Abstract
In the green alga, Chlamydomonas, chloroplast DNA is maternally transmitted to the offspring. We previously hypothesized that the underlying molecular mechanism involves specific methylation of maternal gamete DNA before mating, protecting against degradation. To obtain direct evidence for this, we focused on a DNA methyltransferase, DMT1, which was previously shown to be localized in chloroplasts. The full-length DMT1 protein with a molecular mass of 150 kD was expressed in insect cells, and its catalytic activity was determined. In vitro assays using synthetic DNA indicated methylation of all cytosine residues, with no clear selectivity in terms of the neighboring nucleotides. Subsequently, transgenic paternal cells constitutively expressing DMT1 were constructed and direct methylation mapping assays of their DNA showed a clear nonselective methylation of chloroplast DNA. When transgenic paternal cells were crossed with wild-type maternal cells, the frequency of biparental and paternal offspring of chloroplasts increased up to 23% while between wild-type strains it was ∼3%. The results indicate that DMT1 is a novel type of DNA methyltransferase with a nonselective cytosine methylation activity, and that chloroplast DNA methylation by DMT1 is one of factors influencing maternal inheritance of chloroplast genes.
PRESENCE of 5-methylcytosine (m5C) as a minor base in DNA was established as early as 1951 (Wyatt 1951). The ratio of m5C to the total bases varies among organisms, ranging between <0.25% in bacteria up to 7% in plants (Hall 1971), and its physiological functions have long been a matter of debate. In bacteria, m5C and N6-methyladenine act to protect the host DNA from cleavage by restriction enzymes (Kuhnlein and Arber 1972). Their contribution to these systems, referred as restriction modification, has been shown to be catalyzed by DNA methyltransferases in combination with corresponding restriction endonucleases in many species, including Escherichia coli (Smith and Kelly 1984).
In the case of eukaryotes, it is only relatively recently that relationships between m5C and epigenetic control of gene expression have been established (Bird 2002). Currently m5C is considered to play a key role in silencing gene expression by altering DNA and/or chromatin structures (Bird 2002) and various disorders due to defective DNA methylation including cancer development with abnormal methylation patterns have been described (Robertson and Wolffe 2000). Transcriptional gene silencing caused by RNAi in plants is also considered to be associated with DNA methylation (Jones et al. 2001).
The m5C entities are located almost exclusively in CpG doublets in mammalian DNA, whereas they are found in CpG, CpHpG, and CpHpH (H is A, T, or C) in plant DNA (Oakeley and Jost 1996). The methylation of these sequences is catalyzed by distinct DNA methyltransferases, which transfer the methyl group of S-adenosyl-l-methionine (AdoMet) to the 5-position of the cytosine residues. In mammalian cells, Dnmt1 and Dnmt3 are thought to methylate CpG for maintenance and de novo generation, respectively (Okano et al. 1998). In plants, genes encoding three different types of DNA methyltransferases have so far been identified: methyltransferase type 1 (MET1), chromomethylase (CMT), and domains-rearranged methyltransferase (DRM), these being proposed as responsible for CpG, CpHpG, and CpHpH methylation, respectively (Cao et al. 2000). Proteins belonging to the MET1 group have been partially characterized from several plant species, but biochemical evidence regarding the latter two is limited. Recently, we have identified and characterized a tobacco enzyme belonging to the DRM family and found that it is a de novo cytosine methyltransferase, which actively excludes the CpG sequence (Wada et al. 2003). However, exact biological functions of each plant DNA methyltransferase remain to be determined.
Organelle genes in many eukaryotes are maternally transmitted to the offspring (Birky 2001). Their biological significance, however, is not completely understood, although some advantages have been proposed. For example, uiniparental inheritance reduces the spread of cytoplasmic parasites and selfish organelle DNA (Birky 1995). Similarly, since parasitic DNA must be excluded from an egg cell, paternal DNA, which is regarded as an invader, is actively degraded (Joder et al. 1997). Another idea is that, since organelle DNA encodes proteins essential for survival, its recombination must be minimized to inhibit the formation of cytoplasmic heterozygotes (Sager 1972). In the case of selective degradation of DNA, a mechanism in which degraded DNA is utilized as a source of nucleotides during starvation has been proposed (Sears and Van Winkle-Swift 1994).
The mechanisms of uniparental inheritance are diverse (Birky 2001). In mammals, dominance of maternal genes in mitochondria may be due simply to the abundance of mitochondria in an egg cell, several thousandfold over that in sperm (Gyllenstein et al. 1991; Birky 1995). However, an active mechanism to eliminate paternal genes has also been postulated (Shirata et al. 1998). In plants, paternal genes in plastids are actively excluded, being degraded during male gametophyte development or double fertilization (Messing and Grossniklaus 1999).
This is also the case for the green alga Chlamydomonas, a unicellular soil microorganism with a simple sexual life cycle, showing segregation by mating type (mt), denoted by plus (mt+) and minus (mt−)(Sager 1972). Upon mating, transmission of nuclear genes obeys Mendelian laws of inheritance, whereas that of chloroplast genes is maternal through mt+ (Sager 1972). The underlying molecular mechanism has been hypothesized to involve a restriction-modification system, in which preferential DNA methylation in maternal genes is critical (Sager and Lane 1972). Alternatively, methylated genes have been suggested to be selectively replicated in germinating zygotes, resulting in apparent maternal inheritance (Umen and Goodenough 2001). In either case, methylation of chloroplast DNA is proposed to play an essential role. In a previous study, we isolated a gene encoding an mt+-specific chloroplast-resident DNA methyltransferase, DMT1 (CrMET1), and proposed it to be the key element in maternal inheritance of chloroplast genes (Nishiyama et al. 2002). In this report, we provide direct evidence for this, documenting its enzymatic properties and an increase in exceptional progeny from transgenic paternal cells expressing DMT1 from a transgene.
