Abstract
The outer surface protein C (ospC) locus of the Lyme disease bacterium, Borrelia burgdorferi, is at least an order of magnitude more variable than other genes in the species. This variation is classified into 22 ospC major groups, 15 of which are found in the northeastern United States. The frequency distributions of ospC within populations suggest that this locus is under balancing selection. In multiple-niche polymorphism, a type of balancing selection, diversity within a population can be maintained when the environment is heterogeneous and no one genotype has the highest fitness in all environments. Genetically different individuals within vertebrate species and different vertebrate species constitute diverse environments for B. burgdorferi. We examined four important host species of B. burgdorferi and found that the strains that infected each species had different sets of ospC major groups. We found no variation among conspecific hosts in the ospC major groups of their infecting strains. These results suggest multiple niches create balancing selection at the ospC locus.
BALANCED polymorphisms within populations can be used to understand the dynamics of natural selection. Most of the genetic polymorphisms observed in natural populations are neutral or nearly neutral alleles that are dominated by genetic drift and carry little information about the ecological interactions or the selection pressures a population faces (Kimura and Ohta 1971; Ohta 1992). However, the few loci with multiple alleles and long coalescence times are generally maintained by balancing selection (Charlesworth et al. 1997; May et al. 1999). Investigations of these loci can increase our understanding of the dynamics of natural selection and the ecological interactions driving selection in natural populations. We propose that the polymorphism at the outer surface protein C (ospC) locus of Borrelia burgdorferi sensu stricto, the etiological agent of Lyme disease, will provide insight into the mechanisms of selection in natural populations.
Lyme disease is the most prevalent vector-borne disease in the United States and Europe (Johns et al. 2001). B. burgdorferi s.s. (Spirochaetaceae) is an obligate internal parasite that is vectored between vertebrate hosts by hard-bodied ticks of the Ixodes ricinis complex (Burgdorfer et al. 1989; Lane et al. 1991; Bosler 1993; de Silva and Fikrig 1997; Shih and Chao 2002). To persist in nature, B. burgdorferi s.s. must remain in this zoonotic cycle because neither the vertebrates nor the vectors can transmit B. burgdorferi s.s. from mother to offspring (Magnarelli et al. 1987; Burgdorfer et al. 1988). I. scapularis, the exclusive vector for B. burgdorferi s.s. in the northeastern United States, feeds a maximum of three times during its two-year life cycle, once at each of the three non-egg life stages (Wilson and Spielman 1985; Anderson 1989; Bosler 1993). The proliferation of B. burgdorferi s.s depends strongly on when, during the year, each I. scapularis subadult stage actively seeks a bloodmeal. Small mammals and ground-foraging birds become infected with B. burgdorferi s.s. in the early summer when infected nymphs, the second non-egg life stage, feed on them. Larvae, the first non-egg life stage, of the succeeding tick cohort, then feed on these infected hosts in the late summer and become infected before they molt to nymphs, renewing the cycle (Anderson 1989; Bosler 1993). Transmission of B. burgdorferi s.s. is enhanced because the nymphal season occurs before the larval season, increasing the probability a larva feeds upon a B. burgdorferi s.s.-infected animal (Lane et al. 1991). Adult I. scapularis need not be considered in the life history strategy of B. burgdorferi s.s. because the adults generally parasitize a different set of vertebrate species than do the subadults, effectively removing adult ticks from the zoonotic cycle (Anderson 1989).
B. burgdorferi s.s. expresses ospC when it migrates from the midgut of the tick to the salivary glands during tick feeding and for the first 10 days after it enters the vertebrate host (Fingerle et al. 1995; Schwan et al. 1995; Montgomery et al. 1996; Schwan and Piesman 2000). OspC is one of the first and most heavily targeted borrelial antigens by the vertebrate immune system (Wilske et al. 1986, 1993; Dressler et al. 1993; Fung et al. 1994; Margolis et al. 1994; Stevenson and Barthold 1994; Engstrom et al. 1995). The immune response poses a strong selective pressure on ospC, as the immune system quickly eliminates B. burgdorferi s.s. with antigenic OspC epitopes (Preac-Mursic et al. 1992; Gilmore et al. 1996). It has been suggested that the selection pressure from the vertebrate immune response is responsible for the ospC polymorphism (Qiu et al. 1997; Wang et al. 1999).
ospC is a highly polymorphic single-copy gene with a long coalescence time compared to neutrally evolving genes (Dykhuizen et al. 1993; Jaurisheipke et al. 1993; Masuzawa et al. 1997; Wang et al. 1999; Baranton et al. 2001). The genetic diversity at the ospC locus has been classified into major groups of alleles. An ospC major group is defined as a group of alleles that are different in >8% of their nucleotide sequence from alleles in other major groups (average difference of ∼20%) and <2% different from other alleles in the same major group (average difference of <1%; Wang et al. 1999). No reported sequences are between 2 and 8% different from any other sequence. Of the 22 ospC major groups found worldwide in B. burgdorferi s.s., four are found only in Europe (Seinost et al. 1999) and one has recently been discovered in the southeastern United States (Lin et al. 2002). Of the remaining ospC major groups, 15 occur at fairly even frequencies in every well-sampled population in the northeastern United States (Seinost et al. 1999; Wang et al. 1999; Qiu et al. 2002). For the sake of brevity and clarity, we refer to all strains of B. burgdorferi s.s. with the same ospC major group as an oMG. For example, all strains with an allele from ospC major group A are called oMG A. We reserve the term ospC major group for discussions of the alleles at the genetic locus.
