Abstract
Transposons and transposon-like repetitive elements collectively occupy 44% of the human genome sequence. In an effort to measure the levels of genetic variation that are caused by human transposons, we have developed a new method to broadly detect transposon insertion polymorphisms of all kinds in humans. We began by identifying 606,093 insertion and deletion (indel) polymorphisms in the genomes of diverse humans. We then screened these polymorphisms to detect indels that were caused by de novo transposon insertions. Our method was highly efficient and led to the identification of 605 nonredundant transposon insertion polymorphisms in 36 diverse humans. We estimate that this represents 25–35% of ∼2075 common transposon polymorphisms in human populations. Because we identified all transposon insertion polymorphisms with a single method, we could evaluate the relative levels of variation that were caused by each transposon class. The average human in our study was estimated to harbor 1283 Alu insertion polymorphisms, 180 L1 polymorphisms, 56 SVA polymorphisms, and 17 polymorphisms related to other forms of mobilized DNA. Overall, our study provides significant steps toward (i) measuring the genetic variation that is caused by transposon insertions in humans and (ii) identifying the transposon copies that produce this variation.
TRANSPOSONS and transposon-like repetitive elements collectively occupy an impressive 44% of the human genome sequence (Lander et al. 2001). Alu and LINE (L1) elements alone account for ∼30% of the genome sequence and are the most abundant transposable elements in humans (Lander et al. 2001). Both Alu and L1 also are actively mobile in the genome today and serve as ongoing sources of human genetic variation (Moran et al. 1996; Ostertag and Kazazian 2001; Batzer and Deininger 2002; Brouha et al. 2003; Dewannieux et al. 2003). The remaining transposon-like elements in the genome have some or all of the hallmark features of transposons, such as target site duplications (TSDs), terminal repeats, and/or poly(A) tails, but are not known to remain functional (Smit and Riggs 1996; Smit 1999; Lander et al. 2001).
Alu elements have been actively mobile in primate genomes during the past 65 million years and consequently have expanded to >1 million copies in the human genome today (Batzer and Deininger 2002 and references therein). The earliest Alu elements appear to have been monomeric derivatives of 7SL RNA, and these monomers later gave rise to dimeric Alu elements (Ullu and Tschudi 1984; Slagel et al. 1987; Britten et al. 1988; Jurka and Zuckerkandl 1991). Alu J elements are the oldest dimeric elements in the human genome (Jurka and Smith 1988; Batzer and Deininger 2002). Although these elements were highly active ∼55–65 million years ago, they are thought to have lost the ability to transpose long ago (Jurka and Smith 1988; Batzer and Deininger 2002). Likewise, Alu S elements, which are intermediate in age, are thought to have become inactive at least 35 million years ago (Jurka and Smith 1988; Batzer and Deininger 2002; Johanning et al. 2003). Alu Y elements, in contrast, are the youngest Alu elements in the genome and these elements remain actively mobile today (Batzer and Deininger 2002; Dewannieux et al. 2003). The Alu J, S, and Y families (and their subfamilies) contain a series of hierarchical DNA sequence changes that arose during Alu evolution (Slagel et al. 1987; Jurka and Smith 1988; Batzer and Deininger 2002; Jurka et al. 2002). Each Alu family contains a unique set of diagnostic base changes that can be used to identify copies belonging to that family.
The second most abundant class of transposons in humans, the LINE (L1) elements, are autonomous poly(A) retrotransposons (Ostertag and Kazazian 2001 and references therein). These elements also have reached high copy numbers in the human genome (∼500,000) and collectively occupy ∼17% of the genome sequence (Lander et al. 2001). Like Alu, L1 elements have been actively mobile over a long period of time and have been classified according to their respective ages using specific base changes (Boissinot et al. 2000; Ovchinnikov et al. 2002; Brouha et al. 2003). The oldest L1 elements in the genome have accumulated deleterious mutations that render them inactive. However, younger L1 elements have been identified that remain actively mobile today (Moran et al. 1996; Brouha et al. 2003). These active copies contain two intact open reading frames, ORF1 and ORF2, which encode proteins that are necessary for L1 retrotransposition (Feng et al. 1996; Moran et al. 1996). ORF1 encodes a 40-kD protein with RNA-binding activity (Hohjoh and Singer 1996, 1997a,b; Kolosha and Martin 1997; Martin et al. 2000, 2003; Martin and Bushman 2001), whereas ORF2 encodes a protein with both endonuclease (EN) and reverse transcriptase (RT) activities (Mathias et al. 1991; Feng et al. 1996; Moran et al. 1996; Cost et al. 2002). EN and RT work together in a process known as target-primed reverse transcription (TPRT; Luan et al. 1993) that integrates a newly synthesized L1 cDNA into a DNA target site (Cost et al. 2002). Alu RNA (and other cellular RNAs) can compete for the L1 machinery during the TPRT process, leading to the retrotransposition of these alternative RNAs instead of the normal L1 mRNA (Esnault et al. 2000; Wei et al. 2001; Dewannieux et al. 2003). This “trans” replication mechanism is thought to account for the massive expansion of Alu elements in the human genome and for the existence of processed pseudogenes.
Because Alu and L1 remain actively mobile in the human genome today, they serve as ongoing sources of genetic variation by generating new transposon insertions (reviewed in Ostertag and Kazazian 2001 and Batzer and Deininger 2002). For example, estimates suggest that a new Alu insertion occurs approximately once every 200 live births (Deininger and Batzer 1999). As a consequence, a large number of polymorphic Alu and L1 insertions have accumulated in human populations. Many of these insertions are expected to be genetically neutral and, therefore, would have little or no impact on human phenotypes. However, other insertions (primarily those within genes) have been found to cause altered human phenotypes, including diseases. For example, disease-causing Alu insertions have been observed in the BRCA2 gene (Miki et al. 1996), the glycerol kinase gene (Zhang et al. 2000), and others (Deininger and Batzer 1999). Disease-causing L1 insertions likewise have been observed in at least 14 different genes, causing cancers (Morse et al. 1988; Miki et al. 1992; Liu et al. 1997), hemophilia (Kazazian et al. 1988), muscular dystrophy (Narita et al. 1993), and other diseases. It is likely that additional transposon insertions will be found to affect human phenotypes as well.
As an initial step toward studying the potential phenotypic variation that is caused by Alu and L1 elements, it is necessary to identify all of the polymorphic insertions that exist in human populations. Only a fraction of such insertions have been identified to date, largely because the methods for detecting transposon insertion polymorphisms are labor intensive. Most of the known Alu and L1 insertion polymorphisms have been identified by systematically screening individual element copies in human populations using PCR assays (Carroll et al. 2001; Roy-Engel et al. 2001; Myers et al. 2002; Abdel-Halim et al. 2003; reviewed in Ostertag and Kazazian 2001 and Batzer and Deininger 2002). Transposon display assays also have been used to identify transposon insertion polymorphisms (Sheen et al. 2000; Badge et al. 2003). Although these methods have been useful for identifying polymorphisms, they are not likely to be sufficient on a genome-wide scale to identify all of the polymorphic Alu and L1 copies that exist in human populations. Thus, new and more efficient methods are necessary to identify transposon insertion polymorphisms.
In addition to Alu and L1 elements, some of the remaining transposons and transposon-like elements in the genome also might be polymorphic and, therefore, would contribute to human genetic diversity. Despite the fact that there are many families of such elements in humans (Smit and Riggs 1996; Smit 1999; Lander et al. 2001), no comprehensive studies have been conducted to examine whether these elements are polymorphic or remain actively mobile. As is the case for Alu and L1, such elements would be of interest because they represent sources of human genetic variation and might also cause mutations that lead to human diseases.
In an effort to measure the levels of genetic variation that are caused by human transposons, we have developed an efficient method to broadly detect transposon insertion polymorphisms of all kinds in humans. The method exploits DNA sequencing traces that originally were generated from diverse humans for single-nucleotide polymorphism (SNP) discovery projects (Sachidanandam et al. 2001; International HapMap Consortium 2003). We have developed a computational pipeline that now analyzes these traces to identify transposon insertion polymorphisms. Our study provides significant steps toward (i) measuring the genetic variation that is caused by transposon insertions in humans and (ii) identifying the transposon copies that produce this variation.
MATERIALS AND METHODS
Identifying insertion and deletion candidates using DNA sequencing traces from diverse humans:
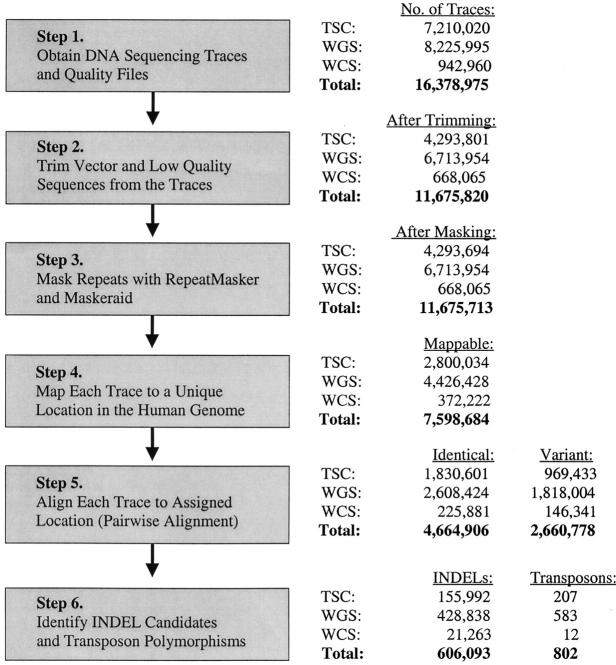
DNA sequencing traces and accompanying quality files were obtained from Cold Spring Harbor Laboratory [traces generated by the SNP Consortium (TSC)] or from the Trace DB archive at the National Center for Biotechnology Information (NCBI). Insertion and deletion (indel) and transposon insertion polymorphisms were identified from these traces using a sequential series of computer programs and databases as outlined in Figure 1. Many of these programs were obtained from NCBI or from other sources as indicated below. Other custom Perl programs were developed for indel and transposon polymorphism discovery as necessary and are available upon request. Most of these programs and databases were installed locally on Dell workstations running Microsoft 2000, XP, or Red Hat Linux operating systems. A 12-CPU Linux cluster also was constructed and utilized for the RepeatMasker and MegaBLAST steps of the pipeline (Figure 1).
Figure 1.—
Computational pipeline for indel and transposon polymorphism discovery. A flow chart of the computational steps that were followed for the discovery of indel and transposon polymorphisms is shown on the left (boxes). A breakdown of the number of traces present at the end of each step is shown on the right. The number of indel and transposon polymorphisms identified is listed at the bottom. Note that the numbers are broken down for each of the three populations examined. TSC, the SNP Consortium traces; WGS, whole-genome shotgun traces; WCS, whole-chromosome shotgun traces.
A total of 16.4 million DNA sequencing traces were processed using the pipeline depicted in Figure 1. The traces first were screened for vector contamination using the VecScreen system developed by NCBI and were trimmed as necessary. Low-quality regions of the traces then were identified and trimmed with a custom Perl program that uses the Phred quality scores in the accompanying quality files to identify such regions (Ewing and Green 1998; Ewing et al. 1998). Our method identified the longest high-quality region of each trace and then trimmed the flanking data upon encountering 5 bases in a row with Phred scores <25. The longest high-quality interval from each trace was chosen for further analysis and the remaining data were set aside. Trimmed traces also were required to have average Phred scores of at least 25 and minimum lengths of 100 bases.
After trimming, each trace then was mapped to a unique location in the human genome sequence (build 33 for the TSC traces and build 34 for the remaining traces). Builds 33 and 34 of the human genome sequence database were obtained from the University of California (Santa Cruz) and installed locally to perform this step (Kent et al. 2002). All known repeats (including all transposons and transposon-like repetitive elements defined in Repbase Volume 7, Issue7; Jurka 2000) first were temporarily masked in the traces using RepeatMasker (version 2001/07/07; A. Smit, unpublished data) and MaskerAid (Bedell et al. 2000). The single longest unmasked “anchor sequence” of the trace then was used to assign each trace to a unique genomic location using MegaBLAST (NCBI). The anchor sequence was required to have a minimum of a 50-base match at 100% identity for a trace to be mapped successfully. Traces with anchor sequences that matched to more than one genomic location with 100% identity, or that did not have a minimum of a 50-base match at 100% identity, were set aside to avoid traces that mapped to duplicated regions of the genome (Bailey et al. 2002). After the traces were successfully mapped to unique genomic locations, they were unmasked and aligned to their assigned genomic locations using the Bl2Seq program (NCBI). The Bl2Seq program allowed for as much as a 16-base gap in the alignments and led to identification of indels as large as 16 bases in length.