MATERIALS AND METHODS
Strains and culture condition:
Chlamydomonas reinhardtii 7003A (mt−) and 7024E (mt+) are resistant to spectinomycin and erythromycin, respectively. Vegetative cells were cultured in Tris/acetate/phosphate (TAP) liquid medium at 28° under continuous light. Gamete cells were induced and grown on Snell's agar plates (Snell 1982) at 21° for 4–6 days under a light/dark cycle of 16/8 hr.
Expression of DMT1:
The full coding sequence of DMT1 (formerly CrMET1; accession no. AB073989) was cloned into a baculovirus transfer vector, pDEST20, as an in-frame fusion protein equipped with a his-tag using the Gateway cloning system (Invitrogen, Carlsbad, CA). The N-teminal-truncated DMT1 (ΔN-DMT1) was cloned into pFastBac HTa (GIBCO BRL, Carlsbad, CA). Each resulting plasmid was transformed into E. coli with the bacmid, in which expression of the recombinant gene is driven by a polyhedron promoter. Insect cells (Sf9) were infected with the recombinant bacmids, and crude extracts were prepared by brief sonication and centrifugation. For enzymatic assay, contaminating genomic DNA from Sf9 cells was removed by partial purification of the protein fraction with nickel-NTA agarose (QIAGEN, Hilden, Germany). Each preparation was subjected to SDS-polyacrylamide gel electrophoresis and stained with Coomassie Brilliant Blue, which showed polypeptides of expected size (100 or 150 kD), along with several additional small polypeptides (data not shown). Electrophoresed samples were also blotted onto cellulose paper (Immobilon-N; Millipore, Bedford, MA) and subjected to immunostaining using antibody against his-tag as described below.
DNA methylation assay:
DNA methylation activity was monitored by incorporation of the 3H-labeled methyl group of AdoMet into recipient DNA substrate as described (Wada et al. 2003) with modification. A 100-μl reaction mixture containing an appropriate amount of crude or partially purified protein, 20 mm Tris-HCl (pH 7.7), 5 mm EDTA, 0.2 mg/ml BSA, 25% (v/v) glycerol, 1 mm DTT, 0.1 mg/ml RNaseA, 2 μm AdoMet (methyl-3H; specific activity 10 Ci/mmol; New England Nuclear Life Science Products, Boston), and substrate DNA was incubated at 37° for an appropriate time period. To determine the substrate specificity, 4.6 μg of partially purified DMT1 was incubated with a synthetic 24-mer oligonucleotide: 5′-ACGATCGTACGATCGTACGATCGT-3′ (for CpG), 5′-ACTGCAGTACTGCAGTACTGCAGT-3′ (for CpWpG, where W is A or T), and 5′-AGCATGCTAGCATGCTAGCATGCT-3′ (for CpWpW). All these sequences are palindromic, which form duplexes. The reaction was stopped by addition of proteinase K (GIBCO BRL) with further incubation at 50° for 1 hr, and substrate DNA was extracted with phenol/chloroform treatment. After precipitation with ethanol, samples were spotted onto diethylaminoethyl paper, which was washed with 0.5 m sodium phosphate, dried, and assessed for radioactivity (Nakano et al. 2000b). Under these experimental conditions, the radioactivity in the absence of the enzyme (background) was ∼1800 cpm, whereas it was minimal 25,000 cpm in the presence of the enzyme. Background values were subtracted from each measured value to estimate the net incorporation. The linearity of the reaction velocity in terms of elapsed time and enzyme amount was confirmed by plotting the initial velocity against five different time points (data not shown).
Methylation mapping:
Total DNA was extracted from vegetative untransformed mt− cells (7003A) and transformed mt− cells (no. 107). The bisulfite treatment was performed as described (Nishiyama et al. 2002), and modified DNA was subjected to first PCR using a sequence-specific primer set (forward, 5′-GTTTTAGAGTTAAAAGAGAAGAATAATGGG-3′; reverse, 5′-CCACCACATAAATCATTCATAAACTACACAACC-3′). Amplified samples were then subjected to second PCR using another primer set (forward, 5′-GGTTTTATAAATAGAAATTAAAGTAGGTGTTGG-3′; the reverse was the same as used for the first amplification). The PCR products were cloned and sequenced as described (Nishiyama et al. 2002).
Transformation:
The plasmid pBle-AR-DMT1-flag was constructed by fusing DMT1 genomic DNA (accession no. AB108536) to a flag-tag in frame, driven by the heat-shock protein 70A (HSP70A) promoter and the ribulose bisphosphate carboxylase/oxygenase 2 (RBCS2) promoter from Chlamydomonas (Harris 1989). The bleomycin-resistant gene cassette was used for selection with zeocin (Invitrogen). The spectinomycin-resistant mt− cells (7003A) were treated with gametelysin at 27° for 30 min and then transformed with pBle-AR-DMT1-flag by electroporation as described (Schroda et al. 2000). The voltage and time constant of the charged electric pulse were 1800 V/cm and 5 msec, respectively. After electroporation, cells were diluted with 8 volumes of TAP-60mm sucrose medium and cultivated at 27° for 16 hr under light. After postcultivation, cells were spread on selection plates containing 3 μg/ml zeocin, which were kept at 27° under light until transformant colonies grew up.
