Abstract
On the basis of synthetic lethality, five genes in Caenorhabditis elegans are known to be redundant with the mec-8 gene, which encodes a protein that contains two copies of an RNA recognition motif (RRM) and affects alternative RNA splicing. The molecular identities of two of the redundant genes, sym-1 and sym-5, were previously reported. The remaining three genes have now been cloned, and their synthetically lethal phenotypes with mec-8 are described in more detail. Animals homozygous for mec-8 and sym-2 loss-of-function mutations die during late embryogenesis. The SYM-2 predicted protein contains three RRMs; we propose that SYM-2 and MEC-8 can substitute for each other in promoting the maturation of the transcripts of a vital gene. Animals homozygous for mutations in mec-8 and in either sym-3 or sym-4 have the same striking defect: they arrest development just prior to or just after hatching with a pharynx that appears fully formed but is not properly attached to the body cuticle. sym-3 encodes a protein of unknown function with orthologs in Drosophila and mammals. sym-4 encodes a WD-repeat protein and may also have orthologs in Drosophila and mammals. We propose that SYM-3 and SYM-4 contribute to a common developmental pathway that is redundant with a MEC-8-dependent pathway.
GENETIC redundancy is one of the major challenges to a thorough understanding of eukaryotic genomes (Thomas 1993; Cooke et al. 1997; Nowak et al. 1997; Hartwell 2004). Genetically tractable organisms provide a means of addressing this issue through the isolation of mutations that confer synthetic phenotypes, phenotypes that depend on mutations in at least two different genes. One of the first systematic approaches for the isolation of synthetically lethal mutations made use of the budding yeast Saccharomyces cerevisiae (Bender and Pringle 1991). A strain was constructed that had a deletion of a gene that is not essential for viability. The strain also had a mitotically unstable plasmid containing a wild-type copy of the gene and an independent marker for assessing inheritance of the plasmid. Segregants that failed to inherit the plasmid were nonmutant phenotypically, but when a mutation in another gene was induced that was synthetically lethal with the mutation in the first gene, inheritance of the plasmid became essential for viability because it complemented the first mutation and thereby prevented the synthetic lethality. Doubly mutant lines were easily identified on the basis of an absolute requirement for inheritance of the plasmid.
Synthetic mutations have also been described in Caenorhabditis elegans. One example involves doubly mutant hermaphrodites that have excess induction of the precursor cells of the vulva, whereas each single mutant has a normal vulva (Ferguson and Horvitz 1989). Another analysis of genetic redundancy in C. elegans began with a serendipitous observation of synthetic lethality between a loss-of-function mutation in the mec-8 gene and certain viable mutations in unc-52. Although there are defects associated with loss-of-function mutations in mec-8, which encodes a putative RNA splicing factor (Lundquist et al. 1996), null alleles of the gene are for the most part homozygous viable (Lundquist and Herman 1994). The defects include a loss of mechanosensation, for which the gene was named (Chalfie and Sulston 1981); aberrant chemosensation; a slight dumpiness in the body; and a partially penetrant cold-sensitive embryonic lethality. Homozygotes for null alleles segregate ∼27% dead embryos at 16° (vs. 1% at 25°), and the arrested embryos exhibit a Pat (paralyzed arrest at the twofold stage of embryogenesis) phenotype (Lundquist and Herman 1994). In contrast, complete lethality results when animals are homozygous for a mec-8 mutation and any of a set of viable mutations in unc-52 (Lundquist and Herman 1994), which encodes a proteoglycan that resembles perlecan (Rogalski et al. 1993). On the basis of severe alleles of the gene, unc-52 is essential for viability. Null alleles prevent proper formation of the myofilament lattice and result in dead embryos with the Pat phenotype (Rogalski et al. 1993; Hresko et al. 1994; Williams and Waterston 1994). The viable alleles of unc-52, on the other hand, are nonsense mutations in certain exons that undergo alternative splicing that is dependent on the mec-8 gene (Lundquist et al. 1996; Spike et al. 2002). The synthetic lethality between mec-8 and one of the viable alleles of unc-52 results from a decreased ability to skip an unc-52 exon that has the nonsense mutation; most mRNAs have the mutant exon and are therefore nonfunctional.
Although the mec-8 and unc-52 genes are not functionally redundant, the striking nature of the synthetic lethality of mec-8 mutations with unc-52 viable mutations motivated a search for other mutations that might be synthetically lethal with a mutation in mec-8, with the goal of uncovering truly redundant genes (as well as other genes that might behave like unc-52). The search was based on a segregation analysis similar to the plasmid screen mentioned above for yeast (Bender and Pringle 1991). A minichromosome, or extrachromosomal array, containing wild-type copies of the mec-8 gene was used to rescue mutant copies of mec-8 present on the chromosomes. The array also contained a segregation marker, permitting assessment of potentially doubly mutant lines that acquire the requirement that the array be inherited for viability. Five sym (synthetically lethal with mec-8) genes were discovered in this search (Davies et al. 1999). With the exception of sym-5, each of the sym mutations lacks a discernible phenotype when mec-8 is wild type, and with the exception of the combination of sym-1 and sym-5, double-mutant combinations of the sym genes with each other do not confer synthetic lethality or other obvious defects.
The molecular identities of sym-1 and sym-5 have been previously reported (Davies et al. 1999). Both encode proteins with leucine-rich repeats, and a functional fusion of SYM-1 with GFP is secreted from the apical surface of the hypodermis. The combination of sym-1 and mec-8 results in dead embryos that have detached body muscles. RNA interference (RNAi) of sym-5 (or sym-1) in a mec-8 mutant background also confers embryonic lethality. For neither sym-1 nor sym-5 is the molecular basis of the synthetic lethality with mec-8 known, but it is reasonable to assume that these genes function, albeit redundantly, in the attachment of body muscle to the cuticle. These genes are presumed to be either redundant with a gene that requires mec-8 for maturation of its transcript or part of a developmental or other pathway that is redundant with another pathway, one of whose genes requires mec-8 for maturation of its transcript.
Because mec-8 affects alternative RNA splicing of the unc-52 gene, an explanation for the pleiotropy of mec-8 mutations is an involvement of mec-8 in the splicing of transcripts from other genes, including genes that function in mechanosensation, chemosensation, and synthesis of the cuticle (interference of which could cause a dumpy shape). An involvement of mec-8 in RNA splicing can also account for the synthetic lethality of mec-8 with the sym genes, which can be interpreted as one of two modes of genetic redundancy. In one possibility, another gene can substitute for mec-8 in the splicing of critical genes. The sym-2 gene described here may be such an example. In a second possibility, genes or genetic pathways that require mec-8 are redundant with genes or pathways that do not require mec-8. If the second possibility exists, as seems likely on the basis of the nature of the sym-1 and sym-5 gene products, which seem to not be involved in RNA splicing, one might expect a variety of genes to be synthetically lethal with mec-8, as described here for sym-3 and sym-4. These genes may act together in a redundant pathway for attachment of the pharynx to the outer body. The search for mutations synthetically lethal with mec-8 therefore may have revealed an unknown pathway for a critical step during development.
MATERIALS AND METHODS
Genes and alleles:
General growth of C. elegans was as described (Brenner 1974). The wild-type strain was N2 var. Bristol. The mec-8 alleles u74, u314, and u456, all of which behave genetically as null alleles (Lundquist and Herman 1994), were used; they have been characterized at the molecular level (Davies et al. 1999). In this article, mec-8 without an allele designation refers to mec-8(u74). The isolation of sym-2(mn617) II, sym-3(mn618) X, and sym-4(mn619) X has been described (Davies et al. 1999). The following additional mutations were used: LGII—dpy-10(e128), rol-6(e187), unc-4(e120), unc-52(e444), mnDf67, mnDf68, mnC1; LGIII—ncl-1(e1865), unc-36(e251); LGX—lon-2(e678), unc-6(e78). Descriptions of the markers, mutations, and rearrangements can be obtained from WormBase (http://www.wormbase.org).
Correlating the sym mutations with the physical map:
The sym mutations had been previously assigned to chromosomal regions by standard genetic mapping (Davies et al. 1999). For the purpose of positional cloning, single-nucleotide polymorphisms (SNPs) between the N2 isolate and CB4856, a Hawaiian strain, were the basis of precise mapping with respect to the physical map (Jakubowski and Kornfeld 1999; Wicks et al. 2001). In the case of sym-2, Rol non-Unc and Unc non-Rol recombinant progeny of rol-6 sym-2 unc-4/Ha hermaphrodites, where Ha designates the homologous chromosome from CB4856, were picked, homozygosed, and scored with respect to their possible synthetic lethality with mec-8. Similarly, Lon non-Unc recombinants were picked among the self-progeny of lon-2 sym-3 unc-6/Ha hermaphrodites, and viable (non-Sym) Unc-3 recombinant self-progeny of mec-8; sym-4 unc-3/Ha hermaphrodites were identified. SNPs that affect restriction-enzyme sites were chosen on the basis of the website http://genome.wustl.edu/projects/celegans/index.php?snp=1 and were assayed on DNA prepared by the polymerase chain reaction (PCR) from single recombinant worms.