A new algorithm also was developed to identify indels that were >16 bp in length. Our strategy was designed to split trace data into two blocks upon encountering a region in the pairwise alignment that no longer matched the query. The first block of sequence that matched was maintained in the correct position, and the nonmatching sequence was moved over as a block, 1 base at a time, until a match was obtained. The Perl program that was developed to accomplish this task moved the nonmatching block until it detected either a perfect alignment or a distance of 10,000 bases (the maximum distance allowed by the program). The 5 bases on each side of an indel candidate were required to have Phred scores of ≥20 to ensure that high-quality bases were being used to locate the indel junctions. Indel candidates were deposited into dbSNP under accession nos. ss8029278–ss8176133, ss8475737–ss8484870, ss14926095–ss15354938, and ss15357378–ss15378640.
Identifying transposon insertion polymorphisms by screening human indels:
Transposon insertion polymorphisms were identified among indels using a custom computer algorithm. First, indels were identified for which at least 80% of the indel sequence was occupied by a known transposon as defined by the definitions of all human transposons and repeats in Repbase (Vol. 7, Issue 7; Jurka 2000). This step was accomplished by querying an Oracle database that stored RepeatMasker output data (and other information) for each indel. Next, selected candidates were examined with a custom Perl program to determine whether potential TSDs were present. Such duplications generally flank transposon insertions and are hallmarks of most transposons (Berg and Howe 1989). Therefore, if an indel was caused by a transposon insertion, it generally would be expected to be flanked by a TSD (one copy of the duplicated sequence actually is contained within the indel itself, since the duplication is created during the insertion of the transposon). Candidate transposon insertions also were screened with a custom Perl program to identify potential poly(A) tails, which are associated with certain retrotransposons. Finally, the genomic contexts of all transposon indel candidates were examined to identify true de novo insertions vs. indels that were caused by deletions or duplications within existing transposon copies. All indels that met at least the first test were inspected and curated manually (see supplemental Table 1 at http://www.genetics.org/supplemental/ for the final curated set). Six hundred and five nonredundant polymorphisms were identified that were caused by de novo transposon insertions (these are listed in the “Alu,” “L1,” “SVA,” and “Other” sections of supplemental Table 1). Another 50 nonredundant polymorphisms were caused by deletions or duplications within existing transposons (these are listed in the “Deletions and duplications” section of supplemental Table 1).
Analysis of transposon subfamilies:
Alu transposon insertions were classified initially using RepeatMasker (A. Smit, unpublished data) and Repbase (Vol. 7, Issue 7; Jurka 2000). Each polymorphic copy also was compared independently to the consensus sequences of all known Alu subfamilies (Repbase Vol. 7, Issue 7; Jurka 2000). To accomplish this goal, all Alu insertions identified were coaligned with the consensus sequences of all Alu subfamilies using the Clustal W program. Key diagnostic bases then were analyzed to further assist with the assignments of these elements to specific subfamilies. Each copy then was compared to the assigned subfamily consensus using Bl2seq (NCBI). In some cases, element copies also were compared to the consensus sequences of several neighboring families. A final assignment was made on the basis of the best match obtained. L1-Hs and L1-P elements were classified initially using RepeatMasker (A. Smit, unpublished data). The L1-Hs elements then were assigned to a given subfamily using the classification system described by Brouha et al. (2003). All other transposons were classified using the RepeatMasker system (A. Smit, unpublished data) and Repbase (Vol. 7, Issue 7; Jurka 2000).
Validation of the computational pipeline by PCR:
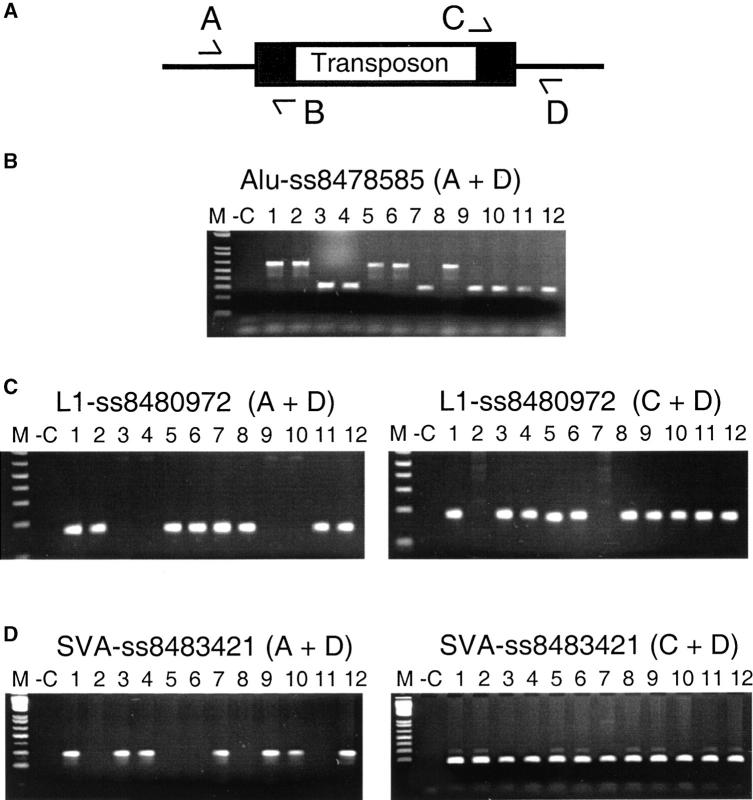
Sixty-one transposon insertions were chosen arbitrarily from the TSC data set and examined by PCR to evaluate the accuracy of our computational predictions (Table 6). PCR assays were designed for each of the 61 polymorphic transposon copies using primers that either flanked (A and D primers) or were located within (B and C primers) a given transposon as depicted in Figure 3. All primers used in these studies are listed in supplemental Table 2 at http://www.genetics.org/supplemental/. A total of 68 PCR assays were designed initially. Seven (10%) of these assays failed due to technical reasons and these assays were abandoned. The remaining 61 assays (90%) yielded band(s) of the expected size(s) and were used to assay 12–24 DNA samples from the Coriell diversity panel (Figure 3 and Table 6). The Coriell diversity panel of 24 DNA samples was obtained from the Coriell Repository, Camden, New Jersey (Collins et al. 1999). Lymphocyte cultures of this panel also were obtained from Coriell and, in some cases, DNA was prepared from these cells. PCR reactions were carried out in 50-μl volumes as described previously (Kimmel et al. 1997). PCR products were run on 1.5% agarose gels and sized using a 1-kb ladder marker (Invitrogen, San Diego).
TABLE 6.
Verification of TSC trace predictions by PCR
| dbSNP no. | Chromosomal locationa | Type of transposon |
Alleles examined |
% golden path allele |
% trace allele |
|---|---|---|---|---|---|
| ss8475858 | Chr1: 35,902,357–35,902,357b | Alu Ya5 | 48 | 4 | 96 |
| ss8475874 | Chr1: 43,270,765–43,271,100 | Alu Y | 46 | 11 | 89 |
| ss8476214 | Chr1: 180,191,288–180,191,688 | Alu Yg6 | 46 | 46 | 54 |
| ss8480173 | Chr2: 184,452,525–184,452,867 | Alu Ya5 | 46 | 4 | 96 |
| ss8481265 | Chr3: 117,596,173–117,596,496 | Alu Ye5 | 44 | 0 | 100 |
| ss8481475 | Chr3: 182,282,816–182,283,143 | Alu Y | 16 | 63 | 38 |
| ss8481677 | Chr4: 36,367,085–36,367,417 | Alu Ya4 | 20 | 30 | 70 |
| ss8481874 | Chr4: 98,569,039–98,569,364 | Alu Ya5 | 44 | 43 | 57 |
| ss8481886 | Chr4: 100,528,683–100,528,998 | Alu Ya5 | 48 | 0 | 100 |
| ss8481943 | Chr4: 127,436,762–127,437,090 | Alu Ya4 | 44 | 11 | 89 |
| ss8482744 | Chr5: 174,741,730–174,742,046 | Alu Y | 14 | 21 | 79 |
| ss8482858 | Chr6: 20,873,358–20,873,677 | Alu Ya5 | 32 | 44 | 56 |
| ss8482986 | Chr6: 52,830,475–52,830,475 | Alu Y | 48 | 96 | 4 |
| ss8483054 | Chr6: 74,416,861–74,417,166 | Alu Ya5 | 36 | 14 | 86 |
| ss8483191 | Chr6: 116,796,384–116,796,702 | Alu Yi6 | 14 | 14 | 86 |
| ss8483704 | Chr7: 94,998,283–94,998,613 | Alu Ya4 | 32 | 22 | 78 |
| ss8484187 | Chr8: 76,176,865–76,177,188 | Alu Ya5 | 48 | 40 | 60 |
| ss8484397 | Chr8: 140,446,315–140,446,646 | Alu Yg6 | 48 | 4 | 96 |
| ss8484534 | Chr9: 30,481,284–30,481,594 | Alu Ya5 | 44 | 20 | 80 |
| ss8484574 | Chr9: 41,460,809–41,461,130 | Alu Ya5 | 46 | 2 | 98 |
| ss8484611 | Chr9: 74,581,632–74,581,947 | Alu Yb8 | 48 | 33 | 67 |
| ss8476640 | Chr10: 53,785,800–53,786,116 | Alu Y | 10 | 50 | 50 |
| ss8477096 | Chr11: 42,428,408–42,428,718 | Alu Yb9 | 20 | 5 | 95 |
| ss8477278 | Chr11: 102,948,539–102,948,851 | Alu Yd8 | 48 | 0 | 100 |
| ss8477519 | Chr12: 45,361,618–45,361,965 | Alu Yf1 | 48 | 0 | 100 |
| ss8477629 | Chr12: 89,348,858–89,349,061 | Alu Ya4/Ya5 | 48 | 4 | 96 |
| ss8478585 | Chr15: 79,108,405–79,108,719 | Alu Ya5 | 48 | 29 | 71 |
| ss8478989 | Chr17: 13,905,127–13,905,415 | Alu Y | 44 | 9 | 91 |
| ss8480426 | Chr20: 11,512,266–11,512,588 | Alu Sx | 26 | 31 | 69 |
| ss8480530 | Chr20: 53,978,074–53,978,387 | Alu Ya5 | 48 | 4 | 96 |
| ss8480534 | Chr20: 55,148,643–55,148,959 | Alu Y | 20 | 10 | 90 |
| ss8480629 | Chr21: 29,522,273–29,522,612 | Alu Yb8 | 46 | 17 | 83 |
| ss8484940 | ChrX: 115,208,464–115,208,782 | Aluc | 20 | 60 | 40 |
| ss8480093 | Chr2: 160,351,662–160,354,192 | L1 Ta-0 | 20 | 10 | 90 |
| ss8480896 | Chr3: 7,322,025–7,322,322 | L1 pre TA | 20 | 40 | 60 |
| ss8480972 | Chr3: 33,524,976–33,525,861 | L1 Ta-1nd | 20 | 60 | 40 |
| ss8481820 | Chr4: 78,694,670–78,694,935 | L1 pre TA | 12 | 33 | 67 |
| ss8482149 | Chr4: 182,572,693–182,574,194 | L1 Ta-1 | 48 | 0 | 100 |
| ss8482285 | Chr5: 24,416,029–24,418,937 | L1 Ta-1 | 18 | 11 | 89 |
| ss8482213 | Chr5: 2,021,152–2,024,339 | L1 Ta-1d | 20 | 50 | 50 |
| ss8483298 | Chr6: 153,060,963–153,064,780 | L1 Ta-1d | 20 | 30 | 70 |
| ss8483034 | Chr6: 66,255,638–66,257,631 | L1 Ta-1 | 20 | 25 | 75 |
| ss8483013 | Chr6: 57,469,905–57,475,956 | L1 PA3 | 48 | 0 | 100 |
| ss8484339 | Chr8: 126,551,711–126,557,723 | L1 Ta-1d/Ta-0 | 20 | 75 | 25 |
| ss8484350 | Chr8: 129,421,743–129,427,855 | L1 Ta-1d | 16 | 6 | 94 |
| ss8484672 | Chr9: 96,187,490–96,188,291 | L1 pre TA | 18 | 11 | 89 |
| ss8477608 | Chr12: 82,346,513–82,347,076 | L1 Ta-1 | 16 | 12.5 | 87.5 |
| ss8477509 | Chr12: 40,524,321–40,525,144 | L1 Ta-1 | 18 | 22 | 78 |
| ss8478229 | Chr14: 49,510,268–49,510,585 | L1 pre TA | 20 | 90 | 10 |
| ss8478193 | Chr14: 38,088,431–38,090,252 | L1 pre TA | 20 | 60 | 40 |
| ss8478558 | Chr15: 68,737,658–68,743,326 | L1 Ta-1d/Ta-0 | 20 | 75 | 25 |
| ss8479344 | Chr18: 50,014,733–50,014,733 | L1 Ta | 18 | 39 | 61 |
| ss8479308 | Chr18: 39,283,540–39,284,523 | L1 Ta-1 | 20 | 20 | 80 |
| ss8480503 | Chr20: 42,710,042–42,711,239 | L1 Ta-1 | 20 | 70 | 30 |
| ss8481522 | Chr3: 195,440,681–195,441,437 | SVA | 48 | 46 | 54 |
| ss8483321 | Chr6: 158,457,569–158,458,368 | SVA | 48 | 17 | 83 |
| ss8483556 | Chr6: 29,793,437–29,796,056 | SVA | 48 | 4 | 96 |
| ss8483421 | Chr7: 10,248,637–10,250,470 | SVA | 48 | 67 | 33 |
| ss8483579 | Chr7: 55,028,493–55,030,810 | SVA | 44 | 32 | 68 |
| ss8478175 | Chr14: 33,497,755–33,499,561 | SVA | 48 | 75 | 25 |
| ss8477420 | Chr12: 13,090,355–13,090,458 | 5S | 48 | 67 | 33 |
Chromosomal coordinates are given for the July 2003 build (build 34) of the human genome sequence (University of California, Santa Cruz; Kent et al. 2002).
