Abstract
Interval mapping of quantitative trait loci (QTL) for resistance to late blight, height, and maturity was performed on a tetraploid full-sib family of potato comprising 227 clones from a cross between a susceptible parent, 12601ab1, and a resistant cultivar, Stirling, which were of similar height and main crop maturity. Thirty-eight AFLP primer combinations provided 585 informative markers, and 23 SSRs proved useful for identifying linkage groups (LGs). A simplex QTL allele was found on LGV of Stirling close to marker STM3179, which was associated with early maturity, short plants, and susceptibility to blight and explained 54.7, 26.5, 26.3, and 17.5% of the variation for maturity, height, tuber blight, and foliage blight. When the residuals from the regressions of foliage and tuber blight on maturity were analyzed, there was no significant effect of a QTL on LGV, but a duplex QTL allele for resistance was found on LGIV of Stirling, which explained 30.7 and 13.6% of the variation for foliage and tuber blight on an additive model. Partial dominance for resistance explained even more of the variation, up to 37.2% for foliage blight. A major gene for blight resistance in Stirling was also mapped to LGXI.
IN recent years, advances in computer technology have led to an increasing interest in extending linkage analysis and quantitative trait locus (QTL) mapping methods from diploid to polyploid plant species, which display complex polysomic inheritance (Doerge and Craig 2000; Luo et al. 2001; Gallais 2003). This is not surprising, given the occurrence and evolutionary significance of autopolyploidy in wild plants and the economic importance of several autopolyploid crop species such as tetraploid alfalfa and potato, hexaploid sweet potato, and octaploid strawberry and sugarcane (Gallais 2003).
Luo et al. (2001) presented a methodology for the construction of a linkage map in an autotetraploid species using either codominant or dominant molecular markers scored on two parents and their full-sib progeny. The methods were applied to a simulated data set and to a small set of SSR (microsatellite) and amplified fragment length polymorphism (AFLP) markers scored in a full-sib population of tetraploid potato. Hackett et al. (2003) then introduced simulated annealing into the methodology for ordering the molecular markers in a linkage group (LG). This approach facilitates the examination of orders close to the optimum to see which marker placings are fixed and to identify the markers whose positions are less certain. The method was tested by computer simulation and application to 30 AFLP and SSR markers on linkage group IV of potato. Once a reliable linkage map is available, the method of Hackett et al. (2001) can be used for QTL interval mapping in a full-sib family of an autotetraploid species derived by crossing two parents. The success of the method was demonstrated in a simulation study but not on real data. The theory in these articles (Hackett et al. 2001, 2003; Luo et al. 2001) was based on the simplest situation that can give tetrasomic inheritance, namely random pairing of four homologous chromosomes to give two pairs of bivalents at meiosis and an orderly meiosis in which all possible diploid gametes are equally likely and viable.
In practice, many departures from this simple situation could occur, in particular: multivalents and double reduction, lack of complete homology between chromosomes and hence departures from random pairing, distorted segregation due to differential fertility and viability, errors in scoring gels, and genuine anomalies that require cytogenetical explanation.
It is, therefore, important to determine if this simple theory can be applied to real data and give useful results. There is also the need to demonstrate in polyploids the advantages of testing for the presence of a QTL at locations between markers over regression models that compare trait means for different phenotypes at a single marker, as done by Meyer et al. (1998) and Bradshaw et al. (1998) in potato and Ming et al. (2001) in sugarcane. Finally, there is the need to show that genetical analysis at the tetraploid level really does provide information about allelic diversity and interactions, which is not available from analysis of diploid relatives or haploids of the tetraploid species.
Quantitative resistance to late blight [Phytophthora infestans (Mont.) de Bary] of potato is an ideal trait for testing the simple theory on real data of economic importance. Late blight is still the most serious disease of potatoes worldwide, particularly in developing countries with poor farming communities in highland regions disproportionately affected (International Potato Center 1997), and durable resistance is a major objective of many potato-breeding programs. Progress in the genetical dissection of quantitative resistance to P. infestans using DNA-based markers in diploid mapping populations has been reviewed by Gebhardt and Valkonen (2001) and Simko (2002). QTL have been found on almost every chromosome and have been repeatedly detected on III, IV, and VI and on V associated with late maturity. The genetical basis of the association between resistance and maturity on V has been analyzed further but not resolved (Collins et al. 1999; Oberhagemann et al. 1999; Visker et al. 2003). Meyer et al. (1998) constructed a partial linkage map of AFLPs in tetraploid potato and associated markers thought to be on chromosome VIII, but now known to be on chromosome IV, with glasshouse-assessed quantitative resistance to foliage blight. Their mapping population consisted of a subset of 94 randomly chosen F1 plants from a cross between the blight-resistant cultivar Stirling and the susceptible Scottish Crop Research Institute (SCRI) breeding clone 12601ab1 (Bradshaw et al. 1995). This article presents and discusses the results of interval mapping of QTL for resistance to foliage and tuber blight, maturity, and canopy height (as an indicator of vigor) on the full set of 227 F1 clones from the 12601ab1 × Stirling population using 38 AFLP primer combinations and 23 SSRs to identify linkage groups of interest.
MATERIALS AND METHODS
Plant material:
The cross 12601ab1 × Stirling was one of a diallellic set of crosses made in 1992 (Bradshaw et al. 1995). The reciprocal cross was made in 1993. Seedlings were raised in a glasshouse during the summer of 1994 and their tubers harvested in mid-September. Although subsequent molecular marker analysis of 272 clones failed to detect any selfs, it did highlight a distinct group of 45 clones from 12601ab1 × Stirling with an unusually low number of paternal bands. As there was no obvious experimental or genetical explanation for the origin of these distinct clones, they were omitted from the linkage and QTL analyses. This left 227 clones, of which 169 and 58 had 12601ab1 and Stirling, respectively, as the female parent. The two parents and the clones were maintained in a glasshouse in 1995 and 1996 and then from 1997 until 2000 at a high-grade seed site (Blythbank Farm, West Linton, Peeblesshire, UK). From 1998 onward, as back up and for convenience, a single plant of each clone was also grown in a glasshouse. In 1998, the tubers for this glasshouse maintenance came from the seed site, whereas in subsequent years they came from the glasshouse-grown plants.
Field assessment of foliage blight resistance in 1998 (226 clones):
The assessment was done at Yonderton Farm, Ayrshire, United Kingdom, on the western seaboard of Scotland where the weather conditions are conducive to the development of blight. The trial had a randomized complete block design with two replicates and two-plant plots. There was one plot of each clone and three of each parent in both replicates. Tubers were planted on 13 May in pairs of drills separated by a third “spreader drill” of the susceptible cultivar King Edward. On July 20, glasshouse-grown plants of cultivar King Edward were inoculated in the laboratory with a zoospore suspension of the complex race 1, 2, 3, 4, 6, 7 of P. infestans. Three days later, on July 23, they were placed in their pots at meter intervals along the spreader drills. The amount of blight present on the two plants in each plot was scored on Malcolmson's 1–9 scale of increasing resistance (Cruickshank et al. 1982) on July 30 and on August 3, 7, 11, and 14, 1998.
The resistance of cultivar Stirling (score 7.50) and susceptibility of clone 12601ab1 (score 2.67) were clear by August 11 (fourth score, fb4) and this date was considered optimal for discriminating between the clones in the population. The area under the disease progress curve (AUDPC) from July 23 until August 14 was also calculated for each plot, following conversion of the 1–9 scale to the percentage of necrotic tissue (100, 90, 80, 70, 60, 40, 25, 10, 0). Subsequent analyses were done both on fb4, for comparison with the glasshouse tests (see later) and with previously reported results (Bradshaw et al. 1999), and on the square root of the AUDPC on the percentage scale (saudpc%) because its distribution was the least skewed of those considered.
Two replicates of the R-gene differentials, with the exception of R9, which was not available, were planted adjacent to the trial in three-plant plots and scored on the same dates. The scores confirmed that the trial had been inoculated with the intended race 1, 2, 3, 4, 6, 7.
