Abstract
Medaka is emerging as a model organism for the study of vertebrate development and genetics, and its effectiveness in forward genetics should prove equal to that of zebrafish. Here, we identify by positional cloning a gene responsible for the medaka i-3 albino mutant. i-3 larvae have weakly tyrosinase-positive cells but lack strongly positive and dendritic cells, suggesting loss of fully differentiated melanophores. The region surrounding the i-3 locus is syntenic to human 19p13, but a BAC clone covering the i-3 locus contained orthologs located at 15q11–13, including OCA2 (P). Medaka P consists of 842 amino acids and shares ∼65% identity with mammalian P proteins. The i-3 mutation is a four-base deletion in exon 13, which causes a frameshift and truncation of the protein. We detected medaka P transcripts in melanin-producing eyeballs and (putative) skin melanophores on embryos and an alternatively spliced form in the non-melanin-producing ovary or oocytes. The mouse p is similarly expressed in gonads, but not alternatively spliced. This is the first isolation of nonmammalian P, the functional mechanism of action of which has not yet been elucidated, even in mammals. Further investigation of the functions of P proteins and the regulation of their expression will provide new insight into body color determination and gene evolution.
MEDAKA, a small freshwater teleost, is a useful vertebrate for genetic and developmental studies, because it reaches reproductive maturity in a short period (∼2 months) and its eggs, which are laid every day, are highly transparent [transparency can be maintained until the adults are fully mature (Wakamatsu et al. 2001)]. Tools for genetic experiments, such as inbred strains (Yamamoto 1975; Hyodo-Taguchi 1980; Shima and Shimada 1994), linkage maps (Naruse et al. 2004), genomic libraries from divergent inbred strains (Matsuda et al. 2001; Kondo et al. 2002), an expressed sequence tag (EST) database (http://mbase.bioweb.ne.jp/dclust/medaka_top.html), and an 8.9× genome sequence database (http://dolphin.lab.nig.ac.jp/medaka/) are publicly available. These tools have allowed the positional or positional-candidate cloning of the genes responsible for spontaneous medaka mutants with curious phenotypes (Tomita 1992). Such studies have provided intriguing insights into development and evolution in various fields, such as eye development (Loosli et al. 2001), scale formation (Kondo et al. 2001), sex determination (Matsuda et al. 2002), and body coloration (Fukamachi et al. 2001, 2004). On-going large-scale ethylnitrosourea mutagenesis (see Furutani-Seiki et al. 2004) and whole-genome sequencing projects will further accelerate these gene-hunting experiments and confirm the medaka as a model organism for genetic and developmental studies of vertebrates.
In medaka, ∼50 body-color mutants exhibit various defects in their chromatophores (Iwamatsu 1997; Kelsh et al. 2004). To date, however, only three genes responsible for these mutants have been isolated, i.e., those encoding tyrosinase, membrane-associated transporter protein (MATP), and somatolactin for the i-, b-, and ci-locus mutants, respectively (Koga and Hori 1997; Fukamachi et al. 2001, 2004). Identification of more genes and functional comparison of these with the corresponding genes in other model organisms (in mouse or zebrafish, which has one or three kinds of chromatophores, respectively, whereas medaka has four) should identify the conserved and divergent aspects of chromatophore-associated genes. This should in turn provide intriguing insights into how genes evolved to achieve such diversity of organisms. In view of these considerations, we undertook to isolate the gene responsible for a medaka pigmentation mutant with one of the most drastic phenotypes, the i-3 mutant.
MATERIALS AND METHODS
Fish:
The i-3 albino strain arose spontaneously from wild-type population around Tottori, Japan, and was kept at Nagoya University (Tomita 1992). We crossed wild-type HNI (Northern inbred) and i-3 (Southern origin) strains, and collected F2 intercross siblings for locus mapping. Phenotypes were determined by microscopy during embryogenesis and reconfirmed after hatching. We fixed the hatched fry in 100% ethanol and isolated their DNA using the PI-50 automated DNA isolation system (Kurabo).
Tyrosinase reaction:
Hatched fry were rinsed in Ringer solution for medaka (Yamamoto 1975) and incubated in reaction mixture [0.06% tyrosine, 0.024% NaHCO3, 80 mm NaCl, and 10 mm iodoacetamide in 30 mm phosphate buffer (pH 7.3)] for 24 hr at 37° (Hishida et al. 1961).
Mapping and chromosome walking:
We amplified the DNA markers by PCR using the following parameters: 94° for 1 min and then 30 cycles of 98° for 20 sec, 60° for 1 min, and 72° for 45 sec. Products were digested with restriction enzymes, if necessary, to detect polymorphisms on agarose or acrylamide gel. We used an HNI-based bacterial artificial chromosome (BAC) genomic library for chromosome walking (Kondo et al. 2002). Hybridization screening was performed using the AlkPhos direct labeling and detection system (Amersham Biosciences), and BAC ends were sequenced directly after the BAC DNA was purified with the Plasmid Midi kit (QIAGEN, Valencia, CA).
Shotgun sequencing:
We used the QIAGEN large-construct kit to isolate BAC129C11 DNA and the TOPO shotgun subcloning kit (Invitrogen, Carlsbad, CA) to construct the library. The inserts were amplified by colony PCR, and the products were directly sequenced in both directions. We assembled the sequence data using Lasergene software (DNASTAR, Madison, WI).
