Abstract
Following the advent of a gene targeting technique in Drosophila, different methods have been developed to modify the Drosophila genome. The initial demonstration of gene targeting in flies used an ends-in method, which generates a duplication of the target locus. The duplicated locus can then be efficiently reduced to a single copy by generating a double-strand break between the duplicated segments. This method has been used to knock out target genes by introducing point mutations. A derivative of this method is reported here. By using different homologous regions for the targeting and reduction steps, a complete deletion of the target gene can be generated to produce a definitive null allele. The breakpoints of the deletion can be precisely controlled. Unlike ends-out targeting, this method does not leave exogenous sequence at the deleted locus. Three endogenous genes, Sir2, Sirt2, and p53 have been successfully deleted using this method.
TWO general strategies can be used to modify a gene of interest by homologous recombination. Either an ends-in or an ends-out targeting strategy is chosen on the basis of the desired outcome. These terms refer to the placement of a DNA double-strand break (DSB) that is used to stimulate recombination. For ends-in targeting, a double-strand break is generated within the target-homologous region of the donor construct. Recombination between the donor and target typically generates a duplication of the target locus. In spite of this duplication, ends-in donors can be designed in several ways to generate mutant alleles of the target locus. Homologous recombination can be achieved using just a fragment of the target gene to generate truncated gene copies. Alternatively, point mutations may be introduced into each of the two copies, generating two mutant copies of the gene. Finally, in a two-step process, a single-copy mutant allele may be generated. All these methods have been used to produce mutants in Drosophila (Rong and Golic 2000, 2001; Rong et al. 2002; Seum et al. 2002; Dolezal et al. 2003; Egli et al. 2003; Elmore et al. 2003; Lankenau et al. 2003; Liu and Kubli 2003; Sogame et al. 2003). One problem with these methods is that, without additional characterization, it is difficult to be absolutely sure that a null allele has been generated.
A deletion of a gene represents a definitive null allele, and a method that uses ends-in gene targeting to generate deletions has been demonstrated in bacteria (Posfai et al. 1999). That method involved two steps that are very similar to the steps used to carry out allelic substitution in Drosophila (Rong et al. 2002). First, ends-in targeting produced an allele carrying duplications of the targeted region. Second, a DNA DSB was generated between the duplicated segments. Repair of this break by the cell is typically accomplished by a mechanism that reduces the duplication to a single copy. By proper design of the donor this reduction event can generate a specific deletion.
We wished to test whether this adaptation of ends-in targeting could be used to generate target gene deletions in Drosophila. We targeted three genes with appropriately designed ends-in donors and succeeded in generating deletions for all three. The results of these experiments are reported here.
MATERIALS AND METHODS
Donor constructs:
All oligonucleotide sequences are shown in Table 1. For subcloning, PCR reactions were all performed using the Herculase system (Stratagene, La Jolla, CA). Template DNA was genomic DNA prepared from the w1118 strain.
TABLE 1.
Sequences of oligonucleotides used
| Primer name | Primer sequence |
|---|---|
| I-SceI-Not1 | 5′-GGCCTAGGGATAACAGGGTAAT-3′ |
| I-SceI-Not2 | 5′-GGCCATTACCCTGTTATCCCTA-3′ |
| I-SceI-R1 | 5′-AATTGTAGGGATAACAGGGTAATC-3′ |
| I-SceI-R2 | 5′-AATTGATTACCCTGTTATCCCTAC-3′ |
| sir2-1 | 5′-TCGAGCTCGCGGCCGCTAGGCGCAACTACGCCAAGTACAATC-3′ |
| sir2-2 | 5′-TCGAGCTCATTTACCTTCATGGTCTCAGCCCCTTC-3′ |
| sir2-3 | 5′-GGTAAATGCGGCCGCTCGTCCAGCTC-3′ |
| sir2-4 | 5′-CCTCTAGATTTTTCCGAACCCAAGCGCGCGCACAC-3′ |
| sir2-5 | 5′-GCTCTAGAGAAGCTGCGAAATCGTAGCCGAG-3′ |
| sir2-6 | 5′-AACTCGAGCGGCCGCACCATTCCTACTTCCAGCTGGTCGCG-3′ |
| sir2-C1 | 5′-CCAAATGGGTGCGAAGCTGACG-3′ |
| sir2-C2 | 5′-GGCCCTCGGCTACGATTTCGCAG-3′ |
| p53-1 | 5′-CGAGCTCGCGGCCGCAACCTCATTTACGGACAATTATGTGAGC-3′ |
| p53-2 | 5′-TCCCCGCGGCAGGCGCATCGCAACGATGAGAC-3′ |
| p53-3 | 5′-CTGCGCGGCCGCCTGTGTCGCG-3′ |
| p53-4 | 5′-CTCTAGACAAAGAGATTAAGCTTTACTTACCGAAC-3′ |
| p53-5 | 5′-CTCTAGACAATCGCCGATGCCAATGTTTATCG-3′ |
| p53-6 | 5′-TGCGGCCGCCTTATTGATTGAGTGTCTGCACGGG-3′ |
| P53B-1 | 5′-TCCTCCGATTCACTGCTTACTCTTAAG-3′ |
| P53B-2 | 5′-TCGCGCTGCGTGGCGAATGGCAAAATC-3′ |
| CG5085-1 | 5′-GGGCCCTCAAGAAGCTTCGGTGCTAAAG-3′ |
| CG5085-2 | 5′-CCTTATCCATTGTGTCGTCTAGAATTC-3′ |
| CG5085-3 | 5′-TAGACCCTAAGATTAGTTAGTAACATCCG-3′ |
| CG5085-4 | 5′-CATTTATTCATTGAAGATTGGCTTTGGC-3′ |
| CG5085-C1 | 5′-GCCCCAGGCTAGTCTAAATAG-3′ |
| CG5085-C2 | 5′-GAAAGAAAGCTCGCGCTATTAG-3′ |
Sir2 construct:
This construct is designed to delete the entire coding region of Sir2 (2.7 kb), from the start codon to a point 67 bp downstream of the stop codon. The PCR product from primers sir2-3 and sir2-4 was ligated into the pBluescript vector (Stratagene) using the XbaI site and the NotI site. An I-SceI site was then inserted at the NotI site by ligating in the double-stranded oligonucleotide produced by annealing of the oligonucleotides I-SceI-Not1 and I-SceI-Not2. The PCR product of primers sir2-1 and sir2-2 was then added to the plasmid at the SacI site. A clone carrying the insert in the correct orientation was selected. The genomic DNA fragment between primers sir2-5 and sir2-6 was then amplified and ligated into the plasmid using the XbaI and XhoI sites. The assembled construct was released from that vector using the NotI sites and transferred into the same site of pTV2 (Rong et al. 2002).