Tuber blight test in 1999 (200 clones):
Seed tubers were planted on June 9, 1999 in 10-cm pots of soil-based compost placed on a glasshouse bench in a two-replicate randomized complete block design. Within each replicate, each clone was represented once, and each parent three times, by plots of eight plants in adjacent pots, although for some clones there were fewer than eight plants. Tubers were harvested on August 17, just as the plants were coming into flower and before the tubers were fully mature. The produce of the eight plants was bulked and, where possible, the 20 largest tubers were selected and placed in a 12.5-cm pot. In practice, the average sample size of the clones was 16.87 tubers, excluding 17 plots, which were treated as missing because they had <7 tubers. The tuber samples were inoculated immediately after harvest by dipping the pots momentarily in a suspension containing 2.5 × 104 zoospores/ml of race 1, 2, 3, 4, 6, 7 of P. infestans. The pots were stored for 48 hr in a constant environment chamber at 15° and 100% relative humidity (RH) and then left to dry on the floor of a glasshouse header house at ambient temperature until scored. The number of infected tubers in each sample was recorded over the period August 30–September 1, ignoring any infections that had penetrated the skin through a wound or the stolon scar. The percentage of infected tubers was calculated (tb%).
Glasshouse foliage blight tests in 2000 (225 clones):
The whole-plant glasshouse test developed by Malcolmson (1976) and Stewart et al. (1983a) was used to identify which clones had inherited Stirling's major R-gene for blight resistance by comparing their reactions to the simple race 1, 4 with those to the complex race 1, 2, 3, 4, 6, 7. The results have already been reported by Stewart et al. (2003) in another context. It proved possible to determine the R-gene status of 215 (137 with and 78 without the R-gene) of the 225 clones assessed. Ten clones were excluded as it was not possible to categorize them with confidence, despite repeating the blight test on them in 2001. The R-gene was mapped as a qualitative trait along with the molecular markers (see later). The scores with the complex race (fbc) on Malcolmson's 1–9 scale of increasing resistance (Cruickshank et al. 1982) were analyzed as a quantitative trait.
Height (225 clones) and maturity (222 clones) assessment:
The population was grown at a ware site in 1999 (Mylnefield Farm, Dundee, United Kingdom) and again in 2000 and 2001 (Gourdie Farm, Dundee). Each year the trial had a randomized complete block design with two blocks and single-drill plots of five tubers spaced 45 cm apart within drills with 75 cm between drills in 1999 and 90 cm in 2000 and 2001. There were enough tubers to include 217 and 162 clones in the first and second blocks in 1999, 219 clones in both blocks in 2000, and 225 clones in both blocks in 2001. Three replicates of each of the two parents were included in every block. The trials were planted on April 26, 1999, May 1, 2000, and May 4, 2001. Fertilizer, herbicide, aphicide, and fungicide (for control of late blight) applications were standard for a ware crop in southeast Scotland.
Canopy height (ht) from the top of the drill was measured (in centimeters) on July 14, 1999, July 13, 2000, and July 24, 2001.
Maturity (mat) was scored on a 1 (all plants in a plot dead) to 9 (all plants still green) scale on August 12, 1999 and August 22, 2000. In 2001, the trial was still green at burn down. The trials were burned down with split applications of sulphuric acid on August 22, 1999, August 27, 2000, and August 19, 2001.
GENSTAT 5 Release 3 (Genstat 5 Committee 1993) was used to analyze the data by the method of residual maximum likelihood with years, clones, and blocks as random effects.
The heritability (h2) of clone means was estimated as
5.47 σ2c/5.47 σ2c + 1.89 σ2cy + σ2 for height and
3.54 σ2c/3.54 σ2c + 1.85 σ2cy + σ2 for maturity,
where σ2c, σ2cy, and σ2 are the variance components for clones, clones × years of interaction, and residual plot-to-plot variation.
Clone means over years were based on the best linear unbiased prediction of clone effects, which are usually smaller than if they had been estimated as fixed effects.
Genomic DNA isolation:
Between 5 and 10 g of young, fully expanded leaves were harvested from glasshouse-grown plants, “flash-frozen” in liquid nitrogen, and stored at −70°. Genomic DNA was extracted from 1 g of frozen leaf material following the protocol set out in the Nucleon Phytopure kit (Nucleon Biosciences, Manchester, UK). The quality and quantity of the DNA samples were evaluated under UV illumination after ethidium-bromide staining of 1% agarose gels.
AFLP assays:
AFLP assays were performed using a modification of the protocol of Vos et al. (1995) as described by Milbourne et al. (1997). The 6-bp cutting enzymes PstI (10 unit/μl) and EcoRI (10 unit/μl) were obtained from Pharmacia (Piscataway, NJ) and GIBCO BRL (Gaithersburg, MD), respectively, and the 4-bp cutting enzyme MseI (4 unit/μl) from New England Biolabs (Beverly, MA). The One Phor All buffer (which is compatible with all enzymes used), T4 DNA ligase, and T4 polynucleotide kinase were purchased from Pharmacia. AFLP markers were derived from 33 PstI/MseI combinations and 5 EcoRI/MseI combinations. The approximate size of each marker was estimated by comparison with the mobility of the bands of the Sequamark 500-bp ladder (fmol sequencing system; Promega, Madison, WI) by linear interpolation. The primer sequences can be deduced from the marker designations. Autoradiograms were scored manually, each band being scored as a locus with a dominant (present) vs. recessive (absent) allele. Intensity differences were observed in the segregating bands, but visual interpretation was not considered reliable enough to assign allelic dosage to a given individual in the population. The 38 primer combinations yielded 585 informative markers, of which 162 were simplex and 50 were duplex in Stirling, 205 and 84 were simplex and duplex in 12601ab1, and 84 were present as simplex markers in both parents (double simplex).
SSR assays:
The forward primer was radiolabeled by mixing 0.1 μm forward primer with 0.04 μm [γ33P]ATP (10 mCi/ml), 1 × T4 polynucleotide kinase buffer, and 0.1 unit T4 polynucleotide kinase (New England Biolabs) to make a final reaction volume of 0.5 μl/assay. This was incubated at 37° for 30 min followed by heating to 65° for 10 min to terminate the reaction.
Twenty-microliter PCR reactions consisted of 50 ng of DNA, 0.2 mm dNTPs (Roche), 0.1 μl Taq DNA polymerase (5 unit/μl, Roche), 0.1 μl unlabeled reverse primer (10 μm), and 0.5 μl labeled forward primer (10 μm). The cycling conditions for PCR using a PE 9700 thermocycler were as follows: 3 min at 94° followed by 35 cycles of 15 sec at 94°, 30 sec at the specific annealing temperature for each primer (see Table 1), 30 sec at 72°, and then 5 min at 72°. Equal volumes of electrophoresis loading buffer (98% deionized formamide, 0.5 mm EDTA pH 8.0, 25 mg/ml bromophenol blue, 25 mg/ml xylene cyanol) were added to the samples, which were then denatured at 95° for 5 min, “snap cooled” on ice, and subjected to 5% denaturing polyacrylamide gel electrophoresis as described previously (Milbourne et al. 1997). Buffer on the anodal side was supplemented with 0.5 m NaOAC to create an ionic gradient, which resulted in better separation of the larger fragments.
TABLE 1.