Loci that are dominated by neutral processes are expected to have few alleles at relatively high frequencies within a population. The ospC major groups violate this expectation, suggesting that the ospC polymorphism is not neutral (Wang et al. 1999). The polymorphism is probably not maintained by a balance between local selection and high migration between populations, as local tick populations are differentiated at the 16S rRNA mitochondrial gene, suggesting a very low migration rate of ticks between populations (Qiu et al. 2002). Balancing selection, on the other hand, is expected to preserve multiple alleles at intermediate frequencies with high sequence diversity in a population (Vekemans and Slatkin 1994), giving them deeper coalescence than neutrally evolving alleles (May et al. 1999; Schierup et al. 2000), as seen at the ospC locus (Wang et al. 1999; Lin et al. 2002; Qiu et al. 2002).
In haploid organisms such as B. burgdorferi s.s., the most likely forms of balancing selection are negative frequency-dependent selection and multiple niche polymorphism. Negative frequency-dependent selection, which occurs in populations when rare genotypes have a selective advantage over common genotypes (Gromko 1977), has been shown to maintain polymorphisms in natural populations, including many host-parasite systems (Thrall et al. 1995; Brunet and Mundt 2000; Uyenoyama 2000; Uyenoyama et al. 2001). Negative frequency-dependent selection caused by a strong secondary immune response to OspC could maintain the ospC polymorphism. Animals cannot clear a B. burgdorferi s.s. infection with an immune response, but the immune memory to OspC protects them from future infections of the same strain (Barthold 1999). Therefore, an animal previously infected by one strain of B. burgdorferi s.s. cannot be reinfected by another strain with the same ospC major group due to a secondary immune response, or immune memory, to OspC. However, those animals are still susceptible to infection by strains with different ospC major groups (Preac-Mursic et al. 1992; Gilmore et al. 1996; Probert et al. 1997; Barthold 1999). Rare oMG's could therefore have a selective advantage in natural systems. With simple negative frequency dependence, all of the oMG's would be at approximately the same frequency at equilibrium. In natural populations, however, oMG's vary ∼20-fold in frequencies (Qiu et al. 1997; Wang et al. 1999). If a secondary immune response to OspC is the only factor preventing B. burgdorferi s.s. from infecting an animal, every host would be infected by all of the oMG's that they had been challenged with during the nymphal season.
Multiple-niche polymorphism, also referred to as diversifying selection, is a conceptually possible model with a long history in the theoretical literature (Levene 1953; Gliddon and Strobeck 1975; Strobeck 1979; Czochor and Leonard 1982). Yet there is very little uncontroversial evidence that this type of selection has maintained polymorphisms in natural systems (Hedrick 1986). Multiple-niche polymorphism can maintain diversity within populations when the environment is heterogeneous and no one genotype has the highest fitness in all environments. The environments B. burgdorferi s.s. may encounter, the different species of vertebrate hosts, may be very heterogeneous. If the fitness of a strain depends upon an interaction between the ospC major group and a factor unique to the host species, the frequency distribution of the oMG's should differ significantly among vertebrate species. Conspecific individual vertebrates, regardless of species, may also be heterogeneous environments for B. burgdorferi s.s. due to variability among individuals and might also vary significantly in the frequency distribution of oMG's. Differences between species and variability within species are not mutually exclusive and both could be operating in this system to maintain the polymorphism at ospC.
In this study we present evidence that the diversity observed at the outer surface protein C locus of B. burgdorferi s.s. is a multiple-niche polymorphism due to differences among host species. The frequency distributions of oMG's are not different among individuals, suggesting that variability among individuals within a species is not creating a heterogeneous environment for B. burgdorferi s.s. These data suggest that interspecies multiple-niche polymorphism is necessary and sufficient to maintain the polymorphism at the ospC locus. However, our data do not reject the possibility that negative frequency-dependent selection is also occurring in this system, but it is neither necessary nor sufficient to explain our data. Negative frequency-dependent selection and multiple-niche polymorphism are not mutually exclusive forces and may both contribute to the creation and maintenance of this polymorphism.
MATERIALS AND METHODS
We collected larval ticks that had fed on four species of vertebrates trapped in a natural system to determine the oMG's that had infected each animal. Because B. burgdorferi s.s. is not transmitted from mother to offspring in I. scapularis, we assumed that every oMG detected in a larva had also infected the host that larva had fed on. We chose to examine engorged larvae, as opposed to testing animal tissues directly, for two reasons. Testing several larvae that have fed upon a vertebrate can provide a quantitative assessment of the variability among conspecifics. In addition, we do not know if all the oMG's that a host is infected with can be detected in a small skin sample. We were able to account for all of the oMG diversity within a host and test for variation among conspecifics and among species by sampling engorged larvae from each host. In most instances, all of the variability within a host was detected by sampling five larvae from that host.