DNA micro-injections and mutational rescue:
For assessing rescue of the sym mutations, strains having the general genotype mec-8 I; ncl-1 unc-36 III; sym; mnEx52[mec-8(+) ncl-1(+) unc-36(+)] were micro-injected (Mello and Fire 1995) with DNA preparations of cosmid clones, provided by A. Coulson (The Wellcome Trust Sanger Institute, Hinxton, UK), that had been selected on the basis of the SNP mapping. Transgenic worms were identified by co-injection of the plasmid pRF4, which confers a dominant rolling behavior (Mello et al. 1991), or co-injection of pTG96, which expresses SUR-5::GFP in nuclei (Yochem et al. 1998). Rescue of the sym mutations was based on the ability of transgenic mothers to produce healthy, Unc-36 progeny. In the absence of rescue, the only viable progeny are fully coordinated, because they must inherit the meiotically unstable array mnEx52, which rescues both unc-36 and mec-8, and therefore rescues the synthetic lethality between mec-8 and the sym mutation. Rescue of the synthetic lethality by a newly formed array that contains wild-type copies of a sym gene changes the pattern of segregation: inheritance of mnEx52 is no longer needed for viability, and viable Unc-36 progeny are observed.
Once positive cosmid clones had been identified for sym-3 and sym-4, the Expand Long Template PCR System (Roche Diagnostics) was used to generate DNA for assessing individual genes for rescue. Wild-type genomic DNA was used as template, and the reaction conditions and the lengths of oligonucleotide primers were according to manufacturer's recommendations. The genomic DNA sequence (C. elegans Sequencing Consortium 1998), accessed via WormBase, was used to design primers. For sym-3, the final primers used were LRB-63 (AAAAAGAGTAGGCAAAGGGCGCAGAGTG) and LRB-64 (GGCTCAAGTGTGACTCGAACTAAAGCTC) and for sym-4, LRB-79 (TTGCTGATGCCGTCATTGACAAGTTCCG) and LRB-80 (GCGGAGACGAGAAGAACAAACTTCATCC). PCR products were micro-injected at ∼15 μg/ml.
RNA manipulations:
Total RNA was isolated from mixed-stage populations using TRIzol (Invitrogen, San Diego). cDNA having the proper 5′ and 3′ ends was generated with the FirstChoice RNA-ligase-mediated rapid amplification of cDNA ends (RLM-RACE) kit (Ambion, Austin, TX). For analyzing cDNA from each of the sym genes, PCR products were generated using a gene-specific primer and a kit-specific primer for the 3′ or 5′ end.
For RNAi, PCR products representing coding regions were generated with pairs of primers that both contained the T7 promoter sequence, and double-stranded RNA (dsRNA) was allowed to form during transcription of the PCR products with a T7 MEGA-script kit (Ambion; Kennerdell and Carthew 1998). For sym-2 (ZK1067.6), the primer sequences were LRB-19 (TAATACGACTCACTATAGGGAATCGGCAAACGTCTCATC), which derives from the first exon, and LRB-20 (TAATACGACTCACTATAGGCCGACTTCTTGATCTTCCAG), which derives from the third exon and is upstream of the region encoding the first RNA recognition motif (RRM). The primers for sym-3 (C54H2.1) were LRB-53 (TAATACGACTCACTATAGGTTCAGCAAAGCTCGTCTTCG) and LRB-54 (TAATACGACTCACTATAGGGTTCAGCCACATAAGACGTC), and for sym-4 (R03E1.1), LRB-42 (TAATACGACTCACTATAGGTACTGCGGATATTCTCGACG) and LRB-43 (TAATACGACTCACTATAGGGTTAGCGAATCCGATTGTCC). After purification with a QIAquick kit (QIAGEN, Chatsworth, CA), dsRNA ranging from 250 to 500 μg/ml was injected into both arms of the gonads of hermaphrodites, which were periodically transferred to new plates for several days to give discrete intervals of egg laying.
DNA sequence analysis:
For detection of mutations, DNA representing all of the exons and the parts of introns immediately flanking the exons was produced from homozygous sym mutants by PCR. After purification with a QIAquick kit (QIAGEN), the DNA sequence of the PCR products was determined directly on both strands by the Advanced Genetic Analysis Center Facility at the University of Minnesota. DNA sequences were compared with the genomic sequence of the wild-type strain, obtained from GenBank, with the BLAST program (Altschul et al. 1997). Because the genomic sequence can contain errors, however infrequent, each mutation was verified by determining the corresponding DNA sequence from SP2141, the parent strain of the sym mutations.
For cDNA analyses, the DNA sequence of RLM-RACE products or of reverse-transcribed PCR products was determined directly. For sym-2, the 5′ and 3′ ends of the mRNA were deduced from RLM-RACE products, but the internal part was determined from PCR products generated from the incomplete cDNA clones yk82b3 and yk479a2, which were provided by Y. Kohara (National Institute of Genetics, Mishima, Japan).
Amino acid sequences were aligned and displayed by means of the PileUp and PrettyBox programs from the Wisconsin Package Version 10.3, Accelrys (GCG), San Diego.
RT-PCR of sym-3(mn618):
RNA was isolated from mixed-stage populations of wild-type worms or from sym-3 homozygotes as described above. cDNA was prepared using the RLM-RACE kit, and PCR products spanning the sixth intron were generated using the cDNA as template. The 5′ primer, LRB-137, had the sequence AAGTCGACGAACCAGTTGTCGAAC and was derived from the 5′ end of the sixth exon. The 3′ primer was the “outer primer” from the RLM kit [the primer that represents the linker added to the poly(A) tail of the cDNA]. After analytical gel electrophoresis and purification with a QIAquick column, the mixtures of PCR products were each directly subjected to DNA sequencing en masse.
RESULTS
The phenotype of the mec-8; sym-2 double mutant:
A strain homozygous for sym-2 lacks an obvious mutant phenotype, but a strain that is heterozygous for sym-2 and homozygous for a loss-of-function mutation in mec-8 produces progeny that arrest growth and development at the twofold stage of embryogenesis (Davies et al. 1999). This stage of arrest has been closely studied, because mutations that affect the structure of body muscles or their attachment to the body cuticle confer an arrest at this stage, usually with paralysis of the embryos, which is termed the Pat phenotype (Williams and Waterston 1994). Because the mec-8; sym-2 double mutants exhibit twitching of body muscles, they have been described as having a mild Pat phenotype (Davies et al. 1999). An example of the synthetic lethality between sym-2 and mec-8 is shown in Figure 1A. The cause of the twofold arrest is unknown.
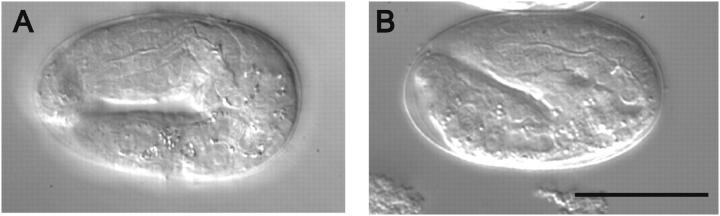
Figure 1.—
The synthetic lethality of mec-8 and sym-2. (A) Nomarski image of an embryo arrested at the twofold stage that segregated from a strain having the genotype mec-8(u74); rol-6 unc-4/sym-2. (B) Image of a developmentally arrested embryo that segregated from a mec-8(u314) homozygote following injection of dsRNA derived from part of the first and third exons and all of the second exon of the sym-2 gene. Injection of mec-8(u74) homozygotes with the same dsRNA produced the same effect. Bar, 25 μm.
RNA interference can be a potent method for depleting a gene product in C. elegans (Fire et al. 1998). It was therefore of interest to determine whether or not sym-2(RNAi) would differentially affect a mec-8 strain vs. the wild-type strain. As indicated below, the sym-2 gene encodes a protein containing copies of an RRM. To minimize the chance of cross-interference, double-stranded RNA was derived from a region of unique DNA sequence outside of the region that encodes the copies of the RRM (see below). Injection of N2 hermaphrodites had no discernible effect on viability, on gross morphology, or on behavior of the progeny. In contrast, injection of mec-8(u74) or mec-8(u314) homozygotes produced striking embryonic lethality (Figure 1B). During the expected peak of inhibition, >95% of the progeny died as embryos; the remaining hatched but were malformed and died in the first larval stage. Thus, RNAi of sym-2 can elicit synthetic lethality with mec-8.
