Abstract
The Drosophila klarsicht (klar) gene is required for developmentally regulated migrations of photoreceptor cell nuclei in the eye. klar encodes a large (∼250 kD) protein with only one recognizable amino acid sequence motif, a KASH (Klar, Anc-1, Syne-1 homology) domain, at its C terminus. It has been proposed that Klar facilitates nuclear migration by linking the nucleus to the microtubule organizing center (MTOC). Here we perform genetic and immunohistochemical experiments that provide a critical test of this model. We analyze mutants in the endogenous klar gene and also flies that express deleted forms of Klar protein from transgenes. We find that the KASH domain of Klar is critical for perinuclear localization and for function. In addition, we find that the N-terminal portion of Klar is also important for function and contains a domain that localizes the protein to microtubules apical to the nucleus. These results provide strong support for a model in which Klar links the nucleus to the MTOC.
NUCLEAR movements are significant to a wide variety of developmental processes. Neural cell migration during human brain development depends on nuclear migration (Morris et al. 1998; Morris 2000; Wynshaw-Borris and Gambello 2001). Migration of the oocyte nucleus from the posterior to the anterior of the oocyte is necessary for determination of the major axes of the Drosophila body plan (van Eeden and St Johnston 1999). In Drosophila embryogenesis, migration of syncytial nuclei to the embryo cortex forms a syncytial blastoderm prior to cellularization (Foe et al. 1993). Nuclei in precursor cells of the vulva and neurons of Caenorhabditis elegans must migrate or the cells die (Malone et al. 1999).
In the Drosophila eye, developmentally regulated migrations of photoreceptor nuclei determine cell shape (Fischer-Vize and Mosley 1994). Drosophila compound eyes are composed of ∼800 identical facets or ommatidia. Each ommatidium has a core of 8 photoreceptors (R cells) arranged in a trapezoid surrounded by a hexagonal lattice of pigment cells. Four apical cone cells secrete the lens. The eye develops during the larval and pupal stages in a cellular monolayer called the eye disc (Wolff and Ready 1993). A wave of morphogenesis, starting at the posterior of the disc, travels anteriorly and leaves behind it rows of assembling facets. The wave front is a physical indentation in the disc called the morphogenetic furrow. All nuclei plunge basally in the furrow and then rise apically in those cells that assemble into facets. The eight R cells assemble first in a fixed order, followed by the cone cells and the pigment cells.
In klar mutants, although the sequence of ommatidial assembly is largely unperturbed, R-cell nuclei fail to rise posterior to the morphogenetic furrow (Fischer-Vize and Mosley 1994). The failure of apical nuclear migration results in misshapen ommatidia and thus global defects in eye morphology. R-cell nuclear migration is a microtubule- and dynein-dependent process. Mutations in the p150 subunit of the dynein-regulator dynactin (Glued mutants) result in defects in R-cell nuclear migration similar to those in klar mutants (Fan and Ready 1997). In addition, mutants in two other Drosophila genes implicated in dynein regulation, Bicaudal-D and DLis-1, have R-cell nuclear migration defects similar to klar mutants (Swan et al. 1999).
Klar is a large (∼250 kD) protein with only one recognizable domain, a KASH (Klar, Anc-1, Syne-1 homology) domain at its C terminus (Figure 1; Mosley-Bishop et al. 1999; Starr and Han 2002). Only two proteins, Klar and C. elegans Anc-1 (homologous to vertebrate Syne-1 and Drosophila Msp-300) are known to contain KASH domains (Mosley-Bishop et al. 1999; Starr and Han 2002). Like Klar, C. elegans Anc-1 functions in nuclear positioning (Starr and Han 2002, 2003). Anc-1 anchors nuclei to actin; the C-terminal KASH domain embeds in the nuclear envelope and an N-terminal actin-binding domain, separated from the KASH domain by a long spectrin-like region, connects the nucleus to actin. Syne-1 has also been shown to associate with the nuclear membrane via its KASH domain (Apel et al. 2000; Zhang et al. 2001; Zhen et al. 2002). In contrast, Drosophila Msp-300 has been detected in the cytoplasm, but not at the nuclear membrane (Volk 1992). Moreover, a role for Msp-300 in nuclear positioning has not been reported (Rosenberg-Hasson et al. 1996). Thus, it is unclear if the KASH domain functions as a nuclear membrane localization signal in all cellular contexts.
Figure 1.—
KASH domain sequence comparisons. The KASH domain amino acid sequences of Klarsicht in fruit flies (Drosophila melanogaster) and mosquito (Anopheles gambiae) are aligned with the KASH domains of Msp-300, Anc-1, and Syne-1, which are homologs in fruit flies and mosquitoes, nematodes (C. elegans), and mice (Mus musculus). The shaded areas indicate transmembrane domains, and the boxed amino acids are identical in all KASH domains. The KASH domains constitute the final amino acids of their respective proteins. Accession numbers for the sequences are as follows: Dm Klarsicht, AAD43129 (Mosley-Bishop et al. 1999); Ag Klarsicht, XP_310059.1 (Anopheles Genome Sequence Consortium, 2003); Dm Msp-300, NP_723065 (Adams et al. 2000); Ag Msp-300, XP_310133 (Anopheles Genome Sequence Consortium, 2003); Ce Anc-1, NP_491353 (Starr and Han 2002); and Mm Syne-1, AAG24393 (Apel et al. 2000).
Klar is perinuclear and is also associated with microtubules apical to the nucleus (Mosley-Bishop et al. 1999; Patterson et al. 2004). The model for Klar function in R cells is that Klar forms a bridge between the nuclear membrane and microtubules (Figure 2; Patterson et al. 2004). According to the model, the KASH domain of Klar is held in the nuclear membrane through indirect interactions with nuclear lamin, an intermediate filament protein that is a major component of the inner nuclear membrane. At the microtubules, Klar is thought to interact with the minus-end-directed motor dynein. Dynein, through its connection to Klar, would walk the nucleus up to the microtubule organizing center (MTOC), which forms apically in differentiating R cells.