Immunostaining:
For identification of DMT1 trangenic cell lines, each transformed cell was cultured in TAP liquid medium at 28° under continuous light and suspended in 2 volumes of lysis buffer consisting of 100 mm Tris-HCl (pH 6.8), 4% SDS, 200 mm DTT, 20% glycerol. After centrifugation, 10 μl of the supernatant was subjected to SDS-polyacrylamide gel electrophoresis, blotted onto cellulose membranes (Immobilon-N; Millipore, Bedford, MA), and immunostained by incubating with a flag-tag monoclonal antibody (1:5000 dilution; Sigma, St. Louis) for 1.5 hr at room temperature. Secondary staining was performed with horseradish peroxidase-conjugated, goat anti-mouse antibody (1:5000 dilution; Dako, Carpinteria, CA).
Partial purification of DMT1 protein:
Methyltransferase was partially purified as described (Sano et al. 1981). Briefly, vegetative cells of 7024E (wild type, mt+), CC1154 (me-1, mt+), and DMT1 transgenic lines 53 and 107 were grown in 2-liter batches in TAP medium, harvested at midlogarithmic phase of growth, and resuspended in 10 ml 50 mm Tris-HCl containing 150 mm NaCl, 1 mm EDTA, and 5% (v/v) glycerol. Cells were disrupted by sonication (1 min at setting 5; Ultrasonic Processor XL, Farmingdale, NY) and subjected to successive centrifugation at 2000 rpm for 10 min and 40,000 rpm for 2 hr. The resulting clear supernatant was dialyzed against 20 mm Tris-HCl containing 20 mm NaCl, 1 mm EDTA, and 5% (v/v) glycerol at 4° for 16 hr, and appropriate aliquots were assayed for methyltransferase activity as described above. The amount of proteins was measured by the method of Bradford (1976).
Mating and scoring exceptional zygotes:
Gametes were suspended in Tsubo mating buffer (1.2 mm HEPES at pH 6.8, 1 mm MgSO4) 2–4 hr after the beginning of the light period. Mating was performed by mixing approximate equal numbers of mt+ and mt− gametes, which were preincubated for 5–6 hr under light (Nishimura et al. 1999). The mating efficiency was checked by observation of the mixed cells stained with SYBR Green I (Nishimura et al. 2002). After 0.5–1 hr of mating, the mixed cell solutions were plated on plates containing 3% agar in 1/5 m medium (Harris 1989). To obtain single zygote colonies, the aliquots were diluted prior to plating and incubated for 1 day at 21° under light, and then allowed to mature for 12–14 days in the dark. After maturation of zygotes, unmated gametes were eliminated by gently scraping the surface of plates with a razor and killed by chloroform treatment. Plates were then kept under light at 25° for germination. Germinated colonies were replated on TAP agar plates, and duplicated copies were prepared on TAP agar plates containing 500 mg/liter spectinomycin or 100 mg/liter erythromycin to score the antibiotic resistance.
RESULTS
Enzymatic properties in vitro:
Enzymatic properties of DMT1 were determined by in vitro assays of the recombinant protein. The full coding sequence of DMT1 was cloned in frame into a baculovirus transfer vector containing a his-tag. A truncated cDNA was also prepared by deleting a sequence of 424 amino acids from the N terminus (Figure 1A). Both were transformed into E. coli with the bacmid (genome of a baculovirus) to undergo recombination, and resulting recombinant bacmids were introduced into Sf9 insect cells. These proteins were then isolated and their sizes confirmed to be 150 and 100 kD for the full-length and truncated samples, respectively (Figure 1, A and B). DNA methylation activity was examined for these proteins using dcm− E. coli genomic DNA as the methyl acceptor. The full-length protein was shown to actively transfer methyl groups to DNA, whereas the truncated one was totally inactive (Figure 1C), indicating the N-terminal region to be critical for recognition of the substrate. The sequence specificity of recombinant DMT1 was then examined using synthetic 24-mer oligonucleotides each containing six sites of CpG, CpWpG, or CpWpW. Results showed that DMT1 almost equally methylates any cytosine residues located in these sequences, suggesting nonselective characteristics (Figure 1D).