Sirt2 construct:
This construct is designed to delete virtually the entire gene, from the fourth codon through the stop codon (1.2 kb). The PCR product from primers CG5085-3 and CG5085-4 was blunt-end ligated into the Klenow-filled EcoRI site of pBluescript. A clone with the insert oriented to place the CG5085-4 primer end next to the SpeI vector site was selected. This plasmid was digested with SpeI and NcoI, removing a small fragment of DNA from the plasmid. These ends were filled with Klenow and then ligated to make an EcoRI site unique in the plasmid. Oligonucleotides I-SceI-R1 and I-SceI-R2 were annealed to generate an I-SceI site and then ligated into the unique EcoRI site in the plasmid. The PCR product of primers CG5085-1 and CG5085-2 was treated with Klenow and kinase and then blunt-end ligated into the plasmid at the EcoRV site, and a plasmid with the desired orientation of the insertion was selected. This construct was removed by digestion with NotI and Acc65I and then ligated into the same sites of pTV2.
p53 construct:
This construct is designed to produce a deletion starting 37 bp upstream of the transcription start point to a point within the last intron (3.5 kb). The last exon still remains after deletion, containing seven codons including a stop codon. The PCR product from primers p53-3 and p53-4 was ligated into pBluescript using the XbaI site and the NotI site. An I-SceI site was then put into the construct by annealing the oligonucleotides I-SceI-Not1 and I-SceI-Not2 and ligating this into the NotI site, destroying the NotI site in the process. The PCR product of primers p53-1 and p53-2 was then added into the plasmid using the SacI and SacII sites. A new NotI site was also carried into the construct by the primer p53-1. The PCR product produced by the primers p53-5 and p53-6 was then cloned into the plasmid using the ApaI and XbaI sites. Another NotI site was introduced with the primer p53-6. The assembled construct was transferred to the NotI site of pTV2 using the flanking NotI sites.
Transformation:
The donor P-element constructs were transformed into flies carrying the w1118 null mutation using standard P-element-mediated transformation (Rubin and Spradling 1983).
Fly stocks and heat shocks:
Fly strains used in this work are described at http://flybase.bio.indiana.edu. Heat shocks were performed in a circulating water bath as previously described (Golic and Lindquist 1989).
The targeting screen:
Targeting crosses were carried out by the rapid targeting method described previously (Rong and Golic 2001). The lines that were nonmosaic in the presence of a constitutive FLP gene, indicating that whs was no longer flanked by FRTs, were kept as putative targeting events. The whs marker was then mapped with respect to the target chromosome. Diagnostic PCR was performed as an initial molecular screen. The structures of the targeted alleles were ultimately analyzed by Southern blotting.
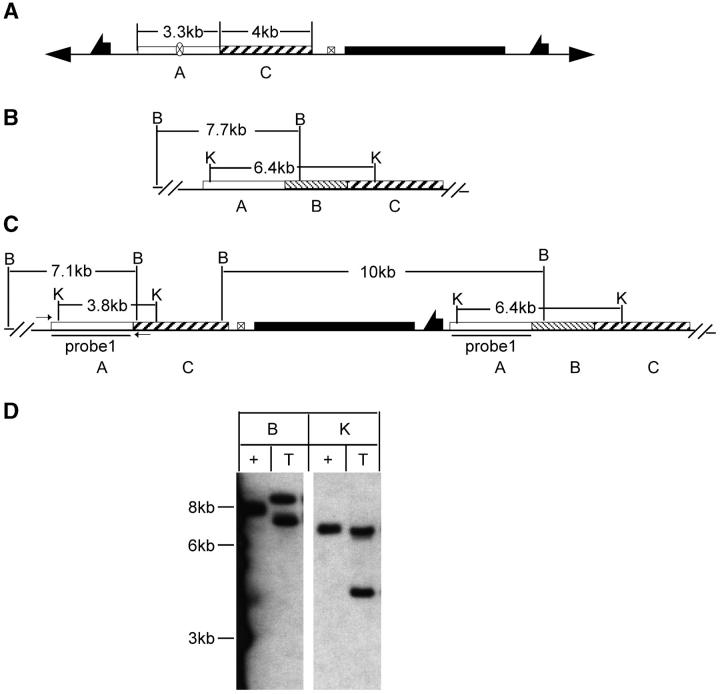
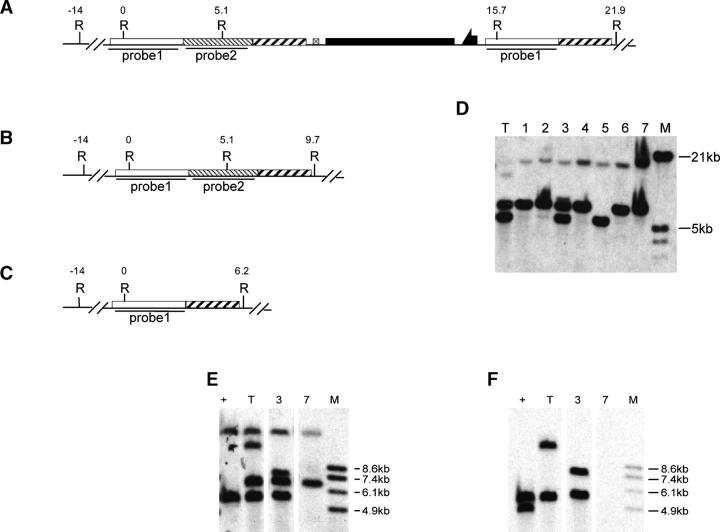
For Sir2, diagnostic PCR was performed on potential target lines using a primer outside of the A region and the other within the C region (Figure 2C). This pair of primers amplifies the 3.3-kb A region in a targeted allele. The endogenous locus gives a 5.9-kb fragment that includes the A and B regions. Nontargeted events should give only the endogenous 5.9-kb product.
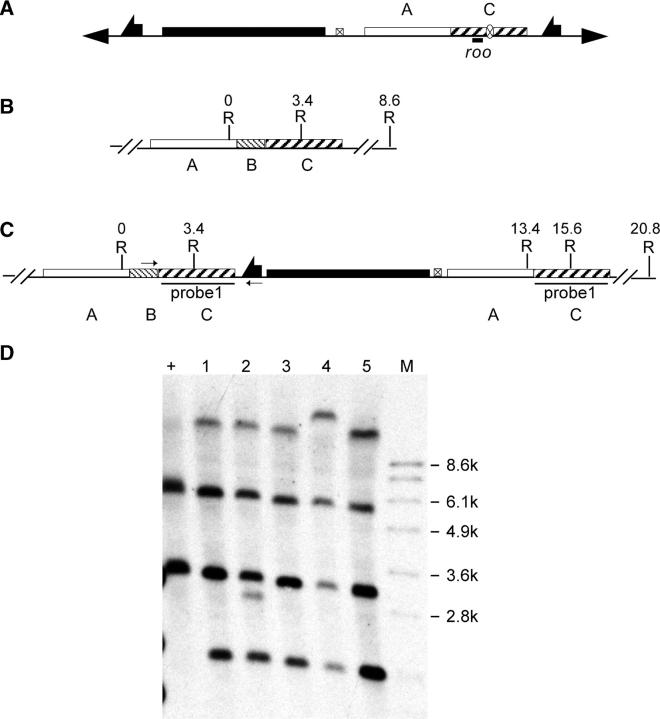
Figure 2.—
Molecular analysis of Sir2 targeting. (A) The donor construct. Restriction sites and fragment sizes are indicated for the wild-type allele (B) and the expected targeted allele (C). Small arrows in C indicate the primers used for diagnostic PCR. Southern blot results are shown in D, with marker sizes indicated at the left. The region used to probe the blot is indicated in C. Plus indicates genomic DNA from the wild type; T, DNA from flies carrying a targeted allele; B, BamHI; K, KpnI.