Summary information on the six SSRs used to identify LGs IV, V, and XI including length in base pairs and optimum annealing temperature
| SSR | Repeat | LG | Sequence | Length (bp) | Annealing temperature |
|---|---|---|---|---|---|
| STM5140 | (AAT)7 | IV | F: GCTATTGTTGCAGATAATACG | 180–190 | 45° |
| (estp104b1) | R: GCCATGCACTAATCTTTGA | ||||
| STM3160 | (AG)25 | IV | F: ACTCAGAACACCGAATGG | 105–127 | 54° |
| R: CTCCACTACTGGTCAAATCC | |||||
| STM3179 | (AT)10 | V | F: ACCATAACCATCATCGTCTAC | 168–185 | 54° |
| R: GATAGTGATTGGTGGTGATTC | |||||
| STM5148 | (GAA)17 | V | F: TCTTCTTGATGACAGCTTCG | 380–440 | 55° |
| R: ACCTCAGATAGTTGCCATGTCA | |||||
| STM5109 | (TA)9 | XI | F: ACAGCTTTACTCTGCTCAACAA | 208–212 | 57° |
| (estp2OB8) | R: TGACGGCGTTATTACAAAC | ||||
| STM5130 | (CGG)5 | XI | F: AAAGTACAGCGAAGATGACGAC | 240–250 | 57° |
| (estp21B15) | R: TTACCTTTGCAACCTTGCC |
Construction of linkage map of markers:
The first stage of the linkage analysis was to identify the most likely dosage for each marker, that is, simplex or duplex for dominant markers present in one of the parents, double simplex or higher dosage for dominant markers present in both parents, or various dosages for multi-allelic markers. The dosage with the highest probability conditional on the observed parent and offspring phenotypes was identified using the approach of Luo et al. (2000). The goodness of fit of the most likely genotype was assessed, and markers for which this was a poor fit were checked and either corrected or excluded as appropriate. All simplex, duplex, double-simplex, and multi-allelic markers were analyzed by cluster analysis to partition them into linkage groups (Luo et al. 2001). Dominant markers present in both parents other than the double-simplex ones were excluded because they are very uninformative about recombination.
For each linkage group, recombination frequencies and LOD scores between every pair of markers were calculated for all possible phases using the EM algorithm, as described by Luo et al. (2001). The recombination frequencies and LOD scores from the phase with the highest likelihood were used to order the markers. A simulated annealing algorithm (Hackett et al. 2003) was used to identify the order with the minimum value of the weighted least squares criterion (Stam 1993) and to calculate map distances between the markers. All of these analyses were done using the TetraploidMap software program of Hackett and Luo (available from authors).
Analysis of trait data for QTL:
A preliminary analysis of the trait data was performed by regressing the trait on the presence/absence scores of each marker in turn. The threshold for declaring a significant association was determined by a permutation test (Churchill and Doerge 1994) to overcome problems of multiple testing. This analysis eliminated markers with no significant effect on the trait and gave an approximate location for QTL affecting blight, maturity, and height.
However, unless a population has been scored for many multi-allelic microsatellite markers that are sufficiently informative to follow the inheritance of the four alleles from each parent individually, regression on a single marker is not very informative. Therefore, linkage groups showing significant associations with a trait were analyzed further using the QTL interval mapping method of Hackett et al. (2001). The first step estimated a linkage map for each group, as described in the previous section, and identified the phase of each marker in the parents. This information was used to reconstruct a graphical genotype for each offspring, to deduce how chromosome segments had been inherited from each parent with the fewest recombinations required to give the offspring's phenotype. From the graphical genotypes, the set of possible genotypes at a putative QTL anywhere on the chromosome was identified and their probabilities calculated. The trait values were then modeled as functions of the possible QTL genotypes for a grid of positions along the chromosome, using a normal mixture model, but rewritten as an iterative weighted regression (Jansen 1992), updating the probabilities of the QTL genotypes for each regression step. An additive model was used for the effects of the QTL alleles at this stage in the analysis. Hence the effects of the QTL alleles from one of the parents were modeled as additive effects as
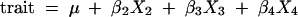 |
where μ is a constant term; β2, β3, and β4 are the effects of the QTL alleles on homologous chromosomes C2, C3, and C4; and X2, X3 and X4 are indicator (1/0) variables showing whether or not each individual has that allele. The effect of the allele on homologous chromosome C1 is fixed at zero to avoid overparameterization of the model.
Once the chromosomal position with the highest likelihood for a QTL was found, alternatives, such as models including dominance effects, were explored.
RESULTS
Phenotypic data:
The phenotypic distributions of clone means are shown in Figure 1 for fb4, saudpc%, tb%, fbc, ht, and mat. There were statistically significant differences (P < 0.001) between clones for all traits. Reciprocal crosses are not distinguished as there were no statistically significant reciprocal differences (P > 0.05). Heritabilities were all high: 0.878 (fb4), 0.853 (saudpc%), 0.870 (tb%), 0.629 (fbc), 0.876 (ht), and 0.916 (mat). The high values for ht and mat reflect the much larger variance component for clones compared with that for the clones × years of interaction, namely 68.65 vs. 15.32 for height and 2.476 vs. 0.160 for maturity. Cultivar Stirling had better resistance to late blight than clone 12601ab1 in its foliage (fb4, saudpc%, and fbc) and tubers (tb%), (P < 0.001 for fb4, saudpc%, and tb% and P < 0.05 for fbc), whereas the two parents had similar heights and maturities, although Stirling was significantly shorter (P < 0.001) than 12601ab1.
Figure 1.—
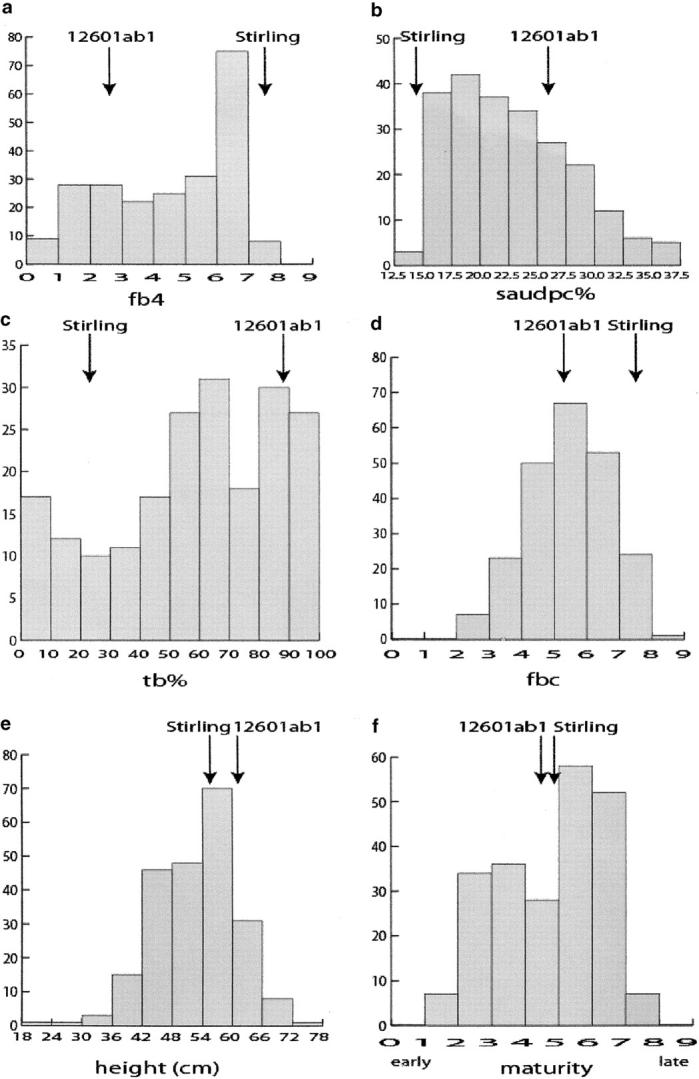
The phenotypic distributions of clone means for (a) fb4 fourth foliage blight score in field on a 1–9 scale of increasing resistance; (b) saudpc% square root of the area under disease progress curve on percentage scale; (c) tb% percentage of blight-infected tubers; (d) fbc foliage blight score for complex race in glasshouse on a 1–9 scale of increasing resistance; (e) height; and (f) maturity.
Correlations among traits:
The correlations among traits are shown in Table 2. All except the correlation between mat and fbc were statistically significant (P < 0.05). Thus, resistance to blight in the foliage (fb4, saudpc%, and fbc) was correlated with resistance in the tubers (tb%) and foliage resistance in the glasshouse (fbc) was correlated with resistance in the field (fb4 and saudpc%). The magnitudes of the correlations with fb4 and saudpc% were very similar as these traits were highly correlated, both being measures of field resistance at Yonderton in 1998, but with scales running in opposite directions.
TABLE 2.
