Abstract
A total of 37 original cDNA libraries and 9 derivative libraries enriched for rare sequences were produced from Chinese Spring wheat (Triticum aestivum L.), five other hexaploid wheat genotypes (Cheyenne, Brevor, TAM W101, BH1146, Butte 86), tetraploid durum wheat (T. turgidum L.), diploid wheat (T. monococcum L.), and two other diploid members of the grass tribe Triticeae (Aegilops speltoides Tausch and Secale cereale L.). The emphasis in the choice of plant materials for library construction was reproductive development subjected to environmental factors that ultimately affect grain quality and yield, but roots and other tissues were also included. Partial cDNA expressed sequence tags (ESTs) were examined by various measures to assess the quality of these libraries. All ESTs were processed to remove cloning system sequences and contaminants and then assembled using CAP3. Following these processing steps, this assembly yielded 101,107 sequences derived from 89,043 clones, which defined 16,740 contigs and 33,213 singletons, a total of 49,953 “unigenes.” Analysis of the distribution of these unigenes among the libraries led to the conclusion that the enrichment methods were effective in reducing the most abundant unigenes and to the observation that the most diverse libraries were from tissues exposed to environmental stresses including heat, drought, salinity, or low temperature.
GENOME projects are progressing at an unprecedented pace for a wide range of species from bacterial to human due to the continuing improvement of high-throughput technologies (http://www.ncbi.nih.gov/Genomes/index.html). However, among higher plants only the two model species, Arabidopsis (Arabidopsis thaliana L. Heynh.) and rice (Oryza sativa L.), have had their genomes completely sequenced. Arabidopsis is economically insignificant but has been the leading model for plant genome research due to its compact size and short generation time, the ease of producing mutations and transgenic plants, its small genome (157 Mb; Bennett et al. 2003), and timely development of core genomic resources, including deep-coverage BAC libraries and a complete genome microarray.
In recent years, rice has come into its own as a model species, representing monocotyledonous plants for genomic research. Rice is the world's largest contributor of calories for direct human consumption and second only to wheat in worldwide production acreage. Rice is a member of the grass family and as such carries far more extensive gene relationships and similarities in genome organization to wheat and other grasses than does Arabidopsis (Gale and Devos 1998). Rice also has reliable transformation systems, a rapidly increasing number of mutants, extensive germplasm collections, and a worldwide network of production-oriented researchers and farmers. With the sequence of the 450 Mb (Bennett and Leitch 1995) rice genome entering the public sector in draft form in 2002 (Goff et al. 2002; Yu et al. 2002) and expected to be finished in 2005, all major cereal genome research efforts now draw heavily from rice as the premier plant genome model.
Progress in genome sequencing for all other plants has lagged behind Arabidopsis and rice. In many cases, the main reason has been the presence of a much larger genome size. For example, bread wheat is an allohexaploid species (Triticum aestivum L., 2n = 6x = 42, AABBDD) with a genome size of 17,300 Mb (Bennett and Leitch 1995), which is 110 and 38 times as large as Arabidopsis and rice, respectively. A further barrier has been that the bulk of the extra genome size is composed of at least 90% repetitive DNA (McCarthy et al. 2002), which complicates genome sequence assembly and discourages investment in comprehensive genome sequencing. Nevertheless, the economic and social relevance of wheat and other crop plants, together with a resourceful and motivated research community, have driven crop plant genomic research. Wheat genome research also benefits from its polyploid nature, which provides opportunities to understand the organization and evolution of genomes that have a history of polyploidy, genome reduction, and effective diploidization.
As an interim alternative to whole-genome sequencing, many research communities have turned to collecting transcript sequences from cDNA library sequencing. Single-end sequences of cDNAs are known as expressed sequence tags (ESTs). EST sequencing has proven to be a powerful approach for gene discovery (Adams et al. 1991, 1993, 1995), amenable to large- or collective small-scale efforts. The number of ESTs has grown exponentially and access to them has improved during the past decade such that, as of April 2004, there were >20 million EST sequence accessions in the National Center for Biotechnology Information (NCBI) dbEST database (http://www.ncbi.nlm.nih.gov/dbEST). This included nearly 1 million ESTs from hexaploid wheat and its near relatives in the tribe Triticeae (Hordeum vulgare L., diploid and tetraploid Triticum species, Secale cereale L., and Aegilops speltoides Tausch.).
The work on cDNA libraries summarized here reflects the recognition by a consortium of U. S. and international wheat researchers that large-scale EST sequencing was the most practical first step in the development of extensive knowledge of wheat genes and the hexaploid wheat genome. As described in the accompanying articles in this issue (Hossain et al. 2004; Lazo et al. 2004; Linkiewicz et al. 2004; Miftahudin et al. 2004; Munkvold et al. 2004; Peng et al. 2004; Randhawa et al. 2004; Conley et al. 2004; Qi et al. 2004), physical mapping efforts that combined a unique set of wheat deletion stocks (Qi et al. 2003) with EST-based probes facilitated comparative genome analyses between wheat and other cereal species. When this National Science Foundation (NSF)-funded project was initiated in the fall of 1999, only 8 wheat EST sequences were publicly available. By the end of the project in 2003, the project had generated and deposited in the NCBI dbEST some 117,000 EST sequences. This work has been accompanied by other national and international efforts providing additional wheat EST sequences, bringing the dbEST total for wheat to some 500,000 ESTs, the most for any plant species. With the advantage of a retrospective analysis this study examines the EST data set produced by this project to assess the efficacy of the materials and methods chosen to produce cDNA libraries. Our cDNA libraries were produced from a wide range of tissues and stages of development in combination with different growth conditions and genotypes. To address the issue of redundant sequencing of highly abundant cDNAs, several libraries were produced using “normalization” or “subtraction” methods.
MATERIALS AND METHODS
Plant materials:
The Chinese Spring genotype of wheat was the primary source of tissues for cDNA library construction. In addition, five other hexaploid wheat genotypes, including BH1146, Brevor, Butte 86, Cheyenne, and TAM W101, were used for some libraries. Also, four wheat-related species, T. monococcum L. (genome AA), T. turgidum L. (genomes AABB), Ae. speltoides (genomes SS), and S. cereale (genomes RR), were used to recover genes related to specific traits of developmental stages that were not readily accessible using hexaploid wheat. We report on ESTs from 37 different primary, unenriched cDNA libraries and 9 enriched libraries derived from 6 of these primary (parent) libraries (Tables 1 and 2). Sources of plant materials are further described below in the details of library construction.
TABLE 1.
