Abstract
Myosin VIIs provide motor function for a wide range of eukaryotic processes. We demonstrate that mutations in crinkled (ck) disrupt the Drosophila myosin VIIA heavy chain. The ck/myoVIIA protein is present at a low level throughout fly development and at the same level in heads, thoraxes, and abdomens. Severe ck alleles, likely to be molecular nulls, die as embryos or larvae, but all allelic combinations tested thus far yield a small fraction of adult “escapers” that are weak and infertile. Scanning electron microscopy shows that escapers have defects in bristles and hairs, indicating that this motor protein plays a role in the structure of the actin cytoskeleton. We generate a homology model for the structure of the ck/myosin VIIA head that indicates myosin VIIAs, like myosin IIs, have a spectrin-like, SH3 subdomain fronting their N terminus. In addition, we establish that the two myosin VIIA FERM repeats share high sequence similarity with only the first two subdomains of the three-lobed structure that is typical of canonical FERM domains. Nevertheless, the ∼100 and ∼75 amino acids that follow the first two lobes of the first and second FERM domains are highly conserved among myosin VIIs, suggesting that they compose a conserved myosin tail homology 7 (MyTH7) domain that may be an integral part of the FERM domain or may function independently of it. Together, our data suggest a key role for ck/myoVIIA in the formation of cellular projections and other actin-based functions required for viability.
MYOSIN VIIAs are actin-based motor proteins essential for a variety of biological processes (Chen et al. 1996; Hodge and Cope 2000; Yamashita et al. 2000; Berg et al. 2001; Redowicz 2002; Tzolovsky et al. 2002; Ahmed et al. 2003 and references therein). In vertebrates, they play a key role in sensory perception: defects in myosin VIIA lead to deafness and blindness in humans, retinal defects and deafness in mice, and aberrant auditory and vestibular function in zebrafish. The cellular basis of these phenotypes suggests that the defects are the consequence of aberrant actin cytoskeleton function. Moreover, the tissue-specific expression pattern of myosin VIIA correlates well with the phenotypes observed. Biochemical experiments on purified or recombinant proteins show that the myosin VIIAs have plus (barbed) end-directed motor activity on actin filaments and a characteristic actin-activated ATPase activity (Udovichenko et al. 2002; Inoue and Ikebe 2003). Structurally, the myosin VIIA heavy chain is well conserved and the various domains provide for motor- and cargo-binding functions. The myosin VIIA head and neck are composed of a conserved N-terminal region (∼60 amino acids), a characteristic motor domain, and four to five isoleucine-glutamine (IQ) motifs that bind calmodulin (Cheney and Mooseker 1992; Todorov et al. 2001) and/or specific light chains. The myosin VIIA tail begins with a short sequence predicted to form an alpha-helical coiled-coil that may contribute to dimerization. The remainder of the tail consists of a tandem repeat of myosin tail homology 4 (MyTH4) domains and partial four-point 1, ezrin, radixin, and moesin (FERM) domains (see below) that are separated by an SH3 subdomain and are thought to mediate dimerization and binding to other proteins or cargo.
Myosin VIIAs are part of a myosin subfamily that is conserved phylogenetically in metazoa and amoebozoa (Dictyostelium) but is lacking in sequenced fungal and plant species. The subfamily includes myosin VIIA, myosin VIIB, myosin VII (from species that do not have distinct myosin VIIA and myosin VIIB genes, see below and discussion), myosin X, and myosin XV. In vertebrates and insects, the closely related myosin VIIB heavy chain is encoded by distinct, myosin VIIB genes (see references above and Chen et al. 2001) that have not been characterized extensively. The fly myoVIIB gene is encoded by a transcription unit at polytene location 28B. The VIIB heavy chains share clear sequence similarity throughout the heavy chain, but lack a region predicted to form a coiled-coil. No pathologies associated with myosin VIIB have so far been discovered in any species. Caenorhabditis elegans and Dictyostelium discoideum have a single myosin VII heavy chain gene (not distinct VIIA and VIIB forms). MyoI encodes the D. discoideum myosin VII: it is essential for the initial steps of cell adhesion that contribute to phagocytosis, cell-cell interactions, translocation across a substrate, and the formation of filopodia (Titus 1999; Tuxworth et al. 2001). Interestingly, recent analysis of vertebrate myosin VIIA mutants suggests that it is not required for the early adhesion events in phagocytosis (Gibbs et al. 2003). Nevertheless it may play an important role in linking adjacent steriocilia in hair cells (Kussel-Andermann et al. 2000). Thus far, no mutants are available for the worm hum-6/myoVII. Myosin X and XV, like members of the myosin VII subfamily, include tails with one or more FERM domains. Flies have a myosin XV encoded by a transcription unit at polytene location 10A, but they do not have a myosin X, which may have some overlap in function with myosin VIIs in vertebrates (Yamashita et al. 2000; Berg et al. 2001; Tuxworth et al. 2001; Tzolovsky et al. 2002).
The crinkled (ck) locus has been studied intermittently for the past 70–80 years (Gubb et al. 1984; Ashburner et al. 1999), and genomic sequence analysis suggested that it was likely to encode myosin VIIA (Ashburner et al. 1999). A mutation in ck was first identified by Bridges in the 1930s, but the allele was lost (Bridges and Brehme 1944). Detailed studies on the region around Adh identified a number of alleles in the l(2)br27 complementation group with phenotypes very similar to those described by Bridges for ck, so ck and l(2)br27 were deemed allelic (Gubb et al. 1984). A number of phenotypes attributable to mutations at the ck locus have been described [other synonyms for ck are listed in FlyBase (FlyBase Consortium 2003)]. Severe mutations in ck are lethal or semilethal, with a small fraction of homozygous [ck/ck or hemizygous, ck/Df(ck−)] flies reaching adulthood (<0.5–5%, so-called “escapers”). Adult escapers of these lethal alleles show common expressivity of characteristic defects that include stubby microchaetae; short, multiple setae that are frequently branched; short aristae that are more highly branched than normal; and wavy and crumpled wings. In the context of specification of asymmetric cytoskeletal organization for planar polarity, crinkled suppresses both frizzled gain-of-function and disheveled loss-of-function mutations (Winter et al. 2001). More recently, ck homozygotes were shown to have aristae that are abnormal in morphology (He and Adler 2002).
Here we formally demonstrate that the myosin VIIA heavy chain is encoded by the ck locus (Ashburner et al. 1999). We demonstrate that the ck/myoVIIA transcript is differentially spliced and show that protein is present at a low level throughout development and in different body parts. We establish that severe alleles die as embryos and document new phenotypes in escapers that are consistent with disruption of actin cytoskeletal dynamics. In addition, we propose a homology model for the structure of the ck/myoVIIA head that indicates that the N-terminal 55 amino acids constitute a spectrin-like SH3 subdomain comparable to that seen in myosin-II heavy chains. Finally, we use sequence comparisons to show that only the first two of the three lobes of each of the FERM domains is well conserved with other FERM domain proteins. Sequences that follow lobe 2 are highly conserved among myosin VIIs and show only weak sequence similarity with other proteins. We refer to these sequences as a MyTH7 domain.
MATERIALS AND METHODS
Fly husbandry and stocks:
Flies were raised and crosses were performed at 22° or 25° on standard yeast-cornmeal-agar medium using standard methods (Roberts 1998). EP(2)2051 was obtained from Janos Szidonya at the Szeged stock center and BG00682 from the Baylor/Berkeley Drosophila Genome Project (BDGP) Gene Disruption Project. Other ck alleles came from the stock collection at the Department of Genetics, University of Cambridge, United Kingdom. All other mutations and deletions used in this study are fully described in FlyBase (FlyBase Consortium 2003).
Genetic mapping:
Mutant alleles were mapped to ck by crossing to deletion stocks and by crossing ck alleles inter se. Adult flies prepared for SEM analysis were from crosses of ck alleles to the large chromosomal deletion Df(2L)osp29 (35B1–3; 35E6).
Verification of P(PZ)07130 as a ck allele:
Deletions with one breakpoint at the insertion site of the P element were isolated using the P-element transposase-mediated “male recombination” technique described elsewhere (Preston et al. 1996). Putative deletions were recognized by exchange between the flanking markers dumpy (dp), black (b) and cinnabar (cn), brown (bw). The cross scheme was as follows: male P(PZ)07130, ry+/CyO flies were crossed to females of the genotype dp b cn bw; Dr, D2-3/TM6B; individual male flies of the genotype P(PZ)07130, ry+/dp b cn bw; Dr, D2-3/+ were recovered and crossed to female CyO, dplvI b pr cn bw/Gla flies. Exceptional progeny of the genotype dp b P(PZ)07130/CyO, dplvI b pr cn bw indicated duplications extending proximally or deletions extending distally from the insertion site; those of the genotype P(PZ)07130 cn bw/CyO, dplvI b pr cn bw indicated duplications extending distally or deletions extending proximally from the insertion site.
Identification of Drosophila myosin VIIA:
Standard methods were used for molecular biology throughout this study unless specified (Sambrook et al. 1989). Degenerate primers designed to amplify conserved sequences from myosin heavy chains but not the three fly myosin genes whose sequences were available at the time (muscle myosin II heavy chain, zipper non-muscle myosin II heavy chain, and ninaC myosin III heavy chain) were used to amplify DNA from a fly head cDNA library (Itoh et al. 1985), from an embryonic cDNA library (Brown and Kafatos 1988), and from a Kc0 cell line cDNA library in λgt11 (origin unknown).
ck/myoVIIA cDNA:
A nearly full-length ck/myoVIIA cDNA (LD10736) was identified by the BDGP. cDNA from CsCl prepared plasmid DNA and PCR product derived from genomic DNA isolated from heterozygous or homozygous mutant flies and amplified with custom gene-specific primers (IDT, Coralville, IA or GIBCO BRL, Gaithersburg, MD) were sequenced. Raw sequences were viewed with EditView (Perkin Elmer, Wellesley, MA) and differences between sequences from two templates in the same fly (i.e., from the mutant and balancer chromosomes) gave two peaks at approximately half height. Sequences were assembled and analyzed with Sequencher (Gene Codes, Ann Arbor, MI) or SeqMan (DNAStar, Madison, WI). All differences were verified by sequencing homozygous mutant (identified in the progeny of heterozygotes through the absense of a GFP-marked balancer chromosome) or hemizygous mutant escapers [ckallele/Df(2L)osp29]. Alterations that effected changes in protein coding were verified by sequencing in both directions PCR product amplified from independently isolated genomic DNA. Sequences were compared with Align or MegAlign (DNAStar).
