Abstract
This report describes the rationale, approaches, organization, and resource development leading to a large-scale deletion bin map of the hexaploid (2n = 6x = 42) wheat genome (Triticum aestivum L.). Accompanying reports in this issue detail results from chromosome bin-mapping of expressed sequence tags (ESTs) representing genes onto the seven homoeologous chromosome groups and a global analysis of the entire mapped wheat EST data set. Among the resources developed were the first extensive public wheat EST collection (113,220 ESTs). Described are protocols for sequencing, sequence processing, EST nomenclature, and the assembly of ESTs into contigs. These contigs plus singletons (unassembled ESTs) were used for selection of distinct sequence motif unigenes. Selected ESTs were rearrayed, validated by 5′ and 3′ sequencing, and amplified for probing a series of wheat aneuploid and deletion stocks. Images and data for all Southern hybridizations were deposited in databases and were used by the coordinators for each of the seven homoeologous chromosome groups to validate the mapping results. Results from this project have established the foundation for future developments in wheat genomics.
HEXAPLOID wheat (2n = 6x = 42, Triticum aestivum L.) is one of the world's cornerstone crops, feeds more people than any other crop (∼600 million tons is produced annually), and is the most widely adapted of the major crops, thus offering potential for increased food production. Hexaploid wheat is composed of three genomes (A, B, and D), each of which contains seven pairs of chromosomes, which have been identified and characterized by Sears (1966), who established that there is a strong homoeologous relationship among chromosomes belonging to the three genomes. The wheat genome, while complex, offers a unique opportunity for enhancing our understanding of variation in gene density and evolution between and within plant chromosomes.
Technical complexities in studying the wheat genome include that it is an allohexaploid composed of ∼16,000 Mb of DNA (Arumuganathan and Earle 1991), ∼40 times the size of the rice (Oryza sativa L.) genome. However, even with the large size of this hexaploid genome, the genes within the three component genomes remain largely colinear (Van Deynze et al. 1995). Extensive aneuploid stocks have been developed, including nullisomic-tetrasomic and ditelosomic lines (Sears 1954, 1966; Sears and Sears 1978). The ability of the homoeologous chromosomes of polyploid wheat to buffer losses of chromosome fragments has been shown and developed (Endo 1988, 1990) and a collection of overlapping deletion lines involving all chromosome arms has been accumulated and characterized (Endo and Gill 1996; Qi et al. 2003). The breakpoints of the sequential deletions available for a chromosome arm define physical segments (bins) for that arm. This deletion series offers a unique opportunity to perform bin mapping of all single-dose restriction fragments by their presence or absence in DNA from members of the deletion population.
Expressed sequence tags (ESTs) are short cDNA sequences that serve to “tag” the gene from which the messenger RNA (mRNA) originated and that can serve multiple important uses. Typically, anonymous ESTs are single-pass sequenced to yield a 200–700 bp sequence that can be used to search DNA and protein databases for similar genes (Adams et al. 1991). Information from the search can be used to determine if a specific gene (or sequence motif) has been found in the same or other organisms and if its function has been determined. Until recently the lack of ESTs from species of the Triticeae tribe [wheat, barley (Hordeum vulgare L.), rye (Secale cereale L.)] had been a serious limitation to gene-sequence-based research for wheat. In May 2000, GenBank contained only 9 ESTs for wheat, 86 for barley, and none for rye. A large EST data set was a high priority for large-scale efforts to characterize the wheat genome more fully. An international effort was organized to develop “deep” wheat and barley EST collections. To this end, the International Triticeae EST Cooperative (ITEC) established the goal of generating 300,000 publicly available ESTs each for wheat and barley (see the ITEC website at http://wheat.pw.usda.gov/genome/).
The present and accompanying reports present results of a National Science Foundation-funded project to generate the main U.S. public contribution to the ITEC wheat EST effort, to assemble these ESTs into unique sets as contigs, and to map the EST restriction fragments by Southern hybridization with a subset of nullisomic-tetrasomic and ditelosomic lines and 101 of the deletion lines defining unique deletion bins for each of the 21 wheat chromosomes of the hexaploid wheat genome (http://wheat.pw.usda.gov/NSF/). The characterization of the deletion stocks used in this project was reported by Qi et al. (2003). Data from the EST chromosome deletion maps have been used to analyze the relationships of chromosome recombination rates to chromosome structure and evolution (Akhunov et al. 2003a,b) and the genomic relationships between wheat and rice (Sorrells et al. 2003). Results of mapping these ESTs into the seven wheat homoeologous chromosome groups are presented in this issue in accompanying articles by Hossain et al. (2004), Linkiewicz et al. (2004), Miftahudin et al. (2004), Munkvold et al. (2004), Peng et al. (2004), Randhawa et al. (2004), and Conley et al. (2004), with a summary, genome-wide analysis by Qi et al. (2004). The present report describes the generation of project ESTs, the selection and preparation of unique EST probes for large-scale mapping of wheat genes, the basic rationales and protocols utilized for mapping with wheat aneuploid and deletion stocks, the bioinformatics tools used and developed to coordinate this large multi-institution project, and current methods to access and query the project EST and bin-mapping data.
MATERIALS AND METHODS
EST production and probe preparation
cDNA libraries:
Fifty-one cDNA libraries were either constructed specifically for this project or made available from other sources. Of these, 41 cDNA libraries were used for EST production (Table 1) and for the final contig assembly. The libraries were derived from diverse tissues, stages, and treatments, including root, root tip, seedling, shoot, leaf, spike, seed, endosperm, anther, embryo, and apical meristem. Chinese Spring wheat, the internationally recognized basic research genotype for wheat, was used as the principal mRNA source. When Chinese Spring was not an appropriate choice for specific research aspects, e.g., for specific stress responses, differential development, or ease of tissue isolation, other T. aestivum cultivars such as BH1146, Brevor, Butte 86, Cheyenne, Sumai3, and TAMW101 and related species such as T. monococcum L., T. turgidum L., Aegilops speltoides Tausch, and S. cereale were used. Details of the library preparation and analyses are presented in the accompanying article by Zhang et al. (2004).
