Abstract
In yeast, four-stranded, biparental “joint molecules” containing a pair of Holliday junctions are demonstrated intermediates in the repair of meiotic double-strand breaks (DSBs). Genetic and physical evidence suggests that when joint molecules are resolved by the cutting of each of the two Holliday junctions, crossover products result at least most of the time. The double-strand break repair (DSBR) model is currently accepted as a paradigm for acts of DSB repair that lead to crossing over. In this study, a well-defined mammalian gene-targeting assay was used to test predictions that the DSBR model makes about the frequency and position of hDNA in recombinants generated by crossing over. The DSBR model predicts that hDNA will frequently form on opposite sides of the DSB in the two homologous sequences undergoing recombination [half conversion (HC); 5:3, 5:3 segregation]. By examining the segregation patterns of poorly repairable small palindrome genetic markers, we show that this configuration of hDNA is rare. Instead, in a large number of recombinants, full conversion (FC) events in the direction of the unbroken chromosomal sequence (6:2 segregation) were observed on one side of the DSB. A conspicuous fraction of the unidirectional FC events was associated with normal 4:4 marker segregation on the other side of the DSB. In addition, a large number of recombinants displayed evidence of hDNA formation. In several, hDNA was symmetrical on one side of the DSB, suggesting that the two homologous regions undergoing recombination swapped single strands of the same polarity. These data are considered within the context of modified versions of the DSBR model.
HOMOLOGOUS recombination is the process by which two identical or very similar DNA sequences undergo an exchange of genetic information. Homologous recombination is of fundamental importance in living organisms; in meiosis, it generates genetic diversity that is important for enhancing species diversity, and in mitosis, it aids in the repair of replication errors and other forms of DNA damage, such as double-strand breaks (DSBs). However, homologous recombination is also potentially deleterious; it can generate chromosomal aberrations and lead to the homozygosis of recessive oncogenes seen in some human cancers. There are two major outcomes of homologous recombination: gene conversion, a nonreciprocal form of recombination in which genetic information on a recipient chromosome is replaced with genetic information from a donor chromosome, and crossing over, in which genetic exchange between chromosomes results in the generation of reciprocal products.
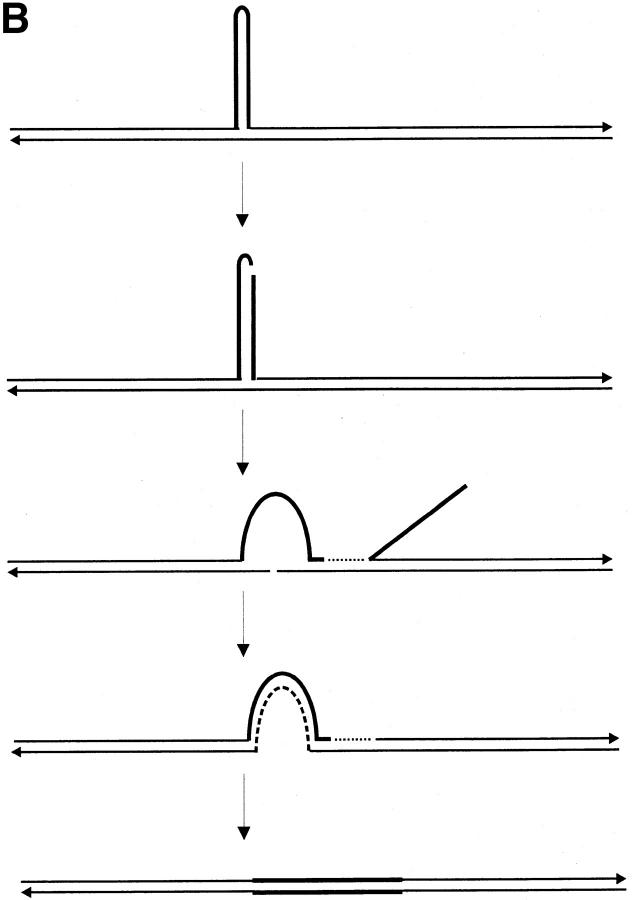
One mechanism for repairing a DSB by homologous recombination is presented in Figure 1. The double-strand break repair (DSBR) model was proposed to explain the repair of a DSB in a sequence insertion (“ends-in”) plasmid during gene targeting in Saccharomyces cerevisiae (Orr-Weaver et al. 1981; Orr-Weaver and Szostak 1983), and features of it are compatible with yeast meiotic recombination (Resnick 1976; Szostak et al. 1983; Schwacha and Kleckner 1994, 1995; Gilbertson and Stahl 1996; Foss et al. 1999; Allers and Lichten 2001; Hunter and Kleckner 2001). The DSBR model is currently accepted as a paradigm for acts of DSB repair that lead to crossing over (Allers and Lichten 2001; Hunter and Kleckner 2001). Following the DSB, the DSBR model proposes that 3′ single-strand tails will be generated by exonuclease activity. In yeast meiosis, 5′–3′ resection of the DNA ends yields 3′ tails that can exceed 1000 nucleotides (nt; Cao et al. 1990; Sun et al. 1991; Bishop et al. 1992). The processing of DSBs into 3′ overhangs is also observed in phage λ (Hill et al. 1997). In mammalian cells, 5′–3′ resection occurs at the ends of transfected DNA (Henderson and Simons 1997) and at meiotic DSBs (Zenvirth et al. 2003). In the DSBR model, 3′ single-strand tails invade a homologous duplex, prime new DNA synthesis, and ligate to the 5′ ends creating the canonical double Holliday junction intermediate. DNA forms consistent with D-loops, and intermediates bearing single and double Holliday junctions have been isolated from meiotic yeast cells (Bell and Byers 1983; Collins and Newlon 1994; Schwacha and Kleckner 1994, 1995; Allers and Lichten 2001; Hunter and Kleckner 2001). The initial strand invasion events are predicted to generate two regions of asymmetric heteroduplex DNA (hDNA), one on each side of the DSB. DNA synthesis from the invading strands generates a gene conversion tract toward the chromosomal sequence on each side of the DSB. Outward branch migration of each Holliday junction may generate symmetrical hDNA on both sides of the DSB (Cunningham et al. 1980; West et al. 1983). The double Holliday junction intermediate is resolved by junction cutting. A crossover results from opposite-sense cleavage of the Holliday junctions and can be of two types. The product illustrated in Figure 1F(i) results from cutting of the noncrossing strands at the junction to the left of the DSB (vertical cutting) and the crossing strands at the junction to the right of the DSB (horizontal cutting), while the product in Figure 1F(ii) results from horizontal-vertical cutting of the same junctions, respectively. In the absence of cellular mismatch repair (MMR), Figure 1F(i) and (ii) illustrate the predicted configuration of hDNA and gene conversion tracts in the two recombinant products (asymmetric hDNA, A; symmetrical hDNA, S; gene conversion toward the chromosomal sequence, GCC). The canonical DSBR model predicts random cleavage of the Holliday junctions, generating the crossover products illustrated in (i) and (ii) with equal frequency. However, evidence suggests that the mode of resolution is nonrandom, with the preferred mode involving the cutting of like strands in one of which there is newly synthesized DNA near the junction [Figure 1F(i); Gilbertson and Stahl 1996; Foss et al. 1999; Baker and Birmingham 2001]. In yeast meiosis, Holliday junction cleavage in the preferred sense is observed in ∼84–89% of crossover events (Foss et al. 1999; Merker et al. 2003). The double Holliday junction intermediate may also give rise to various noncrossover products, depending on whether resolution involves the cleavage of two, one, or no Holliday junctions as explained in Gilbertson and Stahl (1996)(not shown).
Figure 1.—
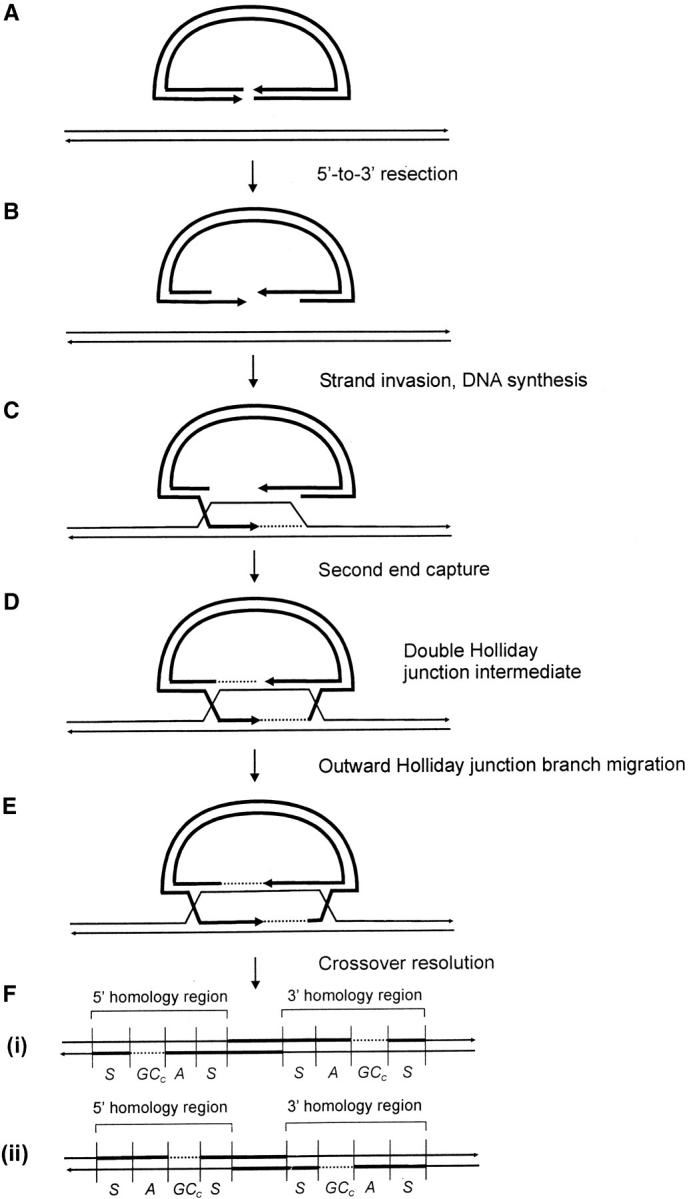
Crossing over according to the canonical DSBR model. The process of homologous recombination that leads to crossing over between a sequence insertion (“ends-in”) gene-targeting vector and the cognate chromosomal locus is depicted. The gene-targeting vector is indicated by the thick lines, while the chromosome is indicated by the thin lines. Dashed lines indicate regions of newly synthesized DNA. Refer to text for details.
If the two recombining DNAs contain sequence differences, one or more mismatches in hDNA might be generated, and these can affect the recombination outcome in several ways. As an aid to describing crossover recombinants that arise according to the gene-targeting scheme in Figure 1, a variation of the nomenclature used to describe meiotic recombination events in eight-spored fungi was adopted (Petes et al. 1991). This is permissible because products that would normally segregate to different chromosomes during meiotic recombination in fungi are preserved in gene targeting with a sequence insertion vector as 5′ and 3′ homologous repeats on the same chromosome (Figure 1). Figure 2 illustrates a sequence insertion vector aligned with the corresponding homologous region on the chromosome. The two sequences differ by a marker pair positioned arbitrarily to the “right” of the DSB. One recombinant type (normal 4:4 segregation) displays the vector-borne (solid circle) marker in one homology region and the chromosomal (open circle) marker in the other. Any pattern that differs from normal 4:4 segregation is referred to as aberrant segregation. Aberrant segregation that results in vector-borne or chromosomal markers in the 5′ and 3′ homology regions is referred to as full conversion (denoted FC) or as half-conversion (HC) if the recombinants contain both vector-borne and chromosomal markers. In yeast and fungi, HC is also referred to as postmeiotic segregation (Esposito 1971; Petes et al. 1991). Full conversions fall into the 6:2 (open chromosomal marker in both 5′ and 3′ homology regions) or 2:6 (solid vector-borne marker in both 5′ and 3′ homology regions) classes, signifying the transfer of information from both strands of a donor molecule to the recipient. Half-conversions fall into the 5:3 or 3:5 classes, in which information from a single strand of the donor molecule is transferred to the recipient. In HC, one position in the recombinant has a sectored genotype, while the nonsectored site may contain either a vector-borne or a chromosomal marker. The aberrant 4:4 class (Ab 4:4) is sectored for both sites in the 5′ and 3′ homology regions, with the vector-borne and chromosomal markers in trans configuration as a result of crossing over.
