Abstract
Cf resistance genes in tomato confer resistance to the fungal leaf pathogen Cladosporium fulvum. Both the well-characterized resistance gene Cf-9 and the related 9DC gene confer resistance to strains of C. fulvum that secrete the Avr9 protein and originate from the wild tomato species Lycopersicon pimpinellifolium. We show that 9DC and Cf-9 are allelic, and we have isolated and sequenced the complete 9DC cluster of L. pimpinellifolium LA1301. This 9DC cluster harbors five full-length Cf homologs, including orthologs of the most distal homologs of the Cf-9 cluster and three central 9DC genes. Two 9DC genes (9DC1 and 9DC2) have an identical coding sequence, whereas 9DC3 differs at its 3′ terminus. From a detailed comparison of the 9DC and Cf-9 clusters, we conclude that the Cf-9 and Hcr9-9D genes from the Cf-9 cluster are ancestral to the first 9DC gene and that the three 9DC genes were generated by subsequent intra- and intergenic unequal recombination events. Thus, the 9DC cluster has undergone substantial rearrangements in the central region, but not at the ends. Using transient transformation assays, we show that all three 9DC genes confer Avr9 responsiveness, but that 9DC2 is likely the main determinant of Avr9 recognition in LA1301.
PLANTS are continuously challenged by a diverse array of pathogens. Resistant plants carry resistance (R) genes that enable them to recognize pathogen strains that carry matching avirulence (Avr) genes, a phenomenon called the gene-for-gene relationship (Flor 1946). In response to pathogen pressure, sophisticated surveillance systems have evolved to maintain and to generate new R genes in plants (Michelmore and Meyers 1998; Hulbert et al. 2001). R genes often occur in clusters, and extensive sequence exchange between homologs can occur through unequal recombination. This leads to novel sequence combinations and possibly to novel R genes. Although many R genes and R-gene clusters have been isolated, the evolution of R-gene clusters in natural populations is still poorly understood.
One of the best-studied pathosystems that follows the gene-for-gene relationship is the tomato-Cladosporium fulvum interaction (Joosten and de Wit 1999; Rivas and Thomas 2002). C. fulvum is a fungal biotrophic leaf pathogen of tomato (Lycopersicon) species. Resistance of tomato cultivars to C. fulvum has been introduced from wild tomato germ plasm. The tomato resistance gene Cf-9 that originates from L. pimpinellifolium (Lp) confers resistance to strains of C. fulvum that secrete the Avr9 protein (Jones et al. 1994). In resistant Cf-9 plants, a hypersensitive response (HR) is mounted at the infection site upon Avr9 recognition, thereby restricting the growth of the fungus. A high proportion of Lp accessions collected from their natural habitat are able to recognize Avr9 (Laugé et al. 2000; Van der Hoorn et al. 2001a), which suggests that the ability to recognize Avr9 may be beneficial to wild tomato plants.
Cf-9 is a member of the Hcr9 (homologs of Cladosporium fulvum resistance gene Cf-9) gene family (Parniske et al. 1997). Hcr9's encode proteins with a stretch of extracellular leucine-rich repeats, a transmembrane domain, and a short cytoplasmic tail that lacks an obvious signaling signature (Joosten and de Wit 1999; Rivas and Thomas 2002). Tomato plants usually carry several clusters of Hcr9's, and up to five homologs per cluster have been reported (Parniske et al. 1997, 1999; Parniske and Jones 1999). The Cf-9 cluster contains five homologs, Hcr9-9A–9E, of which Hcr9-9C is the functional Cf-9 gene (Parniske et al. 1997). Comparative analysis of Hcr9 clusters suggested that point mutation, unequal recombination, gene conversion, gene duplication, and translocation have contributed to the diversification of individual Hcr9's. Orthologous Hcr9's are more similar than Hcr9 paralogs, suggesting that sequence exchange occurs most frequently between orthologs (Parniske and Jones 1999). We previously identified the natural Cf-9 variant 9DC in Lp (Van der Hoorn et al. 2001a). 9DC has the same activity and specificity in conferring HR-associated Avr9 responsiveness as Cf-9 and was suggested to be ancestral to Cf-9 (Van der Hoorn et al. 2001a).
To date, only Hcr9 clusters of different species, clusters with different Cf genes, or clusters located at different chromosomal positions have been compared to study Hcr9 evolution (Parniske et al. 1997, 1999; Parniske and Jones 1999). Since 9DC and Cf-9 are clearly related and both genes confer Avr9 responsiveness (Van der Hoorn et al. 2001a), we investigated the relationship between their respective clusters at the individual and at the population level. Isolation of the 9DC cluster of the Lp accession LA1301, from which the 9DC gene was originally isolated (Van der Hoorn et al. 2001a), provided us with a unique opportunity to compare the Cf-9 and 9DC clusters in detail. Extensive sequence homology between both clusters was revealed. We identified numerous rearrangements in the central region of the clusters that allowed us to conclude that Cf-9 is ancestral to 9DC. Multiple unequal recombination events have resulted in the generation of three 9DC genes in the 9DC cluster, which all confer Avr9 responsiveness.
MATERIALS AND METHODS
Plant material:
Accessions of Lp were donated by the C. M. Rick Tomato Genetics Resource Center of the University of California, Davis (http://tgrc.ucdavis.edu/). The Cf-9 and 9DC genes of all Avr9-responsive L. pimpinellifolium accessions used in this study have been sequenced (Van der Hoorn et al. 2001a). These accessions were collected throughout the natural geographical range of L. pimpinellifolium and therefore represent distinct populations (Van der Hoorn et al. 2001a). The tomato cultivar MoneyMaker (MM) and the near-isogenic line MM-Cf9 (Tigchelaar 1984), which contains the Cf-9 cluster (Parniske et al. 1997), were used as controls. Plants were grown under standard greenhouse conditions. Avr9-responsive plants were selected by injection of leaflets with Avr9 protein (10 μg/ml) and screened for visible necrosis.
DNA manipulations:
DNA manipulations were performed according to standard protocols (Sambrook and Russell 2001). DNA sequence analysis was performed using Lasergene software programs (DNASTAR, Madison, WI). PCRs were performed with AmpliTaq (Perkin-Elmer, Wellesley, MA) or the Expand High Fidelity PCR system (Roche, Basel, Switzerland) for fragments >2 kb. Hybridizations were performed with 32P-labeled probes (Prime-a-gene labeling system, Promega, Madison, WI). Genomic DNA blots were hybridized with either a 9DC ORF probe or a Cf-9-Hcr9-9D (9D) intergenic region (IR) probe, which was obtained by PCR with primers IRF and IRR on a pBluescript II SK− (Stratagene, La Jolla, CA) library clone (see below). Primers were synthesized by Sigma-Genosys (Cambridge, UK). Primer sequences (5′–3′ direction, restriction sites underlined) are: 9AS1, tttttccatgggttgtgtaaaacttata; H5S1, tttttccatggattgtgtagaacttgta; CS1, gccgttcaagttgggtgtt; CS5, tttccaacttacaatcccttc; CS10, aaaccagaagaacctacaatta; CS11, cccccctgcagtcactaatatcttttcttgtgc; DS1, gagagctcaacctttacgaa; DS9, tttttccatgggttgtgtaaaacttgtg; DS12, cccccctgcagtaattaatatcttttcttgtgc; DS13, ggaagagaggttcacttcgta; DS14, ccaagtctaacactatcaacatttc; DCS1, gttcttatcctttaacacccaac; IRF, ctaatgtatacaaagcaacaaacc; and IRR, tgaagttgtgaagggaagc.