TABLE 1.
Wheat EST project cDNA libraries with data on EST productivity and mapping
| Namea | Tissue and conditionb | No. 5′ ESTs |
No. 3′ ESTs |
No. ESTs mappedc |
|---|---|---|---|---|
| TA054XXXd | Anther, meiotic, untreated | 9,139 | — | 0 |
| TA038E1X | Crown, seedling, salt stressed | 943 | 286 | 148 |
| TA016E1X | Crown, seedling, vernalized | 2,286 | 703 | 416 |
| TA012XXX | Embryo, mature, ABA-treated (Brevor) | 2,107 | — | 3 |
| TA049E1X | Embryo, mature, dormant (Brevor) | 2,927 | 148 | 51 |
| TA001E1X | Endosperm, 5–30 DPA (Cheyenne) | 2,728 | 1,216 | 382 |
| TA001E1S | Endosperm, 5–30 DPA, subtracted (Cheyenne) | 269 | — | 0 |
| TA036E1X | Leaf, mature, dehydrated | 641 | 180 | 88 |
| TA027E1X | Leaf, mature, dehydrated (TAMW101) | 905 | 136 | 60 |
| TA031E1X | Leaf, flag, at anthesis, heat stressed | 973 | 341 | 213 |
| TA037E1X | Leaf, seedling, sheath, salt stressed | 964 | 248 | 124 |
| TA008E1X | Root, seedling, etiolated | 4,017 | 453 | 520 |
| TA008E3N | Root, seedling, etiolated, normalized | 4,308 | 1,184 | 764 |
| TA065E1X | Root, seedling, salt stressed | 2,055 | — | 0 |
| TA055E1X | Root, at full tillering, drought stressed | 1,310 | — | 0 |
| TA058E1X | Root, at full tillering, unstressed | 1,025 | — | 0 |
| TA047E1X | Root tip, seedling, unstressed | 959 | 75 | 16 |
| TA056E1X | Root tip, seedling, aluminum stressed | 1,032 | — | 0 |
| TA048E1X | Root tip, 4 days old, aluminum stressed (BH1146) | 991 | 55 | 8 |
| TA059E1X | Seed, whole grain at 3–44 DPA (Butte 86) | 3,649 | — | 0 |
| TA005E1X | Seedling, whole, dehydrated | 795 | 92 | 51 |
| TA007E1X | Seedling, whole, cold stressed | 938 | 301 | 161 |
| TA015E1X | Seedling, whole, heat stressed | 821 | 291 | 146 |
| TA006E1X | Shoot, seedling, etiolated | 2,261 | 264 | 207 |
| TA006E2N | Shoot, seedling, etiolated, normalized | 1,686 | 174 | 122 |
| TA019E1X | Spike, preanthesis | 11,194 | 3,071 | 1,590 |
| TA018E1X | Spike, 5–15 DPA | 2,860 | 709 | 419 |
| TA032E1X | Spike, 5–20 DPA, heat stressed | 1,012 | 309 | 196 |
| TA017E1X | Spike, 20–45 DPA | 1,076 | 177 | 144 |
| TA009XXXe | Spike, mature, challenged with Fusarium graminearum (Sumai3) | 10,287 | 1,077 | 361 |
| TA006G1Xf | Spike, mature, challenged with F. graminearum (Sumai3) | 727 | — | 46 |
| TA066E1X | Mixed tissue (root, leaf, crown, stem, sheath) at maturity, unstressed | 1,404 | — | 0 |
| TM011XXX | Apex, shoot, vegetative (5-week; accession Dv92) | 3,031 | — | 11 |
| TM043E1X | Apex, shoot, early reproductive, 7-week vernalized (accession Dv92) | 2,647 | 930 | 426 |
| TM046E1X | Apex, shoot, 1-month vernalized (accession G3116) | 3,363 | — | 0 |
| TT039E1X | Plant, whole, mature (Langdon-16) | 1,194 | 284 | 157 |
| SC010XXX | Root tip, seedling, aluminum stressed (Blanco) | 1,198 | — | 0 |
| SC013XXX | Root tip, seedling, control (Blanco) | 778 | — | 0 |
| SC024E1X | Anther, mature, unstressed (Blanco) | 4,631 | 1,071 | 468 |
| AS040E1X | Anther, premeiotic, untreated | 2,466 | 804 | 339 |
| AS067E1X | Anther, early premeiotic, untreated | 1,044 | — | 0 |
| Total | 98,641 | 14,579 | 7,637 |
The species source of the library is indicated by the first two letters of the name: TA, T. aestivum; TM, T. monococcum; TT, T. turgidum; SC, S. cereale; and AS, Ae. speltoides.
All of the TA libraries are from the hexaploid wheat Chinese Spring genotype except where indicated otherwise in parentheses; RNA for both Ae. speltoides libraries came from F2 plants from the cross 2-12-4-8-1-1-1-(1) by PI 36909-12-811-(1).
Coordinator-confirmed mapping data as of February 2, 2004.
Library contributed by P. Langridge, University of Adelaide.
Library contributed by G. Muehlbauer, University of Minnesota.
Library contributed by J. Fellers, USDA-ARS, Kansas State University.