Expression analysis:
We used Isogen (Nippon Gene) to isolate total RNA from the organs and embryos of medaka and mouse. First-strand cDNA was synthesized using a 3′ adaptor and ReverTra Ace (Toyobo) following the manufacturer's protocol, and the 5′ adapter was added after a 20-min incubation. The reaction was further incubated for 1 hr and used as the template for RT-PCR. Thirty PCR cycles were applied to all the experiments in Figure 4. RACE was performed by nested PCR using primers complementary to the 3′ or 5′ adapters and specific primers complementary to the translated P sequences. The strongest bands were excised from the agarose gel, purified, subcloned into the pCR4-TOPO vector (Invitrogen), and sequenced. We performed whole-mount in situ hybridization on bg8 embryos by using DIG-labeled partial P riboprobes as described elsewhere (Fukamachi et al. 2001).
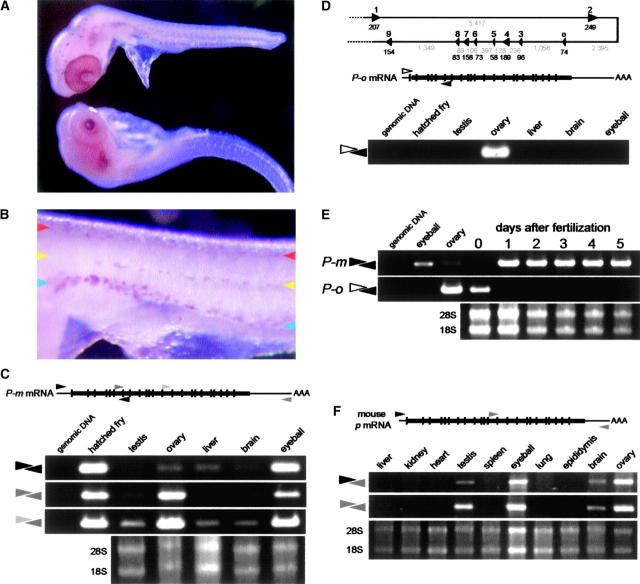
Figure 4.—
Expression of medaka P and mouse p mRNA. (A) Whole-mount in situ hybridization using medaka 6-day bg8 embryos using P riboprobes (top, antisense; bottom, sense). Positive cells are obvious in the eye and dorsal part of the head. (B) Higher magnification of the trunk. P-positive cells are distributed on the dorsal-most part (red arrowhead) and along the lateral line (yellow arrowhead) and the ventral side (blue arrowhead), which corresponds well to positions of three melanophore stripes in wild-type embryos (Kelsh et al. 2004). (C) Expression of medaka P in adult organs. The broad horizontal line and vertical lines in the diagram of P-m mRNA show the translated region and boundaries between exons, respectively. Black, dark gray, and light gray arrowheads indicate the approximate positions of the primers used for RT-PCR. (D) An ovary- (or oocyte-) specifically spliced form of P mRNA (P-o) occurs in medaka. (Top) Diagram shows the genomic structure of the 5′ region of the medaka P locus. Triangles show the positions of exons 1–9 and an ovary-specific exon (o). Lengths of exons and introns are indicated by black and gray, respectively. RT-PCR using a primer complementary to the ovary-specific exon (open arrowhead in the middle diagram) revealed its exclusive expression in the ovary. (E) Expression of medaka P-m and P-o during embryonic development. cDNAs were synthesized from embryos collected 0–5 days after fertilization and RT-PCR was performed using the primers shown in C and D. (F) Expression of mouse p in adult organs. RT-PCR was performed using two sets of primers that amplify the whole cDNA (black and gray arrowheads) or only the 3′ region (gray arrowheads). Strong expression was observed in the eyeball, ovary, and testis. Identical results were obtained with both sets of primers, except that the relative band intensity of the whole cDNA in testis was weak, suggesting the possible loss of the 5′ region of p mRNA in the testis by alternative splicing (see text).
RESULTS
Phenotype of the medaka i-3 mutant:
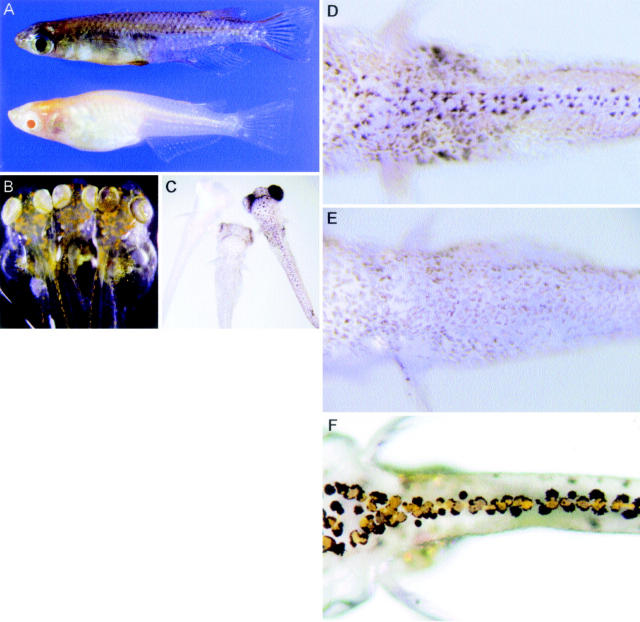
Medaka i-3 is a mutant isolated by Tomita (1992) using spontaneous mutant screening and its phenotype resembles oculocutaneous albinism (OCA) in mammals; i.e., melanin deposition is strongly suppressed over the whole body and eyes (Figure 1, A and B). It is almost indistinguishable from the i-locus mutants that lack the melanin-synthesizing enzyme, tyrosinase (Koga and Hori 1997), but the i-3 and the i (i-1) fry show distinct differences when incubated in tyrosine and iodoacetamide (Figure 1C). Amelanotic melanophores in the i-1 fry were not stained after incubation, indicating a complete lack of tyrosinase. In contrast, the i-3 fry exhibited cells stained brown (due to newly synthesized melanin) distributed over the whole body surface, indicating the presence of active tyrosinase in these cells. However, the i-3 fry do lack the strongly stained and slightly more dendritic cells that are observed on the dorsalmost part of another albino strain (bg8), which lacks MATP/AIM-1 (Fukamachi et al. 2001; Figure 1, C–E). The area in which the strongly tyrosinase-positive cells appear in bg8 fry corresponds to the dorsal melanophore stripe in the wild-type fry (Figure 1F). We observed that the wild-type fry has the weakly tyrosinase-positive cells in the interstripe regions, too (data not shown). Therefore, it seems that (1) whereas the distribution of strongly tyrosinase-positive cells (fully differentiated melanophores) is restricted to the stripe regions of medaka fry, weakly positive cells (putative melanophore precursor cells) are also distributed in the interstripe regions, and (2) the i-3 mutants have only melanophore precursors and lack fully differentiated cells, whereas the bg8 mutants have both cell populations.