The terminal arrest of 80% or more of the embryos from mec-8 mothers treated with sym-2 dsRNA resembled that of the mec-8; sym-2 double mutants that segregate from sym-2 heterozygotes in a mec-8 mutant background (Figure 1), indicating that a significant maternal effect does not exist for sym-2. As an independent assessment of a maternal effect, the progeny of mothers of genotype mec-8(u456); sym-2 unc-4 were examined; the mothers were rare escapers from a mec-8(u456); mnC1dpy-10 unc-52/sym-2 unc-4 strain. [Rare mec-8; sym-2 unc-4 escapers were also recovered from a mec-8(u74); mnC1 dpy-10 unc-52/sym-2 unc-4 strain (Davies et al. 1999).] Virtually all of the progeny of such mothers died at the same stage of embryogenesis as was seen for the sym-2 homozygotes derived from sym-2 heterozygotes in a mec-8 mutant background. A maternal effect therefore seems unlikely.
The failure of mec-8(+); sym-2 animals to exhibit a mutant phenotype might be the consequence of the sym-2 mutation being a weak allele. To test this notion genetically, sym-2 was placed over genetic deficiencies that delete the rol-6 sym-2 let-23 region: sym-2/mnDf67 unc-4 and sym-2/mnDf68 unc-4 progeny of crosses between sym-2 males and mnC1 dpy-10 unc-52/mnDf unc-4 hermaphrodites were readily identified and exhibited wild-type viabilities and fertilities. We conclude that the sym-2 mutation is probably not a weak allele that exhibits synergy with mec-8 mutations. Although RNAi should not be used to infer the null phenotype of a gene, the effective ability of sym-2 dsRNA to produce striking synthetic lethality with mec-8 mutations while not affecting wild-type worms reinforces an interpretation of sym-2 as redundant with mec-8 for a vital function. Strengthening the notion that it is a significant mutation, mn617 affects a tyrosine codon, described in more detail below, that is conserved in the predicted sym-2 gene product from another species, C. briggsae (Figure 2B).
Figure 2.—
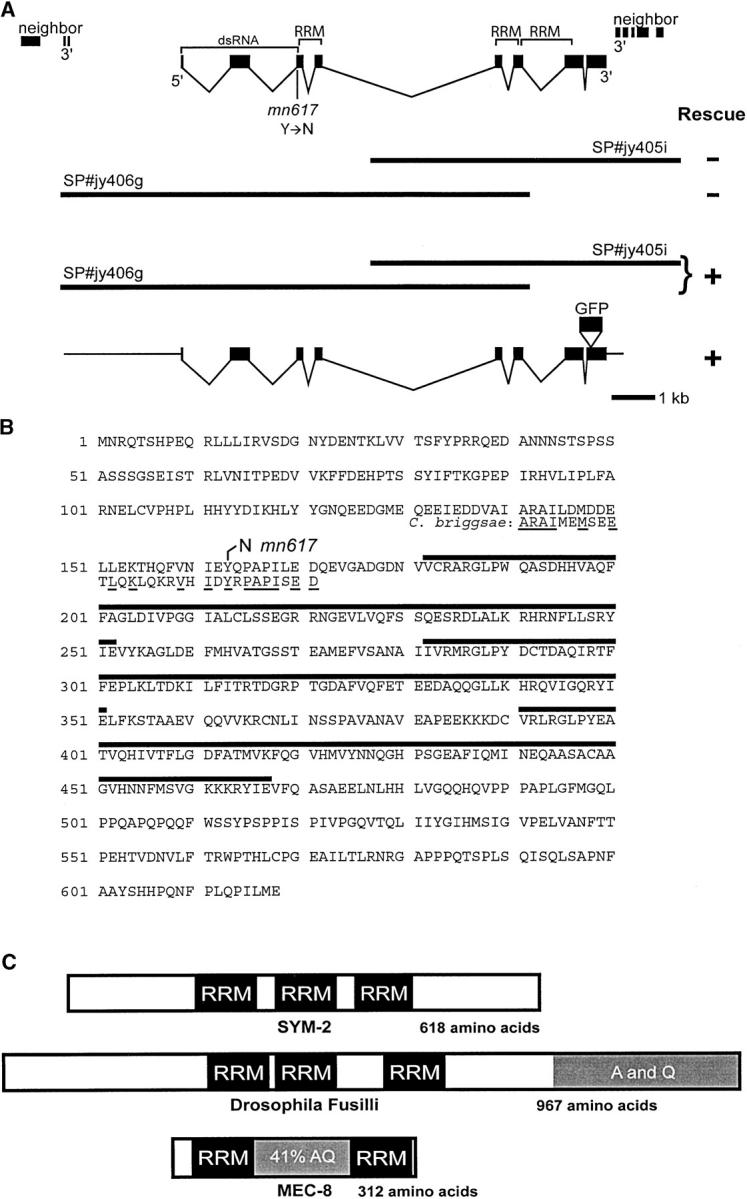
The sym-2 gene and its predicted product. (A) Two subclones of ZK1067 rescued when co-injected but not when individually injected, implicating ZK1067.6 as sym-2. The genomic positions of the eight exons of the gene, determined from cDNA sequences from 5′-RLM-RACE and from 3′-RACE, are indicated relative to the 5′ and 3′ flanking genes, the terminal exons of which are shown at the extreme left and right. The position of the sym-2(mn617) mutation, the regions encoding copies of the RRM motif, and the region used for deriving dsRNA are also indicated. The ability of subclones to rescue the mec-8; sym-2 synthetic lethality is summarized below the schematic of the gene. A schematic of a GFP fusion (SP#jy424b) that rescued sym-2 is also shown. (B) The amino acid sequence predicted for the translation of sym-2. Thick lines over the sequence delimit the three copies of the RNA recognition motif, and the amino acid change associated with mn617 is indicated. Shown beneath parts of the third and fourth lines of the C. elegans sequence is the predicted amino acid sequence, based on data in WormBase, for the region of SYM-2 from C. briggsae that flanks the tyrosine affected by mn617, indicating that the tyrosine is conserved in C. briggsae. Identical amino acids are underlined. (C) A schematic comparison of SYM-2 with the Drosophila hnRNP F-like protein Fusilli (Wakabayashi-Ito et al. 2001) and with MEC-8 (Lundquist et al. 1996), which has a region rich in alanines and glutamines that is not present in SYM-2. The carboxyl-terminal third of Fusilli is also rich in these amino acids.
Identification of the sym-2 gene:
Genetic mapping placed sym-2 to the left of the unc-4 gene on LGII (Davies et al. 1999). To define its position in more detail and to correlate it with the physical map, snip SNP mapping was performed, which placed sym-2 between a DraI SNP on cosmid clone T24B8 and the let-23 gene on cosmid ZK1067. Micro-injection of strain SP2249 [mec-8 I; sym-2 II; ncl-1 unc-36 III; mnEx52[mec-8(+) ncl-1(+) unc-36(+)]] with cosmid DNA from the interval revealed that clone T23G7 obviated the need for mnEx52, the extrachromosomal array that contains mec-8(+), indicating rescue of the sym-2 mutation. Plasmid subclones representing the various genes predicted for T23G7, except for ZK1067.6, were individually tested for rescue without success. In contrast, co-injection—but not single injection—of two overlapping subclones, SP#jy405i and SP#jy406g, efficiently rescued sym-2 (Figure 2). Homologous recombination within the region of overlap should generate an intact ZK1067.6 gene, as seen for other genes such as sur-2 (Singh and Han 1995). Thus, ZK1067.6 is likely to be sym-2.
DNA sequence analysis of ZK1067.6 from a homozygous sym-2 strain revealed a single-nucleotide change relative to the N2 DNA sequence deposited in GenBank. DNA sequencing of the parental strain confirmed that the change is specific to the sym-2 mutant isolate. The nucleotide change, which results in the change of a tyrosine codon to an asparagine codon in the third exon of the reading frame, confirms that ZK1067.6 is sym-2.
The product of sym-2 contains three copies of an RRM:
An analysis of the 5′ and 3′ ends of sym-2 cDNA generated by the RLM-RACE method combined with an analysis of the internal parts of cDNA clones provided by Y. Kohara demonstrated that the mRNA is not trans-spliced and is 2262 or 2269 nucleotides long, the variability arising from different 5′ ends. The donor of the first intron has the sequence GCAAGUUG, in contrast to the much more common sequence GURAGUUU in C. elegans (Blumenthal and Steward 1997). The use of the GC donor has recently been confirmed by new cDNA clones submitted to GenBank and WormBase by Y. Kohara. A GC donor is present in ∼0.6% of the introns of C. elegans, and a small fraction of these are involved in alternative RNA splicing (Farrer et al. 2002). sym-2 appears to fall in the major class, because the cDNA analysis provided no evidence for alternative splicing. Interestingly, sequence data in WormBase indicate that the GC donor is conserved in the sym-2 gene of C. briggsae, but the exon structure of the gene has not been rigorously determined for this organism.