Library construction, clone selection, and sequencing:
Genomic DNA was isolated (Van der Beek et al. 1992) from LA1301 plants homozygous for 9DC and partially digested with Sau3AI. Fragments were cloned into the Lambda FIX vector and packaged using the Lambda FIX II/XhoI Partial Fill-In vector kit (Stratagene) and transfected to E. coli KW251 (Promega). Phages carrying an Hcr9 were identified by hybridization of plaque lift filters with a 9DC ORF probe. On the first phage isolations, a PCR was performed with the 9DC-specific primers DS1 and CS1 (Van der Hoorn et al. 2001a) to identify phages that contain 9DC. Pure phages were obtained after two subsequent screens. Phage DNA was isolated using a plate-lysate method (Sambrook and Russell 2001). The tomato genomic DNA inserts were cloned into the NotI site of pBluescript II SK− (Stratagene). End sequences of clones were determined using the universal M13F and M13R primers. Inserts were subcloned in pBluescript II SK− and sequenced by primer walking (triple-strand coverage, BaseClear, Leiden, The Netherlands).
Agroinfiltration assays:
Individual Hcr9's were amplified by PCR with gene-specific primer pairs 9AS1/CS11 (‘9A’), DS9/DS12 (9DC3) and H5S11/CS11 (‘9E’) using library clones as templates. 9D was amplified from MM-Cf9 genomic DNA with primer pair DS9/DS12. The Hcr9's were cloned in pRH80, sequenced (BaseClear), and subcloned in the binary plasmid pMOG800 (Van der Hoorn et al. 2000), yielding overexpression agroinfiltration constructs. The previously described 9DC overexpression construct (Van der Hoorn et al. 2001a) was used for both 9DC1 and 9DC2 genes. Genomic agroinfiltration constructs were made by subcloning of the NotI inserts of pBluescript II SK− clones into pBIVM2 [a pCGN1548 (McBride and Summerfelt 1990) derivative]. Agroinfiltration assays with Nicotiana tabacum cv. Petite Havana SR1 were performed as described, with use of the pAvr9 and pCf9 constructs (Van der Hoorn et al. 2000), except that Agrobacterium tumefaciens strain GV3101 was used. To test the relative activities of Cf genes, agroinfiltration dilution series were performed as described previously (Van der Hoorn et al. 2001b).
RESULTS
9DC genetics and cluster conservation:
Both the 9DC and Cf-9 genes confer Avr9 recognition in the Lp population. Sequencing of seven alleles of both genes (including the previously isolated Cf-9 allele by Jones et al. 1994) showed only three polymorphism nucleotides in 9DC and none in Cf-9 (Van der Hoorn et al. 2001a). The high DNA sequence homology (99.8%) among 9DC, Cf-9, and 9D suggested that these genes are allelic. Selfings of an F1 of LA1301 and the susceptible cultivar MM-Cf0 showed a 3:1 segregation for HR-associated Avr9 recognition (65:24, χ2 = 0.18, P > 0.67), which indicates that Avr9 recognition is inherited as a monogenic dominant trait in LA1301. LA1301 was crossed to cultivar MM-Cf9 (Tigchelaar 1984) and subsequently backcrossed to MM-Cf0. All 330 BC1 plants responded with an HR upon injection with Avr9 protein, confirming that 9DC is allelic, or very closely linked, to Cf-9 (<1.8 cM, P = 95%) and located at the Milky Way locus (Parniske et al. 1999).
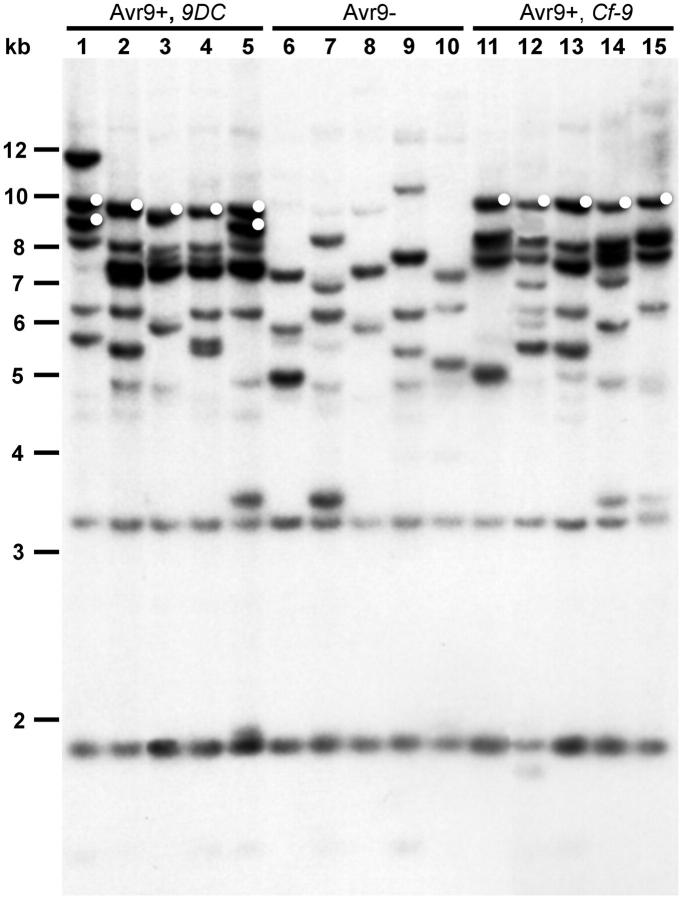
To assess the possible conservation of the 9DC and Cf-9 clusters in the Lp population, genomic DNA blots from a sample of Lp plants were hybridized with the 9DC ORF probe (Figure 1). The Lp plants that carry either sequenced 9DC or Cf-9 alleles represent accessions from geographically distant locations throughout the natural range of Lp (Van der Hoorn et al. 2001a) and therefore represent different local populations. In addition, Lp plants from geographically distinct locations that lack both Cf-9 and 9DC were studied (Figure 1). Similar hybridization patterns may be expected if the Hcr9 clusters are conserved in different Lp plants. However, only some hybridizing fragments appear to be conserved, and both Cf-9- and 9DC-containing Lp plants display variation in their Hcr9 hybridization patterns. Avr9-nonresponsive plants show fewer Hcr9-hybridizing bands, as does the susceptible cultivar MM-Cf0. This suggests that the nonresponsive Lp plants harbor fewer Hcr9's than Avr9-responsive plants, possibly due to a Milky Way cluster with only one or a few Hcr9's, as observed in MM-Cf0 (Parniske et al. 1997).
