Abstract
The focus of this study was to analyze the content, distribution, and comparative genome relationships of 996 chromosome bin-mapped expressed sequence tags (ESTs) accounting for 2266 restriction fragments (loci) on the homoeologous group 3 chromosomes of hexaploid wheat (Triticum aestivum L.). Of these loci, 634, 884, and 748 were mapped on chromosomes 3A, 3B, and 3D, respectively. The individual chromosome bin maps revealed bins with a high density of mapped ESTs in the distal region and bins of low density in the proximal region of the chromosome arms, with the exception of 3DS and 3DL. These distributions were more localized on the higher-resolution group 3 consensus map with intermediate regions of high-mapped-EST density on both chromosome arms. Gene ontology (GO) classification of mapped ESTs was not significantly different for homoeologous group 3 chromosomes compared to the other groups. A combined analysis of the individual bin maps using 537 of the mapped ESTs revealed rearrangements between the group 3 chromosomes. Approximately 232 (44%) of the consensus mapped ESTs matched sequences on rice chromosome 1 and revealed large- and small-scale differences in gene order. Of the group 3 mapped EST unigenes ∼21 and 32% matched the Arabidopsis coding regions and proteins, respectively, but no chromosome-level gene order conservation was detected.
THE success of cultivated wheat (Triticum aestivum L.) as a worldwide food crop can be attributed to its diverse genetic resources, unusually wide adaptation, and broad utility to humankind. Hexaploid wheat is a self-pollinated species composed of three related diploid genomes designated AA, BB, and DD, each of which has seven chromosomes. These three genomes are descendants of extant diploid species that have been widely used as germplasm in genetic studies and crop improvement. The polyploid nature of the wheat genome provides considerable genetic buffering that allows aneuploids and deletions to remain viable and fertile. Sears's (1954) pioneering work in the development of various kinds of aneuploids, such as nulli-tetrasomic and ditelosomic lines, has been instrumental in revealing the complex genome structure of wheat as well as the genetic control of numerous traits. More recently, these aneuploids have been used for mapping isozymes and molecular markers (for a summary see Hart et al. 1993). Wheat deletion lines have also been used extensively for mapping (Delaney et al. 1995; Endo and Gill 1996; Qi et al. 2003). Such lines are genetic stocks with deletions of one or more chromosome segments caused by a gene present on Aegilops cylindrica host chromosome 2C known as a gametocidal gene. In the monosomic condition of wheat chromosomes with the Ae. cylindrica chromosome 2C, this gene causes random breakage and loss of parts of wheat chromosomes (Endo 1988). The terminal deletions can then be isolated in a homozygous condition by self-pollination and cytological examination. More than 436 deletions have been isolated and used for mapping molecular markers in the wheat genome (Endo and Gill 1996).
The homoeologous group 3 chromosomes are among the largest in physical size (Dvořák et al. 1984; Gill et al. 1991). Gene density has been found to be higher at the ends of these chromosomes (Gill et al. 1993; Lukaszewski and Curtis 1993; Moore 2000) with the exception of 3DS where there is an interstitial region with higher gene density than at the end (Qi et al. 2004). A number of important traits are known to be controlled by loci on these chromosomes, including grain yield and seed weight (Berke et al. 1992a,b), kernel color (Sears 1944; Metzger and Silbaugh 1970; Nelson et al. 1995), chromosome pairing (Sears 1982; Dong et al. 2002), seed dormancy (Osa et al. 2003), glume blotch (Stagonospora nodorum) resistance (Ma and Hughes 1995), stem rust (Puccinia graminis f. sp. tritici) resistance (Hare and McIntosh 1979), leaf rust (P. recondita) resistance (McIntosh et al. 1977), and a number of isozymes (Hart et al. 1993; McIntosh et al. 1998).
Among the grasses, comparisons with chromosomes from other species that are related to wheat group 3 indicate that this group is the most conserved in gene content and order. The wheat group 3 chromosomes are most closely related to barley (Hordeum vulgare L.) chromosome 3 (Devos and Gale 1993; Nelson et al. 1995), rye (Secale cereale L.) chromosome 3 (Devos et al. 1992), rice (Oryza sativa L.) chromosome 1 (Devos et al. 1992; Ahn et al. 1993; Kurata et al. 1994; Van Deynze et al. 1995b), maize (Zea mays L.) chromosomes 3 and 8 (Van Deynze et al. 1995b; Wilson et al. 1999), sorghum (Sorghum bicolor L.) chromosome 3 (Whitkus et al. 1992; Klein et al. 2003), and diploid oat (Avena spp.) chromosomes C and G (Van Deynze et al. 1995a). However, recent comparative maps based on DNA sequence analyses reveal considerable rearrangement, insertions, and deletions between wheat and rice (Sorrells et al. 2003; La Rota and Sorrells 2004).
A project funded by the National Science Foundation had the primary goal of studying the structure and function of the expressed portion of the wheat genome by mapping wheat unigenes (http://wheat.pw.usda.gov/NSF). cDNA clones from expressed sequence tags (ESTs) representing unigenes were used for mapping in the wheat genome, using a set of the wheat deletion stocks. More than 100,000 ESTs from various tissues of wheat at different stages of development have been sequenced and wheat cDNA clones mapped by this project to deletion bins as of March 17, 2003, were used for further analysis. Accompanying articles describe the cDNA library development (Zhang et al. 2004) and EST sequencing, unigene assembly, and cDNA probe selection (Lazo et al. 2004). A summary article by Qi et al. (2004) gives an overview of the deletion bin-mapping results at the genome level. The characterization of the deletion lines used in this study was described previously by Qi et al. (2003).
The purpose of this study was to conduct an in-depth analysis of the results for the group 3 chromosomes and relate them to previous knowledge regarding these chromosomes and their relationship to the model species, rice and Arabidopsis thaliana (L.) Heynh. Data, figures, and supporting analyses for this research can be obtained from supplemental online material at (http://wheat.pw.usda.gov/pubs/2004/Genetics).
MATERIALS AND METHODS
Genetic stocks:
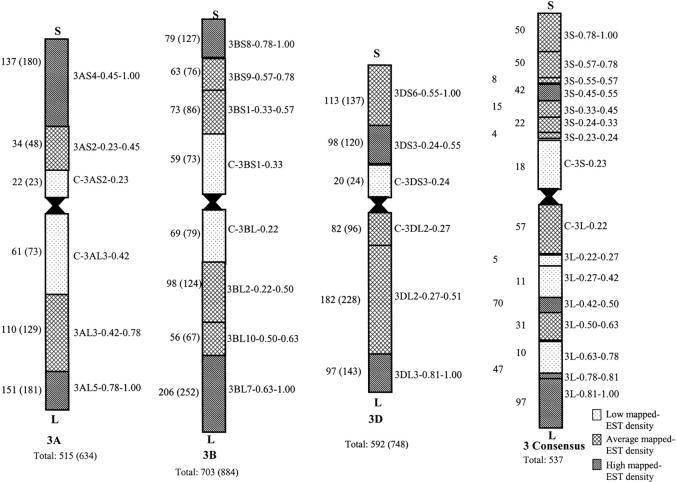
Chromosome bin mapping was performed by hybridizing a cDNA clone corresponding to each EST selected from the unigene set (referred to hereafter as mapped EST) to a Southern blot of DNA from a panel of wheat genetic stocks. The panel was composed of 101 lines with specific regions of chromatin deleted (Endo and Gill 1996; Qi et al. 2003) obtained from B. S. Gill (Kansas State University) and the nulli-tetrasomic and ditelosomic aneuploids (Sears 1954; Sears and Sears 1978) obtained from the U.S. Department of Agriculture (USDA)-Sears collection of wheat genetic stocks (USDA-Agricultural Research Service, University of Missouri). These genetic stocks allowed for the assignment of fragments to bins delineated by the deletion breakpoints. The aneuploid stocks used in the present study included three nullisomic-tetrasomic (NT) lines (N3AT3D, N3BT3D, and N3DT3B) and four ditelocentric lines [Dt3AL (3AS3), Dt3BL, Dt3DS, and Dt3DL] (Sears 1954, 1966; Sears and Sears 1978). Four, six, and four deletions were used to characterize chromosomes 3A, 3B, and 3D, respectively (Figure 1).
