Abstract
The objectives of this study were to develop a high-density chromosome bin map of homoeologous group 7 in hexaploid wheat (Triticum aestivum L.), to identify gene distribution in these chromosomes, and to perform comparative studies of wheat with rice and barley. We mapped 2148 loci from 919 EST clones onto group 7 chromosomes of wheat. In the majority of cases the numbers of loci were significantly lower in the centromeric regions and tended to increase in the distal regions. The level of duplicated loci in this group was 24% with most of these loci being localized toward the distal regions. One hundred nineteen EST probes that hybridized to three fragments and mapped to the three group 7 chromosomes were designated landmark probes and were used to construct a consensus homoeologous group 7 map. An additional 49 probes that mapped to 7AS, 7DS, and the ancestral translocated segment involving 7BS also were designated landmarks. Landmark probe orders and comparative maps of wheat, rice, and barley were produced on the basis of corresponding rice BAC/PAC and genetic markers that mapped on chromosomes 6 and 8 of rice. Identification of landmark ESTs and development of consensus maps may provide a framework of conserved coding regions predating the evolution of wheat genomes.
COMMON wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) has a genome of ∼16 million kilobases per haploid cell, which is 35 times larger than that of rice (Oryza sativa L.) and ∼110 times that of Arabidopsis (Bennett and Smith 1976). It is composed of three genomes, contributed by T. uratu Tum. ex Gand. (A genome), Aegilops speltoides Tausch or an extinct close relative (B genome), and Ae. tauschii Coss. (D genome; McFadden and Sears 1946; Kihara 1954; Nishikawa 1983; Dvořák and Zhang 1990). The order of loci in these three genomes is thought to be colinear except for a 4A–5A–7B translocation, a putative 2B–6B translocation, and two inversions on chromosome 4A (Devos et al. 1995; Mickelson-Young et al. 1995).
A complete series of aneuploid wheat lines missing an entire chromosome or an arm of a chromosome has been developed (Sears 1954). More recently, a system of generating an unlimited number of deletion lines has become available; a chromosome with gametocidal properties from Ae. cylindrica host was used to generate frequent chromosome breaks in the wheat background (Endo 1988). The deletions were isolated in a wheat background as the breaks were caused only in the gametes lacking the alien chromosome. Most of the deletions were from a single break followed by the loss of the chromosome region distal to the breakpoint (Endo 1990). The systematic production of common wheat stocks containing terminal chromosomal deletions of various lengths has been reported and 430 deletion lines involving all 21 wheat chromosomes have been isolated (Endo and Gill 1996). From this collection, deletion lines were selected for the present study that provided extensive coverage of the wheat genome, subdividing it into 159 chromosome bins (Qi et al. 2003).
Physical maps of RFLPs produced using deletion stocks have been reported for the chromosomes of all seven homoeologous groups of hexaploid wheat (Gill et al. 1993; Kota et al. 1993; Hohmann et al. 1994, 1995; Delaney et al. 1995a,b; Mickelson-Young et al. 1995; Gill et al. 1996a,b). Arm-specific physical maps and identification of gene-rich areas and genes controlling phenotypic traits have also been reported (Endo and Mukai 1988; Endo et al. 1991; Endo and Gill 1996; Faris et al. 2000; Sandhu et al. 2001; Weng and Lazar 2002). The physical maps of homoeologous group 6 and group 7 chromosomes and a comparative map of chromosomes 7 of wheat and barley (Hordeum vulgare L.) have been reported (Werner et al. 1992; Hohmann et al. 1995; Weng et al. 2000). Landmark loci, which represent cDNA clones and single- or low-copy genomic DNAs that correspond to highly conserved coding regions, are useful tools in locating orthologous loci across the Triticeae genomes. These regions are of significance in understanding genome evolution among the species of Triticeae. Conserved linkages with similar gene content and gene order have been reported among many related species (Tanksley et al. 1992; Ahn and Tanksley 1993; Sorrells et al. 2003). The high colinearity of molecular markers between wheat and barley genomes at the genetic-map level has been well documented, which will accelerate integrative mapping among species (Devos et al. 1995; Van Deynze et al. 1995; Dubcovsky et al. 1996). Analyzing the degree of linkage conservation and synteny of chromosome segments between the homoeologous group 7 chromosomes of wheat and barley, Hohmann et al. (1995) identified extensive homologies between these chromosomes.
Expressed sequence tags (ESTs) are partial sequences of cDNA clones that correspond to mRNA and facilitate the identification of many genes (Adams et al. 1991). These sequences have been used to develop new molecular markers to analyze genome structure and to discover genes in many organisms, such as human, mouse, rat, Medicago trunculata, maize (Zea mays L.), and rice (Adams et al. 1991, 1995; Hillier et al. 1996; Covitz et al. 1998; Ewing et al. 1999; Marra et al. 1999; Scheetz et al. 2001; Fernandes et al. 2002). Previously, 238 genes with orthologous locations among the three genomes of wheat were identified. Thirty-nine (16.5%) of these genes were localized in the chromosomes of group 7 (McIntosh et al. 2003). Recently, a consortium of scientists (Lazo et al. 2004) identified ∼117,000 ESTs developed from the sequences of cDNAs of different tissues and developmental stages primarily of hexaploid wheat (http://wheat.pw.usda.gov/project).
ESTs from this collection representing wheat unigenes were physically mapped to individual chromosomes/chromosomal intervals using wheat nullisomic and ditelosomic lines and deletion stocks (Sears 1966; Endo and Gill 1996). This study summarizes the mapping of >2000 EST loci to the three homoeologous group 7 chromosomes of wheat, an assessment of conserved loci, and the distribution of mapped EST loci to the chromosome bins defined by the deletion stocks. Patterns of distribution and duplication of loci within and among the group 7 chromosomes of wheat and comparisons with rice and barley genomes were investigated. This is the first report of the mapping of such a large number of ESTs to this homoeologous group, and location of these genes in the rice genome offers the possibility of positioning similar genes across grass genomes.
MATERIALS AND METHODS
Genetic stocks:
In this study various cytogenetic stocks of the hexaploid wheat cultivar Chinese Spring (T. aestivum) were used. These were 24 nullisomic-tetrasomic (NT), 21 ditelosomic (DT), and 101 deletion lines (del) lines (Sears 1954, 1966; Sears and Sears 1978; Endo and Gill 1996). A detailed description of these stocks is provided in Qi et al. (2003). The fraction length (FL) value of each deletion breakpoint identifies the position of the breakpoint from the centromere relative to the length of the complete arm. A bin is defined by two deletion breakpoints and is given a name followed by the arm fraction-length endpoints for which the deletion is diagnostic; e.g., 7AL16-0.86-0.90 designates the region from a breakpoint at 86% of the 7AL arm to one at 90%. These aneuploid and deletion stocks provide a complete coverage of the wheat genome, subdividing it into 159 chromosome bins. All the genetic stocks selected for EST mapping were cytologically and/or molecularly verified by C-banding and Southern hybridization with >500 EST clones (Qi et al. 2003).
EST singletons:
The clones used in this study were developed from cDNA libraries of different tissues and developmental stages of wheat and other related species in the Triticeae tribe (Lazo et al. 2004; Zhang et al. 2004). The cDNA clones were sequenced and clones with unique sequences (unigenes) were used in this study as probes for mapping all homoeologous chromosomes. The distribution of the mapped loci along the chromosomes of wheat genomes approximates mapped-EST distribution in wheat.