Figure 2.—
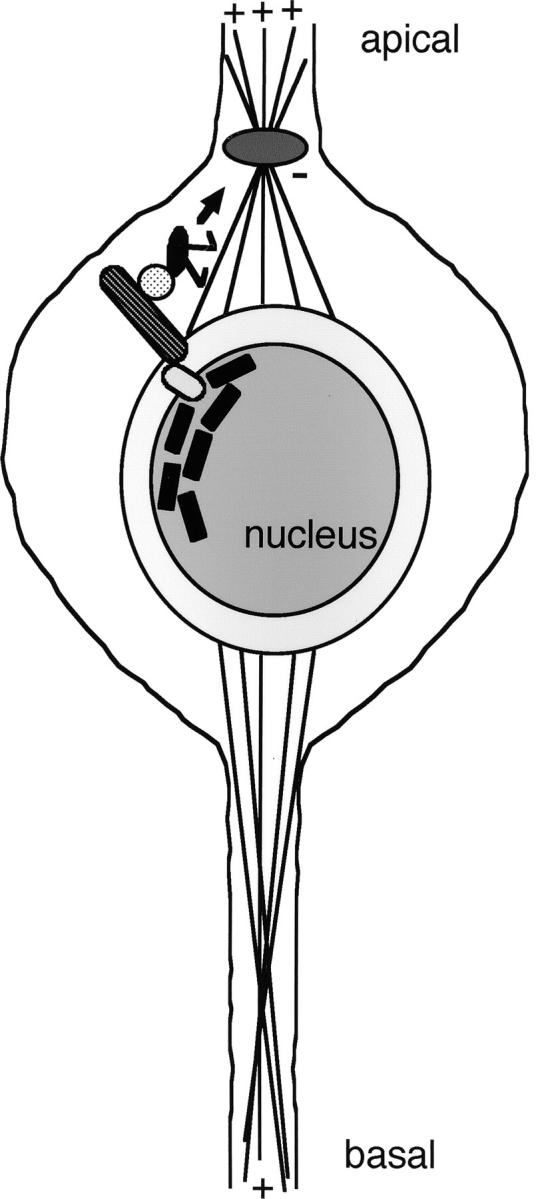
Diagram of the model for Klar function in R-cell nuclear migration. The large hatched bar is Klarsicht, the black rods are lamin, and the dark gray oval is the MTOC. The light gray oval and circle are hypothetical proteins that link Klarsicht with lamin and dynein. The black oval with the legs is dynein and the arrow indicates the direction of its movement, toward the MTOC. Plus and minus indicate plus and minus ends of microtubules.
The model for Klar function derives from experiments with klar and nuclear lamin Dm0 (Lam) mutants (Patterson et al. 2004). Lam mutant eye discs lose perinuclear Klar and have klar-like R-cell nuclear migration defects. Thus, nuclear membrane association of Klar, via Lam, is essential for nuclear migration. Moreover, in klar or Lam mutant discs, MTOCs form normally in R cells but lose their association with the nuclei; the nuclei are basal in the mutants, but the MTOCs remain in their normal apical position. Thus, Klar is required for the connection between the MTOC and the nucleus. The proposed role of dynein is based on the observation that Klar is present on microtubules only apical to the nucleus, toward the minus ends at the MTOC. Also, as mentioned above, dynactin mutants have an R-cell nuclear migration phenotype similar to that of klar (Fan and Ready 1997). Finally, klar is also required for lipid droplet migration in embryos and in this role, there is evidence that Klar regulates dynein (Welte et al. 1998; Gross et al. 2000).
The proposal that Klar connects the nucleus to the MTOC makes several testable predictions about Klar protein. First, Klar is expected to interact with the nuclear membrane via its KASH domain. Second, Klar should have a separate domain for interaction with apical microtubules. Third, both the KASH domain and the microtubule-binding domain should be essential for Klar function. Here, we test these predictions by using genetics and immunocytochemistry. We determine the DNA sequences of six klar point mutations. Next we generate flies with transgenes that express an N-terminal or a C-terminal portion of Klar. We assay the subcellular localization and function of the Klar protein fragments. The results support a model in which Klar links the nucleus with the MTOC.
MATERIALS AND METHODS
Drosophila genetics:
klarmCD4, klarmFQ19, klarmBX4, klarmBX5, klarmBX6, and klarmBX15 are described in Fischer-Vize and Mosley (1994) and Mosley-Bishop et al. (1999). Df(3L)emcE12 (61A-61D3) is described in Lindsley and Zimm (1992). P{w+, elav-Gal4} and P{w+, UAS-6mklarFL} are described in Patterson et al. (2004). P{w+, glrs-6mklarFL} is described in Mosley-Bishop et al. (1999). P{w+, GMR-Gal4} (Freeman 1996) was obtained from the Bloomington Drosophila Stock Center. P-element transformation of w1118 flies was according to standard procedures. All experimental crosses were performed at 25°.
Analysis of eyes:
External eyes were photographed using an Olympus SZX12 microscope and a Kodak DC120 digital camera. Sectioning of retinas was as described in Tomlinson and Ready 1987. Retinal sections were photographed using a Zeiss Axioplan and a Zeiss AxioCam. Larval eye discs were immunostained as follows [see Fischer-Vize et al. (1992) for details]. Discs were dissected in PBS and fixed for 45–55 min in PEMS on ice, washed for 15 min three times in PBST, and then incubated in primary antibody in PBST for 1–2 hr at room temperature. Discs were washed for 5 min three times in PBST and then incubated in secondary antibody in PBST for 1–2 hr at room temperature or overnight at 4°. Discs were then washed for 5 min three times in PBST and mounted in Vectashield (Vector, Burlingame, CA). Prior to mounting, some discs were treated with phalloidin, which was dried and resuspended in PBST (0.1 unit/μl). In Figures 4 and 7, the primary antibody was rat anti-Elav 7E8A10 obtained from the Developmental Studies Hybridoma Bank (DSHB, Iowa City, IA) used at 9:1. The secondary antibody was Cy5-goat-anti-rat (Jackson) used at 1:200. Alexa568-phalloidin (Molecular Probes, Eugene, OR) was used as described by Overstreet et al. (2003). In Figure 6, the primary antibodies were rat anti-Elav (described above) and rabbit anti-myc (ICL Labs; RMYC-45A-2) used at 1:1000. Secondary antibodies were Alexa633-goat-anti-rat and Alexa488-goat-anti-rabbit each used at 1:600. Confocal microscopy was with a Leica TCS SP2. All images were processed using Adobe Photoshop software.