Preparation of antigen:
A fragment of ck/myoVIIA (Arg822 to Ser1130) was cloned into pGEX-6P-1 (Amersham Pharmacia, Piscataway, NJ) using engineered XmaI and XhoI restriction sites. Protein was expressed in Escherichia coli and purified using standard GST methods. Fractions were monitored with SDS-PAGE. Guinea pigs were immunized commercially (Pocono Rabbit Farm, Canadensis, PA) and sera were characterized using immunoblots.
SDS-PAGE and immunoblotting:
Living Drosophila embryos (20), larvae (3), pupae (2), or adults (2) were ground directly into 100 μl of hot SDS-PAGE sample buffer and then boiled for 5 min. Antennae, heads, thoraxes, and abdomens were hand dissected on a dry-ice-cooled aluminum block from flies frozen in liquid N2. Frozen fly body parts were prepared as described above using six heads, six thoraxes, and six abdomens per 100 μl of sample buffer. Samples (10 μl) were resolved by SDS-PAGE on 7.5% acrylamide, 0.75% bis-acrylamide using standard methods (Laemmli 1970). Gels were blotted and blots were processed by standard methods using 5% normal goat serum (NGS) and 5% normal horse serum (NHS) in Tris-buffered saline (TBS) that consists of 20 mm TrisCl (pH 7.5) and 154 mm NaCl with 0.08% Tween for blocking and incubation steps. Probed blots were rinsed in TBS plus 0.1% Tween, developed with luminescent substrate [ELC Plus (Amersham, Piscataway, NJ) or Super Signal West Pico (Pierce, Rockford, IL)], and then exposed to film. Primary guinea pig serum (JDF no. 1515) was diluted 1:1000 and incubated ∼16 hr at 4°. For loading controls antisera was directed against fly nonmuscle myosin-II (no. 656 diluted 1:1000; Kiehart and Feghali 1986) or β-tubulin (E7; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). Secondary antibodies were affinity purified, peroxidase conjugated, rabbit anti-guinea pig, goat anti-rabbit, or goat anti-mouse antibodies (Zymed, South San Francisco, CA) diluted 1:5000 and incubated 1 hr at 22°. Molecular mass standards (10–250 kD) were Precision Plus All Blue prestained (Bio-Rad, Hercules, CA). Exposed films were scanned into Adobe Photoshop (San Jose, CA).
Homology modeling:
We used the SWISS-MODEL automated comparative protein modeling server to generate a three-dimensional (3-D) homology model of the motor domain (Guex and Peitsch 1997). Using the First Approach mode, we submitted to the server the amino acid sequence for ck/myoVIIa and five structures for use as templates: chicken skeletal muscle myosin subfragment-1 (ExPDB 2mysA); D. discoideum MyoE, a myosin I (ExPDB 1lkxC); scallop adductor muscle myosin S1 (ExPDB 1b7tA); D. discoideum myosin II truncated S1 (ExPDB 1vom_); and chicken smooth muscle myosin II motor domain (ExPDB 1br2A). The primary sequences of the motor domain of these five myosins show 42.5, 37.3, 37.2, 36.8, and 36.3% identity, respectively, when aligned with the ck/myo VIIA head. The primary sequence alignment returned by the SWISS-MODEL server was compared to pairwise sequence alignments between ck/myoVIIA and each of the reference structures. After introducing shifts caused by obvious misalignments, the preliminary model was resubmitted to the server for optimization. This process was repeated three more times to generate the model shown in Figures 6 and 7. The model was validated using WHAT IF (Vriend 1990). The “spare part” algorithm employed by SWISS-MODEL searches existing crystal structures solved to better than 2.5-Å resolution to build segments of ck/myoVIIA that correspond to regions that remain unresolved in the template structures and enables the model to be continuous. The model, called “homology model for ck/myoVIIA,” can be downloaded for viewing at the Kiehart Lab website (http://www.biology.duke.edu/kiehartlab/) using Rasmol or SwissPdb Viewer.
Figure 6.—
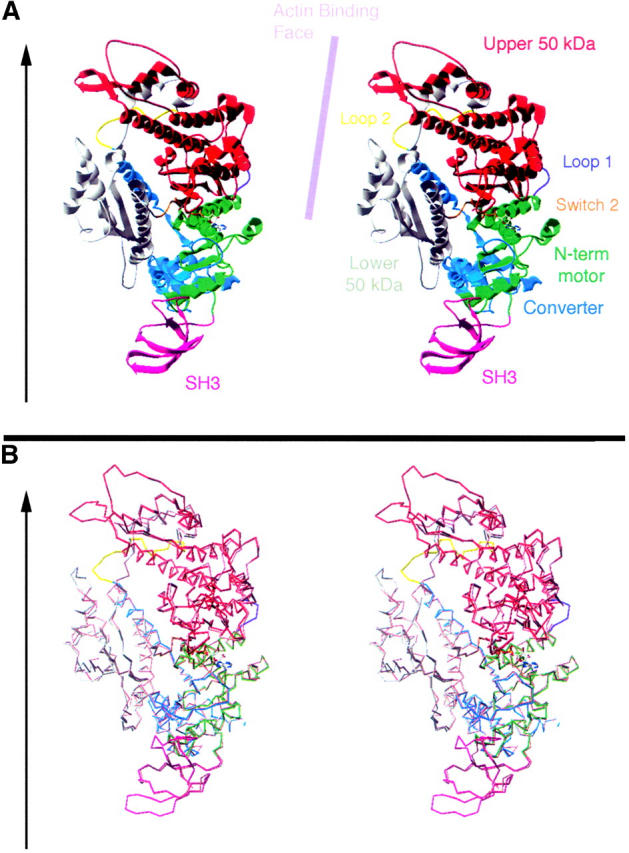
Homology model for the fly ck/myoVIIA motor shows the N-terminal, SH3 subdomain and its fit with the primary reference structure, chicken smooth muscle myosin. (A) A stereoscopic view of a ribbon diagram is color coded to display the model and its relationship to key structural features of the myosin head. Regions include the SH3 subdomain (pink), the N-terminal subdomain of the motor core (green, these first two subdomains correspond to the 25-kD N-terminal fragment of skeletal muscle myosin head), the upper 50-kD subdomain (red), the Switch 2 junction (orange), the lower 50-kD subdomain (gray), the Loop 2 junction (yellow), and the converter (blue, which comprises part of the 20-kD head subfragment). Arrow to the left indicates the approximate orientation of the actin filament to which this myosin orientation would bind and the lilac bar runs parallel to the actin-binding face of the molecule. (B) A stereoscopic view of the α-carbons traces the polypeptide backbone of our model superimposed on that of chicken smooth muscle myosin II (ExPDB 1br2A). The ck/myoVIIA model is shown in the same colors as in A, and the reference is in peach. The fit is good throughout the proposed N-terminal SH3 subdomain (where the reference structure is resolved) and throughout the core of the motor domain. The surface loop in the top left-hand face of the diagram deviates from the reference.
RT-PCR and 5′ RACE:
Total RNA from overnight collections of w1118 embryos was prepared by standard methods and used as template for RT-PCR (using the One-Step RT-PCR kit; QIAGEN, Valencia, CA) or 5′ RACE (using the FirstChoice RLM-RACE kit; Ambion, Austin, TX). ck-specific primers were used for both RT-PCR and RACE and products were TA cloned (QIAGEN PCR cloning kit) into DH5α cells. DNA from randomly picked colonies was prepared by standard methods and subjected to automated sequencing.
Lethal phase analysis:
Brief egg collections were made on grape plates with yeast paste from small population cages by standard methods (Wieschaus and Nüsslein-Volhard 1998). Embryos were dechorionated, transferred to a grid marked on a new plate, and overlaid with a 1:1 mixture of Halocarbon 27 and 700 oils (Sigma/Aldrich, St. Louis). A dissecting microscope was used to select embryos that were undergoing cellularization and/or gastrulation. Hatch rates were determined after 36 hr. Larvae were collected, counted, and transferred to a new food vial. After 5-7 days the number of larvae that had formed pupae was counted.
SEM:
Adult flies were fixed in 70% ethanol for several hours, dehydrated into 100% ethanol, and then critically point dried in CO2 by methods recommended by the manufacturer of the critical point dryer (Ted Pella, Redding, CA). Samples were coated with 60% Au, 40% Pd, with a Hummer V sputter coater (Anatech, Springfield, VA), and then observed with a Philips XL30 ESEM TMP manufactured by the FEI company (Portland, OR). The morphology of samples observed without sputter coating appeared identical to that of coated specimens but was more readily altered as a consequence of beam damage at higher magnifications.
RESULTS
Fly myosin VIIA:
We cloned myosin VIIA from flies using a PCR strategy designed to recover unconventional myosins. We recovered a partial cDNA encoding novel sequences similar to myosin heavy chain motor domains (Chen et al. 1991)—this partial clone became the founding member of the myosin VII subfamily of motor proteins (Cheney et al. 1993; Mooseker and Cheney 1995). Positional cloning of a human Usher syndrome type 1B and of the Shaker defect in mouse characterized a full-length myosin VIIA cDNA and its gene (Chen et al. 1996; Weil et al. 1996; Kelley et al. 1997 and references therein). We recovered additional cDNA and genomic sequences and performed polytene in situ to show that this gene mapped to chromosomal location 35B (not shown). The physical map provided by subsequent sequencing of the Drosophila genome through the region (Ashburner et al. 1999) was compared to the genetic map and indicated that myosin VIIA likely corresponded to the ck gene.
crinkled encodes myoVIIA:
Sequence analysis, reversion analysis, and fine-scale genetic mapping confirm that the myoVIIA transcription unit corresponds to the locus disrupted by ck mutations. An overview of the transcription unit, its relationship to other genes in the region, and to the orthologous transcription units from D. pseudoobscura and Anopheles gambiae appears in Figure 1. The domain structure of ck/myoVIIA appears as a schematic (Figure 1H) and on a sequence alignment with human myosin VIIA and worm myosin VII (Figure 2). An alignment with the two insect orthologs is shown in supplementary Figure 1 (http://www.genetics.org/supplemental/).