At the U.S. Department of Agriculture (USDA)-Agricultural Research Service (ARS) Western Regional Research Center (Albany, CA), ∼117,000 ESTs were produced from 43 cDNA libraries (primarily of wheat) representing a wide range of tissues, developmental stages, and environmental stresses (Lazo et al. 2004). Amplified PCR products for unigenes (inserts) were prepared and sent to 10 mapping laboratories (http://wheat.pw.usda.gov/NSF) for Southern hybridization.
Southern hybridization:
Procedures used for genomic DNA isolation, restriction endonuclease digestion, gel electrophoresis, and DNA gel blot hybridization were as described in Qi et al. (2003) and are available on-line at http://wheat.pw.usda.gov/NSF/project/mapping_data.html. For Southern analysis, genomic DNA was digested with EcoRI. Lambda DNA digested with HindIII and BstEII was used as a size marker. Images of all autoradiographs are available on line at http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi.
Localization of ESTs:
EST loci were assigned to a specific chromosome, arm, and/or deletion bins on the basis of the presence or absence of the restriction fragments in a given set of DNA lanes of a Southern blot (Sears 1954, 1966; Endo and Gill 1996). Details of the mapping procedure can be found at http://wheat.pw.usda.gov/NSF/project/mapping.
Map construction:
The group at North Dakota State University was responsible for analyzing the mapping data for homoeologous group 7 chromosomes (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi). On the basis of the physical size (in micrometers) of chromosomes and chromosome arms and on the relative length of chromosome intervals (bins), the expected number of EST loci for each was calculated (Gill et al. 1991; Endo and Gill 1996). The χ2-test was used to test for randomness of the distribution patterns of EST loci among the chromosomes, chromosome arms, and deletion bins of group 7 of wheat.
The EST probes that hybridized to only three RFLP fragments and mapped across the three genomes were identified. The homoeologous map positions of the loci produced by these probes and those associated with the ancestral translocation involving 7BS were identified on the basis of the overlapping FL values in the bins and were used in the construction of a consensus physical map of group 7.
Those ESTs that hybridized to more than three fragments, many of which mapped onto homoeologous group 7 chromosomes, identified duplicated loci. On the basis of the pattern of duplicated loci, different classes of duplicated regions were evident. Whenever duplicated fragments of a particular EST mapped in the same deletion bin it was considered as an intrabin duplication. Interchromosomal duplication was defined as those events where duplicated loci of a particular EST mapped to chromosome deletion bins other than those in group 7. The consensus duplication was defined for those ESTs whose loci mapped to consensus positions across the chromosomes of homoeologous group 7 as well as to consensus positions across the chromosomes of another homoeologous group. The intra- and interarm duplications were defined for those ESTs whose loci were duplicated into the same or another arm of chromosomes of homoeologous group 7.
EST density:
The proportion of the chromosome for each deletion bin was calculated on the basis of Gill et al. (1991). The physical length, arm ratio data of a chromosome, and proportion of arm missing in chromosome bins were used in calculating the megabase values in deletion breakpoint-defined chromosome bins.
Chi-square statistics to test the homogeneity of EST content distribution along chromosome arms were used. The observed frequencies of ESTs in chromosome bins were tested against the null hypothesis of uniform EST distribution along the chromosome arms. Under the null hypothesis, the expected number of ESTs is proportional to the length of the bin. The distribution of EST loci along the physical length of each missing segment was analyzed by estimating the ratio of the percentage of mapped loci to the relative percentage of missing arm in the deletion breakpoint-defined regions (Weng and Lazar 2002).
Ordering ESTs into chromosome bins and comparison of map positions:
Deletion mapping provides a fast and efficient method of locating many loci within a chromosome bin; however, the order of loci within a bin cannot be determined. A putative order of ESTs can be inferred using in silico comparison to the rice genome sequence as reported by Sorrells et al. (2003). Each mapped EST locus is a unique EcoRI restriction digest signature of known molecular weight, marking a specific expressed segment of an individual chromosome of wheat. We considered only ordering of the ESTs placed on the consensus map. Corresponding rice BAC/PACs were identified by searching the ESTs at http://www.gramene.org/perl/SeqTable and genetically mapped molecular markers corresponding to BAC/PACs were identified. On the basis of centimorgan distances of these markers in different rice chromosomes, the relative order of ESTs in the different chromosome bins was determined, and the map positions were compared. Considering the homoeologous relationship between chromosomes of group 7 of wheat and barley, map positions of barley chromosome 7 markers were compared to those of wheat group 7 markers (Kleinhofs et al. 1993; Hohmann et al. 1995). Corresponding rice BAC/PACs with markers were identified by searching the sequences of RFLP markers of chromosome 7 of barley at http://www.gramene.org/perl/SeqTable (Künzel et al. 2000). Identified BAC/PACs were compared with the BAC/PACs and ESTs previously identified in the wheat consensus map.
RESULTS
Chromosome bin maps of 7A, 7B, and 7D:
Nine hundred nineteen EST probes were mapped in homoeologous group 7 chromosomes of wheat, identifying 2148 loci. Among the ESTs, 528 mapped on 7A and identified 661 loci, 549 mapped on 7B and identified 719 loci, and 613 mapped on 7D and identified 768 loci. The distributions of ESTs without duplication and showing no ambiguous loci in 7A, 7B, and 7D are presented in Figure 1, A, B, and C, respectively. The χ2 analysis indicated a significantly higher number of EST loci mapped into 7D even though it had the smallest physical size.
Figure 1.—
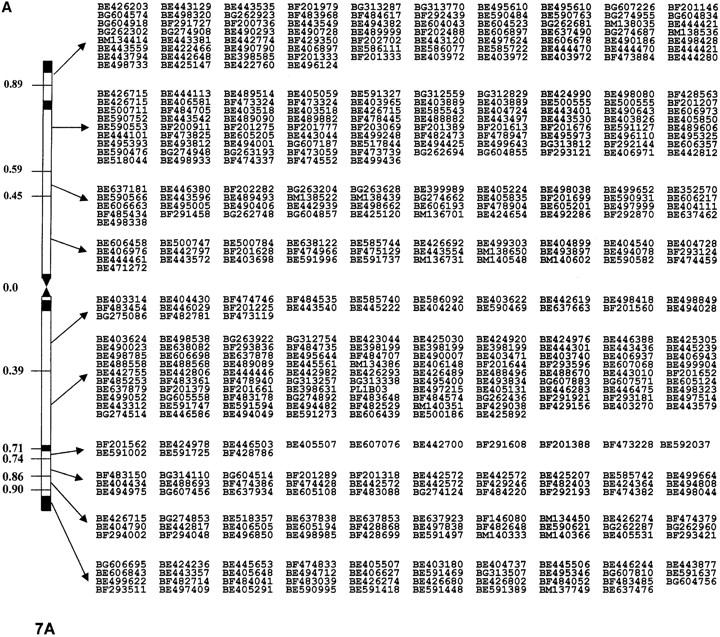

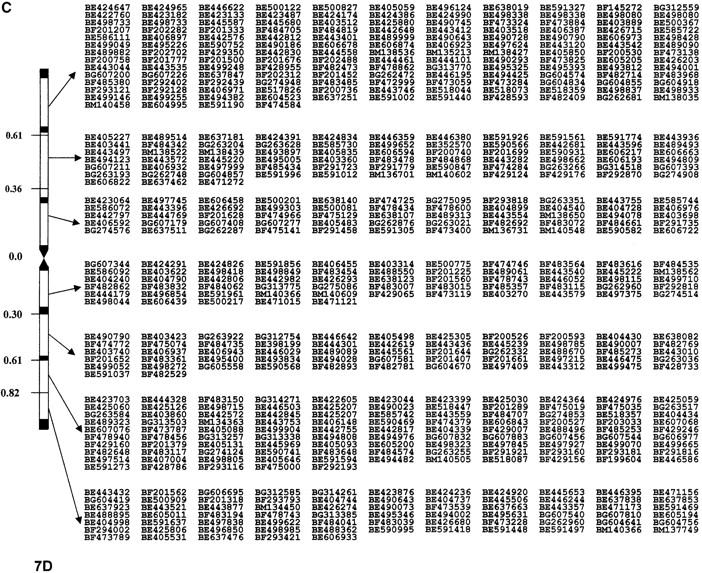
Physical EST maps of chromosomes of group 7 showing distribution of ESTs mapped in different deletion bins. The deletion bins for each chromosome are marked on the left. The intrabin duplicated loci are represented only once per bin. The number of loci per bin is that presented in Table 1 minus the duplications. Information on the exact locus designation and restriction fragment mapped to each bin can be found at http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi. The short arm of each chromosome is oriented toward the top while the long arm is toward the bottom. (A) physical map of chromosome 7A; (B) physical map of chromosome 7B; (C) physical map of chromosome 7D.
