size of the evolved clone over the starting clone. This adjustment was necessary to eliminate intrinsic differences in vegetative fitness of C. neoformans in the four testing environments. The standardization allows the comparison of vegetative fitness differences derived only from newly accumulated mutations. The mean and standard deviation of relative vegetative fitness were calculated for the starting clone and for each of the 64 evolved clones in the four environments.
The data were then used to estimate the genomic mutation rate per mitotic division (Û), the average effect per mutation (â), and the mutational heritability h2m. Since JEC21 is haploid, estimation procedures followed those for haploids as described by Lynch and Walsh (1998). Because phenotypic changes in MA lines were products of both the mutation rate and the effect per mutation, with a given phenotypic change, mutation rate and mutational effects would be in inverse relationship. Current procedures provide estimates for minimum mutation rate (Ûmin) and maximum mutational effects (âmax) (Lynch and Walsh 1998) in haploids:
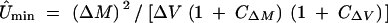 |
and
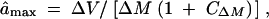 |
where ΔM and ΔV denote estimates of the rates of change of mean and variance, and CΔM and CΔV are squared coefficients of sampling variance (ratios of sampling variance to squared estimates) of ΔM and ΔV, respectively. The mutational heritability h2m was obtained as mutational variance scaled by environmental variance. These functions have been used in mutation accumulation studies in bacteria, plants, and animals (e.g., see a summary in Lynch and Walsh 1998). These mutational parameters were obtained for each of the two MA conditions and in each of the four testing environments.
Genotype-environment interaction of spontaneous mutations:
The vegetative growth data of MA clones were analyzed by three-way factorial analysis of variance (ANOVA). The three factors were MA line, incubation temperature, and medium. Sums of squares were partitioned into sources attributable to the three main effects, three pairwise two-way interactions, one three-way interaction, and the within-treatment random error. The three-way ANOVA was done for the two MA conditions separately. Wilcoxon's signed-rank test was used to compare fitness among environments for the same MA lines. These statistical tests followed the procedures in Sokal and Rohlf (1981), using the Microsoft Excel program.
Comparison between clinical and environmental strains:
The vegetative growth data of environmental and clinical samples were analyzed by two-way ANOVA with each strain as a replicate. The two factors were source of strain and incubation temperature. Sums of squares were partitioned into sources attributable to the two main effects and temperature-strain source interaction. The statistical test followed the procedures in Sokal and Rohlf (1981), using the Microsoft Excel program.
RESULTS
Reduced fitness among derived clones:
To standardize our comparisons, the relative vegetative fitness of the original clone in each of the four environments was scaled to 1 and those of all other evolved clones were adjusted accordingly as a simple ratio of colony sizes between the derived clone and the original clone (the denominator). The relative fitness for each of the derived clones at generation 600 (G600) is presented in Table 1. As shown, after 600 asexual divisions, relative to that of the starting clone, the fitness of all 16 clones had decreased in all four environments (Table 1).
TABLE 1.
Relative fitness of MA lines at generation 600 in various environments
| Testing environments
|
|||||
|---|---|---|---|---|---|
| MA condition | MA line | T25/SD | T25/YEPD | T37/SD | T37/YEPD |
| T25/YEPD | 1 | 0.112 | 0.828 | 0.056 | 0.109 |
| 2 | 0.112 | 0.939 | 0.062 | 0.652 | |
| 3 | 0.710 | 0.899 | 0.595 | 0.516 | |
| 4 | 0.776 | 0.878 | 0.867 | 0.891 | |
| 5 | 0.822 | 0.888 | 0.379 | 0.538 | |
| 6 | 0.934 | 0.888 | 0.303 | 0.636 | |
| 7 | 0.944 | 0.808 | 0.056 | 0.592 | |
| 8 | 0.738 | 0.727 | 0.477 | 0.766 | |
| Mean ±SD | 0.644 ±0.338 | 0.857 ±0.066 | 0.349 ±0.293 | 0.588 ±0.229 | |
| T37/YEPD | 1 | 0.450 | 0.754 | 0.760 | 0.887 |
| 2 | 0.690 | 0.913 | 0.890 | 0.785 | |
| 3 | 0.790 | 0.845 | 0.830 | 0.750 | |
| 4 | 0.910 | 0.894 | 0.910 | 0.781 | |
| 5 | 0.640 | 0.715 | 0.890 | 0.797 | |
| 6 | 0.870 | 0.884 | 0.870 | 0.727 | |
| 7 | 0.440 | 0.541 | 0.150 | 0.852 | |
| 8 | 0.940 | 0.928 | 0.850 | 0.785 | |
| Mean ±SD | 0.715 ±0.195 | 0.809 ±0.132 | 0.768 ±0.254 | 0.795 ±0.052 | |
Fitness of the original starting clone was scaled to 1 in each environment. Each data point represents the mean of 10 measurements
For MA lines maintained at temperature 25° (T25), the highest mean fitness was observed in the same environment where mutations were accumulated (T25/YEPD) and the lowest mean fitness was at a different temperature (37°) and on a different medium, SD (T37/SD). At G600, the most significant drops occurred in MA lines 1, 2, and 7. These clones had only one to two cell divisions under the T37/SD condition (i.e., approximately two to four cells per colony). These three clones could be considered to have conditional lethal mutations in this environment. Lines 1 and 2 also showed a similar growth pattern in the T25/SD environment where only four to eight cells were observed per colony. Coupled to the decline in fitness was divergence among lines. The standard deviation of fitness among MA lines was lowest in the MA environment (T25/YEPD), and much greater in the other three environments (Table 1).