Figure 2.—
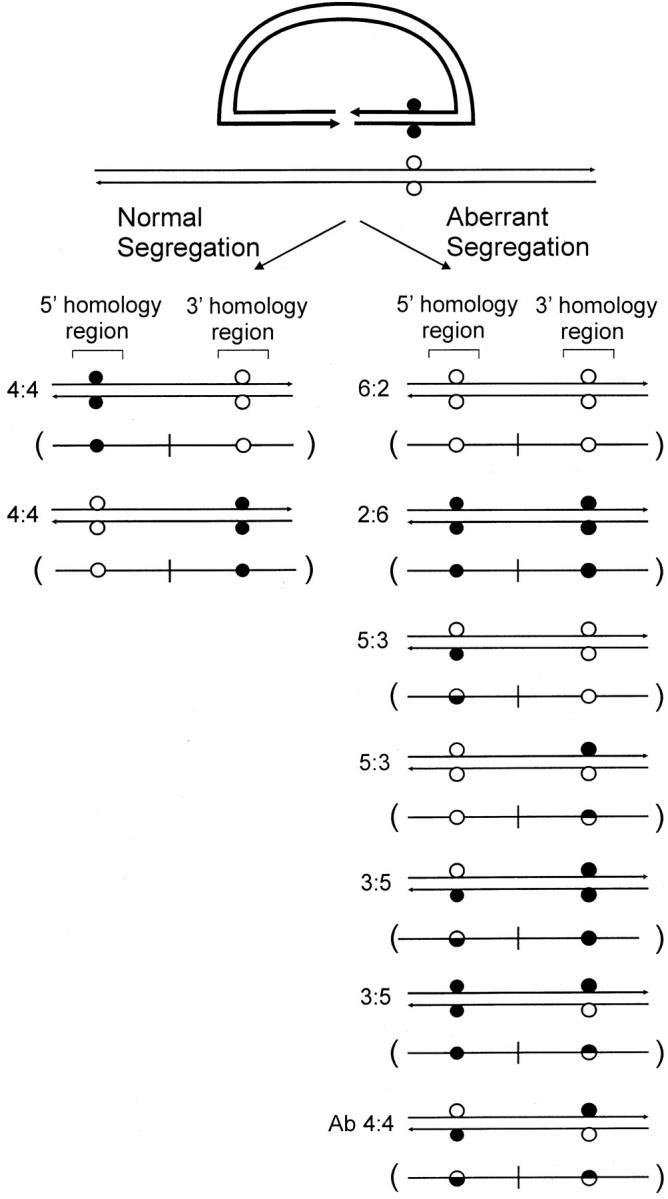
Recombinant segregation patterns. Nomenclature used to describe meiotic recombination events in eight-spored fungi is adopted to characterize equivalent outcomes for gene targeting with a sequence insertion vector. For each segregation class, the double-strand product is represented in a more simplified, single-strand form in parentheses, a scheme adopted in Figures 4–6 and 8. Refer to text for details.
In this study, we exploited a well-defined gene-targeting assay to investigate crossing-over mechanisms in mammalian cells in culture. The sequence insertion plasmid bears ∼7 kb of homology to the target locus, namely, the haploid, chromosomal immunoglobulin μ gene constant (Cμ) region. Within the vector-borne Cμ region of homology, there are three unique restriction enzyme sites for introducing the recombination-initiating DSB, and at regular intervals the vector-borne Cμ region is marked by small palindrome insertions, which are semirefractory to MMR (Nag et al. 1989; Bollag et al. 1992; Donoho et al. 1998; Li and Baker 2000). The palindrome insertions facilitated the detection of hDNA in the recombinants and allowed us to test several features of the crossing-over reaction predicted by the DSBR model.
MATERIALS AND METHODS
Recipient hybridoma and gene-targeting vector:
The gene-targeting system exploits the wild-type murine hybridoma cell line Sp6/HL, which bears a single copy of the trinitrophenyl (TNP)-specific chromosomal immunoglobulin μ heavy chain gene (Köhler and Shulman 1980; Köhler et al. 1982) that serves as the target for homologous recombination with the 12.4-kb sequence insertion vector pTCμEn−4pal. This vector was constructed by inserting a 7016-bp HindIII/EcoRI wild-type Cμ-region fragment into a derivative of pSV2neo (Southern and Berg 1982) in which the 372-bp NsiI/NdeI fragment encompassing the SV40 early region enhancer important in neo expression had been deleted. As shown previously (Bautista and Shulman 1993; Ng and Baker 1998), the enhancer-trap feature enriches for gene-targeting events at the chromosomal immunoglobulin μ-locus. To permit detection of hDNA, the endogenous MfeI, AflII, KpnI, and XhoI sites within the vector-borne Cμ region were replaced by insertion of a 30-bp palindrome (5′-GTACTGTATGTGCGGCCGCACATACAGTAC-3′; Li and Baker 2000). The palindrome was engineered with a unique NotI restriction enzyme site for identification (indicated by underlining). To permit ligation, each palindrome was synthesized with terminal nucleotides appropriate to the endogenous site being replaced (indicated in lowercase italics in the following sequences) and then self-annealed; for palindrome insertion at the MfeI site, the sequence 5′-aattcGTACTGTATGTGCGGCCGCACATACAGTACg-3′ was synthesized, whereas, for insertion at the AflII site, the sequence was 5′-ttaacGTACTGTATGTGCGGCCGCACATACAGTACg-3′. In the case of the KpnI site, the sequence was 5′-GTACTGTATGTGCGGCCGCACATACAGTACgtac-3′, while for XhoI, it was 5′-tcgaGTACTGTATGTGCGGCCGCACATACAGTAC-3′. Insertion of a single palindrome at each site was confirmed by restriction enzyme mapping. Three unique restriction enzyme sites within the vector-borne Cμ region (Esp3I, Eco47III, and SpeI) are available for introduction of the recombination-initiating DSB. With the exception of the small palindrome insertions, the vector-borne and chromosomal Cμ regions are isogenic. Palindrome-containing plasmids were propagated as reported earlier (Li and Baker 2000).Vector construction and plasmid recovery were performed according to standard procedures (Sambrook et al. 1989).
Recovery and characterization of targeted recombinants:
Transfer of vector DNA into the hybridoma cells was performed by electroporation under conditions described previously (Baker et al. 1988). The limited dilution cloning procedures used to recover independent G418-resistant (G418R) recombinants representing the progeny of single G418R cells have been described (Ng and Baker 1999a; Li and Baker 2000). A combination of Southern and PCR analysis was used in recombinant identification as described (Ng and Baker 1999a; Li and Baker 2000). Genomic DNA was prepared according to the procedures of Gross-Bellard et al. (1973). Restriction enzymes were purchased from New England Biolabs (Beverly, MA), Bethesda Research Laboratories (Gaithersburg, MD), and Pharmacia (Piscataway, NJ) and were used in accordance with the manufacturers' specifications. Gel electrophoresis, blotting, 32P-probe preparation, and hybridization were all performed by standard procedures (Sambrook et al. 1989).
The conditions used for PCR have been described (Ng and Baker 1999a). For amplification of the 7459-bp endogenous chromosomal Cμ region, forward and reverse primers 9931FCμ, 5′-GCAAGAGTGAGTAGAGCTGGCTGG-3′, and 17390RCμ, 5′-GGTTCGGTTCTGTCTGCACTACTC-3′, respectively, were utilized. In targeted recombinants, PCR amplification of the 5′ Cμ region was performed with forward and reverse primers 9931FCμ and AB9745, respectively, while for the 3′ Cμ region, the forward and reverse primers were AB15527 and 17390RCμ, respectively. The DNA sequence and binding site of primer AB9745 were reported previously (Li and Baker 2000). The forward primer AB15527 (5′-CCTTGTGGTCAGTGTTCATCTGCT-3′) binds to the coding strand of the vector-borne neo gene, while the reverse primer 17390RCμ binds to the noncoding strand outside the 3′ border of the vector-borne region of homology to the chromosome. DNA sequence analysis was performed on an ABI Prism automated sequencer (model 3100) according to standard procedures.
RESULTS
Experimental system:
The experimental system has been described previously (Baker et al. 1988; Ng and Baker 1999a; Li and Baker 2000). In brief, the haploid chromosomal immunoglobulin μ gene constant (Cμ) region in a mouse hybridoma cell line serves as the target for homologous recombination with a transfected pSV2neo-based enhancer-trap sequence insertion vector linearized within the Cμ region of homology (Figure 3A). The enrichment in crossover gene-targeting events provided by the enhancer-trap insertion vector, together with recovery of recombinants by limited dilution cloning, makes it highly likely that the product(s) of individual crossover reactions are retained for molecular analysis (Ng and Baker 1999a; Li and Baker 2000; Baker and Birmingham 2001). Therefore, all segregation classes in Figure 2 can, in principle, be represented among the recombinants.
Figure 3.—
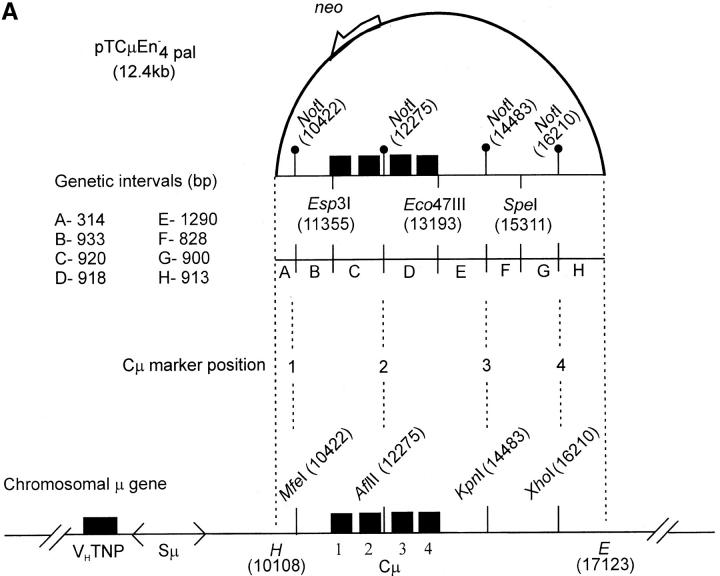
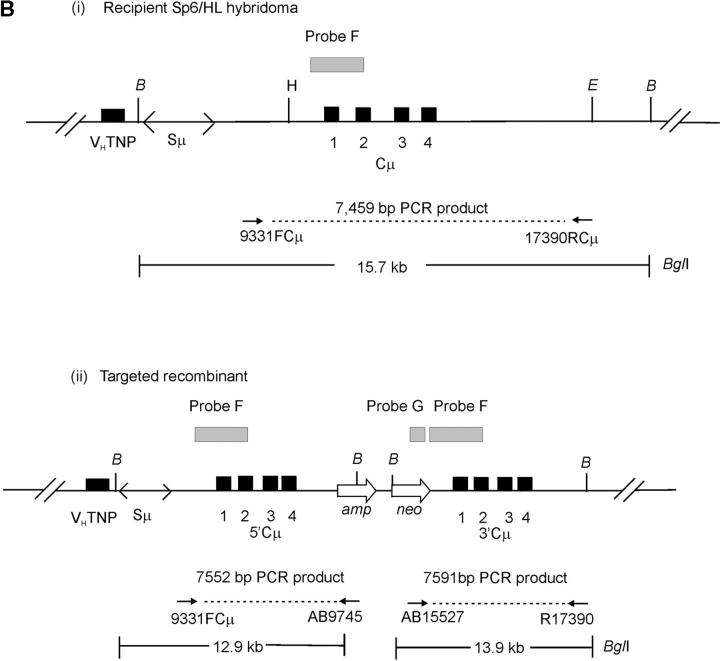
Vector and chromosomal DNA structures. (A) Enhancer-trap gene-targeting vector. The vector pTCμEn−4pal contains a 7016-bp segment of homology to the Cμ region of the mouse chromosomal immunoglobulin μ-gene inserted into a derivative of pSV2neo in which the SV40 early region enhancer has been deleted. The vector-borne Cμ region of homology is interrupted at regular intervals by insertion of a 30-bp palindrome sequence containing a diagnostic NotI restriction enzyme site (Li et al. 1999), which replaces endogenous MfeI, AflII, KpnI, and XhoI sites. Unique Esp3I, Eco47III, and SpeI recognition sequences within the vector-borne Cμ region permit introduction of the recombination-initiating DSB. As the genetic intervals (A–H) indicate, flanking palindrome markers reside approximately equidistantly from each DSB site. The numbering system used to denote restriction enzyme sites is based on the genomic wild-type μ gene sequence (Goldberg et al. 1981; Bilofsky et al. 1986). (B) Chromosomal immunoglobulin μ-gene structures. (i and ii) The structures of the haploid chromosomal μ-gene in the recipient Sp6/HL and targeted recombinant hybridoma cell lines, respectively. Diagnostic bands that can be detected through PCR and Southern analysis are indicated. DNA probe fragments used in Southern analysis include probe F, an 870-bp XbaI/BamHI fragment, and probe G, a 762-bp PvuII fragment from the neo gene of pSV2neo. VHTNP, TNP-specific chromosomal immunoglobulin heavy chain variable region; Sμ, immunoglobulin μ-gene switch region; Cμ, four exons composing the immunoglobulin μ-gene constant region; B, BglI; E, EcoRI; H, HindIII. The diagrams are not drawn to scale.
