Abstract
A genetic screen for Chlamydomonas reinhardtii mutants with copper-dependent growth or nonphotosynthetic phenotypes revealed three loci, COPPER RESPONSE REGULATOR 1 (CRR1), COPPER RESPONSE DEFECT 1 (CRD1), and COPPER RESPONSE DEFECT 2 (CRD2), distinguished as regulatory or target genes on the basis of phenotype. CRR1 was shown previously to be required for transcriptional activation of target genes like CYC6, CPX1, and CRD1, encoding, respectively, cytochrome c6 (which is a heme-containing substitute for copper-containing plastocyanin), coproporphyrinogen III oxidase, and Mg-protoporphyrin IX monomethylester cyclase. We show here that CRR1 is required also for normal accumulation of copper proteins like plastocyanin and ferroxidase in copper-replete medium and for apoplastocyanin degradation in copper-deficient medium, indicating that a single pathway controls nutritional copper homeostasis at multiple levels. CRR1 is linked to the SUPPRESSOR OF PCY1-AC208 13 (SOP13) locus, which corresponds to a gain-of-function mutation resulting in copper-independent expression of CYC6. CRR1 is required also for hypoxic growth, pointing to a physiologically meaningful regulatory connection between copper deficiency and hypoxia. The growth phenotype of crr1 strains results primarily from secondary iron deficiency owing to reduced ferroxidase abundance, suggesting a role for CRR1 in copper distribution to a multicopper ferroxidase involved in iron assimilation. Mutations at the CRD2 locus also result in copper-conditional iron deficiency, which is consistent with a function for CRD2 in a pathway for copper delivery to the ferroxidase. Taken together, the observations argue for a specialized copper-deficiency adaptation for iron uptake in Chlamydomonas.
COPPER is usually utilized in organisms as a cofactor in enzymes or electron transfer proteins that catalyze redox reactions or oxygen chemistry. Therefore, it is an essential micronutrient. However, the potential for Cu ions to participate in Fenton chemistry resulting in the production of reactive oxygen species means that cells must balance the acquisition of the nutrient to saturate but not exceed intracellular copper binding sites. Studies of copper homeostasis have focused on the pathways of copper uptake from the environment, intracellular distribution via chaperones and distributive transporters, and mechanisms for sequestration and detoxification. These works have revealed a variety of assimilatory copper transporters whose function is regulated by copper availability (Dancis et al. 1994; Zhou and Gitschier 1997; Peña et al. 2000). In eukaryotes as well as in compartmentalized bacteria, there are distributive transporters that deliver copper to particular compartments, where it can be assembled into the active site of proteins like Fet3p, a ferroxidase required for iron assimilation, or plastocyanin, required for photosynthesis (Yuan et al. 1995; Askwith and Kaplan 1998; Tottey et al. 2001; Sancenón et al. 2003; Shikanai et al. 2003). Small proteins, called copper chaperones, bind intracellular copper and transfer it to appropriate compartments, in some cases via the distributive transporters (Glerum et al. 1996a,b; Culotta et al. 1997; Klomp et al. 1997; Lin et al. 1997; Valentine and Gralla 1997; Himelblau and Amasino 2000).
An immediate first line of defense to nutrient deficiency is the activation of assimilatory mechanisms, and this is well established for copper as well (Vulpe and Packman 1995; Yamaguchi-Iwai et al. 1997; Labbé and Thiele 1999). On the other hand, much less is known about how cellular metabolism is adjusted to deal with the situation where the amount of copper available is insufficient for the biosynthesis of a full complement of copper proteins. One well-established metabolic adaptation in photosynthetic microorganisms is the substitution under copper-deficient conditions of an iron-containing cytochrome for the abundant blue copper protein plastocyanin (Wood 1978; Sandmann et al. 1983; Merchant and Bogorad 1986). The molecular details of this adaptive switch are best understood in Chlamydomonas. In copper-replete medium, plastocyanin is normally present in Chlamydomonas chloroplasts at a stoichiometry of ∼8 × 106 molecules/cell, but when the cells are grown in copper-deficient medium, Chlamydomonas alters its requirement for copper (Merchant et al. 1991). The organism replaces this abundant copper protein with a catalytically equivalent heme-containing substitute, cytochrome c6 (cyt c6), and in this situation it requires much less copper for growth.
The mechanism by which the switch between plastocyanin and cyt c6 is achieved involves copper-responsive modification of gene expression. The PCY1 gene, encoding plastocyanin, is constitutively expressed irrespective of the copper status of the cell, but under copper-deficient conditions the apoprotein is actively degraded by an as yet unidentified protease in the thylakoid lumen (Li and Merchant 1995; Li et al. 1996). In contrast, the CYC6 gene, encoding cyt c6, is transcribed only in Cu-deficient cells. This occurs via transcriptional activation through copper-responsive elements containing a GTAC core (Quinn and Merchant 1995; Quinn et al. 2000). As Cu availability falls below the amount needed to saturate intracellular copper binding sites (∼9 × 106/cell), a number of cellular responses besides changes in plastocyanin and cyt c6 abundance are coordinately induced, suggesting that a common signal transduction pathway is responsible for handling nutritional copper deficiency.
For instance, a high-affinity copper-uptake system involving a transporter and a cell surface reductase is induced in copper deficiency. On the basis of the fact that the velocity of transport increases 20-fold but the Km remains the same, Hill et al. (1996) concluded that Chlamydomonas must upregulate the same transporter that operates in Cu-replete cells. Proteolysis of apoplastocyanin is observed only in Cu-deficient cells, suggesting that a specific protease may be expressed (Li et al. 1996). Another change is the transcriptional activation of the CPX1 gene, encoding coproporphyrinogen oxidase in the tetrapyrrole biosynthetic pathway (Hill and Merchant 1995; Quinn et al. 1999). The function of upregulation of CPX1 is not known; it has been suggested that it provides for increased flux for heme biosynthesis, perhaps to provide the cofactor for cyt c6. However, the recent discovery of copper-regulated differential accumulation of isoforms Crd1 vs. Cth1 of a chlorophyll biosynthetic enzyme (Moseley et al. 2002b; Tottey et al. 2003) suggests a more fundamental and broader-based connection between Cu and the tetrapyrrole pathway.
In this work we sought to discover components of copper-responsive signal transduction through isolation and genetic classification of mutants that require copper supplementation for growth. We established (i) a specialized copper-deficiency adaptation for high-affinity iron uptake, (ii) that a single pathway controls all known responses to copper deficiency, (iii) that the pathway is required also for copper homeostasis under copper-replete conditions, and (iv) that the previously identified genetic and molecular connection between copper nutrition and hypoxia is physiologically significant.
MATERIALS AND METHODS
Growth conditions:
Chlamydomonas reinhardtii strains were grown in copper-supplemented (6 μm added CuSO4) or copper-deficient Tris-acetate-phosphate (TAP) media as described by Quinn and Merchant (1998). Liquid cultures were grown in an orbital shaker (200 rpm) at 25° under continuous light at an intensity of 50–150 μmol/m2/sec. Agar plates were grown at 22°–25° and 50–100 μmol/m2/sec. For arginine-requiring strains the medium was supplemented with 250 μg/ml arginine. For growth at low oxygen concentrations, cell cultures were bubbled with gas mixtures as follows: 99.8% air and 0.2% CO2 (control) or 98% N2, 1.8% air, and 0.2% CO2 (low oxygen conditions). The output mixture had a total flow of 2.5 liter/min/liter of culture and was filtered through a sterile 0.45-μm syringe filter. Cultures were maintained for 4 days at room temperature (22°–24°) with normal room lighting (5–15 μmol/m2/sec) on a shaker at 100 rpm. The oxygen content of the cultures was measured with a standardized oxygen electrode (Orion Research, Beverly, MA).
Protein analysis:
Soluble and pellet fractions of cells were prepared as described previously (Quinn and Merchant 1998; Quinn et al. 1999). Proteins were separated by discontinuous SDS-PAGE and transferred to polyvinylidene difluoride membranes for immunodetection of plastocyanin, cyt c6, coprogen oxidase, Crd1, Cth1, Fox1, and OEE1. The amounts of samples loaded were normalized on the basis of either cell counts or chlorophyll. Antibody dilutions were 1:2000 for α-plastocyanin, 1:1000 for α-cyt c6, 1:3000 for α-coprogen oxidase, and 1:1000 for α-Crd1, α-Fox1, and α-OEE1.
Analysis of RNA:
RNA isolation and analysis by hybridization was performed essentially as described previously (Quinn et al. 1999; Moseley et al. 2000; La Fontaine et al. 2002). Three to 5 μg of total RNA was loaded in each lane. Hybridization was detected by exposure to PhosphorImager screens (Molecular Dynamics, Sunnyvale, CA). For quantitative real-time PCR, genomic DNA was removed from the total RNA preparation by treatment with RQ1 DNAse (Promega, Madison, WI) according to the manufacturer's instructions. Complementary DNA, primed with oligo(dT), was generated with reverse transcriptase (GIBCO BRL, Gaithersburg, MD) also according to the manufacturer's instructions, and used in the amplification reaction directly after dilution. The amplification reaction was carried out with reagents from the DyNAmo HS Sybr green qPCR kit (MJ Research, Watertown, MA). Each reaction contained 10 μl 2× master mix, 0.3 μm primers, and cDNA corresponding to 5 ng/μl input RNA in the reverse transcriptase reaction. For each gene, the primers were as follows:
CBLP: 5′-GCCACACCGAGTGGGTGTCGTGCG-3′ and 5′-CCTTGCCGCCCGAGGCGCACAGCG-3′;
ATX1: 5′-GGCATCCAGACGGTGAAAAGCG-3′ and 5′-TCACTGCGCCAGCTTTGCCTC-3′;
CTP1: 5′-GGTGTTGACCTGGCTACGCTGTG-3′ and 5′-GCCACCGGTGCCAAGGGAGGAG-3′;
CTP2: 5′-AGCAGCAGCAGCGGGGTGCAG-3′ and 5′-GTCGCTGCCGGCGATGCCGAC-3′.