Figure 1.—
Phenotype of the medaka i-3 mutant. (A) Melanin deposition in the eyes and skin is strongly suppressed in i-3 (bottom) compared with the wild type (top). (B) Phenotype of i-3 fry (middle) is almost indistinguishable from that of the tyrosinase (OCA1) mutant, i-1 (left). Slight deposition of melanin is detectable in the eyes of the MATP/AIM-1/Uw (OCA4) mutant, bg8 (right), but not in i-3 or i-1. Pink-colored pigment cells are leucophores. (C) Tyrosinase reactions using i-1 (left), i-3 (middle), or bg8 (right) fry were completely negative in i-1 and differentially positive in i-3 and bg8. (D and E) Higher magnification of the dorsal parts of bg8 (D) and i-3 (E) after the tyrosinase reaction. Note that strongly positive cells are distributed in a line in bg8 but are absent from i-3. Distribution of these strongly tyrosinase-positive cells in bg8 corresponds to dorsal, lateral, and ventral stripes of fully differentiated melanophores in wild-type fry (F and other data not shown; see Kelsh et al. 2004). All the fry in B–F are 0–1 day after hatching.
Chromosome walking to the i-3 locus:
The i-3 locus was mapped to medaka linkage group 4 (LG4) using M markers (Martinez-Morales et al. 2004). Orthologous analyses of its flanking ESTs indicated that the region around the i-3 locus is syntenic to human 19p13 (Figure 2A). We found the adapter-related protein complex 3 delta (AP3D1) gene on 19p13, which is the gene responsible for mouse mocha mutants and the human pigmentation disorder, Hermansky-Pudlak syndrome (Kantheti et al. 1998). The medaka i-3 mutants, however, do not show any obvious defects other than in pigmentation, so that AP3D1 does not seem to be encoded by i-3. Therefore, we took a positional approach to identifying the i-3 gene.
Figure 2.—
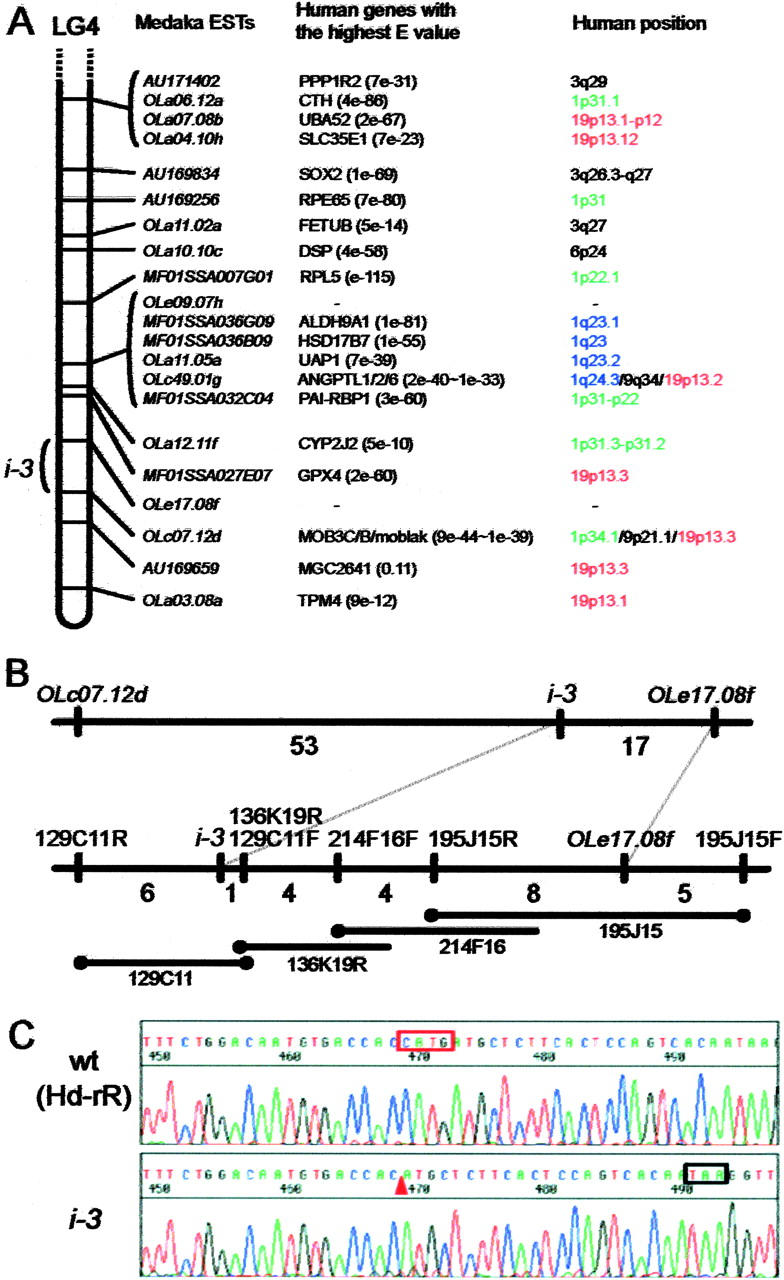
Positional cloning of the medaka i-3 gene. (A) Syntenic relationship between medaka and human. Sequences of ESTs on medaka LG4 were obtained from the M base, and their human orthologs were defined as the genes with the highest E-values on BlastX, except for OLc49.01g and OLc07.12d. The human orthologs of these two ESTs belong to gene families and we detected multiple human genes with high E-values. The three genes with the highest E-values are listed as putative orthologs of them. No orthologous genes were detected for OLe09.07h or OLe17.08f. (B) Chromosome walking from OLe17.08f to the i-3 locus. The numbers between neighboring loci denote the number of recombinants detected among 843 intercross siblings (1686 meioses). BAC ends mapped are indicated by solid circles. BAC 129C11 contains the entire i-3 mutation candidate region. The insert lengths of the BACs are not reflected in the lengths of the bars. (C) The i-3 mutation detected in the 13th exon. Top, Southern inbred Hd-rR is the wild type; bottom, i-3. Four-base deletion and its position in the i-3 allele are indicated by the red box and arrowhead, respectively. The black box shows a novel stop codon created by the frameshift.