The sym-2 gene can encode a primary translation product of 618 amino acids. It contains three copies of the RRM that is present twice in MEC-8 (Figure 2). mn617 is a T-to-A transversion, changing the 163rd codon of the open reading frame from tyrosine to asparagine, 19 codons upstream of the region encoding the first RRM (Figure 2B). In MEC-8, the two copies of the RRM are separated by a region rich in alanines and glutamines (Lundquist et al. 1996). SYM-2, in contrast, lacks regions that are comparably rich in these amino acids (Figure 2C). SYM-2 is most similar to mammalian hnRNP F and H and to Drosophila Fusilli (Figure 2C), each of which has three copies of the RRM (Wakabayashi-Ito et al. 2001). The resemblance with these proteins is confined, however, to the RRM. In fact, outside of the copies of the RRM, there is no striking resemblance of SYM-2 with other protein species in GenBank, including C. elegans proteins.
The phenotype of the mec-8; sym-3 double mutant:
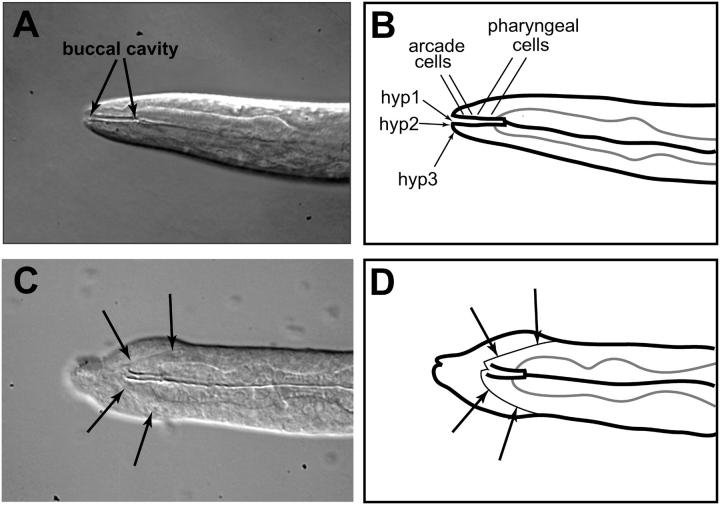
One-quarter of the progeny of a strain heterozygous for mec-8 but homozygous for sym-3 are doubly mutant and die either as late-stage embryos or as L1 larvae. The arrested L1 larvae have been described as having a characteristic swelling of the anterior end, which has been termed a “bulbous nose” (Davies et al. 1999). The arrested embryos and hatchlings have now been examined in more detail with Nomarski optics, revealing an anatomical defect: the anterior end of the pharynx, which appears to be of normal morphology, is not properly attached to the outer body cuticle, precluding the formation of a normal mouth (Figure 3, A–C). Unable to eat, the hatchlings eventually arrest growth and development in the L1 stage.
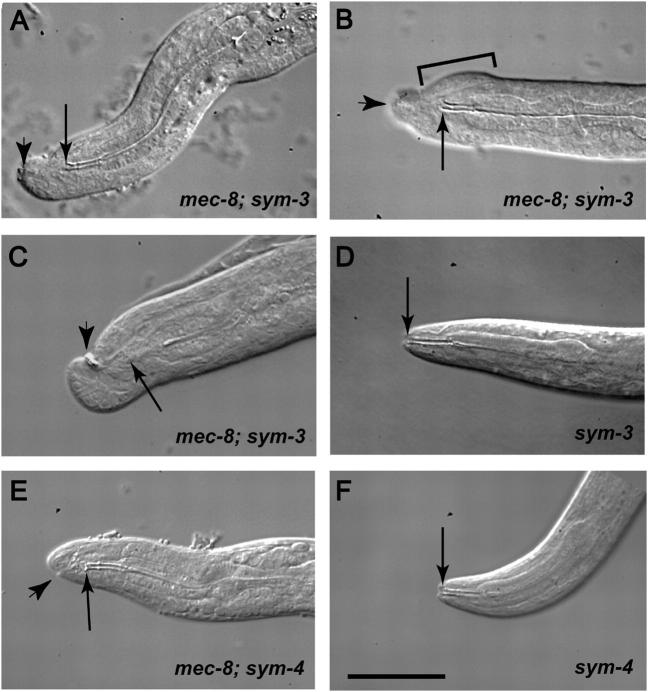
Figure 3.—
The phenotype of sym-3 or sym-4 in combination with mec-8(u74) or mec-8(+). (A–D) Nomarski images of sym-3 homozygotes. (E and F) Nomarski images of sym-4 homozygotes. In A–F, an arrow indicates the anterior end of the buccal cavity of the pharynx. The walls of the cavity appear as parallel lines in these optical sections, which are longitudinal, because of strong refraction of light caused by cuticle that forms the wall of the cavity. (A–C) mec-8; sym-3 double mutants. (A) The anterior end of the pharynx appears to be fully formed but is not properly connected to the anterior tip (arrowhead) of the animal. Overall, the morphology of the pharynx, which is shown in its entirety, appears to be normal. (B) The “bulbous nose” (indicated with a bracket) associated with mec-8; sym-3 (and mec-8; sym-4) double mutants is particularly apparent in this animal. As in A, the pharynx is not properly attached to the body. (C) An example of a malformed, asymmetric anterior end of the body, which is sometimes seen in the double mutant. The arrowhead indicates refraction caused by an accumulation of material. The anterior end of the pharynx (arrow), which is in a different focal plane, is not properly attached to the body. (D) The wild-type phenotype of a sym-3 homozygote in a mec-8(+) background. Note the orifice formed by the proper junction of the anterior end of the pharynx with the body cuticle. (E) The failure of proper attachment of the pharynx to the outer body in a mec-8; sym-4 double mutant. As in C, the anterior end of the body is asymmetric. (F) The wild-type phenotype of a sym-4 homozygote in a mec-8(+) background. Bar (pertains to A–F), 25 μm.
There is a striking uniformity in the degree of posterior displacement of the pharynx; its anterior tip is almost always ∼10 μm from the anterior tip of the animal. Indicating the possibility that a mouth has been formed or at least partly formed, the worms seem to have an indentation of the anterior tip of the body cuticle (Figure 3), and sometimes strings of refractile material (not shown) can be seen between the indentation and the anterior tip of the buccal cavity of the pharynx. Occasionally, the indentation itself has refractile material, and sometimes the indentation is displaced from the center relative to the longitudinal axis of the animal. A worm displaying both of these traits is shown in Figure 3C. The homozygous sym-3 mutation in a wild-type mec-8 background, on the other hand, has been described as conferring no discernible mutant phenotype (Davies et al. 1999). Indeed, the junction of the pharynx with the outer body appears to be completely normal on the basis of Nomarski optics (Figure 3D).
There are several possible explanations for the aberrant morphology of the anterior end. The defect could lie with the pharynx, with the hypodermal cells involved in the attachment of the pharynx, or with both. As a first step toward an understanding, the junction of the hypodermis and the pharyngeal primordium was examined in sym-3(−) and sym-3(+) embryos that segregated from a strain having the genotype mec-8; ncl-1 unc-36; sym-3; mnEx169 [sym-3(+); sur-5::gfp]. The embryos were distinguished by their inheritance of the extrachromosomal array, which expresses SUR-5::GFP in addition to sym-3(+). An abnormal invagination resembling a keyhole is evident at the junction in array-minus embryos (Figure 4A). An array-plus embryo, in contrast, has only a moderate invagination (Figure 4B), which resembles the invagination seen in wild-type N2 embryos (Figure 4C). The array-plus embryo in B, when examined 45 min later, had formed what appeared to be a normal junction with the body: the buccal cavity could be seen at the extreme anterior tip of the embryo. An array-minus embryo, on the other hand, has a pronounced defect later in embryogenesis: the buccal cavity is displaced posteriorly from the anterior end of the embryo, and a channel appears to connect the buccal cavity to the exterior of the body (Figure 4D). It therefore appears that the pharynx is connected to the outer body, which can account for the constant displacement of the pharynx from the anterior tip and for the refractile strings that can sometimes be seen between the buccal cavity and the anterior tip, but the connection is aberrant and perhaps becomes occluded, preventing feeding. One possibility is that the pharynx is abnormally short and thereby compresses the anterior end of the animal, but it is not clear why this would produce a bulbous nose rather than a more uniform compression of the anterior end. Although an aberrancy in the pharynx cannot yet be ruled out, the morphology of the mec-8; sym-3 embryos is perhaps more consistent with an overgrowth of hypodermal tissue that disrupts the formation of a normal mouth.