Interestingly, for MA lines maintained at 37° (T37), the highest mean fitness was observed in the T25/YEPD environment, not in the original MA environment, T37/YEPD. Also different was the environment producing the lowest mean fitness, T25/SD. Overall, the loss of mean fitness was smaller for MA lines maintained at 37° than for those maintained at 25°. Unlike the large differences in mean fitness among testing environments for MA lines maintained at T25/YEPD, testing conditions had relatively minor effects on the average vegetative fitness for MA lines maintained at T37/YEPD. Similar to the conditional lethality of the spontaneous mutations observed above, one line, line 7, from MA condition T37/YEPD had only a few (six to eight) cells per colony, likely the result of conditional lethal mutation(s). The patterns of fitness divergence among MA lines maintained in the T37/YEPD environment were similar to those from MA lines maintained at T25/YEPD. The least divergence was observed in the original MA environment (T37/YEPD). Greater divergences were seen in the other three testing environments (Table 1).
Differences in estimates of mutational parameters:
On the basis of the patterns of fitness decline and the among-line divergence, the genomic mutation rate (Ûmin), mutational effect (âmax), and mutational heritability h2m for fitness were estimated for the two MA conditions in each of the four testing environments. A summary of the results of these estimates is presented in Table 2. Both MA conditions and testing environments showed influences on all three mutational parameters.
TABLE 2.
Estimates of mutational parameters from two mutation accumulation conditions in four testing environments
| Testing condition
|
|||||
|---|---|---|---|---|---|
| MA condition | Mutation parameters | T25/SD | T25/YEPD | T37/SD | T37/YEPD |
| T25/YEPD | Ûmin (×10−3) | 1.815 | 1.127 | 5.662 | 1.982 |
| âmax | 0.253 | 0.007 | 0.147 | 0.085 | |
| h2m (×10−3) | 2.124 | 0.213 | 1.185 | 1.962 | |
| T37/YEPD | Ûmin (×10−3) | 5.334 | 1.253 | 0.702 | 1.363 |
| âmax | 0.057 | 0.029 | 0.156 | 0.014 | |
| h2m (×10−3) | 0.326 | 0.445 | 0.646 | 0.091 | |
The mutational parameters Ûmin, âmax, and h2m are defined in materials and methods.
For MA lines maintained at T25/YEPD, the estimated mutation rate was highest (5.662 × 10−3) under the T37/SD testing condition and lowest (1.127 × 10−3) under the T25/YEPD condition. Interestingly, while the fitness effect per mutation was lowest in the T25/YEPD (0.00656) environment, the highest was in T25/SD (0.25337), not in T37/SD (0.14669). The rank order of mutational heritability estimates was similar to that of mutational effects (Table 2).
For MA lines maintained at T37/YEPD, the estimated mutation rate was highest (5.332 × 10−3) under the T25/SD testing condition and lowest (0.702 × 10−3) under T37/SD. However, the estimated mutational effect was largest in T37/SD. Similar to estimates from MA lines maintained at T25/YEPD, both mutational effects and mutational heritability for vegetative fitness were lowest in the T37/YEPD environment where MA lines were maintained (Table 2).
There are several noteworthy observations regarding estimates from the two MA conditions. First, the testing environments exhibiting the highest estimates of mutation rates were the ones most dissimilar to the original MA conditions (T37/SD for MA lines maintained at T25/YEPD and T25/SD for MA lines maintained at T37/YEPD). Second, the testing conditions exhibiting the lowest mutational effects and mutational heritability were the environments where MA lines were originally maintained. Third, except for MA lines maintained and tested in the T25/YEPD environment, all estimates of mutational parameters were within a 10-fold range of each other. Fourth, except for MA lines maintained and tested in the T25/YEPD environment, estimates of mutational effects and mutational heritability were smaller in MA lines maintained at T37/YEPD than in those maintained at T25/YEPD.