Collection:
Four important host species for B. burgdorferi s.s., Peromyscus leucopus (white-footed mouse), Tamias striatus (eastern chipmunk), Blarina brevicauda (short-tailed shrew), and Sciurus carolinensis (gray squirrels), were trapped at the Institute of Ecosystem Studies in Millbrook, New York, in August 2002 (Schmidt et al. 1999). Captured animals were housed in the Institute of Ecosystem Studies rearing facility for 72 hr in wire mesh cages suspended over shallow water to allow the larval I. scapularis to feed to repletion and drop off. The engorged larvae were collected from the water twice daily into microfuge tubes. Microfuge tubes were stored at −20° until the DNA was extracted. In addition, we collected 188 host-seeking nymphs at the Institute of Ecosystem Studies in July 2002 to assess the frequencies of oMG's in the cohort of nymphs that infected the individuals trapped in this study.
Extraction:
One tick was placed in a microfuge tube and homogenized with a sterile 200-μl pipet tip in 100 μl of a 5% Chelex resin (Bio-Rad, Richmond, CA) solution in water. A total of 900 μl of 5% Chelex was added to each tube (no extra 5% Chelex was added to preparations of the foraging nymphs), which was subsequently vortexed for 30 sec. The tubes were then rocked gently overnight at 56°, vortexed for 30 sec, heated to 95° for 15 min, vortexed for 30 sec, and finally centrifuged at 10,000 rpm. We found that 1 ml of 5% Chelex solution was necessary to reduce the concentration of inhibitors found in engorged larvae to permit successful PCR.
Semi-nested PCR:
A 594-bp fragment of the ospC locus was amplified using semi-nested PCR. The semi-nested PCR protocol is more sensitive than the PCR-based method previously reported (Qiu et al. 2002), although more prone to contamination. PCR contamination was carefully avoided by using barrier pipet tips and negative controls. First, a 617-bp fragment was amplified using primers OC6(+) and OC623(−) (primer sequences are reported in Qiu et al. 2002). Each 50-μl PCR reaction contained 10 μl of the DNA extraction, 50 mm KCl, 10 mm Tris (pH 8.3), 2.5 mm MgCl, 0.5 μm of each primer, dNTPs at 0.2 mm per nucleotide and 1 unit of recombinant Taq. After an initial denaturation at 95° for 1 min, the mixture was run for 30 cycles at 95° for 40 sec, 54° for 35 sec, and 72° for 1 min. The second round of PCR amplifies a 594-bp fragment using OC6(+Fluo) (a fluorescein label was commercially added to the 5′ end) and OC602(−). The reaction conditions for the second round of PCR were identical to the first except 1 μl of first-round PCR product was used as template and the reaction was run for 40 cycles. A total of 5 μl of the second PCR reaction was run on a 1.5% agarose gel with ethidium bromide to assess the presence or absence of B. burgdorferi s.s. in the sample tick. Amplified fragments of different sizes were never detected, suggesting that these reaction conditions are specific for ospC. The PCR products from 66 infected host-seeking nymphs and five infected larvae from each of 15 individuals from each species (10 for S. carolinensis) tested were used in the subsequent reverse line blot procedure.
Reverse line blot:
The reverse line blot is a very effective, high-throughput procedure that is well suited for systems where the alleles are defined and sequencing is not feasible because multiple alleles are present in a sample. As opposed to standard Southern blots, reverse line blot attaches the probe to the membrane, and a labeled test sample hybridizes to the probes. Many of the previously designed probes, including the positive control probe, work well (Qiu et al. 2002) and are used in this study. Probes for ospC major groups G, I, and J were modified to increase signal strength and reduce potential cross-specificity (Table 1). Two new probes that bind ospC major group C were added to include group C in the analysis. ospC major group C appears to be a recombinant of alleles from ospC major groups I, B, and E, negating the possibility of using a probe unique to major group C. We designed two probes for major group C, one that has dual specificity for ospC major groups I and C (OC-C − 7) and one that is specific for major groups E and C (OC-C 182 + 2). With this combination, ospC major group C can be distinguished from all other ospC major groups and its frequency can be estimated. However, ticks that are infected by both ospC major groups I and E will have the same pattern on the reverse line blot membrane as a tick infected with oMG's I, E, and C. The frequencies of oMG's I and E are not affected by this discrepancy, as alleles from major group C do not bind to probes for either ospC major groups I or E. ospC major group C was not present in our samples from the Institute for Ecosystem Studies nor was any tick infected by both oMG's I and E. Every PCR product run on the reverse line blot hybridized to at least one of our ospC major group probes, suggesting that novel oMG's are either not present or in very low frequencies at the Institute for Ecosystem Studies.
TABLE 1.