In cases where both chromosomal coordinates are the same, the insertion occurred in the trace.
Too short to classify.
Figure 3.—
PCR validation studies. The strategy for the PCR validation assays is shown along with some examples of these assays. (A) The locations of the four primers (A, B, C, and D) that were used to evaluate transposon polymorphisms by PCR are shown. (B) A typical Alu PCR assay is shown in which primers flanking the transposon (A and D) are used to determine whether a given Alu copy is present in the genome of an individual. The larger band is produced when the element is present, whereas the smaller band is produced when the element is absent. Lane M, 1-kb marker; −C, negative control lacking template DNA; lanes 1–12, 12 PCR reactions evaluating DNA samples from the Coriell panel. (C) A typical L1 PCR assay is shown in which two PCRs are performed to identify all of the alleles present in the Coriell panel. The assay on the left uses the A and D primers to identify alleles that lack an L1 insertion, whereas the assay on the right uses primers C and D (or A and B) to identify alleles containing the L1 insertion. These two assays are used together to evaluate whether a given allele is homozygous or heterozygous in a given individual. The lanes are the same as for B. For cases in which the L1 element is relatively short (<2 kb), the allele containing the L1 insertion often was detected in the A plus D assay as well. (D) A typical SVA assay is depicted. These assays are performed the same way as the L1 assays (in C above), using two assays to evaluate all SVA alleles present.
Analysis of additional genomic SVA elements:
In addition to the SVA copies identified in the trace experiments, 28 other genomic SVA copies were selected from the human genome sequence using SVA element query sequences and the BLAT program (Kent 2002). These SVA copies were examined by PCR to assess whether they were polymorphic in at least one individual of the Coriell panel. PCR primers were developed to examine the status of each SVA copy as described in Figure 3 and supplemental Table 2. PCR reactions were carried out as outlined above and in Figure 3. An SVA copy was considered to be polymorphic if both alleles (one with and one without the transposon insertion) could be identified at least once. Fifty-nine additional SVA element copies were identified by manual inspection of the first 50 Mb of human chromosome 1 using the University of California, Santa Cruz genome browser (Kent et al. 2002). The genomic regions surrounding all of these SVA copies were compared to the equivalent chimp genomic sequences to determine whether the chimp contained an SVA element at the equivalent position (supplemental Table 1).
RESULTS
A strategy for detecting genetic variation caused by transposon insertions in humans:
Our strategy for detecting transposon insertion polymorphisms in humans involved identifying a large number of indel polymorphisms in human populations and then screening these polymorphisms to identify de novo transposon insertions. We reasoned that this strategy should be successful since transposon insertion polymorphisms are equivalent to insertions and deletions in genomes. Relatively few indels had been identified in human populations prior to our study, despite the fact that indels are abundant in the genomes of model organisms such as Drosophila melanogaster (Berger et al. 2001) and Caenorhabditis elegans (Wicks et al. 2001) and were likely to be abundant in humans as well. Therefore, we began our study by developing new computational methods to discover indel polymorphisms in the genomes of diverse humans (materials and methods).
Our strategy involved mining indels from DNA sequencing traces that previously had been generated for SNP discovery projects. All of the traces used in our study originally were generated at genome centers by resequencing pools of genomic DNA from diverse humans. For example, a set of 7.1 million traces, which originally had been generated by shotgun sequencing the DNA of 24 diverse humans (Sachidanandam et al. 2001), was obtained from TSC. A second set of 8.2 million whole-genome shotgun (WGS) traces, which originally had been generated by shotgun sequencing the DNA of eight unrelated African-American adults (four males and four females from the Baylor Polymorphism Resource; International HapMap Consortium 2003), was obtained from the Baylor and Whitehead Genome Centers. Finally, a much smaller set of 0.9 million whole-chromosome shotgun (WCS) traces, which had been generated by shotgun sequencing chromosome 20-specific libraries from four diverse humans (International HapMap Consortium 2003), was obtained from the Sanger Center. Because these DNA sequencing traces were derived from diverse humans, we expected them to harbor various forms of genetic variation, including indels. We developed a computational pipeline to identify indels within these traces by comparing them to the human genome reference sequence (builds 33 and 34; Figure 1).
A total of 606,093 indel candidates were identified by analyzing 16.4 million traces with our computational pipeline (Figure 1 and materials and methods). The majority of these indels (428,838 or 70.8%) were identified from the WGS traces. An additional 155,992 indels (25.7%) were identified from the TSC traces, and 21,263 indels (3.5%) were identified from the WCS traces. Overall, these indel candidates were distributed throughout the human genome and were found on all 24 chromosomes (data not shown). They ranged in size from 1 to 9969 bp in length and contained a wide array of different DNA sequences. All indel candidates were deposited into dbSNP under the “Devine_lab” handle (http://www.nih.nlm.gov/SNP).
We next developed a computer algorithm to identify indels that were caused by de novo transposon insertions. The method was designed to identify indels for which a single transposon copy and its associated sequences (e.g., its target site duplication) accounted for the indel (see materials and methods). Eight hundred and two transposon insertion polymorphisms were detected with these methods in the three populations examined (Table 1). Four major classes of transposon insertions were identified in these experiments: (i) Alu insertions, (ii) L1 insertions, (iii) SVA insertions, and (iv) insertions of “other” elements.
TABLE 1.
Transposon insertion polymorphisms identified in humans
| TSC | WGS | WCS | |
|---|---|---|---|
| Bases analyzed | 989,283,997 | 2,271,983,242 | 110,411,692 |
| Fraction of genome | 0.30 | 0.69 | 0.033 |
| No. polymorphisms observed (% total)
| |||
| TSC | WGS | WCS | |
| Alu-total | 173 (0.836) | 487 (0.835) | 10 (0.833) |
| Alu Ya5 | 67 (0.324) | 148 (0.254) | 7 (0.583) |
| Alu Yb8 | 30 (0.145) | 132 (0.226) | 1 (0.083) |
| L1-total | 26 (0.126) | 64 (0.110) | 2 (0.167) |
| L1-Ta | 25 (0.121) | 58 (0.099) | 2 (0.167) |
| L1-other | 1 (0.005) | 6 (0.010) | 0 (0) |
| SVA | 6 (0.029) | 25 (0.043) | 0 (0) |
| Other | 2 (0.010) | 7 (0.012) | 0 (0) |
| Total | 207 | 583 | 12 |
Alu insertion polymorphisms were by far the most abundant polymorphisms identified in the three experiments (Table 1). A total of 173 of 207 (83.6%) of the polymorphisms in the TSC data set were Alu insertions. Likewise, 487 of 583 (83.5%) of the polymorphisms in the WGS set were Alu insertions, and 10 of 12 (83.3%) of the polymorphisms in the WCS set were Alu insertions. L1 insertions were the next most abundant polymorphisms identified, representing 12.6 and 11.0% of the TSC and WGS data sets, respectively (Table 1). Although the L1-Ta class was the most abundant subfamily of L1, other non-Ta L1 elements were identified as well (see below). SVA element insertions were the third most abundant class of transposon polymorphisms identified, representing 2.9 and 4.3% of the TSC and WGS data sets, respectively. Finally, the remaining transposon insertion polymorphisms were caused by a miscellaneous collection of low-frequency insertions. These elements were pooled into a single group of other polymorphisms (Table 1).
It is important to note that our measurements were remarkably consistent between the data sets. This was particularly true for the TSC and WGS data sets, which were significantly larger than the remaining WCS set. For example, as noted above, Alu polymorphisms represented ∼83% of the transposon insertions in all three of the populations examined. The percentages of L1 insertions likewise were very similar in these experiments (Table 1). Overall, the TSC and WGS experiments were remarkably similar given the differences in the populations that were used to generate these trace sets (Table 1). Nevertheless, the results were not completely identical between the populations. For example, Alu Ya5 polymorphisms represented 32.4% of the insertions in the TSC population and 25.4% of the insertions in the WGS population (Table 1). Therefore, at least some of these element families might have amplified at slightly different rates in the populations examined.
We next inspected all of the polymorphic transposon insertions from the three populations to determine whether any of the copies were redundant in the three data sets. In fact, 149 polymorphisms were identified in which the same transposon allele was detected more than once in our trace experiments. In most of these cases (115 of 149 or 77.2%), the alleles were detected independently twice (supplemental Table 1). Another 25 of 149 alleles (16.8%) were detected three times and the remaining 9 of 149 alleles (6%) were detected four to six times (supplemental Table 1). These results provide confidence in our method and suggest that at least some of our transposon insertion polymorphisms are present at high frequencies in human populations. To perform additional analyses of these transposons, we developed a nonredundant data set of 605 transposon insertions (supplemental Table 1).
Alu Y insertion polymorphisms:
A total of 505 nonredundant Alu insertion polymorphisms were identified in the three populations of our study, including both full-length and partial Alu insertions (supplemental Table 1 and Table 2). These elements were compared to all known Alu families and were classified to determine which Alu elements were detected in our experiments (materials and methods). The vast majority of our Alu insertions were Alu Y elements, with 500 of 505 (99%) of the insertions falling in this category (supplemental Table 1 and Table 2). Alu Ya5 elements were the most abundant subfamily in our study, representing 33.7% of the insertions (supplemental Table 1 and Table 2). Alu Yb8 polymorphisms also were abundant, representing 25.5% of the insertions (supplemental Table 1 and Table 2). Alu Y, Alu Yc1, and Alu Ya4 elements were present at intermediate levels (between 5.9 and 7.5%), and these three families together represented 19.7% of all nonredundant Alu insertions in our study. Most of the remaining Alu Y-related insertions were present at relatively low levels and were distributed among 15 different Alu Y subfamilies (supplemental Table 1 and Table 2). Notably, Alu polymorphisms were detected from most of the known Alu Y subfamilies, including Alu Ya, Yb, Yc, Yd, Ye, Yf, Yg, and Yi (supplemental Table 1 and Table 2). Moreover, although we did detect several new small groups of Alu Y insertions that might be considered novel subfamilies (see below and Figure 2), no new extended Alu Y families of significant size were detected in our study.