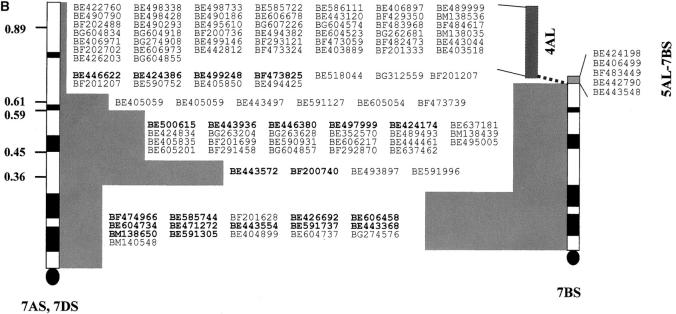
A total of 267 probes mapped to all three chromosomes of homoeologous group 7. Of these, 119 probes were unique, identifying only three loci. These probes were highly conserved among the genomes and could be considered as landmark probes. A consensus physical map of group 7 was developed with 117 (consensus position could not be resolved for two ESTs) of these probes, providing a framework map for chromosome 7 (Figure 2, A and B). An additional 49 probes could be added to this group if the ancestral translocation event involving 7BS was considered. These markers mapped to 7AS, 7DS, and the 5AL-4AL segment derived from 7B (Figure 2B).
Figure 2.—
Consensus map of homoeologous group 7 of wheat including the ancient translocation involving 7BS, 4AL, and 5AL. (A) consensus map of the long arm of group 7; (B) consensus map of 7AS and 7DS (on the left) with landmark probes shared with 7BS (0.00–0.59 interval). Forty-four EST detected loci mapped to 7AS and 7DS (0.59–1.00 interval) and translocated from 7BS to 4AL. A smaller segment of 5 EST detected loci representing an unequal translocation from 5AL to 7BS. Probes in boldface type were ordered on the basis of the corresponding markers mapped on the linkage maps of chromosomes 6 and 8 of rice.
Distribution of the loci and gene density:
All of the 23 deletion breakpoints defined bins among the homoeologous group 7 chromosomes contained different numbers of EST loci (Table 1). A trend for increasing numbers of EST loci mapped from proximal to distal regions of all chromosome arms was observed. All of the centromeric bins except C-7BS1-0.27 contained a significantly lower number of EST loci than expected on the basis of the size of the bins (Table 1). Except for the distal bin of chromosome arm 7BS (7BS1-0.27-1.00), all other distal regions contained relatively higher numbers of EST loci. The relative density of ESTs was expressed as the percentage of mapped loci per unit of physical length for each deletion bin and chromosome arm. For example, bin 7AL1-0.39-0.71 distal to bin C-7AL1 is physically 32% of the arm length and 39% of the loci on the long arm of chromosome 7A (7AL) were mapped to this bin (Table 1). Assuming the physical length of 7AL is 100 units then the ratio of mapped loci per unit arm length is ∼1.22. From Table 1, it is clear that in each arm the density of mapped loci per unit arm length increased from the centromeric region to the distal end, except for the short arm of chromosome 7B. The trend in 7BS can be explained by a double translocation event involving this arm (details presented later). The percentage of mapped loci per unit length among the distal chromosome deletion bins of all chromosome arms varied from 0.71 to 3.75.
TABLE 1.
Chromosome bins with relative physical length, number of loci per bin, and ratio of mapped loci per unit arm length of group 7 of wheat
| Chromosome arm and DNA content (Mb)a |
Chromosome bin | % arm | Mb DNA | No. of locib |
χ2c | % loci | Loci/unit arm length |
|---|---|---|---|---|---|---|---|
| 7AS = 407.53, χ2 = 0.226 | C-7AS8-0.45 | 45 | 183.40 | 36 | 61.81*** | 13 | 0.28 |
| 7AS8-0.45-0.59 | 14 | 57.08 | 46 | 1.52 | 17 | 1.21 | |
| 7AS5-0.59-0.89 | 30 | 122.26 | 108 | 8.09** | 39 | 1.30 | |
| 7AS1-0.89-1.00 | 11 | 44.83 | 84 | 96.24*** | 31 | 2.82 | |
| 7AL = 407.53, χ2 = 0.226 | C-7AL1 | 39 | 158.94 | 31 | 59.95*** | 11 | 0.28 |
| 7AL1-0.39-0.71 | 32 | 130.42 | 113 | 4.24* | 39 | 1.22 | |
| 7AL17-0.71-0.74 | 3 | 12.33 | 16 | 6.05* | 5 | 1.66 | |
| 7AL21-0.74-0.86 | 12 | 48.91 | 34 | 0.024 | 12 | 1.00 | |
| 7AL16-0.86-0.90 | 4 | 16.30 | 45 | 95.03*** | 15 | 3.75 | |
| 7AL18-0.90-1.00 | 10 | 40.75 | 52 | 18.02*** | 18 | 1.80 | |
| 7BS = 360.65, χ2 = 0.57 | C-7BS1-0.27 | 27 | 97.38 | 84 | 2.26 | 32 | 1.18 |
| 7BS1-0.27-1.00 | 73 | 263.28 | 180 | 0.83 | 68 | 0.93 | |
| 7BL = 540.98, χ2 = 0.38 | C-7BL2-0.33 | 33 | 180.01 | 49 | 42.38*** | 14 | 0.39 |
| 7BL2-0.33-0.63 | 30 | 162.29 | 92 | 2.79 | 25 | 0.83 | |
| 7BL7-0.63-0.78 | 15 | 81.15 | 40 | 3.97* | 11 | 0.73 | |
| 7BL10-0.78-1.00 | 22 | 119.02 | 184 | 133.92*** | 50 | 2.37 | |
| 7DS = 346.90, χ2 = 0.620 | C-7DS5-0.36 | 36 | 124.89 | 68 | 18.40*** | 21 | 0.61 |
| 7DS5-0.36-0.61 | 25 | 86.73 | 75 | 0.21 | 24 | 0.96 | |
| 7DS4-0.61-1.00 | 39 | 135.29 | 173 | 20.09*** | 55 | 1.41 | |
| 7DL = 381.59, χ2 = 0.51 | C-7DL5-0.30 | 30 | 114.48 | 72 | 13.18*** | 20 | 0.66 |
| 7DL5-0.30-0.61 | 31 | 118.30 | 80 | 10.02*** | 21 | 0.71 | |
| 7DL2-0.61-0.82 | 21 | 80.14 | 120 | 23.91*** | 33 | 1.57 | |
| 7DL3-0.82-1.00 | 18 | 68.69 | 95 | 12.67*** | 26 | 1.44 |
Relative distributions of loci mapped per chromosome were 7A = 815.06, χ2 = 5.49, *P = 0.05; 7B = 901.63, χ2 = 6.41, **P = 0.01; and 7D = 728.49, χ2 = 22.33, ***P = 0.005.