For Sirt2, diagnostic PCR was initially used to identify targeting events with a strategy similar to that used for Sir2. Only events that showed the expected product in the PCR were selected for Southern blot analysis.
Reduction:
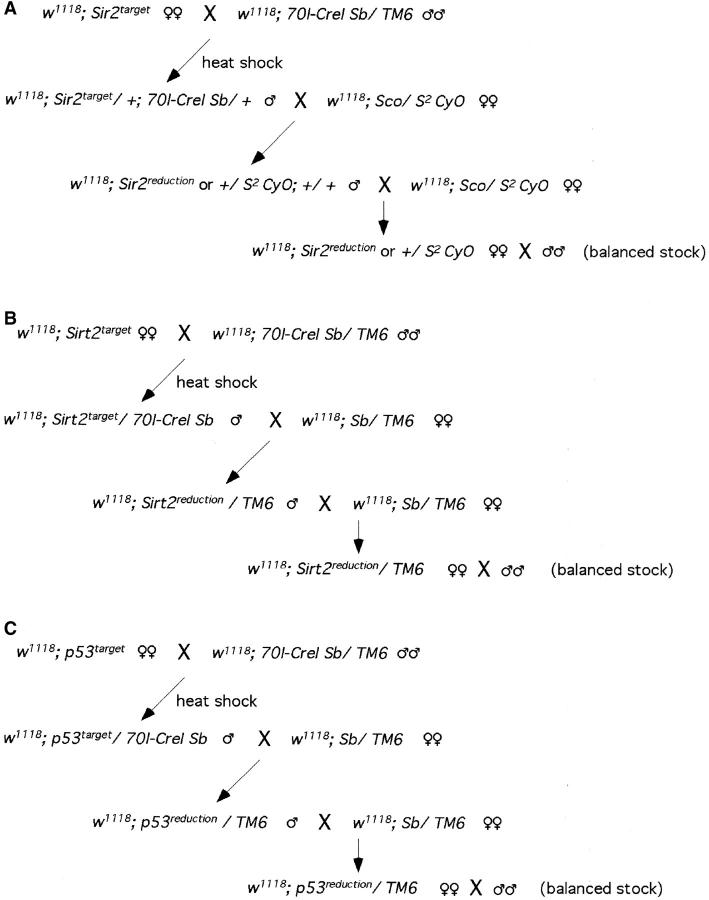
The procedure to reduce the tandem duplications to generate deletion alleles was similar to the reduction procedure described previously (Rong et al. 2002). Flies with selected targeted alleles were crossed to w1118; 70I-CreI Sb/TM6 flies and their progeny were heat-shocked at 36° for 1 hr at 0–2 days of age. Males carrying a targeted allele and 70I-CreI were then individually crossed to w1118 flies carrying an appropriate balancer (S2 CyO for Sir2, TM6 for Sirt2 and p53). Single white-eyed sons with the balancer were isolated from each cross. By backcrossing to the balancer strain, homozygous lines for reduction alleles were generated. DNA from homozygous flies was used for characterization by PCR and/or Southern blot analysis.
Diagnostic PCR and Southern blotting:
All diagnostic PCR reactions used genomic DNA prepared from homozygous flies (males or virgin females). The Ex Taq system (Fisher) was used. PCR conditions were designed according to manufacturers' recommendation.
For Southern blots, genomic DNA was prepared from 15–30 homozygous flies (males or virgin females) for each sample. After overnight digestion by the appropriate restriction enzyme, the DNA was separated by agarose gel electrophoresis and transferred to a nylon membrane. The membrane was probed with Dig-labeled probe (Roche). Conjugated anti-Dig antibody was used to detect hybridization, followed by a chemiluminescence reaction and exposure to X-ray film. In the presentation of results, in some cases uninformative lanes were deleted.
RESULTS
General method:
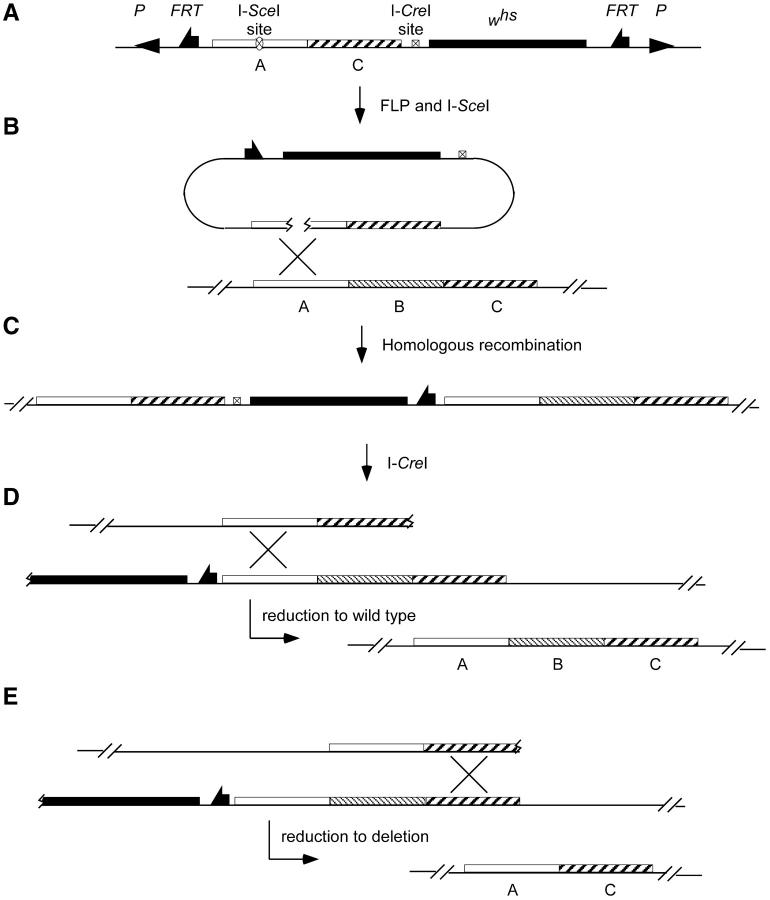
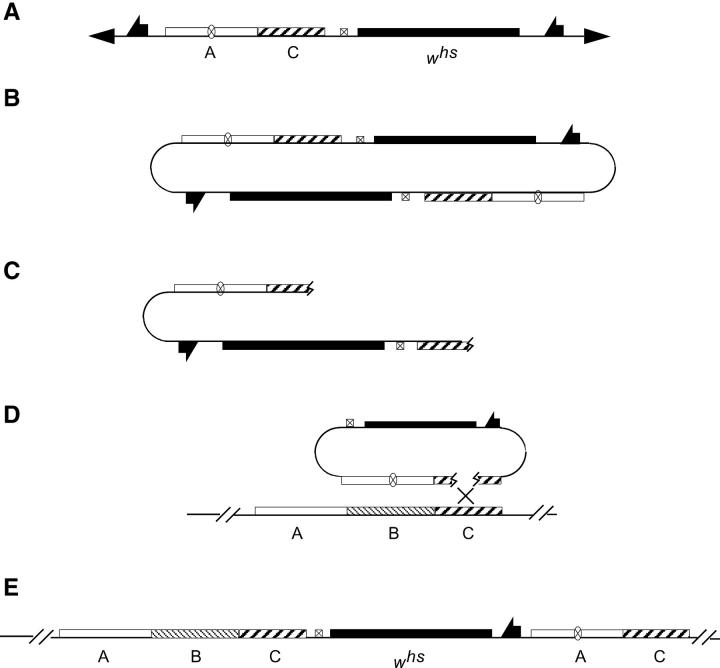
To design an ends-in targeting to generate a deletion, the desired deletion junction is first created in vitro in the donor construct and then it is recapitulated in vivo after targeting. Thus, the first step is to build a donor construct that carries DNA regions from the left and right of the gene that is to be deleted. In Figure 1, these flanking regions are indicated as A and C, and the region to be deleted is indicated as B. The junction of A-C regions in the donor construct corresponds to the breakpoints of the desired deletion. Targeted homologous recombination is directed to one of these flanking regions by placement of the I-SceI recognition sequence that will be used to generate a double-strand break. In Figure 1, the I-SceI site is placed in region A.