Oligonucleotide probes
| Designation | Length | Sequence (5′-3′) |
|---|---|---|
| OC-C182 + 2 | 20 | TGCAAGTAAGGTCTCAACTT |
| OC-C − 7 | 25 | TCCGTTGTTATCTGCCTCATTATCT |
| OC-G + 3 | 19 | GGTGTTGTGATTCGCATCA |
| OC-I + 1 | 24 | GTTTGAAATTAAATATGCTCCTGA |
| OC-J + 7 − 6 | 17 | TTGACCCACTTCAGCAC |
The method for reverse line blotting is as follows: 400–600 thymine residues were added to the 3′ end of all probes using terminal transferase (Roche Molecular Biochemicals, Indianapolis) to enhance sticking to the nylon membrane. Tailed probes were fixed to a 15- × 15-cm nylon membrane (Bio-Rad Zeta-Probe membranes), using a MiniSlot 30 manifold (Immunetics, Cambridge, MA). Forty picomoles of each probe was diluted in 2 ml of TE buffer (10 mm Tris-HCl, 1 mm EDTA, pH 8.0) and evenly applied to individual slots (0.8 mm × 13 cm) created by the manifold. Each slot was subsequently washed twice with 2 ml TE buffer, removed from the manifold, and immediately UV-crosslinked (125 mJ). Checkerboard hybridization followed the protocol described in Qiu et al. (2002). Chemiluminescent detection was performed using the procedure suggested by the manufacturer [digoxygenin (DIG) luminescent detection kit; Roche Molecular Biochemicals], substituting anti-fluorescein-AP, Fab fragments (Roche Molecular Biochemicals) at a 1:1000 dilution for the DIG antibodies. The membrane can be used up to 10 times by stripping the membrane after each use. To reuse a membrane, a boiling 0.1× SSC/0.5% SDS solution was poured over a used membrane and rocked for 15 min. This procedure was repeated two to three times. Membranes were exposed to X-ray film overnight to determine the extent the membrane was stripped. Once prepared, the membrane should be stored at 4° in 2× SSC (0.3 m NaCl, 30 mm sodium citrate, pH 7.0).
Statistical methods:
Differences in the frequency distribution of oMG's among species were assessed using hierarchical log-linear analysis of frequencies (Sokal and Rohlf 1995). If one larva from an individual tested positive for an oMG, that individual was considered infected by that oMG in the statistical analysis. This hierarchical log-linear analysis of frequencies is a three-dimensional contingency table with species, oMG, and counts of presence or absence as the factors. To test for intraspecific variation, we used the number of infected ticks that tested positive for each oMG from each individual in the analysis. In this analysis, the frequency distributions of the oMG's from each individual host are compared in a similar three-dimensional contingency table with conspecific individuals, oMG, and counts of presence or absence as the factors. Intraspecific differences in the number of oMG's that tested positive per infected larva were evaluated using a single-classification ANOVA (Sokal and Rohlf 1995).
RESULTS
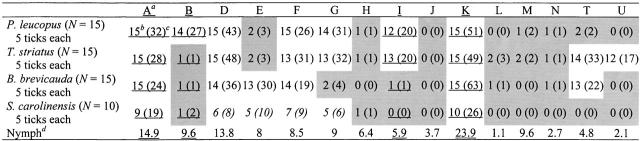
Using PCR and reverse line blotting, we assayed 66 infected host-seeking nymphs from the Institute for Ecosystem Studies and five infected larvae from each of 15 individuals from each vertebrate species except S. carolinensis, of which 10 individuals were assayed, for the presence of each oMG. We determined the percentage of host-seeking nymphs that test positive for each oMG, which is a measure of the total oMG diversity at the Institute for Ecosystem Studies. By testing larvae from animals, we determined both the number of larval ticks from each individual that tested positive for each oMG and the number of conspecific individuals that tested positive for each oMG. Differences in the frequency distributions of oMG's among individuals, among species, and between each species and the host-seeking nymphs were tested statistically.
Host-seeking nymphs:
A total of 188 host-seeking nymphs collected from the Institute for Ecosystem Studies were processed by PCR of the ospC gene to determine the percentage of nymphs infected by B. burgdorferi s.s. Of these, 66 (35%) tested positive for B. burgdorferi s.s. by PCR, similar to what was found in previous studies using a direct immunofluorescence antibody-staining assay (Table 2; LoGiudice et al. 2003). The PCR products from the 66 infected nymphs were subsequently run on a reverse line blot to determine the frequency distribution of oMG's in the cohort of nymphs that infected the animals examined in this study. We detected 15 oMG's at frequencies more even than expected by chance (Table 3), similar to what was observed previously at the Institute for Ecosystem Studies and in other localities in the northeastern United States (Qiu et al. 2002).
TABLE 2.
Percentages of infected ticks
| P. leucopus (%) | T. striatus (%) | B. brevicauda (%) | S. carolinensis (%) | Nymphs (%) | |
|---|---|---|---|---|---|
| Semi-nested PCR | 78 (±2.7) | 55 (±3.5) | 42.6 (±3.1) | 19.2 (±2.5) | 35 |
| LoGiudice et al. (2003) | 92.1 (±1.7) | 55 (±2.5) | 41.8 (±2.6) | 14.7 (±2.3) | 37.6a |
TABLE 3.