The loci presented here are those that were unambiguously assigned to each bin and not to the entire region, chromosome arm, or chromosome.
Significance levels were *P = 0.05, **P = 0.01, and ***P = 0.005.
The megabase content of DNA and gene densities for all deletion breakpoint-defined regions are summarized in Table 1. For example, in the short arm of chromosome 7A, four deletion breakpoint-defined regions were examined, and 274 loci mapped to this arm. Eighty-four loci mapped to bin 7AS1-0.89-1.00, while 108 loci mapped to bin 7AS5-0.59-0.89, and 46 loci mapped to bin 7AS8-0.45-0.59. On average, 72% of all mapped loci in homoeologous group 7 chromosomes were located in the distal regions, and the number of loci mapped in these regions was about seven times higher than that in the centromeric chromosome bins. The highest density of EST loci was observed in bin 7AL16-0.86-0.90 followed by bin 7BL10-0.78-1.00. The megabase contents of these chromosome bins were 16.30 and 119.02, and 45 and 184 loci were mapped in these bins, respectively.
Duplication:
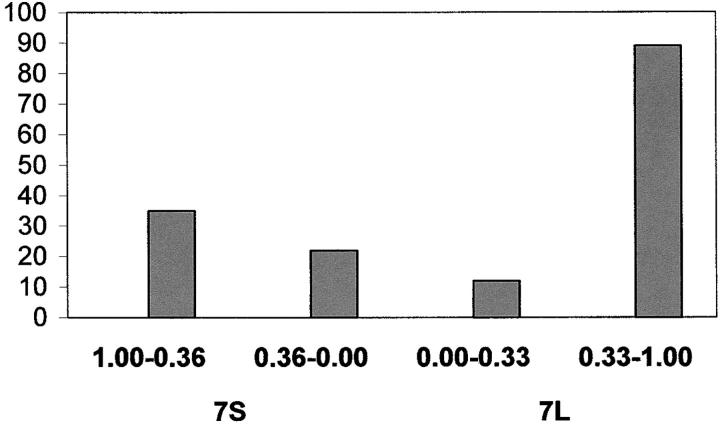
Of the 267 probes mapped into the three genomes of homoeologous group 7 chromosomes, 63 (24%) identified duplicated loci either on the chromosomes of other homoeologous groups or on the same arm or different arms of chromosomes of group 7. Seven of these ESTs identified duplicated loci placed on the consensus regions across the three genomes of groups 3, 5, and 6. Eighty-five (31%) ESTs identified duplicated loci in the same deletion bin where they were placed. The distribution of ESTs producing duplicated loci along the length of the consensus chromosome 7 is presented in Figure 3 and the number of ESTs with each type of locus duplication pattern is presented in Table 2.
Figure 3.—
Pattern of distribution of EST loci duplicated along the length of the arms of consensus chromosome 7. The vertical axis is the number of loci. The horizontal axis divides the short and long arms into approximately a proximal one-third and distal two-thirds portions.
TABLE 2.
Number of ESTs in different duplication patterns
| Duplication pattern | No. of ESTs |
|---|---|
| Intrabin | 85 |
| Interchromosomal | 47 |
| Unique (consensus) duplication | 7 |
| Intra-arm | 5 |
| Interarm | 4 |
7BS > 4AL and 5AL > 7BS translocations:
Of the 919 ESTs mapped to homoeologous group 7, 44 were mapped on the short arms of chromosomes 7A and 7D, but instead of mapping to the short arm of chromosome 7B they mapped to the long arm of chromosome 4A (Figure 2B). Out of these 44 probes 31 mapped to bin 7AS1-0.89-1.00 and 13 mapped to 7AS5-0.59-0.89. All of these 44 probes mapped to the distal bin of 7DS (7DS4-0.61-1.00). These probes also mapped to the distal 41% of the long arm of chromosome 4A. Twenty-nine mapped into bin 4AL4-0.80-1.00, 13 into bin 4AL5-0.66-0.80, and 2 mapped into bin 4AL13-0.59-0.66.
Five of the probes mapped to the long arms of chromosomes 5B and 5D, and instead of mapping to a similar region on chromosome 5A, they mapped to the short arm of chromosome 7B (Figure 2B).
Ordering the EST loci among chromosome bins and comparison of map positions with rice and barley:
Of the 117 probes (excluding those involved in the 7BS translocation) placed on the consensus map of homoeologous group 7, the possible order for 38 probes was determined on the basis of the molecular markers mapped to chromosomes 6 and 8 of rice (Figure 4), on the assumption of retained colinearity of loci among these species (Sorrells et al. 2003). In general, the probes mapping to rice chromosome 6 all mapped distal to the centromere in the consensus wheat chromosome, while all those mapping to rice chromosome 8 mapped near the wheat centromere (Table 3, Figure 4). All the ESTs in interval 0.45–0.59 of the short arm corresponded to rice chromosome 6 except BE424174, which mapped to rice chromosome 8 (Table 3, Figure 4).
Figure 4.—
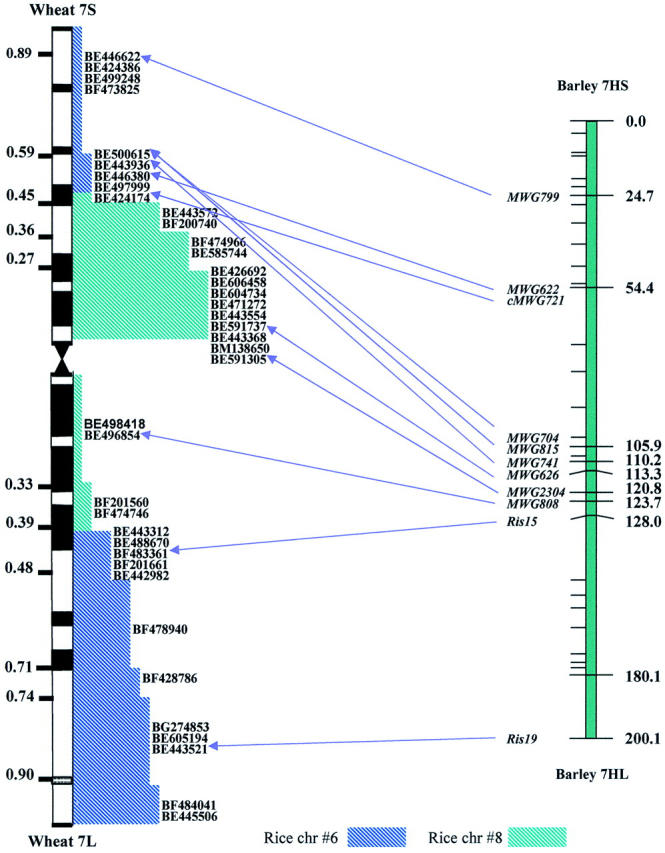
Comparison of map positions of molecular markers of rice and barley with tentatively ordered EST loci on the consensus map of homoeologous group 7. The linkage map of barley chromosome 7, adapted from Künzel et al. (2000), is depicted on the right. The stepwise increases in the height of the colored diagonal lines were used in wheat group 7 to differentiate among the regions.
TABLE 3.