Source materials of cDNA libraries and numbers and characteristics of EST clones
| No. of
|
Most frequent junk |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Library designationa |
Genotype | Tissue | Stageb | Condition | Clones | ESTs | In contigsc |
Unique contigsd | Singles | Unigenes/clone | Unique/clonee | Junkf (%) |
Sourceg | % |
| AS040E1X | F2 from cross | Anther | Premeiotic | 2,570 | 3,359 | 1,045 | 246 | 1,376 | 0.942 | 0.631 | 1.21 | TREP | 0.35 | |
| AS067E1X | F2 from cross | Anther | Premeiotic | 1,068 | 1,068 | 445 | 19 | 425 | 0.815 | 0.416 | 2.20 | TREP | 0.82 | |
| SC010XXX | Blanco | Root tip | Seedling | Aluminum | 1,229 | 1,229 | 361 | 39 | 701 | 0.864 | 0.602 | 0.24 | TREP | 0.08 |
| SC013XXX | Blanco | Root tip | Seedling | 894 | 894 | 265 | 21 | 554 | 0.916 | 0.643 | 0.33 | Chimera | 0.22 | |
| SC024E1X | Blanco | Anther | Developing spike | 6,553 | 7,679 | 1,645 | 893 | 2,976 | 0.705 | 0.590 | 0.98 | rRNA | 0.45 | |
| TA001E1S | Cheyenne | Endosperm | 5–30 DAP | 222 | 222 | 122 | 5 | 70 | 0.865 | 0.338 | 2.20 | Mitochon. | 0.88 | |
| TA001E1X | Cheyenne | Endosperm | 5–30 DAP | 3,111 | 3,111 | 1,222 | 72 | 753 | 0.635 | 0.265 | 2.63 | rRNA | 1.91 | |
| TA001E2N | Cheyenne | Endosperm | 5–30 DAP | 65 | 65 | 36 | 0 | 28 | 0.985 | 0.431 | 90.75 | Sorghum | 90.33 | |
| TA005E1X | Chinese Spring | Root and shoot | Seedling | Drought | 875 | 971 | 540 | 29 | 315 | 0.977 | 0.393 | 1.02 | Chimera | 0.51 |
| TA005E2S | Chinese Spring | Root and shoot | Seedling | Drought | 828 | 828 | 403 | 56 | 220 | 0.752 | 0.333 | 2.36 | E. coli | 0.83 |
| TA006E1X | Chinese Spring | Shoot | Seedling | Etiolated | 2,728 | 3,021 | 1,395 | 132 | 757 | 0.789 | 0.326 | 3.27 | Mitochon. | 1.76 |
| TA006E2N | Chinese Spring | Shoot | Seedling | Etiolated | 1,508 | 1,622 | 530 | 128 | 148 | 0.450 | 0.183 | 2.41 | rRNA | 1.08 |
| TA006E3N | Chinese Spring | Shoot | Seedling | Etiolated | 657 | 715 | 420 | 32 | 214 | 0.965 | 0.374 | 65.79 | Sorghum | 64.07 |
| TA007E1X | Chinese Spring | Root and shoot | Seedling | Cold | 1,033 | 1,333 | 671 | 59 | 444 | 1.079 | 0.487 | 1.91 | TREP | 0.44 |
| TA007E2S | Chinese Spring | Root and shoot | Seedling | Cold | 491 | 534 | 223 | 31 | 206 | 0.874 | 0.483 | 23.82 | E. coli | 19.69 |
| TA007E3S | Chinese Spring | Root and shoot | Seedling | Cold | 491 | 507 | 232 | 24 | 182 | 0.843 | 0.420 | 91.05 | Sorghum | 90.02 |
| TA008E1X | Chinese Spring | Root | Seedling | Etiolated | 3,634 | 4,169 | 2,008 | 169 | 1,193 | 0.881 | 0.375 | 0.95 | rRNA | 0.24 |
| TA008E3N | Chinese Spring | Root | Seedling | Etiolated | 4,450 | 5,317 | 2,245 | 579 | 904 | 0.708 | 0.333 | 2.60 | E. coli | 0.77 |
| TA012XXX | Brevor | Embryo | Dormant seed | ABA | 1,767 | 1,767 | 889 | 8 | 761 | 0.934 | 0.435 | 17.58 | rRNA | 16.23 |
| TA015E1X | Chinese Spring | Root and shoot | Seedling | Heat | 1,339 | 1,652 | 734 | 43 | 630 | 1.019 | 0.503 | 1.73 | Mitochon. | 0.54 |
| TA016E1X | Chinese Spring | Crown | Seedling | Cold | 2,789 | 3,513 | 1,642 | 163 | 1,174 | 1.010 | 0.479 | 1.01 | rRNA | 0.31 |
| TA017E1X | Chinese Spring | Spike | 20–45 DAP | 1,167 | 1,317 | 523 | 32 | 272 | 0.681 | 0.260 | 0.98 | rRNA | 0.38 | |
| TA018E1X | Chinese Spring | Spike | 5–15 DAP | 2,912 | 3,535 | 1,609 | 123 | 1,186 | 0.960 | 0.450 | 0.31 | TREP | 0.23 | |
| TA019E1X | Chinese Spring | Spike | Preanthesis | 12,194 | 15,316 | 5,344 | 790 | 5,698 | 0.906 | 0.532 | 0.65 | TREP | 0.23 | |
| TA027E1X | TAM W101 | Leaf | Full tillering | Drought | 1,333 | 1,486 | 674 | 52 | 499 | 0.880 | 0.413 | 1.78 | Mitochon. | 0.93 |
| TA027E2S | TAM W101 | Leaf | Full tillering | Drought | 991 | 991 | 523 | 14 | 361 | 0.892 | 0.378 | 1.88 | Chloroplast | 0.69 |
| TA031E1X | Chinese Spring | Leaf | Full tillering | Heat | 1,177 | 1,530 | 724 | 70 | 517 | 1.054 | 0.499 | 1.29 | TREP | 0.84 |
| TA032E1X | Chinese Spring | Spike | 5–20 DAP | Heat | 1,180 | 1,506 | 684 | 49 | 571 | 1.064 | 0.525 | 0.86 | rRNA | 0.33 |
| TA036E1X | Chinese Spring | Leaf | Full tillering | Drought | 787 | 981 | 456 | 30 | 376 | 1.057 | 0.516 | 2.29 | TREP | 0.60 |
| TA037E1X | Chinese Spring | Sheath | Sheath | Salt | 977 | 1,242 | 613 | 47 | 360 | 0.996 | 0.417 | 1.90 | rRNA | 0.39 |
| TA038E1X | Chinese Spring | Crown | Crown | Salt | 1,241 | 1,537 | 811 | 54 | 499 | 1.056 | 0.446 | 1.35 | rRNA | 0.45 |
| TA047E1X | Chinese Spring | Root tip | Root tip | 1,157 | 1,210 | 688 | 45 | 278 | 0.835 | 0.279 | 0.98 | E. coli | 0.33 | |
| TA048E1X | BH1146 | Root tip | Root tip | Aluminum | 1,016 | 1,069 | 565 | 32 | 329 | 0.880 | 0.355 | 0.56 | Chimera | 0.37 |
| TA049E1X | Brevor | Embryo | Dormant seed | Dormant | 3,138 | 3,283 | 1,411 | 114 | 990 | 0.765 | 0.352 | 6.12 | rRNA | 4.83 |
| TA055E1X | Chinese Spring | Root | Full tillering | Drought | 1,326 | 1,326 | 686 | 15 | 508 | 0.900 | 0.394 | 1.41 | TREP | 0.74 |
| TA056E1X | Chinese Spring | Root tip | Seedling | Aluminum | 1,208 | 1,208 | 719 | 28 | 321 | 0.861 | 0.289 | 0.66 | Chimera | 0.33 |
| TA058E1X | Chinese Spring | Root | Full tillering | 1,024 | 1,024 | 583 | 8 | 357 | 0.918 | 0.356 | 1.73 | TREP | 0.86 | |
| TA059E1X | Butte 86 | Grain | 3–44 DAP | Various | 3,708 | 3,708 | 1,674 | 61 | 785 | 0.663 | 0.228 | 3.13 | rRNA | 1.49 |
| TA065E1X | Chinese Spring | Root | Full tillering | Salt | 2,129 | 2,129 | 1,153 | 26 | 738 | 0.888 | 0.359 | 1.16 | Chimera | 0.70 |
| TA066E1X | Chinese Spring | All | Full tillering | 1,412 | 1,412 | 767 | 10 | 434 | 0.851 | 0.314 | 0.77 | TREP | 0.28 | |
| TM011XXX | DV92 | Shoot apex | Vegetative | 3,188 | 3,188 | 1,318 | 131 | 1,194 | 0.788 | 0.416 | 2.92 | TREP | 1.83 | |
| TM043E1X | DV92 | Shoot apex | Reproductive | Vernalized | 2,802 | 3,590 | 1,349 | 307 | 1,143 | 0.889 | 0.517 | 3.65 | TREP | 1.88 |
| TM046E1X | G3116 | Shoot apex | Reproductive | Vernalized | 3,471 | 3,471 | 1,308 | 143 | 1,295 | 0.750 | 0.414 | 2.72 | TREP | 1.51 |
| TT039E1X | Langdon-16 | All | Various | 1,203 | 1,472 | 684 | 71 | 471 | 0.960 | 0.451 | 0.67 | rRNA | 0.20 | |
| Total | 89,043 | 101,107 | 16,740 | 5,000 | 33,213 | 0.561 | 0.429 | |||||||
The species source of the library is indicated by the first two letters of the designation: TA, T. aestivum; TM, T. monococcum; TT, T. turgidum; SC, Secale cereale; and AS, A. speltoides. Any enrichment technique applied is indicated by the last character in the designation: N, normalized and S, subtracted.
DAP, days after pollination.
In contigs, number of contigs in which sequences from this library participate. The total is not the sum of the column, but the total number of contigs when considering all of the libraries at once.
Unique contigs, those that are composed of sequences only from this library.
Unique/clone, (unique contigs + singles)/number of clones.
Percentage of library composed of clones other than the intended cDNAs.
TREP, Triticeae Repeat Sequence Database (http://wheat.pw.usda.gov/ggpages/ITMI/Repeats/index.shtml); Mitochon., Mitochondrion.
TABLE 2.