Correlations among the traits of mat, ht, tb%, and foliage blight in the field (fb4 and saudpc%) and glasshouse (fbc) where early maturity has a low score and resistance has a low score for tb% and saudpc% but a high score for fb4 and fbc
| mat | 1.000 | |||||
|---|---|---|---|---|---|---|
| ht | 0.674 | 1.000 | ||||
| tb% | −0.508 | −0.403 | 1.000 | |||
| fb4 | 0.559 | 0.511 | −0.641 | 1.000 | ||
| saudpc% | −0.517 | −0.461 | 0.635 | −0.963 | 1.000 | |
| fbc | 0.033 | 0.157 | −0.274 | 0.459 | −0.455 | 1.000 |
| mat | ht | tb% | fb4 | saudpc% | fbc |
Maturity (mat) was correlated with all of the other traits except fbc. Linear regression was used to partition the trait variance for fb4, saudpc%, and tb% into a component associated with maturity and a residual effect, which can be interpreted as true resistance/susceptibility. The component associated with maturity was statistically significant (P < 0.001) for all three traits and the percentage variance accounted for was 31.1% (fb4), 26.9% (saudpc%), and 25.4% (tb%). While the full range of blight scores could be seen at medium maturity, the association between resistance and late maturity was clear for fb4 and saudpc% and so was the association between early maturity and susceptibility for tb%. The linear regressions are
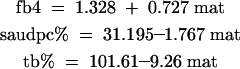 |
Stirling's major R-gene:
Stirling's major R-gene was treated as a qualitative trait and mapped along with the molecular markers. The ratio of 137:78 clones with and without the R-gene was a better fit to a 1:1 ratio (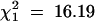 ) than to a 5:1 ratio (
) than to a 5:1 ratio (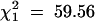 ), but a statistically significant (P < 0.001) departure from expectation. Nevertheless, it was concluded that a single copy of the R-gene was present in cultivar Stirling. It mapped to linkage group XI at 79 cM on a marker map running from EAACMCAG_91.0 at 0 cM to PGTAMCAC_600.0 at 122 cM, with the SSR markers STM5130 and STM5109 at 36 and 60 cM, respectively. The closest marker in simplex coupling was PCTMCAC_153.0, 16 cM away at 95 cM, and this marker had the highest chi-square (72.39) of those tested for association with the R-gene and also displayed distorted segregation (125:83,
), but a statistically significant (P < 0.001) departure from expectation. Nevertheless, it was concluded that a single copy of the R-gene was present in cultivar Stirling. It mapped to linkage group XI at 79 cM on a marker map running from EAACMCAG_91.0 at 0 cM to PGTAMCAC_600.0 at 122 cM, with the SSR markers STM5130 and STM5109 at 36 and 60 cM, respectively. The closest marker in simplex coupling was PCTMCAC_153.0, 16 cM away at 95 cM, and this marker had the highest chi-square (72.39) of those tested for association with the R-gene and also displayed distorted segregation (125:83, 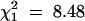 , P < 0.01).
, P < 0.01).
Marker-trait regressions:
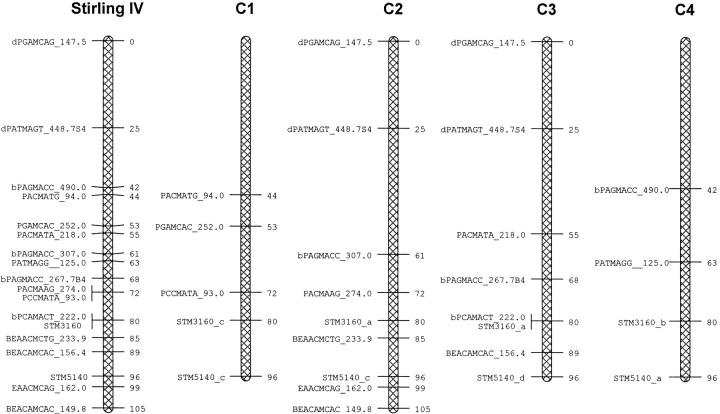
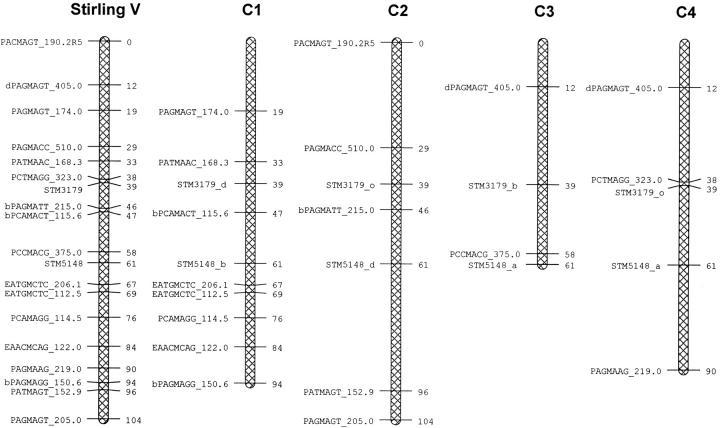
Regression of the trait data on the presence/absence of each marker identified 14 markers that were significantly associated with one or more of the traits according to a permutation test (with |t| >3.8; Table 3). All of these markers were present only in the Stirling parent. Linkage analysis showed that these markers belonged to two linkage groups, identified as linkage groups IV and V by the presence of microsatellite markers and AFLPs shared with a diploid reference map referred to as cross SH × RH (Isidore et al. 2003). The maps of these linkage groups, with their four homologs, are shown in Figures 2 and 3. The simplex allele STM5148_b on C1 of linkage group V of Stirling was also present in 12601ab1 and did not segregate. The percentage variance of the traits mat, ht, tb%, fb4, saudpc%, and fbc that was accounted for by the regression on each of the 14 markers is shown in Table 3. Mat and ht are both highly associated with the markers from linkage group V, but not with those from linkage group IV, and fbc is associated only with the markers from linkage group IV. Both field measures of foliage blight, fb4 and saudpc%, show associations with markers from both groups. Tuber blight (tb%) is associated with markers from linkage group V according to the permutation test, but also has a weak association with markers from linkage group IV.
TABLE 3.
Percentage trait variance accounted for by regression on marker phenotypes
| Trait
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Marker | Dosage | Linkage group |
mat | ht | tb% | fb4 | saudpc% | fbc |
| PAGMAGT_174.0 | S | V | 25.4 | 14.9 | 12.6 | 10.8 | 9.2 | NS |
| PATMAAC_168.3 | S | V | 38.9 | 24.8 | 18.4 | 17.2 | 15.7 | NS |
| STM 3179_d | S | V | 41.5 | 21.3 | 20.0 | 12.1 | 11.9 | NS |
| STM 5148_a | D | V | 7.9 | 6.4 | 1.7 | 2.2 | NS | NS |
| EATGMCTC_206.1 | S | V | 15.0 | 12.8 | 4.2 | 6.1 | 5.7 | NS |
| EATGMCTC_112.5 | S | V | 14.8 | 12.2 | 2.2 | 6.0 | 5.1 | NS |
| PCAMAGG_114.5 | S | V | 8.8 | 13.8 | 5.4 | 8.0 | 7.0 | NS |
| EAACMCAG_122.0 | S | V | 10.4 | 8.7 | NS | 3.8 | 3.4 | NS |
| PATMAGT_448.7 | D | IV | NS | NS | 2.6 | 7.4 | 6.9 | 1.8 |
| PACMATG_94.0 | S | IV | NS | NS | NS | 3.5 | 3.9 | 9.2 |
| PGAMCAC_252.0 | S | IV | NS | NS | NS | 3.6 | 3.5 | 8.7 |
| PACMAAG_274.0 | S | IV | NS | NS | 2.2 | 7.7 | 9.2 | 6.4 |
| PCCMATA_93.0 | S | IV | NS | NS | 2.7 | 8.6 | 9.5 | 7.6 |
| STM 3160_a | D | IV | NS | NS | NS | 13.3 | 10.8 | 8.1 |
The dosage of each marker is indicated as S (simplex) or D (duplex). NS indicates that the regression is not significant at P < 0.05, while italic type indicates a regression that is significant according to a permutation test (|t| >3.8). Traits are mat, ht, tb%, and foliage blight in field (fb4 and saudpc%) and glasshouse (fbc).