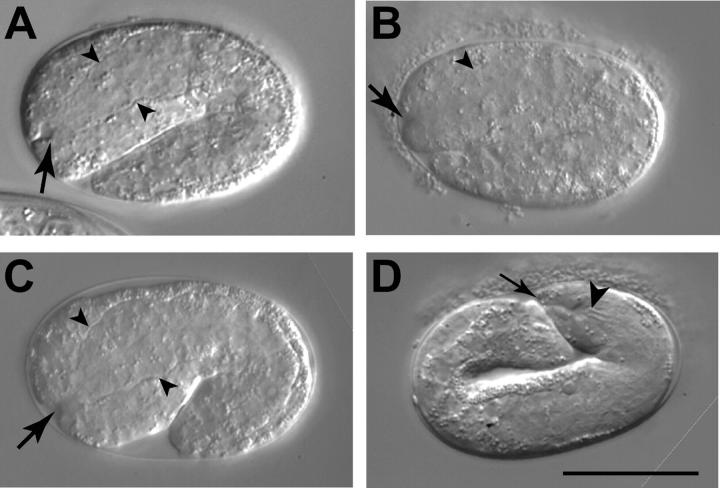
Figure 4.—
Abnormal morphology at the junction of the hypodermis and pharyngeal primordium in embryos. (A) A pronounced invagination (arrow) at the anterior end of a mec-8; sym-3 mutant embryo. The basement membrane of the pharyngeal primordium is evident (arrowheads). (B) An apparently normal invagination (arrow) at the anterior end of a mec-8(−); sym-3(+) sibling of the embryo in A. The focal plane with the maximum invagination is shown. The dorsal part of the pharyngeal primordium can be seen (arrowhead). (C) The invagination of a wild-type N2 embryo. (D) A mec-8; sym-3 embryo later in development exhibiting extreme abnormality at the anterior end. The buccal cavity (arrowhead) is grossly recessed from the anterior tip of the embryo (arrow). A canal may exist between the anterior tip and the buccal cavity. In contrast, the anterior ends of normal embryos at this stage resemble those of the larvae shown in Figure 3, D and F. Bar, 25 μm.
Identification of the sym-3 gene:
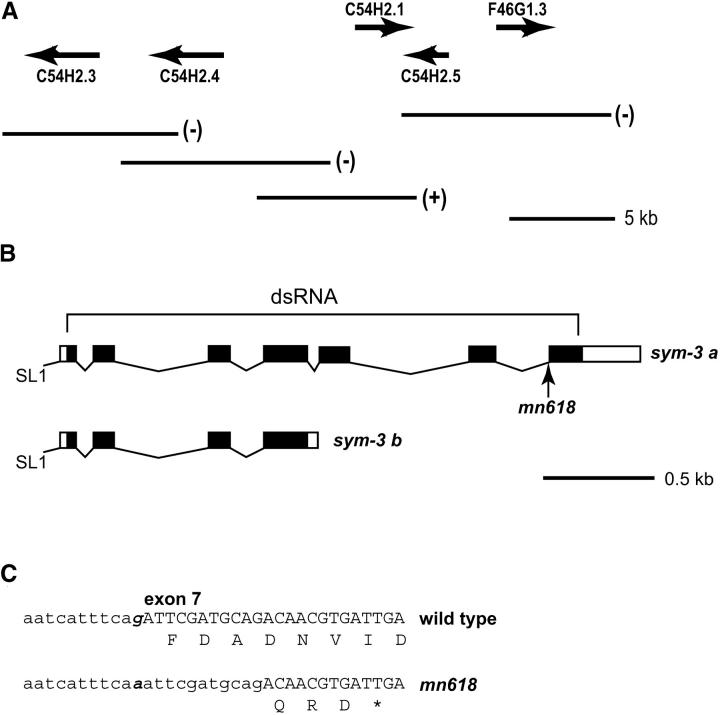
Genetic mapping placed sym-3 between dpy-8 and unc-6 on LGX (Davies et al. 1999). Its position on the physical map has now been investigated by SNP mapping, which has placed the mutation between the cosmid clones W01C8 and F46G11. When cosmid DNA from the interval was tested for rescue by micro-injection of SP2250, a strain of genotype mec-8 I; ncl-1 unc-36 III; sym-3 X; mnEx52[mec-8(+) ncl-1(+) unc-36(+)], C54H2, was shown to rescue fully the synthetic lethality. Injection of long-range PCR products indicated that C54H2.1 is sym-3 (Figure 5A).
Figure 5.—
Identification of the sym-3 gene (C54H2.1). (A) The ability of regions of the cosmid C54H2 (C. elegans Sequencing Consortium 1998) to rescue sym-3 on the basis of micro-injection of long-range PCR products. (B) The relative positions of the exons for the two forms of the gene and the location of sym-3(mn618) in sym-3a are indicated. The exons used for derivation of dsRNA are also indicated. (C) On the basis of DNA sequencing of RT-PCR products, the mn618 mutation of the splice acceptor of exon 7 results in the use of a cryptic acceptor and subsequent truncation of the open reading frame. Intron sequences are shown in lowercase.
An analysis of cDNA produced by the RLM-RACE method indicated that sym-3 produces two mature transcripts. One is 562 nucleotides in length for a predicted protein product, termed SYM-3B, of 158 amino acids; the other has 1231 nucleotides for a product, SYM-3A, of 305 amino acids. The mRNAs have identical 5′ ends, which are trans-spliced to SL1, but the shorter transcript results from alternative 3′ end formation within the fourth intron of the gene relative to the larger mRNA (Figure 5B). The shorter transcript, which is less abundant than the long form, is polyadenylated 12 nucleotides downstream of a sequence AAUGAA, which is the second-most common sequence for polyadenylation in C. elegans (Blumenthal and Steward 1997).
DNA sequence analysis of a homozygous strain has revealed that the sym-3 mutation is a G-to-A transition of the final base of the splice acceptor of the seventh and final exon of sym-3a (Figure 5, B and C). The alteration of the nearly completely conserved G [the consensus for splice acceptors in C. elegans is UUUUCAG (Blumenthal and Steward 1997)] would be expected to have a striking effect on splicing of the primary transcript of the longer species of mRNA. Indeed, an analysis of RT-PCR products generated from a sym-3 strain revealed a difference relative to the wild-type N2 strain. Direct DNA sequencing of the PCR products provided no evidence for use of the mutant splice site. Instead, a cryptic site 11 bp downstream and having the sequence GAUGCAG is used as the splice acceptor (Figure 5C); there was no evidence from the DNA sequencing trace for use of other splice acceptors in the region. The consequence of the cryptic splice is an introduction of three novel amino acids before a stop codon, which would truncate the product by 58 amino acids relative to the longer form of sym-3.
The sym-3 protein has orthologs in Drosophila and mammals:
The sym-3 gene products have an amino acid sequence relationship with proteins of unknown function in insects and mammals. The resemblance encompasses all of the short form, but the long form extends for 147 amino acids beyond the region of the resemblance (Figure 6). The insect and mammalian proteins are longer than SYM-3A, and they resemble each other in the region that extends beyond the end of SYM-3A. They least resemble each other in their middle sections; this region begins where SYM-3A diverges in sequence from each of them and where SYM-3B terminates. The carboxyl-terminal third of the insect or mammalian protein does not have an amino acid resemblance to any predicted gene product in C. elegans; i.e., the insect or mammalian gene is not an obvious fusion of two distinct genes in C. elegans.
Figure 6.—

The amino-terminal half but not the carboxyl-terminal half of SYM-3A is similar to the amino-terminal parts of proteins of unknown function from other species. The amino acid sequences of proteins from Drosophila (GenBank no. NM136259), mosquito (Anopheles gambiae; GenBank XM-318591), and mouse (GenBank NM 153560) are aligned. The proteins, which are shown in their entirety, are longer than SYM-3A and resemble each other outside the region of similarity with SYM-3A. The proteins least resemble each other in their middle parts, which correspond to the carboxyl-terminal half of SYM-3A. The location and consequence of the mn618 mutation are indicated. The fourth row of the alignment includes the final four amino acids predicted for SYM-3B, which is otherwise identical with SYM-3A up to this point.
The sym-3 mutation, which lies carboxyl terminal to the region of resemblance with the orthologs, should truncate the long form of SYM-3 by 58 amino acids. The mutation, however, should not affect the shorter transcript. Injection of mec-8(u314) animals or of the wild-type strain with dsRNA representing all of the sym-3 exons (Figure 5B) caused a small percentage of the progeny of both strains to die as embryos but did not otherwise elicit a strong phenotype and therefore could not be used to address the question of whether or not sym-3(mn618) causes a loss of a nonessential function in a wild-type background or is a mutation that is specific for a mec-8 mutant background.