Sequence processing:
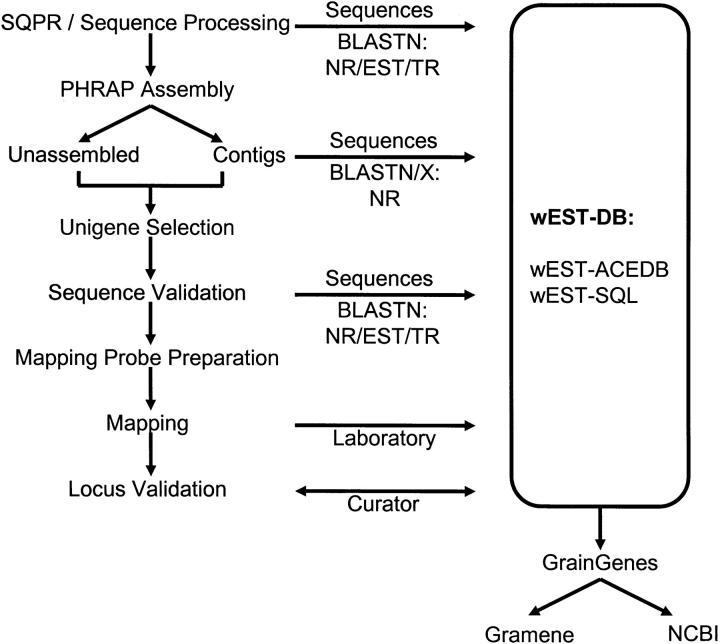
Samples were prepared for DNA isolation and archiving using high-throughput processing methods adapted for 96-well microtiter plate formats. Sequencing was done on an ABI Prism 3700 sequencer (Applied Biosystems, Foster City, CA). The flow of sequence production and analysis is illustrated in Figure 1. Sequence and quality files from trace files were read by the phred program (Ewing and Green 1998; Ewing et al. 1998) using a quality score setting of 20. This allowed the definition of a quality read length (QRL) of <0.01 errors/100 bases. In other words, a QRL was a string of nucleotides read from the sequencing trace file that maintained an acceptable average quality value. From these QRLs, high-quality sequence data were extracted after removing vector ends and short sequences and then filtered for sequences from Escherichia coli, plastids, repetitive sequences, and other sequence anomalies. Also removed were sequences <100 bp, sequences of low complexity, or sequences with >40% of bases with a score below phred 20 within a QRL. A custom web-accessible tracking system, SQPR, was developed and utilized to follow the quality of sequence generation (Lazo et al. 2001). Within SQPR, the phred program was used to read and process trace files. In general, plates that were advanced for further processing required an 80% success rate per 96-well plate run with QRL reads >350 bases in length. Processed sequence data were deposited on the project website (http://wheat.pw.usda.gov/wEST) and into the NCBI dbEST resource using NCBI sequence-submission protocols to annotate sequences.
Figure 1.—
Overview of sequence processing and database entry. The column on the left shows, from top to bottom, the data processing pipeline for EST sequencing and preparation of the unigene set for genome mapping. At all steps of the process, data were entered into wheat EST databases (wEST-DB), accessible from http://wheat.pw.usda.gov/wEST. Archives used for blast comparisons included nonredundant (NR), dbEST (EST), and local Triticeae (TR) databases. Sequences deposited into databases were derived from initial 5′ sequencing, formed assembly contigs, and 5′/3′ validation sequencing.
Contig assemblies:
To determine the unique nature of sequences within the collections, assembly algorithms were applied to the sequence pools using the phrap algorithm (http://www.phrap.org). Assemblies were performed on all sequences advanced through the sequence cleaning process. Phrap parameters (penalty, −5; minmatch, 50; minscore, 100) were set to allow like-sequences with 90% identity over a 100-base length to form contig clusters. Periodic assemblies were performed as sequence pools increased. In addition, assemblies were performed on the cDNA libraries individually to assess the level of library redundancy (http://wheat.pw.usda.gov/NSF/library_redundancy.html). Sequencing within a library continued with the identification of additional unique sequences, the goal being to reduce redundancy. The results from the global assembly of all project ESTs were used to estimate the extent of ESTs unique to a specific library and to estimate the number of unique clusters assembled using the Triticeae sequences. Subassemblies also compared candidate gene clusters between the species represented in the EST pool (bread wheat, durum wheat, diploid wheat, rye, and Ae. speltoides).
Candidate sequences for bin mapping were further screened to eliminate sequences for which matches were found in a series of formatted databases using the blastN algorithm (Altschul et al. 1997). Databases included UniVec (NCBI), E. coli (NCBI, GenBank), plastid and mitochondria (GenBank), rRNA (GenBank), and the Triticeae repetitive (TREP) element collection (Wicker et al. 2002); ESTs with significant matches (E-value <10−20) to the previous databases were removed. All processed EST sequences were compared to the NCBI nonredundant databases by the blastN and blastX algorithms using default settings (Altschul et al. 1997).
Probe selection and preparation:
Probe selection followed a stringent series of steps to assure minimal problems when performing Southern analysis in the mapping labs. ESTs selected for validation were rearrayed into 384-well format, 5′ resequenced to confirm clone identity, then 3′ sequenced to provide additional validation or to eliminate sequences where 5′ sequences did not overlap, but were from the same original sequence. ESTs with 3′ sequence similarity >90% over at least 100 bases were considered as duplicates and not suitable for bin mapping. Inserts were amplified by PCR using vector-based primers flanking the vector cloning site and purified using QIAQuick 96 kits (QIAGEN, Chatsworth, CA) either manually or on a BioRobot 8000 (QIAGEN). An aliquot of the purified PCR products was separated by electrophoresis in a 1% agarose gel, stained with ethidium bromide, and photographed. The bands were sized and quantitated using mass and size standards and Quantity One software (Bio-Rad, Hercules, CA). The amplification was considered successful if one prominent DNA band was generated. Aliquots of the purified PCR products were transferred to new plates and shipped to the mapping laboratories to be used as mapping probes. In some cases validated clones were sent and the mapping laboratory performed their own amplifications of inserts as described in the accompanying articles.