Figure 4.—
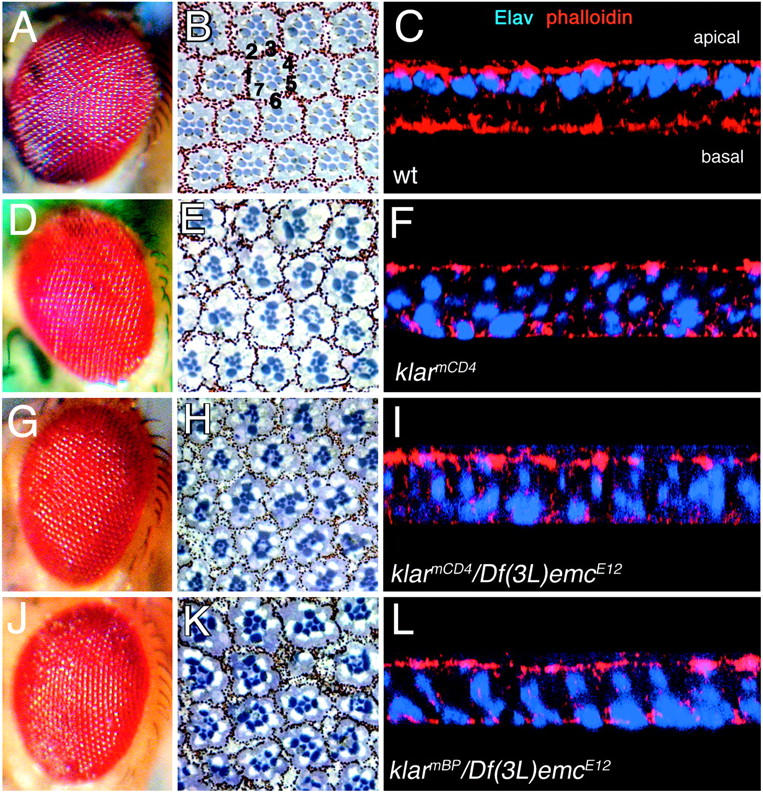
klarCD4 has little or no klar+ function. External adult eyes (A, D, G, J), light microscope images of adult apical retinal sections (B, E, H, K), and confocal microscope images of larval eye discs (C, F, I, L) are shown. The genotypes indicated in C, F, I, and L apply to the entire horizontal row. The black numbers in B indicate the seven R cells (R1–7) visible in apical sections of the retina. Eye color differences in A, D, G, J are due to eye color mutations in the backgrounds, not to the klar alleles. The larval eye discs are labeled with anti-Elav to highlight R-cell nuclei (blue) and phalloidin to highlight cell membranes (red). Five eyes of each genotype were observed and representative data are shown.
Figure 6.—
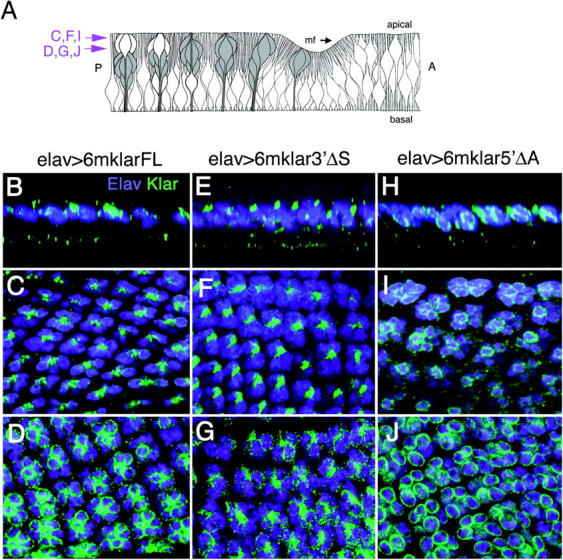
Localization of 6mKlar proteins in wild-type eye discs. (A) A diagram of a Z section through a developing eye disc is shown. The morphogenetic furrow (mf) is moving in the direction of the arrow. A, anterior; P, posterior. Photoreceptor cells are shaded gray; their nuclei and most of the cell cytoplasm migrate apically as the cells differentiate. The unshaded apical cell bodies are cone cells. Purple arrows at the left mark the XY planes of the confocal images indicated (after Tomlinson and Ready 1986). (B–J) Confocal images of eye discs that express the 6mKlar transgene indicated in R cells are shown, labeled with anti-Elav to highlight R-cell nuclei (purple) and anti-myc to reveal 6mKlar proteins. B, E, and H are Z sections, and C, D, F, G, I, J are XY sections. The 6mKlar protein apical to the nuclei is thought to be associated with microtubules because it closely resembles the localization pattern of the microtubule-associated protein Futsch (Patterson et al. 2004).
DNA sequence analysis:
klar alleles were amplified by PCR using as a template total genomic DNA prepared from a single homozygous or hemizygous (in trans to Df(3L)emcE12) fly. Genomic DNA was prepared as described in Chen and Fischer (2000). Several primer pairs (sequences available upon request) and standard PCR conditions were used. The sequences of the PCR products were determined directly using automated fluorometric methods. Sequences were analyzed with MacVector (Accelrys) software.