As shown in Figure 3A, the vector-borne Cμ region contains four 30-bp palindrome insertions containing a diagnostic NotI site (Li and Baker 2000) that replace the endogenous μ gene MfeI, AflII, KpnI, and XhoI sites. In addition, three unique restriction enzyme sites (Esp3I, Eco47III, and SpeI) permit a recombination-initiating DSB to be introduced at different positions within the Cμ region. Small palindromes are useful in recombination studies because they form hairpin loops when encompassed in heteroduplexes with the wild-type sequence (Nag et al. 1989). These structures are semirefractory to MMR in yeast (Nag et al. 1989; Detloff et al. 1991, 1992; Porter et al. 1993) and mammalian cells (Bollag et al. 1992; Donoho et al. 1998; Li and Baker 2000). Thus, a colony derived from a single cell in which hDNA has formed at a particular site during recombination may have a mixed or “sectored” genotype, consisting of two cell populations: one bearing the palindrome marker, and the other, the corresponding endogenous restriction enzyme site. In the vector, a NotI palindrome resides at approximately the same distance to the left and right of each DSB site. Thus, from recombinants derived with each cut vector, information about the disposition of hDNA during repair of the DSB can be obtained.
Two separate electroporations were performed with each linearized vector, and independent G418R transformants arising from the expansion of single G418R cells were recovered by limited dilution cloning in 96-well plates as described previously (Ng and Baker 1999a; Li and Baker 2000). In total, the transfections yielded 3377 independent G418R transformants, of which the first 924 cell lines were analyzed to identify correctly targeted recombinants. Initial screening by PCR eliminated 598 G418R transformants bearing an unmodified chromosomal μ-locus as revealed by the specific 7459-bp fragment using primers 9931FCμ and 17390RCμ [Figure 3B(i); data not shown]. The remaining 326 G418R transformants were examined in Southern analysis of BglI-digested genomic DNA using chromosome and vector-specific DNA probes to positively identify the 5′ and 3′ Cμ-region duplication characteristic of correctly targeted recombinants bearing a single integrated vector copy [Figure 3B(ii); data not shown]. This second screening revealed 65 targeted G418R recombinants. Of these, the diagnostic Cμ duplication [Figure 3B(ii)] was evident in 63 recombinants, which were subjected to genetic marker analysis as described below, while in the other two recombinants, the chromosomal μ-locus contained a tandem Cμ-region triplication indicative of the targeted integration of two vector copies (Ng and Baker 1999b). The remaining 236 cell lines were random G418R transformants bearing an intact chromosomal μ-locus [the 15.7-kb BglI fragment as indicated in Figure 3B(i)], while in 25 G418R transformants, the chromosomal μ-locus was absent according to Cμ probe F binding. Cell lines of the latter category have been observed previously, and although they have not been analyzed in detail, might represent a form of illegitimate integration of the targeting vector into the μ-locus (Baker and Read 1993).
The mean frequency of targeted recombinants among the random G418R transformants was similar for transfections involving the Esp3I-, Eco47III-, and SpeI-cut vectors, namely, ∼6, ∼9, and ∼8%, respectively (data not shown), suggesting that the position of the vector-borne DSB site did not influence homologous recombination. This is not surprising, given that the length of the homologous segments flanking each DSB site (Figure 3A) is above the ∼1 kb reported previously as sufficient for promoting mammalian gene targeting (Shulman et al. 1990; Hasty et al. 1991; Thomas et al. 1992).
Assignment of Cμ-region genetic markers:
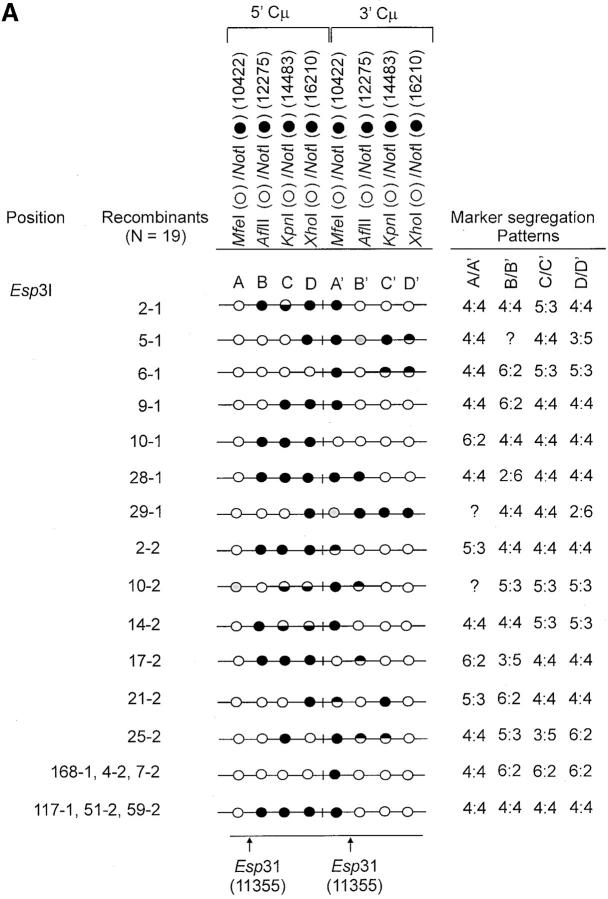
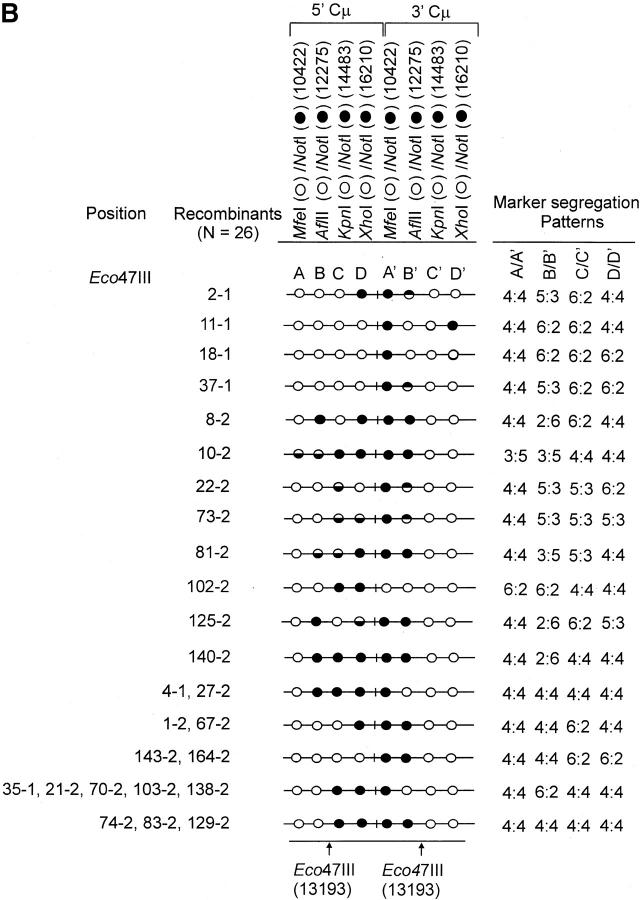
The determination of the Cμ-region genetic markers in the 63 recombinants was performed according to PCR and gel analysis methods described previously (Ng and Baker 1999a). As indicated in Figure 3B(ii), PCR amplification using primer pair 9931FCμ/AB9745 generates a specific 7552-bp product from the 5′ Cμ region while primer pair AB15527/17390RCμ generates a specific 7592-bp product from the 3′ Cμ region. The 5′ and 3′ Cμ-region PCR products were tested for their sensitivity to cleavage with restriction enzymes diagnostic of each endogenous Cμ-region site (MfeI, AflII, KpnI, and XhoI) as well as NotI, specific for the vector-borne palindrome (Figure 3A), and the fragment sizes resolved by standard gel electrophoresis. The Cμ-region marker patterns in the individual recombinants as determined from these digests (data not shown) are presented in Figure 4, A–C. Recombinants recovered from the two separate electroporations are identified by the coding “-1” and “-2”. Nomenclature for describing meiotic recombination in eight-spored fungi was adopted for purposes of characterizing the allele segregation patterns in the 5′ and 3′ Cμ regions in the recombinants. The half-solid circles denoting positions of 5:3 segregation are presented with the vector-borne palindrome sequence on the bottom strand in the 5′ Cμ region and on the top strand in the 3′ Cμ region. This format is consistent with the marker pattern that is predicted from cleavage of the double Holliday junction intermediate in the preferred sense [i.e., resolution of the type shown in Figure 1F(i); Gilbertson and Stahl 1996; Foss et al. 1999; Baker and Birmingham 2001], although it is recognized that a minority of resolutions may generate the product shown in Figure 1F(ii) (Foss et al. 1999; Merker et al. 2003). Note that in seven recombinants [cell lines 5-1, 29-1, and 10-2 generated with the Esp3I-cut vector (Figure 4A) and cell lines 8-1, 9-1, 28-1, and 89-2 generated with the SpeI-cut vector (Figure 4C)], a single position within one of the Cμ regions is devoid of a genetic marker (denoted by a shaded circle). In six of these recombinants, the missing marker made it difficult to categorize segregation on one side of the DSB, and these positions are denoted by a question mark.
Figure 4.—
Genetic marker analysis. The genetic marker patterns are presented for the 5′ and 3′ Cμ-region duplication in independent recombinants derived following transfection with the (A) Esp3I-cut, (B) Eco47III-cut, and (C) Spe-I-cut enhancer-trap vectors. For simplicity, unnecessary details of the recombinant Cμ-region duplication shown in Figure 3B are omitted. The Cμ-region positions that are sensitive to cleavage with one of the chromosomal restriction enzymes (MfeI, AflII, KpnI, and XhoI) are indicated by open circles, while those sites that are cleaved with NotI, diagnostic of the vector-borne palindrome sequence, are denoted by solid circles. The Cμ-region marker positions exhibiting a mixed cleavage pattern with NotI and the corresponding chromosomal restriction enzyme are denoted by half-solid circles (sectored sites). In those instances in which sectored sites resided in both members of the recombinant Cμ-region duplication, the indicated “trans” marker configuration was established through analysis of individual subclones (originating from a single cell) as detailed earlier (Li and Baker 2000). Those Cμ positions devoid of either a chromosomal or a vector marker are indicated by shaded circles. The position corresponding to the DSB site is presented at the bottom of each diagram relative to the genetic markers in the 5′ and 3′ Cμ regions. The segregation class of each pair of allelic markers is based on the nomenclature in Figure 2.