Frequency distribution of oMG's from four species of reservoir host
Interspecific differences in oMG frequencies:
As reported in previous studies that used a different method to detect B. burgdorferi s.s., we found that many larvae are not infected with B. burgdorferi s.s. after feeding on infected animals (Table 2). Moreover, we found that a larva does not carry all the B. burgdorferi s.s. oMG's that infect the host on which the larvae had fed. Therefore, we tested five infected ticks from each of 15 individuals from each host species, except S. carolinensis where 10 individuals were examined, to determine which oMG's had infected each animal. An individual was considered infected by an oMG if any of the larvae that had fed on that individual tested positive for that oMG. In all vertebrate species except S. carolinensis, each oMG was found in either >80% or <15% of the individuals from each species. However, the oMG's that were found in high frequencies differed among the vertebrate species [P < 0.0001, d.f. = 42 (4 × 15 × 2), hierarchical log-linear analysis of frequencies; Table 3].
oMG's A, D, F, and K were found at very high frequencies in all four species. oMG G was found at high frequency in three of the species examined; oMG's E, I, and T were in two species; and oMG's B and U were each found at high frequency in only one species. Five oMG's (H, J, L, M, and N) were found in very low frequencies in all of the species examined. It is highly unlikely that these oMG's were not detected due to methodological problems, because all were found in the host-seeking nymphs. oMG's C and O were found in neither the host-seeking nymphs nor any of the vertebrate species.
Low-frequency oMG's:
Some oMG's were detected in <3 of the 75 larvae examined from a species. We assessed the possibility that these oMG's were falsely detected by repeating the PCR and reverse line blot from these DNA extractions. Of 21 samples retested, 19 of these rare oMG's were absent in a follow-up test. From 232 samples we experimentally determined that the 100-fold dilution of an engorged larva during the DNA extraction reduces the concentration of different oMG's such that 24% are not detected in a single follow-up test. This may be expected, as the number of B. burgdorferi s.s. transmitted from infected vertebrates to feeding larvae is likely to be quite small (Benach et al. 1987). However, low-frequency oMG's were absent in the follow-up PCR products at a much greater rate than expected at the DNA dilution we used (P < 0.0001, d.f. = 1, chi-square). Thus, we conclude either that many of these low-frequency oMG's were false positives or that they are in a much lower density in the engorged larvae than oMG's that commonly infect a vertebrate species. We discuss possible biological causes of these low-frequency oMG's later in this article.
Distribution of oMG's among conspecific hosts:
All 5 larvae from some animals tested negative for oMG's for which most of their conspecific hosts tested positive. For two reasons all 5 larvae may test negative for an oMG even if the host carries it. First, the larvae are infected by only a subset of the oMG's that have infected the animal the larva had fed upon. Every animal, regardless of species, was infected by six to eight oMG's. However, infected larvae carried between one and seven oMG's. On average, larvae that had fed on P. leucopus tested positive for 3.19 (±1.05) oMG's, 3.65 (±1.23) oMG's were found in larvae from T. striatus, 2.73 (±1.05) were in larvae from B. brevicauda, and 1.75 (±0.83) were in larvae from S. carolinensis. Second, the dilution of the larvae for the DNA extraction reduces the density of spirochetes in the PCR reaction so that some oMG's that are in that larva are not amplified. Because an animal may falsely test negative for an oMG by our methods, we calculated the probability that 5 randomly sampled larvae will test negative for an oMG when the host they fed upon is actually infected by that oMG. If all of the individuals in a species are actually infected by an oMG, the probability a randomly sampled larva will test negative for that oMG is equal to the percentage of larvae from that species that actually tested negative for that oMG. For example, 43 of the 75 infected larvae (57%) that had fed on P. leucopus tested negative for oMG A (Table 3). The probability that 5 larvae randomly sampled from P. leucopus will test negative for oMG A is approximately the fraction of larvae testing negative raised to the fifth power (0.575), or 0.06. Thus, we expect that 0.9 (= 0.06 × 15) of the 15 P. leucopus examined will test negative for oMG A, even though they are truly infected by this oMG. The number of individuals from each species expected to test negative for each of the oMG's coincides well with the number of individuals that actually tested negative in our sample (Table 4). Sampling too few larvae can account for the oMG's that were not found in a small percentage of the individuals of a species. However, this does not exclude the possibility of intraspecific variation in resistance to oMG's.
TABLE 4.
The actual and expected number of individuals testing negative for eachoMG
| A | B | D | E | F | G | I | K | T | U | |
|---|---|---|---|---|---|---|---|---|---|---|
| P. leucopus | 0a | 1 | 0 | − | 0 | 1 | 3 | 0 | − | − |
| 0.93b | 1.6 | 0.21 | 1.8 | 1 | 3.2 | 0.05 | ||||
| T. striatus | 0 | − | 0 | − | 2 | 2 | 2 | 0 | 1 | 3 |
| 1.5 | 0.1 | 1 | 0.9 | 3.2 | 0.1 | 0.8 | 4.2 | |||
| B. brevicauda | 0 | − | 1 | 2 | 1 | − | − | 0 | 2 | − |
| 2.2 | 0.6 | 1.2 | 3.5 | 0 | 2.6 | |||||
| S. carolinensis | 1 | − | 4 | 5 | 3 | 5 | − | 0 | − | − |
| 2.3 | 5.7 | 4.9 | 5.3 | 6.6 | 1.2 |
−, oMG's that do not infect the species.
Regular type, number of individuals testing negative in this sample.
Underlined type, number of individuals expected to test negative.
Intraspecific variation:
Most of the animals examined were infected by every oMG that commonly infected that animal's conspecifics. However, differences among individuals could be quantitative, as opposed to qualitative. Thus, the frequency in which each oMG tested positive in the larvae from each individual within a species was examined to quantitatively assess the intraspecific differences within each species. We found that there was no significant difference among conspecifics in any of the species examined [P > 0.85, d.f. = 196 (S. carolinensis d.f. = 126), hierarchical log-linear analysis of frequencies]. In addition, the frequency distributions of the number of oMG's found per infected tick were not significantly different among conspecifics [P > 0.05, d.f. = 14 (S. carolinensis d.f. = 9), single-classification ANOVA].