Figure 1.—
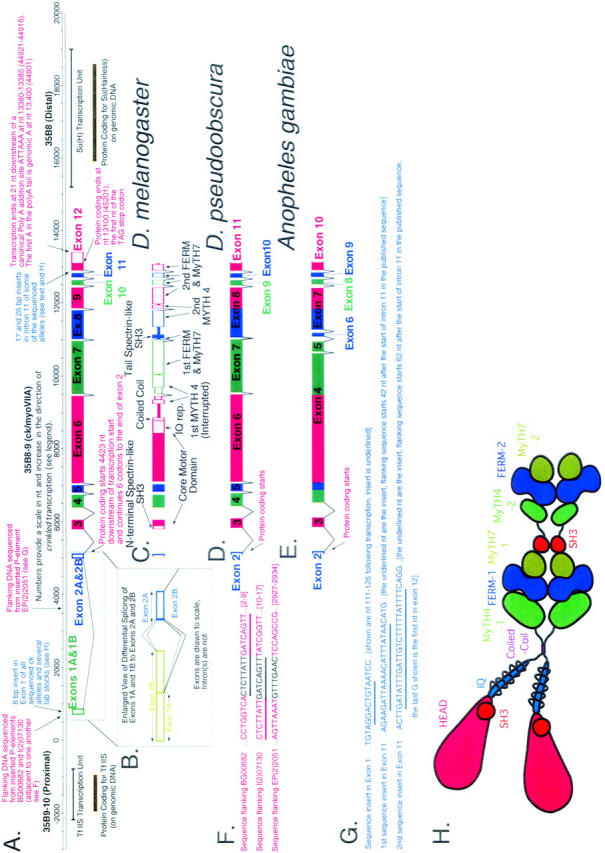
A schematic overview of genomic organization at the crinkled locus at polytene location 35B shows ck/myoVIIA transcription units from Drosophila melanogaster, D. pseudoobscura, and Anopheles gambiae and indicates significant differences in exon/intron structure. (A) Numbers provide a scale in nt and increase in the direction of crinkled transcription (“0” was chosen early in the project at a site predicted to be close to transcription start—its location is therefore arbitrary). Transcription start, identified by 5′ RACE, is at nt 663. This origin corresponds to nt 58301 in accession no. AE003646 from the Drosophila genome project (in which numbers decrease in the direction of ck transcription). Adjacent genes TfIIS and Suppressor of Hairless are also shown. (B) An enlarged view of exons 1 and 2 from D. melanogaster shows differential splicing at the first intron. (C) Domain structure of ck/myoVIIA protein mapped onto the exon/intron structure of the D. melanogaster gene. (D and E) Exons and introns in the D. pseudoobscura and A. gambiae ck/myoVIIA genes are shown (compare to A). These species last shared a common ancestor with D. melanogaster 25 million and 250 million years ago, respectively (Russo et al. 1995; Zdobnov et al. 2002). In D. pseudoobscura, a single exon 8 replaces exons 8 and 9 of D. melanogaster. In contrast, in A. gambiae, the exons corresponding to the melanogaster exons 4, 5, 6, and part of 7 are “fused” into exon 4. Likewise, parts of the melanogaster exons 8 and 9 are fused to make the A. gambiae exon 7. (F) Sequence flanking three P-element-induced alleles of ck. Sequence in black is the 8-bp target sequence that is duplicated upon P insertion. Note that the target sites for BG00682 and P(PZ)07130 are directly adjacent to one another (shared sequence is underlined). (G) Insertional polymorphisms in D. melanogaster exon 1 and intron 11. (H) Schematic of the ck/myoVIIA protein outlines the overall structure of the protein and is color coded using the scheme in Figure 2. The schematic is drawn approximately to scale: the area of each “domain” is roughly proportional to the number of amino acids in that domain. Contact between dimerized heavy chains is shown at the coiled-coil region, the FERM domains, and the tail SH3 domain because those domains are thought to mediate protein-protein interactions. It is important to note that no evidence, either for or against such interactions, exists. Similarly, the MyTH4 and MyTH7 domains may also contribute to intradimer interactions. Rings drawn around the IQ motif region represent light chains.
Figure 2.—
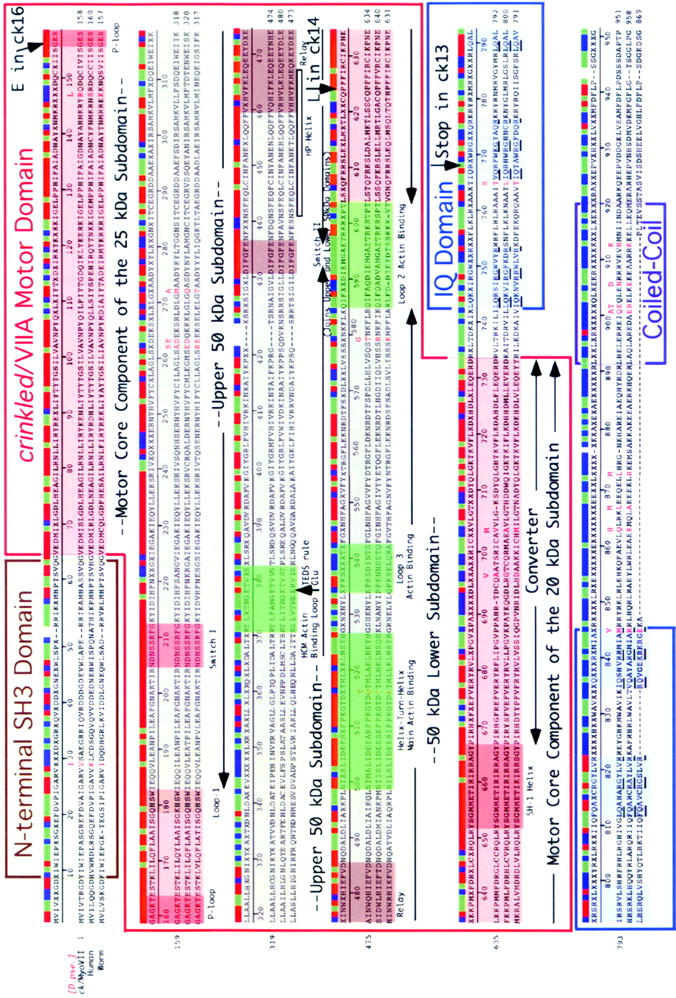
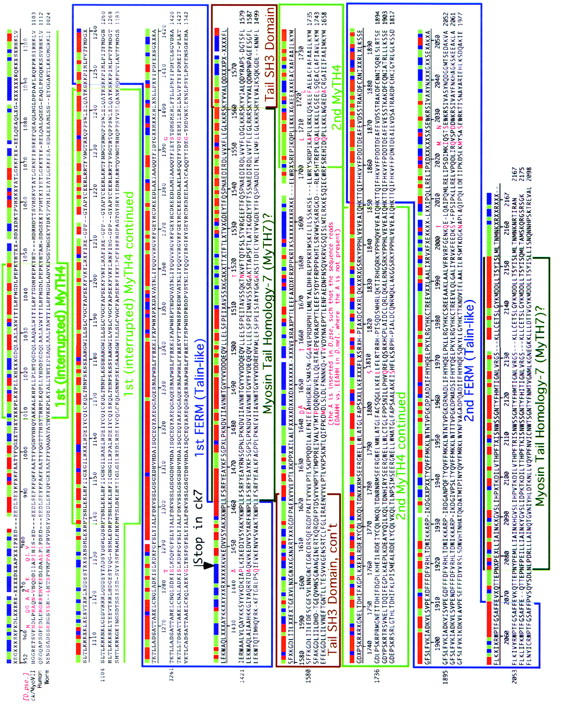
Outlines of the domain structure of ck/myosin VIIA are superimposed on a sequence alignment that compares ck/myoVIIA from D. melanogaster and D. pseudoobscura to myosin VIIA from humans and hum6/myoVII from C. elegans. Numbers indicate residue number in D. melanogaster. Domains were identified by searching the conserved domain database with reverse position-specific BLAST (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and/or correspond to the position of comparable domains previously identified in human myosin VIIA (Chen et al. 1996). Additional features are described in the text.
Lesions in the ck/myoVIIA open reading frame characterize ck mutations:
Sequence analysis of genomic DNA purified from hemizygous escaper adults [ck7, ck13, ck14, and ck16/Df(ck−)] show nonsense mutations (premature stop codons) in ck7 (Leu1445Stop, truncates upstream of the first FERM domain) and ck13 (Arg768Stop, truncates in the light chain-binding IQ domain; see Figure 2). In addition, we found missense mutations in ck14 and ck16 that alter highly conserved sequences in the C-terminal, 20-kD subdomain of the myosin motor (Pro684Leu) and in the P-loop, polyphosphate binding sequence, GESGAGKT (Gly-156-Glu; discussed below), respectively. Our sequencing effort also identified two insertional polymorphisms in various ck and control stocks that we sequenced (Figure 1G).
The locations of two P-element insertional mutations near transcription start [P(PZ)07130, BG00682] and a third P-element insert in the middle of intron 1 [EP(2)2051], all of which fail to complement ck alleles, are shown in Figure 1F. In trans with the large ck− deletion Df(2L)osp29, they show characteristic ck phenotypes.