Figure 1.—
The scheme for producing deletions. (A) The donor P element carrying regions from the left and the right of the gene to be deleted. (B) FLP and I-SceI excise the donor DNA from the chromosome and generate a double-strand break within its A region, stimulating recombination with the endogenous A region. (C) The expected structure of the allele produced by recombination between the donor DNA and the target locus. Then, in a second step, I-CreI is used to produce a double-strand break to the left of the whs marker gene. (D) If the chromosomal break is repaired by recombination within region A, the whs marker will be lost and a wild-type allele is produced. (E) If repair occurs by recombination within region C, the result will be a deletion of region B.
After transformation of the donor, targeting is carried out in the manner that has been described (Rong and Golic 2000, 2001). Expression of FLP recombinase and I-SceI endonuclease is used to excise the donor DNA from its chromosomal location and generate a double-strand break that stimulates recombination in region A. Homologous recombination between donor and target is expected to generate a targeted allele with the structure shown in Figure 1C. The targeted allele has a duplication of regions A and C flanking the whs gene (a hypomorphic allele of the white gene), while region B is present as a single copy to the right side of the whs gene. A recognition site for the rare cutting endonuclease I-CreI is included within the targeting vector and lies next to the whs gene. In a second step, I-CreI expression can generate a double-strand break at the I-CreI site in a targeted allele. Such a double-strand break lying between repeated sequences is frequently repaired by single-strand annealing, causing recombination between flanking homologous regions (Rong and Golic 2000; Preston et al. 2002; Rong et al. 2002; Lankenau et al. 2003). The duplication is reduced to a single copy, with the DNA segment between the repeated sequences removed. These reduction events can be readily detected by loss of the whs gene. When reduction occurs by recombination within region A, a wild-type allele will be recovered (Figure 1D). However, recombination may also occur within region C to generate a deletion of region B (Figure 1E). Molecular strategies such as PCR or Southern blotting can be used to detect deletion alleles.
Sir2 targeting and deletion:
Three independent transformants of the Sir2 (CG5216) deletion donor were used for targeting (Table 2). We recovered 20 putative targeting events from 184 vials. In 10 of those lines the whs gene mapped to chromosome 2, where the endogenous Sir2 gene resides; the remaining 10 were discarded. One was confirmed as a targeting event by both PCR (primers as in Figure 2C) and Southern blot analysis (Figure 2D) and was used later in the reduction step to generate the deletion. To determine whether the remaining 9 potential targeting events on chromosome 2 were targeted to the Sir2 locus, they were genetically mapped with respect to the first verified targeted allele. Females that were heterozygous for the verified allele and each remaining allele (all carried the whs gene) were testcrossed to w1118 males to score meiotic recombination. Three of nine testcrosses produced white recombinants, indicating that the whs gene in these lines was integrated at a locus other than Sir2; they were not targeted. The other six testcrosses gave only white+ progeny, indicating that their whs gene is located at Sir2 and they are, in all probability, targeted events. These were also tested by diagnostic PCR (primers shown as arrows in Figure 2C; data not shown). Four of the seven targeted alleles, including the previously verified allele, produced the expected PCR product. Although the other three lines were likely produced by recombination of the donor with the target locus, they probably have more complex arrangements than the predicted structure diagrammed in Figure 2C.
TABLE 2.
Targeting frequency forSir2
Donor, the designation for the three transformant lines used in this study; location, chromosomal location for each of the P elements; vials, number of vials screened for each P-element insertion; T, number of targeted events recovered from each donor; NT, number of nontargeted events recovered; PCR+, number of events giving positive diagnostic PCR results (see text for details).
Targeting was confirmed by recombination except for one line (see text for details).
In these crosses only one-half of the tested females carried the donor element; we have corrected by multiplying the number of vials tested by 0.5.
This allele was used for reduction.
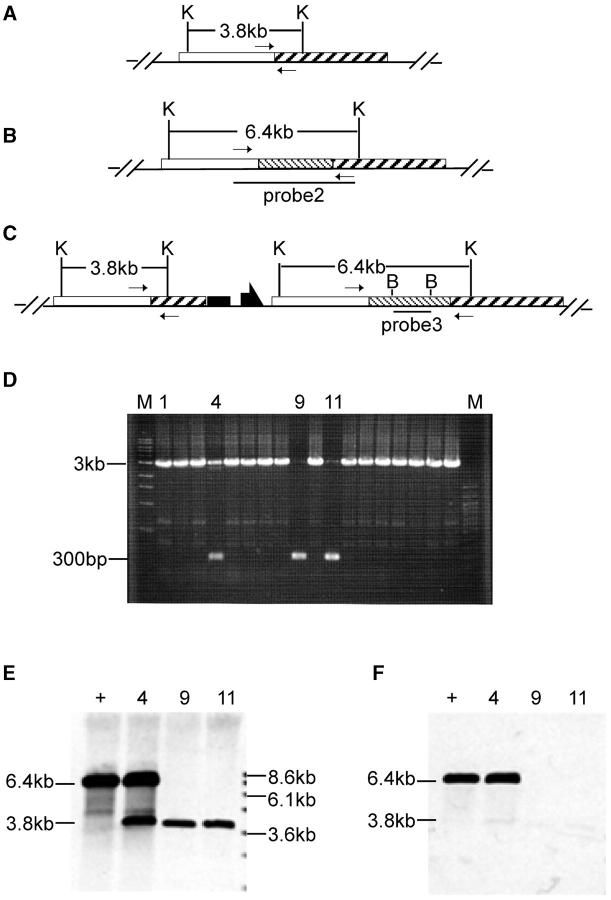
To recover a Sir2 deletion, flies with the targeted allele were crossed to w1118; 70I-CreI Sb/TM6. The progeny of this cross were heat-shocked at age 0–2 days (36°, 1 hr), to induce I-CreI expression. Stubble (Sb) sons carrying the targeted allele were crossed to w1118; Sco/S2 CyO females individually. From each cross, a single w Cy Sb+ son was picked, and balanced or homozygous stocks were established for chromosome 2 (Figure 3A). Because less than half of the chromosomes collected in this scheme will carry a reduced version of the targeted allele (the father's unmarked homolog will be found in the majority of white progeny), we used a PCR test for rapid initial screening. Twenty-two independent homozygous lines were obtained from the targeted allele of Sir2. Diagnostic PCR was performed on DNA from homozygous flies using the primers Sir2-C1 and Sir2-C2 flanking the A-C junction (as indicated in Figure 4, A, B, and C). The desired deletion allele of Sir2 should generate only a PCR product of 300 bp (Figure 4A). The wild-type allele should produce a PCR product of 3 kb (Figure 4B). Most lines exhibited the 3-kb product (Figure 4D), but three lines produced the 300-bp band. Among these, one line gave both 300-bp and 3-kb products. This line apparently resulted from a deletion of whs sequence only, which can result from exonucleolytic enlargement of the I-CreI-generated cut followed by nonhomologous end joining (NHEJ; as indicated in Figure 4C). Two lines produced only the 300-bp product and appear to be the desired deletion. Although lanes 9 and 11 of the gel in Figure 4D show a small amount of the 3-kb band, further PCR tests showed that this resulted from contamination (not shown). To more completely characterize these three lines we performed genomic Southern blotting (Figure 4E). We first used probe 2 (produced by the primers Sir2-C1 and Sir2-C2 from wild-type allele; Figure 4B), which detects the Sir2 gene and flanking regions. The blot was then stripped and rehybridized with probe 3 (a 1.7-kb BamHI fragment from the center of probe 2; Figure 4C) that recognizes only the Sir2 region. The results confirmed the conclusions from the PCR test. The 3.8-kb breakpoint fragment was present in the putative deletion lines (lanes 9 and 11) but Sir2 was absent. We conclude that these lines represent the expected deletion of Sir2.