The phenotype of the mec-8; sym-4 double mutant:
The mec-8; sym-4 double mutant resembles the mec-8; sym-3 double mutant in having a bulbous nose (Davies et al. 1999). As for sym-3, there is no overt phenotype when mec-8 is wild type. The double-mutant combination of sym-3 and sym-4 in a wild-type mec-8 background also is without a phenotype (Davies et al. 1999). Closer examination of mec-8; sym-4 segregants has revealed L1 larvae and late-stage embryos with improperly attached pharynges as described above for sym-3 (Figure 3E). As for sym-3 in the mec-8 mutant background, there is a constant displacement of the anterior tip of the buccal cavity from the anterior tip of the animal, and an asymmetry in the indentation of the anterior tip of the animal relative to the central axis is sometimes seen (Figure 3E). In contrast, the junction of the pharynx and outer body appears to be normal for the sym-4 mutation when mec-8 is wild type (Figure 3F). The similarity of the improperly attached pharynx associated with the mec-8; sym-3 or mec-8; sym-4 double mutant strengthens the notion that sym-3 and sym-4 are involved in a common function or pathway.
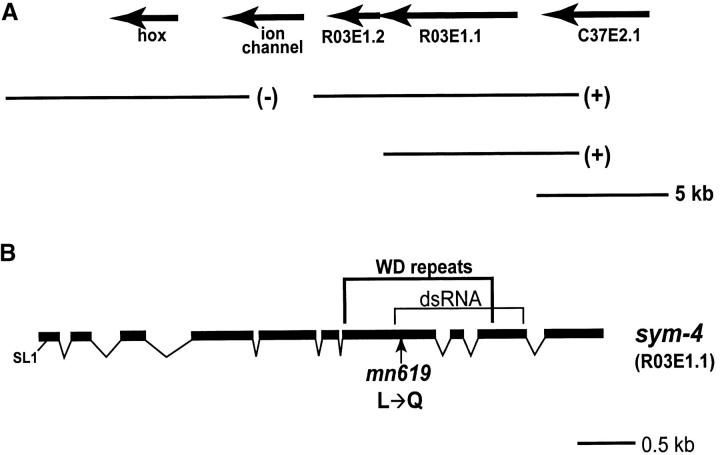
Identification of the sym-4 gene:
Linkage of sym-4 to unc-3 on the right arm of LGX has been previously demonstrated (Davies et al. 1999). A correlation with a small region of the physical map has now been achieved by snip SNP mapping, placing the mutation between cosmids C33G3 and C11H1. A clone, R03E1, from the interval fully rescues sym-4. On the basis of micro-injection of long-range PCR products corresponding to genes on the cosmid, sym-4 is R03E1.1 (Figure 7A), a conclusion reinforced by DNA sequencing of sym-4(mn619), which is a T-to-A transversion, creating a missense mutation in the seventh exon (Figure 7B). The mn619 change is not in the parental strain SP2141; DNA sequencing of the region from this strain showed a complete agreement with the N2 sequence submitted to GenBank by the C. elegansSequencing Consortium (1998). [A second allele of sym-4, mn620, has been reported (Davies et al. 1999); DNA sequencing has shown that it has a change identical to that of mn619, raising the likelihood that the mutants were not isolated independently of each other.]
Figure 7.—
Identification of the sym-4 gene (R03E1.1). (A) The position of the genes for the rescuing cosmid R03E1 (C. elegans Sequencing Consortium 1998) and the ability of long-range PCR products to rescue sym-4. (B) A schematic of the exons of the gene and the location of the sym-4(mn619) mutation. The region of the gene that encodes the seven WD repeats and the exons used for derivation of dsRNA are indicated.
The product of sym-4 contains WD repeats:
Analysis of cDNA by RLM-RACE has shown that the major form of the sym-4 mRNA derives from 10 exons (Figure 7B). The 5′ ends of this mRNA fall into two general classes. Approximately one-half are SL1 trans-spliced 23 nucleotides upstream of the presumed AUG start codon. The other class contains mRNAs that are not trans-spliced and have variable 5′ ends. The 5′ end of the major species is 64 nucleotides upstream of the AUG, with other species having even longer 5′ ends. There are also minor, uncharacterized forms of the mRNA that have 5′ ends that start within exon 4 of the major form. Although sym-4 has variation in the 5′ ends of its mRNA, there is no evidence for alternative splicing of the primary transcript or for alternative 3′-end formation.
The predicted product of the sym-4 gene has 1043 amino acids and contains a region in the carboxyl portion that has seven tandem copies of the WD motif. The sym-4 mutation changes a leucine to a glutamine in the third WD repeat. Although genes encoding WD proteins are common in eukaryotes, a stretch of WD repeats in SYM-4 has a striking resemblance to a stretch of tandem WD repeats in a human protein and a Drosophila protein, both of unknown function (Figure 8). The third and fourth repeats in particular are similar among the species, and a codon for leucine corresponding to the one altered by the sym-4 mutation is present in these other species in the midst of this strong resemblance. Although the human and fly proteins are similar to SYM-4 both in size and in having their WD repeats confined to their carboxyl-terminal halves, their amino-terminal portions have no sequence resemblance to SYM-4. In fact, the amino terminal half of SYM-4 lacks amino acid sequence resemblance to other proteins in the databases, including C. elegans proteins.
Figure 8.—

A comparison of the carboxyl-terminal half of SYM-4 with the carboxyl-terminal halves of WD-repeat proteins of unknown function from human (GenBank accession no. NP-061918) and from Drosophila melanogaster (NP 651742). The seven predicted WD repeats are designated. Note that strong resemblance extends over several adjacent copies of the WD motif and that the mn619 mutation affects a conserved amino acid in the region of strong resemblance.
As for sym-3, injection of dsRNA derived from sym-4 failed to elicit a phenotype in wild-type worms or in worms homozygous for mec-8(u314). The region used for the analysis of RNAi should be significant with respect to function of the protein, as it encodes many of the WD motifs and includes the site that has undergone the sym-4 mutation, as shown in Figure 7. Not all phenotypically mutable genes, however, are susceptible to RNAi in C. elegans (Fire et al. 1998; Kamath and Ahringer 2003). Some of these genes function in the nervous system, and others encode stable proteins (Kamath and Ahringer 2003), but it is safe to say that the cause of insusceptibility is not known in every case. That the general conditions used for sym-3 and sym-4 are otherwise effective can be seen from the striking effect of the sym-2 dsRNA on mec-8 worms. Also, it was possible to elicit phenotypes for other genes on the sym-3 and sym-4 cosmids. For example, injection of wild-type worms with dsRNA from R03E1.2 results in swollen, rod-like progeny that die in the first larval stage. The analysis here did not make use of rrf-3 worms, which have increased sensitivity to RNAi (Simmer et al. 2003), but rrf-3 mutants are not completely wild type. In any case, it remains unknown whether sym-4(mn619) is a loss-of-function mutation in an otherwise wild-type background. The possibility remains that it may have only a reduction of function, which is apparent, albeit dramatically so, only in a mec-8 mutant background.
DISCUSSION
The molecular cloning of sym-2, sym-3, and sym-4 completes the initial analysis of a set of five genes that were identified by their synthetic lethality with mutations in the mec-8 gene (Davies et al. 1999). Although redundant with mec-8, the products of sym-2, sym-3, and sym-4 are distinct from each other and from the products of the previously described sym-1 and sym-5 genes, which encode proteins with leucine-rich repeats (Davies et al. 1999). The variety of genes found to be redundant with mec-8 probably reflects in part an involvement of its product in the splicing of transcripts from several genes that may not have a structural or functional relationship with each other. This is a straightforward conclusion based on the pleiotropy of mec-8 mutations themselves, indicating an involvement in distinct functions, ranging from mechanosensation and chemosensation to body shape. The almost certain involvement of mec-8 in the processing of gene transcripts required for these functions makes general conclusions about the extent of genetic redundancy uncertain. Nevertheless, mec-8 and the five sym genes illustrate the potential extent of genetic redundancy: the distinct phenotype of sym-1 or sym-5 in a mec-8 mutant background vs. sym-3 or sym-4 implies that at least two different pathways or processes are redundant with at least two pathways or processes that require mec-8. Because the isolation of sym mutations is probably not saturated, additional pathways that are redundant or partly redundant with mec-8-dependent pathways may yet be discovered.
The classic category of genetic redundancy is composed of pairs of genes that encode similar products that can apparently substitute for each other, at least under laboratory conditions. One of the first discoveries of this nature was the histone genes of S. cerevisiae (Rykowski et al. 1981). Redundant, or partly redundant, genes that encode similar products are also well known in C. elegans. Among the examples are lin-12 and glp-1 (Lambie and Kimble 1991); ksr-1 and ksr-2 (Ohmachi et al. 2002); npc-1 and npc-2 (Sym et al. 2000); and akt-1 and akt-2 (Paradis and Ruvkun 1998). Because the products of both sym-2 and mec-8 have copies of an RNA recognition motif, these genes may also fall in this category. sym-2 and mec-8 are, in fact, 2 of 100 genes in C. elegans that encode RRM proteins (C. elegans Sequencing Consortium 1998). As possibly illustrated by sym-2 and mec-8, redundancy within this family, which might reflect in part an increase in fitness due to greater accuracy and efficiency of having more than one gene involved in a particular aspect of RNA maturation, might obscure forward and reverse genetic analyses of these processes, and the analysis of doubly or even triply mutants (or RNAi combinations) may thus be needed to infer function for many of the RRM genes.