Figure 2.—
Marker map of linkage group IV of Stirling showing the four homologous chromosomes (C1–C4) combined and separately.
Figure 3.—
Marker map of linkage group V of Stirling showing the four homologous chromosomes (C1–C4) combined and separately.
Interval mapping:
Interval mapping was used to examine the effects of the QTL on linkage groups IV and V of Stirling. The effects of the QTL alleles from the Stirling parent were initially modeled as additive effects. As no significant associations between traits and alleles from the 12601ab1 parent were detected by regression analysis, these effects were not included in the model. The locations of the maxima of the likelihood profiles and the estimates of QTL parameters are shown in Tables 4 and 5 for linkage groups V and IV, respectively.
TABLE 4.
Interval mapping for Stirling linkage group V
| Trait | Location (cM) |
R2 (%) | Constant | Allele 2 | Allele 3 | Allele 4 |
|---|---|---|---|---|---|---|
| mat | 36.0 | 54.7 | 1.2 (0.23) | 2.3 (0.18) | 2.2 (0.17) | 2.5 (0.18) |
| ht | 34.0 | 26.5 | 39.7 (1.55) | 7.2 (1.22) | 8.6 (1.13) | 9.3 (1.19) |
| tb% | 44.0 | 26.3 | 104.1 (5.67) | −30.6 (4.45) | −26.2 (4.20) | −31.6 (4.22) |
| fb4 | 44.0 | 20.3 | 1.9 (0.40) | 1.8 (0.31) | 1.7 (0.29) | 2.1 (0.30) |
| saudpc% | 44.0 | 17.5 | 29.8 (1.07) | −4.2 (0.83) | −4.3 (0.77) | −5.0 (0.80) |
Location, the location of the maximum of the likelihood profile; R2, the trait variance accounted for; Constant, the constant terms in the interval mapping model; Allele 2, Allele 3, and Allele 4, effects of the different alleles (relative to the constant). Standard errors are shown in parentheses. Traits are mat, ht, tb%, and foliage blight (fb4 and saudpc%).
TABLE 5.
Interval mapping for Stirling linkage group IV
| Trait | Location (cM) |
R2 (%) | Constant | Allele 2 | Allele 3 | Allele 4 |
|---|---|---|---|---|---|---|
| tb% | 72.0 | 10.6 | 72.8 (6.14) | −16.3 (4.68) | −15.6 (4.74) | 2.5 (4.67) |
| fb4 | 72.0 | 24.1 | 3.0 (0.38) | 1.8 (0.29) | 1.8 (0.29) | 0.1 (0.29) |
| saudpc% | 72.0 | 24.5 | 27.7 (0.99) | −5.1 (0.75) | −4.5 (0.76) | −0.4 (0.75) |
| fbc | 66.0 | 28.7 | 4.4 (0.23) | 1.3 (0.17) | 1.2 (0.18) | 0.2 (0.18) |
| r_tb% | 72.0 | 13.6 | 9.9 (5.20) | −13.9 (3.96) | −12.7 (4.00) | 5.6 (3.95) |
| r_fb4 | 74.0 | 31.5 | −1.6 (0.31) | 1.7 (0.23) | 1.6 (0.24) | −0.1 (0.24) |
| r_saudpc% | 74.0 | 30.7 | 4.4 (0.82) | −4.8 (0.62) | −4.1 (0.63) | −0.1 (0.62) |
Trait names prefixed with r_ indicate the residuals after regression on maturity. Location, location of the maximum of the likelihood profile; R2, the trait variance accounted for; Constant, the constant terms in the interval mapping model; and Allele 2, Allele 3, and Allele 4, effects of the different alleles (relative to the constant). Standard errors are shown in parentheses. Traits are tb% and foliage blight in the field (fb4 and saudpc%) and glasshouse (fbc).
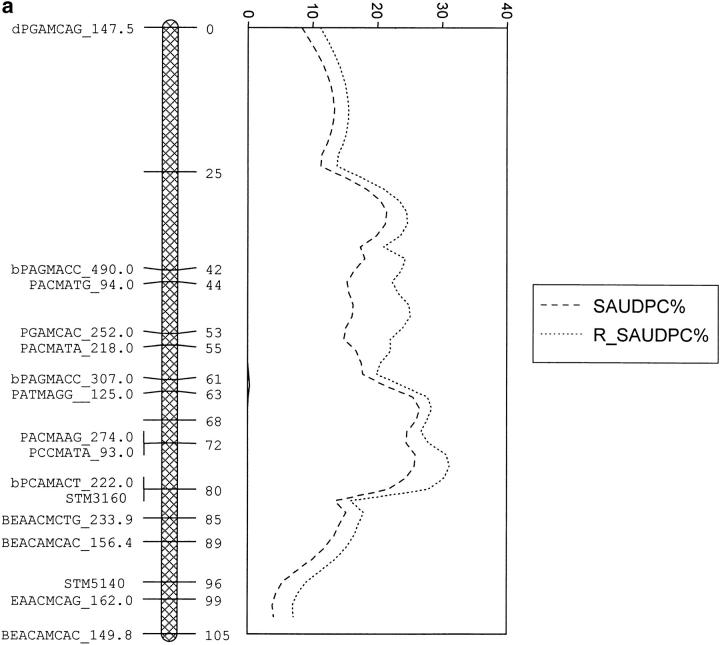
For linkage group V, the QTL for saudpc%, fb4, tb%, ht, and mat mapped to similar positions and explained from 17.5 to 54.7% of the variation, which was more than the best individual marker (Table 3). The likelihood profiles for mat and saudpc% are shown in Figure 4b. For each trait the effects of alleles 2, 3, and 4 were similar in size and sign, suggesting that the QTL on linkage group V is a simplex allele on homolog C1 with a significantly different effect from those on the other three homologs; i.e., the genotype is Qqqq and gives rise to two gametes, Qq and qq. The allele Q on homolog C1 is associated in the offspring with earlier maturity (3.53 vs. 5.87 where 9 is late), shorter height (48.07 vs. 56.43 cm), and susceptibility to blight in the tubers (74.63% vs. 45.17%) and foliage as measured by fb4 (3.77 vs. 5.63 where 9 is resistant) and saudpc% (25.3 vs. 20.8). There was no evidence for a QTL affecting fbc on linkage group V. When the residuals from the regression of tb%, fb4, and saudpc% on mat were analyzed, there was no significant effect of a QTL on linkage group V, thus providing evidence for a QTL having a direct effect on maturity, which in turn affects the response to blight.
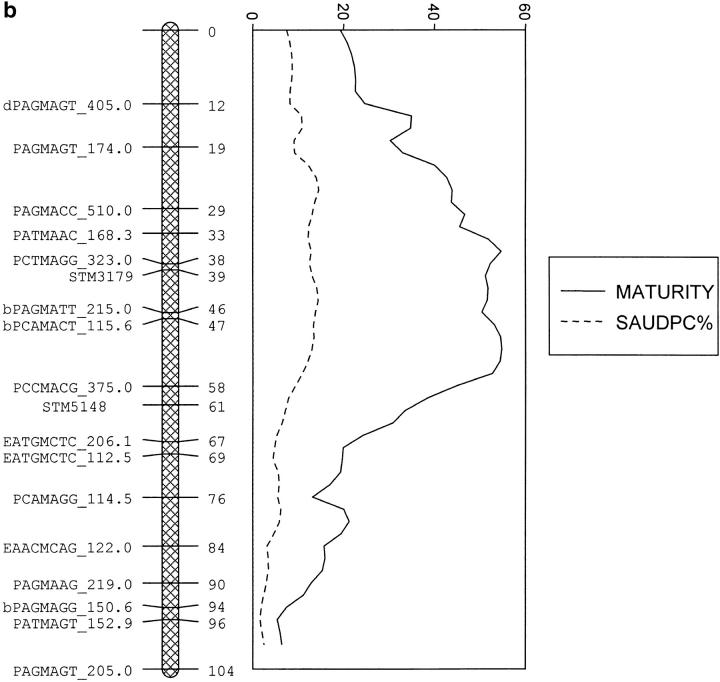
Figure 4.—
The likelihood profiles for (a) foliage blight score before and residuals after regression on maturity (saudpc% and r_saudpc%) on linkage group IV of Stirling and (b) maturity and foliage blight (saudpc%) on linkage group V of Stirling.