Figure 3.—
The crossing schemes for I-CreI-mediated reduction at (A) Sir2, (B) Sirt2, and (C) p53.
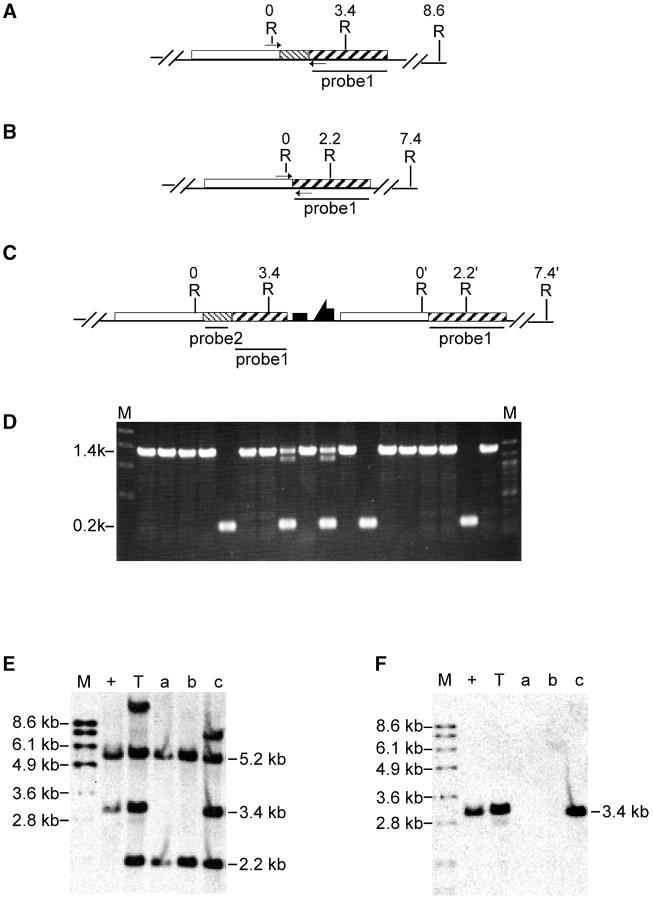
Figure 4.—
Molecular analysis of Sir2 reduction. Restriction sites are shown for (A) the deletion allele, (B) the wild-type allele, and (C) a targeted allele in which part of whs has been deleted by NHEJ. Small arrows indicate the locations of primers used for the diagnostic PCR shown in D. In D, M indicates molecular weight markers with sizes indicated at the left and specific lanes are numbered for reference in E and F. (E and F) Southern blotting of KpnI-digested DNA from wild type (+) and from the reduction alleles represented in lanes 4, 9, and 11. E was probed with probe 2 (indicated in B) and then stripped and reprobed with probe 3 (shown in F). K, KpnI; B, BamHI.
Sirt2 targeting and deletion:
We used five donor lines for targeting of Sirt2 (CG5085) and recovered 42 potential targeting events (Table 3). The whs marker of the potential targeting events was mapped. Surprisingly, only 14 of 42 lines mapped to chromosome 3, where the target gene is located. In previous gene targeting performed in our lab, we have not observed such a high frequency of nontargeted events (Rong et al. 2002). To find an explanation for the high frequency of nontargeted events, we carried out a BLAST search, comparing the sequence of the donor DNA with the complete genome. A 429-bp sequence in the donor matches part of the transposon roo (Figure 5A). This sequence is found in all the major chromosome arms. The presence of this moderately repetitive DNA in the donor might account for the higher frequency of nontargeted events, possibly by directing recombination to other sites in the genome.
TABLE 3.
Targeting frequency forSirt2
| Donor | Location | Vials | FLP nonmosaic |
On 3 | PCR+ |
|---|---|---|---|---|---|
| 1 | 2 | 118 | 8 | 1 | 0 |
| 2 | 2 | 131 | 5 | 3 | 1a |
| 3 | 2 | 54 | 20 | 2 | 1 |
| 4 | 3 | 63 | 4 | 3 | 1 |
| 5 | 3 | 68 | 5 | 5 | 2 |
Donor, location, and vials have the same meaning as in Table 2. FLP nonmosaic, putative targeted events; see text for details. On 3 refers to the events from the previous column that mapped to chromosome 3. PCR+, diagnostic PCR with positive results. See text for details.
This line was used for reduction step.
Figure 5.—
Molecular analysis of Sirt2 targeting. (A) The donor P-element construct, with the location of the roo element indicated. Restriction sites and coordinates, in kilobases, are given for (B) the wild-type allele and (C) the expected targeted allele. In C the small arrows indicate the locations of primers used for diagnostic PCR. The region used as a probe for Southern blotting is indicated. (D) Southern blotting of EcoRI-digested genomic DNA. “+” indicates wild type; lanes 1–5 indicate targeted alleles. Lanes 1, 3, and 5 represent the expected event. R, EcoRI.
An even more likely explanation might be that these nontargeted events simply represent cases in which the donor was cut by I-SceI, a cut end was degraded so that one FRT was removed, and then the ends were rejoined. The loss of one FRT would prevent subsequent excision by FLP and would allow the element to pass the initial screen, which simply looks for a lack of white mosaicism in the presence of FLP. The mapping of the nontargeted events is consistent with this explanation. The location of whs was determined for 26 of the potential targeting events obtained from donor elements inserted on chromosome 2. At least 15 of these 26 events were still located on chromosome 2 and 4 more were most likely on 2, because they were not on X or 3. Thus, the majority of events had not moved from the starting chromosome, as predicted by the hypothesis that most of the nontargeted events simply represent loss of an FRT at the original location. In cases such as this, it might be advantageous to initially screen for a change in linkage of the donor, rather than use the mosaic test we employed.
These explanations could be tested by examining the insertion sites of the nontargeted events, but since our main concern here was generating deletions of Sirt2, we did not carry out such an examination. In either case, the results are probably attributable to the sequence of this donor construct, since we have not observed such a high frequency of nontargeted events when targeting other genes.