Fine-scale genetic mapping of P(PZ)07130 to ck:
Fine-scale genetic mapping shows that P(PZ)07130 maps to ck and not to adjacent loci. We isolated ck deletions using the P-element transposase-mediated male recombination technique (Preston et al. 1996). These deletions usually retain the original insertion, extend either distally or proximally from the insertion site, and are recognized by exchange between flanking markers. From 14,545 progeny we selected 28 independent recombinants between the flanking markers (14 dp b and 14 cn bw). The 14 dp b recombinants could be either deletions extending distally or duplications extending proximally. Two were deletions affecting loci distal of ck. They were both lethal over severe ck alleles. The 14 cn bw recombinants could be either deletions extending proximally or duplications extending distally. Two were deletions affecting loci proximal of ck. These had very weak ck phenotypes over ck alleles [similar to the original P(PZ)07130]. Two further recombinants did not affect other loci but were weak ck alleles. Thus, the P elements map molecularly to myosin VIIA sequence and genetically to the ck locus.
Reversion analysis:
To investigate further the relationship between P(PZ)07130 and the ck locus, we performed reversion analysis. Of 54 transposase-induced excisions, 34 reverted to wild type and complemented severe ck alleles: they are likely the consequence of precise P-element excisions. In contrast, 20 excisions failed to complement other ck alleles, 17 of which had a phenotype stronger than the original P-insertion allele. These lines were likely the consequence of small deletions that removed part of the ck transcription unit. None of these more severe alleles were lethals, nor were they deletions of adjacent loci as demonstrated genetically. The P(PZ)07130 insertion is not the cause of lethality in this chromosome: hemizygotes of this chromosome [in trans to Df(2L)osp29, a deficiency that removes the ck locus] or trans-heterozygotes with severe ck alleles are viable, suggesting that the lethality is due to another lesion on the chromosome.
Phenotype of developmental arrest:
To evaluate when ck/myoVIIA function is required in development, we investigated when the most severe ck/myoVIIA mutant animals die as hemizygotes. Nearly all ck13 mutants die as embryos and most ck7 mutants die as larvae, suggesting an acute need for ck/myoVIIA function in both stages. We have not yet identified any morphological defects that correlate with these lethal phenotypes. All combinations of ck alleles yield some adult escapers, demonstrating that ck is not absolutely essential for viability. Nevertheless, for all intents and purposes, ck is essential: all emerging adults show a variety of morphological defects (described below) and fail to live very long. We have tested a small number of escapers for fertility and find that hemizygous males and females of the severe ck alleles are not fertile (ck7, ck13, and ck16). In all, 15 EMS-induced alleles of ck, plus 4 insertional alleles (3 P-element insertions and 1 due to the insertion of the complex element TE36) have been characterized genetically. Their hemizygous viabilities vary between 0.1 and 20% (the 3 P-element alleles are all weak by this criterion), but all hemizygous escapers have a typically crinkled phenotype.
Phenotypes of escapers:
To understand the function of ck/myoVIIA better, we examined hairs, bristles, and aristae in escaper, hemizygous ck flies by scanning electron microscopy (SEM, Figures 3 and 4). We confirmed that these ck/myoVIIA mutant flies had wispy aristae (previously described as “feathery,” Figure 4) and had aberrant wing hairs (setae) and bristles (chaetae) as described previously (Gubb et al. 1984; He and Adler 2002).
Figure 3.—
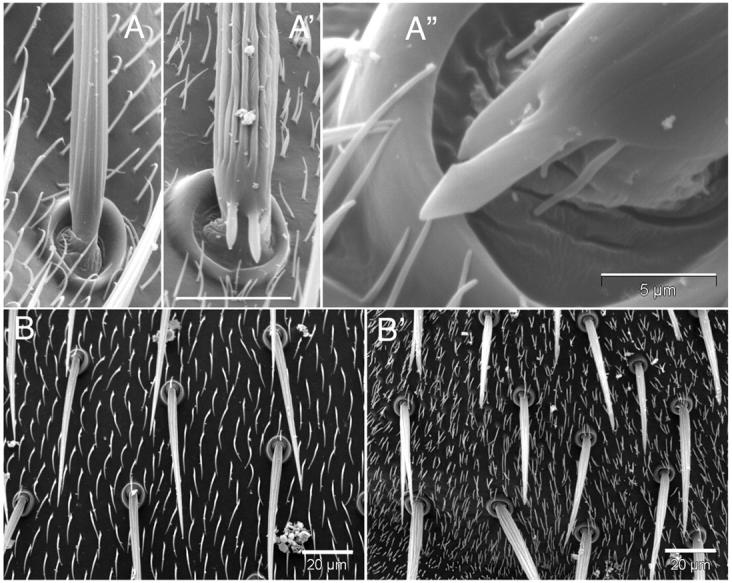
Mutations in ck/myoVIIA alter the morphology of setae, micro- and macrochaetae, and the number of and distribution of setae on the thorax. (A) Deep ridges in and aberrant projections from the shaft of the macrochaetae on the thorax of ck13/Df flies (A′ and A′′, compare to control, A). Surrounding setae are shorter and more numerous. Bar in A′, for A and A′, 20 μm. Bar in A′′, 5 μm. (B) Microchaetae are sometimes branched and setae are short and more numerous on the thorax of ck13/Df mutant (B′) vs. control (B) flies. Bars, 20 μm.
Figure 4.—
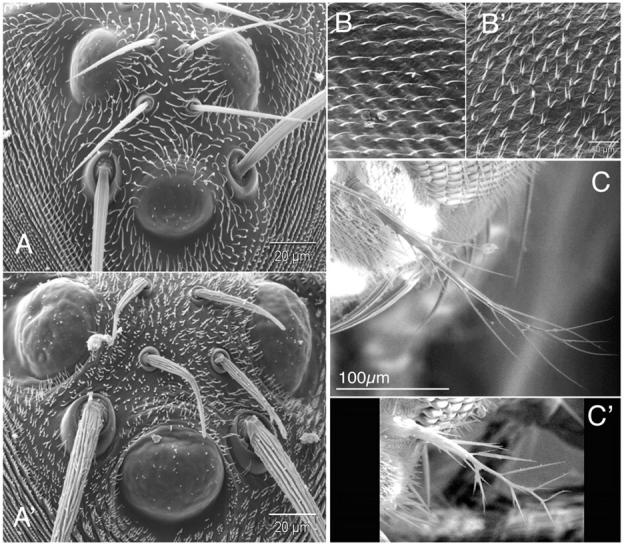
Defects in ck/myoVIIA disrupt the morphology and distribution of setae, microchaetae, and macrochaetae on the head and wing and alter the morphology of the aristae. (A) Macrochaetae, microchaetae, and setae near the ocelli on control (A) and ck13/Df mutant (A′) flies. Microchaetae are twisted and bent, and micro- and macrochaetae have abnormally deep and irregular grooves. Multiple setae also characterize mutant vs. control flies. Bars in A and A′, 20 μm. (B) Wing hairs on ck13/Df mutant (B′) flies show a multiple wing hair (setae) phenotype (vs. control, B). Bar in B′, for B and B′, 20 μm. (C) Aristae on ck13/Df mutants (C′) are shorter and more highly branched than controls (C). Bar in C, for C and C′, 100 μm.
We observed four additional phenotypes (Figure 3). First, SEMs show that escapers have odd projections emanating from the base of macrochaetae on their thoraxes, perpendicular to the long axis of the bristle (Figure 3, A′ and A′′). Second, both microchaetae and macrochaetae are stubby, branched, frequently twisted (Figure 4, A vs. A′), and not uniformly (or smoothly) tapered. These bristle morphologies appear related to the third phenotype: deep grooves that are curved and fused characterize ck mutant bristles (compare macrochaetae in Figure 3, A vs. A′ and A′′, and microchaetae in Figure 4, A vs. A′).
Finally, we observe multiple setae (hairs), with as many as six to eight per cell on all body parts (vs. one setum per cell in wild type; compare setae in Figure 3, A and B to A′, A′′, and B′, as well as in Figure 4, A to A′). The defect in hair structure seen on the wings and the rest of the mutant's body is strikingly different. Wing hairs tend to split into two to three hairs that are frequently branched near their tips and are more slender than their wild-type counterparts. Moreover, they are more like wild type over wing veins. In contrast, on the abdomen, thorax, legs, and head, hairs are far more numerous (five to eight per cell), much shorter, and less likely to branch. These aberrant phenotypes suggest that ck/myoVIIA plays a crucial role in positioning actin bundles during bristle and hair morphogenesis.
ck/myoVIIA is widely expressed:
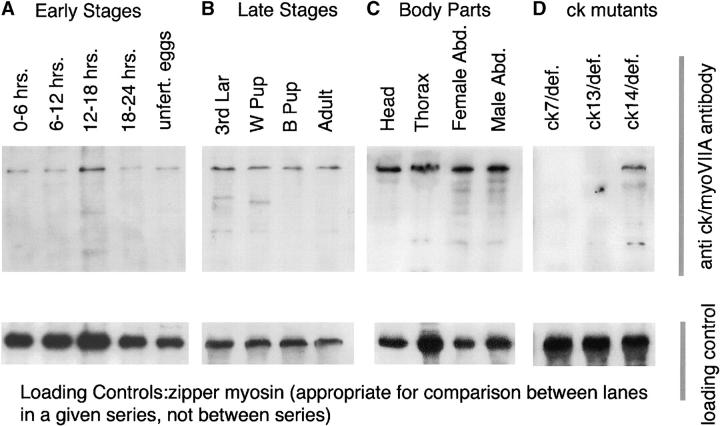
Antiserum directed against unique sequences in ck/myoVIIA was used to evaluate ck/myoVIIA expression by immunoblots. ck/myoVIIA is maternally loaded (is present in unfertilized eggs) and its abundance remains relatively constant, at a low level, throughout development (Figure 5, A–D, and data not shown). It reacts strongly with a single band, consistent in size with the protein product predicted from cDNA sequence. Lower molecular mass bands are likely breakdown products—following incubation of extracts at room temperature, the abundance of the 250-kD band decreases and the lower bands increase (data not shown). The serum fails to detect ck/myoVIIA protein in adult escapers of ck7 and ck13 (Figure 5, both with premature stop codons), thereby verifying specificity of this antiserum. Because the antiserum was raised against an internal fragment of ck/myoVIIA, it is possible that in the ck13 allele a stable, N-terminal fragment of protein is made and retains partial function. Our data suggest that no ck7 protein is made (however, see discussion regarding the severity of ck13 vs. ck7 alleles based on phenotype of arrest).