Figure 1.—
Graphical representation of the mapped ESTs and loci and mapped-EST densities by deletion bin for each of the group 3 chromosomes and the consensus chromosome bin map. Areas of low and high mapped-EST density had χ2 P < 0.01. Numbers without parentheses correspond to the number of ESTs mapped to the bin of the chromosome as a whole. Numbers within parentheses represent the total restriction fragments (loci) mapped to the bin. A more detailed summary of mapped ESTs and corresponding loci statistics can be found in supplemental online material (http://wheat.pw.usda.gov/pubs/2004/Genetics).
Chromosome bin mapping:
Mapping was conducted in 10 labs using similar protocols. Procedures used for probe selection, DNA isolation, EcoRI endonuclease digestion, gel electrophoresis, Southern hybridization, scoring, and confirmation were as described by Lazo et al. (2004). For homoeologous group 3, 996 ESTs representative of 987 unigenes (http://wheat.pw.usda.gov/NSF/progress_mapping.html) were mapped on the deletion lines, resulting in 2266 mapped restriction fragments, referred to hereafter as loci, on the three homoeologous chromosomes and were used for the remainder of the analyses. Loci were placed into a bin by relating the absence of a band in a particular deletion line to the portion of the chromosome distal to the breakpoint. The regions between adjacent breakpoints were referred to as bins.
EST distribution statistics:
Validated ESTs with at least one group 3 map position, and related confirmed loci mapping to a specific deletion bin, were used for analyzing mapped EST and locus distributions. Loci mapped to a chromosome, chromosome arm, or a portion of a chromosome arm containing more than one deletion bin were excluded from the analyses. Chi-square (χ2) analysis was used to examine EST and locus distributions between group 3 chromosomes by comparing observed and expected values for each individual chromosome against the observed and expected values for the rest of the group. In all cases statistical significances for EST density did not differ from those for locus density so probability values were listed for EST statistics except where noted. Expected EST and locus values were calculated from the total number of ESTs and loci mapped to the group and weighted by physical chromosome size (Gill et al. 1991). Comparisons between long and short arms were based on the number of ESTs and loci mapped to the chromosome with expected values weighted by chromosome arm physical size (Gill et al. 1991). χ2 analyses of EST and locus density along the group 3 chromosomes used observed values against expected values calculated by multiplying the total ESTs or loci for the arm by the deletion bin fraction length (FL; Endo and Gill 1996), assuming equal distribution over a chromosome arm. In all χ2 distribution analyses P < 0.01 was considered significant. Supplementary information can be obtained from supplemental online material at (http://wheat.pw.usda.gov/pubs/2004/Genetics).
Development of the group 3 consensus chromosome bin map:
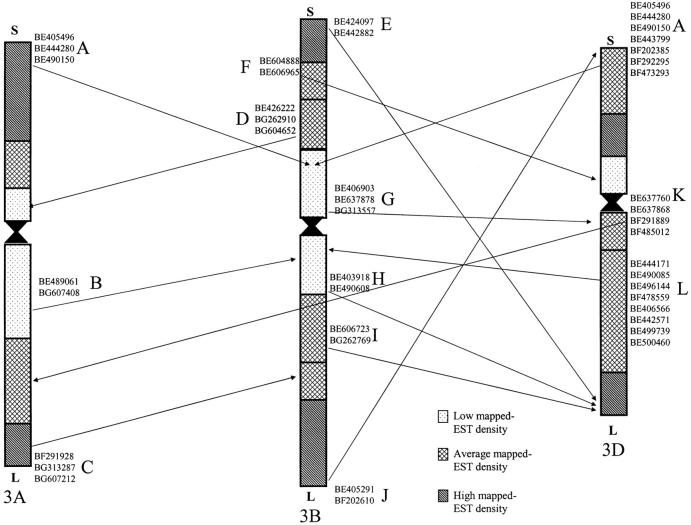
To develop the group 3 consensus chromosome bin map those ESTs detecting two or more validated loci on different group 3 chromosomes were obtained. These data were then combined in a pairwise fashion, in all combinations of chromosomes, and ordered on the basis of corresponding deletion bin order. The finished pairwise comparisons were then condensed into a single nonredundant deletion bin consensus map. Those ESTs detecting loci mapping to nonoverlapping bins on homoeologous chromosomes were flagged and termed an anomaly. The original deletion mapping Southern hybridization film images for each anomalous probe were checked and either confirmed or corrected by the mapping lab. ESTs mapped to two overlapping deletion bin loci and one anomalous locus were placed into the consensus bin corresponding to the overlapping loci. In those instances where an EST had only two conflicting loci, the EST was grouped with any similar anomalies and, if possible, ordered by rice chromosome 1 blastN best-hit order. Those anomalies that could not be positioned were placed into a consensus bin corresponding to one of the deletion bin loci (Figure 2). ESTs that mapped to overlapping intervals across two or more chromosomes were used to define the consensus bins. For example, wheat EST BF202444 mapped to the deletion bins 3AS4-0.45-1.00, 3BL1-0.33-0.57, and 3DS6-0.55-1.00, indicating a consensus map position between the fraction lengths 0.55 and 0.57 (3S-0.55-0.57) on the short arm. The spreadsheets used for assembling the consensus map and detecting anomalies can be found in supplemental online material (http://wheat.pw.usda.gov/pubs/2004/Genetics).
Figure 2.—
Anomalies of EST distribution detected by two or more loci. The GenBank accession numbers for the ESTs detecting the anomalies are included next to the appropriate arrow. The 12 anomalies were given letter codes (A–L). The spreadsheet used for the detection of anomalies can be found in supplemental online material (http://wheat.pw.usda.gov/pubs/2004/Genetics).
Gene ontology analyses:
The gene ontology (GO) database and its set of associated protein sequences were downloaded in May 2003 from http://www.geneontology.org and used for sequence comparison of consensus unigene sequences containing mapped ESTs. Local blastX results against the GO protein database (E-value < 10−5) were parsed and used to link to GO numbers corresponding to functional gene families. Hits to the GO database were counted under classifications at level 2 of the biological process category and level 3 of the molecular function category. Classifications were selected on the basis of relevance and all other classifications were combined and named “other”; therefore, each matched unigene was classified on both molecular function and biological activity according to the GO database standard (Figure 3). Comparisons were made between group 3 mapped-EST unigenes and the entire mapped unigene set using χ2 analyses. More detailed information can be found in supplemental online material at (http://wheat.pw.usda.gov/pubs/2004/Genetics).