Figure 1.—
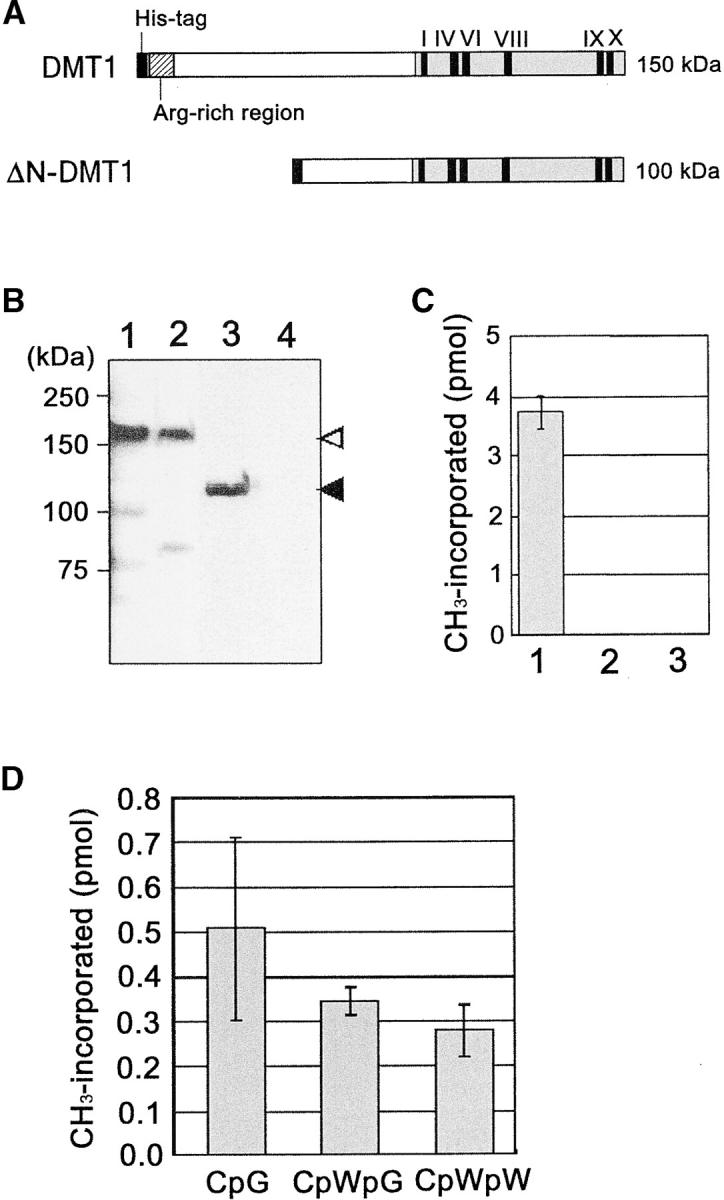
In vitro methylation activity of DMT1. (A) Schematic of full-length DMT1 (DMT1) and N-terminal truncated DMT1 (ΔN-DMT1) harboring the his-tag at the N terminus. The conserved motifs in the C-terminal catalytic domain (I–X) and the Arg-rich region are indicated. ΔN-DMT1 lacks 424 amino acids of the N terminus. (B) In vitro expression of DMT1 proteins. Protein solutions were fractionated on SDS-PAGE and subjected to immunostaining with a his-tag antibody. Samples are crude extract of DMT1 (lane 1), partially purified DMT1 (lane 2), crude extract of ΔN-DMT1 (lane 3), and crude extract of nontransformed Sf9 cells (lane 4). Protein size is indicated on the left, and the expected positions for DMT1 and ΔN-DMT1 are indicated by open and solid arrowheads, respectively. (C) Activity analysis of de novo methylation. The reaction mixture containing 10 μl of crude extract, [3H]AdoMet (specific activity 7.7 × 104 cpm/pmol), and 7 μg of dcm− E. coli genomic DNA was incubated for 1.5 hr. Samples were the crude DMT1 (lane 1), crude ΔN-DMT1 (lane 2), and crude extracts from nontransformed cells (lane 3) as the control. Values are means of triplicate experiments and error bars represent the standard deviation. (D) Substrate specificity of DMT1. Reaction mixtures containing 4.6 μg of partially purified DMT1 and [3H]AdoMet were incubated for 1.5 hr with 4.9 μm (in terms of methylatable C) of synthetic 24-mer oligonucleotides containing six sites of CpG, CpWpG, or CpWpW (W is A or T). Values are means of triplicate measures and error bars represent the standard deviations.
Construction of transgenic lines:
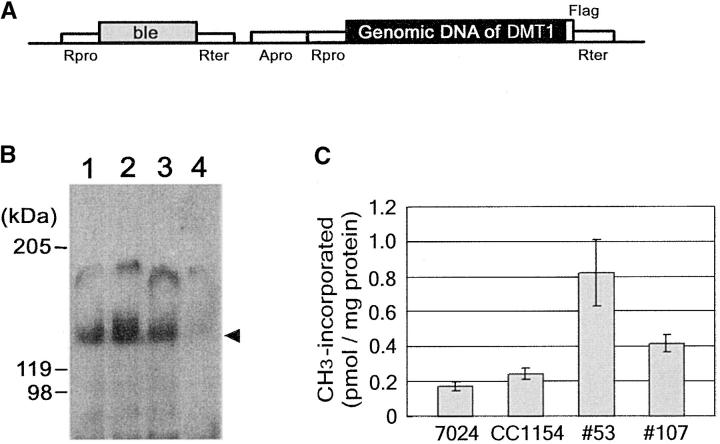
DMT1 genomic DNA was fused to the flag-tag in frame, driven by promotors derived from heat-shock protein 70A and Chlamydomonas ribulose bisphosphate carboxylase/oxygenase 2 (Figure 2A). The resulting plasmid was electrically transferred into spectinomycin-resistant mt− cells (strain 7003A), which were then selected for bleomycin resistance with zeocin. Antibiotic-resistant transformants were again screened for expression of DMT1 by immunostaining with a flag-tag antibody, and three stable lines expressing the DMT1 protein of 150 kD were finally obtained (Figure 2B). The methyltransferase activity of expressed protein was determined using crude preparations (Figure 2C). Vegetative cells of 7024E (wild type, mt+) showed low activity with a methyl group transfer of 0.16 pmol/mg protein. This activity could be attributable to a methyltransferase with a low molecular mass (60 kD), which is not involved in chloroplast DNA methylation (Sano et al. 1981). The CC1154 mutant line (me-1, mt+) with a constitutive methylation in chloroplast DNA (Nishiyama et al. 2002) exhibited a 1.5-fold higher activity than the control did. DMT1 transgenic lines 53 and 107 also showed high activity, as much as 4- and 2.5-fold more, respectively, than the control level (Figure 2C). Results indicated that the transgene produced an active enzyme, of which activity was high enough to methylate chloroplast DNA in planta.