Figure 5.—
Immunoblots demonstrate that ck/myoVIIA is expressed throughout fly development, at comparable levels in head, thorax, and male and female abdomens, and is altered in ck mutant animals. (A) Early stages. Expression is comparable in unfertilized eggs and in timed, 6-hr embryo collections. Abundance appears to be somewhat increased at 12–18 hr, but there is also an increase in the loading control (zipper myosin, bottom, and tubulin, not shown). (B) Later stages show an essentially constant level of protein throughout development (comparable to the level in embryos, not shown). (C) Body part blot shows equivalent amounts of ck/myoVIIA in all body parts tested. The wavy band in the thorax sample is due to the high abundance of muscle myosin that migrates just below the ck/myoVIIA in SDS-PAGE. (D) Blots of ck7/Df and ck13/Df show no immunoreactive band in these animals, consistent with premature stop codons that truncate the protein or cause message instability.
ck/myoVIIA transcription unit:
We sequenced a nearly full-length ck cDNA and aligned it with Drosophila melanogaster genomic sequence (Release 3). Exon-intron boundaries and the overall organization of the transcription unit are shown in Figure 1 and supplemental Figure 1. ck lies ∼1.8 kb proximal of the Suppressor of Hairless transcription unit and 1.4 kb distal to the TfIIS transcription unit on the left arm of the second chromosome. It is encoded by a 12.8-kb transcription unit [transcription start to poly(A) addition site] that includes 12 exons and 11 introns (Figure 1 and Table 1). It makes a 7.0- to 7.2-kb mature transcript, is transcribed in a proximal to distal direction, and is differentially spliced. Due to the large size of the first two introns, the first 0.2–0.4 kb of cDNA (depending on splicing) spans 5.4 kb of genomic DNA. The remainder of the cDNA is more compactly organized, such that the 6784 bp of cDNA spans 7419 bp of genomic DNA and is interrupted by 9 introns of close to minimal length. The structure of the transcription unit and the protein that it encodes is detailed in supplemental material at http://www.genetics.org/supplemental/. Remarkably similar ck/myoVIIA proteins (see below) are encoded by orthologous genes in both D. pseudoobscura and the mosquito, A. gambiae (Holt et al. 2002), but the distribution of exons and introns is not preserved (Figure 1).
TABLE 1.
Exon-intron boundaries in insectck/myoVIIA
|
D. melanogaster
|
D. pseudoobscura
|
A. gambiae
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon, intron no. |
Start | End | Length | Protein no. (nt) |
Length (nt) |
Exon, intron no. |
Start | End | Length (nt) |
D. melanogaster equivalent |
Protein no. (nt) |
Length (nt) |
Exon, intron no. |
Start | End | Length (nt) |
D. melanogaster equivalent |
Protein no. (nt) |
Length (nt) |
| Exon 1 | 663 | 1,011 | 349 | Exon 1 | |||||||||||||||
| Intron 1 | 1,012 | 4,993 | 3,982 | Intron 1 | |||||||||||||||
| Exon 2 | 4,994 | 5,102 | 109 | Exon 2 | Exon 2 | ||||||||||||||
| Exon 2 ORF |
5,085 | 5,102 | 18 | 1–6 | 6 | Exon 2 ORF |
5,083 | 5,100 | 18 | Exon 2 | 1–6 | 6 | Exon 2 ORF |
4,320 | 4,337 | Exon 2 | 1–6 | 6 | |
| Intron 2 | 5,103 | 5,781 | 679 | Intron 2 | 5,101 | 5,841 | 741 | Intron 2 | Intron 2 | ||||||||||
| Exon 3 | 5,782 | 6,042 | 261 | 7–93 | 87 | Exon 3 | 5,842 | 6,102 | 261 | Exon 3 | 7–93 | 87 | Exon 3 | 10,004 | 10,264 | 261 | Exon 3 | 7–93 | 87 |
| Intron 3 | 6,043 | 6,372 | 330 | Intron 3 | 6,103 | 6,522 | 420 | Intron 3 | Intron 3 | 10,265 | 10,333 | 69 | Intron 3 | ||||||
| Exon 4 | 6,373 | 6,722 | 350 | 94–210 | 117 | Exon 4 | 6,523 | 6,872 | 350 | Exon 4 | 94–210 | 117 | Exon 4 | 10,334 | 14,463 | 4,130 | Exons 4, 5, 6, part of 7 |
94–1,472 | 1,379 |
| Intron 4 | 6,723 | 6,788 | 66 | Intron 4 | 6,873 | 6,940 | 68 | Intron 4 | Intron 4 | 14,464 | 14,528 | 65 | |||||||
| Exon 5 | 6,789 | 6,989 | 201 | 211–277 | 67 | Exon 5 | 6,941 | 7,141 | 201 | Exon 5 | 211–277 | 67 | |||||||
| Intron 5 | 6,990 | 7,053 | 64 | Intron 5 | 7,142 | 7,201 | 60 | Intron 5 | |||||||||||
| Exon 6 | 7,054 | 9,448 | 2,395 | 278–1,075 | 798 | Exon 6 | 7,202 | 9,596 | 2,395 | Exon 6 | 278–1,075 | 798 | |||||||
| Intron 6 | 9,449 | 9,504 | 56 | Intron 6 | 9,597 | 9,660 | 64 | Intron 6 | |||||||||||
| Exon 7 | 9,505 | 10,955 | 1,451 | 1,076–1,559 | 484 | Exon 7 | 9,661 | 11,111 | 1,451 | Exon 7 | 1,076–1,559 | 484 | Exon 5 | 14,529 | 14,692 | 164 | End of 7 | 1,473–1,560 | 88 |
| Intron 7 | 10,956 | 11,016 | 61 | Intron 7 | 11,112 | 11,174 | 63 | Intron 7 | Intron 5 | 14,693 | 14,760 | 68 | |||||||
| Exon 8 | 11,017 | 11,803 | 787 | 1,560–1,821 | 261 | Exon 8 | 11,175 | 12,530 | 1,356 | Exons 8, 9 | 1,560–2,012 | 450 | Exon 6 | 14,761 | 14,995 | 235 | Part of 8 | 1,561–1,607 | 47 |
| Intron 8 | 11,804 | 11,862 | 59 | Intron 8 | 12,531 | 12,589 | 59 | Intron 9 | Intron 6 | 14,996 | 15,061 | 66 | |||||||
| Exon 9 | 11,863 | 12,428 | 566 | 1,822–2,010 | 189 | Exon 7 | 15,062 | 16,283 | 1,222 | End of 8, part of 9 |
1,608–2,015 | 408 | |||||||
| Intron 9 | 12,429 | 12,490 | 62 | Intron 7 | 16,284 | 16,367 | 84 | ||||||||||||
| Exon 10 | 12,491 | 12,637 | 147 | 2,011–2,058 | 48 | Exon 9 | 12,590 | 12,736 | 147 | Exon 10 | 2,013–2,059 | 48 | Exon 8 | 16,368 | 16,514 | 147 | Exon 10 | 2,016–2,063 | 48 |
| Intron 10 | 12,638 | 12,705 | 68 | Intron 9 | 12,737 | 12,805 | 69 | Intron 10 | Intron 8 | 16,515 | 16,579 | 65 | |||||||
| Exon 11 | 12,706 | 12,826 | 121 | 2,059–2,099 | 41 | Exon 10 | 12,806 | 12,926 | 121 | Exon 11 | 2,060–2,100 | 41 | Exon 9 | 16,580 | 16,700 | 121 | Exon 11 | 2,064–2,104 | 41 |
| Intron 11 | 12,827 | 12,895 | 69 | Intron 10 | 12,927 | 12,996 | 70 | Intron 11 | Intron 9 | 16,701 | 16,779 | 79 | |||||||
| Exon 12 | 12,896 | 13,439 | 544 | Exon 11 ORF |
12,997 | 13,200 | 204 | Exon 12 | 2,101–2,168 | 68 | Exon 10 | 16,780 | 16,987 | 208 | Exon 12 | 2,105–2,173 | 68 | ||
| Exon 12 ORF |
12,896 | 13,098 | 203 | 2,100–2,167 | 68 | ||||||||||||||
ck/myoVIIA protein organization:
All the D. melanogaster ck/myoVIIA transcripts include a 6501-bp open reading frame that encodes a 250-kD protein (Figures 1 and 2). The size of this ORF is consistent with the ck/myoVIIA band seen on immunoblots (Figure 5). The relationship between ck/myoVIIA, its ortholog from humans (61.7% identical), the single myosin VII found in C. elegans (58.8% identical), and its orthologs from other insects is shown in sequence alignments (Figure 2 and supplemental Figure 1). A consensus sequence indicates the shared amino acid if two or more sequences match, an x if there is no match, and a blank if there is a gap. A corresponding bar color codes the alignment: regions of sequence identity are shown in red (perfect match), sequence similarity is in green (two of the three residues match), no match is in blue, and a gap is blank. Boxed sequences indicate various domains that are shared between ck/myoVIIA and other proteins. Sequence motifs in the myosin head are shaded and labeled and are based on detailed modeling of the 3-D structure of the ck/myoVIIA head described below. Also shown in red text, in and above the alignments, are the 40 amino acids that distinguish the D. melanogaster ck/myoVIIA from the D. pseudoobscura ck/myoVIIA (the amino acid shown above the alignment is the one from pseudoobscura, the two proteins are 98.2% identical). For comparison, the A. gambiae protein is 88.1% identical (supplementary Figure 1). Hot spots for amino acid substitutions exist in several locations.