The reaction conditions were: 95° for 15 min, followed by cycles of 95° for 10 sec, 65° for 10 sec, 72° for 30 sec up to a total of 35 cycles. The fluorescence was measured at each cycle at 72° and 83°. The 2-ΔΔCT method was used to analyze the data (Livak and Schmittgen 2001). For visualizing the relative abundance of each transcript, product was removed from each set of reactions for a particular primer pair at a cycle where all samples showed product increase in the exponential phase and was separated by electrophoresis (2% agarose). The size of the product was estimated against markers (λDNA digested with PstI).
Strains and genetic analysis:
The wild-type strain used was CC-125. Insertional mutants were generated in the CC-425-pCU4 background (arg2 cw15 mt+; Quinn and Merchant 1995) and were backcrossed at least twice to wild-type or arginine auxotrophic strains (arg2 derived from CC-425 or arg7 derived from CC-1931). The suppressor of pcy1-ac208 strain (sop13) was isolated by growing the parental pcy1-ac208 strain on Cu-supplemented minimal medium plates in continuous light. Fast-growing colonies were chosen and screened by immunoblot analysis for cyt c6 expression in +Cu medium. Mating of strains of opposite mating types and dissection of zygotes was performed as described by Harris (1989). Complementation analysis was performed by generating vegetative diploids according to the method of Harris (1989), using arg2 and arg7 double mutants with crd1, crd2, crr1, and pcy1-ac208sop13 strains.
Measurements of fluorescence kinetics:
Room temperature chlorophyll fluorescence induction kinetics were measured using an open FluorCam detector (Photon Systems Instruments, Brno, Czech Republic). Fluorescence emissions were recorded from either liquid cultures or colonies of cells on plates after dark adaptation periods of at least 5 min, using an actinic light intensity of ∼60 μE/m2/sec for 2–3 sec.
Insertional mutagenesis:
To mutagenize cells, the glass bead method (Kindle 1990) was used to insert the plasmid pSP109 containing the ble gene that confers resistance to Zeocin (Stevens et al. 1996). The strain used for transformation was CC-425 transformed with pCU4 (Quinn and Merchant 1995). In the pCU4 construct the copper responsive CYC6 promoter drives the arylsulfatase encoding gene ARS2. [This strain was used because we had initially intended to use the reporter for screening among the transformants for regulatory mutants (no reporter gene expression in −Cu medium), but arylsulfatase expression varied too much post-transformation, and the strategy had to be abandoned.] After transformation, the cells were plated on +Cu TAP plates. After 5–7 days transformants were transferred to a new +Cu plate (50 transformants/plate) and after an additional 5–7 days these colonies were transferred to two sets of new plates, one set with and one set without copper. After two more transfers to +Cu or −Cu plates the transformants were scored for differences in appearance, growth, and fluorescence induction kinetics.
RESULTS
Three loci, CRR1, CRD1, and CRD2, are involved in copper nutrition signaling:
In previous work we had defined a nutritional copper signaling pathway on the basis of the differential accumulation of cyt c6 and plastocyanin that function in photosynthetic electron transfer (Merchant and Bogorad 1987a; Quinn and Merchant 1995). To identify additional players in the pathway, we screened colonies of insertionally mutagenized cells (see materials and methods) for copper nutrition-dependent growth phenotypes and photosynthetic function (see Moseley et al. 2000). Of 7568 Zeocin-resistant colonies tested, we identified 11 strains that displayed copper-conditional phenotypes, and these were subsequently categorized as carrying mutations in regulatory or target genes on the basis of phenotypic analysis (described below) and named copper response regulator (crr) or copper response defect (crd) strains, respectively. Genetic complementation and recombination analyses separated the mutants into three complementation groups: CRR1, represented by allelic strains crr1-1 and crr1-2; CRD1, represented by 8 strains (of 8 crd1 mutants identified in this screen plus another 4 from a separate screen, 9 were analyzed by recombination and complementation tests and shown to be allelic; Moseley et al. 2000); and CRD2, represented by a single strain, crd2-1 (Table 1).
TABLE 1.
Complementation and recombination analysis ofcrr andcrd strains
| crr1-1 | crr1-2 | crd1-1 | crd1-5 | crd2-1 | |
|---|---|---|---|---|---|
| crr1-1 | − | −a | ND | + | + |
| crr1-2 | 0/57 (17)b | − | ND | ND | ND |
| crd1-1 | 12/24 (6) | ND | − | ND | ND |
| crd1-5 | 80/200 (random spores) | ND | − | + | |
| crd2-1 | 15c/83 (19) | ND | 27/56 (16) | ND | − |
ND, not done.
The results of complementation tests are shown above the diagonal (top and right). A plus sign indicates that the vegetative diploids displayed wild-type growth and fluorescence induction kinetics on −Cu TAP medium, i.e., positive complementation, whereas a minus sign indicates that the diploids displayed mutant growth and photosynthesis phenotypes, i.e., negative complementation.
The results of recombination tests are shown below the diagonal (bottom and left). The numerator refers to the number of recombinant progeny, while the denominator indicates the total progeny scored. The number in parentheses indicates the number of zygotes from which the spores originated.
Only wild-type spores were scored as recombinants, since it was not possible to distinguish crd2-1crr1-1 double mutants from crr1-1 single mutants using growth and fluorescence analysis.
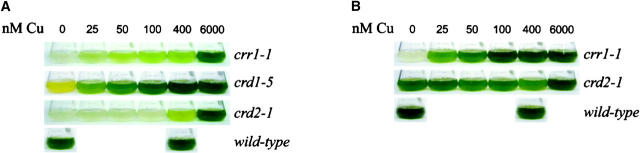
CRR1:
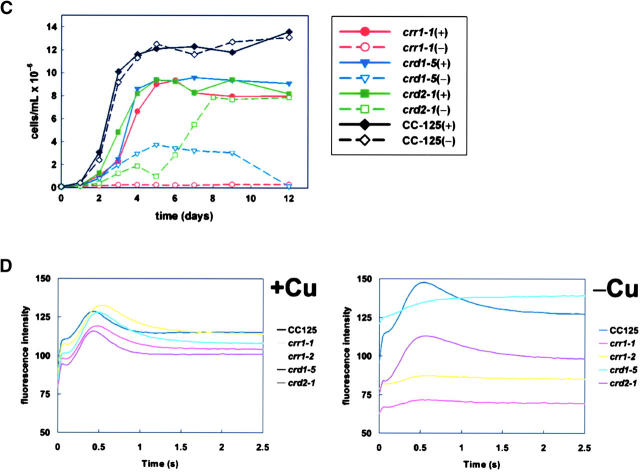
Copper-deficient crr1 cultures grow poorly compared to wild-type cells (Figure 1, A–C). A normal growth rate of crr1 is evident only upon provision of excess copper (see 6 μm sample, Figure 1A), and severely copper-depleted crr1 cells never reach the cell densities of wild-type cultures, indicating that the strain is copper limited for growth (see 0 and 25 nm samples, Figure 1B). In previous work we noted that complete repression of the CYC6 gene, even in cultures with high cell density (1–2 × 107 cells/ml), occurred at 400 nm medium copper (Hill et al. 1991; Merchant et al. 1991), so the higher requirement for medium copper in crr1 suggests that the CRR1 locus controls aspects of copper homeostasis besides transcriptional activation of CYC6. Fluorescence induction and decay kinetics indicate a defect in photosynthetic electron transfer in −Cu crr1 cells, but when crr1 strains are grown in excess copper, photosynthesis is normal (Figure 1D). This can be explained if −Cu crr1 strains lack cyt c6 and if +Cu crr1 strains can accumulate plastocyanin, and indeed this is the case (Figure 2B).
Figure 1.—
Growth phenotypes of candidate mutants. (A) Appearance of cultures after growth for 5 days [wild type (CC125), crr1-1, and crd2-1] or 6 days (crd1-5), in TAP medium supplemented with the indicated Cu concentrations. (B) Appearance of the same wild-type, crr1-1, and crd2-1 cultures after 13 days of growth. (C) Plot of cell density over time for wild-type (CC125), crr1-1, crd1-5, and crd2-1 cultures over 12 days of growth in TAP medium supplemented with 6 μm (+) or 0 μm (−) Cu. (D) Chlorophyll fluorescence induction curves from wild-type vs. mutant strains grown on solid TAP medium, with (+, 6 μm) or without (−, 0 μm) Cu.