For linkage group IV, the QTL likelihood profiles for tb%, fb4, saudpc%, and fbc all peaked at similar positions and explained between 10.6 and 28.7% of the variance, which was considerably more than the best individual marker (Table 3). The residuals from the regressions on maturity of the first three of these traits (r_tb%, r_fb4, and r_saudpc%) also peaked at similar locations and explained a higher proportion of the trait variance due to the elimination of the variance associated with the maturity QTL on linkage group V. The likelihood profiles for saudpc% and r_saudpc% are shown in Figure 4a. For every trait the effect of allele 4 was not significantly different from zero, and hence from allele 1, whereas the effects of alleles 2 and 3 were significantly different from zero, but not from each other; i.e., the genotype of the QTL is QQqq and gives rise to three gametes, QQ, Qq, and qq. There were no significant effects of linkage group IV on mat or ht.
Alternative QTL models:
Stepwise regression was used to explore different models for the QTL effects on linkage group IV, using the estimated QTL genotypes for each individual, weighted by their probabilities at the peak of the likelihood profile. Table 6 shows the mean and standard error associated with the six possible QTL genotypes for r_saudpc% and fbc. For fbc, no improvement could be found on the additive model in which Qq is intermediate between QQ and qq. In contrast, with r_saudpc%, improvements were found on the additive model, which accounted for 30.7% of the variance. A model in which Q was completely dominant to q (i.e., Qq equals QQ) accounted for 35.3% of the variance. However, the mean trait value for individuals with QQ (Q23) was lowest (most resistant) and including a separate term for this genotype explained 37.2% of the variance. Hence, there is evidence that the possession of two copies of the resistance allele confers more resistance than a single copy; i.e., dominance is incomplete. Similar results were obtained for the other traits (tb%, fb4, saudpc%, r_tb%, and r_fb4) with the exception of fbc, which has already been mentioned.
TABLE 6.
Mean and SE of r_saudpc% and fbc foliage blight scores associated with each QTL genotype in offspring of Q1234 (= QQqq) for linkage group IV and comparison with additive model in Table 5
| QTL genotype of gametes |
Proposed model |
Mean (SE) for r_saudpc% |
Additive model for r_saudpc% from Table 5 |
Mean (SE) for fbc |
Additive model for fbc from Table 5 |
|---|---|---|---|---|---|
| Q23 | −2.7 (0.62) | −4.50 | 7.0 (0.19) | 6.9 | |
| Q12 | −1.6 (0.64) | −0.40 | 5.7 (0.19) | 5.7 | |
| Q24 | −1.0 (0.55) | −0.50 | 5.8 (0.17) | 5.9 | |
| Q34 | −0.6 (0.57) | 0.20 | 5.8 (0.17) | 5.8 | |
| Q13 | −0.3 (0.61) | 0.30 | 5.5 (0.18) | 5.6 | |
| Q14 | 5.9 (0.58) | 4.30 | 4.7 (0.16) | 4.6 |
Combining the two linkage groups:
Finally, as fb4, tb%, and mat are commonly measured in the potato-breeding program at SCRI, it was considered worthwhile to determine how much of the variation in fb4 and tb% could be explained by the QTL on linkage groups IV and V. Regression of fb4 and tb% on both mat and a dominant effect of a duplex allele at about position 72 cM on linkage group IV accounted for 56.3% of the variance in fb4 and 39.6% of the variance in tb%. The effect of the R-gene locus on linkage group XI was not significant.
DISCUSSION
The QTL interval mapping method of Hackett et al. (2001) allowed a more sophisticated genetical analysis of quantitative resistance to late blight than was possible in the earlier study of the same population by Meyer et al. (1998). Furthermore, inclusion of data on height and maturity, as well as on resistance to tuber blight and a field assessment of foliage resistance, resulted in more information on the genetics of blight resistance and the relative importance of various effects. Finally, the use of a simple and a complex race of P. infestans in glasshouse tests allowed Stirling's R-gene to be mapped to chromosome XI. As R7 has been mapped to this chromosome (El-Kharbotly et al. 1996), and the R7 differential was both a maternal and a paternal great-grandparent of Stirling, it is a likely candidate for Stirling's R-gene, but this needs confirmation once appropriate races of P. infestans are available. Although the analysis of Stewart et al. (2003) provided evidence of clones with the R-gene having higher and lower scores (i.e., more resistance) for fb4 and tb% than clones without the R-gene, the effects were not large (4.96 vs. 4.36 with standard error of difference (SED) of 0.285 and 54.4% vs. 66.2% with SED of 3.97%) and were not significant in the combined regressions on linkage groups IV, V, and XI. For comparison, the estimated means for genotypes QQqq and qqqq at the QTL on linkage group IV were 5.82 and 2.45 for fb4 and 48.8 and 81.4% for tb%.
The limitations of the genetical analysis were those imposed by inadequate genome coverage with appropriate and informative molecular markers and the number of QTL alleles and hence genotypes that occurred and could be distinguished in a full-sib family. Thirty-eight AFLP primer combinations and an initial set of 23 SSRs (which mapped to linkage groups I, IV, V, and XI) failed to produce a complete linkage map of markers (i.e., 12 linkage groups, each comprising four homologous chromosomes) in contrast to our experience in diploid potatoes where 20 AFLP primer combinations and 18 SSRs were adequate with one minor exception (Bryan et al. 2002). Hence, interval mapping was confined to linkage groups in which all four homologous chromosomes had been identified and associations between single markers and traits had been found. In future, the aim is to identify and apply interval mapping to all four homologous chromosomes of all 12 linkage groups. More codominant markers are needed for this purpose and so is more research on the clustering method used to search for linkage groups under polysomic inheritance where the allelic content of pairs of loci affects the information about the maximum-likelihood estimate of the recombination frequency. It may then be possible to detect QTL of smaller effect.
The advantages of the QTL interval mapping method were seen on linkage group V and to an even greater extent on linkage group IV. For mat, ht, and blight (tb%, fb4, and saudpc%), the QTL allele of interest on V was shown to be present in single copy and linked in coupling to seven markers positioned from 19 to 84 cM—more markers than might have been expected by chance with 162 simplex markers and 48 chromosomes, which may reflect polymorphism on LGV from past introgressions. Postulating a QTL between two markers always explained more of the trait variance than either marker alone; for example, the SSR marker allele STM3179 explained 41.5% of the variation in mat compared with 54.7% for the QTL. On linkage group IV, it was concluded that Stirling had two copies of the resistance allele at the QTL. The only two duplex coupling markers were at 25 and 80 cM, and the latter (STM3160_a) was difficult to score and sensitive to errors, being a marker that segregated 5:1. As a consequence, the postulated QTL explained considerably more of the trait variance than either of these markers, including 10.6% for tb%. Likewise, it explained considerably more of the trait variance than the markers at 72 cM, PCCMATA_93.0 and PACMAAG_274.0, both simplex markers associated with susceptibility and resistance, respectively, and postulated to be very close to the QTL. With chromosomal segregation, the difference between the two marker-class means (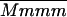 −
− 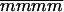 ) for a duplex QTL is 1/3(1 − 2r) (
) for a duplex QTL is 1/3(1 − 2r) (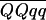 −
− 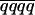 ) with M and Q in coupling and 1/3(1 − 2r) (
) with M and Q in coupling and 1/3(1 − 2r) (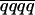 −
− 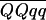 ) with M in coupling with q. Hence, one-third of the difference between the two QTL genotypes is the maximum that can be detected with simplex markers, and this occurs when the recombination frequency (r) is zero. In the earlier work of Meyer et al. (1998), a duplex AFLP marker, which they called p75m48 = 5, was found, which explained 31.6% of the phenotypic variance in blight scores but, as with STM3160_a, it was difficult to score and reproducible results were not achieved in this study. Nevertheless, it is reassuring that the interval mapping analysis gave a similar conclusion about the location of the QTL for foliage blight resistance on linkage group IV.