We tested all the putative targeted alleles on chromosome 3 by PCR with primers that would detect the insertion of an FRT near region B (arrows shown in Figure 5C). PCR with these two primers is expected to produce a band of ∼3.5 kb with the targeted allele template. Nontargeted events and wild-type alleles should give no amplification. Five of the 14 lines produced the PCR product indicative of a targeted allele (data not shown). Southern blot analysis then identified three lines with the expected structure (Figure 5D, lanes 1, 3, and 5). One of these lines was chosen to generate the deletion.
After I-CreI-mediated reduction of the targeted allele, 42 white lines were recovered. All were tested by PCR using DNA from homozygotes as template (Figure 6, A, B, and C). The wild-type allele should generate a 1.4-kb band, which was seen in most lines; 8 lines produced only the 200-bp band that is diagnostic for the deletion (partial results shown in Figure 6D). These lines were further analyzed by Southern blotting, and the expected deletion of Sirt2 was confirmed in all 8 (examples as in Figure 6, E and F, lanes a and b). In addition, two reduction lines produced both the wild type and the deletion PCR products and appear to result from NHEJ repair of the I-CreI double-strand break, accompanied by partial loss of whs sequence. This conclusion was also confirmed by Southern blotting: when the top 10-kb band in the targeted allele contains the whs gene, the band in the NHEJ event is reduced in size (an example is shown in Figure 6E, lane c compared to lane T).
Figure 6.—
Molecular analysis of Sirt2 reduction. The expected structures of (A) the wild-type allele, (B) the deletion allele, and (C) an allele produced by NHEJ repair of the I-CreI break are indicated. EcoRI sites and their coordinates, in kilobases, are indicated. Small arrows indicate locations of primers used for diagnostic PCR. The regions used as probes for Southern blotting are also indicated. (D) Diagnostic PCR. (E and F) Southern blot results of EcoRI-digested DNA for selected alleles probed with probe 1 (E) and stripped and reprobed with probe 2 (F). M, molecular weight markers; +, wild-type allele; T, targeted allele; a and b, the expected deletion alleles; c, NHEJ reduction allele.
p53 targeting:
The targeting of p53 was not straightforward because the expected initial product of targeting was not recovered. Nonetheless, after extensive characterization of one targeted allele, it was possible to generate the desired deletion. Six transformants of the donor were used (Table 4). Five potential targeting events were recovered. These lines were subjected to diagnostic PCR (arrows shown in Figure 7B). The predicted targeting event should give a 4-kb product. However, none of the five lines produced the expected band (not shown). Moreover the five lines did not show the same eye color, suggesting differences in the chromosomal environment of the whs gene in the different lines. Indeed, one of the lines was mapped to the X chromosome, but two did map to chromosome 3, where the p53 gene is located. These lines appear to be the result of integration of the donor at the p53 locus (see below).
TABLE 4.
Targeting frequency forp53
| Donor | Location | Vials | FLP nonmosaic |
T | NT |
|---|---|---|---|---|---|
| 1 | X | 14 | 0 | 0 | 0 |
| 2 | 3 | 14a | 0 | 0 | 0 |
| 3 | 3 | 14a | 1 | 0 | 1 |
| 4 | 3 | 13a | 2 | 2b | 0 |
| 5 | 2 | 10a | 1 | 0 | 1 |
| 6 | 3 | 14a | 1 | 0 | 1 |
Donor, location, vials, and FLP nonmosaic have the same meaning as in Table 3. T, targeted events; NT, nontargeting events.
Only half of the females counted carried the donor construct, and the number of vials was adjusted by 0.5.
Both lines gave positive mosaicism upon I-CreI induction (see text for details).
Figure 7.—
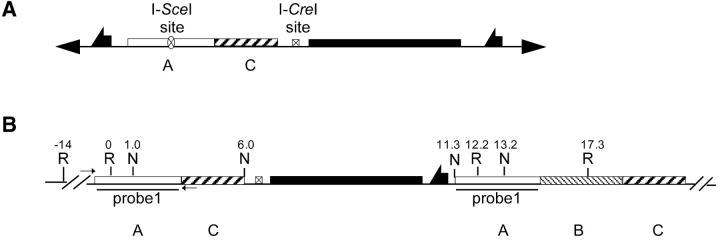
Expected results of p53 targeting. (A) The donor P element. (B) The expected structure of the targeted allele. Restriction sites and coordinates, in kilobases, are given. Small arrows indicate sites of primers used for diagnostic PCR. The region used as a probe for Southern blotting (Figure 8) is shown. R, EcoRI; N, NotI. The EcoRI at the 0 position is a combination of three sites very close to each other.
To differentiate those lines that represent integration at the target locus from nontargeted events we used an I-CreI-based genetic test. In flies carrying a targeted allele, the I-CreI expression causes a high degree of white mosaicism in the eye because DSB repair frequently deletes the whs gene. On the other hand, if the extrachromosomal donor inserts at a nonhomologous region of the genome, the whs marker is not flanked by duplicated regions. Degradation of the cut ends followed by NHEJ is the expected mechanism for loss of whs in this case, and the frequency of loss is much lower (Rong and Golic 2003). Accordingly, the frequency of white mosaicism induced by I-CreI may be used to distinguish targeted and nontargeted events, with targeted alleles expected to show a much greater degree of mosaicism.
Flies carrying a potential targeted allele and the 70I-CreI gene were heat-shocked at age 0–2 days (36°, 1 hr), and the flies that eclosed were scored for white mosaicism in the eye. All flies carried the w1118 null allele on their X chromosomes, so the whs insertion was the only source of w+ product. Although all five lines exhibited significant degrees of mosaicism, two lines stood out by showing more obvious and extensive white patches than the other three lines. These two lines were likely to carry targeted alleles. The molecular results presented below ultimately confirm that at least one of these alleles resulted from targeted integration of the donor.
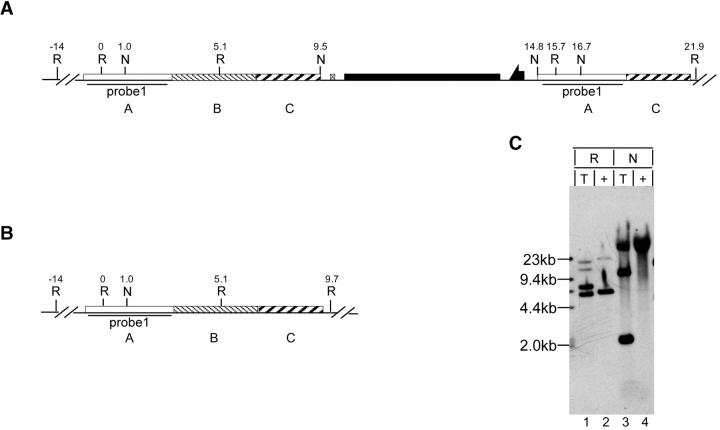
Southern blot analysis of these two lines revealed that neither had the predicted targeting structure shown in Figure 7B. The structure of the allele that was used in the subsequent reduction step was examined in detail (Figure 8). We deduced that the targeted p53 allele has the structure diagrammed in Figure 8A, with the A-B-C segment lying to the left of the whs gene and the A-C segment lying to its right. This is the reverse of what was expected. Using probe 1 (Figure 8C) to examine EcoRI-digested genomic DNA, the wild-type allele should and does give a prominent band at 5.1 kb and a band of lesser intensity at 14 kb. If targeting had occurred as expected (Figure 7B) we should see the same prominent band of 5.1 kb and the lesser band of 14 kb, with another strong band at 12 kb. However, if targeting produced the structure with A-B-C on the left (Figure 8A), then we should see prominent bands of 5.1 and 6.2 kb and lesser bands of 10.6 and 14 kb. This appears to match the pattern shown by the EcoRI digest of the targeted allele. NotI digestion of the wild-type allele generates two large bands of undetermined sizes. With the targeted alleles, NotI should generate additional 5.0- and 1.9-kb bands (plus the two large bands of undetermined size) if the structure has A-C on the left (Figure 7B), but added bands of 8.5 and 1.9 kb (plus the large bands) if the structure has A-B-C on the left (Figure 8A). The results closely match the latter, with one band of ∼9 kb and another of 2 kb.