Figure 2.—
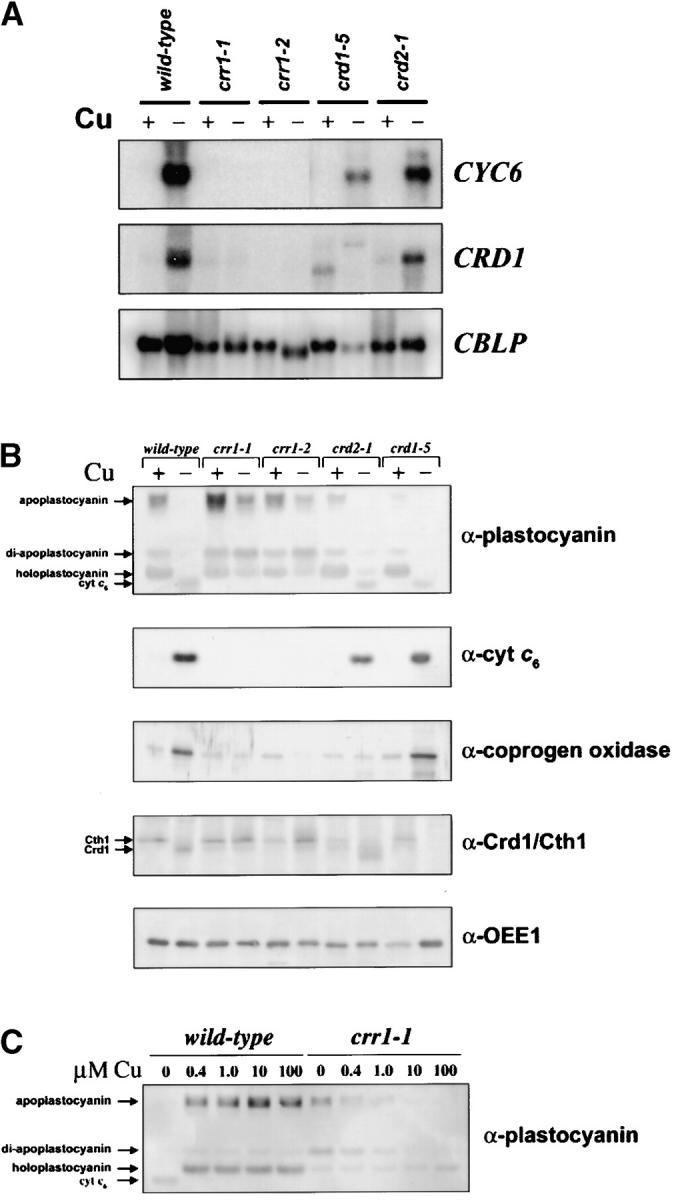
Biochemical phenotypes of crr and crd strains. (A) RNA blot analysis comparing expression of Cu-responsive genes in wild-type vs. mutant cells. Three micrograms of total RNA was loaded in each lane. RNA was prepared from cultures grown in TAP medium supplemented with 6 μm (+) or 0 μm (−) Cu. −Cu crr1-1 and crr1-2 cultures were transferred to Cu-free medium once; all other −Cu cultures were transferred at least twice. CYC6 encodes cyt c6; CRD1 encodes a component of the protoporphyrin IX monomethylester cyclase, formerly known as the COPPER RESPONSE DEFECT 1 gene; and CBLP encodes a Chlamydomonas G-protein β-subunit-like protein, used here as a loading control. (B) Immunoblots to compare accumulation of Cu-responsive proteins in wild-type vs. mutant cells. The migration positions of apo-, di-apo-, and holoforms of plastocyanin are indicated with arrows in the first panel, as is the position of cyt c6, which cross-reacts with the anti-plastocyanin antibody. In the fourth panel, arrows indicate the positions of the Cth1 and Crd1 gene products, which are both recognized by the anti-Crd1 antiserum. (C) Immunoblot showing the accumulation of cyt c6, apo-, di-apo-, and holoplastocyanin in wild-type cells compared to crr1-1 cells grown in TAP medium with the indicated concentrations (micromolar) of Cu.
CRD1:
The CRD1 gene is now known to encode one of two isoforms of a component of the aerobic Mg protoporphyrin IX monomethylester cyclase in chlorophyll biosynthesis: the second isoform was named CTH1 for copper target homolog (Moseley et al. 2002b; Pinta et al. 2002; Tottey et al. 2003). CTH1 is expressed in +Cu cells, while in −Cu cells CRD1 is expressed coordinately with CYC6 and CPX1 (Figure 2, A and B; Moseley et al. 2000, 2002b). The crd1 mutants are accordingly chlorotic in copper-deficient medium, but are rescued by provision of nutritionally relevant amounts of copper with complete rescue occurring at 400 nm copper (Figure 1A). We note that this is the same amount of copper that is required to turn off the CYC6 and CPX1 genes and to support holoplastocyanin biosynthesis to its maximum abundance, which is consistent with CRD1 being a target in the same pathway. The loss of chlorophyll proteins in crd1 in −Cu medium contributes to loss of photosynthetic capacity, evident from comparison of the fluorescence rise and decay kinetics of crd1 strains to those of the wild type (Figure 1D), and hence at high light intensity the crd1 strains are growth compromised in −Cu medium.
CRD2:
The crd2 strain initially showed a more severe growth phenotype in copper-deficient medium compared to crr1 (Figure 1, A and C), but was clearly distinct from crr1 because all cultures eventually reached a high cell density (Figure 1B) and photosynthetic function was independent of copper nutrition status. On this basis, we concluded that copper deficiency limits the growth rate of the crd2 strain, but for crr1 strains, copper deficiency limits the capacity for growth, and the crd2 mutation affects an aspect of metabolism other than photosynthesis.
The CRR1 locus encodes a copper homeostasis regulator required not only for the expression of CYC6, but also for degradation of apoplastocyanin and the biogenesis of plastocyanin and a plasma membrane ferroxidase:
Transcriptional activation:
To distinguish CRR1 as a regulatory locus vs. CRD1 and CRD2 as target genes on the basis of biochemical criteria, we analyzed the expression of the CYC6 gene, the prototypical copper-deficiency target in Chlamydomonas, and also other copper-deficiency targets like CPX1 and CRD1 (Quinn et al. 2002) by RNA blot hybridization (Figure 2A) and immunoblotting (Figure 2B). We noted that both crr1 strains failed to turn on CYC6 transcription under −Cu conditions, and they also fail to activate CPX1 and CRD1 beyond the basal level maintained under copper-replete conditions (Figure 2A) resulting in no cyt c6 in extracts from −Cu cells and only basal abundance of coprogen oxidase (Figure 2B; Hill and Merchant 1995; Moseley et al. 2002b), suggesting that CRR1 is a regulatory locus. Nevertheless both crr1-1 and crr1-2 do respond to other nutrient deficiencies (Quinn et al. 2002; data not shown), indicating that they are capable of gene expression even though they are severely growth compromised. On the other hand, CYC6 is expressed normally in crd2-1 and in all crd1 strains tested, and the CRD1 gene is expressed normally in crd2-1 (Figure 2A; data not shown). These data are compatible with a previously proposed regulatory role for CRR1 in determining the copper-deficiency-induced transcriptional activation of CYC6, CPX1, and CRD1.
Apoplastocyanin degradation:
Immunoblot analysis revealed that the crr1 strains also fail to downregulate plastocyanin abundance in copper deficiency, and most of this protein is found in the apoform (Figure 2B). Li et al. had suggested that the degradation of the plastocyanin polypeptide in −Cu cells required a −Cu-induced protease (Li and Merchant 1995; Li et al. 1996): the accumulation of plastocyanin in crr1 indicates that this protease is yet another target of the CRR1 locus. Plastocyanin regulation occurred normally in crd1 and crd2 mutants.
Holoplastocyanin formation:
Interestingly, in the fully copper-supplemented situation as well, the crr1 strains accumulate more apoplastocyanin relative to wild type. If copper is titrated into copper-deficient medium, we note that the plastocyanin biosynthetic pathway is saturated at ∼400 nm Cu for wild-type cells but requires much more for the crr1-1 strain (Figure 2C), suggesting that copper delivery to the site of holoplastocyanin biosynthesis—either uptake from the medium or distribution to the chloroplast—is dependent on CRR1.
Plasma membrane ferroxidase function is compromised:
We wondered whether the crr1 mutation affected the accumulation of other copper-containing proteins in Chlamydomonas, such as a plasma membrane ferroxidase, encoded by FOX1 (Herbik et al. 2002; La Fontaine et al. 2002). Immunoblot analysis showed that ferroxidase accumulation was also compromised in the crr1 strain (Figure 3C). Loss of ferroxidase occurs probably at the level of protein synthesis or protein stability because the abundance of FOX1 mRNA is not reduced in −Cu vs. +Cu crr1 (Figure 3B). Since the ferroxidase is proposed to function in a high-affinity iron-uptake pathway, we wondered whether compromised ferroxidase biosynthesis would result in poor iron nutrition.