Myosin head domain:
The myosin VIIA head, extending from the N terminus through the motor domain, shows remarkable sequence identity with its human myosin VIIA ortholog and the single myosin VIIs from C. elegans and D. discoideum. There is considerable sequence identity to heads from other myosin superfamily members, including the class I and II myosins, for which crystal structures are available (36–43% identity in the myosin head). This prompted us to generate a 3-D homology model of the ck/myo VIIA head (Figures 6 and 7). The model satisfies most known physical constraints for well-resolved X-ray structures, with a Ramachandran Z-score of −1.687, no Ramachandran outliers, and an average packing Z-score of −1.008. Thus it is a useful working model for the structure of ck/myo VIIA in the absence of a crystal structure. Our confidence in the model is low in the regions, usually variable loops, that are poorly or not resolved in the template structures. The overall topology of our model is similar to that of myosin I and II heads, yet distinct differences are likely to account for the unique properties of myosin VIIA.
Figure 7.—
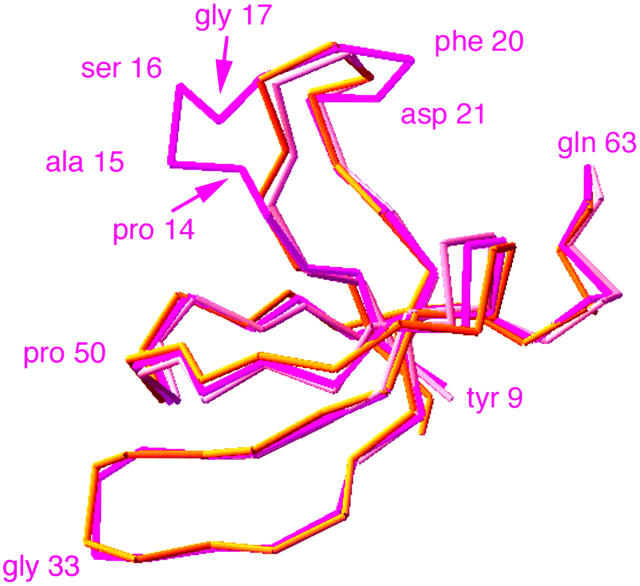
The α-carbon backbone of the N-terminal SH3 subdomain predicted by our model and compared to scallop muscle myosin II (chrome yellow) and chicken smooth muscle myosin (peach). The fit is excellent except where there are additional residues in ck/myoVIIA compared to the reference structures (three additional residues at Pro-14 to Gly-17 and one additional residue at Phe-20 to Asp-21). Shown are ck/myoVIIA residues Tyr-9 to Gln-63, chicken smooth muscle myosin Leu-34 to Ser-84, and scallop muscle myosin II Ser-37 to Glu-83.
ck/myoVIIA has an N-terminal SH3 subdomain:
Like myosin II, the ck/myoVIIA head predicted by our homology model has a N-terminal spectrin-like SH3 subdomain formed by amino acid residues Tyr 9 to Gln 63 (Figure 6). This feature of myosin VIIA has previously been overlooked in the primary sequence alignments used to evaluate domain structure. In myosin II, this region forms a structural unit independent of the rest of the head, similar to the SH3 subdomain of spectrin (Rayment et al. 1993; Dominguez et al. 1998; overall structure of the myosin head reviewed in Geeves and Holmes 1999; Houdusse and Sweeney 2001). The primary sequence of this subdomain is well conserved between ck, its human ortholog, and the worm myosin VII (but not the Dictyostelium myosin VII).
Motor domain:
Our model predicts the traditional division of the myosin head into distinct subdomains, seen in the structures of all myosin heads solved to date and based on limited proteolytic cleavage of myosin II into N-terminal 25-kD, middle 50-kD, and a C-terminal 20-kD fragments. As expected, it suggests that the motor core of the ck/myoVIIA head, which includes the nucleotide-binding pocket (γ-phosphate-binding P-loop of the 25-kD subdomain) and switch I and II of the upper 50-kD subdomain, is conserved in structure. Regions of sequence divergence correspond mainly to flexible surface loops, some of which participate in actin binding, and “hinges” or “joints” between conserved elements of the motor core (Figure 1). Finally, the C-terminal subdomain comprises the “converter,” believed to amplify the relatively small conformational changes in the motor core to drive movement of the lever arm that extends through the neck (and includes the light-chain binding, IQ repeats). The conformations of the joints and of the converter in our model are characteristic of the ADP·Pi-bound state or state II (Houdusse et al. 1999). Our homology model allows interpretation of the structural ramifications of amino acid replacements in ck/myoVIIA mutations (see discussion).
ck/myoVIIA tail and the myosin tail homology (MyTH7) domain:
Most of the remainder of the protein is remarkably similar to its human ortholog and worm homolog. A sequence predicted to form a coiled-coil is considerably shorter than the corresponding region in vertebrate myosin VIIAs and even in vertebrates the role of this region in dimerization has been called in question (Inoue and Ikebe 2003). Following the putative coiled-coil region, two modules, each of which contains a MyTH4 domain and a FERM domain, appear in a tandemly repeated fashion (Figures 1H and 2, supplemental Figure 1). The two modules are separated by an SH3 domain (distinct from the spectrin-like SH3 domain at the very N terminus of the ck/myoVIIA heavy chain). Since original alignments were performed (e.g., Chen et al. 1996), the crystal structures of the ezrin, radixin, and moesin FERM domains showed that FERM domains have a cloverleaf like structure, consisting of three subdomains or lobes, each containing ∼100 amino acids (Pearson et al. 2000; Hamada et al. 2003; Smith et al. 2003 and references therein). Only the first ∼200 amino acids (i.e., the first two lobes) of the ck/myoVIIA FERM domain are well conserved with FERM domains from other proteins. Nevertheless, sequences immediately following these two partial FERM domains are highly conserved among myosin VIIAs, myosin VII, and myosin VIIBs. Indeed, among these subclasses of motor proteins, these stretches are as conserved as or more highly conserved than the two lobes of the FERM domain that directly precede them (vs. the Anopheles, human, and C. elegans ck/myoVIIAs; see Table 2). Due to this remarkable conservation, we refer to the ∼100-amino-acid stretch following ck/myoVIIA FERM 1 and the ∼75-amino-acid stretch following FERM 2 as MyTH7 domains (see Figure 1H and discussion).
TABLE 2.
Sequence conservation among protein domains in theck/myoVIIA tail
| Domain protein | MyTH4-2 (%) | FERM-1 lobes 1 and 2 (%) |
MyTH7-1 (%) | FERM-2 lobes 1 and 2 (%) |
MyTH7-2 (%) |
|---|---|---|---|---|---|
| Anopheles VIIA | 87/95 | 91/97 | 97/98 | 89/97 | 97/98 |
| Human VIIA | 60/76 | 72/86 | 72/86 | 72/85 | 90/96 |
| C. elegans VII | 59/73 | 63/81 | 64/79 | 54/70 | 56/80 |
| Dictyostelium VII | 26/48 | 28/52 | 22/42 | 19/42 | 23/45 |
| Fly VII B | 40/56 | 41/61 | 30/54 | 42/66 | 50/69 |
| Fly XV | 32/43 | 47/61 | NA | 26/51 | 26/46 |
Entries indicate percentage identity/percentage similarity, allowing conservative substitutions. Numbers were generated with BLASTP using a BLOSUM 62 matrix. The fly myoXV has a single FERM domain.
Conserved sequence 3′ of the poly(A) addition site:
3′ of the site at which poly(A) is added, a sequence that is remarkably conserved between D. melanogaster and D. pseudobsura (94.2% identical over 119 bp) suggests that there may be a gene that encodes a micro-RNA (Ambros 2003; Lai et al. 2003). This sequence is not conserved in A. gambiae and shows less secondary structure than many of the micro-RNAs identified through genomic strategies in D. melanogaster (Lai et al. 2003). Experimental analysis will be required to evaluate the significance of the conservation in this region.
DISCUSSION
Here, through a combination of molecular and genetic analysis, we provide formal proof that the fly myosin VIIA heavy chain is encoded by the crinkled locus. In addition, we establish that two severe alleles (molecular nulls that cannot encode more than a fraction of the myosin VIIA motor domain) die as embryos and larvae, although all alleles show a small number of escapers, animals that can survive to adulthood without zyogotically encoded ck/myoVIIA. These escapers are severely compromised—we have been unable to set up a homozygous stock. We demonstrate that the ck/myoVIIA protein is present at low abundance throughout development. While we have been unable to identify a function for ck/myoVIIA that is required for embryonic and larval viability, we document phenotypes that confirm and extend older observations on ck phenotypes: bristles and hairs (chaetae and setae) have aberrant morphologies and/or distributions.
We used the highly conserved sequence among different myosin VIIs to investigate the structure of ck/myoVIIA in two ways. First, we generated a homology model of the ck/myoVIIA head on the basis of solved structures of myosin I and IIs and show that myosin VIIAs have a heretofore unnoticed N-terminal, spectrin-like SH3 subdomain. We used the model to hypothesize the effect of specific amino acid substitutions that characterize sequenced mutants. In addition, we compared the sequence of the melanogaster ck/myoVIIA tail to its orthologs from another D. pseudoobscura, mosquito, and humans and to a myosin VII homolog from worms. We identified two highly conserved protein repeats that are shared by myosin VIIs, VIIAs, and VIIBs. We refer to these conserved repeats as MyTH7 domains. One possibility is that the two MyTH7 domains fold to form a specialized FERM lobe 3 subdomain. Another alternative is that the MyTH7 domain forms a distinct structure that folds and functions independently of the first two lobes of the FERM domain. Clearly, both structure and structure/function analysis of the ck/myoVIIA tail will be required to distinguish among these possibilities. Together our studies provide an essential step in the characterization of this motor protein in flies.