TABLE 2.
Nonredundant transposon insertion polymorphisms
| Polymorphisms | |
|---|---|
| Alu (505 total) | |
| Alu S | |
| Alu Sc-derived | 1 |
| Alu Sp-derived | 1 |
| Alu Sq-derived | 1 |
| Alu Sx-derived | 1 |
| Alu Ya | |
| Alu Ya1 | 4 |
| Alu Ya2 | 1 |
| Alu Ya4 | 27 |
| Alu Ya4-derived | 3 |
| Alu Ya5 | 170 |
| Alu Ya5a2 | 11 |
| Alu Ya8 | 2 |
| Alu Yb | |
| Alu Yb3a1 | 1 |
| Alu Yb3a1-derived | 1 |
| Alu Yb3a2 | 1 |
| Alu Yb3a2-derived | 5 |
| Alu Yb8 | 125 |
| Alu Yb8-derived | 4 |
| Alu Yb9 | 15 |
| Alu Yc | |
| Alu Yc1 | 38 |
| Alu Yc2 | 2 |
| Alu Yd | |
| Alu Yd8 | 4 |
| Alu Ye | |
| Alu Ye2 | 1 |
| Alu Ye2-derived | 1 |
| Alu Ye5 | 15 |
| Alu Ye5-derived | 3 |
| Alu Yf | |
| Alu Yf1 | 1 |
| Alu Yg | |
| Alu Yg6 | 12 |
| Alu Yi | |
| Alu Yi6-derived | 8 |
| Alu Y | |
| Alu Y | 22 |
| Alu Y-derived | 10 |
| Too short to classify | |
| Alu Ya4/5 | 1 |
| Alu Ya5/8 | 5 |
| Alu Yc | 1 |
| Alu Y | 6 |
| Alu | 1 |
| L1 (65 total) | |
| L1 Hs | |
| Ta-0 | 8 |
| Ta-1 | 12 |
| Ta-1nd | 3 |
| Ta-1d | 12 |
| Pre-TA(acg/a) | 4 |
| Pre-TA(acg/g) | 9 |
| Hs (unclassifiable) | 1 |
| Ta (unclassifiable) | 7 |
| Ta-1d/Ta-0 | 3 |
| L1-P | |
| L1 PA2 | 5 |
| L1 PA3 | 1 |
| SVA (39 total) | |
| From traces | 28 |
| Other genomic | 11 |
| Other (7 total) | |
| DNA/Mariner | 1 |
| LTR/ERVK | 2 |
| U2 RNA | 1 |
| U5 RNA | 1 |
| 5S rDNA | 2 |
Figure 2.—
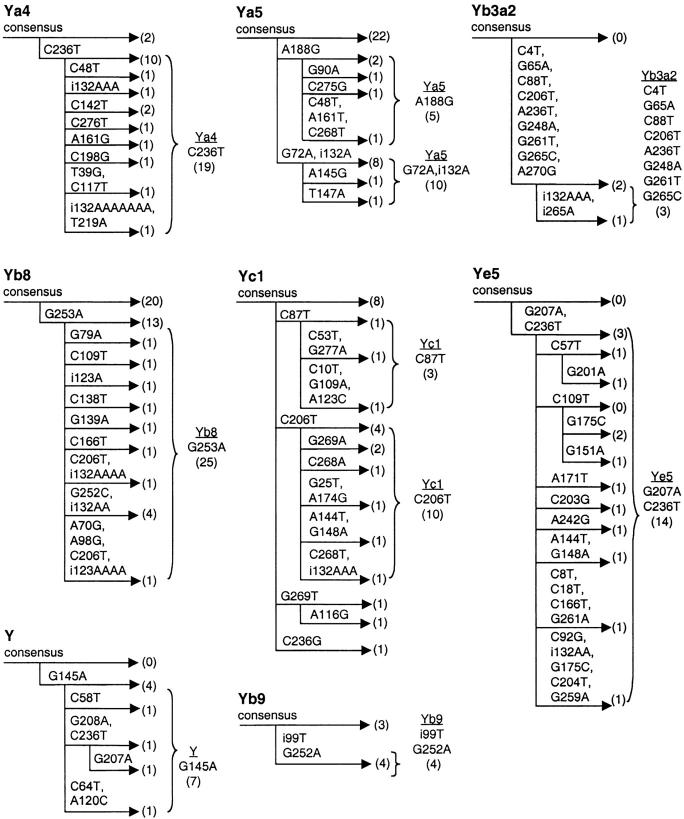
Proposed new evolutionary lineages of Alu. For each Alu subfamily, the number of polymorphic copies retaining the subfamily consensus sequence is compared to groups sharing one or more base pair changes (in parentheses). In several cases, the majority of polymorphic copies of a given subfamily diverge from the subfamily consensus by a few shared changes. An evolutionary progression can be inferred (from left to right) in which new shared base changes appear to have been acquired. The 10 proposed novel evolutionary groups are indicated by braces to the right of the groups. The total number of elements within the group is indicated in the parentheses. These data suggest that a significant number of Alu insertions in the genome can serve as new source genes to produce offspring elements. Only elements that are at least 80% full length are represented.
As outlined above, Alu Ya5 and Alu Yb8 insertions were the most abundant Alu elements in our data sets. Carroll et al. (2001) previously demonstrated that these two Alu subfamilies were highly polymorphic in human populations. In fact, they estimated that 25% of Alu Ya5 elements and 20% of Alu Yb8 elements were polymorphic in at least one individual of a panel of 80 diverse humans (Carroll et al. 2001). On the basis of their copy number estimates for these two elements, we can predict that at least 660 Alu Ya5 insertion polymorphisms and 370 Alu Yb8 insertion polymorphisms should exist in human populations. We found a total of 170 nonredundant Alu Ya5 insertions and 129 nonredundant Alu Yb8 insertions in our study (supplemental Table 1 and Table 2). Only 8 of these polymorphic insertions (4 Alu Ya5 and 4 Alu Yb8) were identified by Carroll et al. (2001). Therefore, 291 of 299 (98.6%) of our Alu Ya5 and Alu Yb8 polymorphisms had not been detected previously. Similar results were obtained with the remaining Alu classes, indicating that our method efficiently identified a large number of novel Alu insertion polymorphisms in human populations.
Polymorphic ancient Alu elements:
In addition to Alu Y elements, we also identified four polymorphic copies of older Alu S elements (Table 2). Two of these examples (Alu ss14941867 and Alu ss8480425) were intact, full-length Alu S insertions with all of the expected features of Alu retrotransposition events, including poly(A) tails and target site duplications (supplemental Table 1). Two additional examples of 5′-truncated or otherwise fragmented copies of Alu S also were identified (supplemental Table 1 and Table 2). One of these insertions (ss14931773) was a 5′-truncated Alu Sc element with a perfect target site duplication. The second insertion (ss15143442) was an Alu Sq element that was truncated at both the 5′ and the 3′ ends and lacked a target site duplication altogether (supplemental Table 1). It is not clear how this second Alu polymorphism was formed. One possibility is that it was caused by an endonuclease-independent mechanism of retrotransposition involving partial Alu RNA templates. Both Alu and L1 elements are known to use an endonuclease-independent mechanism that does not generate target site duplications surrounding the newly transposed copy (Morrish et al. 2002; Abdel-Halim et al. 2003). Perhaps this older Alu Sq element was mobilized by the L1 machinery using this alternative mechanism.
The fact that we identified four ancient Alu S insertion polymorphisms indicates that at least some of the Alu S copies are likely to have retained the ability to transpose long after the majority of Alu S elements became transpositionally inactive. This is most probable for the intact Alu copies discussed above (ss14941867 and Alu ss8480425). These copies do not appear to have been caused by gene conversion events and have estimated ages of 7–23 million years, suggesting that they are younger than most of the Alu S elements (supplemental Table 1). Prior to our study, only the Alu Y elements were thought to be polymorphic in humans, whereas the older Alu S, Alu J, and Alu monomers were thought to have only fixed alleles in human populations. Recent evidence from Johanning et al. (2003) showed that at least some Alu Sx elements appear to have transposed later than previously estimated (∼35 million years ago); however, Alu S insertion polymorphisms were not detected in humans prior to our study.
Sequence variation within polymorphic copies of Alu indicates patterns of Alu evolution:
Significant DNA sequence variation was noted within the polymorphic Alu insertions identified in our study. In most cases, a given element copy could be placed unambiguously within a known Alu subfamily using key diagnostic base changes (materials and methods). Nevertheless, a large number of additional single- and multiple-base changes were noted in these elements relative to their respective consensus sequences (supplemental Table 1). Of particular interest were small groups of Alu elements that clearly belonged to a given element family, but differed from the consensus by one or more shared base changes. Since CpG changes occur independently at a high frequency, it was possible that some of these groups were caused by independent changes at CpG hotspots. However, at least 10 of these groups possessed shared base changes at non-CpG sites (or had unusually high frequencies of a given CpG change along with additional shared changes). Most of these groups also showed evidence for the progressive accumulation of shared mutations. In these cases, a single base change was shared by an initial subset of the elements and additional shared changes appear to have been acquired later. We propose that these groups represent novel evolutionary lineages of Alu elements that are defined by these new base changes (Figure 2). These data suggest that a significant number of Alu insertions go on to serve as new source genes for small numbers of additional retrotransposition events (Deininger et al. 1992).
L1 insertion polymorphisms:
Although most of the ∼500,000 L1 copies in the haploid human genome have accumulated deleterious mutations that render them inactive, some of the younger L1 copies remain actively mobile today (Moran et al. 1996; Brouha et al. 2003). These younger copies belong to the L1-Hs (Human-specific) family of elements (Brouha et al. 2003). The Hs family has been subdivided further into the Ta-0, Ta-1, Ta-nd, and Ta-d subfamilies on the basis of the presence or absence of specific nucleotide changes within the L1 sequence (Boissinot et al. 2000; Ovchinnikov et al. 2002; Brouha et al. 2003). Reflective of their younger ages, L1-Hs elements are highly polymorphic in human populations (Sheen et al. 2000; Ovchinnikov et al. 2001; Myers et al. 2002; Badge et al. 2003; Brouha et al. 2003).
We identified 65 nonredundant L1 insertion polymorphisms in our study (supplemental Table 1 and Table 2). Each of these L1 elements ended in a poly(A) sequence and was flanked by a typical L1 target site duplication (supplemental Table 1). We classified these elements using the system described by Brouha et al. (2003) and found that most of the copies belonged to Ta subfamilies of L1 elements. In fact, elements belonging to the L1 Ta-0, Ta-1, Ta-nd, and Ta-d subfamilies were identified along with some older pre-Ta elements (supplemental Table 1 and Table 2). These results are consistent with the observation that 13 of the 14 L1 insertions that have been found to cause human diseases were L1-Ta elements and the remaining element was a pre-Ta element (reviewed in Moran 1999). Our results also are consistent with previous studies demonstrating that L1-Ta elements are highly polymorphic (Sheen et al. 2000; Ovchinnikov et al. 2001; Myers et al. 2002; Brouha et al. 2003). Although some of our L1 insertion polymorphisms were identified previously, most were unique to our study (supplemental Table 1).
Interestingly, we also identified six polymorphic copies of older L1-P insertions, including five polymorphic L1PA2 insertions and a single L1PA3 insertion (supplemental Table 1 and Table 2). Therefore, in addition to L1-Hs elements, older L1-P elements also are polymorphic in humans. In fact, these elements collectively accounted for 9.2% of the L1 insertion polymorphisms in our study (Table 2). Thus, the spectrum of L1 elements that cause human genetic variation, and perhaps human disease, is broader than previously established (Ovchinnikov et al. 2002). Moreover, since a high level of polymorphism is associated with active transposons, our results suggest that at least some of the L1-P elements in humans and chimps might also remain actively mobile today.