Figure 3.—
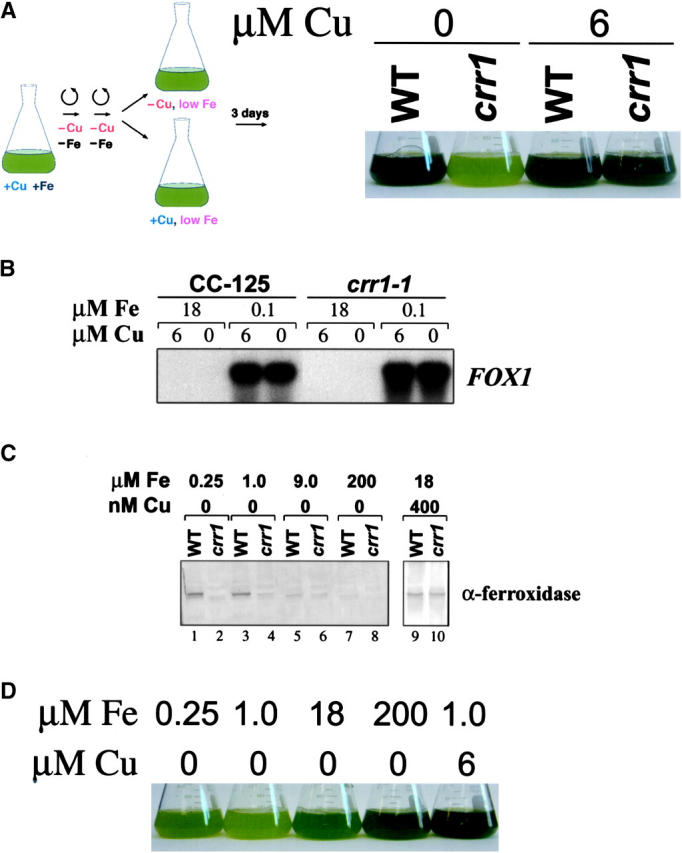
Acclimation responses to Fe deficiency in wild-type vs. crr1 strains. (A) Appearance of wild-type and crr1-1 cultures grown in +Fe (18 μm) and +Cu (6 μm), washed with −Fe −Cu TAP and grown for 3 days in 1 μm Fe TAP with or without 6 μm added Cu, as indicated. crr1-1 cells grown without copper contained ∼50% of wild-type chlorophyll. (B) RNA blot analysis comparing FOX1 mRNA expression in CC-125 (wild type) and crr1-1 mutant strains. CC-125 or crr1-1 cells were grown in TAP medium containing the indicated concentrations of Fe and Cu. Five micrograms of total RNA was loaded per lane. (C) Immunoblots demonstrating accumulation of ferroxidase in wild-type and crr1-1 cells, grown in TAP medium with the indicated concentrations of Fe and Cu. (D) Appearance of crr1-1 cultures grown for 6 days in TAP medium with the indicated concentrations of Fe and Cu, demonstrating partial rescue of the −Cu growth phenotype with excess Fe.
Cells of crr1 were transferred from Cu-enriched, iron-replete (18 μm) medium to copper-enriched vs. copper-depleted, iron-deficient (1 μm) medium. The wild-type culture maintains growth and chlorophyll accumulation, but the crr1 strain displays classic iron-deficiency symptoms (Moseley et al. 2002a). Growth of crr1 is reduced, and the strain is chlorotic (Figure 3A; 1.29 pg chlorophyll/cell in −Cu crr1 relative to 2.31 pg/cell in +Cu crr1 in one representative experiment). Conversely, if crr1 is transferred to copper-deficient medium containing excess iron (200 μm) where the cellular requirement for the ferroxidase is greatly reduced (Figure 3C, compare lanes 1 and 2 vs. 7 and 8), its copper-conditional growth phenotype is suppressed (Figure 3D). We conclude that poor iron nutrition contributes to the growth phenotype of crr1 strains in −Cu medium, possibly because of malfunction of the ferroxidase biosynthetic pathway.
We conclude that CRR1 is a key regulator of nutritional copper homeostasis: it is required in −Cu cells for (i) upregulation of the accumulation of cytochrome c6, coprogen oxidase, and the Crd1 cyclase isoform as indicated previously and (ii) downregulation of plastocyanin via activation of a protease and the Cth1 isoform of the cyclase by transcriptional occlusion of the proximal CTH1 promoter (Cullen et al. 1984; Moseley et al. 2002b). CRR1 is also required in +Cu cells for maintenance of holoplastocyanin and in −Cu cells for maintenance of holoferroxidase.
CRR1 is required for growth under hypoxic conditions:
We noted previously (Moseley et al. 2000, 2002b; Quinn et al. 2000, 2002) that each of the Cu-responsive genes is regulated also by hypoxia—CYC6, CPX1, and CRD1 show increased expression as oxygen is depleted and, in the case of CTH1, the extended 3-kb nonfunctional (with respect to synthesis of the cyclase isoform) transcript is expressed at the expense of the functional 2-kb version. As little as a 50% reduction in the O2 content of the medium is sufficient to upregulate CPX1 and CRD1 expression, and this is mediated via the Crr1 pathway (Quinn et al. 2002). To assess the physiological significance of the hypoxic activation of Crr1 target genes, the impact of the crr1 mutation was tested by assessing growth under oxygen limitation (Figure 4A). Indeed the crr1 mutants are growth compromised in low O2 relative to air. We know that the growth defect is not a secondary effect of hypoxia on copper uptake because hypoxic cells show normal accumulation of holoplastocyanin (Figure 4B), which requires copper delivery to the thylakoid lumen.
Figure 4.—
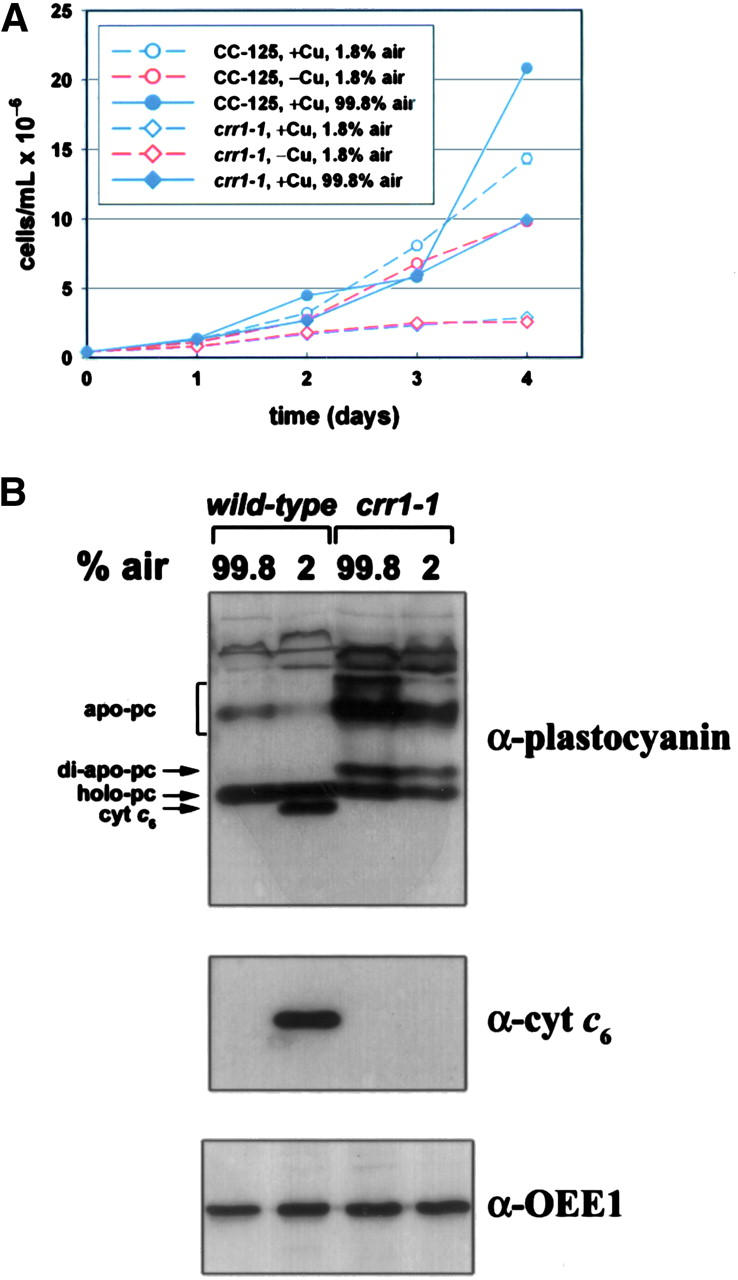
Hypoxic growth of wild type vs. the crr1 strain. (A) Wild-type (CC-125) and crr1-1 strains were grown in TAP medium with 6 μm Cu (blue lines) or without Cu (red lines), bubbled with gas mixtures containing the indicated amounts of air (99.8% vs. 1.8%). All mixtures contained 0.2% CO2, and the balance of the 1.8% air mixture was made up with N2 (98% N2). (B) Immunoblot analysis of plastocyanin and cyt c6 accumulation in soluble protein fractions from wild-type and crr1-1 strains grown for 4 days in 6 μm Cu with the indicated amounts of air. Detection of OEE1 was used to demonstrate equal loading.
Iron-deficiency symptoms and impaired ferroxidase accumulation in crd2 strains:
In Saccharomyces cerevisiae and also in mammalian systems, copper deficiency results in secondary iron deficiency owing to the involvement of a multicopper oxidase in iron assimilation (Askwith and Kaplan 1998). Copper-deficient wild-type Chlamydomonas cells show no symptoms of iron deficiency even though they accumulate much less ferroxidase relative to copper-replete cells (La Fontaine et al. 2002). Therefore, we proposed that copper-deficient Chlamydomonas must use an alternate or modified mechanism for iron uptake relative to copper-replete cells so that its growth is not compromised by reduced ferroxidase abundance, and this idea is supported by the increased iron requirement of the crr1 mutant strains (Figure 3D). Therefore, iron metabolism was a pathway of interest in the context of deciphering the basis of copper-conditional growth phenotypes.
crd2 strains are iron deficient:
We noted during biochemical characterization of our mutant strains that the pattern of migration of iron-containing enzymes Crd1 and Cth1 in the crd2-1 mutant was not normal (Figure 2B, compare crd2 to wild type), but resembled the migration pattern seen in −Fe wild-type cells (J. L. Moseley, unpublished observations). This suggested to us that CRD2 might indeed be involved in an iron-uptake pathway in copper-deficient cells.