Figure 1.—
High-stringency blot of EcoRV-digested genomic DNA of a sample of L. pimpinellifolium and reference genotypes, hybridized with a 9DC ORF probe. Lanes 1–5: LA1301, LA0114, LA1629, LA1637, and LA1659 [Avr9-responsive (Avr9+), all contain sequenced 9DC alleles (Van der Hoorn et al. 2001a)]; lanes 6–10: MM-Cf0, LA0400, LA1279, LA1590, and LA2852 [Avr9-nonresponsive (Avr9-), contain neither 9DC nor Cf-9]; lanes 11–15: MM-Cf9, LA1585, LA1614, LA1687, and LA2653 [Avr9-responsive (Avr9+), all contain sequenced Cf-9 alleles (Van der Hoorn et al. 2001a)]. Fragments marked with a white dot also hybridize to the Cf-9-9D IR region probe. A DNA size ladder (in kilobases) is shown on the left.
Isolation and assembly of the 9DC cluster:
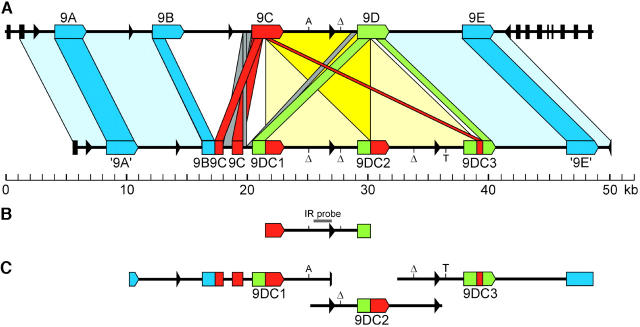
LA1301 was chosen to molecularly characterize the 9DC cluster, since this was also the genotype from which 9DC was originally isolated (Van der Hoorn et al. 2001a), and it exhibits a strong Avr9 response. A genomic phage library with an approximate fivefold genome coverage was made from LA1301. Thirteen phages were selected on the basis of hybridization with a 9DC ORF probe and PCR selection with 9DC-specific primers. The inserts were subcloned in plasmids. Restriction fingerprinting, restriction hybridization, end sequencing of the inserts and AFLP-based fingerprinting of the clones (M. J. D. de Kock, R. Van der Hulst, P. J. G. M. de Wit and P. Lindhout, personal communication) of these plasmids did not result in an unambiguous contig. This suggested that either 9DC sequences are present at two closely linked loci or sequence duplications are present within the 9DC cluster. Hybridization of a DNA blot of BglII-digested clones with a 9DC probe enabled us to form a contig. However, this contig obscured possible duplications within the 9DC cluster. Therefore, BglII subclones (0.7–6.4 kb) were made of selected inserts, which were sequenced by primer walking to prevent sequence assembly artifacts due to repetitive sequences. By sequencing multiple subclones, an 8.7-kb near-perfect direct repeat was detected, which interfered with contig building by conventional methods. A clone containing an insert of 16.5 kb, which almost encompassed the two complete 8.7-kb repeat regions, frequently exhibited recombination in an Escherichia coli rec− strain, which resulted in loss of part of the insert. The size of the remaining insert suggested that one repeat region was lost due to this recombination (data not shown). A similar case of recombination in an E. coli clone leading to loss of part of a Cf gene cluster was previously described for the Cf-2 cluster, which also contains repeated sequences (Dixon et al. 1996). With the inclusion of the 8.7-kb repeat, an unambiguous contig of the 9DC cluster was assembled that consists of 44,539 bp (Figure 2A). The overall organization of the cluster, including the 8.7-kb repeat, has been verified by restriction fingerprinting of individual library clones and a LA1301 DNA blot hybridization using 14 different restriction enzymes and a 9DC ORF probe (data not shown). The near-perfect 8.7-kb repeat includes two 9DC genes with identical coding sequences (9DC1 and 9DC2, sequences described as 9DC in Van der Hoorn et al. 2001a) and a third gene (9DC3), which is similar to 9DC1 and 9DC2. We initially isolated clones containing one of the three 9DC genes, as PCR with 9DC-specific primers resulted in the same product for all three 9DC genes. We conclude that the final contig based on these clones encompasses the complete 9DC cluster for two reasons. First, the 9DC cluster harbors several LipoxygenaseC (LoxC; Heitz et al. 1997) exons, which are thought to have coduplicated with Hcr9's (Parniske et al. 1997). The identity and orientation of the LoxC exons located at the termini of the 9DC cluster correspond to those found at the termini of the Cf-4 and Cf-9 clusters (Parniske et al. 1997). Second, the 9DC cluster harbors orthologs of the most distal Hcr9's from the Cf-9 cluster (‘9A’ and ‘9E’; see Figure 2A).
Figure 2.—
Schematic of the relationships between the Cf-9 and 9DC clusters and of several genomic DNA fragments used in this study. (A) Relationships between the Cf-9 and 9DC clusters. The Cf-9 cluster was previously described (Parniske et al. 1997). Colored arrowed boxes represent complete Hcr9's; colored rectangular boxes represent Hcr9 pseudogenes. Cf-9 (9C)-like sequences are depicted in red; 9D-like sequences in green; other Hcr9 sequences in blue. All Hcr9's and Hcr9 fragments in the Cf-9 and 9DC clusters are in the 5′-3′ orientation. Black arrows and bars represent LipoxygenaseC exons, and the arrows indicate the polarity of transcription of the 3′-exon. Boxes connecting the Cf-9 and 9DC clusters indicate orthologous regions. Note that in the central part of the 9DC cluster an 8.7-kb repeat is present that is almost identical to a region in the Cf-9 cluster (see also B). These regions are connected by light yellow boxes, which overlap in the dark yellow triangle. A and T indicate single polymorphic nucleotides in these regions; “Δ” represents a single nucleotide deletion. (B) The 8.7-kb DNA sequence fragment that is present once in the Cf-9 cluster and as a direct repeat in the 9DC cluster. This fragment includes the 3′-half of Cf-9, the Cf-9/9D intergenic region, and the 5′-half of 9D. The position of the Cf-9-9D IR probe is indicated by a gray horizontal bar above the fragment. (C) Genomic fragments containing one of the three 9DC genes that were cloned in a binary expression vector for agroinfiltration studies to determine their ability to confer Avr9 responsiveness.