SVA insertion polymorphisms are abundant in humans:
The human SVA element is a transposon-like repetitive element that was first identified within the RP gene on human chromosome 6 (Shen et al. 1994). The authors of this original report proposed that SVA represented a composite retrotransposon that contains two previously identified elements (SINE-R and Alu) as well as a variable nucleotide tandem repeat (VNTR) region. Although the authors of this study had no evidence that their proposed element was actively mobile, they suggested that SVA is a retrotransposon because it ended in a poly(A) tail and was flanked by an apparent target site duplication (Shen et al. 1994). Strichman-Almashanu et al. (2001) later estimated that the haploid human genome contains approximately ∼5000 copies of the SVA element.
We identified 28 nonredundant SVA insertion polymorphisms in our trace experiments (supplemental Table 1, Tables 2 and 3). These insertion polymorphisms have all of the hallmark features of retrotransposon insertions. In each case: (i) both empty and SVA-occupied sites were identified in different humans, (ii) the newly inserted SVA copy ended in a poly(A) tail, and (iii) each inserted copy was precisely flanked by a new target site duplication (Table 3). These copies ranged in size from 396 to 2806 bp in length, with the shorter elements lacking 5′ ends due to truncation, or lacking internal VNTR repeats (supplemental Table 1 and Table 3). The target site duplications of all insertions closely resembled (in both length and sequence) the target site duplications of Alu and L1 (Table 3).
TABLE 3.
SVA insertion polymorphisms identified in trace experiments
| SVA name (nearest gene) |
Chromosomal locationa | dbSNP no. | Indel size | Element size |
Target site duplication sequence (5′ to 3′) |
|---|---|---|---|---|---|
| CLIC4-A | Chr1: 24,509,856–24,512,677 | ss15143842 | 2,822 | 2,806 | AAAAATAAAAAATCAA |
| LRIG2 | Chr1: 112,910,067–112,912,521 | ss15142943 | 2,455 | 2,438 | AAAAACACATATTTGCG |
| PPP2R5A | Chr1: 209,527,634–209,529,114 | ss14942498 | 1,481 | 1,463 | AACAATTCACTTCATCTT |
| SSB | Chr2: 170,844,052–170,846,495 | ss15142807 | 2,444 | 2,433 | GAAAATAATGA |
| XRCC5 | Chr2: 217,292,473–217,295,040 | ss15143464 | 2,568 | 2,555 | AAGAACACATGGC |
| AFURS1 | Chr3: 195,440,681–195,441,437 | ss8481522 | 757 | 749 | AAGACTTC |
| KLHL3 | Chr5: 137,098,795–137,100,118 | ss15143086 | 1,324 | 1,308 | AAAATATTACCTCCCT |
| HLA-F | Chr6: 29,793,437–29,796,056 | ss8483556 | 2,620 | 2,601 | AAAAGAAAGACCCAAGCCT |
| HLA-G | Chr6: 30,005,583–30,007,340 | ss15143277 | 1,758 | 1,744 | AAGAATTGAGGAGC |
| SLC17A5 | Chr6: 74,365,117–74,367,305 | ss14142874 | 2,189 | 2,181 | AAAAATGG |
| SERAC1 | Chr6: 158,457,569–158,458,368 | ss8483321 | 800 | 784 | GAAAAATGAACATATC |
| LOC90637 | Chr7: 929,403–931,992 | ss15143254 | 2,590 | 2,576 | AAAACTTAAGAGTG |
| BC002644 | Chr7: 10,248,637–10,250,470 | ss8483421 | 1,834 | 1,817 | AAAGAAAAGAGGTTTAA |
| EGFR | Chr7: 55,028,493–55,030,810 | ss8483579 | 2,318 | 2,303 | TAAAAGCACATTGCA |
| AQP3 | Chr9: 33,413,362–33,414,654 | ss15142745 | 1,293 | 1,055 | AAGAATCTAGTTTTT |
| BC036431 | Chr9: 79,781,468–79,784,045 | ss15143735 | 2,578 | 2,563 | TAAAATGGCTCTAGC |
| RAD23B | Chr9: 105,413,293–105,415,295 | ss15143222 | 2,003 | 1,990 | AATTATTATTATT |
| PRMT3 | Chr11: 20,533,487–20,535,487 | ss15143629 | 2,001 | 1,988 | AATACAGAAATGT |
| CNOT2 | Chr12: 68,881,132–68,883,862 | ss15143419 | 2,731 | 2,713 | AAAAAAGTATGACACTTC |
| EPSTI1 | Chr13: 41,337,810–41,339,462 | ss14938190 | 1,653 | 1,638 | GAAAATCAGCTGGAG |
| FLJ12577 | Chr13: 47,749,416–47,751,876 | ss15143296 | 2,461 | 2,325 | AAACAAAAACAGT |
| C14orf24 | Chr14: 33,497,755–33,499,561 | ss8478175 | 1,807 | 1,792 | AAGACTTACGAATAG |
| PRPSAP2 | Chr17: 18,991,720–18,992,744 | ss15143022 | 1,025 | 1,010 | AAGAAAGAACAAGTT |
| PRKCA | Chr17: 64,815,159–64,816,230 | ss15143644 | 1,072 | 1,057 | AAAAAATGTTTTAAG |
| ZNF137 | Chr19: 57,787,819–57,789,420 | ss15143783 | 1,602 | 1,586 | AAAAATACAAAATTAG |
| AK09261 | Chr19: 58,380,999–58,383,780 | ss15143831 | 2,782 | 2,774 | AAAAAAAA |
| C20orf100 | Chr20: 43,361,555–43,361,953 | ss14942665 | 399 | 396 | TAA |
| AK026502 | Chr22: 25,492,631–25,494,356 | ss15143832 | 1,726 | 1,716 | AGAGGTTAAG |
Chromosomal coordinates are given for the July 2003 build (build 34) of the human genome sequence (University of California, Santa Cruz; Kent et al. 2002). All of the elements shown have poly(A) tails at their 3′ ends. Additional data are provided in supplemental Table 1 and dbSNP (http://www.ncbi.nlm.nih.gov/SNP/). Chr, chromosome.
To further confirm that SVA insertion polymorphisms were indeed abundant in human populations, we developed PCR assays to individually examine 28 additional genomic copies of SVA (materials and methods). These copies were identified arbitrarily from the ∼5000 copies in the human genome by searching the human genome database with SVA element query sequences. A total of 11 of 28 (39%) of the copies tested were found to be polymorphic for insertion in at least one individual of the Coriell panel (Collins et al. 1999; Table 4). Thus, together with the 28 SVA polymorphic copies identified in the trace experiments (Table 3), we have identified a total of 39 independent SVA insertion polymorphisms in human populations. These insertion polymorphisms have all of the sequence features of bona fide SVA retrotransposition events (supplemental Table 1, Tables 3 and 4). These results indicate that not only Alu and L1, but also a third transposon, SVA, is highly polymorphic in human populations (supplemental Table 1, Tables 1–4). Collectively, our results indicate that Alu, L1, and SVA provide the bulk of genetic variation that is caused by transposon insertion polymorphisms in humans.
TABLE 4.
Analysis of additional genomic SVA elements
| SVA name (nearest gene) |
Chromosomal locationa | Element size |
Target site duplication sequence (5′ to 3′) |
Polymorphic in Coriell panel? |
|---|---|---|---|---|
| EIF4G3-A | Chr1: 20,610,706–20,613,260 | 2,544 | TAAAAATTCAT | No |
| SPATA6 | Chr1: 48,217,230–48,220,014 | 2,773 | AAAAGAAAAACC | No |
| CASP8 | Chr2: 202,349,192–202,351,983 | 2,782 | AAGAATTTGA | Yes |
| PLOD2 | Chr3: 147,134,530–147,136,524 | 1,976 | AAAGAAAATGTGGCATATA | Yes |
| CD38 | Chr4: 15,542,595–15,544,984 | 2,374 | GAAAAGCAGCAAGCC | No |
| RAP1B | Chr5: 75,550,653–75,552,974 | 2,305 | AAAAATTAAAAAAACT | Yes |
| Neurestin | Chr5: 167,266,427–167,268,968 | 2,528 | GAAAACAACGTCAA | No |
| POLH | Chr6: 43,594,478–43,596,505 | 2,015 | AAGATTCTTTCAC | No |
| PCMT1 | Chr6: 150,137,472–150,139,466 | 1,930 | GAAAACAGCCA | No |
| LOC90693 | Chr7: 23,417,744–23,420,774 | 2,865 | AAGACTGTCCCCTGC | No |
| FLJ14117 | Chr7: 100,561,431–100,563,976 | 2,529 | AAAAATACAAAAATTGG | Yes |
| TNKS | Chr8: 9,551,502–9,553,345 | 1,829 | GAAAATTCTTTTCTT | Yes |
| FLJ10871 | Chr8: 28,759,637–28,761,987 | 2,260 | AGAAAAATGTAGACATA | No |
| MELK | Chr9: 36,604,406–36,606,089 | 1,669 | CAAAAATAATTTTTT | No |
| PBX3 | Chr9: 123,916,920–123,919,948 | 2,361 | GAAAAGATCA | No |
| HHEX | Chr10: 94,098,024–94,100,358 | 2,320 | GAGAGATGGGATGTG | No |
| ITPR2 | Chr12: 26,828,444–26,830,665 | 2,209 | AAAAATGGAGAAT | No |
| SNRPF | Chr12: 94,736,050–94,738,795 | 2,735 | AAAACTGTGGA | No |
| BRMS1 | Chr14: 34,435,361–34,437,930 | 2,553 | TAAATACCTACGAGTAG | No |
| SPTB | Chr14: 63,350,428–63,353,581 | 2,686 | GAAAATTCTT | Yes |
| PRPSAP2 | Chr17: 18,991,705–18,992,729 | 1,010 | AAGAAAGAACAAGTT | Yes |
| RNF135 | Chr17: 29,451,155–29,453,977 | 2,808 | GAAATAATTAAAAATC | Yes |
| CPX-1 | Chr20: 2,798,148–2,801,409 | 3,247 | AAAAGAACTTGATTT | Yes |
| REM | Chr20: 30,802,228–30,805,279 | 3,036 | AAGATTTGTTTCTTTT | No |
| SLC2A11 | Chr22: 22,520,488–22,523,059 | 2,556 | GAAAAAAAATTAACCT | Yes |
| GPR24 | Chr22: 39,383,157–39,385,673 | 2,503 | AAAACAAACAAACA | Yes |
| UTX | ChrX: 43,944,720–43,947,538 | 2,808 | TTATCAAATGA | No |
| OPHN1 | ChrX: 66,412,207–66,414,450 | 2,232 | TTTTAAACTTTT | No |
Chromosomal coordinates are given for the July 2003 build (build 34) of the human genome sequence (University of California, Santa Cruz; Kent et al. 2002). All of the elements shown have poly(A) tails at their 3′ ends. Additional data are provided in supplemental Table 1.
The recent completion of a draft sequence for the chimpanzee genome allowed us to determine whether the SVA element also is present in the chimp genome. We manually inspected the SVA copies listed in Table 3 and found that 27 of 28 (96.4%) of these copies were absent from the equivalent positions of the chimp genome (supplemental Table 1). The remaining copy appeared to be present, but had only partial sequence coverage in the chimp genome sequence. Therefore, most of these 28 polymorphic SVA copies appear to have been generated relatively recently in humans, at a point in time following the divergence of chimps and humans. However, since these 28 copies were selected on the basis of the fact that they were polymorphic in humans, it was possible that these copies were not representative of all SVAs in the human genome. Therefore, we arbitrarily selected 87 additional copies from the human genome to determine whether they were present in the chimp genome. Twenty-eight of these copies are listed in Table 4, and 59 additional copies were identified in the first 50 Mb interval of human chromosome 1, for a total of 87 copies (supplemental Table 1). Our analysis revealed that only 11 of these 87 SVA copies (12.6%) were precisely present at the equivalent positions of the chimp genome (supplemental Table 1). Seven additional copies had partial sequence coverage in the chimp genome and thus also appeared to be present at equivalent positions. Therefore, the available evidence indicates that ∼18 of the 87 SVA copies (20.7%) are likely to be present at equivalent positions of the human and chimp genomes. The remaining 69 SVA copies (79.3%) were completely absent from the chimp genome sequence. In a few cases, the chimp genome completely lacked sequence coverage in the area of the element, so it is unclear whether the SVA is truly absent in these cases (supplemental Table 1). However, in most of these 69 cases, the SVA element and 1 of the 2 copies of the target site duplication were precisely absent from the chimp genome (supplemental Table 1). Taken together, these data indicate that ∼20.7% of SVA insertions in the human genome were generated prior to the evolutionary divergence of chimps and humans, and up to ∼79.3% of the remaining SVA insertions were generated after the divergence of these species. Thus, SVA is a relatively young transposon that has expanded in the human genome during the past several million years. At least some of the SVA copies appear to have been mobilized very recently, suggesting that SVA might also remain actively mobile in humans and chimps today.