Figure 2.—
Construction of transgenic DMT1 cell lines. (A) Schematic of the plasmid construct used for transformation of Chlamydomonas mt− cells. DMT1 genomic DNA (GenBank/EMBL/DDBJ accession no. AB108536) was fused to the flag-tag in frame at the C terminus and cloned into pBluescript KS−. Promotors were derived from HSP70A (Apro) and RBCS2 (Rpro), and the terminator from RBCS2 (Rter). The antibiotic marker was the bleomycin-resistant gene (ble). (B) Expression of DMT1-flag protein in transformed mt− cells. Chlamydomonas mt− cells (7003A) were transformed with pBle-AR-DMT1-flag and selected with zeocin. Crude cell extracts from three transformed cell lines (nos. 53, 107, and 120; lanes 1–3, respectively) and untransformed cells (7003A, lane 4) were subjected to SDS-PAGE and to immunostaining with a flag-tag antibody. The sizes of proteins are indicated on the left, and the expected position of DMT1 is indicated by the solid arrowhead. (C) Methyltransferase activity assay. Crude cell extracts were prepared from vegetative cells of 7024E (wild type, mt+), CC1154 (me-1, mt+), and DMT1 transgenic lines 53 and 107. Reaction mixtures containing 50 μl of crude extract, [3H]AdoMet (specific activity 1.2 × 104 cpm/pmol), and 2 μg of poly(dI/dC) synthetic substrate were incubated for 1.5 hr. Values are means of triplicate measures and error bars represent the standard deviations.
Methylation mapping:
Methylation sites were then determined by the methylation mapping of a 585-base region of the chloroplast RBCL gene in both wild-type and transgenic mt− vegetative cells. Experimentally, native DNA was isolated, modified by the bisulfite method, amplified with PCR, and cloned. The distribution of m5C was then determined by sequencing nine randomly isolated clones (Figure 3A). The conversion efficiency of cytosines into uracil was directly estimated by aligning sequences between positions 250 and 299 and found to be nearly complete, as all cytosines in wild-type mt− were converted into thymine after PCR (Figure 3A). When DNA originating from transgenic cells (no. 107) was similarly treated and aligned, many cytosines remained intact (Figure 3A), suggesting them to be originally methylated. Notably, methylation does not occur uniformly in the population. Some molecules are heavily methylated while others are not, consistent with our previous observations (Nishiyama et al. 2002). The frequency of methylated sites over total methylatable sites in DNA from transgenic cells was directly estimated by plotting m5C in CpG, CpHpG, and CpHpH of the examined sequence (Figure 3B). The results showed that methylation occurred nonselectively at all sites at frequencies of 30.1% for CpG, 35.3% for CpHpG, and 30.3% for other sites (Figure 3B). These values are consistent with our previous findings for native mt+ gamete cells (36.8, 27.6, and 35.5%, respectively; Nishiyama et al. 2002), suggesting that DMT1 from the transgene is equally as active as the native enzyme. The nearest neighbor frequencies of nucleotides located at the 3′ end of m5C were 36.4, 25.9, and 29.7% for CpA, CpC, and CpT, respectively, suggesting no selectivity for the doublet. These results indicate DMT1 to be essentially a nonselective methyltransferase in terms of the methylation site.
Figure 3.—
Methylation mapping. (A) Identification of m5C. After bisulfite treatment, a 585-base region of RBCL gene was amplified with PCR and cloned, and sequence was determined. C and m5C in the original sequence were converted into T and C, respectively. The original nucleotide sequence at positions 250–299 of wild-type mt− vegetative cells without bisulfite treatment is shown at the top with shading of the methylatable cytosines. The converted bases are indicated by T or C (shaded) in each clone, originating from bisulfite-treated wild-type mt− vegetative cells (7003A) or transgenic mt− vegetative cells (107). The clone number is given on the left. (B) Distribution and frequency of m5C were estimated for 11 clones. Histograms represent the percentage of m5C over the total cytosines (vertical axis) at positions containing CpG (top), CpHpG (middle), and CpHpH (bottom) sequences (H is A, C, or T) in the top strand (horizontal axis). The numbers of methylatable CpG, CpHpG, and CpHpH sequences were 23, 21, and 67, respectively, in the 585-bp DNA fragment tested. Note that not all these sites are shown.
Increase in exceptional progeny:
The frequency of paternal and biparental inheritance of chloroplast genes was then analyzed by crossing transgenic mt− to wild-type mt+ cells (Figure 4). Crossing was performed using erythromycin-resistant mt+ (wild-type 7024E) and spectinomycin-resistant mt− (wild-type 7003A and transgenic lines 53 and 107) with the antibiotic-resistance genes in the chloroplast genomes. After mating, ∼100 zygote colonies in each experiment were scored for mt+-uniparental by erythromycin resistance, for biparental by both spectinomycin and erythromycin resistance, and for mt−-uniparental by spectinomycin resistance (Figure 4). The results revealed that, while the frequency of spontaneous biparental progeny from wild-type crossing was 3 out of 101 (3%), paternal and biparental progeny from transgenic (no. 107) crossing were increased to 23 out of 98 (23.5%; Table 1). The no. 53 line also showed a high frequency at 20% exceptional offspring (Table 1).