Figure 3.—
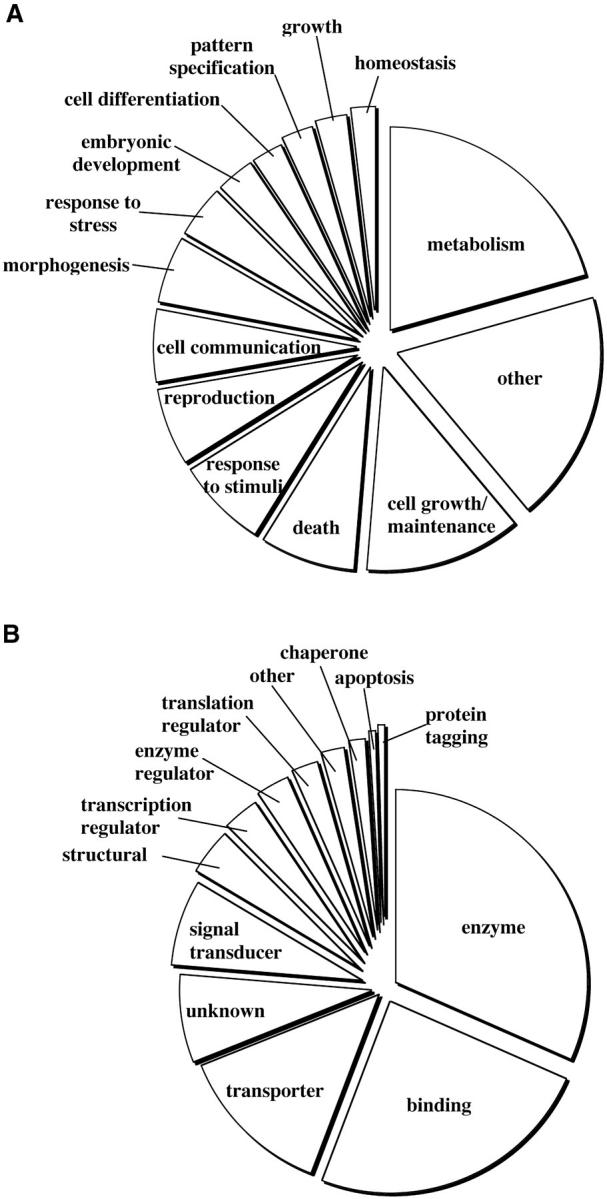
Proportional display of genes mapped on the group 3 chromosomes for various biological functions (A) and molecular activities (B). A more detailed summary of the gene ontology classifications for all mapped ESTs and the group 3 ESTs can found in supplemental online material (http://wheat.pw.usda.gov/pubs/2004/Genetics).
Comparative analysis of wheat group 3 with rice (O. sativa ssp. japonica) and A. thaliana:
A description of the wheat/rice comparison methods, detailed figures, and significance thresholds have been previously described (Sorrells et al. 2003; La Rota and Sorrells 2004). Figure 4 was constructed by matching ESTs in the consensus deletion map with their best hit on rice chromosome 1. The group 3 consensus chromosome bin map was displayed as twice the size of the rice physical map for readability. The wheat consensus chromosome bins are approximately proportional to estimated arm fraction length to the extent allowed by the EST content. All ESTs within the consensus bins were ordered on the basis of their relative best-hit order in rice and evenly distributed throughout the bin. A more detailed figure is in supplemental online material (http://wheat.pw.usda.gov/pubs/2004/Genetics).
Figure 4.—
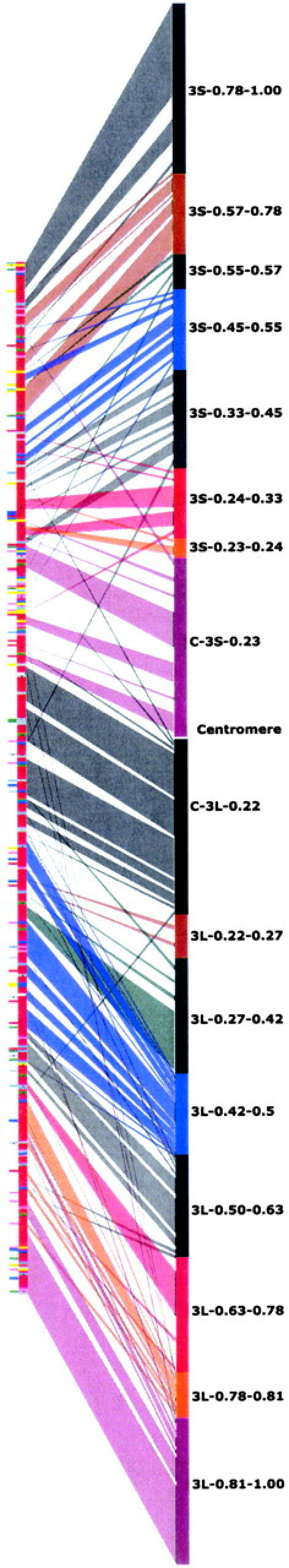
Depiction of the ordered rice chromosome 1 physical map (left) and correspondences with the wheat group 3 consensus chromosome bin map (right). The color coding corresponds to the blastN hits from the mapped wheat unigene set. The red coloring signifies a significant match to wheat group 3, whereas all other colors signify a best match to a different wheat chromosome group (Sorrells et al. 2003; La Rota and Sorrells 2004). The color coding is different for each bin within a chromosome arm but is repeated from the short arm to the long arm. Lines drawn between the rice and wheat group 3 maps show the relative location of each EST in rice and wheat. Thick sections represent a group of EST unigene blastN matches from the same consensus bin without interruption by one or more mapped-EST contig hits from a different consensus bin. Thin lines correspond to single wheat mapped-EST unigene blastN hits where the nearest neighbors are from different consensus bins. A more detailed figure is displayed in supplemental online material (http://wheat.pw.usda.gov/pubs/2004/Genetics).
The Arabidopsis coding region and protein sequences were obtained from the NCBI database and formatted into individual blast databases utilizing the NCBI blast tools (ftp.ncbi.nih.gov). Unigene sequences containing mapped ESTs were masked for known Poaceae repeats (TREP at http://wheat.pw.usda.gov/ggpages/ITMI/Repeats/index.shtml) and compared against the Arabidopsis coding sequence database using the blastN algorithm and against the Arabidopsis protein database using the blastX algorithm (Altschul et al. 1990). Summary statistics were calculated as previously described with the exception of an E-value cut-off of >0.001 (Sorrells et al. 2003; La Rota and Sorrells 2004). Using the summary statistics, only those matches with >70% identity over alignments of 100 bases or more were considered for the blastN alignments and those matches with >40% identity over 33 amino acids were considered for the blastX alignments. The blastX hits were also filtered by taking the total query length and subtracting the total alignment length. Negative numbers indicate a possible repeat or duplicate domain match and were used to selectively filter false hits not removed by truncation at the other threshold criteria. χ2 analyses were used to test the significance of hit distribution across the Arabidopsis genome. Expected hit values were calculated by taking the number of coding regions per chromosome divided by the total coding regions and multiplied by the total number of best hits. Statistical analyses were completed for all of the mapped-EST unigenes and only group 3 best hits.
RESULTS
Distribution of EST loci among the homoeologous group 3 chromosomes:
Of the 5762 informative mapped ESTs in the March 17, 2003, subset, 996, accounting for 2266 loci, were mapped to the homoeologous group 3 chromosomes. Of the mapped ESTs 515, 703, and 592 were mapped to chromosomes 3A, 3B, and 3D and these respectively identified 634, 884, and 748 loci. An overall mean of 2.28 EST loci were mapped to each group 3 chromosome with independently calculated means of 1.23 for 3A, 1.26 for 3B, and 1.26 for 3D. On the basis of χ2 analyses, significantly fewer ESTs and loci mapped to 3A (P < 0.001) than would be expected on the basis of chromosome physical size and significantly more ESTs were mapped to 3D (P < 10−5). The number of mapped ESTs with a location on 3B was not significantly different from that expected. The corresponding mapped-locus density (restriction fragments per relative chromosome length) was 0.88 for 3A, 1.02 for 3B, and 1.11 for 3D. Across the group 3 chromosomes 894 loci were mapped to the short arms and 1372 loci to the long arms, corresponding to significantly different (P < 0.001) mapped-locus densities of 0.92 and 1.06 per chromosome arm, respectively.