Genotype-environment interactions of spontaneous mutations:
The ability of a genotype to modify phenotypic expression in response to different environmental conditions is referred to as phenotypic plasticity (Scheiner 1993). Traditionally, phenotypic plasticity is described by the norm of reaction—a plot of measurements for the same trait in different environments. The difference between measurements in different environments is called environmental sensitivity. Not all genotypes respond similarly to the same environmental signals. The variation in response among genotypes is termed genotype-environment interaction. In this study, the genotypic differences are caused exclusively by spontaneous mutations accumulated during the course of the experiment. Therefore, the genotypic components of genotype-environment interactions are from spontaneous mutations.
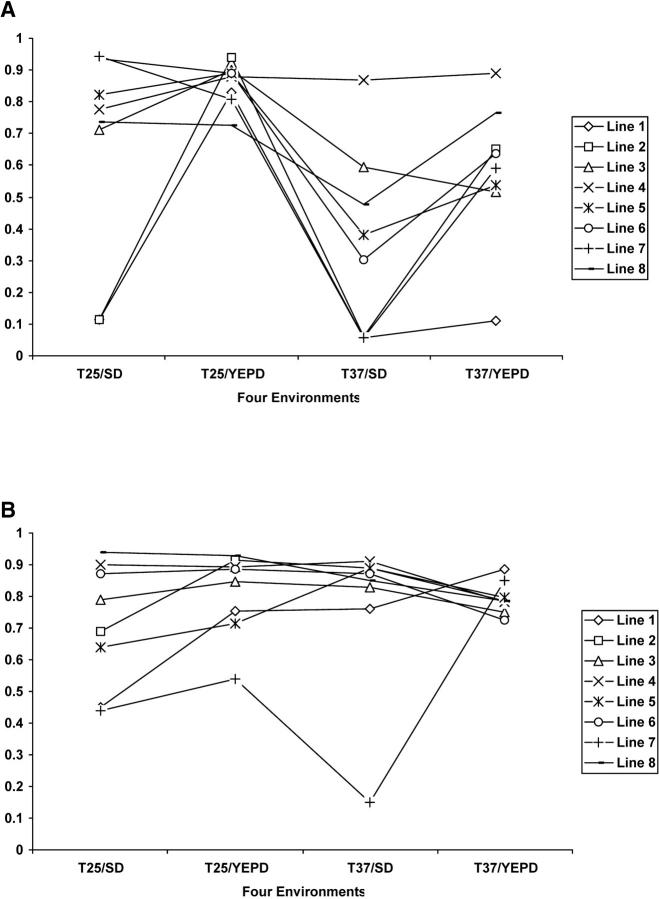
The representative norms of reaction among MA lines in the four environments are shown in Figure 2. Figure 2A shows the eight MA lines maintained at T25/YEPD and Figure 2B shows those maintained at T37/YEPD. If there were no genotype-environment interactions, the MA clones should have parallel reaction norm lines and the crossing of reaction norm lines should not occur. However, as shown in Figure 2, crossing of reaction norm lines was common for MA clones derived from both MA conditions. This result is consistent with the presence of genotype-environment interactions of the spontaneous mutations accumulated in this study.
Figure 2.—
Relative mean fitness of mutation accumulation lines grown under four different conditions, shown on the x-axis from left to right: (i) 25° on SD medium; (ii) 25° on YEPD medium; (iii) 37° on SD medium; (iv) 37° on YEPD medium. Only clones from G600 are shown here. (A) MA lines maintained at 25° on YEPD medium. (B) MA lines maintained at 37° on YEPD medium. All MA lines correspond to those in Table 1.
Statistical significances of genotype-environment interactions of these spontaneous mutations were determined on the basis of three-way ANOVA tests. Results of the ANOVA tests are summarized in Table 3. For single-factor analysis, there was a significant contribution by MA lines under both MA conditions (Table 3). Similarly, the two media (YEPD and SD) showed significant differences in the observed fitness reductions of spontaneous mutations, with cultures grown on the nutrient-poor medium SD showing significantly lower fitness than those grown on the nutrient-rich medium YEPD. Mutations accumulated under different MA conditions reacted differently to testing temperatures. Specifically, mutations accumulated at T25/YEPD showed a significant contribution of temperature to the total fitness variance. However, no significant contribution was observed for temperature for mutations accumulated at T37/YEPD (Table 3).
For bifactor analyses, statistically significant interactions affecting vegetative fitness were observed between MA lines and temperature and between MA lines and medium (Table 3). Interactions between temperature and medium were not statistically significant for mutations accumulated in either MA environment. However, mutations accumulated in both environments showed significant three-way interactions among MA lines, temperature, and medium (Table 3).