Gene ontologies
Each sequence was also searched against the UniProt database (Release 1.5, TrEMBL, Swiss-Prot, and PIR at http://www.ebi.ac.uk/uniprot) resources (Apweiler et al. 2004) using blastX, and best matches (E-value <10−10) were compared to terms of the Gene Ontology (GO) Consortium. Using GO/UniProt comparison tables, candidate GO assignments were predicted on the basis of EST matches to the UniProt reference sequences. Categories were assigned on the basis of biological, functional, and molecular annotations available from GO (http://www.geneontology.org/).
Bioinformatics
Throughout the process from the generation of ESTs to the mapping of a subset of them, data were transferred sequentially to several relational databases (http://wheat.pw.usda.gov/wEST), culminating with the GrainGenes database (http://wheat.pw.usda.gov) from which they were further distributed to additional public databases such as NCBI and Gramene (Figure 1). As sequences were processed, background information was stored using a MySQL-based relational database as a laboratory information management system, hosting information to keep track of clone information, clone production, DNA isolation, sequencing queues, and storage archives.
A series of perl script programs, termed sweeping steps, were used to trim down sequence files to limit the final output to cleaned sequence data (Table 2). Files were generally handled on a run-by-run basis to keep track of clone-library associations for each of the sequences. The sequence was marked up to identify vector-spanning regions of the sequence by cross-match (http://www.genome.washington.edu/UWGC/protocols) and the identified regions were read by the processing script to further trim down the sequence. The blastN/X reports from the screening steps were parsed using perl scripts to upload information into the online databases. ESTs with no or only poor matches to the nonredundant databases were subjected to further comparison using blastN against the NCBI dbEST collection. Information parsed from blast reports included all matched sequences, the blast score, and the E-value (E-value < 10−4), and sequence alignment values. Additional information included tagging the matched database sequence to specific plant taxa, concentrating on species of grasses. All project data are accessible at http://wheat.pw.usda.gov/wEST.
TABLE 2.
Sequence editing steps and nomenclature involved in trimming DNA sequence files
| Step | Sequence nomenclature | ||||
|---|---|---|---|---|---|
| 1. Shred (SQPR) | 0064_H10_O20ZS_80.ab1 | 1032 | 5 | 596 | ABI |
| 2. Sweep1.pl | 0064_H10_O20ZS_80.ab1 | 596 | 0 | 596 | ABI |
| 3. Cross_match | |||||
| 4. Sweep2.pl | 0064_H10_O20ZS_80.ab1 | 552 | 0 | 552 | ABI |
| 5. Add lab designator | WHE0064_H10_O20ZS | ||||
| 6. Submit to GenBank | BE423505 | ||||
Nomenclature was developed to facilitate tracking and data interlinks. Sequence names allowed for tracking plate origin, well position, and primers used in the sequencing reaction. The original name from the sequencing run was changed to reflect action at each processing step (Table 2). For example, from line 1 of Table 2, the sequence 0064_H10_O20 is a random cDNA picked into well O20 of a 384-well deep-well microtiter plate for original growth. This 384-well plate was named “0064-0067” to indicate that, when expanded to 96-well plates for sequencing, it generated 96-well plates numbered 64, 65, 66, and 67. H10 is the well of this clone in plate 64, and O20 the well in the original 384-well plate. The letter Z indicates the end of clone information, and S indicates the primer used in sequencing. Subsequent information included the original length of read (1032 bp), the start position of the QRL (fifth nucleotide), the length of the QRL maintaining phred 20 quality (596 bp), and the type of trace file read by the phred program (from an ABI sequencer). After steps 3 and 4, removing vector sequences, the QRL was 552 bp in length. At this point, a laboratory designator was added (WHE, in this case). Laboratory designators are an internationally recognized system to identify clones via three to four letters. The list of designators can be found at http://wheat.pw.usda.gov/ggpages/Lab.Designators.html. On submission to GenBank, the sequence received its GenBank accession designation following NCBI protocol.
EST mapping
Plant materials:
The collection of stocks in the Chinese Spring background used for mapping EST-specific restriction fragments consisted of a set of 21 nullisomic-tetrasomic lines (Sears 1954, 1966), 24 ditelosomic lines (Sears and Sears 1978), and 101 chromosome deletion lines (Endo and Gill 1996; Qi et al. 2003), which make possible the mapping of fragments to chromosomes, chromosome arms, and subarm locations (bins), respectively. The bin designations were based on the observed fractional position of the bounding deletion breakpoints in the chromosome arm as determined by cytogenetic observation; this set of deletion stocks delineates 159 distinct chromosome bins (Qi et al. 2003). Deletion stocks were obtained from the Wheat Genetics Resource Center, Department of Plant Pathology, Kansas State University, Manhattan, Kansas. The nullisomic-tetrasomic and ditelosomic aneuploids (Sears 1954; Sears and Sears 1978) were obtained from both the Wheat Genetics Resource Center and the USDA-Sears collection of wheat genetic stocks (USDA-ARS/University of Missouri).
Seeds of the aneuploid and deletion stocks were shipped to the 10 mapping laboratories (Table 3 and http://wheat.pw.usda.gov/NSF). Plants were grown in greenhouses and DNA samples were isolated following protocols established in those laboratories.
TABLE 3.