Recombinants displaying normal 4:4 segregation on both sides of the DSB:
In 8 of the 63 recombinants, specifically, 117-1, 51-2, and 59-2 generated with the Esp3I-cut vector (Figure 4A), 74-2, 83-2, and 129-2 generated with the Eco47III-cut vector (Figure 4B), and 1-1 and 24-1 generated with the SpeI-cut vector (Figure 4C), the 5′ Cμ region is characterized by a chromosomal marker(s) (open circles) to the “left” of the DSB and a vector-borne marker(s) (solid circles) to the “right” of the DSB, while the reverse marker pattern is evident in the 3′ Cμ region. To a first approximation, these marker configurations are consistent with the possibility of a crossing-over event at or near the vector-borne DSB. The preservation of vector-borne and chromosomal markers in the 5′ and 3′ Cμ repeats in the recombinants resembles that observed when meiotic chromosomes segregate in the absence of recombination, and therefore we refer to this pattern as normal 4:4 marker segregation (Figure 2). The remaining 55 recombinants bore marker patterns that differed from normal 4:4 segregation. The main features of these aberrant segregations are described in the next sections.
Full conversion tracts are predominantly one-sided:
Our data reveal that FC events in the direction of the unbroken chromosomal sequence (6:2 segregation) are frequently observed on only one side of the DSB (i.e., they are predominantly one-sided). Although the interpretation of recombinants is complicated somewhat by the multiple markers, the following relations are suggested from the information presented in Figure 4, A–C: 6:2 on one side of the DSB and 3:5 on the other were observed 1/63 or 0.02% of the time; 6:2 on one side of the DSB and 2:6 on the other were observed 2/63 or 0.03% of the time; 6:2 on one side of the DSB and 5:3 on the other were observed 7/63 or 0.11% of the time; 6:2 on the two sides of the DSB were observed 7/63 or 0.11% of the time; 6:2 on one side of the DSB and 4:4 on the other were observed 20/63 or 0.32% of the time. Thus, FC emanating from the DSB is suggested in ∼59% of the recombinants. However, only a small fraction of FC events resulted in co-conversion of markers on the two sides of the DSB as evidenced by the low frequency (0.11/0.59 = 0.19) of the 6:2, 6:2 segregation class. In the remaining ∼81% of the convertants, FC tracts are one-sided, and ∼67% (0.32/0.48) display normal 4:4 marker segregation on the other side of the DSB. Of the several recombinants displaying this particular asymmetry, a dramatic example is illustrated by the group of recombinants 168-1, 4-2, and 7-2 generated with the Esp3I-cut vector (Figure 4A), in which FC occurs for all markers to the “right” of the DSB (a distance spanning ∼6 kb) without extending another 933 bp to the “left” of the DSB to co-convert the vector-borne palindrome marker. The FC events are not restricted to the “right of the DSB, since recombinants generated with the Eco47III- and SpeI-cut vectors reveal 6:2 segregation events that span markers to the “left” of the DSB.
Designation of hDNA tracts:
Another notable feature are the patterns of hDNA formation in the recombinants. Before presenting these results, we list several simplifying assumptions that were used in assigning hDNA tracts in the recombinants: (1) recombination is initiated by the vector-borne DSB at the unique Esp3I, Eco47III, or SpeI sites; (2) assimilation of single strands into the unbroken recipient chromosome occurs on both sides of the DSB and proceeds in a continuous fashion beginning from the 3′ end as depicted in the DSBR model (Figure 1); (3) mismatches involving the small palindrome are equally correctable; (4) palindrome markers are sufficiently close to each DSB site to permit detection of hDNA on the two sides of the DSB; and (5) Holliday junction resolution to yield the crossover product is nonrandom. A sizable majority of resolutions display a bias in favor of cutting-like strands in one of which there is newly synthesized DNA near the junction (Gilbertson and Stahl 1996; Foss et al. 1999; Baker and Birmingham 2001; Merker et al. 2003).
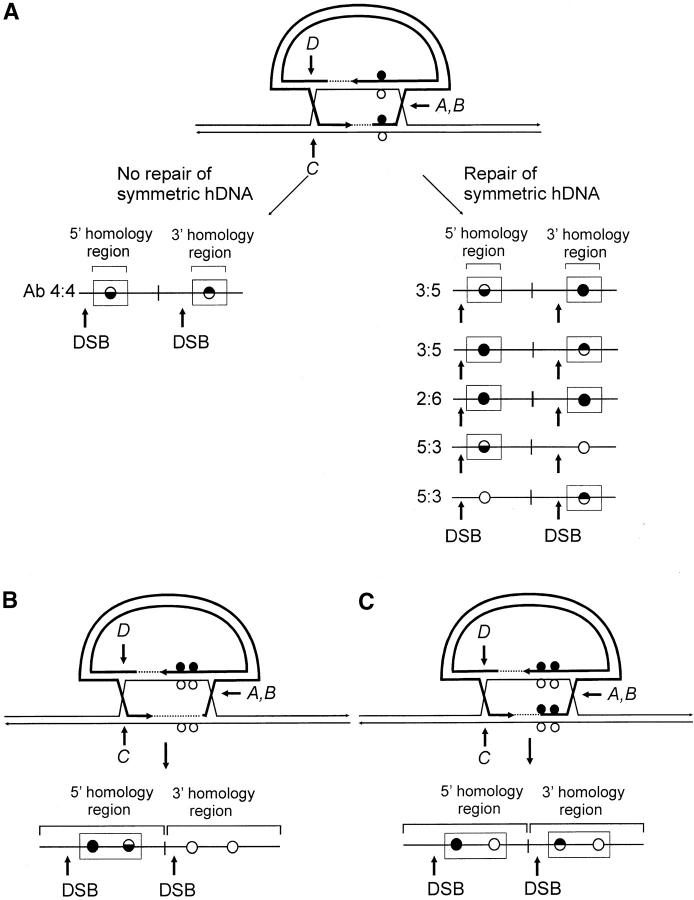
The following additional criteria were used in defining the presence and position of hDNA in the recombinants. As a visual aid to interpretation, Figure 5 presents various possible outcomes of recombination in the case where markers are positioned arbitrarily to the “right” of the DSB. As shown in Figure 5A, the failure to repair mismatches in symmetrical hDNA results in Ab 4:4 marker segregation in the 5′ and 3′ homology regions. In the event of repair, other outcomes are possible. A signature of repair acting on symmetrical hDNA is the appearance of 3:5 and 2:6 marker segregation, in which equivalent positions in the two homology regions contain genetic information from the vector. Another example, 5:3 segregation, denotes formation of hDNA, but it need not be symmetrical. The context of particular repair events can also constitute evidence of hDNA formation. One situation is the case in which a marker repaired in the direction of the vector-borne palindrome resides between a sectored site and the DSB. This pattern suggests that a hDNA tract has spanned the distance from the DSB to the sectored site, as illustrated for the 5′ homology region in the recombinant in Figure 5B. Marker positions B and C in the 5′ Cμ region of recombinant 2-1 generated with the Esp3I-cut vector (Figure 4A) are one example of this pattern; they provide support for the formation of a continuous hDNA tract beginning at or near the DSB and terminating between markers C and D. A second case is where a chromosomal marker is separated from the DSB by a sectored site or a site that has undergone repair in the direction of the vector-borne palindrome. These patterns are suggestive of continuous hDNA tracts spanning the markers (Figure 5C). An example is recombinant 25-2 generated with the Esp3I-cut vector, where continuous hDNA tracts are likely to have spanned marker positions C and D in the 5′ Cμ region and marker positions B′–D′ in the 3′ Cμ region. A third case suggested from the DNA sequencing results presented below can include a Cμ-region position that is devoid of a genetic marker (as denoted in Figure 4A by the shaded circles in recombinants 5-1, 29-1, and 10-2 generated with the Esp3I-cut vector and in Figure 4C in recombinants 8-1, 9-1, 28-1, and 89-2 generated with the SpeI-cut vector).
Figure 5.—
Marker patterns indicative of heteroduplex DNA formation. (A) Marker patterns expected in recombinants in the case in which single mismatches reside in symmetrical hDNA. (B and C) Recombination outcomes in cases in which hDNA tracts contain more than a single mismatch. Marker positions encompassed in hDNA are denoted by enclosure in rectangles. The mismatches are positioned arbitrarily to the “right” of the DSB.
Disposition of hDNA tracts during homologous recombination:
On the basis of the above criteria, a summary of the recombinants in Figure 4, A–C, whose generation is highly likely to have involved formation of a hDNA intermediate is presented in Figure 6, A–C (hDNA is denoted by enclosure of relevant Cμ-region marker positions in rectangles). The results suggest that hDNA formation is frequent, occurring in 11/19 (∼58%) of the recombinants generated with the Esp3I-cut vector, 12/26 (∼46%) of the recombinants generated with the Eco47III-cut vector, and 10/18 (∼56%) of the recombinants generated with the SpeI-cut vector.