Putative order of wheat ESTs based on corresponding rice BAC/PAC and genetic markers
| EST | Chromosomal intervala |
Rice BAC/PAC |
Genetic markers |
Rice chromosome |
Map position (cM) in rice |
|---|---|---|---|---|---|
| Short arm of wheat consensus chromosome 7 | |||||
| BE426692 | 0.00–0.27 | AP003892 | RM308 | 8 | 104.8 |
| BE606458 | 0.00–0.27 | AP004163 | R2027 | 8 | 108.9 |
| BE604734 | 0.00–0.27 | AP005439 | R2027 | 8 | 108.9 |
| BE471272 | 0.00–0.27 | AP004464 | R2027 | 8 | 108.9 |
| BE443554 | 0.00–0.27 | AP003914 | R2662 | 8 | 110.3 |
| BE591737 | 0.00–0.27 | AP004163 | RM3376 | 8 | 114.4 |
| BE443368 | 0.00–0.27 | AP003912 | RZ572 | 8 | 117.0 |
| BM138650 | 0.00–0.27 | AP003928 | S1570 | 8 | 117.0 |
| BE591305 | 0.00–0.27 | AP004044 | R662 | 8 | 117.3 |
| BF474966 | 0.27–0.36 | AP004765 | RZ926 | 8 | 100.4 |
| BE585744 | 0.27–0.36 | AP005509 | R2118 | 8 | 103.6 |
| BE443572 | 0.36–0.45 | AP003892 | RM149 | 8 | 103.7 |
| BF200740 | 0.36–0.45 | AP004761 | S10655 | 8 | 102.4 |
| BE500615 | 0.45–0.59 | AP005619 | R2171 | 6 | 50.4 |
| BE443936 | 0.45–0.59 | AP003524 | P127 | 6 | 61.6 |
| BE446380 | 0.45–0.59 | AP004728 | S2570 | 6 | 64.7 |
| BE424174 | 0.45–0.59 | AP003928 | R662 | 8 | 117.3 |
| BE497999 | 0.45–0.59 | AP003626 | R1559 | 6 | 88.9 |
| BE446622 | 0.61–0.89 | AP004806 | R2291 | 6 | 10.2 |
| BE424386 | 0.61–0.89 | AP002069 | R845 | 6 | 13.5 |
| BE499248 | 0.61–0.89 | AP002536 | RZ2 | 6 | 25.6 |
| BF473825 | 0.61–0.89 | AP003458 | L688 | 6 | 32.1 |
| Long arm of wheat consensus chromosome 7 | |||||
| BE498418 | 0.00–0.30 | AP004591 | C770 | 8 | 13.7 |
| BE496854 | 0.00–0.30 | AP004656 | S1461 | 8 | 38.0 |
| BF201560 | 0.30–0.39 | AP005441 | R2736 | 8 | 39.7 |
| BF474746 | 0.30–0.39 | AP005441 | R2736 | 8 | 39.7 |
| BE443312 | 0.39–0.48 | AP003574 | S20459S | 6 | 65.8 |
| BE488670 | 0.39–0.48 | AP004745 | R3188 | 6 | 66.7 |
| BF483361 | 0.39–0.48 | AP004729 | S10555 | 6 | 78.8 |
| BF201661 | 0.39–0.48 | AP003941 | RM1340 | 6 | 82.9 |
| BE442982 | 0.39–0.48 | AP005446 | C30378S | 6 | 87.5 |
| BF478940 | 0.61–0.71 | AP003771 | S14023 | 6 | 114.3 |
| BF428786 | 0.71–0.74 | AP005192 | L655 | 6 | 106.3 |
| BG274853 | 0.74–0.90 | AP003635 | C556 | 6 | 105.6 |
| BE443521 | 0.74–0.90 | AP003568 | C358 | 6 | 98.6 |
| BE605194 | 0.74–0.90 | AP004744 | E4392S | 6 | 105.6 |
| BF484041 | 0.90–1.00 | AP005750 | S11239 | 6 | 123.1 |
| BE445506 | 0.90–1.00 | AP004685 | C607 | 6 | 124.6 |
The intervals of each consensus chromosome arm are presented from proximal to distal.
Eleven molecular markers mapped on chromosome 7 of barley identified corresponding EST probes mapped to consensus chromosome 7 of wheat, eight of which maintained the same relative order in both genomes. Three EST probes (BE500615, BE446380, and BE424174) mapped in reverse order into the 0.45–0.59 region on the short arm of wheat chromosome 7 (Figure 4).
DISCUSSION
Chromosome bin maps of 7A, 7B, and 7D:
A total of 661, 719, and 768 loci were mapped to wheat chromosomes 7A, 7B, and 7D, respectively. On the basis of the relative sizes of these homoeologous chromosomes (7B > 7A > 7D), significantly higher numbers of EST loci were mapped to 7D followed by 7B and 7A (Figure 1, A, B, and C, respectively), which was in agreement with the findings of Qi et al. (2003). The reduced number of loci on 7B might be explained by the reciprocal translocation, where unequal size fragments were exchanged between 4AL, 7BS, and 5AL. There are 44 7BS-specific ESTs on 4AL and 5 5AL-specific ESTs translocated to 7BS. Due to this uneven exchange, 7BS had the lowest number of EST loci as compared to other short arms of homoeologous group 7 chromosomes (Table 1).
Distribution of loci and gene density:
Physical RFLP maps produced using deletion stocks have been reported for each of the seven homoeologous chromosome groups and for some chromosome arms (Werner et al. 1992; Kota et al. 1993; Hohmann et al. 1994, 1995; Delaney et al. 1995b; Mickelson-Young et al. 1995; Endo and Gill 1996; Gill et al. 1996a,b; Faris et al. 2000; Sandhu et al. 2001; Weng and Lazar 2002). In most cases the numbers of markers analyzed were relatively low, and a majority of the markers were of unknown function or were genomic probes. The highest number of clones used in mapping the three homoeologous chromosome 7's was 111, and only 21 of these were cDNA probes (Hohmann et al. 1995).
A higher marker density was generally observed in the distal regions as compared to the proximal regions of chromosome arms. Akhunov et al. (2003a) analyzed the distribution of EST loci in all chromosomes of the wheat genome using a subset of this project's mapped EST database and determined that in each arm the density of mapped loci increases from the centromeric region to the distal end. The distribution of mapped loci in our study along the arms of group 7 chromosomes supports that pattern.
The highest density of EST loci, as revealed by χ2 and ratio of percentage of mapped loci per unit arm length, was observed in bin 7AL16-0.86-0.90, which agrees with Hohmann et al. (1995). The second-highest density of EST loci was observed in 7BL10-0.78-1.00. The estimated megabase content of bin 7AL16-0.86-0.90 is 16.30 and 45 loci were mapped in this bin; therefore, on average, 1 EST locus was mapped for every 362 kb in this region.
The difference in distribution of recombination along the chromosome length means that the amount of DNA per centimorgan varies depending on the location of the gene on a chromosome. A higher density of EST loci in distal regions was correlated with a higher rate of recombination. Conversely, a lower density of EST loci in proximal regions was correlated with a lower rate of recombination (Akhunov et al. 2003a). Thus, the majority of genes in wheat appear to be located in the high-recombination areas, allowing for effective development and use of map-based cloning strategies to clone genes of interest.
Consensus map:
Because many ESTs detected three orthologous loci among the three homoeologous group 7 chromosomes, and the loci appear to be colinear, it was possible to construct a consensus chromosome deletion bin map of homoeologous group 7. The consensus map provides a detailed resolution of the relative positions of mapped orthologous loci. Because of the colinearity of these loci across the chromosomes of group 7 and a lack of duplication anywhere else in the wheat genome, we identified these 166 loci as landmark markers for this homoeologous group.