SYM-2 resembles Drosophila Fusilli and mammalian hnRNP F and H in having three copies of the RRM. The mammalian proteins affect alternative splicing of several genes (Chen et al. 1999; Chou et al. 1999), and thus it seems reasonable that SYM-2 may also affect alternative splicing, as discussed above in the context of mec-8. Genetic interactions between Fusilli and Cactus indicate that Fusilli functions downstream of the EGF receptor for proper dorsal-ventral polarity of the embryo, but its requirement is more general, because homozygotes have a more severe phenotype, an arrest of growth at the end of embryogenesis (Wakabayashi-Ito et al. 2001). It is not known whether or not mec-8 and sym-2 are required redundantly downstream of let-23, the EFG receptor in C. elegans (Aroian et al. 1990). The combination of mec-8 and sym-2 confers embryonic lethality, whereas progeny that are homozygous for a null allele of let-23 arrest growth later in development, during the first larval stage (Aroian and Sternberg 1991; Koga and Ohshima 1995), but a maternal contribution of let-23 could mask a requirement for the gene during embryogenesis.
Although the most straightforward notion is that mec-8 and sym-2 are required redundantly for the splicing of pre-mRNA from one or more vital genes, sym-2 and mec-8 also illustrate the potential complexity of genetic redundancy and how this complexity can obfuscate genomic analyses. It is also possible, for example, that mec-8 is required for the splicing of a transcript from a gene that is redundant with another gene that requires sym-2 but not mec-8. The gene that requires mec-8 need not be related in sequence to the gene that requires sym-2. The genes also need not be related in function: they could act at different steps in two separate developmental or biochemical pathways that are redundant with each other. Because the pathways could be redundant within the same cell or tissue, expression of both RRM genes within the same cell cannot distinguish the mechanism of redundant function. Adding to the potential complexity, the possibility should be considered that sym-2 affects an aspect of RNA maturation other than splicing.
It is unlikely that sym-3 and sym-4 encode factors that are interchangeable with MEC-8 for RNA splicing. Our interpretation of these genes is that they are part of a developmental pathway for proper attachment of the pharynx to the body, but this pathway is redundant with a pathway that has a gene that requires mec-8 for maturation of its transcript. That sym-3 and sym-4 may be part of the same pathway is inferred from the similarity of their phenotypes in a mec-8 mutant background and from an absence of phenotype for the sym-3 sym-4 double mutant when mec-8 is wild type. Because redundant genes sometimes encode similar products, as we have seen above, one might search the genome for genes that resemble sym-3 or sym-4 and thereby identify candidate genes in a possibly redundant pathway. The C. elegans genome contains 90 genes that encode proteins with WD repeats (C. elegans Sequencing Consortium 1998). Although none seems to be an obvious paralog of sym-4 on the basis of amino acid sequence, one of these genes could be interchangeable with it. sym-3, on the other hand, has no obvious counterpart in the C. elegans genome, and the region that is conserved with mammals and Drosophila is not found in any other predicted gene product in C. elegans. sym-3 and sym-4 may be required together for a function in a pathway, but in a redundant pathway, a single gene may suffice for this function. It is also possible that sym-3 is required for expression of sym-4 (or vice versa), but the gene that is redundant with sym-4 does not require sym-3 for its expression.
The junction of the pharynx with the body cuticle in wild-type worms involves the cells of the buccal cavity of the pharynx and the hypodermal cell hyp1, which in turn is connected to hyp2 (Wright and Thomson 1981; Sulston et al. 1983). A significant feature of the phenotype arising from the combination of sym-3 or sym-4 with mec-8 is that the buccal cavity of the pharynx is of normal length and morphology (Figure 3). If the cells of the cavity are truly wild type, then the pathology might lie with the hypodermal cells involved in forming the mouth or with other hypodermal cells near the junction of the pharynx with the outer body. Perhaps these cells undergo hypertrophy or hyperplasia, thereby producing a bulge, the bulbous nose, of the anterior end of the animal and a partial severing of the pharynx from the anterior end, such that there is a nearly constant distance of the buccal cavity from the anterior end. The refractile strings of material that appear to connect the buccal cavity to the tip may represent a partially severed connection. Although it has been difficult to assess the aberrancy with Nomarski optics or with DAPI staining of nuclei, that overgrowth may exist is perhaps hinted at by an examination of the mec-8; sym-3 double mutant shown in Figure 3B. There appears to be a tapering of an interior part of the body toward the anterior tip of the mutant (Figure 9). The size and shape of this interior part resemble the size and shape of the anterior end of a normal worm, and the apparent overgrowth of the anterior tip of the mutant is peripheral to this tapered part.
Figure 9.—
Possible overgrowth of tissue at the anterior end of a mec-8; sym-3 double mutant. (A) The normal junction of the pharynx with the outer body as seen in the sym-3 single mutant of Figure 3D. The anterior and posterior extremes of the buccal cavity are indicated. (B) A schematic indicating the normal locations of the apical parts of the epithelia (Wright and Thomson 1981; Sulston et al. 1983) at the junction of the pharynx and the anterior tip of the body. The cuticle of the body, of the buccal cavity, and of the pharynx is indicated by a thick black line. The basement membrane of the pharynx is shaded. (C) The mec-8; sym-3 double mutant shown in Figure 3B. The arrows indicate a line of refractivity that tapers in the anterior direction beneath what appears to be an overgrowth of hypodermal material at the anterior end. (D) A schematic of the bulbous nose in C, with a thin black line indicating the tapered refractivity. Reminiscent of a normal anterior end, the thin line appears to be associated with the buccal cavity.
The apparent overgrowth of material at the anterior end raises the possibility that sym-3 and sym-4 are involved in redundant, negative regulation of the number, size, or ploidy of hypodermal cells near or within the junction with the pharynx. If this hypothesis is true, this would be another case of redundant, negative regulation in C. elegans: the SynMuv genes can be divided into two redundant classes of negative regulators of the vulval fates of certain hypodermal cells (Ferguson and Horvitz 1989). Redundant negative regulation may reflect the potential for inappropriate gene expression or for imprecise control of cell division to lead to decreased fitness for metazoa. Cancer, for instance, is to a large extent a perturbation of negative regulation. Regulation in metazoa is thought in general to arise by inhibition of existing cellular processes (Kirschner and Gerhart 1998), and redundancy may be one means by which inhibitions are efficiently imposed: there may be selective pressure for increasing the efficiency of inhibition of a particular process by the evolution of an additional negative regulator. Initially, only partial redundancy may exist with respect to the genes encoding these distinct regulators, but in time complete genetic redundancy may result. Implicit in this argument is that redundant regulators may have independent origins and therefore may not resemble each other molecularly. LIN-35, the retinoblastoma tumor suppressor (Lu and Horvitz 1998), may be an example of this in C. elegans: it participates redundantly with unrelated proteins in negative regulation of cell division during development (Boxem and van den Heuvel 2001, 2002; Fay et al. 2002, 2003). We believe that the majority of cases in which a gene is redundant with a gene that encodes a dissimilar product will be explained by participation in negative regulation of a particular process. (In this argument, we are distinguishing true genetic redundancy from additive or synergistic “sickness,” an overwhelming of an organism caused by a combination of silent or mildly detrimental mutations in unrelated processes.)
Although an improperly attached pharynx is associated with other mutations in C. elegans, the phenotype of the mec-8; sym-3 or mec-8; sym-4 double mutant has not, to the best of our knowledge, been described before. In these other examples, which include the innexin gene inx-3 (Starich et al. 2003) and the double-mutant combination of lin-35 and ubc-18 (Fay et al. 2003), the pharynx usually does not extend as far anteriorly as described herein, and refractile strings between the anterior end of the pharynx and the anterior end of the body are not apparent. Thus, the sym screen may have defined a new defect in attachment of the pharynx to the body cuticle. If sym-3 and sym-4 are involved in negative regulation of a developmental pathway, similar regulation may exist in other animals on the basis of possible orthologs in insects and mammals.