Distribution pattern of mapped ESTs and loci along the group 3 chromosomes:
The EST distributions followed similar patterns within chromosomes 3A and 3B. On the basis of physical size of the deletion bins, mapped-EST and mapped-locus content was significantly greater than expected for the distal bins of both the short and long arms (P < 0.01), less than expected for the proximal bins (P < 0.01), and not significantly different for the central bins (Figure 1). Chromosome 3D had a slightly different pattern than the other two group 3 chromosomes, in that the bins containing a greater-than-expected (P < 0.001) number of mapped ESTs were the central bin of the short arm and the distal bin of the long arm. The proximal bin of the short arm had fewer than expected mapped ESTs and loci (P < 10−7), while the distal bin of the short arm and the proximal and central bins of the long arm did not differ significantly from that expected. The consensus map provided a higher resolution analysis for the group 3 chromosomes. The consensus bin 3S-0.45-0.55 contained a greater than expected number of mapped ESTs and loci while C-3S-0.23 had significantly fewer (P < 0.01). The remainder of the short arm of the consensus map did not significantly differ from that expected. The long-arm consensus map had a central bin, 3L-0.42-0.50, and two distal-most bins, 3L-0.78-0.81 and 3L-0.81-1.00, with greater than expected mapped ESTs and loci (P < 10−5). Three bins toward the middle of the chromosome, 3L-0.22-0.27, 3L-0.27-0.42, and 3L-0.63-0.78, had fewer than expected mapped ESTs and loci (P < 0.01). The remaining bins of the long arm were not significantly different.
Anomalies:
Of the 537 ESTs in the group 3 consensus map, 72 disagreed in map position between two or more homoeologues, corresponding to 44 different anomalies. Of the 44 different anomalies 32 were detected by a single EST, as well as 6 by 2, 3 by 3, 1 by 4, 1 by 7, and 1 by 8 ESTs (Figure 2). Not included in these numbers were 4 ESTs that uncovered a discrepancy in fraction lengths between bins C-3AS2-0.23, 3AS2-0.23-0.45, C-3DS3-0.24, and 3DS3-0.24-0.55. That discrepancy was likely due to inaccurate fraction length estimates or differences in intergenic spaces between chromosomes 3A and 3D. To correct for the difference, the breakpoint at 0.23 in 3A was changed to 0.24 and the breakpoint 0.24 in 3D was changed to 0.23 in the consensus map. The names were not changed in the individual chromosome bin maps to retain the original nomenclature.
Group 3 mapped-EST additional loci:
The 996 ESTs mapped to the group 3 chromosomes accounted for 765 additional loci located in the remaining six chromosome groups. A significantly greater than expected number of loci was mapped to homoeologous groups 4 and 6 (P < 0.01), but the number of loci mapped to groups 1, 2, 5, and 7 agreed with expected random distribution. No significant differences were found among the numbers of these loci mapped to each genome for these other six groups.
Known gene locations:
At least 38 genes affecting morphological and biochemical traits (Table 1) are located on group 3 chromosomes. Among the more important genes are 10 disease resistance genes, viviparous 1, ATPase, sphaerococcum factor, and Hessian fly resistance. Actual and estimated chromosome bin locations were compiled and are shown in Table 1.
TABLE 1.
Morphological and biochemical genes located on group 3 chromosomes and their chromosome bin locations if known
| Chromosome arm or bin name
|
|||||
|---|---|---|---|---|---|
| Gene name | Symbol | A | B | D | Reference maps |
| ATPase1 | C-3BS1-0.33 | Boyko et al. (1999) | |||
| ATPase2 | C-3BL2-0.22 |
Li et al. (1999), Nelson et al. (1995) |
|||
| β-(1-3)-Glucanase | Glb3 | 3AL5-0.78-1 | 3BL7-0.63-1 | 3DL3-0.81-1 | Li et al. (2001), this study |
| b-ZIP class DNA binding protein | Embp | 3DL-0.78-1 |
Delaney et al. (1995), Koba and Tsunewaki (1978) |
||
| Chitinase-1 | Cht1a | 3AL-0.42-0.78 | Li et al. (1999) | ||
| Dihydroflavanol reductase | A1 | 3AL3-0.42-0.78 | C-3BL2-0.22 | C-3DL2-0.27 | This study |
| Esterase-1 | Est-1 | 3DS-0.39-0.55 | Boyko et al. (1999) | ||
| Esterase-5 | Est-5 | 3AL5-0.78-1 | 3BL7-0.63-1 | 3DL3-0.81-1 |
Devos and Gale (1993), Boyko et al. (1999) |
| Glutamic oxaloacetic transaminase |
Got-D3 | 3DL-0.31-0.50 | Boyko et al. (1999) | ||
| Heat-shock protein 17.3 | hsp17.3 | C-3AS2-0.23 | Boyko et al. (1999) | ||
| Hexokinase-1,2 | Hk | 3BS | 3DS | Liu and Gale (1989) | |
| Hybrid chlorosis | Ch2 | 3DL | Koba and Tsunewaki (1978) | ||
| Malic enzyme | Mal-1 | 3DS-0.56-1 | Devos and Gale (1993) | ||
| NADH dehydrogenase | Ndh | 3A | 3BL7-0.63-1.00 | 3DL2-0.27-0.81 | This study |
| Pairing homoeologous | ph2 | 3DS |
Sears (1982), Dong et al. (2002) |
||
| Peroxidase | Per-A3 | 3AL | 3BL | 3DS | Bosch et al. (1987) |
| Phenylalanine ammonia lyase | Pal | 3BS-0.57-1a | Li et al. (1999) | ||
| Phosphodiesterase-1 | Pde | 3A | 3B | 3D | Wolf et al. (1977) |
| Powdery mildew | Pm13 | 3BS-0.87-1 | Boyko et al. (1999) | ||
| Proline rich protein | Prp | 3AL5-0.78-1 | Li et al. (1999) | ||
| Pseudo-black chaff | Pbc | 3B | Kuspira and Unrau (1958) | ||
| Reaction to Mayetiola destructor | H24 | 3DL3-0.81-1 | Ma et al. (1993) | ||
| Reaction to Phaeosphaeria nodorum | Snb1 | 3A | Ma and Hughes (1995) | ||
| Reaction to Puccinia graminis | Sr12 | 3B | Sheen and Snyder (1964) | ||
| Reaction to P. graminis | Sr2 | 3BS-0.78-1 | Hart et al. (1993) | ||
| Reaction to P. graminis | Sr24 | 3DL2-0.27-0.81 |
Hart et al. (1993), Boyko et al. (1999) |
||
| Reaction to P. graminis | Sr27 | 3AS | 3BS | Marais and Marais (1994) | |
| Reaction to P. graminis | Sr35 | 3AL-0.78-1 | Hart et al. (1993) | ||
| Reaction to P. recondita | Lr24 | 3DL-0.78-1 | Boyko et al. (1999) | ||
| Reaction to Puccinia recondita | Lr27 | 3BS-0.78-1 | Nelson et al. (1997) | ||
| Reaction to P. recondita | Lr32 | 3DS-0.87-1 |
Autrique et al. (1995), Boyko et al. (1999) |
||
| Red kernel color | R1 | 3AL5-0.78-1 | 3BL7-0.63-1 | 3DL3-0.81-1 | Nelson et al. (1995) |
| Sphaerococcum factor | s1 | C-3AS2-0.023 | C-3BS1-0.33 | C-3DS3-0.24 | Salina et al. (2000) |
| Suppressor of gametocidal activity |
Igc1 | Tsujimoto and Tsunewaki (1985) |
|||
| Triosphospate isomerase | Tpi-A1 | 3AS | 3BS | 3DS | Pietro and Hart (1985) |
| Virescent | v | 3BS-C-0.33 | Hart et al. (1993) | ||
| Vivpary 1 | vp1 | 3AL-0.42-0.63 | Bailey et al. (1999) | ||
| wsu(dor4) | dor4 | C-3AS2-0.23 | Boyko et al. (1999) | ||
For those genes that were mapped by linkage analysis, bin location was inferred from linked markers mapped on deletion lines.