Figure 8.—
Molecular analysis of p53 targeting. (A) The structure that was deduced for the targeted allele. (B) The wild-type allele. Restriction sites and coordinates, in kilobases, are indicated, along with the region used as a probe for Southern blotting. (C) Southern blot of genomic DNA digested with EcoRI or NotI. T, targeted allele; +, wild-type allele; R, EcoRI; N, NotI.
Although the structure of the allele is not as expected, results from many labs have shown that it is not rare for targeted alleles to have unexpected arrangements of donor components (Rong and Golic 2000; Dolezal et al. 2003; Elmore et al. 2003; Gong and Golic 2003; Lankenau et al. 2003). In any case, the targeted allele was still suitable for generating a deletion of p53.
Seven I-CreI-mediated reduction lines were obtained from this targeted allele. All were viable as homozygotes. They were examined by Southern blotting after EcoRI digestion using region A as probe (probe 1, Figure 9, A, B, and C). Five of these seven reduction alleles exhibited the 6.2-kb band expected for a p53 deletion (Figure 9D, lanes 1, 2, 4, 6, and 7); one appeared to be wild type (lane 5), and one appeared to be the result of NHEJ that deleted at least part of the whs marker, but left both flanking regions (lane 3). The wild-type allele, the targeted allele, and two of the reduction alleles were reexamined in a second blot, where the filter was first hybridized with probe 1 (Figure 9E) and then stripped and rehybridized with probe 2, representing p53 (Figure 9F). These results show, first, that the targeted allele clearly alters the endogenous p53 allele, confirming that the donor had integrated at p53. Second, it confirms that the event represented by lane 3 is indeed a partial deletion of the region carrying whs. And, finally, these results show that the allele represented by lane 7 is a deletion because it lacks any p53 hybridization signal.
Figure 9.—
Molecular analysis of p53 reduction alleles. The structures of (A) targeted, (B) wild-type, and (C) deletion alleles are indicated. (D) Southern blot of EcoRI-digested genomic DNA probed with probe 1. T, targeted allele; 1–7, reduction alleles; M, molecular weight markers. (E) Selected alleles, corresponding to the lane numbers in D, were examined in a second Southern blot of EcoRI-digested genomic DNA using probe 1. The blot was then stripped and reprobed with probe 2 (shown in F).
DISCUSSION
Previous work using ends-in targeting showed that point mutations could be introduced to specifically mutate a target gene (Rong and Golic 2001; Rong et al. 2002; Seum et al. 2002; Dolezal et al. 2003; Egli et al. 2003; Elmore et al. 2003; Lankenau et al. 2003; Liu and Kubli 2003). However, even when nonsense mutations are introduced there is some potential for the production of a partially functional peptide or translational read-through to produce a full-length protein. When a null allele is desired a deletion of the gene would be preferable. The work presented here demonstrates that ends-in targeting can be adapted to make such precise deletions.
The desirability of this type of targeting depends on both the ease of donor construction and the efficiency of targeting and producing the deletion in vivo. The ends-in deletion donor is generally no more difficult to construct than a donor for introducing a point mutation. For deletions, the left and right flanks must be cloned, and then an I-SceI site must be introduced. To introduce a point mutation the gene must be cloned, the desired point mutation must be generated within that clone, and an I-SceI site must be inserted. The effort to generate either type of ends-in donor is probably about equivalent. Moreover, targeting with these deletion donors showed efficiencies ranging from one event per 26 vials (for Sir2) to one event per 87 vials, which is well within the range previously reported for the introduction of point mutations (Rong et al. 2002). Reduction to produce a deletion allele was also quite effective. In the case of Sir2, after exposure to I-CreI, ∼10% of the chromosomes tested carried the desired deletion. This would no doubt have been higher had we marked the chromosomes so that the targeted allele could have been distinguished from its homolog. With Sirt2 and p53, where the chromosomes were marked, the frequencies of reduction to deletion were ∼20% and ∼70%, respectively. Thus, each step of the procedure seems to be as effective in producing deletion alleles as it was for the introduction of point mutations.
In the accompanying article (Gong and Golic 2004, this issue), it is reported that ends-out targeting can be used to make specific gene deletions in Drosophila. The use of ends-out targeting to produce deletions may be more straightforward than the method we report here. The cloning is slightly easier because the I-SceI sites are already contained within the vector for ends-out targeting, and generating the deletion in vivo requires only one step, not the two needed for ends-in deletions. However, if a very precise deletion is desired, ends-in targeting is preferable. The ends-out method replaces the target gene with a whs marker. Although Cre recombinase (Siegal and Hartl 1996) may subsequently be used to remove the marker, a lox site remains at the target site. The ends-in procedure can generate a clean, precise deletion, with no exogenous sequence remaining at the breakpoints. If there is any concern that the locus to be deleted is haplo-insufficient for viability (which is rare; Lindsley et al. 1972), then this ends-in method may be preferable to the generation of deletions by ends-out targeting. In the simplest case, if the deletion of a particular gene caused lethality it would appear as a failure to achieve targeting by the ends-out deletion method. With the ends-in method the targeted allele could be recovered because it should still carry a functional copy of the gene. However, at the reduction step, recovery of the wild-type arrangement only and never the deletion would provide an indication of haplo-insufficiency.
We produced only small deletions in this work, ranging from 1.2 to 3.5 kb. It may be possible to generate much larger deletions by this method: in bacteria, deletions of up to 62 kb have been produced (Posfai et al. 1999). Gong and Golic produced a large deletion (∼47 kb) by ends-out targeting, so by one of these deletion methods at least, it is possible to generate large deletions.
In the reports of the two-step method for allelic substitution, it was concluded that single-strand annealing was the likely mechanism for reduction of the tandem duplication following the I-CreI-induced break (Rong and Golic 2001; Rong et al. 2002; Lankenau et al. 2003). The results obtained here tend to confirm that conclusion. When a DSB is generated, a 5′-3′ single-strand exonuclease proceeds bidirectionally from the break (Maryon and Carroll 1989). For the targeted allele drawn in Figure 1, the exonuclease must pass through the whs marker gene to the right before reaching the A region. The A region to the left of the break may also become single stranded at about the same time because it lies at a similar distance from the I-CreI site. (This will, of course, depend on the size of the B and C regions.) Annealing of the A regions and reduction to the wild-type A-B-C structure is likely to be favored. On the other hand, if the A-B-C structure lies next to the I-CreI site and the A-C structure is on the other side of whs, then reduction by annealing of single-stranded A regions is likely to favor the deletion. We observed exactly this with p53, where the arrangement of the duplication was A-B-C adjacent to the I-CreI site and A-C to the other side of whs. Although with p53 the production of this structure was unintentional, this result could likely be achieved more directly by placing the I-SceI site of the donor within the C region, rather than in the A region as was shown in Figure 1.