To test this idea, we assessed the effect of iron nutrition on the copper-conditional growth phenotype of crd2-1. As noted previously, copper nutrition status does not change the threshold for establishment of iron-deficiency symptoms in the wild-type strain (Figure 5A). In copper-replete medium, the iron-dependent growth of crd2-1 is indistinguishable from wild type, but in copper-deficient medium, crd2-1 is growth compromised at 18 μm medium iron, which is considered iron replete for the wild type (Moseley et al. 2002a). The slow growth phenotype of crd2-1 in −Cu medium can be further exacerbated by decreasing iron supplementation to 1 μm or can be suppressed completely by provision of excess iron to 200 μm (Figure 5A). To test whether crd2-1 cells were internally iron deficient, we analyzed the expression of FOX1 and FTR1 that encode components of the high-affinity iron-uptake pathway (La Fontaine et al. 2002) and FEA1 that encodes a periplasmic iron-uptake component (Rubinelli et al. 2002; M. Allen and S. Merchant, unpublished observations). In the wild-type strain, the expression of these genes is independent of copper (Figure 5B) but is dependent on iron nutrition (La Fontaine et al. 2002; J. A. del Campo and S. Merchant, unpublished observations). We found that in crd2-1, FOX1, FTR1, and FEA1 are all upregulated relative to wild type, and the magnitude of the upregulation is dependent on the degree of copper deficiency (Figure 5B), suggesting that crd2-1 cells are experiencing iron deficiency. This is compatible with the slow growth phenotype of crd2-1, i.e., as if the strain were nutrient limited.
Figure 5.—
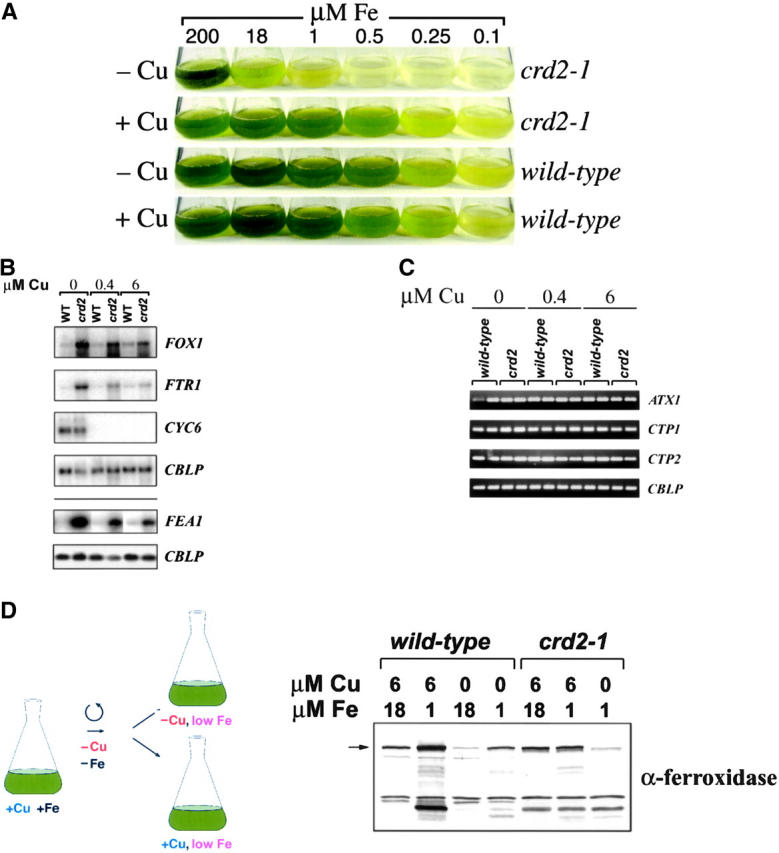
Acclimation responses to Fe deficiency in wild-type vs. crd2 strains. (A) Growth of crd2-1 vs. wild type in +Cu (6 μm) and −Cu (0 μm) TAP with the indicated amounts of added Fe. (B) RNA blot analysis of genes involved in Fe uptake and distribution in wild type and crd2-1 grown in TAP medium with normal Fe supplementation (18 μm) and with the indicated concentration of added Cu. The FEA1 panel and the bottom CBLP panel are separated from the other panels by a line to indicate that these RNA blots were performed with different RNA samples. Three micrograms of total RNA was loaded per lane. CBLP expression is used to demonstrate equal loading of the samples, and CYC6 expression is used as a marker for copper deficiency. (C) Quantitative RT-PCR analysis of the samples described in B. Ten microliters of product was removed from the amplification reaction and analyzed by electrophoresis to authenticate the product on the basis of size and to visualize the difference in the abundance of the specific mRNA in the input RNA sample. Samples were removed at the following cycles: CTP1, 27; CTP2, 27; ATX1, 30; and CBLP, 19. For the experiment shown, CTP1 is twofold induced in the low copper crd2 samples relative to the saturating copper sample. (D) Immunoblot analysis of ferroxidase accumulation in wild-type and crd2-1 cells grown in +Fe (18 μm) and +Cu (6 μm), washed with −Fe −Cu TAP, and grown for 24 hr in 1 μm Fe TAP with or without 6 μm added Cu, as indicated. The position of the ∼140-kD ferroxidase protein is indicated with an arrow. Higher mobility bands, which are stoichiometrically more prevalent in −Cu wild-type cells or in crd2 mutant cells, are likely to be degradation products. Degradation may be more pronounced under these conditions because of a different conformation of the protein when the copper sites are not fully loaded (Hellman et al. 2002).
Compromised ferroxidase biosynthesis:
Since crr1 showed impaired ferroxidase accumulation (Figure 3C), we wondered whether crd2-1 might also be affected in ferroxidase function. Copper- and iron-replete crd2-1 or wild-type cells were transferred to iron-deficient medium to induce ferroxidase accumulation and tested after 24 hr. Copper-supplemented wild-type and crd2-1 cells accumulated ferroxidase to comparable levels but copper-deficient crd2-1 cells did not (Figure 5D). Since we know that the abundance of FOX1 and FTR1 mRNA is high in crd2-1 cells, the lower amount of ferroxidase in −Cu crd2-1 must result from slower synthesis or decreased stability. We conclude that the delayed growth of −Cu crd2-1 cultures results from slower assimilation of iron because of reduced function of Fox1/Ftr1, and we suggest that CRD2 encodes a function required for high-affinity iron uptake especially in a situation where copper is limiting the operation of the multicopper oxidase pathway. Note that the effect of crd2 is specific for ferroxidase biogenesis whereas crr1 strains are affected in the accumulation of plastocyanin and also ferroxidase (Figures 2B and 3C).
One possibility is that CRD2 might encode a component that facilitates Fox1/Ftr1 biogenesis, and its function is more important when copper, which is required for Fox1 biogenesis in the secretory pathway, is scarce within this compartment (La Fontaine et al. 2002). This would be analogous to the increased expression of CPX1 in −Cu cells. Another possibility is that Crd2 is functionally redundant with another molecule whose expression is decreased in copper deficiency. This would be analogous to the Crd1/Cth1 situation.
The biogenesis of a plasma membrane ferroxidase, Fet3p, in S. cerevisiae is proposed to require a cytoplasmic copper chaperone, Atx1, to deliver copper from an assimilatory transporter to a distributive P-type ATPase in the secretory pathway (Yuan et al. 1995; Lin et al. 1997). The phenotype of crd2 was strikingly similar to that of yeast mutants affected in copper loading of Fet3p. For example, the yeast atx1 and ccc2 strains are iron deficient and can be rescued for growth by either copper or iron supplementation. The release of a draft Chlamydomonas genome (http://www.jgi-doe.gov) allowed us to identify two candidate distributive transporters, CTP1 [related to Arabidopsis AHM7 and RAN1 (Hirayama et al. 1999) and mammalian ATP7A/WND (Bull et al. 1993; Chelly et al. 1993; Mercer et al. 1993; Tanzi et al. 1993; Vulpe et al. 1993)] and CTP2 (related to Arabidopsis PAA1; Shikanai et al. 2003). We tested the expression of ATX1, CTP1, and CTP2 by quantitative RT-PCR but could not account for the phenotype on the basis of loss of expression of one of these genes in crd2 relative to the wild type (Figure 5C). The expression of CTP1 was increased slightly in proportion to the decrease in medium copper, which is expected because the gene is regulated by iron deficiency in Chlamydomonas (S. Tottey and S. Merchant, unpublished observations) like the homologous yeast gene (Yamaguchi-Iwai et al. 1996; Lin et al. 1997). The organization of the CTP1 and CTP2 genes was also intact in crd2 as assessed by amplification with gene-specific primers (data not shown).