Figure 6.—
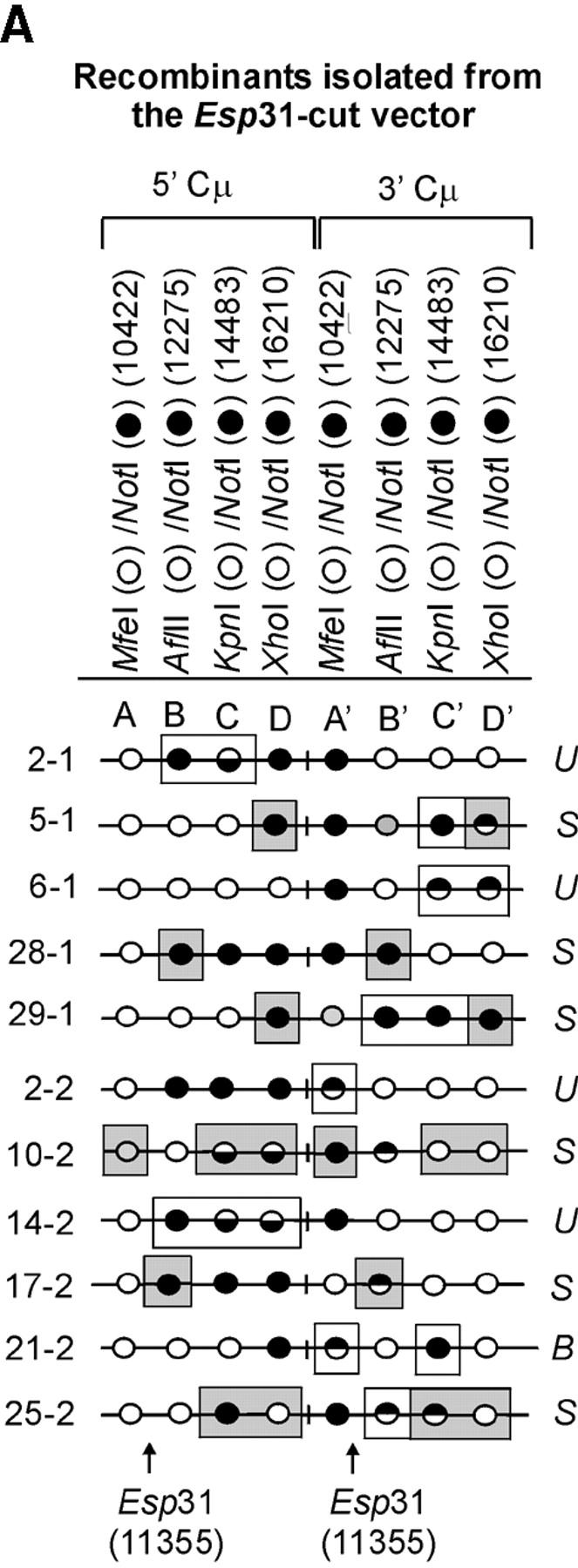
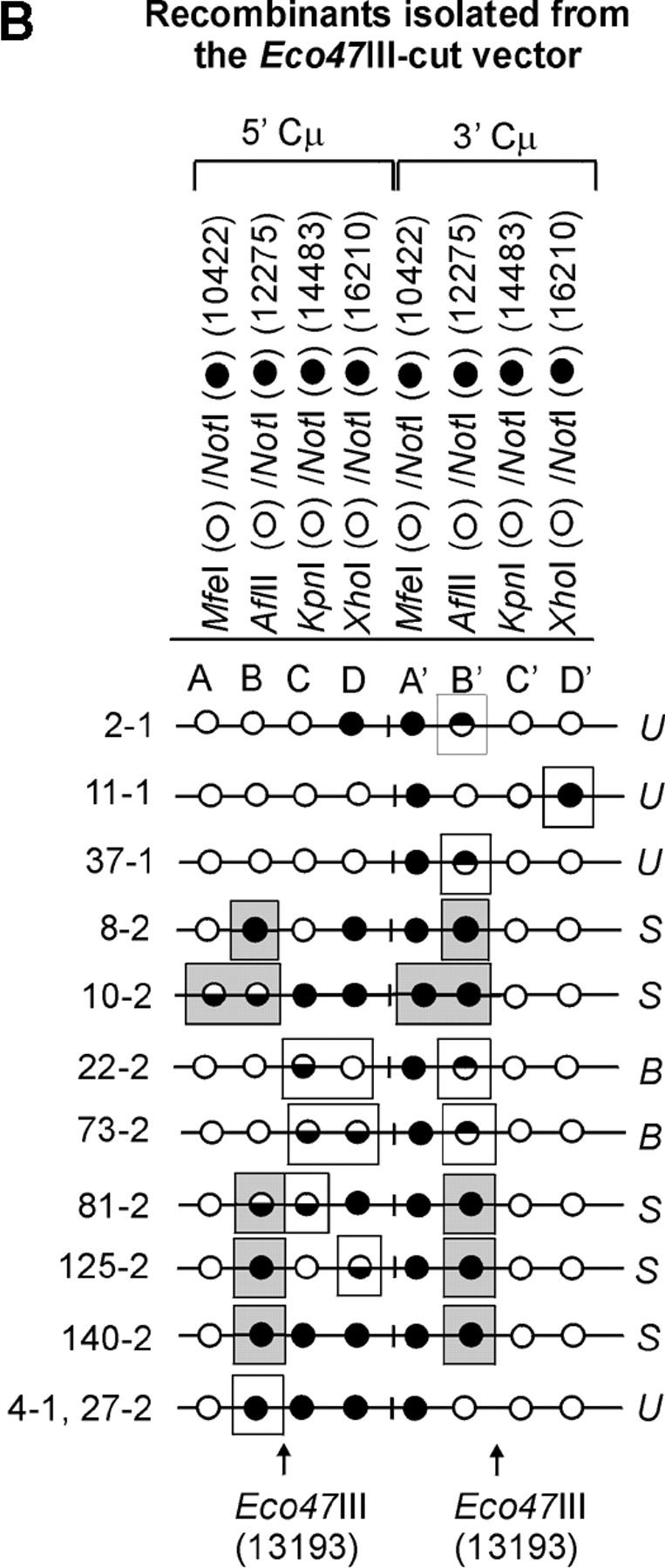
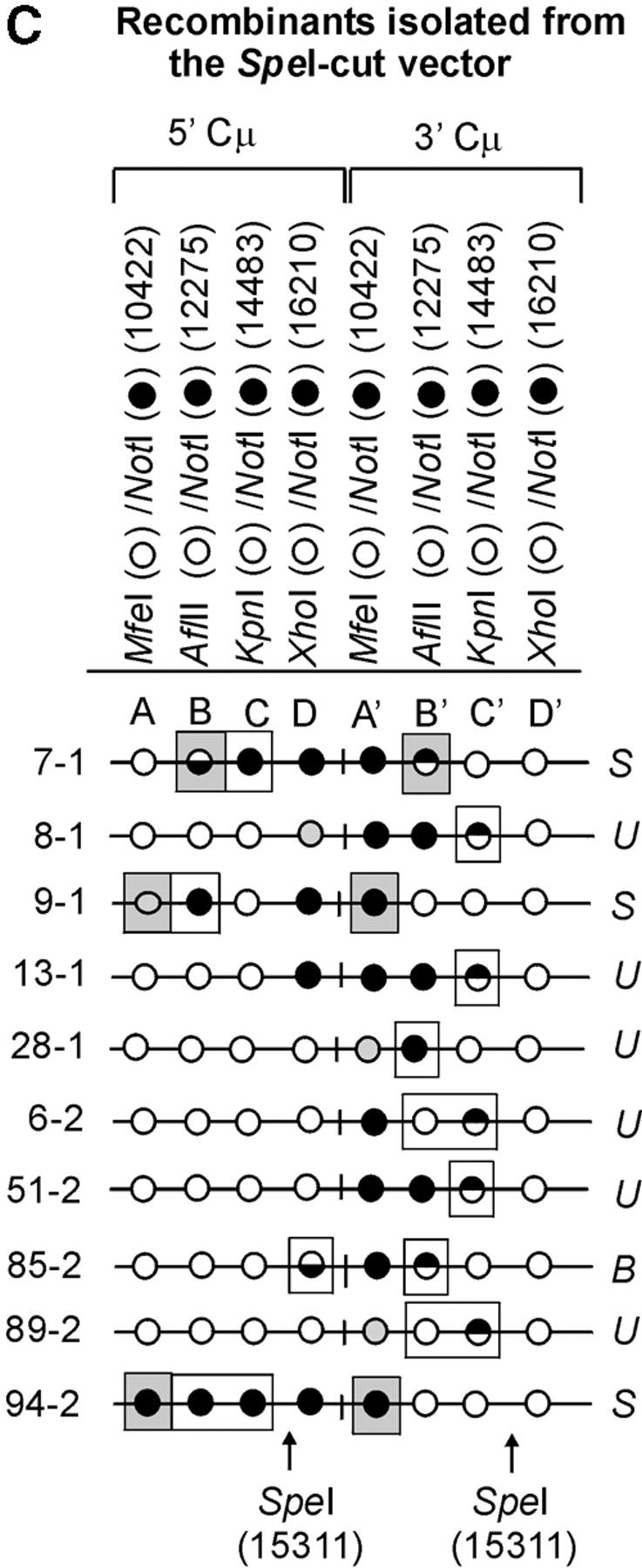
Heteroduplex DNA formation in the recombinants. A subset of recombinants generated with the (A) Esp3I-cut, (B) Eco47III-cut, and (C) SpeI-cut enhancer-trap vectors in which the Cμ-region marker patterns are consistent with formation of hDNA is shown. In the recombinants, minimum tracts of hDNA are indicated by enclosure of relevant Cμ-region marker positions in rectangles. Marker positions that are deemed to reside in symmetrical hDNA are shaded. U, unidirectional hDNA; S, symmetrical hDNA; B, bidirectional hDNA.
The formation of hDNA about the DSB was also extensive. In several recombinants generated with the Esp3I-cut vector (recombinants 5-1, 6-1, 29-1, 10-2, 14-2, and 25-2), the terminal marker located ∼6 kb to the “right” of the DSB was encompassed within hDNA. Essentially the reverse pattern was observed in recombinants generated following transfection with the SpeI-linearized vector (recombinants, 9-1, 28-1, 89-2, and 94-2). When the DSB was positioned in about the middle of the vector-borne region of homology at the Eco47III site, so as to provide ∼3 and ∼4 kb of homology to the “left” and “right” of the DSB, respectively, hDNA tracts were observed to encompass genetic markers at the boundaries of these segments in some recombinants (11-1, 10-2, 22-2, 73-2, and 125-2).
In 14/63 (∼22%) of the recombinants, hDNA tracts are unidirectional in that they reside in one of the Cμ regions either to the “left” or to the “right” of the DSB (recombinants are denoted by the coding “U” in Figure 6). Unidirectional hDNA formation was evident among recombinants generated with each of the cut vectors. The results hinted at the more frequent distribution of unidirectional hDNA to the “right” and “left” of the DSB in recombinants generated with the Esp3I- and SpeI-cut vectors, respectively, which would be consistent with the larger extent of homology available on those sides of the cut vectors. Nevertheless, the expectation that vector cutting at Eco47III, which yields approximately equivalent homology on the two sides of the DSB, would yield equal proportions of events was not fulfilled, although in all cases it is clear that the number of events available for analysis is low.
Bidirectional events (denoted by the coding “B” in Figure 6) are defined as those involving two tracts of 5:3 segregation, one affecting a marker(s) on one side of the DSB in one Cμ region and a second affecting a marker(s) on the opposite side of the DSB in the other Cμ region (i.e., two HC events: 5:3, 5:3 segregation). A notable feature of the present data is the low frequency of this recombinant class. As is evident from Figure 6, a clear 5:3, 5:3 segregation pattern is observed in only 3/63 or ∼5% of the recombinants, namely, 22-2 and 73-2 generated with the Eco47III-cut vector and 85-2 generated with the SpeI-cut vector. In one recombinant (21-2 generated with the Esp3I-cut vector), formation of bidirectional hDNA is also suggested by the particular 3′ Cμ-region marker pattern, namely, the sectored site at position A′ and the site at position C′ that has been repaired to the vector-borne palindrome. In the case of the Esp3I- and SpeI-cut vectors, it might be argued that reduced homology to the “left” and “right” of the DSB, respectively, lowers the efficiency with which the flanking palindrome marker is incorporated into hDNA on one side of the DSB. However, a low frequency of double HC events even among recombinants generated by vector linearization in about the middle of the Cμ region at the unique Eco47III site argues against this possibility.
In a substantial subset of the recombinants (14/63 or ∼22%), one or more pairs of allelic Cμ-region markers display evidence of symmetrical hDNA, suggesting that the two Cμ regions undergoing homologous recombination have swapped single strands of the same polarity. In Figure 6, recombinants in which symmetrical hDNA is suggested are denoted by the coding “S,” and the relevant allelic marker positions are indicated by shading. Evidence for symmetrical hDNA is revealed in 6/19 (∼32%) of the recombinants generated with the Esp3I-cut vector (recombinants, 5-1, 28-1, 29-1, 10-2, 17-2, and 25-2), 5/26 (∼19%) of the recombinants generated with the Eco47III-cut vector (recombinants, 8-2, 10-2, 81-2, 125-2, and 140-2), and 3/18 (17%) of the recombinants generated with the SpeI-cut vector (recombinants, 7-1, 9-1, and 94-2). Furthermore, in recombinant 10-2 generated with the Esp3I-cut vector and in recombinants 81-2 and 125-2 generated with the Eco47III-cut vector, the marker patterns suggest that hDNA has formed on two sides of the DSB in both Cμ regions participating in recombination.
Table 1 summarizes the minimum amount of symmetrical hDNA formation in the 14 recombinants indicated above. In some cases, symmetrical hDNA appears to span one pair of allelic markers producing short tracts (∼0.9 kb). In other recombinants, symmetrical hDNA encompasses two adjacent allelic pairs spanning distances of as much as ∼3 kb. In other instances, symmetrical hDNA tracts span markers over a distance of ∼5 kb on one side of the DSB, and in the case of recombinant 10-2 generated with the Esp3I-cut vector, encompass markers spanning a distance of ∼6 kb on two sides of the DSB.
TABLE 1.
Symmetrical hDNA tracts
| Site of DSB | Recombinant | Minimum estimate of symmetrical hDNA tract length (kb) |
|---|---|---|
| Esp3I | 28-1, 17-2 | 0.9 |
| 5-1, 29-1, 25-24.9 | 4.9 | |
| 10-2 | 5.8a | |
| Eco47III | 8-2, 81-2, 125-2, 140-2 | 0.9 |
| 10-2 | 2.8 | |
| SpeI | 7-1 | 3.0 |
| 9-1, 94-2 | 4.9 |
hDNA spans marker positions on both sides of the DSB in the 5′ and 3′ Cμ regions of homology.