Figure 4.—
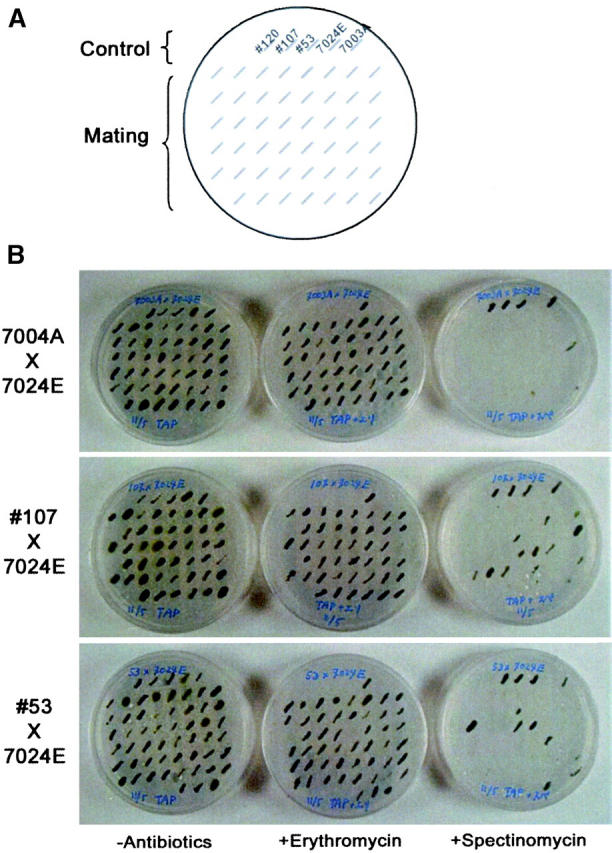
Effects of DMT1 on transmission of chloroplast genes. Each cross was performed using strains with chloroplast antibiotic markers: erythromycin and spectinomycin resistance for mt+ and mt−, respectively. Zygote colonies with serial numbers as indicated were plated; incubated for 4–10 days; and scored for mt+ uniparental colonies by resistance against erythromycin, for biparental colonies by spectinomycin and erythromycin resistance, and for mt− uniparental colonies by spectinomycin resistance. (A) The positions of strains loaded on the plate below. The control lane illustrates colonies from left to right of transgenic lines 120, 107, and 53 and wild-type lines 7024E (mt+) and 7003A (mt−), respectively. The mating lane shows the position of colonies from each crossing. (B) Crossing results. Crossings between 7024E (mt+) and 7003A (wild-type mt−), between 7024E (mt+) and 107 (transgenic mt−), and between 7024E (mt+) and 53 (transgenic mt−) are shown in the top, middle, and bottom, respectively. Culture medium contained no antibiotics (left), erythromycin (middle), or spectinomycin (right) as indicated at the bottom.
TABLE 1.
Effects of Chlamydomonas DMT1 on segregation of mating types
| Progeny
|
|||||
|---|---|---|---|---|---|
| Parents: Crossinga |
Total scored | Uniparental (mt+) (%) |
Biparental (%) |
Uniparental (mt−) (%) |
Exceptional (%) |
| 1. 7024E × 7003A | 101 | 98 (97.0) | 3 (3.0) | 0 (0) | 3 (3.0) |
| 2. 7024E × 53 | 105 | 84 (80.0) | 18 (17.1) | 3 (2.9) | 21 (20.0) |
| 3. 7024E × 107 | 98 | 75 (76.5) | 21 (21.4) | 2 (2.1) | 23 (23.5) |
Crossing was performed as described in the Figure 4 legend, and data were pooled from several crosses to score ∼100 independent progeny colonies. Crossing 1 was performed between wild-type mt+ having erR (7024E) and mt− having specR (7003A); crossings 2 and 3 were performed between the wild-type 7024E and transgenic mt− lines, nos. 53 and 107, respectively.
DISCUSSION
This article documents evidence that the chloroplast-resident DNA methyltransferase, DMT1, catalyzes nonselective methylation of cytosine residues and that resulting methylation of chloroplast DNA is responsible for its persistence in Chlamydomonas offspring.
The experiments showed that DMT1 (CrMET1) possesses nonselective and de novo methylation activity toward cytosine residues. This is in marked contrast to the findings for plant DNA methyltransferases so far reported, which were classified into three groups, but all showing substrate specificity. The majority of enzymes that have been characterized to date belong to the MET1 group, whose target is strictly limited to the CpG doublet (Finnegan and Kovac 2000). Putative proteins belonging to the CMT and DRM groups have a chromodomain and rearranged motifs in their catalytic domains, respectively, and genetic analysis of mutant lines has suggested that they are responsible for maintenance of cytosine methylation at CpHpG and de novo methylation at CpHpH sites, respectively (Cao et al. 2000; Henikoff and Jacobsen 2001). However, we have recently found that tobacco DRM (NtDRM1) methylates almost any cytosines regardless of the flanking nucleotides at both 5′ and 3′ ends, except for the CpG doublet (Wada et al. 2003). This finding suggested that there is no strict sequence specificity between CMT and DRM and that they may redundantly act to regulate methylation of CpHpG and CpHpH (Cao et al. 2000).