HERV-K and other examples of mobilized DNA:
In addition to the three most abundant groups of insertion polymorphisms in this study (Alu, L1, and SVA), we also identified several classes of less abundant insertion polymorphisms that were caused by mobilized DNA. For example, two human endogenous retrovirus K (HERV-K) insertion polymorphisms were identified in our trace experiments (Tables 2 and 5). One of these HERV-K copies was a 969-bp solo LTR element that was flanked by a perfect 5-bp target site duplication (Table 5). The other HERV-K element was a full-length 9462-bp copy that was flanked by a perfect 6-bp target site duplication (Table 5). This full-length copy had intact LTRs at its termini and four intact open reading frames capable of encoding homologs of the retroviral Gag, protease, Pol, and Env proteins (supplemental Table 1 and data not shown). We also identified four examples of mobilized small cellular RNAs, including two polymorphic copies of 5S rDNA and single examples of mobilized U2 and U5 RNA (Table 5). All four of these polymorphic insertions were flanked by L1-like target site duplications, strongly suggesting that these RNAs were mobilized by the L1 machinery. However, none of these mobilized elements contained a poly(A) tail, perhaps suggesting that the poly(A) sequences on template RNAs are not strictly required for the TPRT process (Boeke 1997; Roy-Engel et al. 2003). Finally, we identified a single example of a Mariner dependent-1 (Made1) insertion with an inverted repeat structure that was flanked by a perfect 5-bp target site duplication (Table 5).
TABLE 5.
Insertion polymorphisms generated by other forms of mobilized DNA
| Element type | Chromosomal locationa | dbSNP no. | Indel size |
Element size |
Target site duplication sequence (5′ to 3′) |
|---|---|---|---|---|---|
| 5S rDNA | Chr3: 12,171,534–12,171,588 | ss14942367 | 55 | 40 | GAAAGGTGAAAAGGA |
| HERV-K (LTR) | Chr8: 7,342,808–7,352,275 | ss15143090 | 9,468 | 9,463 | AAAGGT |
| U5 RNA | Chr9: 195,440,681–195,441,437 | ss14936914 | 76 | 60 | GAGAATCCTGGGTTCT |
| 5S rDNA | Chr12: 13,090,355–13,090,458 | ss8477420 | 104 | 90 | GCAAGTGAACATTT |
| HERV-K (LTR) | Chr12: 54,013,482–54,014,455 | ss14933585 | 974 | 969 | GCTAT |
| U2 RNA | Chr13: 108,834,029–108,834,078 | ss14938045 | 50 | 35 | GAAACTGCGAATCCA |
| Made1 (Mariner) | Chr20: 1,037,602–1,037,680 | ss14935893 | 79 | 74 | GCAAA |
Chromosomal coordinates are given for the July 2003 build (build 34) of the human genome sequence (University of California, Santa Cruz; Kent et al. 2002). All of the above elements lack poly(A) tails. Additional data are provided in supplemental Table 1 and dbSNP (http://www.ncbi.nlm.nih.gov/SNP/).
Validation studies:
Several lines of evidence suggested that our computational methods were highly accurate. For example, we detected 149 redundant transposon polymorphisms in the three data sets (supplemental Table 1). Therefore, our methods independently detected identical transposon insertion polymorphisms using totally different traces from different populations. Moreover, we also detected a number of Alu and L1 insertion polymorphisms that had been detected in previous studies with totally different methods. Nevertheless, as outlined below, we also conducted a systematic validation study to further evaluate the accuracy of our computational pipeline (Figure 3; Table 6; materials and methods).
Sixty-one transposon insertion polymorphisms were selected from the TSC data set to conduct this validation study (Table 6). We focused on the TSC data set because DNA was available for the entire panel of 24 diverse humans used in that study (Collins et al. 1999). Since all DNA traces in that experiment were derived from only these 24 individuals (Sachidanandam et al. 2001), any transposon polymorphism that was predicted from the TSC data set also should be found by PCR in at least 1 of these 24 individuals. If the polymorphism was not found in any of the 24, then we would know with certainty that our bioinformatics prediction was incorrect. PCR assays were developed for all 61 of the selected transposon insertion polymorphisms and 12–24 members of the Coriell panel were evaluated to determine whether the polymorphism could be verified. In all 61 cases, the allele predicted by the trace was confirmed in at least 1 of the 24 individuals of the panel (Figure 3 and Table 6). Therefore, our methods produced a 100% success rate for this arbitrarily selected sample of 61 TSC polymorphisms, indicating that our methods are highly accurate.
In five cases, we detected only the allele predicted by the trace in the panel of 24 individuals, and the allele predicted by the reference human genome sequence was not detected by PCR (Table 6). The most likely explanation for these results is that the person(s) represented by the reference human genome sequence had rare “private” transposon insertions at these positions that were absent from the majority of humans. We (and others) have observed similar results with SNPs identified from the TSC traces (Sachidanandam et al. 2001; Tsui et al. 2003), and Myers et al. (2002) have reported similar results with private alleles of transposon insertions. In cases where they have been examined, these private alleles have been verifiable in the DNA clones that were used to sequence the human genome (Myers et al. 2002).
Estimating the number of transposon insertion polymorphisms in humans:
Our study provided a unique opportunity to measure the levels of variation that are caused by transposon insertions in humans. Because our methods utilized DNA sequencing traces, it was possible to determine exactly how many bases of the human genome were sampled for a given trace experiment. In the case of the TSC experiment, 989,283,997 bp were sampled (equivalent to 30% of the haploid human genome). Similarly, 2,271,983,242 bp were sampled in the WGS experiment, equivalent to ∼69% of the human genome. Therefore, it was possible to normalize the data from these experiments to a genome size of 3.3 billion base pairs (100%) to estimate the total number of transposon insertion polymorphisms that were present in the average haploid genome in our study. By doubling these estimates, we determined that the average (diploid) human in our study harbored ∼1283 Alu insertion polymorphisms, 180 L1 polymorphisms, 56 SVA polymorphisms, and 17 other polymorphisms (Table 7). The TSC and WGS populations gave estimates that generally differed by less than twofold, with the WGS giving a higher estimate. Given that the WGS data were generated from eight African-Americans, these results are consistent with the observation of higher levels of genetic diversity in African populations (International HapMap Consortium 2003).
TABLE 7.
Average polymorphism frequencies in humans (diploid)
| No. polymorphisms per average diploid human in group (calculated from Table 1)
|
|||
|---|---|---|---|
| Element | TSC | WGS | Average |
| Alu-total | 1,153.9 | 1,411.3 | 1,282.6 |
| Alu Ya5 | 446.9 | 428.9 | 437.9 |
| Alu Yb8 | 200.1 | 382.5 | 291.3 |
| L1-total | 173.4 | 185.5 | 179.5 |
| L1-Ta | 166.8 | 168.1 | 167.5 |
| L1-other | 6.7 | 17.4 | 12.1 |
| SVA-total | 40.0 | 72.5 | 56.3 |
| Other | 13.3 | 20.3 | 16.8 |
| No. element copies in the human genome
| |||
| Element | Haploid | Diploid | Reference |
| Alu-total | 1,100,000 | 2,200,000 | Lander et al. (2001) |
| Alu Ya5 | 2,640 | 5,280 | Caroll et al. (2001) |
| Alu Yb8 | 1,852 | 3,704 | Caroll et al. (2001) |
| L1-total | 500,000 | 1,000,000 | Lander et al. (2001) |
| L1 (Ta) | 520 | 1,040 | Myers et al. (2002) |
| SVA-total | 5,000 | 10,000 | Strichman-Almashanu et al. (2001) |
| Average polymorphism frequencies per average diploid human in group (no. polymorphisms listed above/total copies in genome listed above) | |||
| Element | TSC | WGS | Average |
| Alu-total | 0.00052 (1/1,923) | 0.00064 (1/1,563) | 0.00058 (1/1,724) |
| Alu Ya5 only | 0.085 (1/11.8) | 0.081 (1/12.3) | 0.083 (1/12.0) |
| Alu Yb8 only | 0.054 (1/18.5) | 0.103 (1/9.7) | 0.079 (1/12.7) |
| L1-total | 0.00017 (1/5,882) | 0.00019 (1/5,263) | 0.00018 (1/5,556) |
| L1-Ta only | 0.160 (1/6.3) | 0.162 (1/6.2) | 0.161 (1/6.2) |
| SVA-total | 0.0040 (1/250) | 0.0073 (1/137) | 0.0057 (1/175) |
We also used our data to estimate the total number of common transposon insertion polymorphisms that are present in human populations. We detected 26% of the 660 polymorphic insertions of Alu Ya5 estimated to exist in human populations by Carroll et al. (2001). Similarly, we identified 35% of the 370 polymorphic insertions of Alu Yb8 estimated to exist in humans by Carroll et al. (2001). We also identified 25% of the 234 polymorphic L1-Ta insertions predicted by Myers et al. (2002). Thus, using the Caroll et al. and Myers et al. studies to calibrate our study, we conclude that we are detecting between 25 and 35% of a given class of transposon insertion polymorphisms. Therefore, given that we identified 605 nonredundant transposon polymorphisms, we estimate that human populations harbor a total of 1730–2420 common transposon insertion polymorphisms (for an average estimate of 2075).
Polymorphism frequencies for Alu, L1, and SVA:
Since we recovered data on Alu, L1, and SVA insertion polymorphisms using a single method, we could calculate the relative polymorphism frequencies for these elements (Table 7). To perform these calculations, we compared the number of polymorphisms that were identified for each transposon to the genomic copy numbers for each element (Table 7). We found that the average polymorphism frequency for all copies of L1 was the lowest, at 0.00018 (one polymorphic L1 insertion per 5556 copies in the genome; Table 7). The average polymorphism frequency for all genomic Alu copies likewise was relatively low at 0.00058 (one polymorphic insertion per 1724 copies in the genome). The average polymorphism frequency for SVA, in contrast, was an order of magnitude higher, at 0.0057 (one polymorphic insertion per 175 copies). Therefore, according to this analysis, SVA is the most polymorphic element in humans and is likely to be one of the youngest elements to expand in the human genome. Nevertheless, when the L1-Ta, Alu Ya5, and Alu Yb8 subfamilies were examined separately from all L1 and Alu copies, these younger L1 and Alu subfamilies had even higher polymorphism frequencies than SVA (Table 7). The L1-Ta subfamily, for example, with ∼1040 copies in the diploid genome, has the highest polymorphism frequency at 0.161 (one insertion per 6.2 copies; Table 7). The Alu Ya5 and Alu Yb8 subfamiles likewise have much higher polymorphism frequencies than all genomic Alu elements (Table 7). Therefore, the most active subfamiles of L1 and Alu have the highest polymorphism frequencies, followed by SVA. Like these other elements, SVA might also harbor extremely polymorphic subfamilies that remain to be discovered but are as polymorphic for insertion as the L1-Ta, Alu Ya5, and Alu Yb8 subfamilies. Alternatively, SVA might have a uniformly lower rate of polymorphism but collectively produces relatively high levels of genetic variation through its higher copy number (SVA has almost 10 times the number of L1-Ta copies). Either of these models would account for the relatively high levels of genetic variation that are caused by SVA insertion polymorphisms (Tables 1 and 7).