Encompasses more than one deletion bin.
Gene content:
Of the 5655 unigene sequences containing mapped ESTs, 1637 (29%) significantly matched at least one protein sequence in the GO database. At least one significant protein match was found for 341 of 987 unigenes mapped to group 3 (35%). Mapped-EST unigenes were classified according to either their biological (Figure 3A) or their molecular function (Figure 3B). Class sizes of those mapped on the group 3 deletion lines and the whole genome were not statistically different for either biological function or molecular function. Metabolism was the largest category of biological function for GO matches to both group 3 and the whole genome, followed by the cell growth and maintenance and response to external stimulus categories. Most of the reported proteins had either enzymatic or binding activity molecular function.
Wheat-rice comparison:
Analysis of the blastN results from the mapped group 3 unigenes against the rice genome indicated that the group 3 chromosomes share the highest level of homoeology with rice chromosome 1 (Figure 4). Of the 537 ESTs used in the consensus map, 232 belonged to unigenes that shared significant similarity with portions of rice chromosome 1, 81 matched a rice sequence on another chromosome, and the remaining 215 ESTs did not significantly match any rice genome sequence to date. We found that 59% of group 3 mapped-EST unigenes showed homology to rice.
Wheat-Arabidopsis comparison:
BlastN comparison of the 5655 mapped-EST unigenes against the Arabidopsis coding region database revealed 1182 (21%) significant matches. When only mapped-EST unigenes for group 3 were considered, 204 out of 988 (20.6%) significantly matched an Arabidopsis coding region. The blastX comparison of all mapped-EST unigenes against the Arabidopsis protein database returned 1799 (32%) matches for all mapped-EST unigenes and 313 (32%) for wheat group 3. The number of unigene matches per Arabidopsis chromosome did not significantly differ from that expected, on the basis of the estimated coding region content per chromosome, for wheat group 3 or total mapped-EST unigenes with either blastN or blastX analyses.
DISCUSSION
Large-scale sequencing of ESTs and generation of a high-density chromosome bin map for hexaploid wheat are an important research tool with applications for molecular marker development (Eujayl et al. 2002; Somers et al. 2003), comparative genomics (Sorrells et al. 2003; La Rota and Sorrells 2004), and genome evolutionary studies (Akhunov et al. 2003a,b). These EST chromosome bin maps will provide invaluable information for physical map construction and sequencing of gene-rich regions of the genome. In addition to the general use of a dense EST chromosome bin map, detailed analysis of individual chromosome groups provides important information to researchers with a particular interest in a chromosome or a homoeologous group of chromosomes.
Distribution of EST loci among the group 3 chromosomes:
The distribution of mapped ESTs and loci among the group 3 chromosomes generally followed the pattern observed for the wheat genomes as a whole (Qi et al. 2004). Chromosome 3B had the most mapped ESTs and loci followed by 3D and then 3A. The locus density (restriction fragments per relative chromosome length) was highest for 3D (1.11) and lowest for 3A (0.88) with 3B in the middle (1.02), following the general trend for all groups (Qi et al. 2004). The higher mapped-EST density (1.06) on the long arms of group 3 chromosomes, compared to the low density (0.92) on the short arms, also followed the general trend for other homoeologous groups (Qi et al. 2004). The close relationship of unique mapped ESTs to loci mapped across the group 3 chromosomes suggested that gene duplication may not be responsible for the increased map data attributed to 3B. The most frequent explanation for the EST mapping discrepancies relates to differences in chromosome size. While size differences do exist among the group 3 chromosomes (Dvořák et al. 1984; Gill et al. 1991), they do not correlate with mapped-EST content, invalidating this simple explanation. It is more likely that the evolutionary history of hexaploid wheat was responsible for the observed differences. An in-depth discussion of this possible explanation can be found in Qi et al. (2004). The mean for number of EST loci mapped to the group 3 chromosomes was found to be 2.28. Assuming that each of the homoeologous chromosomes contains the same gene content one would expect the mean to be near 3. Many factors may have contributed to the lower-than-expected mean, including Southern hybridization technical error, comigrating restriction fragments, and actual sequence loss or divergence between genomes.
Distribution pattern of mapped ESTs and loci:
The distribution of mapped ESTs and loci among the chromosome bins of 3A and 3B matched the general trend observed for most other chromosomes with dense regions at the distal ends of the chromosome arms and sparse regions at the proximal ends of the chromosome arms (Akhunov et al. 2003a,b; Qi et al. 2004). Chromosome 3D was an exception to the trend, with a dense region toward the center of the short arm (Figure 1).
The more detailed group 3 consensus deletion map suggested that mapped-EST density may be more localized than what was indicated by the individual chromosome deletion maps. The region of high mapped-EST density was reduced to only 10% of the short arm in the consensus chromosome bin map. These data may change the overall view of mapped-EST density for chromosome 3A. Though the distal chromosome bin 3AS4-0.45-1.00 appeared to be densely populated with mapped ESTs, the densely mapped consensus deletion bin 3S-0.45-0.55, and the presence of average mapped-EST density in the more distal consensus bins, suggested that the more proximal 10% of the 3AS4 bin was responsible for much of the high distal arm density. The actual mapped-EST density for chromosome arm 3AS may more closely resemble the high density in the middle of the 3DS arm, rather than the high distal density of 3BS, but was masked by the large size of the 3AS4-0.45-1.00 chromosome bin. The lower density of mapped RFLPs at the centromeric and telomeric ends of the short arms of 3A and 3D has been reported previously (Ma et al. 2001). A more central high-density region in 3AS would also explain the average gene densities in the distal short arm consensus bins. Though 3B has a region of higher mapped-EST density at the end of the short arm, the averaging effects of constructing the consensus map and the large amount of telomeric heterochromatin on 3BS abolish its significance. The lower mapped-EST density of the proximal consensus small arm chromosome bin confirmed the pattern seen in all of the group 3 chromosomes.
The long arm of the consensus chromosome bin map also provided a more detailed analysis of the mapped-EST density for group 3. Unlike the proximal 3AL and 3BL deletion bins, the consensus proximal chromosome bin was characterized by average mapped-EST density, resembling the proximal bin of 3DL. The difference between the individual chromosome bin maps and the consensus map was likely due to the differential contribution of ESTs mapped to the proximal bin C-3AL3-0.42 and the higher level of heterochromatin on 3BL. It is likely that the mapped-EST density of the proximal 3AL bin was closer to average in the proximal half and sparse in the more distal half. This hypothesis was supported by the existence of a mapped-EST-deficient region in the two consensus chromosome bins corresponding to the distal half of C-3AL3-0.42. It is also likely that the more proximal one-third of the middle chromosome bin of 3DL (3DL2-0.27-0.81) was less densely populated with mapped ESTs. This was supported by the existence of the very dense consensus deletion bin 3L-0.42-0.50, which suggested that a small section in the middle of the long arm might have a higher density of mapped ESTs than that evidenced by the individual chromosome bin maps. Interestingly a region of sparse mapped-EST density, consensus bin 3L-0.63-0.78, was evident in the distal half of the long arm of the consensus map. This region was characterized by either average or high mapped-EST density in the individual chromosome bin maps.