Comparison between environmental and clinical strains:
A summary of statistics of vegetative fitness for clinical and environmental samples of C. neoformans is presented in Table 4. Overall, strains in both samples grew significantly faster at 37° than at 25°. At 25°, the environmental sample showed a significantly higher vegetative fitness (30.7% growth advantage) than the clinical sample (t = 3.358, d.f. = 78, P < 0.01). At 37°, while the clinical sample showed a mean advantage of 23.4% over the environmental sample, the difference was not statistically significant (t = 0.658, d.f. = 78, P > 0.10), due to large variations among strains within each sample. In both testing environments, the environmental sample showed a greater coefficient of variation (i.e., standard deviation/mean) than the clinical sample, with the smallest coefficient of variation found for the clinical sample incubated at T37/YEPD (0.096/0.637 = 0.15) and the largest for the environmental sample incubated at T25/YEPD (0.091/0.251 = 0.36).
TABLE 4.
Summary of vegetative fitness comparisons between clinical and environmental samples ofC. neoformans at two different temperatures, 25° and 37°
| Testing environment
|
|||
|---|---|---|---|
| Population sample | No. of strains | T25/YEPD | T37/YEPD |
| Environmental | 45 | 0.251 ± 0.091 | 0.516 ± 0.181 |
| Clinical | 35 | 0.192 ± 0.057 | 0.637 ± 0.096 |
| Two-way ANOVA table | |||
| Source of variation | d.f. | Mean square | F |
| A: Sample source (clinical vs. environmental) | 1 | 0.0003 | 0.0205NS |
| B: Incubation temperature (25° vs. 37°) | 1 | 1.1662 | 79.877*** |
| C: A × B Interaction | 1 | 4.1382 | 284.02*** |
| D: Within subgroup (error term) | 156 | 0.0146 | |
Data are presented as mean ± standard deviation of colony sizes in millimeters. ***P < 0.001; NS, not significant.
Due to the averaging effects of the two temperature conditions (Table 4), no statistically significant difference was found between the clinical and the environmental samples when results from the two temperatures were combined in the analysis. In contrast, significant interaction was found between source of strain and incubation temperature (Table 4). The analysis indicated that, overall, the sample from clinical sources grew faster at 37° but slower at 25° than did the environmental strains.
DISCUSSION
In this study, an MA experiment was performed to investigate mutational parameters and genotype-environment interactions of spontaneous mutations for fitness in the human pathogenic yeast C. neoformans. Sixteen MA lines were maintained on the rich medium YEPD, with 8 lines at each of two temperature conditions (25° and 37°). After ∼600 mitotic divisions, all 16 lines showed reduced vegetative fitness. The degree and patterns of reduction in vegetative growth varied between MA conditions, among MA lines within an MA condition, and among testing environments that differed in temperature and medium. Significant genotype-environment interactions were detected for the newly accumulated spontaneous mutations. To my knowledge, this study provides the first estimates of mutational parameters for vegetative fitness in human pathogenic fungi. This is also the first study demonstrating that mutation accumulation conditions can have a significant influence on the types of spontaneous mutations being accumulated.
Estimates of mutational parameters:
The estimated genomic mutation rates (Ûmin) of 0.702 × 10−3–5.662 × 10−3 (an approximately eightfold difference) for vegetative fitness were similar to those determined using the same mutation accumulation technique for a variety of fitness traits in microbes, plants, and animals (Mukai 1964; Keightley and Caballero 1997; Drake et al. 1998; Lynch and Walsh 1998; Vassilieva and Lynch 1999; Zeyl and de Visser 2001). For example, Ûmin was 1.7 × 10−4 in the bacterium E. coli (Kibota and Lynch 1996), 0.02–0.6 for egg-adult viability in Drosophila (Mukai 1964; Drake et al. 1998), 0.0024–0.054 for fitness traits in A. thaliana (Schultz et al. 1999), and 0.003–0.060 for various life-history traits in the model nematode C. elegans (Keightley and Caballero 1997; Vassilieva and Lynch 1999). Unlike the present study, previous studies accumulated mutations under one environmental condition (typically a favorable environment) and examined fitness components typically in the same environment or a different environment that allowed greater sensitivity to detect deleterious mutations (e.g., Zeyl and de Visser 2001). In this study, two MA conditions and four testing environments were used to estimate mutational parameters.
Variation in the average effect per mutation (âmax) was also observed between MA conditions and among the four testing environments. The âmax estimates showed a greater range (0.007–0.253, an ∼36-fold difference) than that of genomic mutation rate (8-fold; see above). The variation in estimates of âmax can be attributable to differences in MA conditions and testing environments, with contributions from both testing temperature and medium (Table 2). Despite such a large variation, the estimates here were generally similar to those in other species. For example, the âmax for viability in D. melanogaster was on average 0.06 in homozygotes (Mukai et al. 1964), 0.1 for total fitness in A. thaliana (Schultz et al. 1999), 0.012 for vegetative growth rate in E. coli (Kibota and Lynch 1996), and 0.217 for overall vegetative fitness in S. cerevisiae (Zeyl and de Visser 2001).