Features of the 9DC cluster:
The 9DC cluster contains five full-length Hcr9's (Figure 2A). Most striking is the presence of three 9DC genes. The 9DC1 and 9DC2 coding sequences are completely identical and are likely the result of a duplication within the 9DC cluster. 9DC3 is identical to 9DC1 and 9DC2 for the first 1635 nucleotides, encompassing the proposed recombination site within 9DC (Van der Hoorn et al. 2001a). The remaining 3′-part has five nucleotide differences when compared with the corresponding region in 9D (Parniske et al. 1997), and the sequence downstream of 9DC3 is nearly identical to that downstream of 9D, indicating that the 3′-part of 9DC3 indeed has a 9D-like origin. Both ‘9A’ and ‘9E’ differ by only 11 bp from their respective orthologs Hcr9-9A and Hcr9-9E from the Cf-9 cluster (Parniske et al. 1997), including a single nucleotide deletion that leads to a premature stopcodon in ‘9E’.
In addition to the complete Hcr9's found in the 9DC cluster, several Hcr9 fragments are present. The 1026-bp ‘9B’ part of the ‘9B9C’ fragment (Figure 2A) is 91% identical and orthologous to Hcr9-9B of the Cf-9 cluster. This ‘9B’ fragment is directly followed by a Cf-9 (9C) fragment (position 206–821 in Cf-9), 831 bp of the Cf-9 promoter directly preceding the Cf-9 gene, and a second Cf-9 fragment that comprises the first 821 nucleotides of the Cf-9 ORF. The two Cf-9 fragments in the 9DC cluster (Figure 2A) carry the same nucleotide difference when compared with Cf-9 itself. The second Cf-9 fragment is followed by a Cf-9 promoter fragment that starts at the same point as the first Cf-9 promoter fragment and merges into the 9D promoter. This chimeric promoter precedes 9DC1. The sequences upstream of ‘9A’ and downstream of ‘9E’ and the ‘9A’-‘9B9C’ and ‘9DC3’-‘9E’ IRs are homologous to the corresponding regions in the Cf-9 cluster (Figure 2A). The 6-kb IR between 9DC1 and 9DC2 and the IR between 9DC2 and 9DC3 each differ by only a single nucleotide from the IR between Cf-9 and 9D (Parniske et al. 1997) and by a single nucleotide from each other (Figure 2A).
Conservation of Cf-9, 9D, and three 9DC genes in Avr9-responsive L. pimpinellifolium plants:
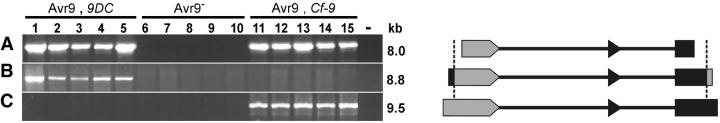
The high sequence conservation in the IRs in the 9DC and Cf-9 clusters and coding sequences of the three 9DC genes and Cf-9 indicates a clear relationship between the Cf-9 and 9DC clusters. Moreover, it suggests that the duplications in the 9DC cluster are recent events. The near-identical 8.7-kb sequence in the Cf-9 and 9DC clusters consists of the 3′-terminal half of Cf-9, the IR between Cf-9 and 9D, and the 5′-terminal half of 9D (Figure 2B). This 8.7-kb conserved sequence could provide a means to further unravel the evolutionary relationships between the 9DC genes and Cf-9. Therefore, we decided to study the occurrence of this region in Lp by PCR, using the same selection of plants used in the hybridization experiment (Figure 1). We designed a set of specific primer pairs, based on the sequences of the three 9DC genes, Cf-9, and 9D. With these primer pairs, products can be obtained only if specific combinations of the above-mentioned genes are present (Figure 3). All 9DC- and Cf-9-containing genotypes appear to carry at least one tandem repeat, which comprises the 3′-half of Cf-9, the Cf-9/9D IR, and the 5′-half of 9D, whereas no PCR fragments were obtained from Avr9-nonresponsive plants (Figure 3A). All 9DC genotypes carry at least one 9DC1-9DC2 or 9DC2-9DC3 direct repeat, including the Cf-9/9D IR. This repeat is not present in either Cf-9-containing or Avr9-nonresponsive genotypes (Figure 3B). Only the Cf-9 genotypes carry a Cf-9-9D repeat, including the Cf-9/9D IR (Figure 3C). Restriction fingerprinting of the PCR products confirmed that all fragments obtained from a specific primer pair are similar and compose the Cf-9/9D IR (data not shown). In addition, control PCR experiments showed the presence of 9D only in all Cf-9 genotypes and the presence of 9DC3 only in all 9DC genotypes. Conversely, Cf-9 is present in none of the 9DC genotypes (Van der Hoorn et al. 2001a; data not shown).
Figure 3.—
Presence of specific tandem repeats of the three 9DC genes or Cf-9 and 9D in a sample of L. pimpinellifolium and reference genotypes, as detected by PCR. Genotypes used for DNA templates are as shown in Figure 1. Lanes 1–5: LA1301, LA0114, LA1629, LA1637, and LA1659 [Avr9-responsive (Avr9+), all contain 9DC]; lanes 6–10: MM-Cf0, LA0400, LA1279, LA1590, and LA2852 [non-Avr9-responsive (Avr9-), contain neither 9DC nor Cf-9]; lanes 11–15: MM-Cf9, LA1585, LA1614, LA1687, and LA2653 [Avr9-responsive (Avr9+), all contain Cf-9]; “—,” water control. (Left) PCR products were size separated on 0.7% agarose gels. Fragment sizes were estimated using a DNA size marker (not shown). (Right) Schematics of the structure of the PCR products. Cf-9 sequences are shaded; 9D sequences are solid. The vertical dashed lines indicate the boundaries of the 8.7-kb fragment as shown in Figure 2B. (A) PCR products obtained with primers CS10 and DS13. CS10 anneals to the 3′-part of both 9DC and Cf-9, and DS13 to the 5′-part of all 9DC genes and 9D. A PCR product (8.0 kb) is obtained if a 9DC1-9DC2, a 9DC2-9DC3 direct repeat, or a Cf-9-9D tandem repeat is present. (B) PCR products obtained with primers DCS1 and CS1, which both anneal to the proposed recombination site (Van der Hoorn et al. 2001a) within all three 9DC genes. A PCR product (8.8 kb) is obtained only when a 9DC1-9DC2 or a 9DC2-9DC3 direct repeat is present. The 17- and 15-bp fragments adjoining the 8.7-kb fragment are not drawn to scale. (C) PCR products obtained with primers CS5 and DS14. CS5 anneals to the 5′-part of Cf-9, and DS14 to the 3′-part of 9D. A PCR product (9.5 kb) is obtained only when a Cf-9-9D tandem repeat is present.