An animal may not be infected by an oMG simply because it was never fed upon by a nymph that was infected by that oMG. To control for this possibility, we calculated the probability that any individual we trapped in August 2002 had been exposed to each of the oMG's during the nymphal season. The probability an animal has not been exposed to an oMG is equal to the frequency of that oMG in the host-seeking nymphal population raised to the number of nymphs that have bitten that individual. The oMG frequencies were estimated from the cohort of host-seeking nymphal ticks that infected the small mammals sampled in this study (Table 3). We calculated the probability that an individual T. striatus or an individual P. leucopus we caught had been exposed to each of the oMG's using the average number of nymphs per individual. We estimated that an average of 60 nymphs fed on an individual P. leucopus, and 250 nymphs fed on T. striatus, during the nymphal host-seeking season. These data were collected in May, June, and July 2002 by counting the number of nymphs on the head of a live animal (R. S. Ostfeld, unpublished data). Visual counts are well correlated with, and are a conservative estimate of, the actual number of nymphs on an individual (R. S. Ostfeld, personal communication), leading to a conservative estimate of the probability of exposure. Nymphal burden data are lacking for S. carolinensis and B. brevicauda, although S. carolinensis are parasitized by a greater number of nymphs than T. striatus in August, and B. brevicauda are parasitized by a fewer number than P. leucopus (Table 5). These data suggest that almost all animals had been exposed to all oMG's, so that lack of exposure is not the cause of individuals testing negative (Table 6).
TABLE 5.
Average tick burden per individual when trapped
| Larvae | Nymphs | |
|---|---|---|
| P. leucopus | 38.97 | 0.58 |
| T. striatus | 34.8 | 1.22 |
| B. brevicauda | 57.47 | 0.06 |
| S. carolinensis | 49.03 | 2.56 |
TABLE 6.
The probability an individual has been challenged by each of theoMG's
| A (%) | B (%) | D (%) | E (%) | F (%) | G (%) | H (%) | I (%) | J (%) | K (%) | L (%) | M (%) | N (%) | T (%) | U (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
P. leucopus (60/yr)a |
>99 | >99 | >99 | 99 | 99 | >99 | 98 | 97 | 89 | >99 | 47 | >99 | 79 | 94 | 72 |
|
T. striatus (250/yr)a |
>99 | >99 | >99 | >99 | >99 | >99 | >99 | >99 | >99 | >99 | 93 | >99 | >99 | >99 | 99 |
An estimation of the number of nymphs per year on each individual.
DISCUSSION
The frequency distributions of oMG's differ significantly among vertebrate species, suggesting species-specific rejection of certain oMG's. Each oMG was detected in either >80% or <15% of the individuals from a species (Table 3). The individuals that tested negative for common oMG's likely tested negative because too few larvae were examined. Low-frequency oMG's were usually found in only 1 or 2 of the 75 larvae sampled from the entire species and many can be attributed to false positives. Members of a host species appear to be uniformly resistant or susceptible to any given oMG. Similarly, only four oMG's, A, B, I, and K, can establish an infection in humans (Seinost et al. 1999). We believe that these other vertebrate species, like humans, can be infected only by certain oMG's.
Interspecies multiple-niche polymorphism:
Our results demonstrate that none of the supposed principal reservoir host species at our study site (Schmidt et al. 1999) were infected by all of the 15 oMG's present in host-seeking nymphs. Each vertebrate species was infected by different subsets of oMG's (Table 3). Thus, the strain that an individual can and cannot reject is determined by an interaction between the ospC major group, or a closely linked gene, and features unique to each species. We propose that variation in the ability of oMG's to infect different vertebrate species is the primary selective force that maintains the ospC polymorphism.
The inability to infect a vertebrate species due to ospC may have at least two possible causes. Differences in the function of the OspC protein among oMG's may determine whether a spirochete can establish an infection. The function of OspC is not known, but it has been speculated that it is an adhesin that binds to vertebrate connective tissue and may be important in establishing an infection. Alternatively, the interspecific variation in the vertebrate immune system may provide resistance to infection by certain oMG's. A vertebrate immune system that quickly eliminates one amino acid sequence may not recognize a highly divergent sequence in the early stages of B. burgdorferi s.s. infection, allowing the later oMG to establish an infection. Further investigations of the interaction between OspC and the vertebrate hosts are necessary to test these hypotheses.
Conceivably, balancing selection acts not on the ospC locus, but on a closely linked locus. With the possible exception of genes on a prophage-like plasmid (Stevenson et al. 1998; Eggers and Samuels 1999; Eggers et al. 2000; Stevenson and Miller 2003), linkage disequilibrium is very high in B. burgdorferi s.s. (Dykhuizen et al. 1993; Balmelli and Piffaretti 1996; Dykhuizen and Baranton 2001), lending support to this hypothesis. Such a linked gene would have to be at least as variable as ospC to produce the patterns seen in this and previous studies (Wang et al. 1999; Qiu et al. 2002). However, in a recent study of 17 highly polymorphic loci in 22 B. burgdorferi s.s. isolates, only two to four alleles were detected at all loci except ospC, of which 11 major groups were found (Qiu et al. 2004). Thus, we believe that ospC is most likely the locus under balancing selection. Regardless of the target of selection, our data suggest that variation among host species is maintaining the polymorphism at the ospC locus.