Landmark loci, presented here as EST clones, could correspond to highly conserved regions and could be a helpful tool in the allocation of orthologous loci across Triticeae genomes and will be useful in genetic mapping of orthologous genes. These regions may be of significance in understanding genome evolution among Triticeae species by analyzing chromosome structural rearrangements, recombination hot spots, suppression of recombination, and gene distribution, duplication, and elimination events in the genome. Hohmann et al. (1995) designated 10 landmark RFLP loci, 5 each for the short and long arms of consensus chromosome 7. They suggested that these loci could be useful in targeting specific genes to specific regions of consensus chromosome 7. In our study 117 loci were colinear across the homoeologous group, and 68% (81/117) were mapped into the region close to the centromere. These loci mapped to the proximal region are possibly conserved over large evolutionary distances and could be linked to, or possibly represent, critical genes that necessitated their presence in the genomes during the establishment of polyploid species (Akhunov et al. 2003b). Because of their evolutionary importance we believe that these 117 loci should be present in closely related species.
Duplication:
There are duplicated loci (paralogous) on almost all of the RFLP linkage and physical maps of Triticeae species reported to date (Anderson et al. 1992; Hohmann et al. 1994; Nelson et al. 1995; Marino et al. 1996; Weng et al. 2000; Weng and Lazar 2002). In a mapping study of the T. monococcum L. genome, Dubcovsky et al. (1996) identified >30% intra- or interchromosomal duplications. In our study, 24% of the ESTs mapping to group 7 identified duplicated loci either on the chromosomes of other homoeologous groups or on the same arm or different arms of chromosomes of group 7. Eighty-five (31%) ESTs identified duplicated loci in the same deletion bin (intrabin) and these could have resulted from the internal cut site of EcoRI within a locus. The observed rate of duplication does not reflect the total duplicated loci of the wheat genome since we mapped only the chromosomes of homoeologous group 7. In an effort to map physically 6421 ESTs in the rice genome, Wu et al. (1998) reported only 2.4% duplicated loci. Hence, there appears to be an order of magnitude more of duplicated loci per gene motif within the homoeologous group 7 chromosomes of wheat than in the small genome of rice. The growth or shrinkage of the plant genomes has been attributed to the growth and shrinkage of repeated nucleotide sequences (Bennetzen 2002; Sanmiguel et al. 2002); however, the growth of the wheat genomes also appears to have been accompanied by the concomitant accumulation of dispersed gene duplications. The similar proportions of duplicated loci to overall genome size of wheat and rice suggest that the accumulation or deletion of repeated sequences and genes could have been coupled and controlled by a common mechanism.
The duplicated loci tend to be located in the distal regions of chromosome arms (Figure 3), whereas the landmark loci were mostly proximal (Figure 2). The distribution of duplicated loci across the wheat genomes was highly correlated with the recombination rates along hexaploid wheat chromosome arms and along chromosome arms in diploid species of the Triticum-Aegilops alliance (Dvořák et al. 1998; Akhunov et al. 2003a). This relationship has been attributed to either hitchhiking of neutral loci with genes selected by natural selection or linkage of neutral loci to mildly deleterious genes not favored by natural selection (Charlesworth 1994). In both scenarios, there is a greater chance for a neutral locus and, by extension, for polymorphism for a duplicated locus, to be eliminated if it is in a low-recombination region than if it is in a high-recombination region. Therefore, it could be suggested that polymorphisms for neutral locus duplications are expected to survive and become fixed preferentially in high-recombination regions. Identification of duplicated regions between homoeologous group 7 and homoeologous groups 3, 5, and 6 of wheat implied that these duplications existed prior to polyploidization (Qi et al. 2003).
7BS > 4AL and 5AL > 7BS translocation:
On the basis of the location of structural genes on chromosomes 4BL, 4DL, and 5AL (Ainsworth et al. 1983), and of endosperm peroxidase on 4AL, 7AS, and 7DS (Kobrechel and Fillet 1975), Naranjo et al. (1987) proposed a double translocation, 4AL to 5AL, 5AL to 7BS, and 7BS to 4AL, in the genome of Chinese Spring wheat. Anderson et al. (1992) analyzed these translocations by RFLP analysis using genomic probes and supported the translocations proposed by Naranjo et al. (1987). Werner et al. (1992) reported a segment of the short arm of chromosome 7B had been translocated to the long arm of chromosome 4A and suggested that ∼20% of the distal region of the 4AL chromosome was derived from a translocation of 7BS. In the present study, we identified loci corresponding to 44 probes mapped to bins 7AS1-0.89-1.00, 7AS5-0.59-0.89, 7DS4-0.61-1.00, 4AL4-0.80-1.00, 4AL5-0.66-0.80, and 4AL13-0.59-0.66 (Figure 2B). Even if we consider that a portion of the chromosomal region between 4AL5-0.66-0.80 and 4AL13-0.59-0.66 was mapped by the loci of these probes, at least 34% of the 4AL chromosome arm at the distal region was derived from a distal translocation event involving 7BS.
Anderson et al. (1992) supported the proposed translocation between 5AL and 7BS by assigning a 5AL-specific fragment of the probe BCD87 to chromosome 7BS. We examined the probes mapped to 7BS and identified loci corresponding to five probes (Figure 2B) that mapped distal on 7BS, 5BL, and 5DL. We could not identify the position of the translocated segment on 7BS because the linear orders of these probes are not known. Our analysis supported the proposed translocation between 5AL and 7BS although the chromosomal segment involved in this translocation appears much smaller than the translocation between 7BS and 4AL, which is in agreement with Jiang and Gill (1994).
Ordering EST loci and comparison of map position with rice and barley:
Homoeology between wheat and rice genomes was first studied by Ahn et al. (1993) followed by Kurata et al. (1994) and Van Deynze et al. (1995) at the macro level. Sorrells et al. (2003) compared rice and wheat genomes at the micro/DNA sequence level. All those studies indicated that rice chromosomes 6 and 8 are homoeologous with Triticeae group 7 chromosomes. Of the 117 ESTs located on the group 7 consensus map, 38 were located to rice BAC/PACs with corresponding genetic markers, and 11 of the BAC/PACs correspond with the sequence of RFLP markers mapped to chromosome 7 of barley (Table 3 and Figure 4). The terminal regions (100.4–118.9 cM) of the long arm of rice chromosome 8 corresponded with the centromeric region (0.0–0.59) of the long arm of consensus chromosome 7 of wheat (Figure 4). The short arm region (13.7–39.7 cM) of rice chromosome 8 corresponded with the centromeric region of the long arm (0.0–0.39) of consensus chromosome 7 of wheat. About 39% of the distal region of the long arm of consensus chromosome 7 corresponded with a 26-cM region of the long arm of chromosome 6 of rice, indicating a putative homoeologous relationship of genes involved in these regions of wheat and rice. The present study identified putative regions of gene content conservation between the wheat group 7 consensus chromosome and rice chromosomes 6 and 8. Orthology of these loci with rice suggests a possible ancestral origin of these loci and that their presence precedes the divergence of the wheat and rice lineages. Hence the proximal low-recombination region of wheat chromosomes could be a region of evolutionary conservation, which is in agreement with the findings of Akhunov et al. (2003a) and Sorrells et al. (2003). Using the rice genome as a template one can predict colinearity with the wheat genomes; however, microsynteny studies have suggested that, in most cases, colinearity will need to be verified at the DNA sequence level (Han et al. 1999; Bennetzen and Ramakrishna 2002). The ordering of mapped ESTs within chromosome bins would be an important enhancement for the wheat/rice comparative analysis.