DISCUSSION
The spectrum of mobile DNA in humans:
In an effort to measure the overall levels of genetic variation that are caused by human transposons, we have developed a new method to broadly detect transposon insertion polymorphisms of all kinds in humans. Our strategy was highly efficient and led to the identification of 605 nonredundant transposon insertion polymorphisms in 36 diverse humans. Since the majority of these insertion polymorphisms had not been identified previously, our method was highly successful at discovering novel transposon polymorphisms. In fact, we estimate that our collection of 605 polymorphisms represents ∼25–35% of all common transposon insertion polymorphisms in human populations (see below). Our strategy, in principle, now could be used to identify all of the common transposon insertion polymorphisms that exist in human populations. Together with all previously identified Alu and L1 insertion polymorphisms, our 605 insertions provide significant progress toward this goal. Approximately 20 million additional human traces (beyond the 16.4 million used here) currently are available from SNP discovery projects, and more traces are being generated by ongoing SNP discovery projects daily. Another ∼17 million chimp traces are available that could be used to identify transposon insertion polymorphisms in the chimp genome relative to the human genome. Thus, our method is likely to be useful in humans as well as other organisms.
Unlike previous strategies, our polymorphism discovery strategy yielded data regarding the relative levels of genetic variation from all classes of transposons. The three most abundant classes of insertion polymorphisms in our study were Alu, L1, and SVA insertions. Although Alu and L1 insertion polymorphisms were expected to occur at high frequencies, no studies had been conducted previously to measure the polymorphism frequency of the SVA element or the remaining elements in the human genome. Therefore, our method has revealed that three transposons, Alu, L1, and SVA, are highly polymorphic in humans, and that these three elements together provide the bulk of genetic variation that is caused by transposon insertions in humans (Tables 1 and 7). Our data also indicate that few, if any, insertion polymorphisms exist for the remaining classes of elements in the human genome.
Most human transposon families are not highly polymorphic:
As mentioned above, an interesting finding of our study is that many transposon families in humans have not generated insertion polymorphisms to any great extent in recent history. Although Alu, L1, and SVA account for a little more than 30% of the human genome sequence, a total of 44% of the genome sequence is occupied by transposons and transposon-like repetitive elements. Therefore, ∼14% of the human genome is occupied by essentially extinct transposon-like families that contain mostly (or totally) inactive, fixed transposon alleles. Our study does not necessarily indicate that these elements are completely inactive, since we have sampled only 36 human genomes. Therefore, it is likely that we have identified only the most polymorphic classes of elements in the genome, and we may have missed polymorphic copies that occur at lower frequencies within smaller families. Such elements would be of great interest, and we do not rule out the existence of these elements, particularly since the heterochromatic regions of the human genome remain unsequenced. For example, our data indicate that elements such as HERV-K have been mobile recently enough to generate polymorphic insertions in human populations (Table 5). Although such elements do not generate a great deal of genetic variation, they would be of great interest if at least some of the polymorphic copies have retained the ability to function as autonomous retrotransposons. Additional studies will be required to determine whether the full-length HERV-K copy discovered in this study (ss15143090, Table 5) remains actively mobile today.
Alu and L1 insertion polymorphisms:
Our results regarding Alu element polymorphisms generally are in good agreement with a large number of previous studies examining the young Alu Y subfamilies of the human genome (reviewed in Batzer and Deininger 2002). Because we detected all Alu subfamilies with a single method, we also were able to measure the relative levels of variation that are caused by each subfamily (Tables 1, 2, and 7). Consistent with previous studies, we found that Alu Ya5 and Alu Yb8 insertion polymorphisms are highly abundant in humans. We also found that Alu Y, Alu Yc1, and Alu Ya4 insertion polymorphisms are moderately abundant in human populations (Table 2). The remaining Alu Y insertions were less abundant and were distributed among 15 different Alu Y subfamilies (Table 2). Our results further indicate that additional polymorphic Alu Y families of any significant size are not likely to exist in humans. Finally, we unexpectedly found a small number of ancient Alu S insertion polymorphisms in humans (Table 2).
Our results regarding L1 element polymorphisms likewise are in good agreement with previous studies examining L1-Hs insertion polymorphisms in humans (Sheen et al. 2000; Ovchinnikov et al. 2001; Myers et al. 2002; Badge et al. 2003; Brouha et al. 2003). However, we also found that older L1-P (primate) insertions represent a significant source of human genetic variation (Ovchinnikov et al. 2002). In fact, L1-P insertions represented close to 10% of all L1 insertion polymorphisms in our study (Table 2). Because we detected all L1 subfamilies with a single method, it was possible to assess the relative levels of variation that were caused by each subfamily and integrate these values with all other elements in the genome (Tables 1, 2, and 7).
SVA is highly polymorphic in humans:
Since the initial discovery of the SVA element, a number of retrotransposon-like insertions have been reported that might have been caused by SVA retrotransposition events (Hassoun et al. 1994; Kobayashi et al. 1998; Rohrer et al. 1999). One of the best candidates in this regard is a 3-kb retrotransposon insertion in the Fukutin gene that was reportedly responsible for 70% of the Fukuyama type muscular dystrophy in Japan (Kobayashi et al. 1998). The element described in that report has some of the features of a de novo SVA insertion; however, it was not referred to as an SVA element in that article, and the sequence of the insertion was not provided (Kobayashi et al. 1998). Two additional retrotransposon insertions also have been referred to as SVA elements by Ostertag and Kazazian (2001) and Ostertag et al. (2003). In one of these cases, the element was reported in the original study to be a SINE-R element rather than an SVA element (Rohrer et al. 1999). Since SINE-R is itself a retrotransposon and also a component of the SVA element, the insertion could be a SINE-R element or a truncated SVA. In the final case, no SVA sequences were actually present within the DNA insertion that was identified (Hassoun et al. 1994), and the inserted DNA segment was proposed to have been mobilized by a 3′-transduction event sponsored by an adjacent SVA element (Ostertag et al. 2003).
We now provide clear evidence for the existence of at least 39 de novo SVA element insertions in humans (Tables 3 and 4). For these 39 insertions: (i) both empty and SVA-occupied sites were identified in the human genome in different individuals, (ii) the SVA copies ended in poly(A) tails, and (iii) the inserted copies were precisely flanked by new target site duplications. Thus, our study indicates that SVA insertion polymorphisms are highly abundant in humans. In fact, SVA insertion polymorphisms provide about one-third the level of genetic variation that is caused by L1 insertions (Tables 1 and 7). Finally, our data also indicate that SVA has amplified independently and perhaps at different rates in the genomes of humans and chimps. Approximately 79% of the SVA insertions in the human genome are absent from the equivalent positions of the chimp genome, indicating that these insertions occurred relatively recently in human history (within the past ∼6 million years).
Since a high rate of polymorphism is a hallmark feature of an active transposon, our data suggest that SVA might be actively mobile in the human genome today. Because SVA does not encode any obvious proteins of its own (it lacks substantial open reading frames), it is likely to be a nonautonomous element that relies upon another transposon for its own transposition. Several aspects of our SVA insertions suggest that they might be mobilized by L1 elements in trans by the same mechanism that mobilizes Alu elements (Dewannieux et al. 2003). For example, the target site duplications of our SVA insertions closely resemble those of Alu and L1 elements in length and sequence (Tables 3 and 4). Our SVA insertions also have poly(A) tails, indicating that they are poly(A) retrotransposons (supplemental Table 1). Finally, many of our polymorphic SVA insertions had 5′-truncations that were similar to the 5′-truncations of L1 elements. Given these similarities to Alu and L1 elements, SVA is likely to be mobilized in trans by L1-encoded proteins (Esnault et al. 2000; Ostertag and Kazazian 2001; Wei et al. 2001; Dewannieux et al. 2003; Ostertag et al. 2003). Therefore, it appears that all three classes of highly polymorphic elements in our study (Alu, L1, and SVA) were generated by the L1 retrotransposition machinery in cis or in trans.
Estimating the levels of variation caused by transposon insertions in human populations:
Our method provided a unique opportunity to measure the levels of genetic variation that are caused by transposon polymorphisms in humans. The measure of variation that is most commonly reported for transposons is the percentage of copies that are polymorphic in at least one individual of a population (Sheen et al. 2000; Carroll et al. 2001; Ovchinnikov et al. 2001, 2002; Roy-Engel et al. 2001; Myers et al. 2002; Abdel-Halim et al. 2003; reviewed in Ostertag and Kazazian 2001 and Batzer and Deininger 2002). This approach is useful from the viewpoint of assessing whether a given transposon family is polymorphic in populations; however, it tends to overestimate the levels of genetic variation that are caused by transposons. This is because high-frequency and low-frequency alleles are counted equally with this type of measurement. In contrast, we measured the polymorphism rates in a manner that included the allelic frequencies of the transposon alleles. High-frequency alleles are encountered more often than rare alleles in our trace experiments, and therefore our method naturally takes into account the allelic frequencies of the transposon insertions. Thus, with this approach, we have been able to estimate the levels of genetic variation that are caused by transposon insertions in populations. As a consequence of factoring in the allelic frequencies, our estimates for polymorphism rates are four- to fivefold lower than those reported previously. For example, 25% of the Alu Ya5 copies previously were reported to be polymorphic in at least one individual of a population of 80 humans (Carroll et al. 2001). We now estimate that ∼6.8% of the Alu Ya5 copies are polymorphic in the average human of our study (Table 7). Likewise, 45% of the L1-Ta element copies previously were reported to be polymorphic in at least one member of a large population (Myers et al. 2002), whereas we now calculate that ∼13.6% of the L1-Ta copies are polymorphic in the average human of our study (Table 7).
We also generated an estimate of the total number of common transposon insertion polymorphisms that exist in human populations. We generated this estimate by comparing the number of insertion polymorphisms discovered in our study for a given element such as Alu Ya5 to the total number expected. For example, Carroll et al. previously had predicted that ∼25% of the 2640 genomic Alu Ya5 copies were polymorphic for insertion in at least one member of a population of 80 diverse humans (Carroll et al. 2001). Therefore, since we identified 170 Alu Ya5 insertion polymorphisms, we determined that we had identified 26% of all expected Alu Ya5 insertion polymorphisms in humans. By calibrating our study with several of these previous studies, we estimate that our 605 transposon insertions represent between 25 and 35% of all common transposon insertion polymorphisms in human populations. Therefore, on the basis of these comparisons, human populations are estimated to harbor between 1730 and 2420 common transposon insertion polymorphisms (for an average of 2075). Together with our 605 polymorphisms, less than half of these polymorphisms have been identified to date, indicating that additional efforts will be required to identify the full set of polymorphic transposon insertions (i.e., the “transposon insertion polymorphome”) in humans.
Together with previous studies, our analysis indicates that SNPs, indels, and transposon insertion polymorphisms represent significant sources of genetic variation in humans. Human populations are estimated to harbor ∼10 million common SNPs (Judson et al. 2002), ∼2 million common indels (our unpublished data), and ∼2000 common transposon insertion polymorphisms (this study). Therefore, with 10 million bases of variation, SNPs account for the majority of common human genetic variation, followed by indels and then transposon insertion polymorphisms. On the other hand, if we assume that the average transposon polymorphism in humans is ∼500–1000 bp in length, then the total amount of variation caused by common transposon insertions is 1–2 million base pairs (equivalent to 10–20% of the base pair variation caused by SNPs). Thus, in terms of the number of base pairs, common transposon insertions cause significant levels of human genetic variation. Moreover, humans also are likely to harbor >10 million rare private transposon insertions (cases in which only one or a few individuals have the insertion). Therefore, transposon insertion polymorphisms cause significant levels of human variation. A number of studies now have shown that SNPs, indels, and transposon insertions all may cause serious phenotypic changes when positioned at critical sites within genes (Collins et al. 1987; Kazazian et al. 1988; Sachidanandam et al. 2001). Nevertheless, a comprehensive map of genetic variation that integrates SNPs, indels, and transposon insertions currently is lacking. A fully integrated map that includes all forms of genetic variation will be necessary to efficiently identify genetic polymorphisms that influence human phenotypes and diseases.