To confirm the PCR results and to study the structure of the 9DC and Cf-9 clusters in the Lp plants, we designed a specific Cf-9/9D IR probe (Figure 2B). This probe was hybridized to the DNA blots used for surveying Hcr9 diversity in Lp (Figure 1). Hybridizing fragments were found only in genotypes that carry 9DC or Cf-9 (Figure 1). The fragments that hybridized to the Cf-9/9D IR probe are a subset of those hybridizing with the 9DC ORF probe (Figure 1). Of the four 9DC genotypes that we sampled in addition to LA1301, only LA1659 showed an IR hybridization pattern identical to LA1301. The upper band in LA1301 represents the 9DC2-9DC3 IR, and the lower band the 9DC1-9DC2 IR. This indicates that both a 9DC1-9DC2 and a 9DC2-9DC3 repeat are present, as in LA1301. The remaining three 9DC genotypes carry only a 9DC2-9DC3 repeat. All Cf-9 genotypes have a hybridization pattern identical to that of MM-Cf9, in which only hybridization of the Cf-9/9D IR is observed. Combined with the PCR data, these results indicate that all sampled Cf-9 genotypes contain the same Cf-9-9D tandem repeat. These data also show that, although the overall structure of the Hcr9 clusters is not conserved (Figure 1), at least the genes directly downstream of 9DC and Cf-9 and the IRs between these genes are conserved in the Lp population.
Activity of individual 9DC genes:
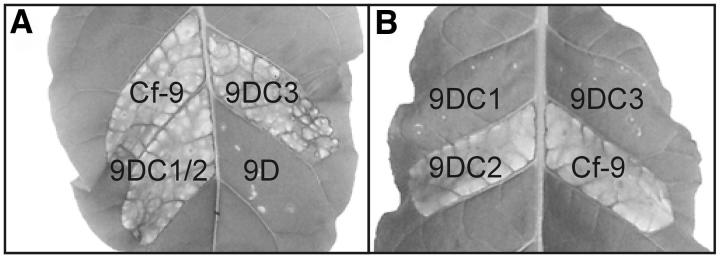
Single amino acid changes in Cf proteins have no, or only minor, effects on their activity, whereas multiple changes can lead to a drastic reduction in activity (Van der Hoorn et al. 2001b; Wulff et al. 2001). 9DC and Cf-9 have the same activity in conferring Avr9 responsiveness, although they differ in 61 amino acids (Van der Hoorn et al. 2001a). Compared with 9DC1 and 9DC2, 9DC3 has 33 nucleotide differences, resulting in only 23 amino acid substitutions. Therefore, 9DC3 may confer Avr9 responsiveness as well. This was tested by co-agroinfiltration of 9DC3 and Avr9. First, the activity of 9DC3 was tested in an agroinfiltration assay in tobacco with Hcr9's under control of the 35S promoter (Van der Hoorn et al. 2000). In this assay, in addition to 9DC1 and 9DC2, 9DC3 is active as well (Figure 4A), as cells in the infiltrated leaf section undergo a typical HR (Van der Hoorn et al. 2000). However, in dilution assays the activity of 9DC3 is reduced four- to eightfold as compared to 9DC and Cf-9 (data not shown). As expected, 9D from the Cf-9 cluster (Figure 4A) and the ‘9A’ and ‘9E’ homologs from the 9DC cluster did not confer Avr9 responsiveness (data not shown). An agroinfiltration assay in which Hcr9's are overexpressed cannot distinguish the intrinsic activities of 9DC1 and 9DC2, which have identical coding sequences. Genomic constructs of the three 9DC genes (Figure 2C), which likely reflect the intrinsic activity of Hcr9's in tomato plants, were therefore generated. Surprisingly, only the genomic 9DC2 construct conferred Avr9 responsiveness in an agroinfiltration assay with Avr9 (Figure 4B). Therefore, 9DC2 is likely the main determinant of Avr9 recognition in Lp LA1301.
Figure 4.—
Combined agroinfiltration of Avr9 under control of the 35S promoter and Hcr9's under control of the 35S or their native promoter. (A) Agroinfiltration of 35S-driven constructs containing the 9DC genes, Cf-9 or 9D. 9DC1/2 represents the identical 9DC1 and 9DC2 coding regions. (B) Agroinfiltration assay of constructs containing 9DC1, 9DC2, or 9DC3 under control of their native promoter; 35S-driven Cf-9 was used as a control. Expression of an Hcr9 that confers Avr9 responsiveness results in visible necrosis. Both photographs were taken 7 days after infiltration.
DISCUSSION
Numerous R genes have been cloned in the past decade and it appears that they frequently occur in clusters (Takken and Joosten 2000; Hulbert et al. 2001; Martin et al. 2003). However, evolution of R-gene clusters at the population level is still poorly understood. Several studies report on the structural rearrangements in R-gene loci (reviewed in Michelmore and Meyers 1998; Hulbert et al. 2001). Unequal recombination, gene conversion, point mutation, duplication, and translocation all contribute to the generation of novel R genes. The discovery of the 9DC gene is an example of a recent event leading to a novel R gene (Van der Hoorn et al. 2001a). Cf-9 and 9DC are related by intragenic recombination, differ in 61 amino acids, but have a similar specificity and activity in conferring Avr9 responsiveness. Here we describe the isolation of the complete 9DC cluster from Lp LA1301 and a detailed comparison with the previously isolated Cf-9 cluster (Parniske et al. 1997). We conclude that several unequal recombination events in the Cf-9 cluster, including two intragenic recombinations, have resulted in three 9DC genes. Surprisingly, all three genes confer Avr9 responsiveness when overexpressed, but only 9DC2 is active under control of its native promoter. Furthermore, we discuss and reconstruct the evolution of the 9DC cluster and show that, in contrast to what had been suggested previously (Van der Hoorn et al. 2001a), Cf-9 is ancestral to 9DC.