Features of normalized and subtracted cDNA libraries
| Normalized/subtracted library designation |
Parent library | Method for preparing single-stranded DNAa |
Driver cDNA |
C0t value (sec-mole/liter) |
Parent library titer |
Enriched library titer |
|---|---|---|---|---|---|---|
| TA001E2N | TA001E1Xb | 2 | PCR-amplified DNAs from library TA001E1X | 5 | 1.2 × 105 | 3.5 × 107 |
| TA006E2N | TA006E1X | 1 | PCR-amplified DNAs from library TA006E1X | 2.5 | 1.1 × 106 | 2.4 × 109 |
| TA006E3N | TA006E1X | 2 | PCR-amplified DNAs from library TA006E1X | 5 | 3.5 × 105 | 4.1 × 107 |
| TA008E3N | TA008E1X | 2 | PCR-amplified DNAs from library TA008E1X | 5 | 1.9 × 107 | 9.0 × 109 |
| TA005E2S | TA005E1X | 2 | Pooled equal amount of PCR-amplified DNAs from nonstressed seedling shoot library TA006E1X and seedling root TA008E1X |
100 | 4.0 × 104 | 8.1 × 106 |
| TA007E2S | TA007E1X | 2 | Pooled equal amount of PCR-amplified DNAs from nonstressed seedling shoot library TA006E1X and seedling root TA008E1X |
100 | 2.4 × 104 | 4.0 × 106 |
| TA007E3S | TA007E1X | 2 | Pooled equal amount of PCR-amplified DNAs from nonstressed seedling shoot library TA006E1X and seedling root TA008E1X |
50 | 5.0 × 104 | 5.5 × 105 |
| TA027E1S | TA027E1X | 2 | 1st stranded cDNAs from non-stressed tissues | 100 | 8.3 × 106 | 2.4 × 105 |
The two methods are described in Preparation of single-stranded phagemid DNA for normalization in materials and methods.
Not included here is an additional subtracted library, TA001E1S, which was a collection of ESTs derived from TA001E1X identified by a different method (see materials and methods).
Bacterial strains:
Standard bacterial strains used for cDNA library construction are stated in the descriptions below for each library. Strain TJC121 was produced during this project to circumvent growth problems associated with strain SOLR (Stratagene, San Diego). The relevant genotype of strain TJC121 is thi-1 metA28 Δ(gpt-lac)5 rpsL150 lamB20::Tn5 hsdR2 Zjj202::Tn10 recA938::Tn9 [F′proAB lacZΔM15 traD36]. The Δ(gpt-lac)5 mutation in combination with the truncated lacZΔM15 gene on the F′ element make this strain suitable for “blue/white” screening for β-galactosidase activity. The lamB20 mutation makes this strain phage λ-resistant. Presence of the F′ element results in F-pilus formation and therefore receptivity to phage fd-like phagemid particles. The lack of a suppressor tRNA mutation renders the strain resistant to glnV44-dependent helper fd phage, which includes Stratagene's ExAssist helper phage. For unknown reasons, strain TJC121 does not rapidly settle to the bottom of culture tubes in liquid culture, which is a problem with SOLR (Stratagene) and some other commonly used phagemid plating strains. In addition, strain TJC121 has excellent survival on the surface of refrigerated petri dishes, possibly because TJC121 carries only the recA mutation rather than additional defects in recombination, repair, and replication pathways. The rpsL15 mutation causes streptomycin resistance and transposons Tn5, Tn10, and Tn9 cause resistance to kanamycin, tetracycline, and chloramphenicol, respectively. Some caution must be exercised with strain TJC121 to maintain the F′ element by selection for proline auxotrophy since the traD36 mutation greatly reduces conjugal transfer and, therefore, F− segregants may persist once they arise in mixed cultures grown in rich medium. Complete details on the construction of strain TJC121 and its properties relative to other strains will be described elsewhere (T. J. Close and R. D. Fenton, unpublished results).
Unenriched cDNA library production methods
Library list:
A total of 44 libraries are summarized in Table 1. Only three (SC010XXX, pSPORT1; SC013XXX, pSPORT2; and TA012XXX, pGAAD10) were based on a vector system other than λZAP (Stratagene), a consistency that facilitates analyses of the enrichment procedures (see below) that were used. Of the 41 λZAP-based libraries, 32 were original, unenriched libraries and 9 resulted from enrichment procedures starting with 6 libraries among the original 32. Two additional λZAP libraries were produced from Chinese Spring wheat by the authors (fully expanded leaf and early preanthesis spike) but were not sequenced due to the expectation of high redundancy with sequences from other libraries. More complete details on the composition of each library listed in Table 1 can be viewed using the HarvEST:Wheat software (http://harvest.ucr.edu) by selecting either the “stringent” or the “relaxed” assembly of “NSF Wheat.” Somewhat different summaries of information on these libraries are also available through GrainGenes, The Institute for Genomic Research (TIGR), and NCBI.
λZAP libraries:
For most libraries, plant tissues were collected, snap frozen, shipped on dry ice or stored at −80°, and then ground in liquid nitrogen prior to RNA extraction by University of California (UC) Riverside authors. Libraries AS040E1X, TA036E1X, TA037E1X, TA038E1X, and TT039E1X were produced at UC Riverside as a training activity, involving D. Zhang, E. D. Akhunov, P. Kianian, C. Otto, and K. Simons in addition to UC Riverside authors D. W. Choi, R. D. Fenton, A. Chin, and T. J. Close. Libraries AS067E1X, TA065E1X, and TA066E1X were produced by E. D. Akhunov in the Dvořák lab at UC Davis, and libraries TM011XXX, TM043E1X, and TM046E1X were produced by V. Echenique in the Dubcovsky lab at UC Davis. In some instances, total RNA was shipped to T. J. Close frozen in water or as an ethanol precipitate. For one library (TM011XXX), fresh tissues were ground in the presence of RNA extraction buffer and sand. For most libraries, total RNA was prepared by the hot phenol procedure described by Verwoerd et al. (1989). Other RNA extraction methods included TRIZOL (GIBCO BRL, Grand Island, NY; TM011XXX, TM043E1X, TM046E1X), CsCl gradient fractionation (TA047E1X, TA048E1X, TA056E1X, TA058E1X), Plant RNeasy (QIAGEN, Valencia, CA; TA055E1X), or phenol and LiCl (Altenbach 1998) followed by Plant RNeasy (TA001E1X, TA059E1X). For most libraries, poly(A) RNA was purified using the PolyATract mRNA Isolation System (Promega, Madison, WI). One library (TA001E1X) was produced by Stratagene, following poly(A) purification using an oligo(dT) column. cDNAs with EcoRI on the 5′-end and XhoI on the 3′-end were synthesized using the ZAP-cDNA synthesis kit (Stratagene). For most libraries, cDNAs >0.5 kb were selected by size fractionation via gel filtration. For some libraries, cDNAs were instead passed through a SizeSep 400 Spun Column (Amersham Biosciences, Piscataway, NJ) and then directionally cloned into the Uni-ZAP XR or, in one case (TM011XXX), the ZAP-Express vector (Stratagene). Following ligation to vector, recombinants were packaged using GigaPack III Gold packaging extract (Stratagene).
Prior to EST sequencing and normalization or subtraction, the primary λcDNA libraries were mass excised in vivo using the host strain XL1-Blue-MRF′ and the helper phage ExAssist (Stratagene) to produce pBluescript phagemid populations. In most cases, 1 × 106 pfu of Uni-ZAP λphage were used for mass excision of phagemid DNA and the multiplication by sibling phagemid production did not exceed 300-fold, although there were exceptions (see supplemental online material 1 at http://wheat.pw.usda.gov/pubs/2004/Genetics). In general, mass excision was performed at 37° for 3 hr with a high ratio of recipient cells to primary λ-phage and a high multiplicity of infection of helper phage to the same host cells. Cultures were centrifuged to create a cell pellet and supernatant, and the supernatant was heated at 70° for 20 min to create a “low amplification” phagemid population. For some libraries, a “high amplification” phagemid population was also produced by resuspending the cell pellet in 40 ml of fresh LB medium, continuing growth at 37° for an additional 16–20 hr, centrifugation to form a pellet and supernatant, and then heating of the supernatant at 70° for 20 min. Titers were determined using SOLR (Stratagene) or TJC121 host cells.
Other unenriched libraries:
Library SC010XXX was produced by G. E. Butler and J. P. Gustafson using SalI and NotI cloning sites in the vector pSPORT1 and the Escherichia coli host strain DH12S. Library SC013XXX was produced by G. E. Butler and J. P. Gustafson using SalI and NotI cloning sites in the vector pSPORT2 and the E. coli host strain DH12S. Library TA012XXX was produced by CLONTECH (Palo Alto, CA) for M. K. Walker-Simmons using a combination of random and oligo(dT) primers in vector pGAD10 and the E. coli host strain DH12S.
Evaluation of the quality of unenriched libraries:
For about one-half of the λZAP libraries, the average insert size was determined by restriction enzyme digestion of 36 randomly chosen clones and by plating phagemid transfectants of SOLR or TJC121 on medium containing X-GAL. The results consistently revealed a maximum of one or two “empty” clones, an average insert size of ∼1400 bp, and a frequency of dark blue colonies in the range of 3–5%, so this type of analysis was discontinued. More indicative measurements of fluctuations in library quality were the frequency and type of contamination observed among EST sequences (Table 1, “Junk %”). In general, there was a strong positive relationship between the size of the primary λ-library and the frequency of clones that carried fragments of the E. coli or phage λ-genome, which were assumed to be low-level contaminants inherent in all the cloning systems that were used. Other library contaminants were rRNA or fragments of the plant genome (detectable as sequences that were completely or nearly completely masked by a Triticeae repeat sequence data set).