Animals homozygous or hemizygous for severe ck mutants almost all die as embryos or in early larval stages. Nevertheless, a small fraction of so-called escapers emerge as adults, indicating that those individuals that persist through an acute, early period in ontogeny, during which ck/myoVIIA is all but essential, can survive through the remaining stages of development. Such escapers have defects in the distribution and morphology of hairs and the morphology of bristles all over their bodies, fail to live very long, and are infertile. Together, these observations demonstrate that myosin VIIA plays a more extensive role in flies than in vertebrates, where defects in myosin VIIA cause aberrant sensory perception but appear to have little or no effect on viability per se (of course from a Darwinian perspective, most vertebrates whose hearing and vision are defective will be far from fit). A simple explanation for this discrepancy may be differences in the pattern of expression of myosin VIIA in flies and vertebrates. In flies, the distribution of mutant phenotypes, immunoblot analysis, and preliminary antibody and RNA in situ studies (data not shown) all point to an expression pattern of ck/myoVIIA that is widespread, if not “ubiquitous.” In contrast, the tissues affected by defects in vertebrate myosin VIIA, cochlea, retina, lung, and testis are commensurate with an expression pattern that is restricted to these tissues and the kidneys. Previous investigators hypothesized that a lack of phenotype in kidney might be attributed to redundant function supplied by additional myosin superfamily members (Hasson et al. 1995). In addition to the widespread role for ck/myoVIIA in epithelial cell function (demonstrated by defective patterns of setae on all body parts), it is interesting to note that myosin VIIA also plays a role in fly sensory perception: there are defects in macrochaetae formation (bristles are sensory structures in flies) and ck/myoVIIA is required for hearing in flies (S. V. Todi and D. F. Eberl, personal communication and Todi et al. 2003). By comparing the function of ck/myoVIIA in embryos—where lethality due to specific effects on sensory cells is unlikely—to the function of ck/myoVIIA in sensory perception, we may well gain insight into the chemomechanical constraints that define the niche in which this motor protein functions.
Another explanation for different roles of myosin VIIA in flies and vertebrates may be redundancy that allows other myosins to compensate for defects in myosin VIIA in vertebrates but not in flies. Indeed, humans, mice, and flies have genes that encode distinct myosin VIIBs and all have a myosin XV, a more distantly related myosin whose tail nevertheless includes two MyTH4 domains and a single FERM domain and is therefore more closely related to this class than to other myosin classes (Yamashita et al. 2000; Berg et al. 2001; Tzolovsky et al. 2002). Interestingly, while defects in myosin VIIB have not been described or associated with disease loci, defects in myosin XV cause deafness in humans and mice, suggesting that myosin VIIA and XV may have some (although clearly not completely) overlapping function (reviewed in Friedman et al. 1999; Redowicz 2002). Overall, sequence comparisons between vertebrate and Drosophila myosin VIIs and XVs suggest that all three of these myosin heavy chain subfamily members were apparently present in the last common ancestor of these organisms (0.6 and 1.2 billion years ago; Benton and Ayala 2003). At this time our observations cannot distinguish between a model in which a common set of functions is performed by a subset of myosin motors or that evolution has called upon fly ck/myoVIIA to perform functions that are distinct from those required in vertebrates. A complete understanding of how these FERM domain myosins contribute to biological function may require strategies designed to affect all three loci simultaneously. The unique molecular genetic tools available in flies, our ability to compare mutagenized ck/myoVIIA function in vitro and in vivo, and the identification of two other myosins in the ck/myoVIIA subfamily (VIIB and XV) suggest that analysis of myosin VIIA function in this system will be particularly rewarding. Moreover, comparing myosin VIIA function in flies and vertebrates to its function in systems that have a single myosin VII isoform may provide further insight into the evolution of this subfamily of the myosin motors.
Our observations indicate that ck/myoVIIA plays an important role in positioning the actin prehairs and bundles that give rise to bristles and hairs. The grooves in bristles are formed during development by bundles of actin filaments that function as struts during bristle formation (see Tilney et al. 2003 and references therein). The multiple body and wing setae phenotype observed suggests that ck/myoVIIA contributes to the distribution and or integrity of microvillus-like prehairs that have been best studied in wings (Wong and Adler 1993; Turner and Adler 1998). Turner and Adler (1998) observed that the multiple wing hair phenotype of ck mutants could be phenocopied by low doses of cytochalasin. Why the absence of a motor protein should mimic the effects of a drug that presumably inhibits F-actin assembly remains a mystery. Analysis of the ontogeny of the ck/myoVIIA phenotype during hair development and analysis of epistasis with other genes that participate in the process may well provide insight into the mechanisms by which ck/myoVIIA functions in this process.
Our homology model of the ck/myoVIIA head facilitates analysis of the defects caused by various missense mutations. In ck14, Pro-624 is replaced by Leu. Pro-624 is located in the 20-kD subdomain, N-terminal to the SH-1 helix and the converter. This proline residue is conserved in the representative myosin VIIs shown in Figure 2, other myosin VIIs for which sequence in the region is available, and in 120 of the 143 myosins shown on the Myosin Home Page (http://www.mrc-lmb.cam.ac.uk/myosin/trees/trees.html). Its replacement with Leu is expected to alter the trajectory of the polypeptide backbone. In addition, it is expected to affect the stereochemistry of the hydrophobic interface between the 20-kD subdomain and the HP helix that extends into the relay element (Figure 2). This interface contributes to the rigidity of the relay that is crucial to the positioning of the converter and, as a consequence, to the overall ability of the ck/myoVIIA motor domain to produce movement.
The ck16 lesion disrupts the phosphate-binding loop that is shared by myosins and other polyphosphate-binding proteins by replacing Gly-156 with Glu. Our model predicts that the disruption of this highly conserved GESGAGKT sequence may stabilize the loop against conformational changes, which would be highly deleterious to motor activity.
The model also makes interesting predictions regarding the detailed structure of the ck/myoVIIA motor domain. For example, it confirms that the junctions between the 25-kD and the upper 50-kD subdomains (loop 1, Gly-178 to Trp-182) and between the lower 50-kD and the 20-kD subdomains (loop 2, Ile-586 to Pro-602) are both short and compact. Loop 1 affects the rate of ADP release by Dictyostelium myosin II (Murphy and Spudich 1998), while loop 2 affects the actin-binding affinity and maximum ATPase rate of this myosin (Murphy and Spudich 1999). These loops vary considerably in length and composition in different myosins, giving rise to functional differences between myosins from different classes and species. The model allows the intelligent engineering of site-directed mutations to probe the function of these loops in ck/myoVIIA. Although the elucidation of the precise orientation of residues will require the high resolution of X-ray crystal structures of the myosin head in various nucleotide states and appropriate EM studies of the acto-myosin VIIA complex, the homology-modeling approach demonstrates its utility in the interpretation of molecular lesions in mutant ck/myoVIIAs and suggests interesting targets for future functional studies.
Analysis of the two nonsense mutations suggests that in these alleles a small fragment of the ck/myoVIIA protein is synthesized. The ck13 mutation (hemizygous animals die as embryos) is more severe than the ck7 mutation (hemizygous animals die as larvae). ck13 encodes an open reading frame that ends in the middle of the ck/myoVIIA IQ domain and is 677 codons shorter that the ck7 open reading frame. We would expect that mRNA instability, due to nonsense-mediated decay (Wagner and Lykke-Andersen 2002; Gatfield et al. 2003), would render both alleles equivalent in severity, yet they show different lethal phases. One possible explanation is that the longer fragment has residual ck/myoVIIA that can substitute, in part, for wild-type function. Alternatively, the longer mutant protein may better stabilize the maternal load of wild-type ck/myoVIIA, thereby increasing ck/myoVIIA activity in the ck7 mutant flies. Our data do not distinguish between the two possibilities.
Our studies provide the data that establish that crinkled encodes fly myosin VIIA, detail the structure of the ck transcription unit, identify the molecular lesion in four ck alleles, establish the phenotype of arrest of severe ck alleles, and document new defects in setae and chaetae morphology in ck escapers. Homology modeling and sequence comparisons identify a spectrin-like SH3 domain comparable to that seen in myosin IIs and show highly conserved sequences that we have dubbed MyTH7 domains. Together, they provide essential groundwork for additional studies on the function of this important motor molecule in development and morphogenesis.
Acknowledgments
We thank Suzanna Lewis at the Berkeley Drosophila Genome Project; Dick Cheney (University of North Carolina, Chapel Hill, NC); Dan Eberl and Sokal Todi (University of Iowa); Meg Titus (University of Minnesota, Minneapolis, MN); and Mariano Garcia-Blanco, Robin Wharton, Rick Fehon, and members of the Kiehart lab for helpful discussion, comments on our manuscript, and encouragement. We also thank Tim Karr for advice on Wolbachia and how to cure our stocks of this pathogen (O'Neill, S. L., and T. L. Karr, 1990, Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature 348: 178–180.). We appreciate assistance from Amanda Boury and Leslie Eibest and funding from the National Science Foundation (for the environmental SEM, DBI-0098534), from the National Institutes of Health (GM33830), and from Pfizer Global Research and Development for a Summer Undergraduate Research Fellowship for M.K.C. Work in Cambridge was supported by a Medical Research Council program grant to M. Ashburner, D. Gubb, and S. Russell.
References
- Ahmed, Z., S. Riazuddin and E. Wilcox, 2003. The molecular genetics of Usher syndrome. Clin. Genet. 63: 431–444. [DOI] [PubMed] [Google Scholar]
- Ambros, V., 2003. MicroRNA pathways in flies and worms. Growth, death, fat, stress, and timing. Cell 114: 269. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., S. Misra, J. Roote, S. E. Lewis, R. Blazej et al., 1999. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: the Adh region. Genetics 153: 179–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton, M. J., and F. J. Ayala, 2003. Dating the tree of life. Science 300: 1698–1700. [DOI] [PubMed] [Google Scholar]
- Berg, J. S., B. C. Powell and R. E. Cheney, 2001. A millennial myosin census. Mol. Biol. Cell 12: 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges, C. B., and K. S. Brehme, 1944 The Mutants of Drosophila melanogaster. Pub. 552, Carnegie Institute, Washington, DC.