Reconstruction of the evolution of the 9DC cluster:
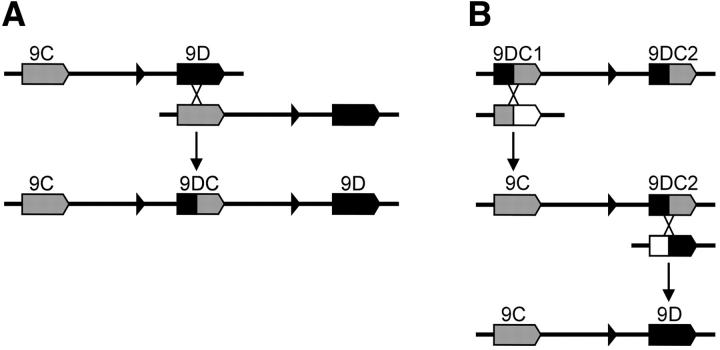
On the basis of previous data, it was initially assumed that 9DC is ancestral to Cf-9 (Van der Hoorn et al. 2001a). Without knowledge of the flanking regions of 9DC, this could be explained by a single intragenic unequal recombination between 9DC and an unidentified homolog, which gave rise to both Cf-9 and 9D. However, we now show that an 8.7-kb region of the Cf-9 cluster that comprises the 3′-half of Cf-9, the Cf-9/9D IR, and the 5′-half of 9D is duplicated in the 9DC cluster (Figure 2, A and B). If the sequence in the Cf-9 cluster is ancestral, a single intragenic unequal recombination event between 9D and Cf-9 explains the generation of the first 9DC gene and its flanking IR sequences (Figure 5A). Conversely, two independent unequal recombinations should have occurred at the same site in the middle of two 9DC genes to create both Cf-9 and 9D from 9DC1 and 9DC2 and two other, unknown Hcr9's (Figure 5B). As this is very unlikely, the sequence in the Cf-9 cluster should represent the ancestral state. Furthermore, identification of Cf-9 alleles in the distantly related tomato species L. hirsutum confirms that Cf-9 indeed is an ancient gene (M. Kruijt, D. J. Kip, M. H. A. J. Joosten, B. F. Brandwagt and P. J. G. M. de Wit, unpublished results).
Figure 5.—
Schematic of the two possible ancestral relationships among Cf-9 (9C), 9D, and 9DC. Arrowed boxes represent complete Hcr9's; Cf-9 sequences are shaded; 9D sequences are solid; open boxes, unknown Hcr9 sequences; solid triangles, LipoxygenaseC exons. Diagonal crossed lines, (unequal) recombination points; arrows point toward one of the possible recombination products. All recombinations are shown in a homozygous genotype. (A) A single intragenic recombination between 9D and Cf-9 could result in the 9DC gene. (B) The Cf-9 and 9D genes can be generated only from two 9DC genes via two independent unequal recombinations at identical positions within each of these 9DC genes. Recombination of 9DC1 with an unknown Hcr9 that carries 5′ Cf-9-like sequences would result in Cf-9. Recombination between 9DC2 and an unknown Hcr9 that carries 3′ 9D-like sequences would generate 9D.
The recent generation of 9DC from Cf-9 and 9D and the isolation of both the Cf-9 (Parniske et al. 1997) and the 9DC clusters allow a detailed reconstruction of the evolution of the 9DC cluster. A model that most likely represents the evolutionary events that have created the 9DC cluster is presented, although alternative explanations cannot be excluded. The termini of the Cf-9 and 9DC clusters are similar, and all major rearrangements have occurred in the central region of the clusters (Figure 2A). A single intragenic unequal recombination event in the central region of the Cf-9 cluster (Figure 6A, cluster 1) would give rise to a cluster with Cf-9, a single 9DC gene, and 9D (Figure 6A, cluster 2). This cluster harbors two identical Cf-9/9D IRs and would therefore be prone to further intragenic unequal recombination due to mispairing of individual homologs. Indeed, a second 9DC copy was generated (Figure 6A, cluster 3). Two scenarios may explain the presence of a third 9DC gene in some of the 9DC clusters (Figure 6, A and B). In scenario A (Figure 6A), 9DC3 was generated before 9DC1. Intragenic recombination between the second 9DC gene and the 9D ortholog of cluster 3 (Figure 6A) generated a cluster that contains the 9DC2 gene and 9DC3 (Figure 6A, cluster 4A), which is present in three of the L. pimpinellifolium genotypes studied here. A final unequal recombination event generated a cluster that contains the two identical 9DC1 and 9DC2 genes and 9DC3 (Figure 6A, cluster 5A), which is present in two of these L. pimpinellifolium genotypes, including LA1301. Alternatively, in scenario B (Figure 6B), all genotypes first accumulated the three 9DC genes, and some have subsequently lost 9DC1 or 9DC2. A recombination point in cluster 3 (Figure 6B) different from that shown in scenario A resulted in a cluster that contains three identical 9DC genes (Figure 6B, cluster 4B). Intragenic recombination between the third 9DC gene and the 9D ortholog of cluster 4B generated a cluster that contains the two identical 9DC1 and 9DC2 genes and 9DC3 (Figure 6B, cluster 5B), which is present in two of the L. pimpinellifolium genotypes studied here, including LA1301. A final unequal recombination event within cluster 5B resulted in loss of 9DC1 and in generation of cluster 6B, which is present in three of the L. pimpinellifolium genotypes studied here.
Figure 6.—
Schematic of two scenarios for the formation of the central region of the 9DC cluster, which involve unequal recombinations due to mispairing of individual Hcr9's. Arrowed boxes represent complete Hcr9's; Cf-9 (9C) sequences are shaded and 9D sequences are solid. Solid triangles, LipoxygenaseC exons. Diagonal crossed lines indicate (unequal) recombination points; arrows point toward one of the possible recombination products. All recombinations are shown in a homozygous genotype. Only the positions of the intragenic recombination sites that generated the first of the two identical 9DC genes and 9DC3 are known exactly. The other recombination sites may be located anywhere within the colinear sequence stretches and are drawn at arbitrary positions within these stretches. Boxed LA numbers to the left of clusters indicate the L. pimpinellifolium accessions that contain these clusters. To the right, numbers indicate the different clusters. The first three clusters are identical in both scenarios. (A) Scenario A, in which 9DC3 was generated prior to 9DC1. (B) Scenario B, in which 9DC3 was generated after 9DC1. (Recombinations that may lead to deletion of Cf-9 or to the formation of the Cf-9 fragments present in the 9DC cluster of LA1301 are not indicated.)
In all five tested 9DC genotypes, Cf-9 and 9D have been lost. 9D was lost through unequal recombination with 9DC2, yielding 9DC3 (Figure 6, A and B). The sequences upstream of 9DC1 that compose the ‘9B9C’ fragment, the two Cf-9 fragments, and the two Cf-9 promoter sequences can be explained only by numerous recombinations. These at least constitute truncation of Cf-9 due to unequal recombination with the Cf-9 promoter; duplication of this Cf-9 fragment-Cf-9 promoter sequence; fusion of the first Cf-9 fragment with an Hcr9-9B ortholog, which would yield the ‘9B9C’ fragment; and fusion of the second Cf-9 promoter fragment with the 9DC1 promoter, which would yield the short chimeric Cf-9/9D promoter.