) with M in coupling with q. Hence, one-third of the difference between the two QTL genotypes is the maximum that can be detected with simplex markers, and this occurs when the recombination frequency (r) is zero. In the earlier work of Meyer et al. (1998), a duplex AFLP marker, which they called p75m48 = 5, was found, which explained 31.6% of the phenotypic variance in blight scores but, as with STM3160_a, it was difficult to score and reproducible results were not achieved in this study. Nevertheless, it is reassuring that the interval mapping analysis gave a similar conclusion about the location of the QTL for foliage blight resistance on linkage group IV.
The interval mapping method was also superior in giving estimates of the three QTL genotypes QQqq, Qqqq, and qqqq, as opposed to only the differences between two dominant marker classes for a marker at an unknown distance from the QTL. In other words, QTL effects and location are not confounded and genotypes QQqq and Qqqq can be distinguished. With only 50 duplex markers in this study, and 45 in the previous one, it would appear fortuitous to find a marker such as p75m48 = 5 closely linked in coupling with a duplex QTL (i.e., MQ/MQ/mq/mq) because there are 12 linkage groups in potato, and for each group there are six possible pairs of chromosomes.
Multi-allelic codominant markers are desirable in potato for greater precision in linkage and QTL analysis (Hackett et al. 2001; Luo et al. 2001). The six SSR markers shown in Table 1 were used primarily to identify the three linkage groups of interest by reference to a standard diploid population, SH × RH (Isidore et al. 2003). None of them allowed recognition of all four homologous chromosomes in a parent, nor had the five SSRs on IX used in the example in Luo et al. (2001). For the 11 SSRs, a total of two to four alleles were found with one to three present in each parent. Hence the use of SNP haplotypes, with the possibility of measuring allele dosages in both parents and progeny, is worth exploring for the future and so is developing more SSRs (Ghislain et al. 2004). It is probably not worthwhile to add to the population size beyond the 227 of this study to increase the number of chiasmata sampled and hence the accuracy of QTL location. It is the QTL of large effect, which can be detected in a study of this size, which are the candidates for marker-assisted selection and map-based cloning. Likewise, just two replicates of each clone were sufficient for high heritabilities of clonal means in all tests and more replicates would appear unnecessary. The limitations of the genetical analysis imposed by the number of QTL alleles, and hence genotypes that occurred and could be distinguished in a full-sib family, were manifest with the QTL on V for maturity. Only two genotypes segregated, Qqq′q′ and qqq′q′, and it is not known how QQq′q′ and Qqq′q′ would compare. Hackett et al. (2001) demonstrated that a maximum of six main effects, 13 biallelic interactions, 12 triallelic interactions, and 4 tetra-allelic interactions can be fitted to the 36 genotypes in a full-sib family arising from the cross Q1Q2Q3Q4 × Q5Q6Q7Q8, but a large population would be required to obtain good estimates of the means. Furthermore, given the number of SSR alleles found, it seems unlikely that eight QTL alleles would, in practice, segregate in a full-sib family. Nevertheless, mapping in tetraploids has provided additional information on allelic diversity and interactions to mapping in diploid relatives and the results are directly relevant to a tetraploid breeding program.
Although the QTL for ht, mat, and blight on V mapped to slightly different locations, it is assumed that there is a single locus with allelic variation for a physiological trait that affects height and maturity and indirectly affects resistance/susceptibility to blight. Likewise, it is assumed that there is a single QTL on IV affecting foliage resistance and tuber blight resistance in the glasshouse and field. However, tight linkage of separate genes cannot be ruled out.
Although cultivar Stirling and clone 12601ab1 had similar foliage maturity scores of 4.95 and 4.61, respectively, the former segregated for maturity and a wide range of variation was seen in the offspring, with 54.7% of the variation explained by a QTL close to marker STM3179 on linkage group V. Hence the QTL allele for early maturity in Stirling must be negated by the other allele (triplex) achieving maturity later than that of clone 12601ab1. Collins et al. (1999) and Visker et al. (2003) also found a QTL of large effect on maturity close to the same marker on V, which explained 62.3% (averaged over 3 years) and 84.0%, respectively, of the variation in their diploid populations. The effect of maturity on blight was marked as can be seen by the linear regressions of fb4, saudpc%, and tb% on mat. Stirling is a maincrop variety and the later-maturing clones that were found in the population are undesirable. Five first early cultivars were included in the yield trials as controls and had a mean maturity score of 2.525 over the 2 years for which the trait was scored. The linear regressions predict increased susceptibility to foliage and tuber blight with earlier maturity, with changes in fb4 from 4.93 to 3.16, in saudpc% from 22.45 to 26.73, and in tb% from 55.77 to 78.23% as maturity changes from 4.95 to 2.525.
When the residuals from the regression of fb4, saudpc%, and tb% were analyzed, there was no significant effect of a QTL on LGV, thus providing evidence for a QTL having a direct effect on maturity, which in turn affects the response to blight. In the absence of maturity scores, one would interpret the blight data in terms of a QTL on V explaining 26.3, 20.3, and 17.5% of the variation in tb%, fb4, and saudpc%. Visker et al. (2003) also found a QTL for foliage resistance to late blight, which appeared to map at the same position as the QTL for maturity on V and which explained 41% of the variation in blight scores. However, when they adjusted the late blight scores for foliage maturity, the effect of blight resistance on V was halved but not eliminated. Hence, they could not rule out two closely linked genes, one affecting blight and maturity and the other affecting only blight resistance. Collins et al. (1999) found in their diploid population that the most significant associations for foliage and tuber blight, maturity and vigor, were with marker alleles on V in the region of STM3179 that originated from the male (susceptible) parent. Susceptibility to foliage blight was associated with early maturity and lack of vigor, but with resistance to tuber blight, the latter result was the opposite from this study. The QTL explained 29.8% (averaged over 3 years) of the variation in foliage blight scores. While it is known that resistance to late blight depends on the physiological age of the plant (Stewart et al. 1983a,b, 1996), further research is required to explain the association between early foliage maturity and tuber susceptibility to blight found in this study with glasshouse-grown potato tubers. In the meantime, it is important in breeding for blight resistance to assess foliage maturity and to explore any relationships with foliage and tuber resistance.
The resistance QTL on LGIV was not associated with late maturity and hence is extremely valuable in a breeding program, but bearing in mind that two copies of the allele give more resistance than only one. Likewise the QTL for foliage resistance on LGIII reported by Visker et al. (2003) was independent of maturity. Although the QTL for blight resistance on LGIV and maturity on LGV explained only 56.3 and 39.6% of the variation in foliage and tuber blight scores, it was encouraging for prospects for molecular breeding to find a QTL of large effect for quantitative resistance, which had traditionally been equated with polygenic resistance. Flanking markers on LGIV allowing the measurement of allele dosage would be very useful. Marker-assisted selection in the seedling generation in the glasshouse could have a major impact on potato breeding because all of the genetical variation from hybridization has been released, yet six generations of expensive clonal selection in the field and special disease and quality tests are usually required to find potential new cultivars.
Lastly, there is the question of whether or not a more elaborate and sophisticated theory is required. The theory of Luo et al. (2001) and Hackett et al. (2001) is based on the simplest situation that can give tetrasomic inheritance, namely random pairing of four homologous chromosomes to give two pairs of bivalents at meiosis. In practice, many departures from this simple situation could occur, in particular: multivalents and double reduction; lack of complete homology between chromosomes and hence departures from random pairing; distorted segregation due to differential fertility and viability; errors in scoring gels (although gels should be scored independently by more than one person and any disagreements that cannot be resolved treated as missing data); and genuine anomalies requiring cytogenetical explanations. Statistical geneticists have made a start with multivalents in the linkage analysis of autotetraploids (Wu et al. 2001) and will no doubt model most if not all of the other situations and find the results intellectually satisfying. How useful such theory will be in practice remains to be seen, bearing in mind that the linkage map is usually a route to the QTL analysis rather than an end in itself, and diploid linkage maps are available. In the meantime, this article shows that simple theory can be applied to real data to give useful results.