The amino acid sequence of the catalytic domain of DMT1 resembles that belonging to the MET1 group, implying a preference for CpG sites. However, an in vitro assay using synthetic oligonucleotides and purified enzyme preparations suggested that DMT1 efficiently methylates any cytosines in CpG, CpWpG, and CpWpW sites. The direct methylation mapping of chloroplast DNA, methylated in vivo by the transgenic DMT1, also revealed that the target site for methylation was apparently random, showing methylation activity toward any cytosines regardless the flanking nucleotides. Approximately 30% of cytosines at any site were found to be equally methylated. In this context, DMT1 rather resembles NtDRM1, which also shows a nonspecific feature (Wada et al. 2003). A critical difference between the two, however, is that the latter actively excludes CpG (Wada et al. 2003), whereas the former does not. This suggests that DMT1 has the flexible sequence-specific recognition mechanism for methylation targeting.
To date, eukaryotic DNA methyltransferases have been proposed to be involved in control of gene expression and/or gene silencing (Bird 2002). In plants, for example, MET1 suppression results in abnormal phenotypes in transgenic Arabidopsis and tobacco plants, suggesting DNA methylation to be critical during development (Finnegan et al. 1996; Nakano et al. 2000a). Members of CMT and DRM have been suggested to function in gene silencing of retrotransposons and epigenetics, respectively (Cao et al. 2000; Lindroth et al. 2001; Cao and Jacobsen 2002; Tompa et al. 2002). In these cases, the site of methylation appears to be important for maintaining the methylation pattern throughout cell division and/or temporarily methylating DNA upon necessity. In contrast, the nonselective recognition of cytosines, in terms of the neighboring nucleotides, by DMT1 suggests that it is not the site of methylation that is important but rather the methylation itself of chloroplast DNA, conferring protection against degradation. In this context of nonselectivity and self-defense functions, DMT1 might be considered to exemplify a fourth type of plant DNA methyltransferase.
The involvement of DMT1 in self-defense of DNA was directly demonstrated by physiological experiments showing that transgenic mt− with hypermethylated gamete DNA by DMT1 exhibits biparental and paternal inheritance at >20% frequency or with an 8-fold higher frequency than in controls. This observation is essentially consistent with findings for a spontaneous mt− mutant, mat-1, which exhibits hypermethylation in gamete DNA and biparental inheritance at a 20-fold higher frequency than in controls (Sager et al. 1981). A possible explanation for the difference in actual frequency is that excess methylation due to constitutive expression of DMT1 throughout the growth period might be detrimental to cells. Indeed, we observed transgenic vegetative and gamete cells exhibiting a detectable level of nuclear DNA methylation and slow growth rate with pale greening (our unpublished observation). This is in clear contrast to mat-1, in which methylation occurs only in chloroplast DNA at the period of gametogenesis and zygote formation (Sager et al. 1981; Nishiyama et al. 2002). Since the extent of methylation greatly varies among populations (Nishiyama et al. 2002), perhaps cells with relatively low methylation are naturally selected for mating.
Degradation of chloroplast DNA from the mt− parental cell begins 45–60 min after mating and is completed within 10 min (Nishimura et al. 2002). Concomitantly, activity of a novel DNase, localized in mt− chloroplasts and activated within 60–90 min after mating, was identified. This nuclease, designated MDN, is proposed to be developmentally controlled, being activated only in mt+ gametes, migrating into mt− to degrade its chloroplast DNA (Nishimura et al. 2002). DNA methylation specific to mt+ chloroplast DNA can provide the explanation of how mt+ chloroplast DNA remains intact during and after mating. To our knowledge, this may be the first example of a eukaryotic restriction-modification system, initially proposed by Sager and Lane (1972).
Acknowledgments
We thank Arthur B. Pardee and Ann Goodman (Harvard Medical School) and Melanie Ehrlich (Tulane University School of Medicine) for helpful discussion and valuable comments. We are also grateful to Yoshihiro Matsuda (Kobe University) for generous gifts of the strains 7003A and 7024E, Kimiyo Nakamura (Nara Institute of Science and Technology) for technical assistance, and Malcolm A. Moore (Intermal, Nagoya) for critical reading of the manuscript. This work was supported by a grant for the Research for the Future Program (JSPS-RFTF001604) from the Japan Society for the Promotion of Science.
This paper is dedicated to the late Professor Ruth Sager, who first proposed the restriction-modification model for maternal inheritance in Chlamydomonas.
Sequence data from this article have been deposited with the EMBL/DDBJ/GenBank Data Libraries under accession no. AB108536 (DMT1 genomic sequence). [The genome annotation rule of Chlamydomonas was determined to strictly use the three-letter designation and strongly suggested not to use the preface including Cr. The Chlamydomonas gene encoding a chloroplast-resident DNA methyltransferase was initially designated as CrMET1 (C. reinhardtii methyltransferase 1; Nishiyama et al. 2002). However, on the basis of the above-mentioned rule, it is renamed as DMT1 in this article.]
References
- Bird, A. P., 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16: 6–21. [DOI] [PubMed] [Google Scholar]
- Birky, C. W., 1995. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl. Acad. Sci. USA 92: 11331–11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky, C. W., 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35: 125–148. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of dye-binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Cao, X., and S. E. Jacobsen, 2002. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12: 1138–1144. [DOI] [PubMed] [Google Scholar]
- Cao, X., N. M. Springer, M. G. Muszynski, R. L. Philips, S. Kaeppler et al., 2000. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. Natl. Acad. Sci. USA 97: 4979–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E. J., and K. A. Kovac, 2000. Plant DNA methyltransferases. Plant Mol. Biol. 43: 189–201. [DOI] [PubMed] [Google Scholar]
- Finnegan, E. J., W. J. Peacock and E. S. Dennis, 1996. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93: 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllenstein, U., D. Wharton, A. Josefsson and A. C. Wilson, 1991. Paternal inheritance of mitochondrial DNA in mice. Nature 352: 255–257. [DOI] [PubMed] [Google Scholar]
- Hall, R. H., 1971 The Modified Nucleosides in Nucleic Acids. Columbia University Press, New York.