Mispairing of R-gene homologs is known from several R-gene families (Hulbert et al. 2001). Inter- and intragenic recombinations give rise to novel R-gene homologs and novel combinations of R-gene homologs and thus contribute to diversity at R-gene loci. However, if the unequal recombination rate within an R-gene cluster is too high, this may lead to homogenization of the R-gene sequences. Consistent with this idea, it was previously suggested that sequence exchange between orthologous Hcr9's occurs more frequently than that between paralogs (Parniske and Jones 1999). However, the generation of three 9DC genes from a cluster that contained only a single 9DC gene by mispairing of and unequal recombination among homologs is unprecedented and suggests that the initial 9DC cluster was unstable due to the presence of the 8.7-kb repeat. In contrast, Cf-9 was found to be very stable in a homozygous background, whereas the meiotic stability of Cf-9 was dramatically reduced in a Cf-4/9 heterozygous background (Parniske et al. 1997). This indicated that unequal mispairing in Hcr9 clusters in a homozygous background is rare, which can be explained by diverged IRs that prevent mispairing of homologs and subsequent homogenization of the homologs. However, the repetitive IR structure of the initial 9DC cluster (Figure 6A) appears to be prone to mispairing, which resulted in unequal recombination and the generation of three 9DC genes. L. pimpinellifolium is a facultative outcrosser (Rick et al. 1977), and therefore the 9DC cluster could be present in heterozygous plants, which would have increased the frequency of mispairing. Therefore, sequence exchange by inter- and intragenic recombination and gene conversion among the 9DC genes and homologs that occupy orthologous positions may lead to further Hcr9 sequence homogenization and a decrease of Hcr9 variation at the MW locus.
The termini of the 9DC cluster (5′ of the ‘9B’ fragment and 3′ of 9DC3) are similar to those of the Cf-9 cluster (Figure 2A). The ORFs of the ‘9A’ and ‘9E’ genes each differ from their orthologs Hcr9-9A and Hcr9-9E of the Cf-9 cluster by only 11 bp, whereas the intergenic regions show a higher proportion of nucleotide differences, as well as some insertions and deletions. In contrast, the 8.7-kb repeat regions in the 9DC cluster, including IRs of almost 6 kb, differ by only one or two nucleotides from the corresponding region in the Cf-9 cluster, which suggests that the formation of the three 9DC genes in the 9DC cluster is a relatively recent event. It further suggests that gene conversion and/or intergenic recombination have occurred at the termini of the Cf-9 and 9DC clusters. However, the termini of the Cf-9 cluster may not be ancestral to those of the 9DC cluster, but more likely represent variation in the Hcr9-9A-like and Hcr9-9E-like homologs in the L. pimpinellifolium population.
Activity of Hcr9's mediating Avr9 recognition in L. pimpinellifolium:
In an agroinfiltration assay, all three 9DC genes confer Avr9 responsiveness under control of the 35-S promoter, although the activity of 9DC3 is four- to eightfold reduced when compared to Cf-9, 9DC1, and 9DC2. Previously, only the two Cf-2 genes from the Cf-2 cluster were found to have the same function in conferring resistance to C. fulvum strains that express the Avr2 protein (Dixon et al. 1996; Luderer et al. 2002), and therefore the three 9DC genes that share the same function represent a unique situation among all known C. fulvum resistance gene clusters. In an agroinfiltration assay with genomic constructs, however, only 9DC2 conferred Avr9 responsiveness. Thus, 9DC2 is likely the main determinant of Avr9 recognition in Lp LA1301. Since 9DC1 and 9DC2 have identical downstream sequences, but 9DC1 has a promoter region of only 797 bp, the observed difference in activity may be attributed to a lower expression level of 9DC1. Unfortunately, this cannot be verified in LA1301 plants, as both genes and their 5′- and 3′-untranslated regions do not contain any polymorphic nucleotides that would enable a discriminatory RT-PCR analysis. 9DC3 has the same promoter as 9DC2, but the terminators of these two genes differ considerably. Therefore, its inactivity when expressed by agroinfiltration under its native promoter may be explained by a combination of the lower activity of the 9DC3 protein and a lower 9DC3 expression level. The agroinfiltration assay with genomic constructs, however, is less sensitive compared to that with overexpression constructs and therefore does not exclude that, in addition to 9DC2, 9DC1 and/or 9DC3 may also be active in Avr9 recognition upon C. fulvum infection of LA1301. Unfortunately, the intolerance of LA1301 to the high humidity used in the standard infection assay prevented successful C. fulvum infections.
Avr9 recognition in tomato:
All isolates of C. fulvum collected to date originate from commercially grown tomatoes. At least one of these strains can overcome the resistance provided by the Cf-9 cluster without an apparent loss of pathogenic fitness (Laugé et al. 1998). Moreover, Avr9 gene replacement did not affect the pathogenic fitness of C. fulvum in greenhouse infection assays (Marmeisse et al. 1993). This suggests that, at least in greenhouse assays, Avr9 may be dispensable. However, Avr9 recognition is present in a high proportion of Lp plants and based on the highly conserved Cf-9 and 9DC genes (Laugé et al. 2000; Van der Hoorn et al. 2001a). Avr9 recognition is also present in several other tomato species and functional Cf-9 alleles have also been identified in the distantly related tomato species Lycopersicon hirsutum (M. Kruijt, D. J. Kip, M. H. A. J. Joosten, B. F. Brandwagt and P. J. G. M. de Wit, unpublished results). This suggests that in wild tomato plants Hcr9's that confer Avr9 recognition may have been maintained by selection. In addition to Cf-9 itself, the Cf-9 cluster also harbors the partial resistance gene Hcr9-9B (Parniske et al. 1997; Laugé et al. 1998; Panter et al. 2002). Although the 9DC cluster does not harbor a complete Hcr9-9B ortholog, it is possible that the other 9DC cluster homologs encode novel resistance specificities. The selective advantage of such a 9DC cluster may explain the prevalence of 9DC over Cf-9 in the Lp population (Van der Hoorn et al. 2001a).
To further study the evolutionary forces that drive the evolution of Cf gene clusters, it would be highly interesting to collect natural C. fulvum strains from wild tomato plants and characterize both the Avr and the R genes. This would add greatly to our knowledge of natural selection and co-evolution in plant-pathogen populations (Bergelson et al. 2001; DeMeaux and Mitchell-Olds 2003; Thrall and Burdon 2003). It may also shed light on the relative importance of the different C. fulvum Avr factors within the C. fulvum population in a natural situation, including Avr9.
Our present study enabled a unique detailed reconstruction of the evolution of a single R-gene cluster at the species level and has shown that unequal recombination can have a major impact on the evolution of R-gene clusters. A great challenge in the near future will be to study R-gene clusters on an even larger scale by using novel R-gene cluster fingerprinting methods.