These findings are in good agreement with the in situ hybridization studies described by Ma et al. (2001). The most proximal two bins on the consensus map long arm appeared to contain high mapped-EST densities, in agreement with the individual chromosome bin maps. The more detailed consensus map uncovered more defined regions of high and low mapped-EST density, some of which were inconsistent with single chromosome bin analysis. Examination of the distribution of mapped ESTs at higher resolution would likely uncover a more complex pattern of mapped-EST density. The estimates of mapped-EST density were sensitive to size and breakpoint accuracy of deletion bins. Error in the size of an individual deletion bin could contribute to larger or smaller consensus bin sizes, subsequently altering the significance and the perceived density.
Anomalies:
The presence of nonoverlapping map positions for an EST in the chromosome bin maps of the homoeologues was termed an anomaly. Anomalies can be the result of actual biological events such as chromosomal rearrangements, transposition, and gene duplication; technical errors in Southern blots, such as the inability to score all restriction fragments; or a function of both in the case of probe cross-hybridization (i.e., gene family members). The existence of an anomaly that was evidenced by a single EST (32 of 44 for group 3) is questionable and more likely due to a technical error, although it is possible that by chance no other mapped EST detected such an anomaly. Anomalies that contain two or more mapped ESTs with the same location pattern (12 of 44) are more likely to be actual products of biological events and are displayed in Figure 2. Out of 12 multi-EST anomalies, 3 were between chromosomes 3A and 3B, 1 was between 3A and 3D, 7 were between 3B and 3D, and 1 was where 3B differed from both 3A and 3D. The greater number of anomalies detected between 3B and 3D was possibly due to the greater number of loci mapped to these two chromosomes.
Anomaly L was first considered to be caused by inaccurate bin lengths (Figure 2). After further examination it was found that correcting the bin length estimates would also create an anomaly in the reverse orientation. The inability to correct the anomaly by correcting bin lengths substantiates the presence of a rearrangement. Anomaly L was the largest anomaly observed with eight mapped ESTs detecting its presence. The nature of this anomaly and its presence in chromosomal areas of low to medium mapped-EST density suggest that the rearrangement between chromosomes 3B and 3D may encompass a large physical distance, assuming mapped-EST density corresponds to relative gene density in the region. Anomaly A was the second-largest anomaly detected with seven mapped ESTs detecting a rearrangement between chromosomes 3B and 3D, three of which detected the same rearrangement between chromosomes 3A and 3B.
Two anomalies, E and J, may both be evidence of the same rearrangement. Two ESTs, BE424097 and BE442882, were mapped to the telomeric bins of chromosome arms 3BS and 3DL. Two additional ESTs, BE405291 and BF202610, were mapped to the telomeric bins of 3BL and 3DS. The observed pattern indicates that a rearrangement possibly occurred within chromosome 3B or 3D and reciprocally exchanged DNA at the telomeric ends of the long and short arms. Another possible explanation would be that a duplication between the telomeric ends of the long and short arms occurred in both chromosomes but only one differing locus was identified in each chromosome.
It is possible that other putative anomalies arose through some forms of duplication that were not detected in the available data. Many of the mapped-EST hybridization films had several additional restriction fragments that could not be mapped because of background, lack of polymorphism between the genomes, or weak signal. The inability to map the additional restriction fragments would lead to an underestimation of duplication. The resolution of the deletion bins also limited the detection of gene duplications within a single bin. In cases where more than one restriction fragment for a particular EST was mapped to the same bin, at least a small percentage would be due to within-bin gene duplication. It was difficult to estimate the percentage of duplication because several of the mapped restriction fragments would be due to within-gene restriction sites. Approximately 13% of the mapped-EST unigenes contained at least one EcoRI restriction site, not including sites within introns, accounting for more than one-half of the ∼23% of ESTs detecting more than one locus on each of the group 3 chromosomes.
Group 3 mapped-EST additional loci:
The ESTs mapped to group 3 also accounted for 765 additional mapped loci throughout the rest of the genome. The presence of these additional loci could be due to cross-hybridization with closely related gene family members, gene duplication, or errors in scoring due to uncharacterized secondary deletions within the deletion stocks. The presence of a significantly greater number of additional loci was detected for the group 4 and group 6 chromosomes. The increased number of loci on group 4 and group 6 could be due to a higher rate of independent duplications between group 3, 4, and 6 chromosomes; concentrations of closely related sequences; or simply chance. However, no large-scale duplication between group 3 and the group 4 and 6 chromosomes was observed.
Wheat-rice comparison:
It has been previously demonstrated that the wheat group 3 chromosomes shared the highest homology with rice chromosome 1 (Devos et al. 1992; Ahn et al. 1993; Kurata et al. 1994; Van Deynze et al. 1995b). The comparisons presented in this study build upon previous sequence-based alignments (Sorrells et al. 2003; La Rota and Sorrells 2004) and are illustrated in Figure 4. Blocks of color represent the consensus sequence matches to the rice genome sequence from a single consensus bin, uninterrupted by matches to another bin. Lines represent a single match flanked by matches from different consensus bins. Blocks of conserved order were present along the lengths of both chromosomes (e.g., 3S-0.78-1.00, C-3L-0.22, and 3L-0.81-1.00), but order rearrangements were apparent between many adjacent consensus bins (e.g., between 3S-0.33-0.45 and 3S-0.24-0.33 and between 3L-0.27-0.42 and 3L-0.42-0.50). The consensus bin 3L-0.42-0.50 was found to be densely populated with mapped ESTs (Figure 1). Because of the high density in this bin, the rearrangement between wheat and rice may involve a large number of genes, possibly complicating the comparative use of rice sequence in this region. The ability to use rice sequence in this area depends on whether the rearranged observed blocks have undergone additional internal rearrangements that are not detectable at this resolution. The observed larger rearrangements between adjacent bins show that the colinear relationship of rice chromosome 1 and the group 3 chromosomes of wheat may be more complicated than previously thought. The fact that many of the adjacent rearrangements include several mapped ESTs suggests that substantial blocks of conserved gene order exist and still provide a basis for useful genome comparisons.
In addition to the larger rearrangements between adjacent consensus bins, numerous single ESTs matched sequences on the other rice chromosomes. In fact, the wheat homoeologous group 3 and rice chromosome 1 comparison may actually be more complex than that shown in Figure 4. Most blocks of conserved order span regions of rice chromosome 1 with matches to wheat ESTs mapped to other homoeologous groups. Also, it was assumed that the EST order within consensus bins would be similar to the order in rice. It is possible that additional rearrangements between rice and wheat were also present within consensus bins but could not be detected. Despite the simplifying assumptions, this wheat group 3 comparative map is the most detailed comparison to date.
Wheat-Arabidopsis comparison:
In the comparison between the group 3 EST unigenes and the Arabidopsis genome sequence, no significantly conserved genome structure was detected. This finding was not unexpected as rice and Arabidopsis also do not share any significant genome structure at the resolution afforded by the EST chromosome bin maps (Devos et al. 1999; Liu et al. 2001).