Identification of the 10 mapping laboratories and their designators and examples of EST locus identifiers derived from the laboratory designator and a GenBank EST accession number
| Laboratory
|
|||
|---|---|---|---|
| Investigator | Institution | Designator | Example of locus identifiera |
| J. A. Anderson | University of Minnesota | UMN | UMW134BE604741-4 |
| J. Dubcovsky | University of California, Davis | UCW | UCW169BE483747-1 |
| J. Dvořák | University of California, Davis | UCD | UCD141BE478737-1 |
| B. S. Gill | Kansas State University | KSU | KSU002BE406357-1 |
| K. S. Gillb | University of Nebraska/Washington State University | UNL | UNL002BF474204-1 |
| J. P. Gustafson | USDA-ARS, University of Missouri | UMC | UMC015BE426301-4 |
| S. F. Kianian | North Dakota State University | NDS | NDS051BE606901-8 |
| N. L. V. Lapitan | Colorado State University | CSU | CSU001BE426257-1 |
| H. T. Nguyenc | Texas Tech University/University of Missouri | TTU/UMW | TTU047BE405648-2 |
| M. E. Sorrells | Cornell University | CNL | CNL014BE591757-2 |
The first three characters are the laboratory designator; the next three characters are the rearrayed plate number; the two letters and six numbers are the GenBank accession number of the EST used as probe; and the number after the dash is the number of the band detected in the autoradiograph, starting with the largest as number 1.
Near the end of the project, K. S. Gill relocated from the University of Nebraska to Washington State University; however, his laboratory designator was not changed.
Approximately midway through the project, H. T. Nguyen relocated from Texas Tech University to the University of Missouri and his laboratory designator changed accordingly.
Diagrammatic representations of the bins, including the position of the breakpoints for homoeologous group chromosomes, can be found at http://wheat.pw.usda.gov/wEST/binmaps/. The mapping data and autoradiogram images are accessible on line from http://wheat.pw.usda.gov/westsql.
Deletion mapping:
All genomic DNA isolations, restriction endonuclease digestions, gel electrophoresis, DNA gel blot hybridizations, and EST analyses were uniformly carried out in each of the 10 mapping laboratories. The cDNA clone corresponding to each EST selected from the unigene set was hybridized to membranes of genomic DNA from each aneuploid and stock digested with EcoRI and blotted onto five membranes of 30 lanes each. λDNA, digested with HindIII and BstEII, was used as a size marker. All five membranes were used in each single hybridization reaction (http://wheat.pw.usda.gov/NSF). Procedures used for genomic DNA isolation, restriction endonuclease digestion, gel electrophoresis, and DNA gel blot hybridization were as described in Qi et al. (2003) and Akhunov et al. (2003a), unless otherwise noted, and are available on line at http://wheat.pw.usda.gov/NSF/project/mapping_data.html.
To provide uniformity in screening mapping data, a template for labeling lanes for each of the five membranes was used by all 10 laboratories for the images generated by autoradiography. A standard template and guidelines for reporting mapping data and laboratory assessments of the results was also provided. A world-wide-web interface (WWW) was used to facilitate the uploading of mapping data and image files by the mapping labs. Project laboratory designators and rearrayed probe plate numbers were added to EST GenBank accession numbers to identify the mapped loci (Table 3). Multiple restriction fragments were resolved by numbering autoradiograph bands in order, starting with 1 for the largest detected fragment.
The large scale of this project, wide geographic distribution of laboratories, and critical need for accuracy in scoring autoradiographs were addressed by a threefold scoring of each EST. Each hybridization profile was analyzed twice in the mapping laboratory where it was produced and uploaded to the project website where it was scored again by the coordinators assigned to each of the seven homoeologous groups. All scoring and accompanying comments were compiled through the WWW interface. Conflicts were resolved by further communication and, if necessary, joint examination of original blots. Only confirmed data were used for analyses. Data for ESTs for which map positions have not been accepted remain as “unconfirmed,” and resolution of these data is ongoing. The project's homoeologous chromosome group coordinators are N. L. V. Lapitan, Colorado State University, group 1; J. A. Anderson, University of Minnesota, group 2; M. E. Sorrells, Cornell University, group 3; J. P. Gustafson, USDA-ARS, University of Missouri, group 4; J. Dubcovsky, University of California, Davis, group 5; K. S. Gill, Washington State University, group 6; and S. F. Kianian, North Dakota State University, group 7.
RESULTS AND DISCUSSION
EST generation:
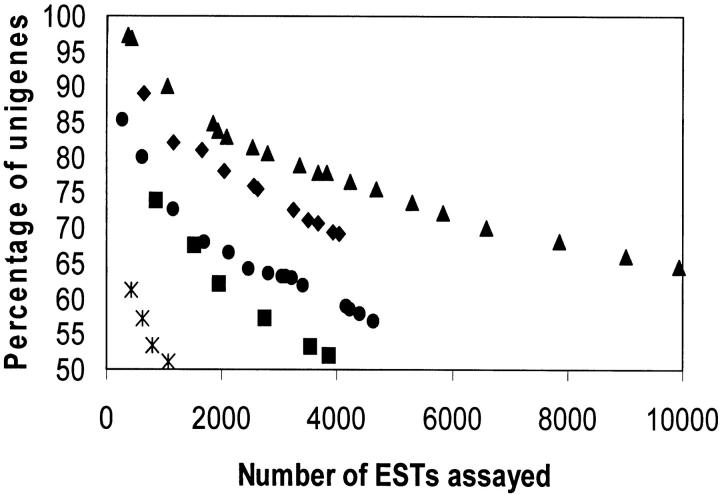
Approximately 99,000 5′ EST sequences were produced from the 41 different libraries and assembled as a pool for probes for EST bin mapping (Table 1). Results of sequencing from each library were regularly monitored to assess success at generating novel sequences. Figure 2 shows examples of such monitoring for five libraries. The library from spikes, 20–45 days past anthesis (DPA; TA017E1X), showed only a small number of new sequences even by 1000 ESTs, consistent with the mRNA source from maturing seeds whose main protein synthetic activity is completion of grain fill. The endosperm library (TA00E1X) continued generating new sequences well beyond that of the maturing spike library since the endosperm library was a pool of endosperm at stages ranging from early endosperm development through maturing endosperm. An example of a library that continued to produce a relatively high percentage of new sequences after deeper sequencing was the preanthesis spike library (TA019E1X).