Acknowledgments
We thank Matthieu Joosten and Maarten Koornneef for critically reading the manuscript; Roger Chetelat (Tomato Genetic Resource Center, Davis, CA) for providing the Lp seeds; Kees Spelt for providing the pBIVM2 vector; Maarten de Kock for help with the AFLP-based clone fingerprinting method; Pim Lindhout for helpful discussions; Mart Berns, Bert Essenstam, Ronald Janssen, and Henk Smid for excellent plant care; and Diana Kip for technical assistance. This study was financially supported by the Dutch Foundation for Applied Sciences.
Sequence data from this article have been deposited with the GenBank Data Libraries under accession no. AY569331.
References
- Bergelson, J., M. Kreitman, E. A. Stahl and D. Tian, 2001. Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- De Meaux, J., and T. Mitchell-Olds, 2003. Evolution of plant resistance at the molecular level: ecological context of species interactions. Heredity 91: 345–352. [DOI] [PubMed] [Google Scholar]
- Dixon, M. S., D. A. Jones, J. S. Keddie, C. M. Thomas, K. Harrison et al., 1996. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84: 451–459. [DOI] [PubMed] [Google Scholar]
- Flor, H. H., 1946. Genetics of pathogenicity in Melampsora lini. J. Agric. Res. 73: 335–357. [Google Scholar]
- Heitz, T., D. R. Bergey and C. A. Ryan, 1997. A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol. 114: 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S. C., C. A. Webb, S. M. Smith and Q. Sun, 2001. Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39: 285–312. [DOI] [PubMed] [Google Scholar]
- Jones, D. A., C. M. Thomas, K. E. Hammond-Kosack, P. J. Balint-Kurti and J. D. G. Jones, 1994. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793. [DOI] [PubMed] [Google Scholar]
- Joosten, M. H. A. J., and P. J. G. M. de Wit, 1999. The tomato-Cladosporium fulvum interaction: a versatile experimental system to study plant-pathogen interactions. Annu. Rev. Phytopathol. 37: 335–367. [DOI] [PubMed] [Google Scholar]
- Laugé, R., A. P. Dmitriev, M. H. A. J. Joosten and P. J. G. M. de Wit, 1998. Additional resistance genes against Cladosporium fulvum present on the Cf-9 introgression segment are associated with strong PR protein accumulation. Mol. Plant Microbe Interact. 11: 301–308. [Google Scholar]
- Laugé, R., P. Goodwin, P. J. G. M de Wit and M. H. A. J. Joosten, 2000. Specific HR-associated recognition of secreted proteins from Cladosporium fulvum occurs in both host and non-host plants. Plant J. 23: 735–745. [DOI] [PubMed] [Google Scholar]
- Luderer, R., F. L. W. Takken, P. J. G. M. de Wit and M. H. A. J. Joosten, 2002. Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45: 875–884. [DOI] [PubMed] [Google Scholar]
- Marmeisse, R., G. F. J. M. Van den Ackerveken, T. Goosen, P. J. G. M. de Wit and H. W. J. Van den Broek, 1993. Disruption of the avirulence gene avr9 in two races of the tomato pathogen Cladosporium fulvum causes virulence on tomato genotypes with the complementary resistance gene Cf9. Mol. Plant Microbe Interact. 6: 412–417. [Google Scholar]
- Martin, G. B., A. J. Bogdanove and G. Sessa, 2003. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61. [DOI] [PubMed] [Google Scholar]
- McBride, K. E., and K. R. Summerfelt, 1990. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14: 269–276. [DOI] [PubMed] [Google Scholar]
- Michelmore, R. W., and B. C. Meyers, 1998. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8: 1113–1130. [DOI] [PubMed] [Google Scholar]
- Panter, S. N., K. E. Hammond-Kosack, K. Harrison, J. D. G. Jones and D. A. Jones, 2002. Developmental control of promoter activity is not responsible for mature onset of Cf-9B-mediated resistance to leaf mold in tomato. Mol. Plant Microbe Interact. 15: 1099–1107. [DOI] [PubMed] [Google Scholar]
- Parniske, M., and J. D. G. Jones, 1999. Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc. Natl. Acad. Sci. USA 96: 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., K. E. Hammond-Kosack, C. Golstein, C. M. Thomas, D. A. Jones et al., 1997. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91: 821–832. [DOI] [PubMed] [Google Scholar]
- Parniske, M., B. B. H. Wulff, G. Bonnema, C. M. Thomas, D. A. Jones et al., 1999. Homologues of the Cf-9 disease resistance gene (Hcr9s) are present at multiple loci on the short arm of tomato chromosome 1. Mol. Plant Microbe Interact. 12: 93–102. [DOI] [PubMed] [Google Scholar]
- Rick, C. M., J. F. Fobes and M. Holle, 1977. Genetic variation in Lycopersicon pimpinellifolium: evidence of evolutionary change in mating systems. Plant Syst. Evol. 127: 139–170. [Google Scholar]
- Rivas, S., and C. M. Thomas, 2002. Recent advances in the study of tomato Cf resistance genes. Mol. Plant Pathol. 3: 277–282. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001 Molecular Cloning: A Laboratory Manual, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Takken, F. L. W., and M. H. A. J. Joosten, 2000. Plant resistance genes: their structure, function and evolution. Eur. J. Plant Pathol. 106: 699–713. [Google Scholar]
- Thrall, P. H., and J. J. Burdon, 2003. Evolution of virulence in a plant host-pathogen metapopulation. Science 299: 1735–1737. [DOI] [PubMed] [Google Scholar]
- Tigchelaar, E. C., 1984. Collections of isogenic tomato stocks. Rep. Tomato Genet. Coop. 34: 55–57. [Google Scholar]
- Van der Beek, J. G., R. Verkerk, P. Zabel and P. Lindhout, 1992. Mapping strategy for resistance genes in tomato based on RFLPs between cultivars: Cf9 (resistance to Cladosporium fulvum) on chromosome 1. Theor. Appl. Genet. 84: 106–112. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R. A. L., F. Laurent, R. Roth and P. J. G. M. de Wit, 2000. Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4-Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13: 439–446. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R. A. L., M. Kruijt, R. Roth, B. F. Brandwagt, M. H. A. J. Joosten et al., 2001. a Intragenic recombination generated two distinct Cf genes that mediate AVR9 recognition in the natural population of Lycopersicon pimpinellifolium. Proc. Natl. Acad. Sci. USA 98: 10493–10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn, R. A. L., R. Roth and P. J. G. M. de Wit, 2001. b Identification of distinct specificity determinants in resistance protein Cf-4 allows construction of a Cf-9 mutant that confers recognition of avirulence protein Avr4. Plant Cell 13: 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff, B. B. H., C. M. Thomas, M. Smoker, M. Grant and J. D. G. Jones, 2001. Domain swapping and gene shuffling identify sequences required for induction of an Avr-dependent hypersensitive response by the tomato Cf-4 and Cf-9 proteins. Plant Cell 13: 255–272. [DOI] [PMC free article] [PubMed] [Google Scholar]