Although the wheat homoeologous group 7 map is based on consensus physical maps that combine deletions from 7A, 7B, and 7D chromosomes that differ in size and in amount of heterochromatin, the results obtained by comparing them to the barley chromosome 7 linkage map are in good agreement with those reported by Werner et al. (1992), Hohmann et al. (1995), and Künzel et al. (2000). Except for the reverse order of four ESTs mapped into the 0.45–0.59 regions on the group 7 consensus map of wheat, the relative positions of all markers and ESTs were maintained between wheat chromosome 7 and barley chromosome 7 (Figure 4). The reverse order of these ESTs corresponding to the molecular markers mapped on rice chromosome 6 identified a minor inversion of the similar region of group 7 chromosomes that has been reported by Hohmann et al. (1995). Thus, these results suggest the ordering of EST probes and map position in comparison of wheat chromosomes of homoeologous group 7 with chromosomes of rice and barley.
The localization and distribution of EST loci into bins along the homoeologous group 7 chromosomes directly reflects the distribution of genes and gene-rich regions of this group in wheat. Identification of landmark probes and putative map positions in rice and barley genomes suggested a detailed analysis of ESTs mapped in the wheat genome could provide valuable information in mapping and identification of genes across grass genomes.
Acknowledgments
We thank S. S. Maan for his valuable guidance throughout this project. This material is based upon work supported by the National Science Foundation under cooperative agreement no. DBI-9975989.
References
- Adams, M. D., J. M. Kelley, J. D. Gocayne, M. Dubnick, M. H. Polymeropoulos et al., 1991. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 252: 1651–1656. [DOI] [PubMed] [Google Scholar]
- Adams, M. D., A. R. Kerlavage, R. D. Fleischmann, R. A. Fuldner, C. J. Bult et al., 1995. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377: 3–17. [PubMed] [Google Scholar]
- Ahn, S. N., and S. D. Tanksley, 1993. Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. USA 90: 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. N., J. A. Anderson, M. E. Sorrells and S. D. Tanksley, 1993. Homoeologous relationships of rice, wheat and maize chromosomes. Mol. Gen. Genet. 241: 483–490. [DOI] [PubMed] [Google Scholar]
- Ainsworth, C. C., M. D. Gale and S. Bird, 1983. The genetics of β-amylase isozymes in wheat. I. Allelic variation among hexaploid varieties and intrachromosomal gene locations. Theor. Appl. Genet. 66: 39–49. [DOI] [PubMed] [Google Scholar]
- Akhunov, E. D., A. W. Goodyear, S. Geng, L. L. Qi, B. Echalier et al., 2003. a The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 13: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov, E. D., A. R. Akhunova, A. M. Linkiewicz, J. Dubcovsky, D. Hummel et al., 2003. b Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc. Natl. Acad. Sci. USA 100: 10836–10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J. A., Y. Ogihara, M. E. Sorrells and S. D. Tanksley, 1992. Development of a chromosomal arm map for wheat based on RFLP markers. Theor. Appl. Genet. 83: 1035–1043. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., and J. B. Smith, 1976. Nuclear DNA amounts in angiosperms. Trans. R. Soc. Lond. Ser. B Biol. Sci. 274: 227–274. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J., 2002. Opening the door to comparative plant biology. Science 296: 60–63. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and W. Ramakrishna, 2002. Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol. Biol. 48: 821–827. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., 1994. Plant self-incompatibility. The key to specificity. Curr. Biol. 4: 545–546. [DOI] [PubMed] [Google Scholar]
- Covitz, P. A., L. S. Smith and S. R. Long, 1998. Expressed sequence tags from a root-hair-enriched Medicago truncatula cDNA library. Plant Physiol. 117: 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, D., S. Nasuda, T. R. Endo, B. S. Gill and S. H. Hulbert, 1995. a Cytologically based physical maps of the group-2 chromosomes of wheat. Theor. Appl. Genet. 91: 568–573. [DOI] [PubMed] [Google Scholar]
- Delaney, D., S. Nasuda, T. R. Endo, B. S. Gill and S. H. Hulbert, 1995. b Cytologically based physical maps of the group-3 chromosomes of wheat. Theor. Appl. Genet. 91: 780–782. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., J. Dubcovsky, J. Dvořák, C. N. Chinoy and M. D. Gale, 1995. Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor. Appl. Genet. 91: 282–288. [DOI] [PubMed] [Google Scholar]
- Dubcovsky, J., M.-C. Luo, G.-Y. Zhong, R. Bransteiter, A. Desai et al., 1996. Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics 143: 983–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák, J., and H. B. Zhang, 1990. Variation in repeated nucleotide sequence sheds light on the phylogeny of the wheat B and G genomes. Proc. Natl. Acad. Sci. USA 87: 9640–9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák, J., M. C. Luo and Z. L. Yang, 1998. Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics 148: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T. R., 1988. Induction of chromosome structural changes by a chromosome of Aegilops cylindrical L. in common wheat. J. Hered. 79: 366–370. [DOI] [PubMed] [Google Scholar]
- Endo, T. R., 1990. Gametocidal chromosomes and their induction of chromosome mutations in wheat. Jpn. J. Genet. 65: 135–152. [Google Scholar]
- Endo, T. R., and B. S. Gill, 1996. The deletion stocks of common wheat. J. Hered. 87: 295–307. [Google Scholar]
- Endo, T. R., and Y. Mukai, 1988. Chromosome mapping of a speltoid suppression gene of Triticum aestivum L. based on partial deletion in the long arm of chromosome 5A. Jpn. J. Genet. 69: 13–19. [Google Scholar]
- Endo, T. R., Y. Mukai, M. Yamamoto and B. S. Gill, 1991. Physical mapping of male-fertility gene of common wheat. Jpn. J. Genet. 66: 291–295. [Google Scholar]
- Ewing, R. M., A. B. Kahla, O. Poirot, F. Lopez, S. Audic et al., 1999. Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res. 9: 950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris, J. D., K. M. Haen and B. S. Gill, 2000. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154: 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, J., V. Brendel, X. Gai, S. Lal and V. L. Chandler, 2002. Comparison of RNA expression profile based on maize expressed sequence tag frequency analysis and micro-array hybridization. Plant Physiol. 128: 896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, B. S., B. Friebe and T. R. Endo, 1991. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34: 830–839. [Google Scholar]
- Gill, K. S., B. S. Gill and T. R. Endo, 1993. A chromosome region-specific mapping strategy reveals gene-rich telomeric ends in wheat. Chromosoma 102: 374–381. [Google Scholar]
- Gill, K. S., B. S. Gill, T. R. Endo and E. V. Boyko, 1996. a Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143: 1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, K. S., B. S. Gill, T. R. Endo and T. Taylor, 1996. b Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 144: 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, F., A. Kilian, J. P. Chao, D. Kudrna, B. Steffenson et al., 1999. Sequence analysis of a rice BAC covering the syntenous barley Rpg1 region. Genome 42: 1071–1076. [DOI] [PubMed] [Google Scholar]