Acknowledgments
Zewei Luo, now at the University of Birmingham, developed the theory and software for linkage analysis while on the Genome Analysis of Agriculturally Important Traits (GAIT) Project. D. Todd and R. N. Wilson provided technical support for the ware trials. This work was financially supported by the Scottish Executive Environment and Rural Affairs Department, the United Kingdom Biotechnology and Biological Sciences Research Council (GAIT Project), and the British Potato Council (Ph.D. studentship to B.P.).
References
- Bradshaw, J. E., H. E. Stewart, R. L. Wastie, M. F. B. Dale and M. S. Phillips, 1995. Use of seedling progeny tests for genetical studies as part of a potato (Solanum tuberosum subsp. tuberosum) breeding programme. Theor. Appl. Genet. 90: 899–905. [DOI] [PubMed] [Google Scholar]
- Bradshaw, J. E., C. A. Hackett, R. C. Meyer, D. Milbourne, J. W. McNicol et al., 1998. Identification of AFLP and SSR markers associated with quantitative resistance to Globodera pallida (Stone) in tetraploid potato (Solanum tuberosum subsp. tuberosum) with a view to marker-assisted selection. Theor. Appl. Genet. 97: 202–210. [Google Scholar]
- Bradshaw, J. E., H. E. Stewart and G. R. Mackay, 1999 New approaches to breeding for late blight resistance: objectives, sources and technology. Proceedings of Global Initiative on Late Blight Conference, March 1999, Quito, Ecuador, pp. 43–46.
- Bryan, G. J., K. McLean, J. E. Bradshaw, W. S. De Jong, M. S. Phillips et al., 2002. Mapping QTLs for resistance to the cyst nematode Globodera pallida derived from the wild potato species Solanum vernei. Theor. Appl. Genet. 105: 68–77. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, A., D. Milbourne, L. Ramsay, R. Meyer, C. Chatot-Balandras et al., 1999. QTL for field resistance to late blight in potato are strongly correlated with maturity and vigour. Mol. Breed. 5: 387–398. [Google Scholar]
- Cruickshank, G., H. E. Stewart and R. L. Wastie, 1982. An illustrated assessment key for foliage blight of potatoes. Potato Res. 25: 213–214. [Google Scholar]
- Doerge, R. W., and B. A. Craig, 2000. Model selection for quantitative trait locus analysis in polyploids. Proc. Natl. Acad. Sci. USA 97: 7951–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kharbotly, A., C. Palomino-Sanchez, F. Salamini, E. Jacobsen and C. Gebhardt, 1996. R6 and R7 alleles of potato conferring race-specific resistance to Phytophthora infestans (Mont.) de Bary identified genetic loci clustering with the R3 locus on chromosome XI. Theor. Appl. Genet. 92: 880–884. [DOI] [PubMed] [Google Scholar]
- Gallais, A., 2003 Quantitative Genetics and Breeding Methods in Autopolyploid Plants. INRA, Paris.
- Gebhardt, C., and J. P. T. Valkonen, 2001. Organization of genes controlling disease resistance in the potato genome. Annu. Rev. Phytopathol. 39: 79–102. [DOI] [PubMed] [Google Scholar]
- Genstat 5 Committee, 1993 GENSTAT 5 Release 3 Reference Manual. Clarendon Press, Oxford.
- Ghislain, M., D. M. Spooner, F. Rodríguez, F. Villamón, J. Núñez et al., 2004. Selection of highly informative and user-friendly microsatellites (SSRs) for genotyping of cultivated potato. Theor. Appl. Genet. 108: 881–890. [DOI] [PubMed] [Google Scholar]
- Hackett, C. A., J. E. Bradshaw and J. W. McNicol, 2001. Interval mapping of quantitative trait loci in autotetraploid species. Genetics 159: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett, C. A., B. Pande and G. J. Bryan, 2003. Constructing linkage maps in autotetraploid species using simulated annealing. Theor. Appl. Genet. 106: 1107–1115. [DOI] [PubMed] [Google Scholar]
- International Potato Center, 1997 Medium-Term Plan 1998–2000. International Potato Center, Lima, Peru.
- Isidore, E., H. Van Os, S. Andrzejewski, J. Bakker, I. Barrena et al., 2003. Toward a marker-dense meiotic map of the potato genome: lessons from linkage group I. Genetics 165: 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. C., 1992. A general mixture model for mapping quantitative trait loci by using molecular markers. Theor. Appl. Genet. 85: 252–260. [DOI] [PubMed] [Google Scholar]
- Luo, Z. W., C. A. Hackett, J. E. Bradshaw, J. W. McNicol and D. Milbourne, 2000. Predicting parental genotypes and gene segregation for tetrasomic inheritance. Theor. Appl. Genet. 100: 1067–1073. [Google Scholar]
- Luo, Z. W., C. A. Hackett, J. E. Bradshaw, J. W. McNicol and D. Milbourne, 2001. Construction of a genetic linkage map in tetraploid species using molecular markers. Genetics 157: 1369–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolmson, J. F., 1976. Assessment of field resistance to blight (Phytophthora infestans) in potatoes. Trans. Br. Mycol. Soc. 67: 321–325. [Google Scholar]
- Meyer, R. C., D. Milbourne, C. A. Hackett, J. E. Bradshaw, J. W. McNicol et al., 1998. Linkage analysis in tetraploid potato and association of markers with quantitative resistance to late blight (Phytophthora infestans). Mol. Gen. Genet. 259: 150–160. [DOI] [PubMed] [Google Scholar]
- Milbourne, D., R. C. Meyer, J. E. Bradshaw, E. Baird, N. Bonar et al., 1997. Comparison of PCR-based marker systems for the analysis of genetic relationships in cultivated potato. Mol. Breed. 3: 127–136. [Google Scholar]
- Ming, R., S-C. Liu, P. H. Moore, J. E. Irvine and A. H. Paterson, 2001. QTL analysis in a complex autopolyploid: genetic control of sugar content in sugar cane. Genome Res. 11: 2075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhagemann, P., C. Chatot-Balandras, R. Schafer-Preg, D. Wegener, C. Palomino et al., 1999. A genetic analysis of quantitative resistance to late blight in potato: towards marker-assisted selection. Mol. Breed. 5: 399–415. [Google Scholar]
- Simko, I., 2002. Comparative analysis of quantitative trait loci for foliage resistance to Phytophthora infestans in tuber-bearing Solanum species. Am. J. Potato Res. 79: 125–132. [Google Scholar]
- Stam, P., 1993. Construction of integrated genetic linkage maps by means of a new computer package: Joinmap. Plant J. 3: 739–744. [Google Scholar]
- Stewart, H. E., P. H. Flavelle, D. C. McCalmont and R. L. Wastie, 1983. a Correlation between glasshouse and field tests for resistance to foliage blight caused by Phytophthora infestans. Potato Res. 26: 41–48. [Google Scholar]
- Stewart, H. E., D. C. McCalmont and R. L. Wastie, 1983. b The effect of harvest date and the interval between harvest and inoculation on the assessment of the resistance of potato tubers to late blight. Potato Res. 26: 101–107. [Google Scholar]
- Stewart, H. E., R. L. Wastie and J. E. Bradshaw, 1996. Susceptibility to Phytophthora infestans of field- and glasshouse-grown potato tubers. Potato Res. 39: 283–288. [Google Scholar]
- Stewart, H. E., J. E. Bradshaw and B. Pande, 2003. The effect of the presence of R-genes for resistance to late blight (Phytophthora infestans) of potato (Solanum tuberosum) on the underlying level of field resistance. Plant Pathol. 52: 193–198. [Google Scholar]
- Visker, M. H., L. C. Keizer, H. J. Van Eck, E. Jacobsen, L. T. Colon et al., 2003. Can the QTL for late blight resistance on potato chromosome 5 be attributed to foliage maturity type? Theor. Appl. Gen. 106: 317–325. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Van De Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. S., R. Wu, C-X. Ma, Z-B. Zeng, M. C. K. Yang et al., 2001. A multivalent pairing model of linkage analysis in autotetraploids. Genetics 159: 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]