Figure 2.—
Diminishing number of unigene ESTs with increasing number of ESTs generated. As libraries were sequenced, the total from each library was assessed for the number of new sequences generated. Shown are plots for five libraries as percentages of unigenes vs. total number of ESTs generated at each increment of EST generation and analysis: ♦, root (TA008E1X); •, anther (SC024E1X); ▴, preanthesis spike (TA019E1X); ▪, endosperm (TA001E1X); and *, 20–45 DPA spike (TA017E1X).
As part of unigene validation and probe selection, all candidate ESTs were 3′ sequenced, and a total of 14,579 3′ sequences were generated for a project total of 113,220 ESTs deposited in GenBank. The first ESTs were deposited in GenBank in July 2001, representing the first large-scale deposition of wheat ESTs to GenBank and the major contribution of the United States to the ITEC wheat EST goal. As of January 2004, there were 577,538 wheat, 377,074 barley, and 9194 rye ESTs in GenBank. Wheat currently ranks first among all plants (and fourth among all organisms), with barley third. Pooling the ESTs of wheat and barley with those of the other species of Triticeae yields ∼977,000 ESTs available to research for candidate gene searches, hybridization probes, or other uses, made possible by the close genetic relationships among these species. This is by far the largest such DNA sequence resource for any plant except where the entire genome has been or is being sequenced (e.g., Arabidopsis and rice).
EST classification:
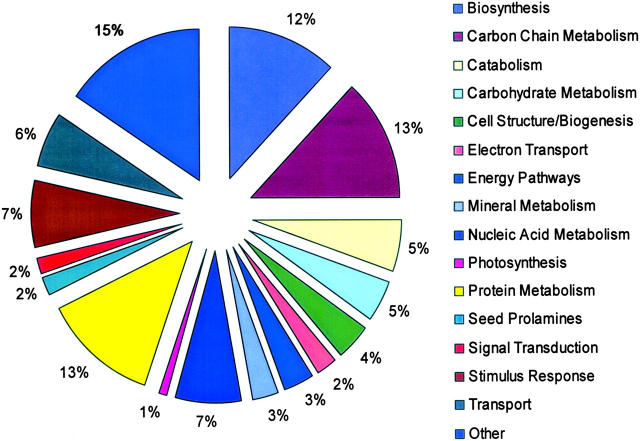
All project ESTs, contigs, and mapped probes and their associated blast annotations are available at http://wheat.pw.usda.gov/wEST. Through such annotation, ESTs can be assigned putative functions on the basis of matches to known sequences. Many different presentations of such data are possible, and in many cases even a single EST can be assigned multiple classifications; e.g., one could have multiple enzyme functions, be associated with a particular organelle, and confer disease resistance. Figure 3 shows distribution of this project's ESTs on the basis of the Gene Ontology sorting and includes classifications of those project ESTs having a significant GO assignment, plus the wheat prolamines not currently covered in GO (gliadin and glutenin classes of wheat seed proteins, polypeptides known for high proline and glutamine content).
Figure 3.—
EST annotation by GO and manual assignments. ESTs with GO assignments are collated and plotted as a percentage of GO-annotated ESTs plus ESTs manually assigned to the gliadin and glutenin (prolamine) classes of wheat seed storage proteins. Some classifications were pooled to reduce the number of classes.
Unigene sets and probe selection:
The project's final contig assembly yielded 18,876 contigs and 23,034 singletons from 116,739 ESTs. A total of 25,310 ESTs representing unigenes were advanced for the probe validation process.
At each step of the validation and insert preparation process there was sample attrition due to the failure to grow cultures, obtain DNA, validate 5′ sequence, obtain 3′ sequence, obtain nonduplicative 3′ sequence, or yield an appropriate PCR product. Even with only a small percentage of failures at each step, only 43% of the original ESTs advanced for probe generation passed all stages of validation and preparation. In cases where ESTs that failed to validate were members of a contig, another contig member was chosen for the next round of probe preparation. Those that passed (13,635) were distributed to the 10 mapping laboratories for physical mapping into chromosome deletion bins.
Mapping:
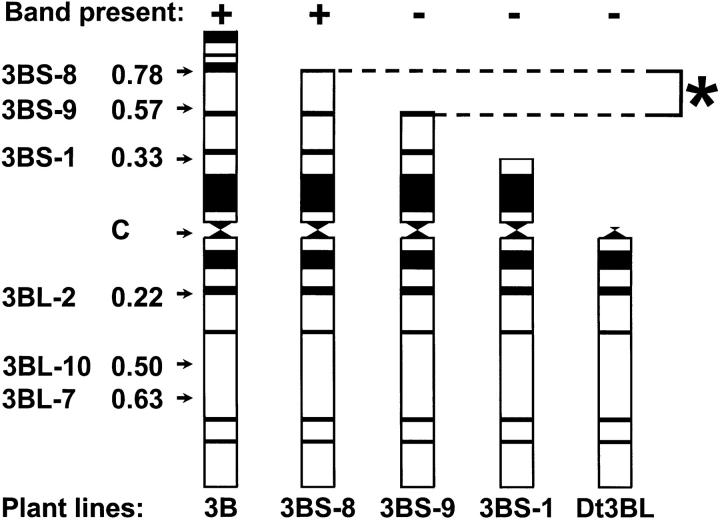
EST fragments were allocated to a given chromosome bin according to the presence or absence of a hybridization fragment in the deletion line. In the example in Figure 4, a probe identified a restriction fragment located on chromosome arm 3BS because the band is missing in the ditelosomic 3BL lane (Dt3BL, missing the 3BS arm). The position of the band was further resolved as being in the deletion bin (indicated by the asterisk in Figure 4), because the band was present in the lane for deletion line 3BS-8, but not in the lanes for deletions 3BS-9 or 3BS-1.