Mutational heritability h2m estimates for vegetative growth in C. neoformans (range 0.091–2.124, more than a 20-fold difference) are also similar to those for a variety of traits in several species (for a summary, see Houle et al. 1996). For example, h2m ranged from 0.38 × 10−3–13.5 × 10−3 for abdominal bristle number and 0.13 × 10−3–0.54 × 10−3 for viability in D. melanogaster; and 1.79 × 10−3–5.57 × 10−3 for a variety of morphological and life history traits in rice (Oryza sativa; Houle et al. 1996).
The decline of vegetative fitness in asexual clones over time is consistent with the observation in fungi that asexual clones cannot persist indefinitely. For >20% of the 70,000 or so identified species of fungi, sexual reproduction has not been found or detected. Interestingly, limited phylogenetic studies have identified that most of these “asexual” species have close relatives capable of sexual reproduction (e.g., Geiser et al. 1996). This result is consistent with the rapid mutational meltdown of asexual fungal clones. The results here provide quantitative estimates for the rates of fitness decline and how environmental conditions can influence the relative fitness of evolving asexual clones (see also below).
Genotype-environment interactions of spontaneous mutations accumulated at T25/YEPD:
Significant interactions were observed between spontaneous mutations accumulated in MA lines and the two testing environmental factors, temperature and medium. In general, the MA lines had greater relative vegetative fitness in the environment T25/YEPD where mutations were accumulated. How did such differences arise? While the exact mechanisms are not known, there are several possibilities. The first possibility is that strains of C. neoformans are typically cultured at ∼25° in the laboratory. Such a practice may have allowed preadaptation of the starting clone to the 25° environment. As described in materials and methods, JEC21 was generated in the early 1990s (Kwon-Chung et al. 1992). Since then, because of its superior mating ability, JEC21 and its isogenic mating partner JEC20 (MATa) have been passed to many clinical and research laboratories as standard tester strains for determining the mating types of unknown C. neoformans cultures. The stock culture used in this study was obtained from Joe Heitman and Tracy Moore of Duke University in 1997 and has since been stored in a −80° freezer. Because the number of transfers and mitotic divisions for JEC21 in different laboratory environments is impossible to trace, the effects of this potential preadaptation to T25/YEPD on the current results cannot be adequately assessed.
The second possibility is that different environments could detect different mutations with some environments detecting a larger number of mutations and/or larger fitness effects of these mutations. For example, the differences in vegetative fitness between the two environments, T25/SD and T25/YEPD, were due to nutrient availability in the medium. SD medium contained only an inorganic nitrogen source (ammonium sulfate) but no organic nitrogen source (no amino acids and nucleotide bases) while YEPD medium had abundant organic compounds, including all amino acids and nucleotide bases, in abundant supply. Therefore, the greater reduction in vegetative fitness on SD medium than on YEPD medium in six of the eight lines (Table 1, MA lines 1, 2, 3, 4, 5, and 8) was likely due to mutations deleterious to the synthesis of organic compounds such as amino acids and nucleotide bases. The remaining two lines, 6 and 7, had a slightly different pattern. Fitness reduction in these two lines was greater in the T25/YEPD environment than in the T25/SD environment. This reversed pattern could be due to mutations in transport of organic compounds. While extensive transport systems were required for the acquisition of organic nitrogenous compounds from YEPD medium, such systems were not required for growth in SD medium as there were no organic nitrogenous compounds in the SD medium. Similarly, the differences between T25/YEPD and T37/YEPD in reduction of vegetative growth for MA lines were due to temperature differences. Lines from the T25/YEPD MA condition showed significantly greater reductions in vegetative growths on T37/YEPD than on T25/YEPD. Of the two parameters, Ûmin and âmax, âmax was over 10 times greater in the T37/YEPD testing environment than in the T25/YEPD environment while Ûmin estimates were similar in these two environments (less than a twofold difference). This pattern suggested that âmax, the per mutation effect, had a greater contribution than Ûmin, the genomic mutation rate, to the difference in fitness declines between T25/YEPD and T37/YEPD environments. Large-scale gene expression analysis of the evolved clones grown in different testing environments could help identify the mutations that contributed the significant genotype-environment interactions.
The third possibility is intracolony selection. In this scenario, the protocols of mutation accumulation are biased in favor of retaining mutations having little or no effect on the specific MA condition. This hypothesis implies that spontaneous mutations were not accumulated freely and that selection within a colony must operate during each growth cycle. Such intracolony selection would reduce the frequency of mutations with large deleterious effects on the MA condition.
It has been assumed that intensive genetic drift such as that applied here—one random individual colony per growth cycle—should minimize selection and allow mutations to accumulate freely. Given the significant differences in fitness among testing environments for the same MA lines, and if intracolony selection was the primary cause, current mutation accumulation protocols must have led to significant underestimates in genomic mutation rates, mutational effects, and/or mutational heritability. To correct such biases, different MA conditions and a variety of testing environments should be conducted for more robust estimates of mutational parameters.