We evaluated 1686 meioses using 843 F2 siblings and detected 17 recombinations between the i-3 locus and the nearest EST, OLe17.08f (Figure 2B). Because the strains we used for crossing (Northern HNI and Southern i-3) are extremely polymorphic [i.e., ∼5% of genomic sequences are single nucleotide polymorphisms (SNPs) or allele-specific nucleotides], we could map any BAC end sequence to confirm its position during chromosome walking and use these as probes for library screening, as long as they were nonrepetitive. Whether or not the BAC end is repetitive can be determined in silico before in vitro hybridization using 8.9× medaka genome sequence data (see above). Taking advantage of these techniques, we proceeded with very efficient and reliable chromosome walking until we reached the i-3 locus.
First, we isolated an OLe17.08f-positive BAC clone (195J15) with an insert of ∼150 kb and identified the polymorphisms at both its ends (195J15F and 195J15R). We detected 5 recombinations between 195J15F and OLe17.08f and 8 recombinations between 195J15R and OLe17.08f (Figure 2B). Therefore, we did not need to walk toward 195J15F but only toward 195J15R. This result also revealed the enhanced recombination frequency in this region; i.e., 1 cM was estimated to be 200 kb, which is much more frequent than the average value of 1 cM = 600 kb (Naruse et al. 2004). Recombination in male meioses was expected to be more frequent than this, because linkage maps on our database, M base (http://mbase.bioweb.ne.jp/dclust/medaka_top.html), indicate a recombination frequency around this region about sixfold higher in males than in females. Therefore, sex-dependent enhancement of meiotic recombination seems to occur in this region, which is similar to the situation around the b locus on LG12, although recombination is enhanced in the female in that case (Fukamachi et al. 2001). We continued chromosome walking by mapping every BAC end used for library screening (Figure 2B; Table 1) and crossed the i-3 locus at the fourth BAC from OLe17.08f after walking ∼1.4 cM (23 recombinations/1686 meioses).
TABLE 1.
BAC-end-based DNA markers closely linked to thei-3 locus
| Marker name | Primer sequence | Product size (bp) | Polymorphism |
|---|---|---|---|
| 195J15F | f: 5′-AAGCTTTACAAGATTCAAAGATTCTTCG-3′ r: 5′-CTCAGTTATGGGGGCCAACG-3′ |
450 | DdeI |
| 195J15R | f: 5′-ATGTCAACATCTGTGGCAAAATTAG-3′ r: 5′-GGATTAGATTTATCTGCGCTTTTCC-3′ |
663 | DdeI |
| 214F16F | f: 5′-GGATTGACAGAAACGAATTGCA-3′ r: 5′-AGATCGAAACTGCTTTTCATTAGAGTTT-3′ |
610 | AseI |
| 136K19R | f: 5′-GACTGAAAACTGCTACTCTCTCTCTGC-3′ r: 5′-AAGCTTTCTCAGCACTGTGCAC-3′ |
463 | PvuII |
| 129C11R | f: 5′-GCCACAGAAATGTAACTGCAAATC-3′ r: 5′-GAACATTAAGTTTACCTCTGTAAACATTTTTG-3′ |
572 | HNI specific |
| 129C11F | f: 5′-AAGCTTCCTGCTACGCCCTAA-3′ r: 5′-CCCATTCATGGCTGGAGG-3′ |
734 | Ins/del (or MnlI) |
The marker name, sequences of forward and reverse primers, product size of the Northern wild-type strain (HNI), and polymorphisms detected between HNI and the Southern mutant (i-3) are indicated. Ins/del, length polymorphism.