Figure 4.—
Mapping loci to chromosome bins using aneuploid stocks. Shown is the mapping of a locus on chromosome 3BS from the data for probe BE406607, band 3 (Figure 1a in Qi et al. 2004). The five diagrams represent the chromosome 3B configurations in five stocks represented as C-banded chromosomes. Heterochromatic regions are shown as solid and C indicates the centromere. Breakpoint names and positions on the respective chromosome arms are shown on the left. The complete 3B chromosome is on the left. The next three are deletion lines named for the deletion breakpoint involved. The last (Dt3BL) is the ditelocentric 3BL line (containing a pair of 3B chromosomes missing the short arm). The presence or absence of a hybridizing band from the sample as detected by autoradiography is indicated by + or − in the “Band present” row. The presence of the band in the first two stocks, but not in the latter three, indicates that it maps to the bin marked by the asterisk (bin 3BS9-0.57-0.78).
March 17, 2003, was selected as a cutoff date for defining a subset of entered and verified mapping data for subsequent use in the in-depth analyses presented in the accompanying articles in this issue. At that time, the combined work of the 10 mapping labs had produced and verified 4485 mapped ESTs (http://wheat.pw.usda.gov/NSF/progress_mapping.html). Mapping data submission and verification continued from that time and as of February 2, 2004, 8318 ESTs had been mapped and 7757 of them verified.
Data access:
All submitted data files were uploaded into databases accessible through Internet connections using WWW browsers. Two database formats were utilized: ACEDB (Durbin and Thierry-Mieg 1991), which had many of the display modules already included, and a MySQL relational database (wEST-SQL), for which recent tool developments have added flexibility in the display of data. Data structure facilitated subsequent incorporation into the publicly available database resources of GrainGenes (Matthews et al. 2003; http://wheat.pw.usda.gov).
Each of the databases provides links among the sequenced ESTs, assembled contig sets, candidate gene identities of the ESTs and contigs, and mapped arm and bin locations. Within the wEST-SQL database (http://wheat.pw.usda.gov/wEST), there are access points for ESTs, contigs, and mapping data. The query resources for the wEST-SQL database provide access from many entry points, including cDNA libraries, ESTs, contigs, and mapping data, as well as the opportunity to submit raw SQL queries.
A blast analysis capability was developed for comparing a user-supplied sequence to all project ESTs, ESTs by library, contigs, and mapped contigs. This capability has been expanded to include all GenBank Triticeae ESTs separately, which can be blast searched by cultivar, contig assemblies from international collaborations, full-length sequence modeled for a barley Affymetrix chip, Triticeae genera, rice and Arabidopsis sequences, the GrainGenes TREP data set for Triticeae repetitive DNAs, and other custom sets to total >100 sequence databases (http://wheat.pw.usda.gov/wEST/blast/). Additional sets were added as requested by the user community.
Project mapping data are primarily available by SQL query through the public version of the database. A query for a specific probe by accession name provides access to mapped locus information by chromosome and bin location with links to 5′ and 3′ sequences. The adaptable queries can be limited to specific genomes, chromosomes, chromosome groups, bins, scored aneuploid and deletion lines, sequence candidate identities, and mapping laboratory searches. Links are provided to associated probe data, including links to the wEST-ACEDB and Generic Genome Browser displays, which is an experimental application being developed by the Generic Model Organism Database project (Stein et al. 2002; http://www.gmod.org).
Although originally developed for this project, the wEST MySQL site and database have been incorporated into the GrainGenes suite of databases and resources. As shown in Figure 1, the project data have flowed into GrainGenes and then to additional public databases. Selected information from this wheat mapping project was distributed into Gramene (Ware et al. 2002), allowing comparison of the alignments of wheat sequences to rice and other grass species. Reciprocal links have been established where appropriate. Access to project data has also been developed at the NCBI website (http://www.ncbi.nlm.nih.gov/). Additional information about access to data is available from the supplemental online material at http://wheat.pw.usda.gov/pubs/2004/Genetics.
Mapped wheat loci:
As of February 2, 2004, 8318 ESTs had been mapped with 7637 of them verified, yielding almost 40,000 scored loci. The subset of mapped and verified ESTs (4485 as of March 17, 2003) used for the analyses in the accompanying articles, after validation and removal of duplications, yielded 16,093 loci mapped to the Chinese Spring aneuploid and deletion stocks with a distribution by genome of 5173 (A), 5774 (B), and 5146 (D). With analyses on a homoeologous chromosome group basis, the total is 15,843 loci with 2212, 2600, 2266, 2236, 2338, 2043, and 2148 loci mapped for chromosome groups 1–7, respectively. The difference in the two totals is due to the fact that analysis by genome can include ESTs with loci that may have mapped to a chromosome or a chromosome arm only but could not have been mapped to a bin. The number of ESTs for a homoeologous group, totaled from a bin-by-bin analysis, will exclude these, yielding a total less than that obtained when analysis is by genome. More detailed presentation of these results is provided in Qi et al. (2004).
Acknowledgments
This material is based upon work supported by the National Science Foundation Cooperative Agreement no. DBI-9975989, the United States Department of Agriculture, Agricultural Research Service Current Research Information System Project no. 5325-21000-010-00D at Albany, California, and the 11 universities participating in this collaborative effort.