Enriched library production methods
Normalized libraries (those depleted of abundant classes of cDNAs) and subtracted libraries (those depleted of abundant classes of cDNAs found in a library from the same type of tissue given a different treatment) were produced using the procedure of Soares and Bonaldo (1998) with modifications for the λUni-Zap cDNA library system (Table 2). The procedures are briefly described below.
Normalization
Preparation of single-stranded phagemid DNA for normalization:
A volume of 10 ml of a “low-amplification” phagemid population was combined with TJC121 cells at a cells-to-phage-mid ratio of 10:1, incubated at 37° for 15 min, and then added to 100 ml of LB broth containing 70 μg/ml ampicillin and 30 μg/ml kanamycin in a 2-liter flask. The flask was shaken for 3 hr at 37° to an OD600 of 0.3–0.4.
Method 1:
The VCSM13 helper phage (Stratagene) was then added to the culture at a MOI of 10:1 (phage to cell) and the culture was grown at 37° with vigorous shaking for 2 hr. The kanamycin concentration was then increased to 70 μg/ml and the culture was grown at 37° for 18 hr, centrifuged at 10,000 × g for 20 min, and the supernatant transferred into a fresh tube, followed by vacuum filtration through a 0.2-μm sterile filter. The filtrate was then used to prepare single-stranded (ss)-plasmid DNA as described by Vieira and Messing (1987).
Method 2:
TJC121 cells harboring double-stranded (ds)-phagemid DNA were pelleted by centrifugation at 1000 × g for 10 min and ds-phagemid DNA was extracted using the Wizard Plus Midipreps Kit (Promega). A 50-ng portion of ds-phagemid DNA was then electroporated into XL1-Blue-MRF′ cells using a CELL-PORATOR (GIBCO BRL). The culture was incubated at 37° in SOC medium for 1 hr, transferred to 100 ml of 2× YT broth containing 70 μg/ml ampicillin, and then grown at 37° to an OD600 of 0.2. The VCSM13 helper phage (Stratagene) was then added at a MOI of 10:1 (phage to cell), the culture was grown at 37° with vigorous aeration for 2 hr, kanamycin was added to 70 μg/ml, the culture was grown at 37° for an additional 8 hr, and then the supernatant was processed to yield ss-plasmid DNA as in method 1. The ss-plasmid DNA was treated with PvuII restriction enzyme for 2 hr at 37° and then purified through a hydroxyapatite (HAP) chromatography column (Bio-Rad, Hercules, CA) as per Soares and Bonaldo (1998).
Preparation of driver DNA for normalization:
A Taq PCR core kit (QIAGEN) was used for PCR amplification of cDNA inserts according to the manufacturer's instructions, with Q-solution added. Oligonucleotides SK (5′-CGCTCTAGAACTAGTGGATC-3′) and T7 (5′-TAATACGACTCACTATAGGGA-3′) (Stratagene) were used as primers and 1 ng of ss-phagemid DNA to be normalized was used as template. The PCR reaction was performed at 94° for 3 min followed by 20–25 cycles of 94° for 1 min, 56° for 2 min, and 72° for 3 min. PCR products were purified using a QIAGEN PCR purification kit (QIAGEN).
Reassociation hybridization for normalization:
In a 0.5-ml siliconized centrifuge tube the following components were present in 9 μl of 50% formamide: 50 ng of ss-plasmid DNA, 500 ng of driver DNA, and 10 μg each of the 5′ blocking oligonucleotides (5′-ACTCGAGGGGGGGCCCGGTACCCAATTCGCCCTATAGTGAGTCGTATTAC-3′), the 3′ blocking oligonucleotide (5′-CGCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGAATT-3′), and the poly(A) tail-blocking oligonucleotides [(dA)30]. The 5′ and 3′ blocking oligonucleotides were added to block the 5′ vector region at the T7 primer site and the 3′ vector region at the SK primer site. The oligonucleotide [(dA)30] was used to block regions in cDNA corresponding to the poly(A) tail of mRNA. The mixture was overlaid with 20 μl of mineral oil and heat denatured at 80° for 3 min, 1 μl of 10× hybridization buffer [1.2 m NaCl, 0.1 m Tris (pH 8.0), 50 mm EDTA, 10% SDS] was added, and the reassociation hybridization reaction was performed at 30° for a sufficient time to reach the calculated C0t value of 2.5 or 5 sec-moles/liter. The remaining ss-plasmid DNA after hybridization was isolated using a HAP chromatography column, followed by concentrating and desalting as described by Soares and Bonaldo (1998).
Conversion of ss-plasmid DNA into ds-plasmid DNA and transformation:
The ss-plasmid DNA recovered from HAP chromatography was converted into partial ds-plasmid DNA by primer extension using the M13 forward primer (5′-GTAAAACGACGGCCAGT-3′) and Sequenase Version 2.0 T7 DNA Polymerase (Amersham Biosciences). The partial duplexes were then electroporated into E. coli DH10B (GIBCO BRL) cells or XL1-Blue-MRF′ cells (Stratagene) using a CELL-PORATOR (GIBCO BRL) and propagated with ampicillin selection. The resulting library was the normalized library.
Subtraction
The procedure to produce all subtracted libraries except TA001E1S was essentially the same as normalization, except for the steps stated below. TA001E1S is a collection of ESTs from clones in the TA001E1X library that were identified by D. Laudencia-Chingcuanco, R. E. Miller, and P. Han in O. D. Anderson's laboratory. These authors used the top 40 genes that were highly expressed in the endosperm (identified from the available ESTs) as probes to hybridize with newly picked clones rearrayed on a nylon filter. Clones that hybridized with the 40 highly expressed genes were not sequenced.
Preparation of driver DNA for subtraction:
For the production of subtracted libraries TA005E2S, TA007E2S, and TA007E3S, cDNA inserts were amplified as driver DNA by PCR from nonstressed cDNA libraries TA006E1X (nonstressed seedling shoot) and TA008E1X (nonstressed seedling root) using the method described above for driver DNA for normalization. Equal amounts of PCR-amplified DNA from these two libraries were pooled. For subtracted library TA027E2S, driver DNA was the first-strand cDNA prepared with a ProSTAR first-strand RT-PCR kit (Stratagene) from the same tissue type from genotype TAMW101 that was used for the stressed library but under normal growth conditions.
Reassociation hybridization for subtraction:
The reassociation hybridization for subtraction followed the same procedure as for normalization except 2.5 μg of driver DNA was used and the hybridization was performed at 30° for a sufficient time to reach the calculated C0t value as 50 or 100 sec-moles/liter.
Source of libraries
Table 1 provides a minimal description of the sources of materials for each library. For most libraries, details on the biological materials used as a source of RNA and the specific roles of each person in library production are displayed within HTML files in the HarvEST:Wheat browser. Related information is also available from the GenBank accessions for ESTs from each library. Abbreviated descriptions of each unenriched library that provided ESTs and two additional libraries produced by our project that were not sequenced [TA025E1X (Chinese Spring unstressed green leaves at the full-tillering growth stage) and TA026E1X (Chinese Spring preanthesis spike)] are available in supplemental online material 2 at http://wheat.pw.usda.gov/pubs/2004/Genetics.
EST sequences
EST sequencing was performed in O. D. Anderson's laboratory from all libraries except TA027E2S, from which EST sequencing was performed by N. Klueva of H. T. Nguyen's lab. The mapping activity of the NSF wheat EST genomics project was supported by sequence processing, assembly, data accessioning and distribution, report generation, and probe distribution by the O. D. Anderson lab as described in Lazo et al. (2004).