Medaka pink-eyed dilution ortholog as a strong candidate for i-3:
BAC 129C11 was shown to contain the entire candidate region for the i-3 mutation because both ends recombined with the i-3 locus (Figure 2B). We then sequenced the insert of 129C11 (∼220 kb) using a shotgun method. Preliminary BlastX analysis of these sequences (∼187 kb in 85 contigs) revealed that BAC 129C11 did not contain genes orthologous to those located on human 19p13, contradicting our former expectation (Figure 2A). Instead, it contained sequences significantly similar to the following human genes (shown by symbols in LocusLink): CYFIP1, NIPA2, GABRG3, OCA2, HERC2, and PEG10, which are all located at human 15q11–13, except for PEG10 (7q21). OCA2 (also called P) is the human ortholog of mouse pink-eyed dilution (p) and is known to be the gene responsible for human OCA type 2 (Rinchik et al. 1993). Because this seemed to be a very likely candidate gene disrupted in the medaka i-3 mutant in terms of its phenotype (Figure 1), we started analysis of this medaka P ortholog.
We first identified the complete open reading frame of medaka P by 3′- and 5′-RACE using embryo cDNA (Figure 3A). The mRNA consists of 24 exons distributed over >40 kb of the genomic region, and the exon-intron boundaries seem to be conserved among medaka, human, and mouse (Figure 3B). Medaka P protein is predicted to consist of 842 amino acids and share ∼65% identity (785 amino acids compared) with human P and mouse p (Figure 3B). A BlastP search also revealed similarity to hoepel1/2 of Drosophila and the arsenical transport proteins of bacteria, with 49 and 33% identity (549 and 508 amino acids compared), respectively. Mammalian P proteins are believed to have 12 transmembrane domains and to act as transporters (Gardner et al. 1992). However, recent web-based programs [SOSUI (Hirokawa et al. 1998), TMHMM (Moller et al. 2001), TMpred (Hofmann and Stoffel 1993), and HMMTOP (Tusnády and Simon 2001)] predict that medaka P has 11, 12, 13, and 14 membrane-spanning regions, respectively. They also predict that human P has 12, 12, 13, and 14, and mouse p has 11, 11, 14, and 14 transmembrane domains, respectively. Thus, the precise secondary structure of the P proteins seems to be unclear.
Figure 3.—

(A) Nucleotide sequence of the medaka P cDNA of the HNI strain. Vertical lines show positions of introns in the genomic DNA. Black boxes show translation initiation and stop codons. Red shows deleted nucleotides in i-3. Nucleotides indicated in light blue are polymorphic in HNI, Hd-rR, and i-3 (gray residues in the 5′-most and 3′-most regions were not compared). Putative poly(A) signal is indicated in green. (B) Alignment of amino acid sequences of P proteins so far isolated, from mouse, human, pig (Sus scrofa), and medaka (Hd-rR). Red letters indicate the residues most commonly conserved at that site in the four species. Boxed residues contain exon-intron boundaries within their codons or are the first amino acids of exons. Light blue underlining shows the untranslated residues from P-o (Figure 4D and see text). Residues with black underlining are substituted with a novel seven amino acids (CSSLQSQ) in i-3. Black circles indicate positions of mutated residues reported in human OCA2 patients. Gray circles and open circles show the positions of polymorphic residues detected in human and medaka, respectively. Three of four residues with open circles (T, V, and D) are substituted into A, L, and E in HNI, respectively, while the third one (S) is substituted into G in i-3. Human mutation and polymorphism information was obtained from the Albinism Database (http://albinismdb.med.umn.edu/) and Kato et al. (2003) and Suzuki et al. (2003).
Polymorphism and mutation analyses of medaka P:
Then we investigated whether the P gene is mutated in the i-3 mutant. We performed RT-PCR using adult eyeball cDNAs of HNI (Northern inbred), Hd-rR (Southern inbred), and the i-3 mutant (Southern origin) and directly sequenced the amplified products. We identified 54 SNPs and 13 allele-specific nucleotides among these loci (3373 bases compared; Figure 3A). Most of them were Northern-Southern polymorphisms, and many fewer polymorphisms were found between the Hd-rR and i-3 strains. Four of these nucleotide polymorphisms cause amino acid substitution (1 of them is i-3 specific), which occurs at residues not conserved among medaka, human, mouse, and pig proteins, indicating that these are not mutagenic but polymorphic differences (Figure 3B). The i-3 mutation was detected as a four-base deletion in exon 13, which would cause a frameshift and the loss of the 394 C-terminal amino acids of the P protein (Figures 2C and 3). This truncation should reduce or abolish the function of medaka P, because the C-terminal region is very highly conserved between medaka and the mammals, indicating its importance for function (Figure 3B). Therefore, we concluded that oculocutaneous albinism in medaka i-3 is caused by this deletion in the medaka pink-eyed dilution ortholog.
P transcripts in medaka and mouse:
We then analyzed the expression of medaka P by in situ hybridization and RT-PCR. Medaka P is strongly expressed in the eyeball of embryos and adults (Figure 4, A and C), where melanin is produced in the choroid membrane and retinal pigment epithelium. We detected P-expressing cells in the skin of embryos, whose distribution pattern is very similar to that of melanophores (Figure 4, A and B). Much weaker expression was detected in non-melanin-producing adult organs (Figure 4C). When we amplified the 3′ region of P, however, we detected additional and even stronger expression in the ovary, although expression of the full-length transcript (inferred from amplification of the 5′ end) was as weak as those in other organs (Figure 4C).