Absence of genetic markers in some Cμ-region positions:
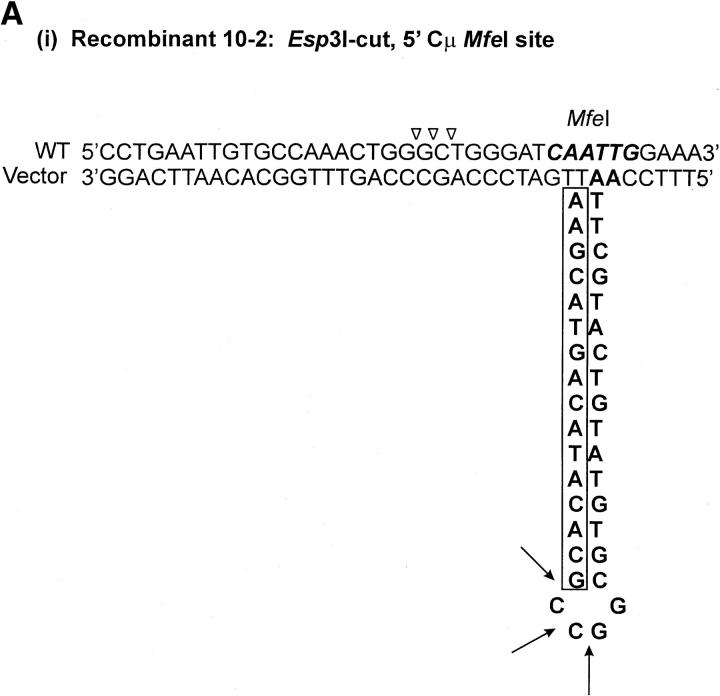
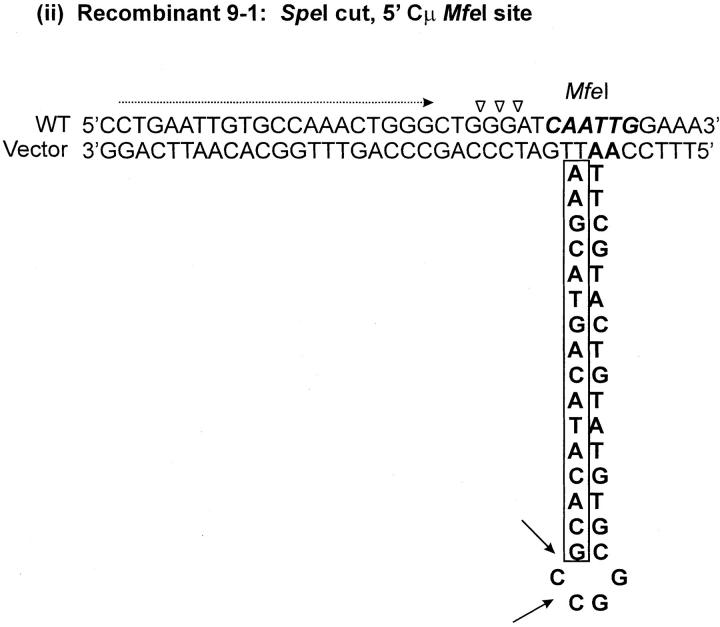
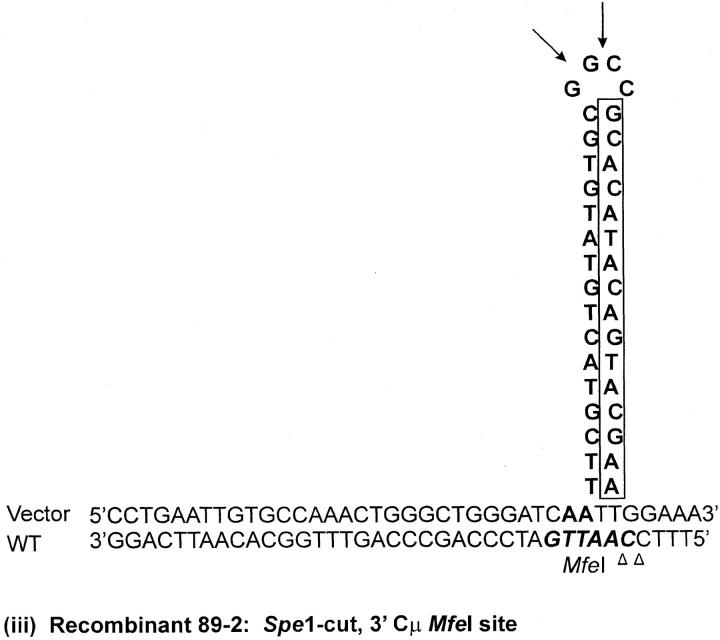
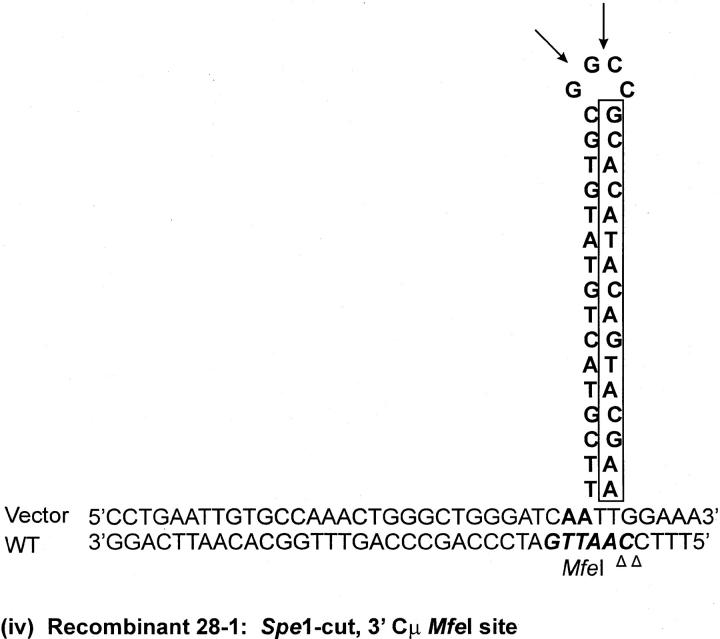
As indicated above, seven recombinants (cell lines 5-1, 29-1, and 10-2 generated with the Esp3I-cut vector and cell lines 8-1, 9-1, 28-1, and 89-2 generated with the SpeI-cut vector) contain a single position within one of the Cμ regions that is devoid of a genetic marker suggesting a mutation at that site (denoted by the shaded circles in Figure 4, A and C). Although the frequency of site loss is low (of the total of 504 Cμ-region marker positions analyzed in the 63 recombinants, only 7/504 or ∼1% were mutant), it was still of interest to investigate the nature of the mutations. In cell lines 9-1 and 10-2, the mutation is located at position 10422 in the 5′ Cμ region, while in cell lines 28-1 and 89-2, the mutation is in the same position in the 3′ Cμ region. Therefore, these mutations were chosen for analysis. Two independent PCR amplifications of genomic DNA spanning the mutation in each recombinant were performed, and a representative clone from each amplification was sequenced on both strands. The independent sequences from each recombinant were equivalent. The results reveal that approximately one-half of the palindrome originally present in the vector-borne Cμ region is deleted in each recombinant (Figure 7). The portion of the palindrome sequence that remains is denoted in boldface type in Figure 7, and the deletion endpoints are summarized at the bottom of the figure. In recombinant 9-1, the flanking sequence 5′-CTGAATTGTGCCAAACTGGG-3′ has been duplicated (illustrated by the tandem, dashed arrows). A proposed mechanism to account for the palindrome deletions in the recombinants is presented below.
Figure 7.—
DNA sequence analysis of mutated Cμ-region positions. Parental DNA consists of the wild-type chromosomal sequence containing the MfeI site and of the corresponding vector-borne sequence in which the MfeI site is replaced by the 30-bp palindrome containing the diagnostic NotI site. The observed sequences spanning the site corresponding to the MfeI/NotI mismatch in the 5′ Cμ region of recombinants 10-2 and 9-1 and the 3′ Cμ region of recombinants 89-2 and 28-1 are indicated. The deletion endpoints that correspond to the observed sequences are also presented.
DISCUSSION
In this study, we exploited a well-defined mammalian gene-targeting assay using a sequence insertion (“ends-in”) vector to test predictions that the DSBR model makes about the frequency and position of hDNA in recombinants generated by crossing over. This was accomplished by replacing endogenous restriction enzyme sites in the vector-borne region of homology with small palindrome insertions, which are semirefractory to MMR and thereby identify hDNA.
The DSBR model (Figure 1) predicts that invasion by the two 3′ ends of the cut vector will generate a crossover recombinant in which asymmetrical hDNA resides on opposite sides of the DSB in the two homology regions (two HC events: 5:3, 5:3 segregation). However, our data do not fully support the DSBR model with respect to this particular prediction. That is, only 3/63 (∼5%) of the crossover recombinants clearly bear the signature of such bidirectional hDNA, namely, recombinants 22-2 and 73-2 generated with the Eco47III-cut vector and recombinant 85-2 generated with the SpeI-cut vector (Figure 4, A–C). Previous studies (Schultes and Szostak 1990; Porter et al. 1993; Gilbertson and Stahl 1996) also noted a lack of doubly unrepaired heteroduplexes among the products of meiotic recombination in S. cerevisiae.
A second interesting feature of the data is that in a large fraction of the recombinants the recombination event appears to have occurred on only one side of the DSB. This is suggested by recombinants displaying unidirectional hDNA formation and by the large number of one-sided FC events (6:2 segregation). In fact, a little over half of the recombinants display FC, and of these, ∼81% are one-sided. A conspicuously high fraction of the one-sided FC events (∼67%) are associated with normal 4:4 segregation of the palindrome marker on the other side of the DSB. The observed unidirectional recombination would seem to be at odds with the canonical DSBR model, which predicts the involvement of the two 3′ ends of the DSB in recombination (Figure 1). The observed “one-sidedness” in recombination might reflect lopsided resection of the DSB ends (Porter et al. 1993) such that one of the palindrome markers is not included in the joint molecule. However, in S. cerevisiae (Alani et al. 1990; Cao et al. 1990; White and Haber 1990; Sun et al. 1991; Bishop et al. 1992), phage λ (Hill et al. 1997), and mammalian cells (Henderson and Simons 1997; Zenvirth et al. 2003), DNA ends are subject to extensive resection of their 3′ termini (perhaps exceeding 1000 nt). In this study, this argues for resection events on both sides of the DSB to normally include the flanking palindrome markers in single-strand DNA. Another possible explanation for one-sided FC events is “lop-sided” double-strand gap formation. However, unless one side of the DSB is somehow protected from degradation, it is difficult to explain how gaps approaching ∼6 kb that would need to form to explain some recombinants (for example, cell lines 168-1, 4-2, and 7-2 generated with the Esp3I-cut vector, and others) could be confined to one side of the DSB. Unidirectionality in homologous recombination has also been observed previously in S. cerevisiae. Using poorly repairable palindrome markers positioned on both sides of a meiotic DSB site at the ARG4 locus, Gilbertson and Stahl (1996) noted a paucity of doubly unrepaired heteroduplexes among the crossover products and a conspicuous excess of one-sided FC events, which were also associated with normal 4:4 segregation on the other side of the DSB. Similarly, Porter et al. (1993) and Merker et al. (2003) observed frequent unidirectional recombination events at the S. cerevisiae HIS4 meiotic recombination hotspot. In considering mechanisms, these authors also did not favor explanations based on “lop-sided” DSB processing.
A third notable feature of our data is that in a large fraction of the recombinants (14/63 or 22%) there was evidence for symmetrical hDNA formation on one side of the DSB. While previous studies have suggested that symmetrical hDNA can form in this system (Li and Baker 2000; Baker and Birmingham 2001), estimates of the extent of its formation have been limited by the restricted distance separating the flanking palindrome markers from the DSB. This information is important because symmetrical hDNA tract length provides a measure of the capacity for Holliday junction branch migration away from the DSB (Figure 1). Our data reveal that symmetrical hDNA tracts span distances ranging from ∼0.9 to ∼6.0 kb (Table 1), suggesting that Holliday junctions form early in the recombination process and are free to branch migrate, converting initial asymmetrical hDNA into symmetrical hDNA (Figure 1E).