- Brown, N. H., and F. C. Kafatos, 1988. Functional cDNA libraries from Drosophila embryos. J. Mol. Biol. 203: 425–437. [DOI] [PubMed] [Google Scholar]
- Chen, T.-L., K. A. Edwards, R. C. Lin, L. W. Coats and D. P. Kiehart, 1991. Drosophila myosin heavy chain at 35B,C. J. Cell Biol. 115: 330a. [Google Scholar]
- Chen, Z. Y., T. Hasson, P. M. Kelley, B. J. Schwender, M. F. Schwartz et al., 1996. Molecular cloning and domain structure of human myosin-VIIa, the gene product defective in Usher syndrome 1B. Genomics 36: 440–448. [DOI] [PubMed] [Google Scholar]
- Chen, Z. Y., T. Hasson, D. S. Zhang, B. J. Schwender, B. H. Derfler et al., 2001. Myosin-VIIb, a novel unconventional myosin, is a constituent of microvilli in transporting epithelia. Genomics 72: 285–296. [DOI] [PubMed] [Google Scholar]
- Cheney, R. E., and M. S. Mooseker, 1992. Unconventional myosins. Curr. Opin. Cell Biol. 4: 27–35. [DOI] [PubMed] [Google Scholar]
- Cheney, R. E., M. A. Riley and M. S. Mooseker, 1993. Phlogenetic analysis of the myosin superfamily. Cell Motil. Cytoskeleton 24: 215–223. [DOI] [PubMed] [Google Scholar]
- Dominguez, R., Y. Freyzon, K. M. Trybus and C. Cohen, 1998. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell 94: 559–571. [DOI] [PubMed] [Google Scholar]
- FlyBase Consortium, 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, T. B., J. R. Sellers and K. B. Avraham, 1999. Unconventional myosins and the genetics of hearing loss. Am. J. Med. Genet. 89: 147–157. [DOI] [PubMed] [Google Scholar]
- Gatfield, D., L. Unterholzner, F. D. Ciccarelli, P. Bork and E. Izaurralde, 2003. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22: 3960–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves, M. A., and K. C. Holmes, 1999. Structural mechanism of muscle contraction. Annu. Rev. Biochem. 68: 687–728. [DOI] [PubMed] [Google Scholar]
- Gibbs, D., J. Kitamoto and D. S. Williams, 2003. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc. Natl. Acad. Sci. USA 100: 6481–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubb, D., M. Shelton, J. Roote, S. McGill and M. Ashburner, 1984. The genetic analysis of a large transposing element of Drosophila melanogaster: the insertion of a w+ rst+ TE into the ck locus. Chromosoma 91: 54–64. [Google Scholar]
- Guex, N., and M. C. Peitsch, 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hamada, K., T. Shimizu, S. Yonemura, S. Tsukita and T. Hakoshima, 2003. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J. 22: 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson, T., M. B. Heintzelman, J. Santos-Sacchi, D. P. Corey and M. S. Mooseker, 1995. Expression in cochlea and retina of the myosin VIIa gene, the gene product defective in Usher syndrome type 1B. Proc. Natl. Acad. Sci. USA 92: 9815–9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B., and P. N. Adler, 2002. The genetic control of arista lateral morphogenesis in Drosophila. Dev. Genes Evol. 212: 218–229. [DOI] [PubMed] [Google Scholar]
- Hodge, T., and M. J. Cope, 2000. A myosin family tree. J. Cell Sci. 113: 3353–3354. [DOI] [PubMed] [Google Scholar]
- Holt, R. A., G. M. Subramanian, A. Halpern, G. G. Sutton, R. Charlab et al., 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- Houdusse, A., and H. L. Sweeney, 2001. Myosin motors: missing structures and hidden springs. Curr. Opin. Struct. Biol. 11: 182–194. [DOI] [PubMed] [Google Scholar]
- Houdusse, A., V. N. Kalabokis, D. Himmel, A. G. Szent-Gyorgyi and C. Cohen, 1999. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell 97: 459–470. [DOI] [PubMed] [Google Scholar]
- Inoue, A., and M. Ikebe, 2003. Characterization of the motor activity of mammalian myosin VIIA. J. Biol. Chem. 278: 5478–5487. [DOI] [PubMed] [Google Scholar]
- Itoh, N., P. Salvaterra and K. Itakura, 1985. Construction of an adult Drosophila head cDNA expression library with lambda gt 11. Dros. Inf. Serv. 61: 89. [Google Scholar]
- Kelley, P. M., M. D. Weston, Z. Y. Chen, D. J. Orten, T. Hasson et al., 1997. The genomic structure of the gene defective in Usher syndrome type Ib (MYO7A). Genomics 40: 73–79. [DOI] [PubMed] [Google Scholar]
- Kiehart, D. P., and R. Feghali, 1986. Cytoplasmic myosin from Drosophila melanogaster. J. Cell Biol. 103: 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussel-Andermann, P., A. El-Amraoui, S. Safieddine, S. Nouaille, I. Perfettini et al., 2000. Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin-catenins complex. EMBO J. 19: 6020–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U. K., 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lai, E. C., P. Tomancak, R. W. Williams and G. M. Rubin, 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker, M. S., and R. E. Cheney, 1995. Unconventional myosins. Annu. Rev. Cell Dev. Biol. 11: 633–675. [DOI] [PubMed] [Google Scholar]
- Murphy, C. T., and J. A. Spudich, 1998. Dictyostelium myosin 25–50K loop substitutions specifically affect ADP release rates. Biochemistry 37: 6738–6744. [DOI] [PubMed] [Google Scholar]
- Murphy, C. T., and J. A. Spudich, 1999. The sequence of the myosin 50–20K loop affects myosin's affinity for actin throughout the actin-myosin ATPase cycle and its maximum ATPase activity. Biochemistry 38: 3785–3792. [DOI] [PubMed] [Google Scholar]
- Pearson, M. A., D. Reczek, A. Bretscher and P. A. Karplus, 2000. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 101: 259–270. [DOI] [PubMed] [Google Scholar]
- Preston, C. R., J. A. Sved and W. R. Engels, 1996. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144: 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment, I., W. R. Rypniewski, K. Schmidt-Base, R. Smith, D. R. Tomchick et al., 1993. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261: 50–65. [DOI] [PubMed] [Google Scholar]
- Redowicz, M. J., 2002. Myosins and pathology: genetics and biology. Acta Biochim. Pol. 49: 789–804. [PubMed] [Google Scholar]
- Roberts, D. B., 1998 Drosophila: A Practical Approach. IRL Press/Oxford University Press, Oxford/New York.
- Russo, C. A., N. Takezaki and M. Nei, 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12: 391–404. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Smith, W. J., N. Nassar, A. Bretscher, R. A. Cerione and P. A. Karplus, 2003. Structure of the active N-terminal domain of Ezrin. Conformational and mobility changes identify keystone interactions. J. Biol. Chem. 278: 4949–4956. [DOI] [PubMed] [Google Scholar]
- Tilney, L. G., P. S. Connelly, L. Ruggiero, K. A. Vranich and G. M. Guild, 2003. Actin filament turnover regulated by cross-linking accounts for the size, shape, location, and number of actin bundles in Drosophila bristles. Mol. Biol. Cell 14: 3953–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus, M. A., 1999. A class VII unconventional myosin is required for phagocytosis. Curr. Biol. 9: 1297–1303. [DOI] [PubMed] [Google Scholar]
- Todi, S. V., Y. Sharma and D. F. Eberl, 2003. Anatomical and molecular design of the Drosophila antenna as a flagellar auditory organ. Microsc. Res. Tech. 63: 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov, P. T., R. E. Hardisty and S. D. Brown, 2001. Myosin VIIA is specifically associated with calmodulin and microtubule-associated protein-2B (MAP-2B). Biochem. J. 354: 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, C. M., and P. N. Adler, 1998. Distinct roles for the actin and microtubule cytoskeletons in the morphogenesis of epidermal hairs during wing development in Drosophila. Mech. Dev. 70: 181–192. [DOI] [PubMed] [Google Scholar]
- Tuxworth, R. I., I. Weber, D. Wessels, G. C. Addicks, D. R. Soll et al., 2001. A role for myosin VII in dynamic cell adhesion. Curr. Biol. 11: 318–329. [DOI] [PubMed] [Google Scholar]
- Tzolovsky, G., H. Millo, S. Pathirana, T. Wood and M. Bownes, 2002. Identification and phylogenetic analysis of Drosophila melanogaster myosins. Mol. Biol. Evol. 19: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Udovichenko, I. P., D. Gibbs and D. S. Williams, 2002. Actin-based motor properties of native myosin VIIa. J. Cell Sci. 115: 445–450. [DOI] [PubMed] [Google Scholar]
- Vriend, G., 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8: 29, 52–56. [DOI] [PubMed] [Google Scholar]
- Wagner, E., and J. Lykke-Andersen, 2002. mRNA surveillance: the perfect persist. J. Cell Sci. 115: 3033–3038. [DOI] [PubMed] [Google Scholar]
- Weil, D., G. Levy, I. Sahly, F. Levi-Acobas, S. Blanchard et al., 1996. Human myosin VIIA responsible for the Usher 1B syndrome: a predicted membrane-associated motor protein expressed in developing sensory epithelia. Proc. Natl. Acad. Sci. USA 93: 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus, E., and C. Nüsslein-Volhard, 1998 Looking at embryos, pp. 179–214 in Drosophila: A Practical Approach, edited by D. B. Roberts. IRL Press/Oxford University Press, New York.
- Winter, C. G., B. Wang, A. Ballew, A. Royou, R. Karess et al., 2001. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105: 81–91. [DOI] [PubMed] [Google Scholar]
- Wong, L. L., and P. N. Adler, 1993. Tissue polarity genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J. Cell Biol. 123: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, R. A., J. R. Sellers and J. B. Anderson, 2000. Identification and analysis of the myosin superfamily in Drosophila: a database approach. J. Muscle Res. Cell Motil. 21: 491–505. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E. M., C. von Mering, I. Letunic, D. Torrents, M. Suyama et al., 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149–159. [DOI] [PubMed] [Google Scholar]