RNA blot analysis:
Eye disc total nucleic acid was prepared as described in Fischer-Vize et al. (1992) and electrophoresed on a 1% formaldehyde gel. NorthernMax (Ambion, Austin, TX) reagents were used according to manufacturer's instructions. The nucleic acid was transferred to positively charged nylon membrane (Ambion) and UV-crosslinked using a Stratalinker 2400 (Stratagene, La Jolla, CA). Biotinylated antisense RNA probes and BrightStar Biodetect (Ambion) were used to detect klar mRNA and 18S RNA. A plasmid, pGEM-5′klar, was constructed for generation of the klar probe as follows. The first 1000 bp of klar cDNA was amplified by PCR using pOT2-klarcDNA (clone LD36052; Open Biosystems) as a template and two primers: 5′-GGCGCGCCATGCACACATGGTTAATA-3′ and 5′-GGTACCTTACGTGGGTGGCGG-3′. The PCR product was ligated in pGEM-T (Promega, Madison, WI) to generate pGEM-5′klar. To generate a the klar probe, pGEM-5′klar was restricted with AscI and transcribed in vitro with SP6 polymerase using the StripEZ RNA kit (Ambion) and biotinylated UTP (Enzo) according to manufacturer's instructions. The 18S RNA probe was synthesized using pTRI-18S (Ambion).
P-element plasmid construction—pUAS-6m3′ΔS:
An ∼5.5-kb AscI-SfoI fragment containing the 6xmyc-tagged N-terminal region of klar was isolated from a plasmid containing 6mklarFL (Patterson et al. 2004). Also, an ∼150-nt SfoI-AscI fragment containing additional coding sequence 3′ to the SfoI site and with a stop codon (TGA) inserted after the codon for P1774 was generated by PCR using a plasmid containing 6mklarFL (Mosley-Bishop et al. 1999) as a template and two primers, 5′-TGTTGTCCAACCACTGCG-3′ (forward) and 5′-GGCGCGCCTCATTCATTCACGGCTCCGTATCGAGGAG-3′ (reverse). The PCR product was ligated into the SmaI site of pBluescript (Stratagene), the insert sequence was determined, and the ∼150-nt SfoI-AscI fragment was isolated. The two fragments were ligated together into the AscI site of pUASgNA, a derivative of pUASg (Rorth 1998) with the NotI site changed to AscI.
pUAS-6m5′ΔA:
Three DNA fragments (1–3) were ligated into the AscI site of pUASgNA. Fragment 1 is an AscI-PacI fragment containing the 6xmyc tag and was generated by PCR using a plasmid containing 6mklarFL as a template and two primers, 5′-GGCGCGCCCAAAATGTTACG-3′ (forward) and 5′-TCATACTCGAGAGGCCTTG-3′ (reverse). The PCR product was ligated into pCRTOPO2.1 (Invitrogen), the insert sequence was determined, and the AscI-PacI fragment was isolated. Fragment 2 is an ∼100-nt PacI-ApaLI fragment containing klar coding sequences starting with the codon for V1860 and a PacI site engineered so that the 6xmyc tag in fragment 1 is in-frame with klar when the PacI sites are ligated. Fragment 2 was generated by PCR using a plasmid containing 6mklarFL as a template and two primers, 5′-GGTCACCGACAGCAATGGTAATC-3′ (forward) and 5′-CGCTCACTTCGTTCTTCAGATGC-3′ (reverse). The PCR product was ligated into pBluescript, the insert DNA sequence was determined, and the PacI-ApaLI fragment was isolated. Fragment 3 was an ∼2-kb ApaLI-AscI fragment of a plasmid containing 6mKlarFL as an AscI fragment.
Protein blot analysis:
Five pairs of eye discs were dissected into 50 μl of 2× Laemmli buffer and boiled for 5 min. One-half of the sample was subjected to SDS-PAGE on a 10% gel (ISC BioExpress). The protein was transferred to PVDF membrane (Immobilon-P; Millipore, Bedford, MA) and the blot was labeled with primary and secondary antibodies using standard procedures. The primary antibodies were anti-c-myc (Santa Cruz Biochemicals; 9E10, sc-40) and anti-β-tubulin (DSHB; mAbE7, against Escherichia coli β-tubulin) each at 1:500. The secondary antibody was HRP-anti-mouse (Jackson) at 1:500. The signal was developed using Amersham reagents (RPN2109D1/2) according to the manufacturer's protocol.
RESULTS
Molecular analysis of klar mutant alleles:
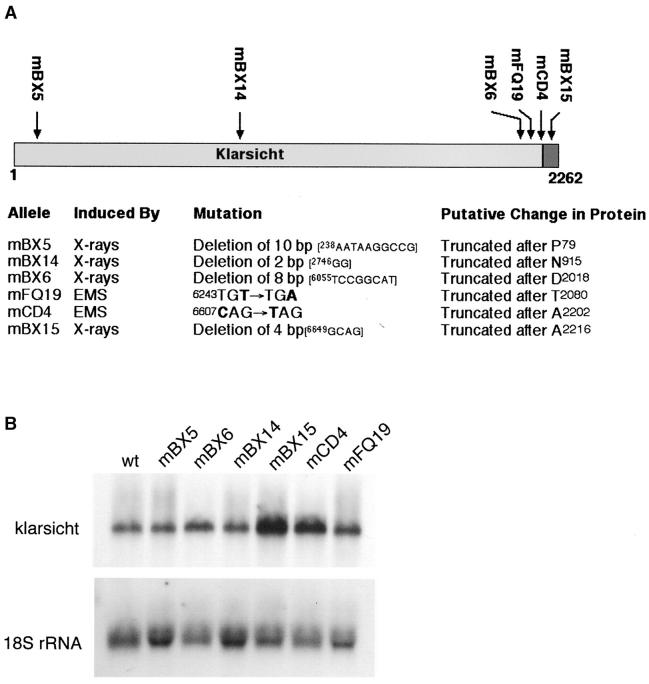
As a first step toward determining which portions of Klar protein are required for its function, we determined the exon DNA sequences of six mutant klar alleles (Fischer-Vize and Mosley 1994). The results of genomic blotting experiments suggested that these alleles contain point mutations (Mosley-Bishop et al. 1999). We find that all six alleles have either nonsense or frameshift mutations in coding exons (Figure 3A). In addition, we performed RNA blotting experiments to detect klar transcripts in eye discs of third instar larvae homozygous for each mutant allele. We find that each mutant klar mRNA is present and at levels similar to those in wild type (Figure 3B).