Differences between MA conditions and implications for C. neoformans life history:
While large differences in fitness reduction among testing environments were observed for MA lines maintained at T25/YEPD, overall those maintained at T37/YEPD showed relatively little variation. In addition, both single-factor effects and interaction effects were weaker for spontaneous mutations accumulated among MA lines maintained at 37° than for those among MA lines maintained at 25°. How could such differences arise and what are the implications for the life history and pathogenicity traits in C. neoformans?
At present, the exact mechanisms for the observed differences between the two MA conditions are not known. One possibility is that genes related to growth at high temperatures in C. neoformans have epistatic and pleiotrophic effects on growth in lower temperature environments and on nutrient acquisition and synthesis pathways. If so, selection pressure exerted by intracolony competition during mutation accumulation at the 37° environment could help maintain the genetic architecture for growth at lower temperatures and a variety of nutrient environments. Interestingly, although statistically not significant, the T25/YEPD testing environment, not the T37/YEPD environment, revealed the least reduction in vegetative fitness of MA clones maintained at T37/YEPD. This result is consistent with the preadaptation hypothesis discussed in the previous section (i.e., the first hypothesis discussed above).
The mutation accumulation experiments reported here were performed under laboratory conditions unlikely to be found in natural environments. Therefore, the mutational estimates are unlikely to correspond exactly to those under clinical or environmental conditions. In their natural habitats, most microorganisms, including C. neoformans, likely experience fluctuations in temperature and nutrient levels. Unfortunately, there is no published estimate of the extent of such fluctuations or their effects on generation times and vegetative fitness in natural habitats. Despite these drawbacks, the results here from laboratory populations of C. neoformans might be relevant to our understanding of the natural history of C. neoformans and, potentially, other microbes as well.
C. neoformans is considered an opportunistic pathogen. The assumed natural reservoirs of C. neoformans are assumed to be in soil, trees, and bird droppings—environments with temperatures typically well below 37° and most of the time probably below 25° (Casadevall and Perfect 1998). Unlike many other fungi, no strain of C. neoformans has been found in large organic composts where temperatures can reach 50°–60°. Indeed, a week-long exposure to 40°–41° environments would kill most strains of C. neoformans (Casadevall and Perfect 1998). In contrast, most strains can grow and maintain viability at 10°–15°. The experiments here showed that in low-temperature environments (such as 25°) the ability to grow at high temperature (such as 37°) could be lost rather quickly. However, the ability to grow at 37° is essential for pathogenesis in mammalian hosts and indeed is the only consensus virulence factor among all human pathogens, fungal pathogens included. Therefore, the data here suggest that extended exposure to high-temperature environments is essential for the evolution and maintenance of this pathogenicity trait in C. neoformans.
There are two candidate high-temperature environments for C. neoformans: (i) in warm-blooded animals and (ii) in microenvironmental niches in soil particles, bird droppings, trees, or other organic matter. These microenvironmental niches could periodically experience high temperatures due to heat generated by the degradation of organic compounds. However, it is unlikely that such high temperatures could be sustained for long periods of time without exceeding 40°–41° and effectively killing the C. neoformans cells.
The possibility that warm-blooded animals might play a greater than expected role in the life history of pathogenic strains of C. neoformans is supported by data from population genetic studies of C. neoformans. Epidemiological surveys have identified a few clones or clonal lineages that dominate the global clinical strains of C. neoformans and have shown that clonal dispersals are common over a wide geographic area (e.g., Brandt et al. 1996; Xu et al. 2000). If pathogenic strains of C. neoformans spent most of their time in soil, trees, and bird droppings and warm-blooded animals were only accidental hosts, it would be expected that geographic populations should diverge from each other rather rapidly. Such divergences should lead to geographic structuring and the lack of clonal dispersal over long distances—phenomena not seen in clinical or environmental populations of C. neoformans (e.g., Brandt et al. 1996; Xu et al. 2000). Indeed, current population genetic surveys are consistent with the hypothesis that warm-blooded animals such as humans and/or other mammalian hosts might act as important reservoirs and selective agents for pathogenic strains. Such a reservoir and the selective pressure for growth at high temperatures could purge nonpathogenic or attenuated strains from human populations, leading to limited genetic diversity, extensive clonality, and long-distance clonal dispersal (also by humans) of these pathogenic strains.