To assess this contradictory result, we performed 5′-RACE using ovary cDNA and identified an alternatively spliced transcript expressed exclusively in the ovary (Figure 4D). This ovary-specific variant (P-o) lacks the first and second exons of melanophore-specific P (P-m) and instead has an extra exon located between the second and third exons (Figure 4D). This ovary-specific exon has several in-frame stop codons and seems to encode no amino acids. Therefore, the protein encoded by P-o is predicted to lack 116 N-terminal amino acids of P-m with no disruption of any of the transmembrane domains predicted by the web-based programs (see above). The P-m and P-o transcripts also exhibit distinct difference in expression during embryogenesis. Whereas P-m is continuously expressed from 1 day after fertilization until hatching (day 5), corresponding to the period of emergence of melanophores in the body, eyes, and yolk sac of embryos, P-o expression was detected only at day 0 (approximately midgastrula stage; Figure 4E), suggesting maternal storage (Figure 4C).
Finally, we analyzed mouse p expression in adult organs to investigate whether there is also an alternative form in the mouse ovary. We detected strong expression of mouse p in the ovary and also in the testis (Figure 4F). Identical results were obtained using primers designed to amplify the full length or only the 3′ region of mouse p mRNA; i.e., mouse p is strongly expressed in the ovary, as in medaka, but it is not alternatively spliced. Isolation of an ovary-specific variant by 5′-RACE (if it exists) seemed to be difficult in this situation. We found no such alternatively spliced mouse p or human P in their respective EST databases. However, further careful investigation might reveal an alternatively spliced form of P in mammals, because a truncated human P has already been reported (Rinchik et al. 1993). Gardner et al. (1992)(Figure 2A) seem to suggest the existence of a shorter form of p in mouse testis (see also Figure 4D herein). Although we and others have observed no anomalies in the reproduction or embryogenesis of the medaka i-3 or mouse p-locus mutants, strong expression of medaka P-o in the ovary and mouse p in the ovary and testis suggests role(s) for the cognate proteins other than in melanogenesis.
DISCUSSION
The colors and patterns on the animal body surface are so diverse, even among closely related species, that pigment cells on the skin (chromatophores) present a fascinating research subject with which to investigate the genetic mechanisms of variation. Studies using >100 mouse coat-color mutants contributed to the isolation of ∼60 relevant genes (see the Coat Color Genes at http://www.cbc.umn.edu/ifpcs/micemut.htm). These surely extended our understanding of the development and regulation of the mammalian chromatophore, the melanocyte. However, little is known about other types of chromatophores in lower vertebrates, which originate developmentally from the “vertebrate-innovating cells” of the neural crest (Shimeld and Holland 2000). Mammals have lost most kinds of chromatophores during the long nocturnal periods experienced by their ancestors, and it should be interesting from an evolutionary standpoint to investigate the fates of the chromatophore-associated genes of lower vertebrates in mammals. Have they all been lost or have some acquired new functions in the development of other neural-crest derivatives?
Common function of P proteins in vertebrate melanogenesis:
Here we elucidated the gene responsible for the medaka i-3 albino mutant and its novel alternatively spliced form expressed in non-melanin-producing cells. There are several working models of the function of mammalian P in melanin biosynthesis, including tyrosine transport (Sidman and Pearlstein 1965; denied by Gahl et al. 1995), tyrosinase routing (Potterf et al. 1998), regulation of melanosomal pH (Puri et al. 2000), melanocyte proliferation and differentiation (Hirobe et al. 2002), glutathione metabolism (Staleva et al. 2002), and tyrosinase processing (Chen et al. 2002; Toyofuku et al. 2002). Our observations suggest that P protein affects tyrosinase activity in and the morphogenesis of melanophores in teleosts (Figure 1, C and E). Because mammalian melanocytes and fish melanophores are homologous but have many different features (e.g., chromatosome movement, nervous control, pheomelanogenesis, etc.), it is possible that the P orthologs do not share identical functions in mammals and other vertebrates (discussed below). However, our results clearly indicate that both the mammalian and the teleost P proteins participate in eumelanogenesis, although further experiments are required to fully elucidate the functional mechanisms.
Role of the nonconserved N-terminal region of the P proteins:
The functions of P in non-melanin-producing cells remain mysterious. In both medaka and mouse, P transcripts are strongly expressed in the ovary (Figure 4, C, D, and F). Moreover, mouse transcripts are also strongly expressed in the testis (Figure 4F). However, neither the medaka i-3 mutant nor the mouse pink-eyed dilution mutant shows obvious defects in fertility or embryonic development under normal breeding conditions, except when these mice are radiation induced and involve large deletions in the neighboring Gabr or Herc2 genes (Nakatsu et al. 1993; Lehman et al. 1998). Furthermore, why the medaka expresses an alternative form of P, instead of P-m, in the ovary, whereas the mouse expresses identical forms, is unclear (Figure 4, C, D, and F). The 116 N-terminal amino acids lost in the P-o protein (if translated) are not conserved either between medaka and the mammals or among the mammals (Figure 3B).
Generally, less conservation indicates that the region is less important in common functions (i.e., eumelanin synthesis in this case), and it has been suggested that the N-terminal region of the P protein might not be functionally critical (Lee et al. 1995). However, a few residues substituted in this region have been reported as the critical mutations in human patients with OCA2 (Figure 3B), suggesting the importance of the N-terminal region of P protein in melanin synthesis (although functional assessment of these mutations is required). Moreover, there must be a positive reason for the retention of this long and nonconserved region across vertebrates and both the splicing variants in medaka; i.e., it is lost in the ovary or oocytes (P-o), but retained in melanophores (P-m) (Figure 3B). Functional assessment of medaka P-m and P-o in melanophores, using a method such as the microinjection of i-3 embryos, should be addressed to investigate the roles of these nonconserved N-terminal residues of the P proteins in melanin synthesis.