- Hillier, L., G. Lennon, M. Becker, M. F. Bonaldo, B. Chiapelli et al., 1996. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 6: 807–828. [DOI] [PubMed] [Google Scholar]
- Hohmann, U., T. R. Endo, K. S. Gill and B. S. Gill, 1994. Comparison of genetic and physical maps of group 7 chromosomes from Triticum aestivum L. Mol. Gen. Genet. 245: 644–653. [DOI] [PubMed] [Google Scholar]
- Hohmann, U., A. Graner, T. R. Endo, B. S. Gill and R. G. Herrmann, 1995. Comparison of wheat physical maps with barley linkage maps for group 7 chromosomes. Theor. Appl. Genet. 91: 618–626. [DOI] [PubMed] [Google Scholar]
- Jiang, I. P., and B. S. Gill, 1994. Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome 37: 717–725. [DOI] [PubMed] [Google Scholar]
- Kihara, H., 1954. Consideration on the evolution and distribution of Aegilops species based on the analyzer-method. Cytologica 19: 336–357. [Google Scholar]
- Kleinhofs, A., A. Kilian, M. A. Saghai Maroof, R. M. Biyashev, P. Hayes et al., 1993. A molecular, isozyme and morphological map of barley (Hordeum vulgare) genome. Theor. Appl. Genet. 86: 705–712. [DOI] [PubMed] [Google Scholar]
- Kobrechel, K., and P. Fillet, 1975. Identification of genomes and chromosomes involved in peroxidase synthesis of wheat seeds. Can. J. Bot. 53: 2326–2344. [Google Scholar]
- Kota, R. S., K. S. Gill, B. S. Gill and T. R. Endo, 1993. A cytogenetically based physical map of chromosome 1B in common wheat. Genome 36: 548–554. [DOI] [PubMed] [Google Scholar]
- Künzel, G., L. Korzun and A. Meister, 2000. Cytologically integrated physical restriction fragment length polymorphism maps of barley genome based on translocation breakpoints. Genetics 154: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata, N., G. Moore, Y. Nagamura, T. Foote, M. Yano et al., 1994. Conservation of genomic structure between rice and wheat. Bio/Technology 12: 276–278. [Google Scholar]
- Lazo, G. R., S. Chao, D. D. Hummel, H. Edwards, C. C. Crossman et al., 2004. Development of an expressed sequence tag (EST) resource for wheat (Triticum aestivum L.): unigene analysis, probe selection and bioinformatics for a 16,000-locus bin map. Genetics 168: 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, C. L., J. C. Nelson, Y. H. Lu, M. E. Sorrells, P. Leroy et al., 1996. Molecular genetic maps of the group-6 chromosomes of hexaploid wheat (Triticum aestivum L). Genome 39: 359–366. [DOI] [PubMed] [Google Scholar]
- Marra, M., L. Hillier, T. Kucaba, M. M. Allen, R. Barstead et al., 1999. An encyclopedia of mouse genes. Nat. Genet. 21: 191–194. [DOI] [PubMed] [Google Scholar]
- McFadden, E. S., and E. R. Sears, 1946. The origin and of Triticum spelta and its free threshing hexaploid relatives. J. Hered. 37: 81–89. [DOI] [PubMed] [Google Scholar]
- McIntosh, R. A., Y. Yamazaki, K. M. Devos, J. Dubcovsky, W. J. Rogers et al., 2003 Catalogue of gene symbols for wheat, Vol. 4, pp. 1–34 in Proceedings of the 10th International Wheat Genetics Symposium, edited by N. E. Pogna, M. Romano, E. Pogna and G. Galterio. Instituto Sperimentale per la Cerealicotura, Rome.
- Mickelson-Young, L., T. R. Endo and B. S. Gill, 1995. A cytogenetic ladder map of the wheat homoeologous group-4 chromosomes. Theor. Appl. Genet. 90: 1007–1011. [DOI] [PubMed] [Google Scholar]
- Naranjo, T., P. Roca, P. G. Goicoechea and R. Giraldez, 1987. Arm homoeology of wheat and rye chromosomes. Genome 29: 873–882. [Google Scholar]
- Nelson, J. C., M. E. Sorrells, A. E. Van Deynze, Y. H. Lu, M. Atkinson et al., 1995. Molecular mapping of wheat: major genes and rearrangements in homoeologous groups 4, 5, and 7. Genetics 141: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, K., 1983 Species relationship of wheat and its putative ancestors as viewed from isozyme variation, pp. 59–63 in Proceedings of the 6th International Wheat Genetics Symposium, edited by S. Sakamoto. Plant Germplasm Institute, Faculty of Agriculture, Kyoto University, Kyoto, Japan.
- Qi, L. L., B. Echalier, B. Friebe and B. S. Gill, 2003. Molecular characterization of a set of wheat deletion stocks for using in chromosome bin mapping of ESTs. Funct. Integr. Genomics 3: 39–55. [DOI] [PubMed] [Google Scholar]
- Sandhu, D., J. A. Champoux, S. N. Bondareva and K. S. Gill, 2001. Identification and physical localization of useful genes and markers to a major gene-rich region on wheat 1S chromosomes. Genetics 157: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmiguel, P., W. Ramakrishna, J. L. Bennetzen, C. S. Busso and J. Dubcovsky, 2002. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A. Funct. Integr. Genomics 2: 70–80. [DOI] [PubMed] [Google Scholar]
- Scheetz, T. E., M. R. Raymon, D. Y. Nishimura, A. McClain, C. W. Roberts et al., 2001. Generation of a high-density rat EST map. Genome Res. 12: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, E. R., 1954 The Aneuploids of Common Wheat. Bull. 572,University of Missouri Agricultural Experiment Station, Columbia, MO.
- Sears, E. R., 1966 Nullisomic-tetrasomic combinations in hexaploid wheat, pp. 29–45 in Chromosome Manipulation and Plant Genetics, edited by R. Riley and K. R. Lewis. Oliver & Boyd, Edinburgh.
- Sears, E. R., and L. M. S. Sears, 1978 The telocentric chromosomes of common wheat, pp. 389–407 in Proceedings of the 5th International Wheat Genetics Symposium, edited by S. Ramanujam. Indian Society for Genetics and Plant Breeding, New Delhi, India.
- Sorrells, M. E., M. La Rota, C. E. Bermudez-Kandianis, R. A. Greene, R. Kantety et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S. D., M. W. Ganal, J. P. Price, M. C. DeVicente, M. W. Bonierbale et al., 1992. High density molecular maps of tomato and potato genomes; biological inferences and practical approaches. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deynze, A. E., J. C. Nelson, E. S. Yglesias, S. E. Harrington, D. P. Braga et al., 1995. Comparative mapping in grasses. Wheat relationships. Mol. Gen. Genet. 248: 744–754. [DOI] [PubMed] [Google Scholar]
- Werner, J. E., T. R. Endo and B. S. Gill, 1992. Towards a cytogenetically based physical map of the wheat genome. Proc. Natl. Acad. Sci. USA 89: 11307–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, Y., and M. D. Lazar, 2002. Comparison of homoeologous group-6 short arm physical maps of wheat and barley reveals a similar distribution of recombinogenic and gene-rich regions. Theor. Appl. Genet. 104: 1078–1085. [DOI] [PubMed] [Google Scholar]
- Weng, Y., N. A. Tuleen and G. E. Hart, 2000. Extended physical maps and a consensus physical map of the homoeologous group-6 chromosomes of wheat (Triticum aestivum L.). Theor. Appl. Genet. 100: 519–527. [Google Scholar]
- Wu, J. Z., N. Kurata, H. Tanoue, T. Shimokawa, Y. Umehara et al., 1998. Physical mapping of duplicated genomic regions of two chromosome ends in rice. Genetics 150: 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., D. W. Choi, S. Wanamaker, R. D. Fenton, A. Chin et al., 2004. Construction and evaluation of cDNA libraries for large-scale expressed sequence tag sequencing in wheat (Triticum aestivum L.). Genetics 168: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]