The importance of a high-temperature environment for the maintenance of growth of C. neoformans at 37° is also supported by vegetative fitness comparisons between clinical and environmental samples. Significant interactions between strain source and temperature were observed. Overall, strains from clinical sources showed a greater vegetative fitness at 37° and a lower fitness at 25° than those from environmental sources. However, it should be noted that, while overall consistent, the magnitude and statistical significance of differences between laboratory populations (Tables 1 and 3) were not identical to those between clinical and environmental populations (Table 4). The lack of statistically significant difference in vegetative fitness between the clinical and the environmental samples (Table 4) at either temperature suggests that the high-temperature environment in humans and/or other mammals might be important for both samples. If the ancestors of the environmental strains had lived only in the environment outside of human or other mammalian hosts, we should expect the environmental strains to lose the ability to grow at high temperature rather quickly. This expectation was not met. Their vigorous growth at high temperature suggests that the environmental strains may have had significant exposure to humans or other mammalian hosts. Results from population genetic studies of environmental samples are also consistent with this hypothesis (see above). At present, aside from the sources prior to the isolation of each strain (patient vs. environment), their previous environments, selection pressure, and the lengths of time in their current environments are unknown for any of these 80 natural strains. In contrast, the spontaneous mutations in laboratory populations were all accumulated under defined conditions over a defined period of time.
It should be emphasized that an extended exposure to warm-blooded animals such as humans does not suggest human-to-human transmission of infectious particles nor does it mean that environmental reservoirs are insignificant. Indeed, natural environments with low temperatures (such as 25°) and low nutrient levels (such as SD with low nitrogen sources) are necessary for mating and sexual reproduction in C. neoformans (Kwon-Chung 1976). Laboratory studies identified that environments with high temperatures (such as 37°) or nutrient-rich medium (such as YEPD) were not conducive for mating and sexual recombination in C. neoformans. Sexual recombination in nature could generate pools of genetically diverse offspring, some of which may have equivalent or greater fitness and pathogenicity than the parental strains. Indeed, sexual recombination in natural environments could help avoid the mutational meltdown that can occur in asexual clones. Recent studies have shown that rare sexual recombination exists in environmental and clinical populations of C. neoformans (Xu et al. 2000; Xu and Mitchell 2003). Given that strains of C. neoformans do not appear to mate at 37°, the recombination events must have occurred in natural environments outside of warm-blooded hosts.
Conclusion:
Using an experimental population approach, this study provided the first estimates of mutational parameters for vegetative growth in human fungal pathogens. The results showed that, with successive population bottlenecks, asexual populations of C. neoformans accumulated mutations deleterious to vegetative fitness. The accumulated spontaneous mutations showed significant genotype-environment interactions. In addition, the two MA conditions (25° and 37°, both on rich medium YEPD) showed significant effects on estimates of mutational parameters, a result suggesting that previous MA experiments using a single MA condition might have produced biased estimates of mutational parameters. Coupled with vegetative fitness comparisons between environmental and clinical samples, the results here suggest that high-temperature environments likely play a much greater role than previously thought in the natural history of pathogenic strains of C. neoformans. Further molecular analysis of mutations accumulated here may help elucidate the nature of the spontaneous mutations influencing vegetative growth in C. neoformans. The approaches presented here may also be applicable for inferring the life history traits of other environmental pathogens of humans and other mammals. At present, comparable experimental investigations are lacking for other human pathogens.
Acknowledgments
I thank Heather Yoell, two anonymous reviewers, and the Associate Editor of Genetics, Mimi Zolan, for comments on this manuscript. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada, the Premier's Research Excellence Award, Genome Canada, the Canadian Foundation for Innovation, and the Ontario Innovation Trust.
References
- Brandt, M. E., L. C. Hutwagner, L. A. Klug, W. S. Baughman, D. Rimland et al., 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 34: 912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall, A., and J. R. Perfect, 1998 Cryptococcus neoformans. ASM Press, Washington, DC.