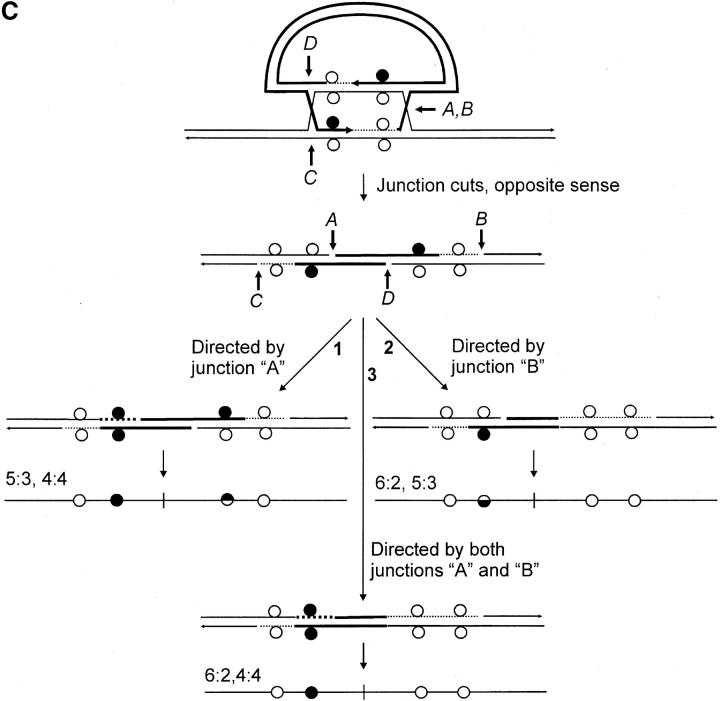
We consider the unique features of the crossover recombinants in this study within the context of three variations of the canonical DSBR model (Figure 1). In the first variation (Figure 8A), the length of heteroduplex formed during recombination is not related to the extent of 5′–3′ DSB resection (Merker et al. 2003). Thus, it is proposed that the initial strand invasion event produces a very short region of heteroduplex on one side of the DSB (usually <200 bp), following which primed DNA synthesis generates a much more extensive length of heteroduplex involving the noninvading strand on the other side of the DSB. In this model, the nature of the constraint imposing a restricted amount of hDNA on the one side of the DSB is not clear. In a second variation (Figure 8B), branch migration in the direction of the newly synthesized DNA generates an intermediate in which the two Holliday junctions and the intervening hDNA are located on the same side of the DSB (Gilbertson and Stahl 1996; Allers and Lichten 2001). The recombinant generated in this event is similar to that depicted in Figure 8A. A third mechanism was proposed by Gilbertson and Stahl (1996) and Foss et al. (1999) to explain one-sided events and their association with normal 4:4 segregation during meiotic recombination at the ARG4 locus in S. cerevisiae. It recognizes that the validity of the DSBR model's prediction of two HC events on opposite sides of the DSB (5:3, 5:3 segregation) holds only in the case in which MMR fails. Thus, in this model, one-sidedness in homologous recombination can be explained through MMR activity. Figure 8C presents this model as it applies to the generation of crossover recombinants of gene targeting in this study. Two proposals that are inherent in the model are:
Each Holliday junction bears a structural asymmetry that biases cleavage of the pair of like strands, one of which contains newly synthesized DNA at that junction. Thus, according to the gene-targeting scheme in Figure 1, resolutions of the type shown in Figure 1F(i) are expected more frequently (Gilbertson and Stahl 1996; Foss et al. 1999; Baker and Birmingham 2001).
MMR is directed not just by discontinuities at the DSB site (referred to as “early” MMR), but also, and perhaps more frequently, by the DNA ends generated through Holliday junction cleavage (referred to as “late” MMR; Alani et al. 1994).
Figure 8.—
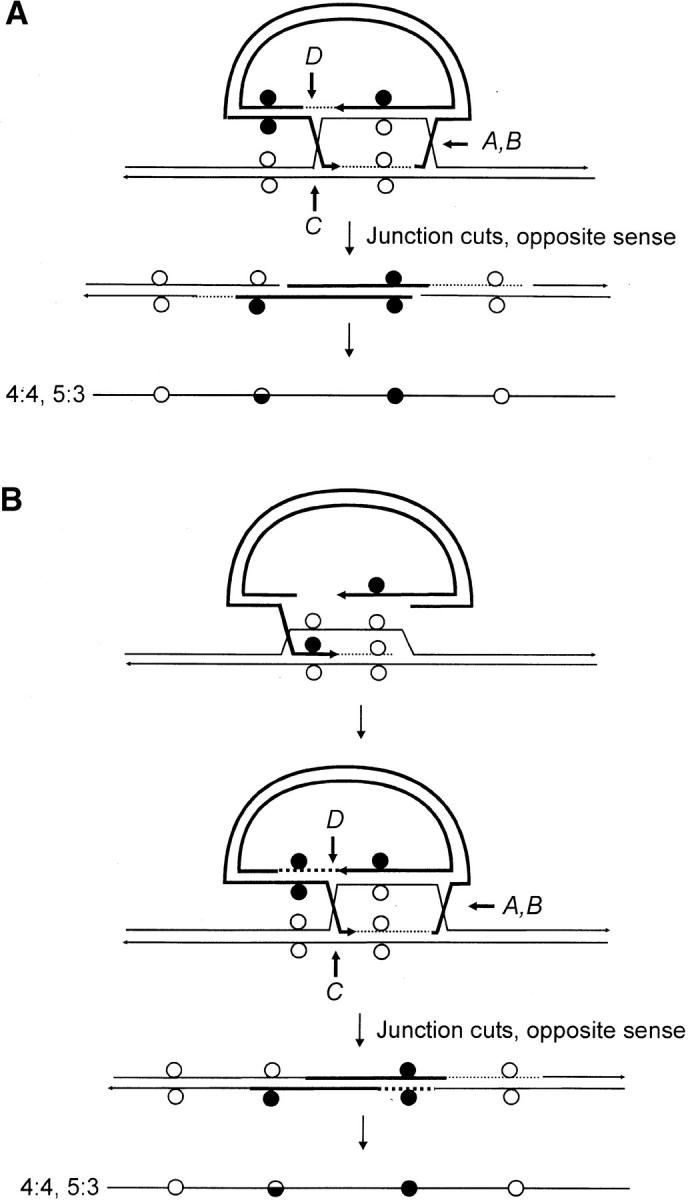
Variations of the canonical DSBR model. (A) The initial strand invasion event produces a very short region of heteroduplex on one side of the DSB, following which primed DNA synthesis generates a much more extensive length of heteroduplex involving the noninvading strand on the other side of the DSB. (B) The initial strand invasion event proceeds according to the canonical DSBR model. However, branch migration in the direction of the newly synthesized DNA generates an intermediate in which the two Holliday junctions and the intervening hDNA are located on the same side of the DSB. In both A and B, alternate sense cleavage generates the crossover product. Initially, the marker segregation pattern is 4:4, 5:3 on the two sides of the DSB. However, the 5:3 marker may be altered through repair to yield a 4:4 or a 6:2 segregation. (C) As in the canonical DSBR model, heteroduplexes form on each side of the DSB, producing an intermediate with two HC events (5:3; 5:3 segregation). Repair activity directed by cutting of the two Holliday junctions in the favored sense (Gilbertson and Stahl 1996; Foss et al. 1999; Baker and Birmingham 2001) can result in restoration and/or conversion events. A repair event issuing from cut junction A results in restoration of normal 4:4 segregation (pathway 1), conversion toward the chromosomal sequence as shown for repair events directed by cut junction B (pathway 2), or both restoration and conversion as shown for repair events directed by cut junctions A and B (pathway 3). Restoration-conversion products also result from repair events directed by junction cuts at C and/or D, and for those involving various combinations of junction cuts at A, B, C, or D, other outcomes are possible (not illustrated).
As shown in Figure 8C, an implicit relationship between crossing over and MMR is predicted when both junctions are cut. Depending on the cut junction and the location of the mismatch with respect to the DSB, MMR will result in conversion toward the vector-borne palindrome as shown in pathway 1 for events directed by cut junction A (in yeast, this would be defined as restoration-type repair, yielding normal 4:4 segregation to the “right” of the DSB); conversion toward the chromosomal sequence as shown in pathway 2 for events directed by cut junction B (in yeast, this would be defined as FC, yielding 6:2 segregation to the “left” of the DSB); or both conversion- and restoration-type repair directed by cut junctions A and B yielding 6:2 and 4:4 marker segregation to the “left” and “right” of the DSB, respectively (pathway 3). Repair events issuing from cut junctions C and/or D would generate similar segregation classes, and other outcomes are possible for combinations of early and late repair events (not illustrated). Junction-directed, restoration-type repair has been suggested as an explanation for the conversion (polarity) gradient observed during meiotic recombination in S. cerevisiae (Petes et al. 1991; Detloff et al. 1992; Gilbertson and Stahl 1996; Foss et al. 1999), although it is recognized that other factors including (1) the length of 3′-end resection (Sun et al. 1991), (2) heteroduplex DNA rejection (Alani et al. 1994; Hillers and Stahl 1999), and (3) differential contributions of heteroduplexes initiated at multiple DSB sites (Merker et al. 2003) may also contribute.
In their simplest forms, the models in Figure 8, A and B, generate recombinants with a HC on one side of the DSB and normal 4:4 marker segregation on the other. In the event of repair, the marker displaying HC may be altered to yield either 4:4 or 6:2 segregation. Thus, in both models, one-sidedness in recombination results from the confinement of hDNA to one side of the DSB, while the marker on the other side of the DSB segregates 4:4. In contrast, the model in Figure 8C provides for a similar efficiency of hDNA formation on the two sides of the DSB, and in the event of early and/or late MMR repair, several segregation patterns are possible. In this regard, our data show that hDNA tracts can be extensive on the two sides of the DSB and that the marker segregation patterns in the recombinants do not always conform to the simple predictions of Figure 8, A and B. We cannot rule out the possibility that all of the above mechanisms participate in the generation of recombinants in this study. However, we suggest that the model in Figure 8C is less constraining, and although not conclusive, would appear to provide an adequate explanation for the recombinants in Figure 4, A–C, including those in which one-sided recombination is associated with normal 4:4 segregation on the other side of the DSB.
In addition, outward branch migration of Holliday junctions positioned on the two sides of the DSB (Figure 1) would provide a suitable explanation for the formation of symmetrical hDNA in our system. In this respect, it of interest to compare and contrast our data with that obtained from studies performed in yeast and fungi. Symmetrical hDNA is detected during meiotic recombination in some fungi, notably Ascobolus (Rossignol et al. 1984) and Sordaria (Kitani et al. 1962). In contrast, symmetrical hDNA would appear to be infrequent, even nonexistent in S. cerevisiae (reviewed in Petes et al. 1991). In fact, in meiotic recombination in S. cerevisiae, considerable gene conversion as well as crossing over are observed within ∼1 kb of the DSB (Orr-Weaver et al. 1988; Schultes and Szostak 1990; Detloff et al. 1992; Smith 2001; Merker et al. 2003). Interestingly, the interval in which recombination appears most frequent coincides with the approximate length of 3′-end resection (Cao et al. 1990; Sun et al. 1991; Bishop et al. 1992). At first glance, this suggests that symmetrical hDNA formation may be disfavored in yeast because Holliday junctions do not usually migrate much farther past their point of formation at the ends of the 3′ single-strand tails. However, locus and/or strain differences may influence hDNA formation (Merker et al. 2003). Also, Stahl and Hillers (2000) suggest that the incidence of symmetrical hDNA may be underestimated in yeast meiosis both as a consequence of it being obscured through junction-directed, restoration-type repair acting on crossover products (Figure 8C) and because the evidence for it is erased through the particular mode that is required to resolve the double Holliday junction intermediate to yield the major noncrossover tetrad class. Although a high frequency of symmetrical hDNA formation is suggested by the data in this study (Figure 6, A–C), in the context of the model in Figure 8C, we also consider it a possibility that more might have been detected had junction-directed repair activities not obscured the evidence.