Figure 3.—
(A) klar mutant allele sequences. The positions of the putative truncations in Klar protein resulting from the DNA lesions in six different mutant klar alleles are shown. Full-length Klar protein is 2262 amino acids (Mosley-Bishop et al. 1999). The solid box at the C terminus is the KASH domain. The numbers adjacent to the DNA sequences affected by the mutations indicate the nucleotide number of the cDNA, where number 1 is the A of the ATG start codon. (B) RNA blots of klar transcripts. Shown is a blot of eye disc total nucleic acid from wild-type (wt) larvae and from larvae homozygous for each of the klar mutant alleles indicated. klar mRNA and 18S rRNA were detected sequentially. The klar band corresponds to at least two mRNAs of similar size (∼8.0–8.5 kb; Mosley-Bishop et al. 1999).
Two alleles, klarmBX5 and klarmBX14, have frameshift mutations in the open reading frame after amino acids 79 and 915, respectively. The protein products of these alleles are difficult to predict. Although the simple expectation is that klarmBX5 and klarmBX14 would produce N-terminal Klar protein fragments, translation reinitiation could result in the production of C-terminal protein fragments. As an antibody that could detect their protein products is not available, we cannot learn much from klarmBX5 or klarmBX14.
Four alleles (klarmCD4, klarmFQ19, klarmBX6, and klarmBX15) have premature stop codons late in the open reading frame (Figure 3A). As a Klar protein truncated somewhat more severely than KlarBX6 is stably produced by a transgene (see below), these four alleles are likely to produce nearly full-length Klar proteins with C-terminal truncations. Notably, klarmCD4 and klarmBX15 should produce Klar protein lacking only the final 60 or 46 amino acids, respectively, which includes most of the KASH domain (Figure 1). These four mutant allele sequences suggest that the KASH domain is important for the function of Klar protein.
klarCD4 has little or no klar+ function:
To explore further the importance of the KASH domain, we asked if klarmCD4, which has a stop codon just prior to the KASH domain, behaves as a strong loss-of-function allele genetically. Toward this end, we compared the mutant eye phenotypes of klarmCD4 homozygotes and hemizygotes (klarmCD4/Df(3L)emcE12). We observed the external eyes and R-cell morphology in sectioned adult eyes and R-cell nuclear positions in developing larval eye discs.
Wild-type external eyes appear smooth and crystalline (Figure 4A). Homozygous klarmCD4 external eyes are subtly rougher than those of wild type (Figure 4D). Sections through the wild-type retina reveal the R cells arranged in a trapezoid. The R cells are identified by their rhabdomeres, light-gathering organelles that project from each R cell into the center of the ommatidium (Figure 4B). In klarmCD4 homozygotes, the R cells are present, largely in their normal positions, but the rhabdomeres are malformed (Figure 4E; Fischer-Vize and Mosley 1994). As the rhabdomeres project out along the entire apical/basal axis of the R cells, the severe cell shape malformations resulting from the lack of an apical nucleus also result in rhabdomere malformations. The positions of the R-cell nuclei in discs were observed by labeling discs with antibodies to Elav, a neural nuclear protein (Robinow and White 1991). In wild-type eye discs, all of the R-cell nuclei are apical (Figure 4C). By contrast, in klarmCD4 eye discs, most of the R-cell nuclei are basal and the remainder are randomly distributed throughout the apical/basal plane (Figure 4F). The eye phenotypes of klarmCD4/Df(3L)emcE12 are qualitatively indistinguishable from those of klarmCD4 homozygotes (Figure 4, D–F and G–I). We also examined the mutant eye phenotypes of klarmBP/Df(3L)emcE12. klarmBP is a translocation that breaks in the middle of the klar coding region and is thus very likely to be null (Fischer-Vize and Mosley 1994; Mosley-Bishop et al. 1999). The phenotypes of klarmBP/Df(3L)emcE12 and klarmCD4 are also indistinguishable (Figure 4, D–F and J–L). We conclude that klarCD4 retains little or no function and thus that the KASH domain is critical to Klar activity.
Transgenes that express 6xmyc-tagged partial Klar proteins:
The results above suggest that the KASH domain is essential for Klar function, perhaps because it localizes Klar to the nuclear envelope. However, we cannot determine the subcellular localization of the mutant Klar proteins. To circumvent this limitation and also to investigate the function of the region of Klar N-terminal to the KASH domain, we generated transgenes that express epitope-tagged partial Klar proteins. Previous results established expression vectors and an epitope tag useful for assays of Klar subcellular localization and for klar function. Localization of wild-type Klar was observed by generating transgenes that express a full-length 6xmyc-tagged Klar (6mKlarFL; Mosley-Bishop et al. 1999; Patterson et al. 2004). When 6mKlarFL is expressed in R cells using a neural-specific elav-Gal4 driver and a UAS-6mklarFL transgene, two distinct aspects of 6mKlarFL localization are observed; 6mKlarFL is detected at the nuclear membrane and also on microtubules apical to the nucleus (Patterson et al. 2004 and see below). When expressed from the GLRS vector, which is active in all cells posterior to the furrow in the eye disc, 6mKlarFL restores significant Klar function to klarCD4 homozygotes (Mosley-Bishop et al. 1999 and see below).