- Harris, E. H., 1989 The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego. [DOI] [PubMed]
- Joder, J. A., C. P. Walsh and T. H. Bester, 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13: 335–340. [DOI] [PubMed] [Google Scholar]
- Jones, L., F. Ratcliff and D. C. Baulcombe, 2001. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11: 747–757. [DOI] [PubMed] [Google Scholar]
- Kuhnlein, U., and W. Arber, 1972. The role of nucleotide methylation in in vitro B-specific modification. J. Mol. Biol. 63: 9–19. [DOI] [PubMed] [Google Scholar]
- Lindroth, A. M., X. Cao, J. P. Jackson, D. Zilberman, C. M. McCallum et al., 2001. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Messing, J., and U. Grossniklaus, 1999 Genomic imprinting in plants, pp. 23–40 in Genomic Imprinting, edited by R. Ohlssen. Springer-Verlag, Berlin. [DOI] [PubMed]
- Nakano, Y., N. Steward, T. Kusano and H. Sano, 2000. a A tobacco NtMET1 cDNA encoding a DNA methyltransferase: molecular characterization and abnormal phenotypes of transgenic tobacco plants. Plant Cell Physiol. 41: 448–457. [DOI] [PubMed] [Google Scholar]
- Nakano, Y., N. Koizumi, T. Kusano and H. Sano, 2000. b Isolation and properties of an S-adenosyl-L-methionine binding protein from the green alga, Chlamydomonas reinhardi. J. Plant Physiol. 157: 707–711. [Google Scholar]
- Nishimura, Y., O. Misumi, S. Matsunaga, T. Higashiyama, A. Yokota et al., 1999. The active digestion of uniparental chloroplast DNA in a single zygote of Chlamydomonas reinhardtii is revealed by using the optical tweezers. Proc. Natl. Acad. Sci. USA 96: 12577–12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, Y., O. Misumi, K. Kato, N. Inada, T. Higashiyama et al., 2002. An mt+ gamete-specific nuclease that targets mt− chloroplasts during sexual reproduction in C. reinhardtii. Genes Dev. 16: 1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, R., M. Ito, Y. Yamaguchi, N. Koizumi and H. Sano, 2002. A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonas maternal gametes. Proc. Natl. Acad. Sci. USA 99: 5925–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeley, E. J., and J.-P. Jost, 1996. Non-symmetrical cytosine methylation in tobacco pollen DNA. Plant Mol. Biol. 31: 927–930. [DOI] [PubMed] [Google Scholar]
- Okano, M., S. Xie and E. Li, 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferase. Nat. Genet. 19: 219–220. [DOI] [PubMed] [Google Scholar]
- Robertson, K. D., and A. P. Wolffe, 2000. DNA methylation in health and disease. Nat. Rev. Genet. 1: 11–19. [DOI] [PubMed] [Google Scholar]
- Sager, R., 1972 Cytoplasmic Genes and Organelles. Academic Press, New York.
- Sager, R., and D. Lane, 1972. Molecular basis of maternal inheritance. Proc. Natl. Acad. Sci. USA 69: 2410–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager, R., C. Grabowy and H. Sano, 1981. The mat-1 gene in Chlamydomonas regulates DNA methylation during gametogenesis. Cell 24: 41–47. [DOI] [PubMed] [Google Scholar]
- Sano, H., C. Grabowy and R. Sager, 1981. Differential activity of DNA methyltransferase in the life cycle of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 78: 3118–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda, M., D. Blocker and C. F. Beck, 2000. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21: 121–131. [DOI] [PubMed] [Google Scholar]
- Sears, K., and K. Van Winkle-Swift, 1994. The salvage/turnover/repair (STOR) model for uniparental inheritance in Chlamydomonas: DNA as a source of sustenance. J. Hered. 85: 366–376. [DOI] [PubMed] [Google Scholar]
- Shirata, H., J.-I. Hayashi, S. Takahama, H. Kaneda and H. Yonekawa, 1998. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 148: 815–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. O., and S. V. Kelly, 1984 Methylases of type II restriction-modification systems, pp. 39–71 in DNA Methylation: Biochemistry and Biological Significance, edited by A. Razin, H. Cedar and A. D. Riggs. Springer-Verlag, New York.
- Snell, W. J., 1982. Study of the release of cell wall degrading enzymes during adhesion of Chlamydomonas gametes. Exp. Cell Res. 138: 109–119. [DOI] [PubMed] [Google Scholar]
- Tompa, R., C. M. McCallum, J. Delrow, J. G. Henikoff, B. Van Steensel et al., 2002. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr. Biol. 12: 65–68. [DOI] [PubMed] [Google Scholar]
- Umen, J. G., and U. W. Goodenough, 2001. Chloroplast DNA methylation and inheritance in Chlamydomonas. Genes Dev. 15: 2583–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, Y., H. Ohya, Y. Yamaguchi, N. Koizumi and H. Sano, 2003. Preferential de novo methylation of cytosine residues in non-CpG sequences by a domains rearranged DNA methyltransferase from tobacco plants. J. Biol. Chem. 278: 42386–42393. [DOI] [PubMed] [Google Scholar]
- Wyatt, G. R., 1951. Recognition and estimation of 5-methylcytosine in nucleic acids. Biochem. J. 48: 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]