The recombinant analysis revealed a low (∼1%) frequency of Cμ-region sites that were devoid of either the NotI-containing palindrome or the corresponding endogenous marker. Sequence analysis of genomic DNA spanning the missing site in cell line 10-2, generated with the Esp3I-cut vector, and cell lines 9-1, 28-1, and 89-2, generated with the SpeI-cut vector, revealed deletion of approximately one-half of the palindrome in each case. Palindromic sequences in double-strand DNA are known to form hairpin structures via cruciform extrusion, which might render the sequence more labile to degradation. However, cruciform formation and/or its persistence is considered poor in the case of palindromes that are <150–220 bp in size (Leach 1994) and would appear to be an unlikely mechanism in the case of the 15-bp palindrome stem loop in this study. Further, in contrast to the one-sided deletion of the palindrome observed here, a spectrum of repair products might be expected in the case of an extruded cruciform, including complete excision of the palindrome or, perhaps, reconstitution of a near-perfect palindrome (Leach 1994). Therefore, we consider it more likely that the deletions arise as the result of improper repair of a mismatch involving the palindrome and the corresponding wild-type sequence in hDNA. If this assumption is correct, the mutation at position 10422 in the 5′ Cμ region in recombinants 9-1 and 10-2 corresponds to a mismatch that would have been created by alignment of the endogenous MfeI site with the vector-borne NotI palindrome, while the mutation in recombinants 28-1 and 89-2 involves the same mismatch at this position in the 3′ Cμ region. As a consequence of crossing over, the vector-borne sequence containing the hairpin would reside on the bottom strand in the 5′ Cμ region of recombinants 10-2 and 9-1 and on the top strand in the 3′ Cμ region of recombinants 28-1 and 89-2 [Figure 9A(i–iv)]. A proposed model to account for the one-sided deletion of the palindrome in each of the recombinants is that of hairpin nicking in the G/C-rich symmetry center and formation of an invasive 3′ end that displaces the nicked strand and undergoes base pairing with complementary microhomology (indicated by the open triangles), followed by DNA synthesis to restore the remaining half of the palindrome stem (Figure 9B). In recombinant 9-1, the copying associated with DNA repair has apparently duplicated a small flanking segment of DNA [indicated by the dashed arrow in Figure 9A(ii)], possibly through replication slippage. We are uncertain as to whether marker loss at these sites results from a localized mismatch repair process that is occasionally aberrant or from an unrelated process altogether. However, hairpin nicking in the symmetry center has been proposed previously as the basis for one-sided deletion in kilobase-sized palindromes in mammalian cells (Collick et al. 1996; Akgün et al. 1997; Lewis 1999; Cunningham et al. 2003) and is consistent with the activity of the SbcCD nuclease in Escherichia coli (Leach 1994).
Figure 9.—
One-sided deletion of palindrome sequences. (A) The proposed hDNA intermediates for the MfeI/NotI mismatch in (i) the 5′ Cμ region in recombinant 10-2, (ii) the 5′ Cμ region in recombinant 9-1, (iii) the 3′ Cμ region in recombinant 89-2, and (iv) the 3′ Cμ region in recombinant 28-1. (B) Hairpin rectification through nicking at the symmetry center followed by repair synthesis. Refer to text for details.
Acknowledgments
We thank members of the Baker laboratory for helpful discussions. The constructive comments of the manuscript reviewers are appreciated. This work was supported by operating grants from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council (NSERC) to M.D.B., an NSERC postgraduate studentship to S.A.L., and an Ontario Graduate Scholarship to R.D.M.
References
- Akgün, E., J. Zahn, S. Baumes, G. Brown, F. Liang et al., 1997. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 17: 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani, E., P. Padmore and N. Kleckner, 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436. [DOI] [PubMed] [Google Scholar]
- Alani, E., R. A. Reenan and R. D. Kolodner, 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers, T., and M. Lichten, 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Baker, M. D., and E. C. Birmingham, 2001. Evidence for biased Holliday junction cleavage and mismatch repair directed by junction cuts during double-strand-break repair in mammalian cells. Mol. Cell. Biol. 21: 3425–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, M. D., and L. R. Read, 1993. Analysis of mutations introduced into the chromosomal immunoglobulin μ gene. Somat. Cell Mol. Genet. 19: 299–311. [DOI] [PubMed] [Google Scholar]
- Baker, M. D., N. Pennell, L. Bosnoyan and M. J. Shulman, 1988. Homologous recombination can restore normal immunoglobulin production in a mutant hybridoma cell line. Proc. Natl. Acad. Sci. USA 85: 6432–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista, D., and M. J. Shulman, 1993. A hit-and-run system for introducing mutations into the immunoglobulin heavy chain locus of hybridoma cells by homologous recombination. J. Immunol. 151: 1950–1958. [PubMed] [Google Scholar]
- Bell, L. R., and B. Byers, 1983. Homologous association of chromosomal DNA during yeast meiosis. Cold Spring Harbor Symp. Quant. Biol. 47: 829–840. [DOI] [PubMed] [Google Scholar]
- Bilofsky, H. S., C. Burks, J. W. Fickett, W. B. Goad, F. I. Lewitter et al., 1986. The GenBank genetic sequence databank. Nucleic Acids Res. 14: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D. K., D. Park, L. Xu and N. Kleckner, 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- Bollag, R. J., D. R. Elwood, E. D. Tobin, A. R. Godwin and R. M. Liskay, 1992. Formation of heteroduplex DNA during mammalian intrachromosomal gene conversion. Mol. Cell. Biol. 12: 1546–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L., E. Alani and N. Kleckner, 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Collick, A., J. Drew, J. Penberth, P. Bois, J. Luckett et al., 1996. Instability of long inverted repeats within mouse transgenes. EMBO J. 15: 1163–1171. [PMC free article] [PubMed] [Google Scholar]
- Collins, I., and C. S. Newlon, 1994. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell 76: 65–75. [DOI] [PubMed] [Google Scholar]
- Cunningham, L. A., A. G. Cote, C. Cam-Ozdemir and S. M. Lewis, 2003. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell. Biol. 23: 8740–8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, R. P., C. Dasgupta, T. Shibata and C. M. Radding, 1980. Homologous pairing in genetic recombination: RecA protein makes joint molecules of gapped circular DNA and closed circular DNA. Cell 20: 223–235. [DOI] [PubMed] [Google Scholar]
- Detloff, P., J. Sieber and T. D. Petes, 1991. Repair of specific base pair mismatches formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff, P., M. A. White and T. D. Petes, 1992. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics 132: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho, G., M. Jasin and P. Berg, 1998. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol. Cell. Biol. 18: 4070–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, M. S., 1971. Postmeiotic segregation in Saccharomyces. Mol. Gen. Genet. 111: 297–299. [DOI] [PubMed] [Google Scholar]
- Foss, H. M., K. J. Hillers and F. W. Stahl, 1999. The conversion gradient at HIS4 of Saccharomyces cerevisiae. II. A role for mismatch repair directed by biased resolution of the recombinational intermediate. Genetics 153: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson, L. A., and F. W. Stahl, 1996. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics 144: 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, G. I., E. F. Vanin, A. M. Zrolka and F. R. Blattner, 1981. Sequence of the gene for the constant region of the μ chain of the Balb/c mouse. Gene 15: 33–42. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard, M., P. Qudet and P. Chambon, 1973. Isolation of high-molecular weight DNA from mammalian cells. Eur. J. Biochem. 36: 32–38. [DOI] [PubMed] [Google Scholar]
- Hasty, P., J. Rivera-Pérez and A. Bradley, 1991. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell. Biol. 11: 5586–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, G., and J. P. Simons, 1997. Processing of DNA prior to illegitimate recombination in mouse cells. Mol. Cell. Biol. 17: 3779–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, S. A., M. Stahl and F. W. Stahl, 1997. Single-strand DNA intermediates in phage λ's Red recombination pathway. Proc. Natl. Acad. Sci. USA 94: 2951–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers, K. J., and F. W. Stahl, 1999. The conversion gradient at HIS4 of Saccharomyces cerevisiae. I. Heteroduplex rejection and restoration of Mendelian segregation. Genetics 153: 555–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, N., and N. Kleckner, 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. [DOI] [PubMed] [Google Scholar]
- Kitani, Y., L. S. Olive and A. S. El-Ani, 1962. Genetics of Sordaria fimicoloa V. aberrant segregation at the G locus. Am. J. Bot. 49: 697–706. [Google Scholar]
- Köhler, G., and M. J. Shulman, 1980. Immunoglobulin M mutants. Eur. J. Immunol. 10: 467–476. [Google Scholar]
- Köhler, G., M. J. Potash, H. Lehrach and M. J. Shulman, 1982. Deletions in immunoglobulin mu chains. EMBO J. 1: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, D. R., 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays 16: 893–900. [DOI] [PubMed] [Google Scholar]
- Lewis, S. M., 1999. Palindromy is eliminated through a structure-specific recombination process in rodent cells. Nucleic Acids Res. 27: 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and M. D. Baker, 2000. Use of a small palindrome genetic marker to investigate mechanisms of double-strand-break repair in mammalian cells. Genetics 154: 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker, J. D., M. Dominska and T. D. Petes, 2003. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 165: 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, D. K., M. A. White and T. D. Petes, 1989. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340: 318–320. [DOI] [PubMed] [Google Scholar]
- Ng, P., and M. D. Baker, 1998. High-efficiency, site-specific modification of the chromosomal immunoglobulin locus by gene targeting in mammalian cells. J. Immunol. Methods 214: 81–96. [DOI] [PubMed] [Google Scholar]
- Ng, P., and M. D. Baker, 1999. a Mechanisms of double-strand-break repair during gene targeting in mammalian cells. Genetics 151: 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, P., and M. D. Baker, 1999. b The molecular basis of multiple vector insertion by gene targeting in mammalian cells. Genetics 151: 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., and J. W. Szostak, 1983. Yeast recombination: the association between double strand gap repair and crossing over. Proc. Natl. Acad. Sci. USA 80: 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., J. W. Szostak and R. J. Rothstein, 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78: 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., A. Nicolas and J. W. Szostak, 1988. Gene conversion adjacent to regions of double-strand break repair. Mol. Cell. Biol. 8: 5292–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., R. E. Malone and L. S. Symington, 1991 Recombination in yeast, pp. 407–521 in The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics, edited by J. R. Broach, J. R. Pringle and E. W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Porter, S. E., M. A. White and T. D. Petes, 1993. Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics 134: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick, M. A., 1976. The repair of double-strand breaks in DNA: a model involving recombination. J. Theor. Biol. 59: 97–106. [DOI] [PubMed] [Google Scholar]
- Rossignol, J. L., A. Nicolas, M. Hamza and T. Langin, 1984. Origins of gene conversion and reciprocal exchange in Ascobolus. Cold Spring Harbor Symp. Quant. Biol. 49: 13–21. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schultes, N. P., and J. W. Szostak, 1990. Decreasing gradients of gene conversion on both sides of the initiation site for meiotic recombination at the ARG4 locus in yeast. Genetics 126: 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha, A., and N. Kleckner, 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Schwacha, A., and N. Kleckner, 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 1–20. [DOI] [PubMed] [Google Scholar]
- Shulman, M. J., L. Nissen and C. Collins, 1990. Homologous recombination in hybridoma cells: dependence on time and fragment length. Mol. Cell. Biol. 10: 4466–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. R., 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35: 243–274. [DOI] [PubMed] [Google Scholar]
- Southern, P. J., and P. Berg, 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1: 327–341. [PubMed] [Google Scholar]
- Stahl, F. W., and K. J. Hillers, 2000. Heteroduplex rejection in yeast? Genetics 154: 1913–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., D. Treco and J. W. Szostak, 1991. Extensive 3′-overhanging, single-stranded DNA associated with meiosis-specific double strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl, 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Thomas, K. R., C. Deng and M. R. Capecchi, 1992. High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol. Cell. Biol. 12: 2919–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, S. C., J. K. Countryman and P. Howard-Flanders, 1983. Enzymatic formation of biparental figure-eight molecules from plasmid DNA and their resolution in E. coli. Cell 32: 817–829. [DOI] [PubMed] [Google Scholar]
- White, C. A., and J. E. Haber, 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth, D., C. Richler, A. Bardhan, F. Baudat, A. Barzilai et al., 2003. Mammalian meiosis involves DNA double-strand breaks with 3′ overhangs. Chromosoma 111: 369–376. [DOI] [PubMed] [Google Scholar]