Conclusions:
The group 3 chromosome bin maps and consensus map provide insight into the structure and organization of the group 3 chromosomes. The high-density maps revealed chromosomal regions of high and low gene density along with putative rearrangements between the genomes. The detailed comparison of the group 3 consensus map with the genomic sequence of rice chromosome 1 unveiled further complexities within the colinearity previously detected between the chromosomes. These analyses provide a resource for understanding the evolution and organization of the wheat genome, for the use of rice as a model genome, and for selective sequencing of gene-rich regions in wheat.
Acknowledgments
This material is based upon work supported by the National Science Foundation under cooperative agreement no. DBI-9975989.
References
- Ahn, S., J. A. Anderson, M. E. Sorrells and S. D. Tanksley, 1993. Homoeologous relationships of rice, wheat and maize chromosomes. Mol. Gen. Genet. 241: 483–490. [DOI] [PubMed] [Google Scholar]
- Akhunov, E. D., A. W. Goodyear, S. Geng, L.-L. Qi, B. Echalier et al., 2003. a The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 13: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov, E. D., A. R. Akhunova, A. M. Linkiewicz, J. Dubcovsky, D. Hummel et al., 2003. b Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc. Natl. Acad. Sci. USA 100: 10836–10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Meyers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Autrique, E., R. P. Singh, S. D. Tanksley and M. E. Sorrells, 1995. Molecular markers for four leaf rust resistance genes introgressed into wheat from wild relatives. Genome 38: 75–83. [DOI] [PubMed] [Google Scholar]
- Bailey, P. C., R. S. McKibbin, J. R. Lenton, M. J. Holdsworth, J. E. Flintham et al., 1999. Genetic map location for orthologous Vp1 genes in wheat and rice. Theor. Appl. Genet. 98: 281–284. [Google Scholar]
- Berke, T. G., P. S. Baenziger and R. Morris, 1992. a Chromosomal location of wheat quantitative trait loci affecting agronomic performance of seven traits using reciprocal chromosome substitutions. Crop Sci. 32: 621–627. [Google Scholar]
- Berke, T. G., P. S. Baenziger and R. Morris, 1992. b Chromosomal location of wheat quantitative trait loci affecting stability of six traits using reciprocal chromosome substitutions. Crop Sci. 32: 628–633. [Google Scholar]
- Bosch, A., C. Vega and C. Benito, 1987. The peroxidase isozymes of the wheat kernel: tissue and substrate specificity and their chromosomal location. Theor. Appl. Genet. 73: 701–706. [DOI] [PubMed] [Google Scholar]
- Boyko, E. V., K. S. Gill, L. Mickelson-Young, S. Nasuda, W. J. Raupp et al., 1999. A high-density genetic linkage map of Aegilopes tauschii, the D-genome progenitor of bread wheat. Theor. Appl. Genet. 99: 16–26. [Google Scholar]
- Delaney, D. E., S. Nasuda, T. R. Endo, B. S. Gill and S. H. Hulbert, 1995. Cytologically based physical maps of the group-2 chromosomes of wheat. Theor. Appl. Genet. 91: 568–573. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., and M. D. Gale, 1993. Extended genetic maps of the homoeologous group 3 chromosomes of wheat, rye, and barley. Theor. Appl. Genet. 85: 649–652. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., M. D. Atkinson, C. N. Chinoy and M. D. Gale, 1992. RFLP-based genetic map of the homoeologous group 3 chromosomes of wheat and rye. Theor. Appl. Genet. 83: 931–939. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., J. Beales, Y. Nagamura and T. Sasaki, 1999. Arabidopsis-rice: Will colinearity allow gene prediction across the eudicot-monocot divide? Genome Res. 9: 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C., R. Whitford and P. Langridge, 2002. A DNA mismatch repair gene links to the Ph2 locus in wheat. Genome 45: 116–124. [DOI] [PubMed] [Google Scholar]
- Dvořák, J., P. E. McGuire and S. Mendlinger, 1984. Inferred chromosome morphology of the ancestral genome of Triticum. Plant Syst. Evol. 144: 209–220. [Google Scholar]
- Endo, T. R., 1988. Induction of chromosomal structural changes by a chromosome of Aegilops cylindrica L. in common wheat. Heredity 79: 366–370. [DOI] [PubMed] [Google Scholar]
- Endo, T. R., and B. S. Gill, 1996. The deletion stocks of common wheat. J. Hered. 87: 295–307. [Google Scholar]
- Eujayl, I., M. E. Sorrells, M. Baum, P. Wolters and W. Powell, 2002. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor. Appl. Genet. 104: 399–407. [DOI] [PubMed] [Google Scholar]
- Gill, B. S., B. Friebe and T. R. Endo, 1991. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34: 830–839. [Google Scholar]
- Gill, K. S., B. S. Gill and T. R. Endo, 1993. A chromosome region-specific mapping strategy reveals gene-rich telomeric ends in wheat. Chromosoma 102: 374–381. [Google Scholar]
- Hare, R. A., and R. A. McIntosh, 1979. Genetic and cytogenetic studies of durable adult-plant resistances in ‘Hope’ and related cultivars to wheat rusts. Z. Pflanzenzüchtung 83: 350–367. [Google Scholar]
- Hart, G. E., M. D. Gale and R. A. McIntosh, 1993 Linkage maps of Triticum aestivum (hexaploid wheat, 2n = 42, genomes A, B, and D) and T. tauschii (2n = 14, genome D), pp. 204–219 in Genetic Maps: Locus Maps of Complex Genomes, edited by S. J. O'Brien. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Klein, P. E., R. R. Klein, J. Vrebalov and J. E. Mullet, 2003. Sequence-based alignment of sorghum chromosome 3 and rice chromosome 1 reveals extensive conservation of gene order and one major chromosomal rearrangement. Plant J. 34: 605–621. [DOI] [PubMed] [Google Scholar]
- Koba, T., and K. Tsunewaki, 1978. Mapping of the s and Ch2 genes on chromosome 3D of common wheat. Wheat Inf. Serv. 45–46: 18–20. [Google Scholar]
- Kurata, N., G. Moore, Y. Nagamura, T. Foote, M. Yano et al., 1994. Conservation of genomic structure between rice and wheat. Nat. Biotechnol. 12: 276–278. [Google Scholar]
- Kuspira, J., and J. Unrau, 1958. Determination of the number and dominance relationships of genes on substituted chromosomes in common wheat Triticum aestivum L. Can. J. Plant Sci. 38: 199–205. [Google Scholar]
- La Rota, M., and M. E. Sorrells, 2004. Comparative DNA sequence analysis of mapped wheat ESTs reveals complexity of genome relationships between rice and wheat. Funct. Integr. Genomics 4: 34–46. [DOI] [PubMed] [Google Scholar]
- Lazo, G. R., S. Chao, D. D. Hummel, H. Edwards, C. C. Crossman et al., 2004. Development of an expressed sequence tag (EST) resource for wheat (Triticum aestivum L.): unigene analysis, probe selection, and bioinformatics for a 16,000-locus bin map. Genetics 168: 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. L., J. D. Faris, J. M. Chittoor, J. E. Leach, S. H. Hulbert et al., 1999. Genomic mapping of defense response genes in wheat. Theor. Appl. Genet. 98: 226–233. [Google Scholar]