The evaluation of libraries reported in the present article was based on the same original EST sequence data, but processed and assembled instead using the HarvEST pipeline (Close et al. 2004; http://harvest.ucr.edu). Unprocessed chromatograms for all EST sequences were provided to T. J. Close and S. Wanamaker by C. C. Crossman, S. Chao, and G. R. Lazo of O. D. Anderson's lab and from N. Klueva. Briefly, the major processing steps were as follows: (1) Phred version 0.020425.c (Ewing and Green 1998; http://www.phrap.org/) was applied to chromatograms to produce sequence and quality files, (2) cross_match version 0.990329 (http://www.phrap.org/) was used to mask cloning system sequences, (3) an in-house script “qvtrim” was used to synchronously remove low-quality regions outside of a sliding window with an average phred quality value of 17, reduce poly(T) or poly(A) ends to a maximum of 17 consecutive T or A nucleotides, and remove residual cloning system sequences, (4) sequences that were <100 bases after steps 1, 2, and 3 were discarded, (5) a filter based on frequency of four-nucleotide repeats was applied to remove additional ESTs that exceeded 100 bases but resulted from poor quality sequencing reactions, (6) orientations were determined using information on sequencing primer, high blastX orientation (default parameters), and presence of a poly(A) or poly(T), (7) blastN searches (Altschul et al. 1997; http://www.ncbi.nlm.nih.gov/BLAST/) were performed to identify contaminant sequences from E. coli, bacteriophage-λ, fungal sources, the repetitive portion of Triticeae genomes [Triticeae Repeat Sequence Database (TREP); http://wheat.pw.usda.gov/ggpages/ITMI/Repeats/index.shtml] or rRNA, (8) several chimera filters, including searches for interior sequences from the cloning system or ESTs that both begin with poly(T) and end with poly(A), were applied to individual EST sequences and chimeras were discarded, (9) assemblies were produced using a special version of CAP3 (Huang and Madan 1999) kindly provided by X. Huang (source date January 9, 2003) and recompiled by S. Wanamaker for the AMD64 processor, (10) contig orientations were determined using the ratio of forward and reverse EST sequences and the orientation of each EST used by CAP3, (11) additional chimera filters, including searches for contigs whose overall orientation cannot be resolved or whose consensus sequence both begins with poly(T) and ends with poly(A), were applied to assembled ESTs, and (12) assembly and chimera removal was repeated several times.
Assembly of the ∼100,000 ESTs by CAP3 using a 64-bit AMD Opteron processor equipped with 8 GB RAM (peak demand ∼2 GB) took ∼2 hr. All information from these processing steps was recorded in a Visual FoxPro database from which the HarvEST:Wheat software is an extraction product. Among the reports that can be generated by the HarvEST:Wheat software, the “Library Summary” is most pertinent to this article. The final column in the Library Summary report shows the portion of each library that is “unique” to it and is reproduced in Table 1. Our definition of “unique to library” is that a sequence from a given library was not assembled by CAP3 with any sequence from any other library.
In an effort to reduce further the impact of contaminations, both inadvertent and inherent to the library construction and enrichment processes, additional comparisons and screenings were done. All individual ESTs and all consensus sequences were periodically compared using blastX to the NCBI translated nonredundant (nr) database (ftp://ftp.ncbi.nih.gov/blast/db), the translated rice genome from TIGR (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules), and the TIGR translated Arabidopsis genome (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/a_thaliana/annotation_dbs). This information was useful for indirect methods of flagging problems. For instance, when it was noted that a very high proportion of unigenes from some enriched libraries was unique to those libraries and that those same libraries were contaminated with sorghum (Sorghum bicolor L.) cDNAs, the following screening sequence was devised to address this issue.
Five filtering steps were applied to EST sequences from libraries suspected of sorghum contamination to minimize consideration of sorghum sequences. Four of these steps were based on blast results (all with default settings), and the fifth was based on assembly using CAP3. All EST sequences from these libraries were compared using blastN with five sources of sequences: (1) sorghum ESTs available from the NCBI dbEST database; (2) EST sequences from all other libraries shown in Table 1; (3) unigene sequences from assembly 31 of HarvEST:Barley; (4) rice coding sequences available from TIGR (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_1.0/all_chrs/all.cds); and (5) the nr database from NCBI, where best hits were defined as “nrTriticeae” if the annotation contained any of the words “Triticum,” “Hordeum,” “wheat,” or “barley.” The “Triticeae BLAST” then consisted of the highest among blast score sources 2, 3, and 5. Filtering step 1 discarded ESTs with a sorghum blast score <150, a rice blast score <150, and a Triticeae blast score <400. Step 2 then discarded ESTs with no sorghum blast score, a rice blast score between 150 and 500, and no Triticeae at least 75 points greater than the rice blast score. Step 3 discarded ESTs with a sorghum blastN score from 150 to 350, but a Triticeae blast score <75 points more than the sorghum blast score. Step 4 discarded ESTs with sorghum blast scores >350 but a Triticeae blast score <50 more than the sorghum blast score. These four steps all eliminated ESTs on the basis of having insufficient confidence to conclude that they are “wheat,” although in fact many must actually be wheat ESTs. Step 5 eliminated ESTs that CAP3 assembled (in a test assembly that added suspect libraries) only with ESTs that were discarded by steps 3 or 4. Supplemental online material 3 at http://wheat.pw.usda.gov/pubs/2004/Genetics contains the blast results from these five filtering steps.
ESTs of putative nonwheat sequence, if identified early in the project pipeline, were not deposited in GenBank and not moved on to the deletion-bin-mapping stage of the project. If ESTs were identified by analysis subsequent to deposition in GenBank, they were accordingly recalled except for 49 putative sorghum probes that were mapped and further annotated to indicate their possible sorghum identity. The fact that an EST was mappable means that either it was actually a wheat sequence or, if it was a sorghum sequence, it represented a region so well conserved that it hybridized with a wheat ortholog and was accordingly a useful marker.
RESULTS
The total number of high-quality ESTs in the final assemblies from the 44 libraries that provided ESTs was 101,107, derived from 89,043 clones. When assembled using the CAP3 “stringent” settings (p = 95, d = 60, f = 100, h = 50), these ESTs comprised 16,740 contigs and 33,213 singletons for a total of 49,953 unigenes (Table 1). When assembled using CAP3 “relaxed” settings (p = 75, d = 200, f = 250, h = 90), these ESTs comprised 16,441 contigs and 20,588 singletons for a total of 37,029 unigenes (see HarvEST:Wheat at http://harvest.ucr.edu). The “stringent” settings achieve more complete isolation of individual paralogs and orthologs than do the relaxed settings. The relaxed assembly was most similar to the assembly discussed in Lazo et al. (2004). The “stringent” assembly used the same CAP3 settings as the barley assembly that was the basis of Affymetrix barley GeneChip content (Close et al. 2004). We refer mainly to the “stringent” assembly in the remainder of this article.
Assessment of the quality of cDNA libraries:
There are two aspects of library quality to consider: (1) how well the library construction methods were accomplished technically, in the sense of the percentage of each library that involved “junk” clones, and (2) to what extent the biological area of interest was represented by the EST sequences obtained. As described above, our goal was to create a diverse collection of ESTs representative of the full spectrum of tissues throughout the wheat life cycle. We began with 37 standard, unenriched libraries and investigated library enrichment strategies that yielded 9 derivative libraries (Table 1). With regard to a functional theme, we concentrated on reproductive development, including a range of environmental stresses that can affect transition to floral development, pollen development, fertilization, grain filling, grain quality, and seed dormancy.
Technical quality of libraries:
Junk clone frequencies are summarized in Table 1, with full details available in supplemental online material 1 at http://wheat.pw.usda.gov/pubs/2004/Genetics. In general, all unenriched libraries were of satisfactory technical quality. Only two (TA0012XXX, TA049E1X) contained >4% junk clones, while 25 contained <2% junk clones. The most common classes of junk clones in the unenriched libraries (and their percentages when highest) were plant genome fragments (TREP-masked; 14 libraries, 0.1–1.9%), rRNA-derived clones (12 libraries, 0.2–16.2%), chimeras (5 libraries, 0.2–0.4%), mitochondrion (3 libraries, 0.5–1.8%), and the E. coli genome (1 library, 0.3%). A clear trend with the λZAP libraries was that the frequencies of E. coli and λ-genome contaminants were diminished in parallel with an increase in the yield of primary λ-phage. Presumably, this was because of a basal level of these contaminants that was diluted by much higher numbers of successfully generated cDNAs. Even the smallest primary library (TM043E1X) had a very tolerable level (1.3%) of these two contaminants.
The frequency of junk clones in enriched libraries was higher than that in the unenriched libraries from which they were derived. This is to be expected when considering that various junk clones, other than rRNA, represent rare sequences. One example was subtracted library TA005E2S, which contained 0.8% E. coli clones, whereas no E. coli clones were found among the sequences from its parent library TA005E1X. Normalized library TA008E3N was another example, with 1.0% E. coli and λ-phage, compared to only 0.2% contamination by these two sources in the parent library TA008E1X. The most dramatic example was subtracted library TA007E2S, which contained 19.7% E. coli and 2.9% phage λ-genome, whereas the most frequent junk in its parent library TA007E1X was 0.4% plant genome fragments. The exceptionally high frequency of bacterial and phage clones in TA007E2S may reflect a bias caused by the amplification steps in this procedure. Three enriched libraries (TA001E2N, TA006E3N, TA007E3S), the latter two of which were produced in an effort to overcome problems with their predecessors (TA006E2N, TA007E2N), suffered from sorghum cDNA contamination, which was a much more severe problem (64–90% of the ESTs were discarded; see supplemental online material 3 at http://wheat.pw.usda.gov/pubs/2004/Genetics). Due to the presence of sorghum cDNA sequences and the filtering that may have discarded a number of wheat sequences, it was not possible to make an equivalent comparison of other classes of junk clones in the residual EST sequences from these three enriched libraries. The number of putative sorghum clones that were hybridized and assigned a genome location amounts to <1% of mapped clones.