References
- Adams, M. D., J. M. Kelley, J. D. Gocayne, M. Dubnick, M. H. Polymeropoulos et al., 1991. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252: 1651–1656. [DOI] [PubMed] [Google Scholar]
- Akhunov, E. D., A. W. Goodyear, S. Geng, L. L. Qi, B. Echalier et al., 2003. a The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 13: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov, E. D., A. R. Akhunova, A. M. Linkiewicz, J. Dubcovsky, D. Hummel et al., 2003. b Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc. Natl. Acad. Sci. USA 100: 10836–10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., A. Bairoch, C. H. Wu, W. C. Barker, B. Boeckmann et al., 2004. UniProt: the universal protein knowledge base. Nucleic Acids Res. 32: D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan, K., and E. D. Earle, 1991. Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol. Biol. Rep. 9: 208–218. [Google Scholar]
- Conley, E. J., V. Nduati, J. L. Gonzalez-Hernandez, A. Mesfin, M. Trudeau-Spanjers et al., 2004. A 2600-locus chromosome bin map of wheat homoeologous group 2 reveals interstitial gene-rich islands and colinearity with rice. Genetics 168: 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin, R., and J. Thierry-Mieg, 1991 A C. elegans database: documentation, code and data available from anonymous FTP servers at ftp.sanger.ac.uk and ncbi.nlm.nih.gov.
- Endo, T. R., 1988. Induction of chromosomal structural changes by a chromosome of Aegilops cylindrica L. in common wheat. J. Hered. 79: 366–370. [DOI] [PubMed] [Google Scholar]
- Endo, T. R., 1990. Gametocidal chromosomes and their induction of chromosome mutations in wheat. Jpn. J. Genet. 65: 135–152. [Google Scholar]
- Endo, T. R., and B. S. Gill, 1996. The deletion stocks of common wheat. J. Hered. 87: 295–307. [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Hossain, K. G., V. Kalavacharla, G. R. Lazo, J. Hegstad, M. J. Wentz et al., 2004. A chromosome bin map of 2148 expressed sequence tag loci of wheat homoeologous group 7. Genetics 168: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo, G. R., J. Tong, R. Miller, C. Hsia, C. Rausch et al., 2001. Software scripts for quality checking of high-throughput nucleic acid sequencers. Biotechniques 30: 1300–1305. [DOI] [PubMed] [Google Scholar]
- Linkiewicz, A. M., L. L. Qi, B. S. Gill, B. Echalier, S. Chao et al., 2004. A 2500-locus bin map of wheat homoeologous group 5 provides new insights on gene distribution and colinearity with rice. Genetics 168: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, D., V. Carollo, G. R. Lazo and O. D. Anderson, 2003. GrainGenes, the genome database for small-grain crops. Nucleic Acids Res. 31: 183–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftahudin, K. Ross, X.-F. Ma, A. A. Mahmoud, J. Layton et al., 2004. Analysis of EST loci on wheat chromosome group 4. Genetics 168: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold, J. D., R. A. Greene, C. E. Bermudez-Kandianis, C. M. La Rota, H. Edwards et al., 2004. Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 168: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J. H., H. Zadeh, G. R. Lazo, J. P. Gustafson, S. Chao et al., 2004. Chromosome bin map of expressed sequence tags in homoeologous group 1 of hexaploid wheat and homoeology with rice and Arabidopsis. Genetics 168: 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L. L., B. Echalier, B. Friebe and B. S. Gill, 2003. Molecular characterization of a set of wheat deletion stocks for using in chromosome bin mapping of ESTs. Funct. Integr. Genomics 3: 39–55. [DOI] [PubMed] [Google Scholar]
- Qi, L. L., B. Echalier, S. Chao, G. R. Lazo, G. E. Butler et al., 2004. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa, H. S., M. Dilbirligi, D. Sidhu, M. Erayman, D. Sandhu et al., 2004. Deletion mapping of homoeologous group 6-specific wheat expressed sequence tags. Genetics 168: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, E. R., 1954. The aneuploids of common wheat. Univ. Mo. Agric. Exp. Stn. Bull. 572: 1–58. [Google Scholar]
- Sears, E. R., 1966 Nullisomic-tetrasomic combinations in hexaploid wheat, pp. 29–45 in Chromosome Manipulations and Plant Genetics, edited by R. Riley and K. R. Lewis, Oliver & Boyd, Edinburgh.
- Sears, E. R., and L. M. S. Sears, 1978 The telocentric chromosomes of common wheat, pp. 389–407 in Proceedings of the 5th International Wheat Genetic Symposium, edited by S. Ramanujam. Indian Society of Genetics and Plant Breeding, New Dehli.
- Sorrells, M. E., M. La Rota, C. E. Bermudez-Kandianis, R. A. Greene, R. Kantety et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L. D., C. Mungall, S. Shu, M. Caudy, M. Mangone et al., 2002. The generic genome browser: a building block for a model organism system database. Genome Res. 12: 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deynze, A. E., J. C. Nelson, E. S. Yglesis, S. E. Harrington, D. P. Braga et al., 1995. Comparative mapping in grasses. Wheat relationships. Mol. Gen. Genet. 248: 744–754. [DOI] [PubMed] [Google Scholar]
- Ware, D., P. Jaiswal, J. J. Ni, X. Pan, K. Chang et al., 2002. Gramene: a resource for comparative grass genomics. Nucleic Acids Res. 30: 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., D. E. Matthews and B. Keller, 2002. TREP: a database for Triticeae repetitive elements. Trends Plant Sci. 7: 561–562. [Google Scholar]
- Zhang, D., D. W. Choi, S. Wanamaker, R. D. Fenton, A. Chin et al., 2004. Construction and evaluation of cDNA libraries for large-scale expressed sequence tag sequencing in wheat (Triticum aestivum L.). Genetics 168: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]