- Li, W. L., J. D. Faris, S. Muthukrishnan, D. J. Liu, P. D. Chen et al., 2001. Isolation and characterization of novel cDNA clones of acidic chitinases and β-1,3-glucanases from wheat spikes infected by Fusarium graminearum. Theor. Appl. Genet. 102: 353–362. [Google Scholar]
- Liu, C. J., and M. D. Gale, 1989. Evidence for the genetic control of hexokinase isozymes by homoeologous group 3 chromosomes in wheat. Cereal Res. Commun. 17: 101–104. [Google Scholar]
- Liu, H., R. Sachidanandam and L. Stein, 2001. Comparative genomics between rice and Arabidopsis shows scant collinearity in gene order. Genome Res. 11: 2020–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewski, A. J., and C. A. Curtis, 1993. Physical distribution of recombination in B-genome chromosomes of tetraploid wheat. Theor. Appl. Genet. 84: 121–127. [DOI] [PubMed] [Google Scholar]
- Ma, H., and G. R. Hughes, 1995. Genetic control and chromosomal location of Triticum timopheevii-derived resistance to Septoria nodorum blotch in durum wheat. Genome 38: 332–338. [DOI] [PubMed] [Google Scholar]
- Ma, X.-F., K. Ross and J. P. Gustafson, 2001. Physical mapping of restriction fragment length polymorphism (RFLP) markers in homoeologous groups 1 and 3 chromosomes of wheat by in situ hybridization. Genome 44: 401–412. [PubMed] [Google Scholar]
- Ma, Z. Q., B. S. Gill, M. E. Sorrells and S. D. Tanksley, 1993. RFLP markers linked to two Hessian fly-resistance genes in wheat (Triticum aestivum L.) from Triticum tauschii (Coss.) Schmalh. Theor. Appl. Genet. 85: 750–754. [DOI] [PubMed] [Google Scholar]
- Marais, G. F., and A. S. Marais, 1994. The derivation of compensating translocations involving homoeologous group 3 chromosomes of wheat and rye. Euphytica 79: 75–80. [Google Scholar]
- McIntosh, R. A., P. L. Dyck and G. J. Green, 1977. Inheritance of leaf rust and stem rust resistances in wheat cultivars Agent and Agatha. Aust. J. Agric. Res. 28: 37–45. [Google Scholar]
- McIntosh, R. A., G. E. Hart, K. M. Devos, M. D. Gale and W. J. Rogers, 1998 Catalogue of gene symbols for wheat, pp. 1–235 in Proceedings of the 9th International Wheat Genetics Symposium, edited by A. E. Slinkard. University Extension Press, University of Saskatchewan, Saskatoon, Canada.
- Metzger, R. J., and B. A. Silbaugh, 1970. Location of genes for seed colour in hexaploid wheat, Triticum aestivum L. Crop Sci. 10: 495–496. [Google Scholar]
- Moore, G., 2000. Cereal chromosome structure, evolution, and pairing. Annu. Rev. Plant Physiol. 51: 195–222. [DOI] [PubMed] [Google Scholar]
- Nelson, J. C., A. E. Van Deynze, E. Autrique, M. E. Sorrells, Y. H. Lu et al., 1995. Molecular mapping of wheat homoeologous group 3. Genome 38: 525–533. [DOI] [PubMed] [Google Scholar]
- Nelson, J. C., R. P. Singh, J. E. Autrique and M. E. Sorrells, 1997. Mapping genes conferring and suppressing leaf rust resistance in wheat. Crop Sci. 37: 1928–1935. [Google Scholar]
- Osa, M., K. Kato, M. Mori, C. Shindo, A. Torada et al., 2003. Mapping QTLs for seed dormancy and the Vp1 homologue on chromosome 3A in wheat. Theor. Appl. Genet. 106: 1491–1496. [DOI] [PubMed] [Google Scholar]
- Pietro, M. E., and G. E. Hart, 1985. The genetic control of triosephosphate isomerase of hexaploid wheat and other Triticeae species. Genet. Res. 45: 127–142. [Google Scholar]
- Qi, L. L., B. Echalier, B. Friebe and B. S. Gill, 2003. Molecular characterization of a set of wheat deletion stocks for use in chromosome bin mapping of ESTs. Funct. Integr. Genomics 3: 39–55. [DOI] [PubMed] [Google Scholar]
- Qi, L. L., B. Echalier, S. Chao, G. R. Lazo, G. E. Butler et al. 2004. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina, E., A. Borner, I. Leonova, V. Korzun, L. Laikova et al., 2000. Microsatellite mapping of the induced sphaerococcoid mutation genes in Triticum aestivum. Theor. Appl. Genet. 100: 686–689. [Google Scholar]
- Sears, E. R., 1944. Cytogenetic studies with polyploidy species of wheat. II. Additional chromosome aberrations in Triticum vulgare. Genetics 29: 232–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, E. R., 1954 The Aneuploids of Common Wheat. Bull. 572, University of Missouri Agricultural Experiment Station, Columbia, MO.
- Sears, E. R., 1966 Nullisomic-tetrasomic combinations in hexaploid wheat, pp. 29–45 in Chromosome Manipulations and Plant Genetics, edited by R. Riley and K. R. Lewis. Oliver & Boyd, Edinburgh.
- Sears, E. R., 1982. A wheat mutant conditioning an intermediate level of homoeologous chromosome pairing. Can. J. Genet. Cytol. 24: 715–719. [Google Scholar]
- Sears, E. R., and L. M. S. Sears, 1978 The telocentric chromosomes of common wheat, pp. 389–407 in Proceedings of the 5th International Wheat Genetics Symposium, edited by S. Ramanujam. Indian Society for Genetics of Plant Breeding, New Dehli, India.
- Sheen, S. J., and L. A. Snyder, 1964. Studies on the inheritance of resistance to six stem rust cultures using chromosome substitution lines of a Marquis wheat selection. Can. J. Genet. Cytol. 6: 74–82. [Google Scholar]
- Somers, D. J., R. Kirkpatrick, M. Moniwa and A. Walsh, 2003. Mining single-nucleotide polymorphisms from hexaploid wheat ESTs. Genome 46: 431–437. [DOI] [PubMed] [Google Scholar]
- Sorrells, M. E., M. La Rota, C. E. Bermudez-Kandianis, R. A. Greene, R. Kantety et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto, H., and K. Tsunewaki, 1985. Gametocidal genes in wheat and its relatives. II. Suppressor of the chromosome 3C gametocidal gene of Aegilops triuncialis. Can. J. Genet. Cytol. 27: 178–185. [Google Scholar]
- Van Deynze, A. E., J. C. Nelson, L. S. O'Donoughue, S. N. Ahn, W. Siripoonwiwat et al., 1995. a Comparative mapping in grasses. Oat relationships. Mol. Genet. Genomics 249: 349–356. [DOI] [PubMed] [Google Scholar]
- Van Deynze, A. E., J. C. Nelson, E. S. Yglesias, S. E. Harrington, D. P. Braga et al., 1995. b Comparative mapping in grasses. Wheat relationships. Mol. Genet. Genomics 248: 744–754. [DOI] [PubMed] [Google Scholar]
- Whitkus, R., J. Doebley and M. Lee, 1992. Comparative genome mapping of sorghum and maize. Genetics 132: 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, W. A., S. E. Harrington, W. L. Woodman, M. Lee, M. E. Sorrells et al., 1999. Can we infer the genome structure of progenitor maize through comparative analysis of rice, maize, and the domesticated panicoids? Genetics 153: 453–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, G., J. Rimpau and T. Lelley, 1977. Localization of structural and regulatory genes for phosphodiesterase in wheat (Triticum aestivum). Genetics 86: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., D. W. Choi, S. Wanamaker, R. D. Fenton, A. Chin et al., 2004. Construction and evaluation of cDNA libraries for large-scale expressed sequence tag sequencing in wheat (Triticum aestivum L.). Genetics 168: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]