Biological quality of libraries:
One measure of EST diversity in each library is the ratio of unigenes per clone. A strong trend was apparent in the unenriched libraries, where the most diverse libraries involved stress treatments and the least diverse were from maturing seed. The nine most diverse unenriched libraries were TA007E1X (seedling, cold treated), TA032E1X (early spike, heat treated), TA036E1X (mature leaf, drought treated), TA038E1X (crown, salt stressed), TA031E1X (flag leaf, heat treated), TA015E1X (seedling, heat treated), TA016E1X (crown, cold treated), TA037E1X (sheath, salt stressed), and TA005E1X (seedling, drought stressed). The three least diverse unenriched libraries were TA001E1X (endosperm), TA059E1X (grain), and TA017E1X (late spike). The numbers of unigenes per clone may exceed 1.0 (Table 1), since some clones were sequenced on both ends and there were many instances of two unjoined “unigenes” from the same clone. The operational definition of unigene in this case was that CAP3 maintained separate sequences.
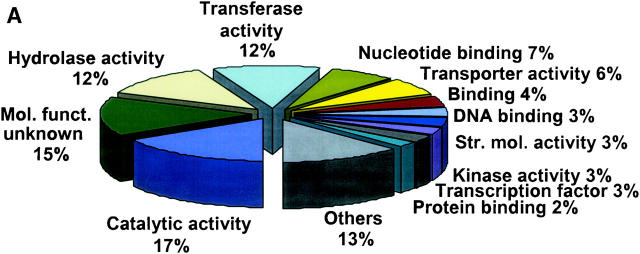
The gene-function diversity of library content is another measure of biological quality. Figure 1A illustrates functional categories of ESTs from the nine most diverse libraries on the basis of comparisons to Arabidopsis genes. The relative sizes of the functional categories in these libraries are very similar to the sizes when the ESTs from all libraries (not shown) are compared to Arabidopsis genes. These nine libraries provided only 12.8% of all clones sequenced (11,398 of 89,043), but contributed 20.0% of the total number of unigenes (9974 of 49,953), 17.6% of unigenes without Arabidopsis blast values of 1e-5 or better (2635 of 15,051), 21.0% of unigenes with Arabidopsis BLAST values of 1e-5 or better (7340 of 34,902), and 37.8% (4105 of 10,859) of all Arabidopsis genes identified by BLAST values of 1e-5 or better.
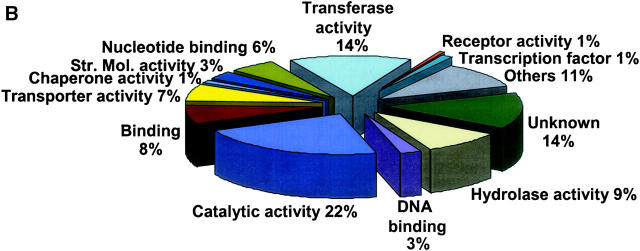
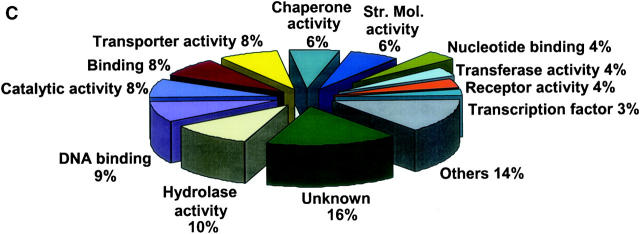
Figure 1.—
Representation of functional categories of genes. Each unigene consensus sequence was compared using blastX with the translated Arabidopsis genome (materials and methods) to identify the most closely related Arabidopsis gene. Those with an E-score 1e-5 or better were listed (one each) and the list analyzed using the gene ontology assignment tools available from The Arabidopsis Information Resource (at http://www.arabidopsis.org/index.jsp) to convert the Arabidopsis gene list to a distribution of gene ontology molecular functions. (A) Unigenes from only the nine most complex libraries. Of these, 73.6% had an Arabidopsis hit with an E-score of 1e-5 or better (7340 of 9974). (B) Unigenes composed of ESTs from two more clones from unenriched library TA027E1X. (C) Unigenes composed of ESTs from two more clones from subtracted library TA027E2S. The category “others” includes a number of additional activities that occur at low frequency and are not very different when comparing the data sets.
By definition, enrichment methods reduce abundant classes of cDNAs likely to be found in unenriched libraries, the potential gain being that rare clones can be more readily accessed. A measure of the biological quality of enriched libraries therefore is the extent to which the initial population of cDNAs has been shifted away from the most abundant classes. Table 3 shows that the most highly represented unigenes in all six of the libraries that served as parents of the enriched libraries were reduced substantially by the enrichment methods that were used. For example, the three most prevalent unigenes in unenriched library TA006E1X encode ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit, together composing 2.0% of all clones in this library (55 of 2728), while the frequency of these three unigenes was only 0.07% (1 of 1508) in ESTs from normalized library TA006E2N and 0% in TA006E3N (Table 3). However, the enrichment in TA006E2N was so extensive that siblings (identical clones that multiplied from a single progenitor during the enrichment) were frequent within this enriched library (data not shown). Because of this excessive bias, an increased C0t value (see materials and methods) was used to produce additional libraries. This modification of the normalization procedure yielded more satisfactory results with normalized library TA008E3N, derived from TA008E3X. All of the subtracted libraries had an increased frequency of novel sequences and clones encoding proteins known to be related to stress responses such as ubiquitin, glutathione S-transferase, pathogen-related protein, peroxisome-type ascorbate peroxidase, subtilisin-chymotypsin inhibitor, RAB15B, and light-inducible protein (Table 4). Together, ESTs from the five subtracted libraries (TA001E1S, TA005E2S, TA007E2S, TA007E3S, and TA027E2S) contributed 1182 unigenes that were not represented by ESTs from any of the other libraries, which is 48.7% of all unigenes (2429) represented by ESTs in these five libraries. Of these 2429 unigenes from these five libraries, 708 do not have an Arabidopsis blastX hit of 1e-5 or better, and 446 of these 708 also do not have a rice blastX hit of 1e-5 or better. Therefore, 18.4% (446 of 2429) of the unigenes from the five subtracted libraries can be considered “novel,” compared with 14.7% (701 of 4764) novel unigenes by this same definition among their four parent libraries (TA001E1X, TA005E1X, TA007E1X, and TA027E1X).
TABLE 3.
Most prevalent unigenes in source library and after normalization or subtraction
| Enriched library
|
Second enriched library |
||||||
|---|---|---|---|---|---|---|---|
| Source library designation | Unigene ID no. | Frequency (%) |
Putative function | Designation | (%) | Designation | (%) |
| TA001E1X | 11501 | 1.06 | Purothionin A-I precursor (T. aestivum) | TA001E2N | 0 | TA001E1S | 0.901 |
| 4446 | 0.932 | γ-Gliadin (T. aestivum) | 0 | 0 | |||
| 14856 | 0.932 | α-Amylase/trypsin inhibitor CM3 precursor | 0 | 0 | |||
| 2093 | 0.836 | Purothionin A-I precursor (T. aestivum) | 0 | 0 | |||
| 15272 | 0.804 | α-Amylase inhibitor Ima1 precursor (T. aestivum) | 0 | 0.45 | |||
| TA005E1X | 2442 | 0.686 | β-Glucosidase (S. cereale) | TA005E2S | 0 | ||
| 14778 | 0.571 | S-Adenosylmethionine decarboxylase precursor (T. aestivum) | 0 | ||||
| 10966 | 0.457 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (T. aestivum) |
0.725 | ||||
| 8073 | 0.457 | Lipid transfer protein 3 (T. aestivum) | 0.121 | ||||
| 2141 | 0.457 | No hit | 0.966 | ||||
| TA006E1X | 10028 | 0.806 | Ribulose bisphosphate carboxylase/oxygenase small subunit (T. aestivum) | TA006E2N | 0.066 | TA006E3N | 0 |
| 2538 | 0.587 | Ribulose bisphosphate carboxylase/oxygenase small subunit (T. aestivum) | 0 | 0 | |||
| 10966 | 0.623 | Ribulose bisphosphate carboxylase/oxygenase small subunit (T. aestivum) | 0 | 0 | |||
| 8073 | 0.587 | Lipid transfer protein 3 (T. aestivum) | 0.199 | 0.152 | |||
| 2635 | 0.55 | α-1,6-Mannosyl-glycoprotein 2-β-N-acetylglucosaminyltransferase (Xenopus laevis) |
0 | 0 | |||
| TA007E1X | 1680 | 0.678 | Glycine-rich RNA-binding protein, low temperature responsive (H. vulgare) |
TA007E2S | 0.204 | TA007E3S | 0 |
| 10966 | 0.484 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (T. aestivum) |
0 | 0.815 | |||
| 1778 | 0.387 | Heat-shock protein cognate 70 (O. sativa) | 0 | 0 | |||
| 16074 | 0.387 | Chlorophyll a/b-binding protein WCAB precursor (T. aestivum) | 0 | 0.204 | |||
| TA008E1X | 3906 | 0.358 | Glutamine-dependent asparagine synthetase 1 (H. vulgare) | TA008E3N | 0.022 | ||
| 5624 | 0.358 | Glutathione S-transferase (T. aestivum) | 0.09 | ||||
| 15768 | 0.358 | Putative calcium-transporting ATPase 8, plasma membrane-type (O. sativa) | 0 | ||||
| TA027E1X | 14030 | 1.725 | No hit | TA027E2S | 0.505 | ||
| 2656 | 0.9 | Ribulose 1,5-bisphosphate carboxylase activase isoform 1 (H. vulgare) | 0 | ||||
| 15313 | 0.75 | Metallothionein (T. aestivum) | 0.505 | ||||
| 7721 | 0.6 | Ribulose bisphosphate carboxylase activase B (T. aestivum) | 0 | ||||
| 4556 | 0.525 | Rubisco activase β-form precursor (Deschampsia antarctica) | 0 | ||||
Unigene ID numbers, unigene frequencies per library, and unigene functions are from HarvEST:Wheat 1.09 (http://harvest.ucr.edu/HWheat109.exe).