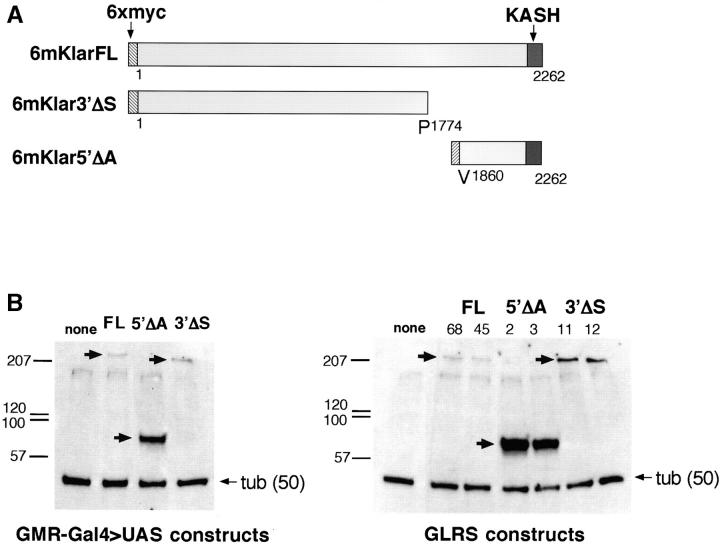
To determine if the N- and C-terminal portions of Klar are differentially required for Klar localization and function, we generated two klar gene constructs that express different 6xmyc-tagged partial Klar proteins; 6mKlar3′ΔS contains the 1774 N-terminal amino acids, and 6mKlar5′ΔA contains the C-terminal 403 amino acids of Klar, which include the KASH domain (Figure 5A). For assays of subcellular localization, transgenes were generated where expression of each construct is controlled by a UAS (UAS-6mklar3′ΔS and UAS-6mklar5′ΔA) and several transformant lines were generated with each. For assays of function, each construct was cloned into the GLRS vector (glrs-6mklar3′ΔS and glrs-6mklar5′ΔA) and several transformant lines were generated.
Figure 5.—
Proteins expressed by klar transgenes. (A) The structures of the complete and partial Klar proteins expressed by transgenes are shown. The hatched bars are 6xmyc epitope tags, and the dark gray bars are KASH domains. The numbers indicate the first and final amino acid of Klar present in each construct. (B) Western blots of larval eye disc extracts showing 6mKlar proteins expressed by three different UAS transgenes (left) and three different GLRS transgenes (right). Two lines of each of the GLRS transgenes are shown and the line numbers are indicated above each lane. Blots were probed with anti-Myc and anti-β-tubulin. 6mKlarFL is predicted to be ∼258 kD, 6mKlar3′ΔS ∼204 kD, and 6mKlar5′ΔA ∼53kD. 6mKlar5′ΔA appears larger than its predicted size. Perhaps the KASH-domain-containing Klar fragment is modified in vivo.
We performed Western blot experiments to determine if 6mKlar protein is expressed stably by each transgene and to identify the transformant lines that express the highest levels of protein. 6mKlar proteins were detected in eye disc protein extracts from larvae with a single UAS transgene expressed by an eye-specific driver (GMR-Gal4) or from larvae with a single GLRS transgene. Anti-myc was used to detect 6mKlar and anti-tubulin was used as a control. One transformant line of each UAS construct and two lines of each GLRS construct with the highest expression levels were chosen for further analysis. The protein blot results for these high-expressing lines are shown in Figure 5B.
Localization of partial Klar proteins in eye discs:
To determine where the partial Klar proteins are located within R cells, eye discs with one copy of a UAS transgene (UAS-6mklarFL, UAS-6mklar3′ΔS, or UAS-6mklar5′ΔA) and one copy of an elav-Gal4 driver transgene were immunostained with anti-myc. The KlarFL protein localizes apically to the R-cell nuclei (Figure 6, B and C) on microtubules and is also perinuclear (Figure 6D; Patterson et al. 2004). We observe that 6mKlar5′ΔA, the C-terminal Klar fragment that contains the KASH domain, retains only one of the two aspects of 6mKlarFL localization; 6mKlar5′ΔA localizes to the nuclear membrane, but not to the apical microtubules (Figure 6, H–J). Conversely, 6mKlar3′ΔS, the N-terminal Klar fragment that lacks the KASH domain, localizes to the apical microtubules, but not to the nuclear membrane (Figure 6, E–G). Thus, distinct domains localize Klar to microtubules and the nuclear envelope.
Ability of partial Klar proteins produced by transgenes to complement the klar mutant eye phenotype:
To determine if the 6mKlar3′ΔS or 6mKlar5′ΔA proteins retain significant levels of Klar function, we tested glrs-6mklar3′ΔS and glrs-6mklar5′ΔA transgenes and glrs-6mklarFL as a control, for complementation of the klarCD4 mutant eye phenotype. We tested for complementation of the nuclear migration defects in eye discs and of the morphological defects in adult eyes. We find that a glrs-6mklarFL transgene rescues both defects significantly (Figure 7, A–C; Mosley-Bishop et al. 1999). By contrast, neither glrs-6mklar3′ΔS nor glrs-6mklar5′ΔA provides significant rescuing activity (Figure 7, D–I). This result is particularly striking given that the partial proteins, especially 6mKlar5′ΔA, are produced at much higher levels than 6mKlarFL (Figure 5B). Although the epitope tag does not affect the function of full-length Klar (Mosley-Bishop et al. 1999), it could affect the function 6mklar5′ΔA. Aside from this caveat, these results indicate that neither the KASH domain nor the N terminus of Klar is sufficient for significant function. Rather, both the N-terminal and the C-terminal Klar fragments are necessary.