Variation among individuals within species:
There is no evidence of individual variation in resistance to different oMG's. Every individual examined tested positive for nearly all of the oMG's that are commonly found in that species (Table 3). Exceptions are likely due to sampling too few larvae (Table 4). However, it is possible that some of the individuals may be resistant to oMG's that commonly infect its conspecifics. The current data suggest that variation among individuals within a species is insignificant and does not affect our conclusions.
Although most apparent occurrences of very rare oMG's appear to result from experimental error, larvae do seem, rarely, to carry oMG's to which the host species they fed upon is seemingly resistant. Three biological processes could be responsible for these observations.
oMG's that do not cause a disseminated infection in humans are found at a low frequency in the skin near the site of nymphal attachment (Seinost et al. 1999). Therefore, larvae that are feeding on a vertebrate host near a feeding nymph, a process known as cofeeding, can become infected by B. burgdorferi s.s. that have not disseminated in the host (Randolph et al. 1996; Patrican 1997; Piesman and Happ 2001; Randolph and Gern 2003).
Larvae that do not complete a bloodmeal will seek a new host (our personal observation), carrying with them the B. burgdorferi s.s. acquired during the first meal, perhaps including strains that are not present in the second host.
Stochastic events such as depression of the host's immune system might account for the rare oMG occurrence.
Because the data suggest that very few larvae are infected by any of these events, none are likely to significantly affect the frequency of the oMG's.
oMG's found in very low frequencies in all vertebrate species examined:
The vertebrate species examined in this study are believed to be the principal hosts for B. burgdorferi s.s. because they support large numbers of larvae (Table 5) that have a high frequency of infection (Table 2; Anderson 1988; Lane et al. 1991; Ostfeld et al. 1996; Schmidt et al. 1999). However, our data suggest that a significant proportion of the infected host-seeking nymphs obtained B. burgdorferi s.s. from other vertebrate host species. For example, oMG M, which does not infect any of these four species, is often detected in host-seeking nymphs. Four other oMG's are not observed in any of these species, but are observed in host-seeking nymphs, albeit at a relatively low frequencies. These oMG's are likely to infect at least one of the many other vertebrate species that I. scapularis subadults parasitize (Anderson 1988).
Negative frequency-dependent selection:
Previous investigators hypothesized that the polymorphism at the ospC locus is maintained by negative frequency-dependent selection caused by a strong secondary immune response to OspC (Wang et al. 1999). A strong secondary immune response to OspC prevents an animal from being infected by an oMG that previously infected the animal (Preac-Mursic et al. 1992; Gilmore et al. 1996; Barthold 1999), but does not prevent infection by strains with other ospC major groups (Probert et al. 1997). This process gives rare oMG's a selective advantage over common oMG's. If the secondary immune response to OspC were the only force rejecting B. burgdorferi s.s., every oMG would be present in most of the animals (Table 6). The data from this study are not consistent with this model, suggesting that negative frequency-dependent selection is neither necessary nor sufficient to maintain the polymorphism at ospC. Negative frequency-dependent selection could play a role within host species. However, a model incorporating only interspecies multiple-niche polymorphism is sufficient to explain the maintenance of the polymorphism.
Multiple-niche polymorphism model:
A model consisting of 15 “ospC niches,” one niche for each oMG, can sufficiently maintain the observed polymorphism in B. burgdorferi s.s. populations. An ospC niche is defined as the assemblage of vertebrate species that are susceptible to an oMG. For instance, P. leucopus is part of the niche for oMG B, while T. striatus is part of the niche for oMG U (Table 3). The fitness of oMG B is much greater than that of oMG U in P. leucopus, while the opposite is true in T. striatus. In fact, the fitness of oMG's U and B are approximately zero in P. leucopus and T. striatus, respectively. The ospC polymorphism persists because no oMG has the greatest fitness in all ospC niches. This model is sufficient to maintain the polymorphism and is the most parsimonious explanation of the data collected in this study and previous studies (Wang et al. 1999; Qiu et al. 2002), but it does not exclude negative frequency-dependent selection as a subsidiary factor.
In localities where B. burgdorferi s.s. is prevalent in host-seeking nymphs, it is likely that every individual from each host species has been challenged by, and is infected with, every oMG that can infect that species (Table 6). Our model suggests that in these areas, the frequency distribution of oMG's in the nymph population is determined by the relative densities of each vertebrate species, the relative number of larvae that feed on individuals from each species, and the percentage of larvae that become infected with each oMG from feeding on that species. We are assuming that other host species not examined are part of some, but not all, of the ospC niches, similar to the four species examined in this study. These species could include some of the 31 species of mammals from seven orders, 49 species of birds from 17 families, and several species of reptiles that I. scapularis has been reported to feed on (Anderson 1988, 1989; Lane et al. 1991).