TABLE 4.
Most prevalent unigenes in subtracted cDNA librariesa
| Library designation |
Unigene ID no. |
Percentage in library |
Putative function | Parent library |
Percentage of parent library |
|---|---|---|---|---|---|
| TA001E1S | 4744 | 1.351 | Low-molecular-weight glutenin (T. aestivum) | TA001E1X | 0.129 |
| 14911 | 1.351 | α/β-Gliadin A-II precursor (T. aestivum) | 0.257 | ||
| 15267 | 1.351 | Succinate dehydrogenase subunit 3 (T. aestivum) | 0.000 | ||
| 738 | 0.901 | Grain softness protein 1a, 15K (clone TSF69) - GSP-1a (T. aestivum) | 0.514 | ||
| 1661 | 0.901 | Probable ribosomal protein S16 (O. sativa) | 0.000 | ||
| TA005E2S | 1521 | 1.329 | No hit | TA005E1X | 0.000 |
| 2141 | 0.966 | No hit | 0.457 | ||
| 1437 | 0.725 | No hit | 0.000 | ||
| 10966 | 0.725 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (T. aestivum) | 0.457 | ||
| 15768 | 0.604 | Putative Calcium-transporting ATPase 8, plasma membrane-type (O. sativa) | 0.000 | ||
| TA007E2S | 1372 | 1.018 | ATP-dependent Clp protease proteolytic subunit, putative (A. thaliana) | TA007E1X | 0.000 |
| 1364 | 0.815 | No hit | 0.000 | ||
| 1385 | 0.815 | CBL-interacting protein kinase 12 (CIPK12; A. thaliana) | 0.000 | ||
| 1387 | 0.815 | No hit | 0.000 | ||
| 1357 | 0.611 | Putative serine/threonine protein kinase (O. sativa) | 0.000 | ||
| TA007E3S | 14030 | 2.444 | No hit | 0.000 | |
| 1857 | 1.833 | RNase S-like protein (O. sativa) | 0.000 | ||
| 897 | 1.018 | Fructose-bisphosphate aldolase | 0.097 | ||
| 10966 | 0.815 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (T. aestivum) | 0.484 | ||
| 1874 | 0.611 | Lipid transfer protein precursor (T. aestivum) | 0.097 | ||
| TA027E2S | 13578 | 1.211 | rab15B protein (T. aestivum) | TA027E1X | 0.000 |
| 54 | 0.605 | rab15B protein (T. aestivum) | 0.300 | ||
| 46 | 0.505 | Putative acid phosphatase (H. vulgare) | 0.450 | ||
| 14030 | 0.505 | No hit | 1.725 | ||
| 15313 | 0.505 | Metallothionein (T. aestivum) | 0.750 |
Unigene ID numbers, unigene frequencies per library, and unigene functions are from HarvEST:Wheat 1.09 (http://harvest.ucr.edu/HWheat109.exe).
Another measure of the effectiveness of the enrichment methods was the extent to which the distribution of molecular functions was changed by the enrichment procedures. For example, Figure 1B shows the distribution of the most abundant clones (with good Arabidopsis hits) in unenriched library TA0027E1X and Figure 1C shows a quite different distribution within subtracted library TA0027E2S. Genes involved in transferase and catalytic activity (housekeeping) were considerably reduced in the subtracted library. In contrast, enriched genes included those encoding DNA-binding proteins, chaperones, structural molecular components, receptors, and transcription factors. A total of 23.0% (270 of 1173) of unigenes from library TA027E1X and 24.2% (214 of 884) from TA027E2S that did not have good Arabidopsis hits were not considered in the analyses shown in Figure 1, B and C. Another interesting measure of the enriched libraries involved the frequency of library-unique clones (Table 1), that is, clones that were in CAP3 unigenes composed solely of ESTs from a given library. Generally, there was not a favorable change in this frequency in the enriched libraries, except in the case of libraries derived from TA001E1X, which, as stated above, had the smallest number of unigenes per clone of all unenriched libraries. Due to the measures that were taken to remove sorghum cDNA contamination in TA001E2N, TA006E3N, and TA007E3S, the percentage of library-unique ESTs in these three libraries in Table 1 may not be accurate.
DISCUSSION
Our EST project began when essentially no Triticeae ESTs had yet been publicly released. As described in Lazo et al. (2004), the main objective of our library construction effort was to provide a stream of materials for EST sequencing that would be sufficient to identify at least 10,000 useable probes for deletion bin mapping. Within this objective, emphasis was given to wheat reproductive development, particularly the end-product—the grain. Therefore, the libraries produced were biased toward factors that influence grain yield and grain quality, especially abiotic stress. While most of the ESTs were derived from random sequencing of standard cDNA libraries, some effort was expended to produce libraries that were depleted of abundant clones and enriched for rare clones.
The normalization and subtraction methods employed substantially reduced the frequency of abundant cDNAs, while increasing the frequency of typically more rare cDNAs encoding transcription factors, receptor-like proteins, and others. In addition, the frequencies of “novel” cDNAs without high BLAST hits were higher in normalized and subtracted libraries than in their parent libraries. These “novel” ESTs seem likely to be more rarely expressed genes on the basis of the rationale that highly and moderately expressed wheat genes were more likely to have been already identified as expressed genes in other organisms (Green et al. 1993; Newman et al. 1994). Also, the subtracted libraries derived from parent libraries involving cold- and drought-stressed tissue were enriched for sequences that have been typically found in response to abiotic stresses (Table 4). The enrichment method that was used to identify rare clones in TA001E1X to generate a collection of ESTs in “library” TA001E1S also reduced the frequency of highly abundant clones, although the small sample size of 222 clones analyzed from this library hampered detailed interpretations. Taken together, the data established the potency of the procedures that were used for eight of the enriched libraries and indicated that the on-filter screening method was effective. A valid question, however, is whether the reduction of redundancy by any method was cost effective. How many additional ESTs could have been sequenced from unenriched libraries, and how many of these would have been rare sequences, for the same expenditures that were used for materials and labor to perform the enrichment methods? At the outset of our project, the balance between savings in sequencing costs and higher expenditures on materials and labor for enrichment methods seemed to favor the enrichment approaches. However, at today's reduced cost of EST sequencing (∼$3.50 per bidirectionally sequenced cDNA), there does not appear to be an advantage of cost efficiency in the production of enriched libraries in newly initiated EST projects. Also, some often-overlooked considerations are that redundancy within EST sequences is necessary to gain confidence in the accuracy of unigene “consensus” sequences and valuable for electronic comparisons of gene expression profiles. Without the advantage of replicate EST sequences, downstream applications that rely on oligonucleotide design and nucleotide polymorphisms can suffer high inefficiencies.
Enrichment methods would seem to be most appropriate for the development of EST collections that reach a stage where only ∼5–10% of randomly sequenced cDNAs reveal library-unique genes. At this diminished rate of novel sequence discovery, one can expect to observe a significant gain from enrichment procedures. As an example, one can calculate the break-even point on a cost of $10,000 for materials and salary necessary to produce high-quality enriched libraries. If the enrichment method increases the discovery rate of library unique genes fivefold to accomplish an increment of 20% (from 5 to 25%), then at a cost of $3.50/bidirectional sequence the expenditure of $10,000 to produce enriched libraries would be recouped from savings in sequencing costs when ∼14,000 clones have been sequenced from the enriched libraries. The break-even point would be a larger number of clones if the sequencing cost per clone were less, outlay for enriched libraries higher, or incremental percentage gain in library-unique clones lower than the values used in this example.