Figure 7.—
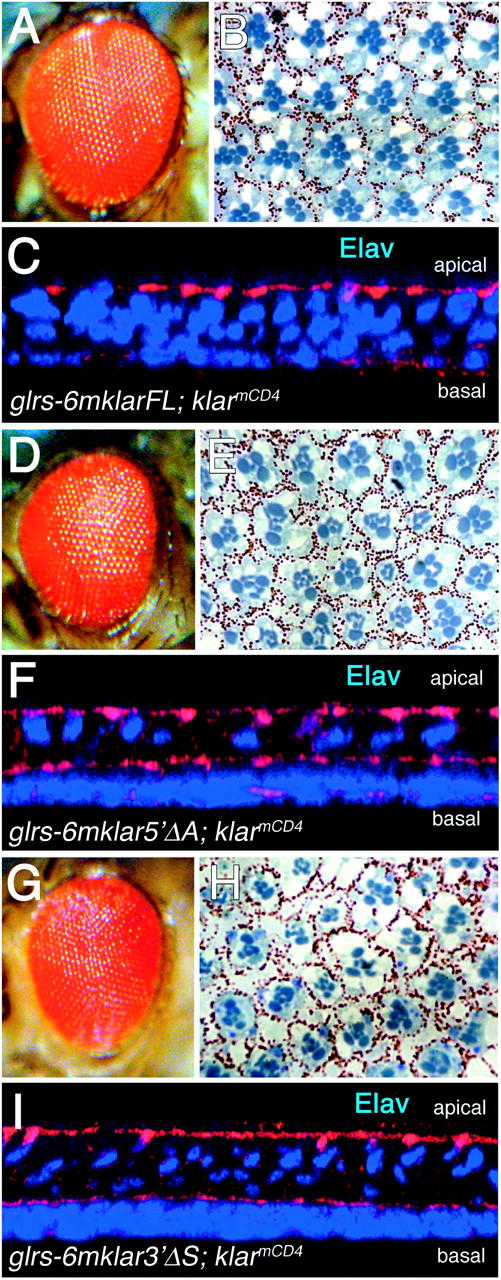
Complementation of klarCD4 by glrs-6mklar transgenes. External adult eyes (A, D, and G), light micrographs of adult retinal sections (B, E, H), and confocal images of larval eye discs (C, F, and I) are shown. Eye discs in C, F, I are labeled with anti-Elav to highlight R cells (blue) and phalloidin to mark cell membranes (red). (A, B, C) Two copies of a glrs-6mklarFL transgene in a klarmCD4 background. (D, E, F) Two copies of a glrs-6mklar5′ΔA transgene in a klarmCD4 background. (G, H, I) Two copies of a glrs-6mklar3′ΔS transgene in a klarmCD4 background. Five eyes of each genotype were observed and representative data are shown.
Mutant eye phenotypes due to Klar protein overexpression:
Overexpression of KlarFL in the eye can result in morphological defects unrelated to nuclear migration (Mosley-Bishop et al. 1999; Patterson et al. 2004). We tested whether expression of the partial Klar proteins 6mKlar5′ΔA or 6mKlar3′ΔS also results in eye morphology defects. None of the GLRS vector transgenes (3 lines of glrs-6mklarFL, 6 lines of glrs-6mklar5′ΔA, or 11 lines of glrs-6mKlar3′ΔS) cause mutant eye phenotypes when present in two copies. Similarly, none of the UAS transgenes, when expression is driven by elav-Gal4, results in a mutant eye phenotype. To boost the expression levels of the UAS transgenes, we used the GMR-Gal4 driver. Each of the 12 UAS-6mklarFL lines tested resulted in roughened external eyes when expressed using GMR-Gal4 (data not shown). By contrast, none of the 11 UAS-6mklar5′ΔA lines produced a phenotype when expressed with GMR-Gal4, and only 1 of 5 UAS-6mklar3′ΔS lines did (data not shown). The failure of 6mKlar5′ΔA overexpression to produce a mutant eye phenotype is particularly striking given that its expression level is considerably higher than that of KlarFL (Figure 5B). We conclude the overexpression phenotype caused by KlarFL is not due solely to the KASH domain or to the microtubule-localization domain, but requires an intact protein.
DISCUSSION
Klar has been proposed to function as a link between the nucleus and MTOC (Patterson et al. 2004). This model predicts that Klar should have discrete domains for nuclear and microtubule attachment and we find that this is the case. When we divide Klar into N-terminal and C-terminal portions, the N terminus localizes to microtubules apical to the nucleus and the C terminus containing the KASH domain is perinuclear. We also show that the KASH domain is important for Klar function. First, we find that four independent klar mutant alleles have nonsense or frameshift mutations that are likely to result in deletion of the C-terminal KASH domain. Second, we show that one of these alleles retains little or no gene activity. Third, a transgene that expresses the N-terminal 1774 amino acids of Klar, which does not include the KASH domain, fails to retain significant klar gene function in vivo. Finally, we also show that the KASH domain alone is insufficient for Klar function. Even when overexpressed, a C-terminal 403-amino-acid Klar fragment that includes the KASH domain does not provide significant Klar function in vivo.
Klar and Anc-1/Syne-1 (and Zyg-12, see below) are unique in that they are held in the nuclear membrane through (probably indirect) interactions with nuclear lamin and they protrude into the cytoplasm. Starr and Han (2002) reported that overexpression of the C. elegans Anc-1 KASH domain results in a dominant negative nuclear anchorage defect, presumably because the KASH domain competes with wild-type Anc-1 for a limited number of docking sites in the nuclear membrane. We were therefore surprised to find that overexpression of the Klar KASH domain does not result in a mutant phenotype. The difference between our results and theirs could be due to technical differences in the two experimental systems. Alternatively, the different results could reflect a mechanistic difference in Anc-1 and Klar function.
Among all of the genomes whose DNA sequences are known, Klar is unique to Drosophila and Anopheles. As nuclear migration and nuclear attachment to the MTOC are universal cellular phenomena, this result is surprising. Other metazoans appear to rely solely on alternative proteins for attaching the nucleus and MTOC and for nuclear migration. One of these proteins is C. elegans Zyg-12. In C. elegans embryos, Zyg-12 attaches nuclear membranes to centrosomes (the MTOC; Malone et al. 2003). Zyg-12 is present at the nuclear membrane and also at centrosomes and the mechanisms proposed for Klar and Zyg-12 function are similar. Yet, the two proteins have no obvious amino acid sequence similarity. Rather, Zyg-12 is a homolog of Drosophila Hook, which in Drosophila is proposed to link organelles other than the nucleus to the cytoskeleton (Kramer and Phistry 1999; Walenta et al. 2001). Hook is expressed in the eye, but a role for it in R-cell nuclear migration has not been reported. Perhaps the variety of mechanisms for connecting the nucleus to the MTOC reflects a requirement for regulation in diverse developmental contexts.