- Charlesworth, B., and N. H. Barton, 1996. Recombination load associated with selection for increased recombination. Genet. Res. 67: 27–41. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., M. T. Morgan and B. Charlesworth, 1990. Inbreeding depression genetic load and the evolution of outcrossing rates in a multilocus system with no linkage. Evolution 44: 1469–1489. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., 1992. Mutation, mean fitness, and genetic load. Oxf. Surv. Evol. Biol. 9: 3–42. [Google Scholar]
- Drake, J. W., B. Charlesworth, D. Charlesworth and J. F. Crow, 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudash, M. R., 1990. Relative fitness of selfed and outcrossed progeny in a self-compatible, protandrous species, Sabatia angularis L. (Gentianaceae): a comparison in three environments. Evolution 44: 1129–1139. [DOI] [PubMed] [Google Scholar]
- Fry, J. D., S. L. Heinsohn and T. F. C. Mackay, 1996. The contribution of new mutations to genotype-environment interaction for fitness in Drosophila melanogaster. Evolution 50: 2316–2327. [DOI] [PubMed] [Google Scholar]
- Geiser, D. M., W. E. Timberlake and M. L. Arnold, 1996. Loss of meiosis in Aspergillus. Mol. Biol. Evol. 13: 809–817. [DOI] [PubMed] [Google Scholar]
- Houle, D., B. Morikawa and M. Lynch, 1996. Comparing mutational variabilities. Genetics 143: 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley, P. D., and A. Caballero, 1997. Genomic mutation rates for lifetime reproductive output and lifespan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 94: 3823–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibota, T. T., and M. Lynch, 1996. Estimate of the genomic mutation rate deleterious to overall fitness in Escherichia coli. Nature 381: 694–696. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1997. Evolutionary genetics of life cycles. Annu. Rev. Ecol. Syst. 28: 391–435. [Google Scholar]
- Kondrashov, A. S., and J. F. Crow, 1991. Haploidy or diploidy: Which is better? Nature 351: 314–315. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., and D. Houle, 1998. Genotype-environment interactions and the estimation of the genomic mutation rate in Drosophila melanogaster. Proc. R. Soc. Lond. Ser. B 258: 221–227. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung, K. J., 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 67: 821–833. [PubMed] [Google Scholar]
- Kwon-Chung, K. J., J. C. Edman and B. L. Wickes, 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60: 602–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, R., 1995. Mutation and conservation. Conserv. Biol. 9: 782–791. [Google Scholar]
- Larraya, L. M., E. Idareta, D. Arana, E. Ritter, A. G. Pisabarro et al., 2002. Quantitative trait loci controlling vegetative growth rate in the edible basidiomycete Pleurotus ostreatus. Appl. Environ. Microbiol. 68: 1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998 Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mackay, T. F. C., and R. F. Lyman, 1998. Polygenic mutation in Drosophila melanogaster: genotype × environment interaction for spontaneous mutations affecting bristle number. Genetica 102/103: 199–215. [PubMed] [Google Scholar]
- Mitton, J. B., and M. C. Grant, 1984. Associations among protein heterozygosity, growth rate, and developmental homeostasis. Annu. Rev. Ecol. Syst. 15: 479–499. [Google Scholar]
- Mukai, T., 1964. The genetic structure of natural populations of Drosophila melanogaster. I. Spontaneous mutation rate of polygenes controlling viability. Genetics 50: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai, T., S. Chigusa and I. Yoshikawa, 1964. The genetic structure of natural populations of Drosophila melanogaster. II. Overdominance of spontaneous mutant polygenes controlling viability in homozygous genetic background. Genetics 50: 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge, L., and N. H. Barton, 1993. Optimality, mutation and the evolution of aging. Nature 362: 305–311. [DOI] [PubMed] [Google Scholar]
- Pringle, A., and J. W. Taylor, 2002. The fitness of filamentous fungi. Trends Microbiol. 10: 474–481. [DOI] [PubMed] [Google Scholar]
- Scheiner, S. M., 1993. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24: 35–68. [Google Scholar]
- Schultz, S. T., M. Lynch and J. H. Willis, 1999. Spontaneous deleterious mutations in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 11393–11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen, G., and J. L. Jinks, 1964. The determination of dikaryotic growth rate in the basidiomycete, Schizophyllum commune: a biometrical analysis. Heredity 19: 629–649. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1981 Biometry: The Principles and Practices of Statistics in Biological Research, Ed. 2. Freeman, New York.
- Steenbergen, J. N., H. A. Shuman and A. Casadevall, 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 98: 15245–15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilieva, L. L., and M. Lynch, 1999. The rate of spontaneous mutation for life-history traits in Caenorhabditis elegans. Genetics 151: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., 1995. Analysis of inbreeding depression in Agaricus bisporus. Genetics 141: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., 2002. Estimating the spontaneous mutation rate of loss of sex in the human pathogenic fungus Cryptococcus neoformans. Genetics 162: 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., and T. G. Mitchell, 2003. Comparative gene genealogical analyses of strains of serotype AD identify recombination in populations of serotypes A and D in the human pathogenic yeast Cryptococcus neoformans. Microbiology. 149: 2147–2154. [DOI] [PubMed] [Google Scholar]
- Xu, J., R. Vilgalys and T. G. Mitchell, 1998. Colony size can be used to determine the minimum inhibitory concentration of fluconazole for pathogenic yeasts. Journal of Clinical Microbiology. 36: 2383–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., R. J. Vilgalys and T. G. Mitchell, 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Molecular Ecology. 9: 1471–1481. [DOI] [PubMed] [Google Scholar]
- Zeyl, C., and G. Bell, 1997. The advantage of sex in evolving yeast populations. Nature. 388: 465–468. [DOI] [PubMed] [Google Scholar]
- Zeyl, C., and J. A. de Visser, 2001. Estimates of the rate and distribution of fitness effects of spontaneous mutation in Saccharomyces cerevisiae. Genetics 157: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]