Acknowledgments
We thank the people at the SNP Consortium, the Sanger Centre, the Baylor Genome Center, and the Whitehead Genome Center for the use of their trace data, and University of California, Santa Cruz for its human genome database. We also thank Shari Corin for helpful advice on this project and for critical review of the manuscript. Finally, we thank Karen Ventii and Summer Goodson for help with some PCR experiments. This work was supported by training grant 2T32GM008490-11 from the National Institutes of Health (E.A.B.), grant 2-80302 from the Emory University Research Council (S.E.D.), grant RSG-01-173-01-MBC from the American Cancer Society (S.E.D.), and grant 1R01HG02898-01A1 from the National Institutes of Health (S.E.D.).
References
- Abdel-Halim, S., G. E. Kilroy, W. S. Watkins, L. B. Jorde and M. A. Batzer, 2003. Recently integrated Alu elements and human genomic diversity. Mol. Biol. Evol. 20: 1349–1361. [DOI] [PubMed] [Google Scholar]
- Badge, R. M., R. S. Alisch and J. V. Moran, 2003. ATLAS: a system to selectively identify human-specific L1 insertions. Am. J. Hum. Genet. 72: 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J. A., Z. Gu, R. A. Clark, K. Reinert, R. V. Samonte et al., 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Batzer, M. A., and P. L. Deininger, 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370–379. [DOI] [PubMed] [Google Scholar]
- Bedell, J. A., I. Korf and W. Gish, 2000. MaskerAid: a performance enhancement to RepeatMasker. Bioinformatics 16: 1040–1041. [DOI] [PubMed] [Google Scholar]
- Berg, D. E., and M. M. Howe, 1989 Mobile DNA. American Society for Microbiology, Washington, DC.
- Berger, J., T. Suzuki, K. A. Senti, J. Stubbs, G. Schaffner et al., 2001. Genetic mapping with SNP markers in Drosophila. Nat. Genet. 29: 475–481. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., 1997. LINEs and Alus—the poly A connection. Nat. Genet. 16: 6–7. [DOI] [PubMed] [Google Scholar]
- Boissinot, S., P. Chevret and A. Furano, 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17: 915–928. [DOI] [PubMed] [Google Scholar]
- Britten, R. J., W. F. Baron, D. Stout and E. H. Davidson, 1988. Sources and evolution of human Alu repeated sequences. Proc. Natl. Acad. Sci. USA 85: 4770–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha, B., J. Schstak, R. M. Badge, S. Lutz-Prigg, A. H. Farbey et al., 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 100: 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, M. L., A. M. Roy-Engel, S. V. Nguyen, A. Salem, E. Vogel et al., 2001. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 311: 17–40. [DOI] [PubMed] [Google Scholar]
- Collins, F. S., M. L. Drumm, J. L. Cole, W. K. Lockwood, G. F. Vande Woude et al., 1987. Construction of a general human chromosome jumping library, with application to cystic fibrosis. Science 235: 1046–1049. [DOI] [PubMed] [Google Scholar]
- Collins, F. S., L. D. Brooks and A. Chakravarti, 1999. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 8: 1229–1231. [DOI] [PubMed] [Google Scholar]
- Cost, G. J., Q. Feng, A. Jacquier and J. D. Boeke, 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21: 5899–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger, P. L., and M. A. Batzer, 1999. Alu repeats and human disease. Mol. Genet. Metab. 67: 183–193. [DOI] [PubMed] [Google Scholar]
- Deininger, P. L., M. A. Batzer, C. A. Hutchison and M. H. Edgell, 1992. Master genes in mammalian repetitive DNA amplification. Trends Genet. 8: 307–311. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., C. Esnault and T. Heidmann, 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41–48. [DOI] [PubMed] [Google Scholar]
- Esnault, C., J. Maestre and T. Heidmann, 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24: 363–367. [DOI] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Feng, Q., J. V. Moran, H. H. Kazazian and J. D. Boeke, 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916. [DOI] [PubMed] [Google Scholar]
- Hassoun, H., T. L. Coetzer, J. N. Vassiliadis, K. E. Sahr, G. J. Maalouf et al., 1994. A novel mobile element inserted in the alpha spectrin gene. Spectrin Dayton. J. Clin. Invest. 94: 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh, H., and M. F. Singer, 1996. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15: 630–639. [PMC free article] [PubMed] [Google Scholar]
- Hohjoh, H., and M. F. Singer, 1997. a Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16: 6034–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh, H., and M. F. Singer, 1997. b Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J. Mol. Biol. 271: 7–12. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium, 2003. The international HapMap project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- Johanning, K., C. A. Stevenson, O. O. Oyeniran, Y. M. Gozal, A. M. Roy-Engel et al., 2003. Potential for retroposition by old Alu subfamilies. J. Mol. Evol. 56: 658–664. [DOI] [PubMed] [Google Scholar]
- Judson, R., B. Salisbury, J. Schneider, A. Windemuth and J. C. Stephens, 2002. How many SNPs does a genome-wide haplotype map require? Pharmacogenomics 3: 379–391. [DOI] [PubMed] [Google Scholar]
- Jurka, J., 2000. Repbase update—a database and an electronic journal of repetitive elements. Trends Genet. 16: 418–420. [DOI] [PubMed] [Google Scholar]
- Jurka, J., M. Krnjajic, V. V. Kapitonov, J. E. Stenger and O. Kokhanyy, 2002. Active Alu elements are passed primarily through paternal germlines. Theor. Popul. Biol. 61: 519–530. [DOI] [PubMed] [Google Scholar]
- Jurka, J., and T. Smith, 1988. A fundamental division in the Alu family of repeated sequences. Proc. Natl. Acad. Sci. USA 85: 4775–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J., and E. Zuckerkandl, 1991. Free left arms as precursor molecules in the evolution of Alu sequences. J. Mol. Evol. 33: 49–56. [DOI] [PubMed] [Google Scholar]
- Kazazian, H. H., C. Wong, H. Youssoufian, A. F. Scott, D. G. Phillips et al., 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332: 164–166. [DOI] [PubMed] [Google Scholar]
- Kent, W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle et al., 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, B., M. Palozzolo, C. Martin, J. D. Boeke and S. E. Devine, 1997 Transposon-mediated DNA sequencing, pp. 455–532 in Genome Analysis: A Laboratory Manual, Vol. 1, edited by B. Birren, E. D. Green, R. M. Myers and P. Hieter. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kobayashi, K., Y. Nakahori, M. Miyake, K. Matsumura, E. Kondo-lida et al., 1998. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature 394: 388–392. [DOI] [PubMed] [Google Scholar]
- Kolosha, V. O., and S. L. Martin, 1997. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. USA 94: 10155–10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Liu, J., M. M. Nau, J. Zucman-Rossi, J. I. Powell, C. J. Allegra et al., 1997. LINE-1 element insertion at the t(11;22) translocation breakpoint of a desmoplastic small round cell tumor. Genes Chromosomes Cancer 18: 232–239. [DOI] [PubMed] [Google Scholar]
- Luan, D. D., M. H. Korman, J. L. Jakubczak and T. H. Eickbush, 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605. [DOI] [PubMed] [Google Scholar]
- Martin, S. L., and F. D. Bushman, 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21: 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. L., J. Li and J. A. Weisz, 2000. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 304: 11–20. [DOI] [PubMed] [Google Scholar]
- Martin, S. L., D. Branciforte, D. Keller and D. L. Bain, 2003. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl. Acad. Sci. USA 100: 13815–13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias, S. L., A. F. Scott, H. H. Kazazian, J. D. Boeke and A. Gabriel, 1991. Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810. [DOI] [PubMed] [Google Scholar]
- Miki, Y., I. Nishisho, A. Horii, Y. Miyoshi, J. Utsunomiya et al., 1992. Disruption of the APC gene by a retrotransposon insertion of L1 sequence in a colon cancer. Cancer Res. 52: 643–645. [PubMed] [Google Scholar]
- Miki, Y., T. Katagiri, F. Kasumi, T. Yoshimoto and Y. Nakamura, 1996. Mutation analysis in the BRCA2 gene in primary breast cancers. Nat. Genet. 13: 245–247. [DOI] [PubMed] [Google Scholar]
- Moran, J. V., 1999. Human L1 retrotransposition: insights and peculiarities learned from a cultured cell retrotransposition assay. Genetica 107: 39–51. [PubMed] [Google Scholar]
- Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis and H. H. Kazazian, 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927. [DOI] [PubMed] [Google Scholar]
- Morse, B., P. G. Rothberg, V. J. South, J. M. Spandorfer and S. M. Astrin, 1988. Insertional mutagenesis of the myc locus by a LINE-1 sequence in human breast carcinoma. Nature 333: 87–90. [DOI] [PubMed] [Google Scholar]
- Morrish, T. A., N. Gilbert, J. S. Myers, B. J. Vincent, T. D. Stamato et al., 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31: 159–165. [DOI] [PubMed] [Google Scholar]
- Myers, J. S., B. J. Vincent, H. Udall, W. S. Watkins, T. A. Morrish et al., 2002. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 71: 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita, N., H. Nishio, Y. Kitoh, Y. Ishikawa and R. Minami, 1993. Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J. Clin. Invest. 91: 1862–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag, E. M., and H. H. Kazazian, Jr., 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35: 501–538. [DOI] [PubMed] [Google Scholar]
- Ostertag, E. M., J. L. Goodier, Y. Zhang and H. H. Kazazian, 2003. Nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 73: 1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov, I., A. B. Troxel and G. D. Swergold, 2001. Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res. 11: 2050–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov, I., A. Rubin and G. D. Swergold, 2002. Tracing the LINEs of human evolution. Proc. Natl. Acad. Sci. USA 99: 10522–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer, J., Y. Minegishi, D. Richter, J. Eguiguren and M. E. Conley, 1999. Unusual mutations in Btk: an insertion, a duplication, and four large deletions. Clin. Immunol. 90: 28–37. [DOI] [PubMed] [Google Scholar]
- Roy-Engel, A. M., M. L. Carroll, E. Vogel, R. K. Garber, S. V. Nguyen et al., 2001. Alu insertion polymorphisms for the study of human genomic diversity. Genetics 159: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel, A. M., A. H. Salem, O. O. Oyeniran, L. Deininger, D. J. Hedges et al., 2003. Active Alu element “A-tails”: size does matter. Genome Res. 12: 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandam, R., D. Weissman, S. C. Schmidt, J. M. Kakol, L. D. Stein et al., 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928–933. [DOI] [PubMed] [Google Scholar]
- Sheen, F. M., S. T. Sherry, G. M. Risch, M. Robichaux, I. Nasidze et al., 2000. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 10: 1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L., L. Wu, S. Sanlioglu, R. Chen, A. R. Mendoza et al., 1994. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and C4B genes in the HLA class III region. J. Biol. Chem. 269: 8466–8476. [PubMed] [Google Scholar]
- Slagel, V., E. Flemington, V. Traina-Dorge, H. Bradshaw and P. L. Deininger, 1987. Clustering and relationships of the Alu family in the human genome. Mol. Biol. Evol. 4: 19–29. [DOI] [PubMed] [Google Scholar]
- Smit, A. F., 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9: 657–663. [DOI] [PubMed] [Google Scholar]
- Smit, A. F. A., and A. D. Riggs, 1996. Tiggers and other DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. USA 93: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strichman-Almashanu, L. Z., R. S. Lee, P. O. Onyango, E. Perlman, F. Flam et al., 2001. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 12: 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, C., L. E. Coleman, J. L. Griffith, E. A. Bennett, S. G. Goodson et al., 2003. Single nucleotide polymorphisms (SNPs) that map to gaps in the human SNP map. Nucleic Acids Res. 31: 4910–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu, E., and C. Tschudi, 1984. Alu sequences are processed 7SL RNA genes. Nature 312: 171–172. [DOI] [PubMed] [Google Scholar]
- Wei, W., N. Gilbert, S. L. Ooi, J. F. Lawler, E. M. Ostertag et al., 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21: 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. A. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., K. M. Dipple, E. Vilain, B. L. Huang, G. Finlayson et al., 2000. AluY insertion (IVS4–52ins316alu) in the glycerol kinase gene from an individual with benign glycerol kinase deficiency. Hum. Mutat. 15: 316–323. [DOI] [PubMed] [Google Scholar]