Acknowledgments
We thank Paul Macdonald and John Sisson for the use of their confocal microscopes. We also thank an anonymous reviewer for helpful comments that improved the manuscript. This work was supported by grants to J.A.F. from the Eye Institute of the National Institutes of Health (RO1 EY13958) and the National Science Foundation (IBN-9808837).
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Apel, E. D., R. M. Lewis, R. M. Grady and J. R. Sanes, 2000. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275: 31986–31995. [DOI] [PubMed] [Google Scholar]
- Chen, X., and J. A. Fischer, 2000. In vivo structure/function analysis of the Drosophila fat facets deubiquitinating enzyme gene. Genetics 156: 1829–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, S.-S., and D. F. Ready, 1997. Glued participates in distinct microtubule-based activities in Drosophila eye development. Development 124: 1497–1507. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize, J. A., and K. Mosley, 1994. marbles mutants: uncoupling cell determination and nuclear migration in the developing Drosophila eye. Development 120: 2609–2618. [DOI] [PubMed] [Google Scholar]
- Fischer-Vize, J. A., G. M. Rubin and R. Lehmann, 1992. The fat facets gene is required for Drosophila eye and embryo development. Development 116: 985–1000. [DOI] [PubMed] [Google Scholar]
- Foe, V. E., G. M. Odell and B. A. Edgar, 1993 Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint, pp. 149–300 in The Development of Drosophila melanogaster, Vol. I, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Freeman, M., 1996. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87: 651–660. [DOI] [PubMed] [Google Scholar]
- Gross, S. P., M. A. Welte, S. M. Block and E. F. Wieschaus, 2000. Dynein-mediated cargo transport in vivo: a switch controls travel distance. J. Cell Biol. 148: 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, H., and M. Phistry, 1999. Genetic analysis of hook, a gene required for endocytic trafficking in Drosophila. Genetics 151: 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992 The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Malone, C. J., W. D. Fixsen, H. R. Horvitz and M. Han, 1999. Unc-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126: 3171–3181. [DOI] [PubMed] [Google Scholar]
- Malone, C. J., L. Misner, N. Le Bot, M.-C. Tsai, J. M. Campbell et al., 2003. The C. elegans Hook protein, Zyg-12, mediates the essential attachment between the centrosome and nucleus. Cell 115: 825–836. [DOI] [PubMed] [Google Scholar]
- Morris, N. R., 2000. Nuclear migration: from fungi to the mammalian brain. J. Cell Biol. 148: 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, N. R., V. P. Efimov and X. Xiang, 1998. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 8: 467–470. [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop, K. L., Q. Li, K. Patterson and J. A. Fischer, 1999. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cell of the Drosophila eye. Curr. Biol. 9: 1211–1220. [DOI] [PubMed] [Google Scholar]
- Overstreet, E., X. Chen, B. Wendland and J. A. Fischer, 2003. Either part of a Drosophila epsin protein, divided after the ENTH domain, functions in endocytosis of Delta in the developing eye. Curr. Biol. 13: 854–860. [DOI] [PubMed] [Google Scholar]
- Patterson, K., A. B. Molofsky, C. Robinson, S. Acosta, C. Cater et al., 2004. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migration of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell 15: 600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow, S., and K. White, 1991. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22: 443–461. [DOI] [PubMed] [Google Scholar]
- Rorth, P., 1998. Gal4 in the Drosophila female germ-line. Mech. Dev. 78: 113–118. [DOI] [PubMed] [Google Scholar]
- Rosenberg-Hasson, Y., M. Renert-Pasca and T. Volk, 1996. A Drosophila Dystrophin-related protein, MSP-300, is required for embryonic muscle morphogenesis. Mech. Dev. 65: 83–94. [DOI] [PubMed] [Google Scholar]
- Starr, D., and M. Han, 2002. Role of Anc-1 in tethering nuclei to the actin cytoskeleton. Science 298: 406–409. [DOI] [PubMed] [Google Scholar]
- Starr, D., and M. Han, 2003. ANChors away: an actin based mechanism of nuclear positioning. J. Cell Sci. 116: 211–216. [DOI] [PubMed] [Google Scholar]
- Swan, A., T. Nguyen and B. Suter, 1999. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1: 444–449. [DOI] [PubMed] [Google Scholar]
- Tomlinson, A., and D. F. Ready, 1986. sevenless: a cell-specific homeotic mutation of the Drosophila eye. Science 231: 400–402. [DOI] [PubMed] [Google Scholar]
- Tomlinson, A., and D. F. Ready, 1987. Cell fate in the Drosophila ommatidium. Dev. Biol. 120: 355–376. [DOI] [PubMed] [Google Scholar]
- van Eeden, F., and D. St Johnston, 1999. The polarization of the anterior-posterior and dorsal-ventral axes during Drosophila oogenesis. Curr. Opin. Genet. Dev. 9: 396–404. [DOI] [PubMed] [Google Scholar]
- Volk, T., 1992. A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development 116: 721–730. [DOI] [PubMed] [Google Scholar]
- Walenta, J. H., A. J. Didier, X. Liu and H. Kramer, 2001. The Golgi-associated Hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 152: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M. A., S. P. Gross, M. Postner, S. M. Block and E. F. Wieschaus, 1998. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell 92: 547–557. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1993 Pattern formation in the Drosophila retina, pp. 1277–1326 in The Development of Drosophila melanogaster, Vol. II, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Wynshaw-Borris, A., and M. J. Gambello, 2001. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 15: 639–651. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., J. N. Skepper, F. Yang, J. D. Davies, L. Hegyi et al., 2001. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 114: 4485–4498. [DOI] [PubMed] [Google Scholar]
- Zhen, Y.-Y., T. Libotte, M. Munck, A. A. Noegel and E. Korenbaum, 2002. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115: 3207–3222. [DOI] [PubMed] [Google Scholar]