The genetic mechanisms that cause variation in animal body colors and patterns are unknown, and the “nonconserved but important domains for function” could be one of these mechanisms. Because mammals have only one kind of chromatophore—the melanocyte—the variations in their body colors and patterns must be generated by the presence or absence of melanocytes (or melanin in the cells) or by switching between black eumelanin and yellow pheomelanin synthesis. The latter has been extensively studied using the agouti and its related mutants (He et al. 2003), but little is known about pattern formation. The P protein is involved in eumelanogenesis but not in pheomelanogenesis, so that pheomelanins in mouse p-locus mutants are only slightly affected (Russell 1949).
Taking these data into consideration, it might be too speculative, but it is possible that the regulation of P-protein expression is involved in eumelanin-pheomelanin switching and that the variable N-terminal regions of P might be responsible for species-specific regulation of switching to form variable patterns by some method of protein-protein (not protein-substrate) interactions. Although little is known about the regulation of P-protein expression, its transcription is upregulated by UVB or melanocortin-1 receptor (MC1R) signaling (Suzuki et al. 2002; Ancans et al. 2003). Further clarification of not only the functions of the P proteins, but also the regulation of their expression should provide interesting insight into body-color determination and variation in vertebrates.
Acknowledgments
We thank K. Naruse of the University of Tokyo for EST information; A. Shimada and S. Takada of the University of Tokyo for fish care; T. Jindo, T. Kitagawa, and H. Takeda for the M markers; and S. Oda of the University of Tokyo for comments on the manuscript. This research was supported by a Grant-in-Aid from the Ministry of Education, Science, Sports, Culture, and Technology (MEXT), Japan (scientific research on priority areas, area number 813).
References
- Ancans, J., N. Flanagan, M. J. Hoogduijn and A. J. Thody, 2003. P-locus is a target for the melanogenic effects of MC-1R signaling: a possible control point for facultative pigmentation. Ann. NY Acad. Sci. 994: 373–377. [DOI] [PubMed] [Google Scholar]
- Chen, K., P. Manga and S. J. Orlow, 2002. Pink-eyed dilution protein controls the processing of tyrosinase. Mol. Biol. Cell 13: 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukamachi, S., A. Shimada and A. Shima, 2001. Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat. Genet. 28: 381–385. [DOI] [PubMed] [Google Scholar]
- Fukamachi, S., M. Sugimoto, H. Mitani and A. Shima, 2004. Somatolactin selectively regulates proliferation and morphogenesis of neural-crest derived pigment cells in medaka. Proc. Natl. Acad. Sci. USA 101: 10661–10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani-Seiki, M., T. Sasado, C. Morinaga, H. Suwa, K. Niwa et al., 2004. A systematic genome-wide screen for mutations affecting organogenesis in Medaka, Oryzias latipes. Mech. Dev. 121: 647–658. [DOI] [PubMed] [Google Scholar]
- Gahl, W. A., B. Potterf, D. Durham-Pierre, M. H. Brilliant and V. J. Hearing, 1995. Melanosomal tyrosine transport in normal and pink-eyed dilution murine melanocytes. Pigm. Cell Res. 8: 229–233. [DOI] [PubMed] [Google Scholar]
- Gardner, J. M., Y. Nakatsu, Y. Gondo, S. Lee, M. F. Lyon et al., 1992. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science 257: 1121–1124. [DOI] [PubMed] [Google Scholar]
- He, L., A. G. Eldridge, P. K. Jackson, T. M. Gunn and G. S. Barsh, 2003. Accessory proteins for melanocortin signaling: attractin and mahogunin. Ann. NY Acad. Sci. 994: 288–298. [DOI] [PubMed] [Google Scholar]
- Hirobe, T., Y. Kawa, M. Mizoguchi, S. Ito and K. Wakamatsu, 2002. Effects of genic substitution at the pink-eyed dilution locus on the proliferation and differentiation of mouse epidermal melanocytes in vivo and in vitro. J. Exp. Zool. 292: 351–366. [DOI] [PubMed] [Google Scholar]
- Hirokawa, T., S. Boon-Chieng and S. Mitaku, 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379. [DOI] [PubMed] [Google Scholar]
- Hishida, T., H. Tomita and T. Yamamoto, 1961. Melanin formation in color varieties of the medaka (Oryzias latipes). Embryologia 5: 335–346. [Google Scholar]
- Hofmann, K., and W. Stoffel, 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. 374: 166. [Google Scholar]
- Hyodo-Taguchi, Y., 1980. Establishment of inbred strains of the teleost, Oryzias latipes. Zool. Mag. 89: 283–301. [Google Scholar]
- Iwamatsu, T., 1997 The Integrated Book for the Biology of the Medaka. University Education Press, Okayama, Japan (in Japanese).