sop13 may be a weak gain-of-function allele of CRR1:
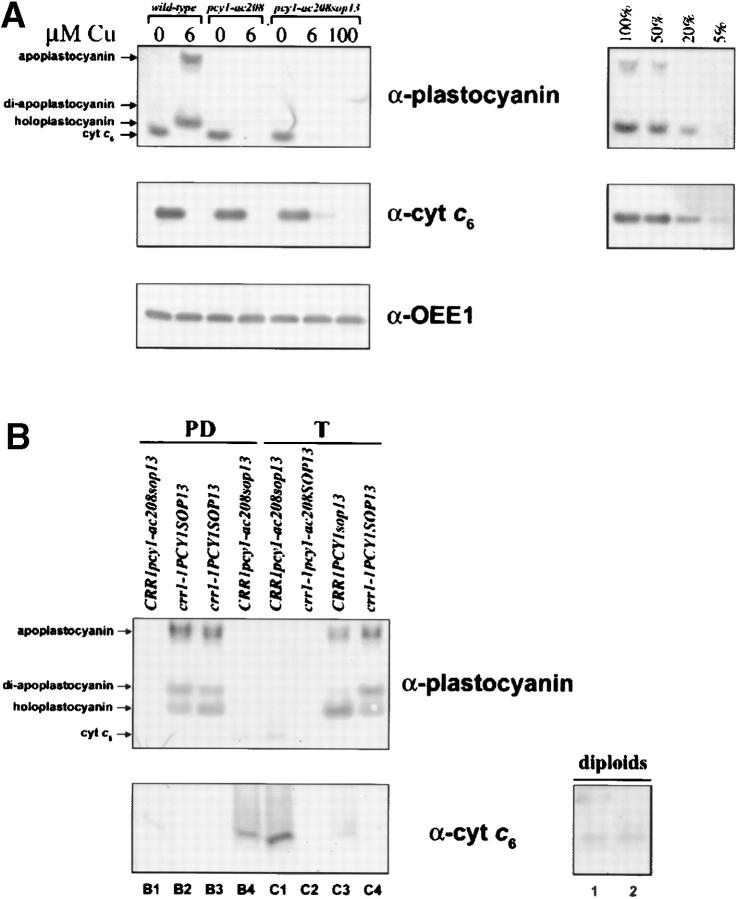
The function of cyt c6 as a substitute for plastocyanin was established definitively when it was shown that the nonphotosynthetic, acetate-requiring phenotype of pcy1-ac208 could be suppressed in copper-deficient medium where CYC6 is expressed (Wood 1978). On this basis, we had selected spontaneous second site suppressors of pcy1-ac208 in which CYC6 was misexpressed in copper-supplemented medium (Merchant and Bogorad 1987b; Li et al. 1996). We considered the possibility that one suppression mechanism might involve mutations at the CRR1 locus that allow copper-insensitive expression of CYC6. To test this model, we isolated new pcy1 suppressor strains that grew photosynthetically on +Cu minimal medium at rates comparable to wild type (see materials and methods). One of these, sop13 (suppressor of pcy1-ac208 13) was chosen for further analysis. Strains pcy1 and pcy1sop13 lack plastocyanin, but in standard TAP medium containing 6 μm copper salts, pcy1sop13 accumulates ∼5–10% of the amount of cyt c6 that is found normally only in the −Cu situation (Figure 6A). This amount can be reduced by provision of excess copper, as noted previously for other suppressed strains (Merchant and Bogorad 1987b).
Figure 6.—
Biochemical and genetic characterization of a pcy1-ac208 suppressor strain. (A) Immunoblot analysis of soluble extracts to compare expression of plastocyanin and cyt c6 in wild type, pcy1-ac208, and a pcy1-ac208 suppressor strain, (pcy1-ac208sop13), grown in TAP medium supplemented with the indicated concentrations of Cu (micromolars). Dilution series of wild-type extracts enables estimation of plastocyanin and cyt c6 abundance. Samples corresponding to equal amounts of chlorophyll were loaded in each lane, and immunodetection of OEE1 was used to demonstrate equal loading. (B) Anti-plastocyanin and cyt c6 immunoblots of +Cu whole cell extracts illustrating phenotypic analysis of two representative tetrads from a cross of crr1-1 and pcy1-ac208sop13. Cyt c6 expression in +Cu cells indicates the presence of the sop13 mutation. Accumulation of cyt c6 in two vegetative diploid strains that are heterozygous for sop13 is also shown.
To test whether the sop13 mutation is linked to the CRR1 locus, we crossed pcy1-ac208sop13 with crr1-1PCY1SOP13 and analyzed the phenotypes of 85 spores from 25 different zygotes, including 8 complete tetrads, and 21 tetrads in total for which the genotype of each spore could be determined. The pcy1-ac208 and sop13 phenotypes were scored by immunodetection of plastocyanin and cyt c6, respectively, in +Cu whole cell extracts (Figure 6B for two representative tetrads) and the crr1 phenotype was scored on the basis of growth on −Cu TAP and fluorescence induction and decay kinetics (not shown). Recombination between CRR1 and SOP13 was not observed (χ2 probability <1% for independent assortment), whereas recombination between CRR1 and PCY1 and PCY1 and SOP13 was. In vegetative diploids that are heterozygous for sop13, we noted that misexpression of cyt c6 is maintained (Figure 6B), indicating that sop13 is a dominant gain-of-function mutation. These data preserve the possibility that sop13 may be a weakly constitutive allele of crr1, although we cannot rule out alternative models such as the possibility that the mutation lies in another component in the pathway that is linked to CRR1. For instance, sop13 could be a cis-acting mutation in the CYC6 promoter, (if CYC6 is linked to CRR1) or it may be a mutation in a linked modifier of CRR1 or in a linked assimilatory copper transporter.
DISCUSSION
CRR1 encodes a regulator of Cu homeostasis under both nutritional deficiency and nutritional sufficiency:
Cytochrome c6 and plastocyanin:
Previously, we showed that Chlamydomonas responds to copper deficiency by changes in gene expression resulting in increased production of an alternate electron transfer catalyst, cyt c6, to take the place of an abundant copper protein, and coprogen oxidase, an oxygen-dependent enzyme in the tetrapyrrole pathway, by transcriptional activation through CuREs associated with the corresponding genes. At the same time, plastocyanin is degraded, presumably to allow redistribution of copper to other copper-containing enzymes. In this work, we define CRR1 as a central regulator of each of these well-characterized responses. The nonphotosynthetic phenotype is attributed to loss of cyt c6, which is essential for the Z-scheme when plastocyanin function is compromised by lack of its cofactor, copper (Figures 1D and 2, A and B).
Heterotrophic growth (Figure 1, A and C) is also affected in −Cu crr1, perhaps because of the requirement for a copper enzyme, cytochrome oxidase, in respiration. Previously (Li and Merchant 1995), we had suggested that plastocyanin degradation was regulated. Plastocyanin is the most abundant copper protein in a photosynthetic cell, and its degradation would remove a major copper sink and in so doing allow copper reallocation to other copper enzyme-containing compartments in the cell. The accumulation of apoplastocyanin in crr1 strains (Figure 2C) confirms that the decreased thermodynamic stability of apo- vs. holoplastocyanin and the increased protease susceptibility (Li and Merchant 1995) is not in itself sufficient for degradation in vivo and that activation of a proteolytic mechanism in −Cu cells is a necessary component of copper homeostasis.
Tetrapyrrole pathway:
The (10- to 20-fold) increased expression of CPX1 in −Cu cells was previously rationalized on the basis of an increased demand for heme owing to production of an abundant (>106 molecules/cell) cytochrome. [Coprogen oxidase is rate limiting in the synthesis of heme from δ-aminolevulinic acid (Andrew et al. 1990)]. In a photosynthetic cell, however, the pathway intermediates downstream of coprogen oxidase are required also for the biosynthesis of chlorophyll, which is a much more abundant tetrapyrrole. The discovery of CRD1 and CTH1 as new Crr1 targets in the chlorophyll biosynthetic pathway suggests a link between copper and tetrapyrrole metabolism whose molecular basis remains to be discovered. The fact that Cth1 cannot fully cover loss of Crd1 function (Moseley et al. 2002b) suggests also additional complexity in the chlorophyll biosynthetic pathway.
Iron metabolism:
We found also that Cu-deficient crr1 strains display pleiotropic symptoms of Fe deficiency (Figure 3A), which we attribute to their reduced capacity for production of a multicopper ferroxidase involved in high-affinity Fe uptake (Figure 3C). Fe deficiency appears to be a major cause of the poor growth of Cu-deficient crr1 strains, since addition of an excess of Fe to the medium improves the growth rate and increases the final cell density of −Cu crr1 cultures (Figure 3D). In S. cerevisiae, nutritional or genetic Cu deficiency results in Fe deficiency owing to the involvement of Fet3p, a multicopper oxidase, in high-affinity Fe uptake (Askwith et al. 1994). However, we find it intriguing that wild-type −Cu Chlamydomonas cells are not Fe deficient to a greater degree than are +Cu cells (as established by monitoring gene expression and chlorosis; La Fontaine et al. 2002). One model is that cellular metabolism might be altered in −Cu cells in a Crr1-dependent pathway to reduce the internal demand for Fe; hence, crr1 strains require more iron compared to the wild type. Another model is that Crr1 may play a role in maintaining an adequate Fe-uptake capacity in Cu-deficient cells. The discovery of CRD2 as a target of nutritional copper signaling and the phenotype of crd2 strains is compatible with this idea. A third possibility is that the reduced capacity for holoferroxidase production in −Cu crr1 strains is simply another consequence of the failure to degrade apoplastocyanin, the presence of which could prevent reallocation of copper from the chloroplast compartment to the secretory pathway where ferroxidase is synthesized. Regardless of the molecular basis, Crr1 clearly plays a role in iron nutrition under copper-deficient growth.
Hypoxia:
Since the same genes were found to be regulated by copper and also oxygen deficiency, one might conclude that low oxygen, perhaps by modification of intracellular steady-state abundance of redox compounds, resulted in coincidental conversion of a thiol containing copper-pathway signaling component from the nonactivating to the activating form. The finding that loss of Crr1 function affects the growth of copper-replete cells under hypoxic relative to well-aerated cells suggests that hypoxic activation of Crr1 targets is physiologically meaningful. We propose that Crr1 function and Crr1 target genes are required for proper adaptation to hypoxic conditions. It is worth noting in this context that HEM13 in S. cerevisiae (encoding coprogen oxidase) is also induced by hypoxia (Zagorec et al. 1988).