In addition, one must carefully consider the limitations and possible pitfalls of enrichment methods. The normalization and subtraction methods that were employed were time consuming and required multiple steps, several of which can potentially be the cause of sibling bias or contamination. The procedures included the following precautionary measures, not all of which were always satisfactory. First, the extent of library amplification during normalization or subtraction procedures was minimized to avoid overrepresentation of short cDNAs or underrepresentation of long cDNAs, because the propagation of cDNA clones tends to vary with plasmid length. Second, PCR-amplified DNA was prepared using reagents that were compatible with high-GC-content templates, again to avoid underrepresentation of some cDNAs. Third, an attempt was made to optimize the reassociation hybridization C0t value. An example of less-than-optimal conditions resulted in the library TA006E2N, which seemed to be so extensively enriched for rare cDNAs compared to the parent library TA006E1X that moderately expressed sequences were underrepresented and a high frequency of siblings of what were originally rare cDNAs was observed. Fourth, and perhaps most important, cautions were taken to avoid cross-contamination between different libraries since only trace amounts of template library DNAs are used to generate enriched libraries. Nevertheless, what must have been initially trace levels of contaminating sorghum cDNAs became the predominant species of cDNA in three enriched libraries. Furthermore, as noted by others (Soares 1997), redundant sequencing of moderately abundant cDNA clones was a persistent problem even for normalized cDNA libraries. Another general issue is that low-abundance cDNAs may be eliminated because of repetitive sequences shared by nonhomologous cDNAs. In retrospect, these various risk factors should be carefully weighed in the decision-making process when allocating resources for cDNA library enrichment methods for gene discovery initiatives. Our project gained significantly from access to rare sequences through the various enriched libraries that were produced, but this path was not a simple one nor was it as bountiful as we imagined it would be.
In summary, our cDNA library production effort far exceeded the goal of providing 10,000 probes suitable for mapping. Furthermore, our interest in major environmental-stress factors that affect reproductive development, including heat, drought, salinity, and low temperature, was fortuitous, since libraries from stressed materials emerged as an efficient source of diverse gene representation.
Acknowledgments
We are grateful to Bento Soares at the University of Iowa for kindly training and helping D.Z. on library normalization and subtraction methods. We also thank C. C. Hsia, Y. Kang, C. J. Rausch, C. L. Seaton, J. C. Tong, C. Londeore, J. Pham, and J. Woo in O. D. Anderson's lab at the U.S. Department of Agriculture-Agricultural Research Service, Albany, California, for helping to sequence clones from all libraries other than TA027E2S, which was sequenced and curated by N.K. We also thank Gary Zank and Tony Vu at the University of California, Riverside, for access to the Beowulf cluster in the Institute of Geoplanetary Physics and Thomas Girke at the University of California, Riverside, Bioinformatics Core Facility for timely advice and helpful derivatives of The Institute for Genome Research rice and Arabidopsis genome annotations and for partnership on the development of a Beowulf cluster in the Genomics Institute. This material is based upon work supported by the National Science Foundation Cooperative Agreement no. DBI-9975989.
References
- Adams, M. D., J. M. Kelley, J. D. Gocayne, M. Dubnick, M. H. Polymeropoulos et al., 1991. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252: 1651–1656. [DOI] [PubMed] [Google Scholar]
- Adams, M. D., A. R. Kerlavage, C. Fields and J. C. Venter, 1993. 3,400 expressed sequence tags identify diversity of transcripts in human brain. Nat. Genet. 4: 256–267. [DOI] [PubMed] [Google Scholar]
- Adams, M. D., A. R. Kerlavage, R. D. Fleischmann, R. A. Fuldner, C. J. Bult et al., 1995. Initial assessment of human gene diversity and expression patterns based on 83 million nucleotides of cDNA sequence. Nature 377(Suppl.): 3–17. [PubMed] [Google Scholar]
- Altenbach, S., 1998. Quantification of individual low-molecular-weight glutenin subunit transcripts in developing wheat grains by competitive RT-PCR. Theor. Appl. Genet. 97: 413–421. [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and psi-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M. D., and I. J. Leitch, 1995. Nuclear DNA amounts in angiosperms. Ann. Bot. 76: 113–176. [Google Scholar]
- Bennett, M. D., I. J. Leitch, H. J. Price and J. S. Johnston, 2003. Comparisons with Caenorhabditis (∼100 Mb) and Drosophila (∼175 Mb) using flow cytometry show genome size in Arabidopsis to be ∼157 Mb and thus 25% larger than the Arabidopsis Genome Initiative estimate of ∼125 Mb. Ann. Bot. 91: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close, T. J., S. Wanamaker, R. A. Caldo, S. M. Turner, D. A. Ashlock et al., 2004. A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol. 134: 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley, E. J., V. Nduati, J. L. Gonzalez-Hernandez, A. Mesfin, M. Trudeau-Spanjers et al., 2004. A 2600-locus chromosome bin map of wheat homoeologous group 2 reveals interstitial gene-rich islands and colinearity with rice. Genetics 168: 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using Phred II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Gale, M. D., and K. M. Devos, 1998. Plant comparative genetics after 10 years. Science 282: 656–659. [DOI] [PubMed] [Google Scholar]
- Goff, S. A., D. Ricke, T. H. Lan, G. Presting, R. Wang et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Green, S., D. Lipman, L. Hillier, R. Waterson, D. States et al., 1993. Ancient conserved regions in new gene sequences and the protein databases. Science 259: 1711–1716. [DOI] [PubMed] [Google Scholar]
- Hossain, K. G., V. Kalavacharla, G. R. Lazo, J. Hegstad, M. J. Wentz et al., 2004. A chromosome bin map of 2148 expressed sequence tag loci of wheat homoeologous group 7. Genetics 168: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., and A. Madan, 1999. CAP3: a DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo, G. R., S. Chao, D. D. Hummel, H. Edwards, C. C. Crossman et al., 2004. Development of an expressed sequence tag (EST) resource for wheat (Triticum aestivum L.): EST generation, unigene analysis, probe selection and bioinformatics for a 16,000-locus bin-delineated map. Genetics 168: 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkiewicz, A. M., L. L. Qi, B. S. Gill, B. Echalier, S. Chao et al., 2004. A 2500-locus bin map of wheat homoeologous group 5 provides new insights on gene distribution and colinearity with rice. Genetics 168: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, E. M., J. Liu, L. Gao and J. F. McDonald, 2002 Long terminal repeat retrotransposons of Oryza sativa. Genome Biol. 3: research0053.1–research0053.11. [DOI] [PMC free article] [PubMed]
- Miftahudin, K. Ross, X.-F. Ma, A. A. Mahmoud, J. Layton et al., 2004. Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold, J. D., R. A. Greene, C. E. Bermudez-Kandianis, C. M. La Rota, H. Edwards et al., 2004. Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 168: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, T., F. Bruijin, P. Green, K. Keegstra, H. Kende et al., 1994. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106: 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J. H., H. Zadeh, G. R. Lazo, J. P. Gustafson, S. Chao et al., 2004. Chromosome bin map of expressed sequence tags in homoeologous group 1 of hexaploid wheat and homoeology with rice and Arabidopsis. Genetics 168: 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L. L., B. Echalier, B. Friebe and B. S. Gill, 2003. Molecular characterization of a set of wheat deletion stocks for using in chromosome bin mapping of ESTs. Funct. Integr. Genomics 3: 39–55. [DOI] [PubMed] [Google Scholar]
- Qi, L. L., B. Echalier, S. Chao, G. R. Lazo, G. E. Butler et al. 2004. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa, H. S., M. Dilbirligi, D. Sidhu, M. Erayman, D. Sandhu et al., 2004. Deletion mapping of homoeologous group 6-specific wheat expressed sequence tags. Genetics 168: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, M. B., 1997. Identification and cloning of differentially expressed genes. Curr. Opin. Biotechnol. 8: 542–546. [DOI] [PubMed] [Google Scholar]
- Soares, M. B., and M. F. Bonaldo, 1998 Constructing and screening normalized cDNA libraries, pp. 49–157 in Genome Analysis: A Laboratory Manual, Vol. 2, edited by B. Birren, E. D. Green, S. Klapholz, R. M. Myers and J. Roskams. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Verwoerd, T. C., B. M. Dekker and A. Hoekema, 1989. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, J., and J. Messing, 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153: 3–11. [DOI] [PubMed] [Google Scholar]
- Yu, J., S. Hu, J. Wang, G. K. Wong, S. Li et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. [DOI] [PubMed] [Google Scholar]