- Kantheti, P., X. Qiao, M. E. Diaz, A. A. Peden, G. E. Meyer et al., 1998. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron 21: 111–122. [DOI] [PubMed] [Google Scholar]
- Kato, A., K. Fukai, N. Oiso, N. Hosomi, S. Saitoh et al., 2003. A novel P gene missense mutation in a Japanese patient with oculocutaneous albinism type II (OCA2). J. Dermatol. Sci. 31: 189–192. [DOI] [PubMed] [Google Scholar]
- Kelsh, R. N., C. Inoue, A. Momoi, H. Kondoh, M. Furutani-Seiki et al., 2004. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech. Dev. 121: 841–859. [DOI] [PubMed] [Google Scholar]
- Koga, A., and H. Hori, 1997. Albinism due to transposable element insertion in fish. Pigm. Cell Res. 10: 377–381. [DOI] [PubMed] [Google Scholar]
- Kondo, M., A. Froschauer, A. Kitano, I. Nanda, U. Hornung et al., 2002. Molecular cloning and characterization of DMRT genes from the medaka Oryzias latipes and the platyfish Xiphophorus maculatus. Gene 295: 213–222. [DOI] [PubMed] [Google Scholar]
- Kondo, S., Y. Kuwahara, M. Kondo, K. Naruse, H. Mitani et al., 2001. The medaka rs-3 locus required for scale development encodes ectodysplasin-A receptor. Curr. Biol. 11: 1202–1206. [DOI] [PubMed] [Google Scholar]
- Lee, S. T., R. D. Nicholls, M. T. Jong, K. Fukai and R. A. Spritz, 1995. Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics 26: 354–363. [DOI] [PubMed] [Google Scholar]
- Lehman, A. L., Y. Nakatsu, A. Ching, R. T. Bronson, R. J. Oakey et al., 1998. A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc. Natl. Acad. Sci. USA 95: 9436–9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosli, F., S. Winkler, C. Burgtorf, E. Wurmbach, W. Ansorge et al., 2001. Medaka eyeless is the key factor linking retinal determination and eye growth. Development 128: 4035–4044. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales, J., K. Naruse, F. Loosli, M. Mitani, A. Shima et al., 2004. Rapid chromosomal assignment of medaka mutants by bulked segregant analysis. Gene 329: 159–165. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., N. Kawato, S. Asakawa, N. Shimizu, Y. Nagahama et al., 2001. Construction of a BAC library derived from the inbred Hd-rR strain of the teleost fish, Oryzias latipes. Genes Genet. Syst. 76: 61–63. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- Moller, S., M. D. Croning and R. Apweiler, 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17: 646–653. [DOI] [PubMed] [Google Scholar]
- Nakatsu, Y., R. F. Tyndale, T. M. DeLorey, D. Durham-Pierre, J. M. Gardner et al., 1993. A cluster of three GABAA receptor subunit genes is deleted in a neurological mutant of the mouse p locus. Nature 364: 448–450. [DOI] [PubMed] [Google Scholar]
- Naruse, K., M. Tanaka, K. Mita, A. Shima, J. Postlethwait et al., 2004. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 14: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterf, S. B., M. Furumura, E. V. Sviderskaya, C. Santis, D. C. Bennett et al., 1998. Normal tyrosine transport and abnormal tyrosinase routing in pink-eyed dilution melanocytes. Exp. Cell Res. 244: 319–326. [DOI] [PubMed] [Google Scholar]
- Puri, N., J. M. Gardner and M. H. Brilliant, 2000. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J. Invest. Dermatol. 115: 607–613. [DOI] [PubMed] [Google Scholar]
- Rinchik, E. M., S. J. Bultman, B. Horsthemke, S. T. Lee, K. M. Strunk et al., 1993. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 361: 72–76. [DOI] [PubMed] [Google Scholar]
- Russell, E. S., 1949. A quantitative histological study of the pigment found in the coat-color mutants of the house mouse. IV. The nature of the effects of genic substitution in five major allelic series. Genetics 34: 146–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, A., and A. Shimada, 1994. The Japanese medaka, Oryzias latipes, as a new model organism for studying environmental germ-cell mutagenesis. Environ. Health Perspect. 102(Suppl.): 33–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimeld, S. M., and P. W. Holland, 2000. Vertebrate innovations. Proc. Natl. Acad. Sci. USA 97: 4449–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman, R. L., and R. Pearlstein, 1965. Pink-eyed dilution (p) gene in rodents: increased pigmentation in tissue culture. Dev. Biol. 12: 93–116. [DOI] [PubMed] [Google Scholar]
- Staleva, L., P. Manga and S. J. Orlow, 2002. Pink-eyed dilution protein modulates arsenic sensitivity and intracellular glutathione metabolism. Mol. Biol. Cell 13: 4206–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, I., T. Kato, T. Motokawa, Y. Tomita, E. Nakamura et al., 2002. Increase of pro-opiomelanocortin mRNA prior to tyrosinase, tyrosinase-related protein 1, dopachrome tautomerase, Pmel-17/gp100, and P-protein mRNA in human skin after ultraviolet B irradiation. J. Invest. Dermatol. 118: 73–78. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Y. Miyamura, J. Matsunaga, H. Shimizu, Y. Kawachi et al., 2003. Six novel P gene mutations and oculocutaneous albinism type 2 frequency in Japanese albino patients. J. Invest. Dermatol. 120: 781–783. [DOI] [PubMed] [Google Scholar]
- Tomita, H., 1992. The lists of mutants and strains of medaka, common gambusia, silver crusian carp, goldfish, and golden venus fish maintained in the laboratory of freshwater fish stocks. Fish Biol. J. Medaka 4: 45–47. [Google Scholar]
- Toyofuku, K., J. C. Valencia, T. Kushimoto, G. E. Costin, V. M. Virador et al., 2002. The etiology of oculocutaneous albinism (OCA) type II: the pink protein modulates the processing and transport of tyrosinase. Pigm. Cell Res. 15: 217–224. [DOI] [PubMed] [Google Scholar]
- Tusnády, G. E., and I. Simon, 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850. [DOI] [PubMed] [Google Scholar]
- Wakamatsu, Y., S. Pristyazhnyuk, M. Kinoshita, M. Tanaka and K. Ozato, 2001. The see-through medaka: a fish model that is transparent throughout life. Proc. Natl. Acad. Sci. USA 98: 10046–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., 1975 An outline of the genetics of the medaka, pp. 154–169 in Medaka (Killifish) Biology and Strains, edited by T. Yamamoto. Keigaku Publishing, Tokyo.