Summary:
We have identified a central component of the signal transduction pathway for Cu and O2 sensing in Chlamydomonas. This factor is implicated in the metabolism of at least three “nutrients” (Cu, Fe, and O2) and reveals a hitherto unanticipated level of integration between nutrient stress acclimation responses.
SOP13 is linked to CRR1 and may be a weakly constitutive crr1 allele:
Genetic analysis demonstrated that the sop13 suppressor of pcy1-ac208 mutation is linked to CRR1. This dominant mutation confers a gain of function, enabling partial induction of CYC6 in +Cu cells, and allowing the accumulation of sufficient cyt c6 to rescue the photosynthesis-minus phenotype of the pcy1-ac208 strain (Figure 6). However, no induction of coprogen oxidase or Crd1 expression or decrease in Cth1 accumulation is observed (data not shown). Unlike cyt c6, which is completely absent in wild-type strains, each of these other proteins is expressed at a significant level in +Cu (Hill and Merchant 1995; Quinn et al. 1999; Moseley et al. 2000, 2002b), and consequently a 5–10% increase or decrease in its abundance would not be discerned in these experiments. Therefore, we retain the model that sop13 is a weak crr1-up allele, which enables leaky expression of the Cu-deficiency responsive gene expression pathway. However, we cannot rule out the model that the sop13 mutation may affect a gene that is tightly linked to CRR1. The possibilities are several: a cis-acting mutation in the CYC6 promoter, a weak loss of function mutation affecting a copper transporter resulting in internal copper deficiency despite plentiful supply in the medium, a weak gain of function of a Crr1-related factor, or a weak gain of function in an upstream or downstream component in the signaling pathway.
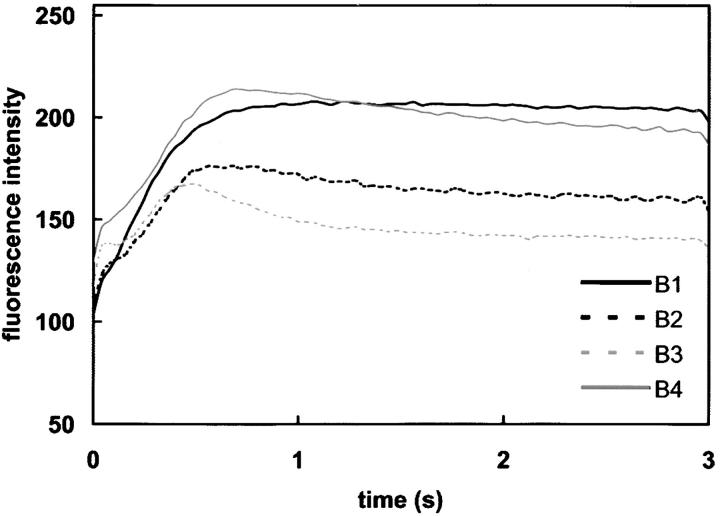
Interestingly, we noted that some pcy1-ac208sop13 spores (16 spores out of 16 different zygotes) did not exhibit wild-type fluorescence on +Cu medium (Figure 7 for a representative tetrad) even though they did express cyt c6, suggesting that the selected phenotype (i.e., photosynthetic growth of pcy1 on +Cu medium) results from a combination of more than one mutation. If misexpression of CYC6 were sufficient to allow photosynthetic growth, all pcy1sop13 strains should be photosynthesis plus. The initial premise that misexpression of CYC6 is sufficient to restore photosynthetic growth to a plastocyanin-minus strain is therefore called into question. We wonder whether a second mutation that changes the composition of PSI or its acceptor side conformation (e.g., at the plastocyanin binding site) may be required for good interaction with cyt c6, which becomes more important when the abundance of cyt c6 is substoichiometric (as is the case in sop13 strains).
Figure 7.—
Photosynthesis phenotypes of crr1 and sop13 strains. Shown are fluorescence rise and decay kinetics of +Cu cells from strains B1–B4 (shown in Figure 6B). B1, CRR1pcy1-ac208sop13; B2, crr1-1PCY1SOP13; B3, crr1-1PCY1SOP13; and B4, CRR1pcy1-ac208sop13.
CRD2 is required for stable accumulation of ferroxidase in −Cu:
The crd2 mutant was isolated as a strain that grew slowly on −Cu medium, but unlike the crr1 strains it did not have a photosynthesis defect (Figure 1). Instead, we have established that the growth phenotype of the crd2 mutant is due to Cu-conditional Fe deficiency (Figure 5). Genes involved in Fe assimilation, such as FOX1, FTR1, and FEA1, are highly induced in −Cu crd2 cells, even at Fe concentrations where the wild type or +Cu crd2 is Fe replete. Like −Cu crr1 strains, the crd2 mutant is unable to synthesize or accumulate normal amounts of ferroxidase under low Fe conditions, suggesting that the defect affects delivery of Cu to apoferroxidase. La Fontaine et al. (2002) showed that at any given Fe concentration, Cu-deficient wild-type cells accumulate only ∼50% of the amount of ferroxidase that is observed in Cu-replete cells; nevertheless, growth rates and the induction of Fe-assimilation-related genes are comparable between +Cu and −Cu. How then are −Cu cells able to keep up with +Cu cells?
One possibility is that ferroxidase is synthesized in excess of the requirement for maximum Fe uptake, and consequently the reduced amount of ferroxidase in −Cu cells is still sufficient to efficiently catalyze Fe assimilation. However, this idea is contrary to the well-accepted dogma that Fe assimilation is very tightly regulated (Eide 1998). An alternative explanation is that there is a Cu-independent “backup” pathway for Fe uptake that is functional in Cu-deficient cells and supplements Fe uptake through the Cu-dependent (ferroxidase-requiring) pathway. Such backup pathways for coping with nutrient deficiencies are prevalent in nature—the replacement of plastocyanin with cyt c6 (Wood 1978), ferredoxin with flavodoxin (Hutber et al. 1977), and Cu- vs. Fe-containing forms of methane monooxygenase (Nielsen et al. 1997) are relevant examples. A third possibility, which is best supported by the evidence presented here, is that the ferroxidase/Ftr1 pathway operates in both +Cu and −Cu, but intracellular Cu distribution must be altered in Cu-deficient cells to ensure that sufficient holoferroxidase is produced to supply the Fe requirements of the cell. CRD2 therefore is likely to encode a Cu-conditional modulator of the Fe-assimilation pathway. A number of candidate genes encoding components of copper homeostasis, including an assimilatory transporter CPT1 and chaperones and distributive transporters CTP1 and CTP2, have been identified in the EST database and the draft genome sequence of Chlamydomonas (La Fontaine et al. 2002; Grossman et al. 2003). While we cannot rule out the possibility that crd2 represents a point mutation or small deletion in one of these genes, we do know the crd2 phenotype does not result from inappropriate expression of the known chaperones and transporters (Figure 5C).
If CRD2 is a modifier of or a bypass mechanism for iron uptake whose function becomes essential in a copper-deficient situation when ferroxidase activity is compromised, it is likely that its expression might be regulated by CRR1. In this case, crr1 mutants would be iron deficient because they fail to activate CRD2 just as crr1 strains are photosynthetically defective because they fail to express CYC6 (Figure 1D). Although we attributed the iron-deficiency phenotype of crr1 to poor copper uptake/delivery and hence loss of ferroxidase function, it is possible that loss of CRD2 expression is another contributing factor to the iron-deficiency and ferroxidase-defective phenotype of crr1 strains.
Acknowledgments
We thank Patrice Hamel and Janette Kropat for helpful comments on the manuscript and for assistance with strains. This work was supported by the National Institutes of Health (grant GM42143). M.E. was supported, in part, by a European Molecular Biology Organization Long-Term Fellowship, J.L.M. was supported in part by the Molecular Biology Ph.D. program and a Dissertation Year Fellowship from the Graduate Division of the University of California (Los Angeles), J.d.C. in part by a postdoctoral fellowship from the Spanish Ministry for Education, and S.T. and Y.K. in part by a University of California Toxic Substances Research and Teaching Program's Lead Campus and Team grant, respectively.
References
- Andrew, T. L., G. R. Pascale and H. A. Dailey, 1990 Regulation of heme biosynthesis in higher animals, pp. 163–200 in Biosynthesis of Heme and Chlorophylls, edited by H. A. Dailey. McGraw-Hill, New York.