One unexpected result was our finding that the majority of events recovered as potential targeting events were not, in fact, targeted. In previous cases from our lab, a majority of potential targeting events were found to be actual targeting events. It is likely that at least part of this frequent nontargeting can be attributed to the specific sequences used in the donors. For instance, the Sirt2 donor carried some moderately repetitive DNA that may have influenced this outcome. Additionally, other investigators and our lab have reported the recovery of more nontargeted than targeted events from certain donor insertion sites (Rong et al. 2002; Seum et al. 2002; Dolezal et al. 2003; Gong and Golic 2004, this issue). Whether our results owe to peculiarities of the donor sequences used or to the A-C donor configuration is unknown.
The initial screening for gene targeting can use movement of the donor from an unlinked location onto the target chromosome (Rong and Golic 2000) or lack of FLP-induced white mosaicism (Rong and Golic 2001). To avoid carrying out large numbers of Southern blots, additional screens were used to further reduce the number of potential candidates. PCR tests successfully identified targeted alleles of Sir2 and Sirt2 (as in Figures 2C and 5C). This was followed by Southern blotting to fully characterize the structure of targeted alleles. Another approach was to examine the frequency of white mosaicism produced by exposure to I-CreI. Alleles with duplicated regions flanking the I-CreI site and whs are expected to produce frequent mosaicism following induction of a DSB (Rong and Golic 2003). Targeted alleles are expected to have such flanking duplications, while nontargeted insertions likely would not and would show less frequent mosaicism. This screen was successful at helping to identify targeted alleles of p53.
The structure of the p53 allele was not as predicted (Figure 8A) and suggests an unusual targeting process had occurred. Although we expected a structure of A-C-whs-A-B-C (Figure 7B), the targeted allele actually had the structure of A-B-C-whs-A-C (Figure 8A). One possible explanation for this targeting event is that two donor molecules (possibly from two sister chromatids) were joined together at an FRT before targeting (Figure 10B). Targeting events resulting from donor dimers have been observed in many cases (Rong and Golic 2000; Dolezal et al. 2003; Gong and Golic 2003; Lankenau et al. 2003). An I-SceI cut at one of the recognition sites was followed by extensive degradation of the ends, exposing the sequence within region C for targeting (Figure 10D). This would generate the recovered allele. A similar event has been proposed to have occurred during the targeting of Nap1 gene (Lankenau et al. 2003). In addition, the second I-SceI site must have been cut and repaired, probably by copying from the sister or homolog, since the NotI sites in both copies of region A are intact, indicating that the I-SceI site has been removed (Figure 8C). The repair event may have occurred before or after targeting.
Figure 10.—
Proposed mechanism for generation of the p53 targeted allele. See text for details.
All the deletion alleles that we produced are viable in homozygous condition. With respect to Sir2, contrary to the report of Rosenberg and Parkhurst (2002), our findings confirm the reports of others (Newman et al. 2002; Astrom et al. 2003) that Sir2 is not a vital gene. For p53, two other null alleles generated by ends-in targeting carrying a point mutation or truncated copies are homozygous viable as well (Rong et al. 2002; Sogame et al. 2003). We also generated the first null allele for the gene Sirt2, and it is also not vital.
This method can be applied to target vital genes as well. Both targeting and reduction steps can be achieved in heterozygous animals, and the deletion that is produced can be maintained in a balanced stock. However, PCR screens to identify the deletion alleles are less useful in such cases. The PCR test we used relied on the absence of the wild-type band in reduction allele homozygotes. In heterozygotes for the deletion/+ or a NHEJ allele/+, both bands are expected to appear. Still, the PCR test can be used to eliminate cases of reduction to wild type, and careful application of Southern blotting can confirm the generation of the expected deletion. Our results show that the method for generating deletions in bacteria that was demonstrated by Posfai et al. (1999) is also useful in Drosophila and probably useful for eukaryotes in general.
Acknowledgments
We thank Scott Hawley and the Stowers Institute for Medical Research for providing support and facilities for a portion of this work and Simon Titen for comments on the manuscript. This work was supported by National Institutes of Health grant GM-065604.
References
- Astrom, S. U., T. W. Cline and J. Rine, 2003. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics 163: 931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal, T., M. Gazi, M. Zurovec and P. J. Bryant, 2003. Genetic analysis of the ADGF multigene family by homologous recombination and gene conversion in Drosophila. Genetics 165: 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli, D., A. Selvaraj, H. Yepiskoposyan, B. Zhang, E. Hafen et al., 2003. Knockout of ‘metal-responsive transcription factor’ MTF-1 in Drosophila by homologous recombination reveals its central role in heavy metal homeostasis. EMBO J. 22: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore, T., R. Ignell, J. R. Carlson and D. P. Smith, 2003. Targeted mutation of a Drosophila odor receptor defines receptor requirement in a novel class of sensillum. J. Neurosci. 23: 9906–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K. G., and S. Lindquist, 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. [DOI] [PubMed] [Google Scholar]
- Gong, W. J., and K. G. Golic, 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W. J., and K. G. Golic, 2004. Genomic deletions of the Drosophila melanogaster hsp70 genes. Genetics 168: 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankenau, S., T. Barnickel, J. Marhold, F. Lyko, B. M. Mechler et al., 2003. Knockout targeting of the Drosophila Nap1 gene and examination of DNA repair tracts in the recombination products. Genetics 163: 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., L. Sandler, B. S. Baker, A. T. C. Carpenter, R. E. Denell et al., 1972. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71: 157–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., and E. Kubli, 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon, E., and D. Carroll, 1989. Degradation of linear DNA by a strand-specific exonuclease activity in Xenopus laevis oocytes. Mol. Cell. Biol. 9: 4862–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, B. L., J. R. Lundblad, Y. Chen and S. M. Smolik, 2002. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics 162: 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai, G., V. Kolisnychenko, Z. Bereczki and F. R. Blattner, 1999. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27: 4409–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C. R., W. Engels and C. Flores, 2002. Efficient repair of DNA breaks in Drosophila: evidence for single-strand annealing and competition with other repair pathways. Genetics 161: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2001. A targeted gene knockout in Drosophila. Genetics 157: 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., S. W. Titen, H. B. Xie, M. M. Golic, M. Bastiani et al., 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, M. I., and S. M. Parkhurst, 2002. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell 109: 447–458. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1983. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 11: 6341–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum, C., D. Pauli, M. Delattre, Y. Jaquet, A. Spierer et al., 2002. Isolation of Su(var)3–7 mutations by homologous recombination in Drosophila melanogaster. Genetics 161: 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal, M. L., and D. L. Hartl, 1996. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogame, N., M. Kim and J. M. Abrams, 2003. Drosophila p53 preserves genomic stability by regulating cell death. Proc. Natl. Acad. Sci. USA 100: 4696–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]