As for sym-1 and sym-5 (Davies et al. 1999), a function for sym-3 and sym-4 may have remained undiscovered for some time had it not been for a search for mutations that are synthetically lethal with mec-8. Moreover, one could not have easily deduced a priori that these genes would be redundant with mec-8. Although the nature of its product makes sym-2 more straightforward, the approach has also facilitated an analysis of this gene. In effect, mec-8 mutations create a sensitized background for isolation of enhancer mutations of various genes whose transcripts require mec-8 for their maturation or of various genes that act in consort with or in the same pathway as genes whose transcripts require mec-8, and the approach using extrachromosomal arrays that rescue mec-8 mutations provides a convenient means of isolating and maintaining these mutations in a mec-8 mutant background. The screen for sym mutations is one of several screens for synthetic mutations in C. elegans that have used extrachromosomal arrays. For instance, the method has been effectively applied to searches for mutations that are synthetically lethal with the C. elegans retinoblastoma gene lin-35 (Fay et al. 2002, 2003). As for the sym genes, the redundant genes identified in these screens often do not resemble each other. As discussed above, one explanation is an increase in fitness due to co-evolution from distinct origins of redundant pathways for negative regulation of certain developmental processes, such as those requiring cessation of the cell-division cycle.
sym-3 and sym-4 are thought to be part of a pathway that is redundant with a pathway that requires mec-8 for splicing of a transcript of a component gene. If this is the case, it should be possible to use the method of extrachromosomal arrays to isolate non-mec-8 mutations that are synthetically lethal with either sym-3 or sym-4 or, expectedly, with both, with the goal of discovering the mec-8-dependent gene and other genes in a pathway that is redundant with a pathway that requires sym-3 and sym-4. In support of the potential effectiveness of this approach, four mutations, defining three or four genes, that are synthetically lethal with sym-1 have been isolated (R. K. Herman, unpublished observations). Because one cannot always predict which genes are redundant with others, this unbiased method offers one of the best ways for enhancing our understanding of genetic redundancy, and the method should add to our understanding of particular processes such as attachment of body muscles to the cuticle in the case of sym-1 or attachment of the pharynx to the body cuticle in the case of sym-3 and sym-4.
Acknowledgments
We thank Katherine Cygnar for help with the SNP mapping of sym-2, Claire Kari for help with the SNP mapping of sym-3, Alan Coulson for generously providing cosmid clones, Yuri Kohara for generously providing cDNA clones for the sym-2 gene, and Jocelyn Shaw for discussions. Some nematode strains were supplied by the Caenorhabditis Genetics Center, which is supported by a contract between the National Institutes of Health (NIH) National Center for Research Resources and the University of Minnesota. Support from the NIH (GM-22387) is gratefully acknowledged.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian, R. V., and P. W. Sternberg, 1991. Multiple functions of let-23, a Caenorhabditis elegans receptor tyrosine kinase gene required for vulval induction. Genetics 128: 251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian, R. V., M. Koga, J. E. Mendel, Y. Ohshima and P. W. Sternberg, 1990. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 348: 693–699. [DOI] [PubMed] [Google Scholar]
- Bender, A., and J. R. Pringle, 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal, T., and K. Steward, 1997 RNA processing and gene structure, pp. 117–145 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- Boxem, M., and S. van den Heuvel, 2001. lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development 128: 4349–4359. [DOI] [PubMed] [Google Scholar]
- Boxem, M., and S. van den Heuvel, 2002. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr. Biol. 12: 906–911. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Chalfie, M., and J. Sulston, 1981. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82: 358–370. [DOI] [PubMed] [Google Scholar]
- Chen, C. D., R. Kobayashi and D. M. Helfman, 1999. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes Dev. 13: 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, M. Y., N. Rooke, C. W. Turck and D. L. Black, 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 19: 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, J., M. A. Nowak, M. Boerlijst and J. Maynard-Smith, 1997. Evolutionary origins and maintenance of redundant gene expression during metazoan development. Trends Genet. 13: 360–364. [DOI] [PubMed] [Google Scholar]
- Davies, A. G., C. A. Spike, J. E. Shaw and R. K. Herman, 1999. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics 153: 117–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer, T., A. B. Roller, W. J. Kent and A. M. Zahler, 2002. Analysis of the role of Caenorhabditis elegans GC-AG introns in regulated splicing. Nucleic Acids Res. 30: 3360–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, D. S., S. Keenan and M. Han, 2002. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, D. S., E. Large, M. Han and M. Darland, 2003. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development 130: 3319–3330. [DOI] [PubMed] [Google Scholar]
- Ferguson, E. L., and H. R. Horvitz, 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Hartwell, L., 2004. Robust interactions. Science 303: 774–775. [DOI] [PubMed] [Google Scholar]
- Hresko, M. C., B. D. Williams and R. H. Waterston, 1994. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J. Cell Biol. 124: 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski, J., and K. Kornfeld, 1999. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics 153: 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., and J. Ahringer, 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J. R., and R. W. Carthew, 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95: 1017–1026. [DOI] [PubMed] [Google Scholar]
- Kirschner, M., and J. Gerhart, 1998. Evolvability. Proc. Natl. Acad. Sci. USA 95: 8420–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, M., and Y. Ohshima, 1995. Mosaic analysis of the let-23 gene function in vulval induction of Caenorhabditis elegans. Development 121: 2655–2666. [DOI] [PubMed] [Google Scholar]
- Lambie, E. J., and J. Kimble, 1991. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112: 231–240. [DOI] [PubMed] [Google Scholar]
- Lu, X., and H. R. Horvitz, 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95: 981–991. [DOI] [PubMed] [Google Scholar]
- Lundquist, E. A., and R. K. Herman, 1994. The mec-8 gene of Caenorhabditis elegans affects muscle and sensory neuron function and interacts with three other genes: unc-52, smu-1 and smu-2. Genetics 138: 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist, E. A., R. K. Herman, T. M. Rogalski, G. P. Mullen, D. G. Moerman et al., 1996. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development 122: 1601–1610. [DOI] [PubMed] [Google Scholar]
- Mello, C., and A. Fire, 1995 DNA transformation, pp. 451–482 in Caenorhabditis elegans: Modern Biological Analysis of an Organism, edited by H. F. Epstein and D. C. Shakes. Academic Press, San Diego.
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak, M. A., M. C. Boerlijst, J. Cooke and J. M. Smith, 1997. Evolution of genetic redundancy. Nature 388: 167–171. [DOI] [PubMed] [Google Scholar]
- Ohmachi, M., C. E. Rocheleau, D. Church, E. Lambie, T. Schedl et al., 2002. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr. Biol. 12: 427–433. [DOI] [PubMed] [Google Scholar]
- Paradis, S., and G. Ruvkun, 1998. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski, T. M., B. D. Williams, G. P. Mullen and D. G. Moerman, 1993. Products of the unc-52 gene in Caenorhabditis elegans are homologous to the core protein of the mammalian basement membrane heparan sulfate proteoglycan. Genes Dev. 7: 1471–1484. [DOI] [PubMed] [Google Scholar]
- Rykowski, M. C., J. W. Wallis, J. Choe and M. Grunstein, 1981. Histone H2B subtypes are dispensable during the yeast cell cycle. Cell 25: 477–487. [DOI] [PubMed] [Google Scholar]
- Simmer, F., C. Moorman, A. M. Van Der Linden, E. Kuijk, P. V. Van Den Berghe et al., 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N., and M. Han, 1995. sur-2, a novel gene, functions late in the let-60 ras-mediated signaling pathway during Caenorhabditis elegans vulval induction. Genes Dev. 9: 2251–2265. [DOI] [PubMed] [Google Scholar]
- Spike, C. A., A. G. Davies, J. E. Shaw and R. K. Herman, 2002. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development 129: 4999–5008. [DOI] [PubMed] [Google Scholar]
- Starich, T. A., A. Miller, R. L. Nguyen, D. H. Hall and J. E. Shaw, 2003. The Caenorhabditis elegans innexin INX-3 is localized to gap junctions and is essential for embryonic development. Dev. Biol. 256: 403–417. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., E. Schierenberg, J. G. White and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Sym, M., M. Basson and C. Johnson, 2000. A model for Niemann-Pick type C disease in the nematode Caenorhabditis elegans. Curr. Biol. 10: 527–530. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., 1993. Thinking about genetic redundancy. Trends Genet. 9: 395–399. [DOI] [PubMed] [Google Scholar]
- Wakabayashi-Ito, N., M. P. Belvin, D. A. Bluestein and K. V. Anderson, 2001. fusilli, an essential gene with a maternal role in Drosophila embryonic dorsal-ventral patterning. Dev. Biol. 229: 44–54. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Williams, B. D., and R. H. Waterston, 1994. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 124: 475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, K. A., and J. N. Thomson, 1981. The buccal capsule of Caenorhabditis elegans (Nematoda: Rhabditoidea): an ultrastructural study. Can. J. Zool. 59: 1952–1961. [Google Scholar]
- Yochem, J., T. Gu and M. Han, 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]