- Askwith, C., and J. Kaplan, 1998. Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci. 23: 135–138. [DOI] [PubMed] [Google Scholar]
- Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li et al., 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76: 403–410. [DOI] [PubMed] [Google Scholar]
- Bull, P. C., G. R. Thomas, J. M. Rommens, J. R. Forbes and D. W. Cox, 1993. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat. Genet. 5: 327–337. [DOI] [PubMed] [Google Scholar]
- Chelly, J., Z. Tumer, T. Tonnesen, A. Petterson, Y. Ishikawa-Brush et al., 1993. Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat. Genet. 3: 14–19. [DOI] [PubMed] [Google Scholar]
- Cullen, B. R., P. T. Lomedico and G. Ju, 1984. Transcriptional interference in avian retroviruses: implications for the promoter insertion model of leukaemogenesis. Nature 307: 241–245. [DOI] [PubMed] [Google Scholar]
- Culotta, V. C., L. W. Klomp, J. Strain, R. L. Casareno, B. Krems et al., 1997. The copper chaperone for superoxide dismutase. J. Biol. Chem. 272: 23469–23472. [DOI] [PubMed] [Google Scholar]
- Dancis, A., D. S. Yuan, D. Haile, C. Askwith, D. Elde et al., 1994. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76: 393–402. [DOI] [PubMed] [Google Scholar]
- Eide, D. J., 1998. The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu. Rev. Nutr. 18: 441–469. [DOI] [PubMed] [Google Scholar]
- Glerum, D. M., A. Shtanko and A. Tzagoloff, 1996. a Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271: 14504–14509. [DOI] [PubMed] [Google Scholar]
- Glerum, D. M., A. Shtanko and A. Tzagoloff, 1996. b SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271: 20531–20535. [DOI] [PubMed] [Google Scholar]
- Grossman, A. R., E. E. Harris, C. Hauser, P. A. Lefebvre, D. Martinez et al., 2003. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryotic Cell 2: 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E. H., 1989 The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego. [DOI] [PubMed]
- Hellman, N. E., S. Kono, G. M. Mancini, A. J. Hoogeboom, G. J. de Jong et al., 2002. Mechanisms of copper incorporation into human ceruloplasmin. J. Biol. Chem. 277: 46632–46638. [DOI] [PubMed] [Google Scholar]
- Herbik, A., C. Bolling and T. J. Buckhout, 2002. The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol. 130: 2039–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K. L., and S. Merchant, 1995. Coordinate expression of coproporphyrinogen oxidase and cytochrome c6 in the green alga Chlamydomonas reinhardtii in response to changes in copper availability. EMBO J. 14: 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K. L., H. H. Li, J. Singer and S. Merchant, 1991. Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J. Biol. Chem. 266: 15060–15067. [PubMed] [Google Scholar]
- Hill, K. L., R. Hassett, D. Kosman and S. Merchant, 1996. Regulated copper uptake in Chlamydomonas reinhardtii in response to copper availability. Plant Physiol. 112: 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau, E., and R. M. Amasino, 2000. Delivering copper within plant cells. Curr. Opin. Plant Biol. 3: 205–210. [PubMed] [Google Scholar]
- Hirayama, T., J. J. Kieber, N. Hirayama, M. Kogan, P. Guzman et al., 1999. RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97: 383–393. [DOI] [PubMed] [Google Scholar]
- Hutber, G. N., K. G. Hutson and L. J. Rogers, 1977. Effect of iron deficiency on levels of two ferredoxins and flavodoxin in a cyanobacterium. FEMS Microbiol. Lett. 1: 193–196. [Google Scholar]
- Kindle, K. L., 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87: 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klomp, L. W., S. J. Lin, D. S. Yuan, R. D. Klausner, V. C. Culotta et al., 1997. Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J. Biol. Chem. 272: 9221–9226. [DOI] [PubMed] [Google Scholar]
- Labbé, S., and D. J. Thiele, 1999. Pipes and wiring: the regulation of copper uptake and distribution in yeast. Trends Microbiol 7: 500–505. [DOI] [PubMed] [Google Scholar]
- La Fontaine, S., J. M. Quinn, S. S. Nakamoto, M. D. Page, V. Göhre et al., 2002. Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryotic Cell 1: 736–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. H., and S. Merchant, 1995. Degradation of plastocyanin in copper-deficient Chlamydomonas reinhardtii. J. Biol. Chem. 270: 23504–23510. [DOI] [PubMed] [Google Scholar]
- Li, H. H., J. Quinn, D. Culler, J. Girard-Bascou and S. Merchant, 1996. Molecular genetic analysis of plastocyanin biosynthesis in Chlamydomonas reinhardtii. J. Biol. Chem. 271: 31283–31289. [DOI] [PubMed] [Google Scholar]
- Lin, S.-J., R. A. Pufahl, A. Dancis, T. V. O'Halloran and V. C. Culotta, 1997. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J. Biol. Chem. 272: 9215–9220. [PubMed] [Google Scholar]
- Livak, K. J., and T. D. Schmittgen, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Mercer, J. F., J. Livingston, B. Hall, J. A. Paynter, C. Begy et al., 1993. Isolation of a partial candidate gene for Menkes disease by positional cloning. Nat. Genet. 3: 20–25. [DOI] [PubMed] [Google Scholar]
- Merchant, S., and L. Bogorad, 1986. Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardtii. Mol. Cell. Biol. 6: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S., and L. Bogorad, 1987. a The Cu(II)-repressible plastidic cytochrome c. Cloning and sequence of a complementary DNA for the pre-apoprotein. J. Biol. Chem. 262: 9062–9067. [PubMed] [Google Scholar]
- Merchant, S., and L. Bogorad, 1987. b Metal ion regulated gene expression: use of a plastocyanin-less mutant of Chlamydomonas reinhardtii to study the Cu(II)-dependent expression of cytochrome c-552. EMBO J. 6: 2531–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S., K. Hill and G. Howe, 1991. Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 10: 1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, J., J. Quinn, M. Eriksson and S. Merchant, 2000. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19: 2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, J. L., T. Allinger, S. Herzog, P. Hoerth, E. Wehinger et al., 2002. a Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21: 6709–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, J. L., M. D. Page, N. P. Alder, M. Eriksson, J. Quinn et al., 2002. b Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, A. K., K. Gerdes and J. C. Murrell, 1997. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol. Microbiol. 25: 399–409. [DOI] [PubMed] [Google Scholar]
- Peña, M. M., S. Puig and D. J. Thiele, 2000. Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275: 33244–33251. [DOI] [PubMed] [Google Scholar]
- Pinta, V., M. Picaud, F. Reiss-Husson and C. Astier, 2002. Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J. Bacteriol. 184: 746–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, J. M., and S. Merchant, 1995. Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7: 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, J. M., and S. Merchant, 1998. Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297: 263–279. [DOI] [PubMed] [Google Scholar]
- Quinn, J. M., S. S. Nakamoto and S. Merchant, 1999. Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J. Biol. Chem. 274: 14444–14454. [DOI] [PubMed] [Google Scholar]
- Quinn, J. M., P. Barraco, M. Eriksson and S. Merchant, 2000. Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275: 6080–6089. [DOI] [PubMed] [Google Scholar]
- Quinn, J. M., M. Eriksson, J. Moseley and S. Merchant, 2002. Oxygen deficiency-responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol. 128: 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinelli, P., S. Siripornadulsil, F. Gao-Rubinelli and R. T. Sayre, 2002. Cadmium- and iron-stress-inducible gene expression in the green alga Chlamydomonas reinhardtii: evidence for H43 protein function in iron assimilation. Planta 215: 1–13. [DOI] [PubMed] [Google Scholar]
- Sancenón, V., S. Puig, H. Mira, D. J. Thiele and L. Peñarrubia, 2003. Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 51: 577–587. [DOI] [PubMed] [Google Scholar]
- Sandmann, G., H. Reck, E. Kessler and P. Boger, 1983. Distribution of plastocyanin and soluble plastidic cytochrome c in various classes of algae. Arch. Microbiol. 134: 23–27. [Google Scholar]
- Shikanai, T., P. Müller-Moulé, Y. Munekage, K. K. Niyogi and M. Pilon, 2003. PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15: 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, D. R., J. D. Rochaix and S. Purton, 1996. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251: 23–30. [DOI] [PubMed] [Google Scholar]
- Tanzi, R. E., K. Petrukhin, I. Chernov, J. L. Pellequer, W. Wasco et al., 1993. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat. Genet. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Tottey, S., P. R. Rich, S. A. Rondet and N. J. Robinson, 2001. Two Menkes-type atpases supply copper for photosynthesis in Synechocystis PCC 6803. J. Biol. Chem. 276: 19999–20004. [DOI] [PubMed] [Google Scholar]
- Tottey, S., M. A. Block, M. Allen, T. Westergren, C. Albrieux et al., 2003. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 100: 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine, J. S., and E. B. Gralla, 1997. Delivering copper inside yeast and human cells. Science 278: 817–818. [DOI] [PubMed] [Google Scholar]
- Vulpe, C., B. Levinson, S. Whitney, S. Packman and J. Gitschier, 1993. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper-transporting ATPase. Nat. Genet. 3: 7–13. [DOI] [PubMed] [Google Scholar]
- Vulpe, C. D., and S. Packman, 1995. Cellular copper transport. Annu. Rev. Nutr. 15: 293–322. [DOI] [PubMed] [Google Scholar]
- Wood, P. M., 1978. Interchangeable copper and iron proteins in algal photosynthesis. Studies on plastocyanin and cytochrome c-552 in Chlamydomonas. Eur. J. Biochem. 87: 9–19. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai, Y., R. Stearman, A. Dancis and R. D. Klausner, 1996. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15: 3377–3384. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai, Y., M. Serpe, D. Haile, W. Yang, D. J. Kosman et al., 1997. Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J. Biol. Chem. 272: 17711–17718. [DOI] [PubMed] [Google Scholar]
- Yuan, D. S., R. Stearman, A. Dancis, T. Dunn, T. Beeler et al., 1995. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc. Natl. Acad. Sci. USA 92: 2632–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorec, M., J. Buhler, I. Treich, T. Keng, L. Guarente et al., 1988. Isolation, sequence, and regulation by oxygen of the yeast HEM13 gene coding for coproporphyrinogen oxidase. J. Biol. Chem. 263: 9718–9724. [PubMed] [Google Scholar]
- Zhou, B., and J. Gitschier, 1997. hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. USA 94: 7481–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]