Annual ospC frequency distribution fluctuations:
The frequency distributions of oMG's in host-seeking tick populations are homogeneous across space within years, but are significantly different among years (Qiu et al. 1997). The data in this study suggest that these dramatic changes in the frequency distributions arise from fluctuations in the population densities of the reservoir hosts, many of which are known to fluctuate dramatically (Goodwin et al. 2001; Ostfeld et al. 2001). Long-term observations on both the frequency distribution of oMG's and the densities of small mammals and birds could be used to further investigate this possibility.
Risk of human Lyme disease:
The data presented in this study support previous claims that humans, like other species, can be infected by only certain oMG's (Seinost et al. 1999). Thus, the risk of human Lyme disease could be reduced by decreasing the percentage of larvae that are infected by human infectious oMG's. P. leucopus is the only species investigated that is infected by all four of the human infectious oMG's. In addition, the density of P. leucopus is high in most areas of the northeastern United States (Allan et al. 2003) and the proportion of larvae infected from P. leucopus is greater than that from any other species investigated (Table 2; LoGiudice et al. 2003). Thus, measures aimed at reducing the relative contribution of B. burgdorferi s.s. from P. leucopus to I. scapularis larvae may significantly reduce the human Lyme disease risk. Reducing the contribution from B. brevicauda should also reduce the human Lyme disease risk, as the larval burden on B. brevicauda is much greater than that on the other species examined (Table 5). Further theoretical and empirical studies are necessary to determine the efficacy of any control measure.
According to the dilution-effect model, maintaining high vertebrate host diversity within a forest community reduces the proportion of infected ticks, as most reservoir hosts infect fewer of the larval ticks that feed on them than do mice (Ostfeld and Keesing 2000a,b). Fragmented and disturbed habitats, such as suburban communities, have low densities of non-mouse reservoir hosts, effectively increasing the proportion of ticks that feed on P. leucopus (Nupp and Swihart 1996; Allan et al. 2003). According to the dilution-effect model, habitat fragmentation will therefore increase the proportion of ticks carrying B. burgdorferi s.s. and will coincidentally increase the proportion of nymphs carrying human infectious strains, both of which increase the risk of human Lyme disease. Thus, maintaining high species diversity by appropriate land management may reduce the risk of human exposure to Lyme bacteria.
Acknowledgments
This study would not have been possible without the help and guidance of Rick S. Ostfeld and the assistance of his field crew at the Institute for Ecosystem Studies. We are grateful to D. J. Futuyma, W. F. Eanes, M. Lerdau, D. M. Stoebel, M. Feldgarden, M. J. Gomes-Solecki, and M. R. Turner for helpful discussion throughout the project and for reading the manuscript. This study benefited from advice from T. J. S. Merritt and T. N. Engstrom. We thank J. F. Anderson and M. Theisen for DNA and strains for positive controls. This study was supported by a Graduate Research Fellowship from the National Science Foundation to D.B. and Public Health Service grant GM60731 to D.E.D. This is contribution 1129 in graduate studies from the Department of Ecology and Evolution, Stony Brook University.
References
- Allan, B. F., F. Keesing and R. S. Ostfeld, 2003. Effect of forest fragmentation on Lyme disease risk. Conserv. Biol. 17: 267–272. [Google Scholar]
- Anderson, J. F., 1988. Mammalian and avian reservoirs for Borrelia burgdorferi. Ann. NY Acad. Sci. 539: 180–191. [DOI] [PubMed] [Google Scholar]
- Anderson, J. F., 1989. Ecology of Lyme disease. Conn. Med. 53: 343–346. [PubMed] [Google Scholar]
- Balmelli, T., and J. C. Piffaretti, 1996. Analysis of the genetic polymorphism of Borrelia burgdorferi sensu lato by multilocus enzyme electrophoresis. Int. J. Syst. Bacteriol. 46: 167–172. [DOI] [PubMed] [Google Scholar]
- Baranton, G., G. Seinost, G. Theodore, D. Postic and D. Dykhuizen, 2001. Distinct levels of genetic diversity of Borrelia burgdorferi are associated with different aspects of pathogenicity. Res. Microbiol. 152: 149–156. [DOI] [PubMed] [Google Scholar]
- Barthold, S. W., 1999. Specificity of infection-induced immunity among Borrelia burgdorferi sensu lato species. Infect. Immun. 67: 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach, J. L., J. L. Coleman, R. A. Skinner and E. M. Bosler, 1987. Adult Ixodes dammini on rabbits: a hypothesis for the development and transmission of Borrelia burgdorferi. J. Infect. Dis. 155: 1300–1306. [DOI] [PubMed] [Google Scholar]
- Bosler, E. M., 1993 Tick vectors and hosts, pp. 18–26 in Lyme Disease, edited by P. K. Coyle. Mosby-Year Books, St. Louis.
- Brunet, J., and C. C. Mundt, 2000. Disease, frequency-dependent selection, and genetic polymorphism: experiments with stripe rust and wheat. Evolution 54: 406–415. [DOI] [PubMed] [Google Scholar]
- Burgdorfer, W., S. F. Hayes and J. L. Benach, 1988. Development of Borrelia burgdorferi in ixodid tick vectors. Ann. NY Acad. Sci. 539: 172–179. [DOI] [PubMed] [Google Scholar]
- Burgdorfer, W., S. F. Hayes and D. Corwin, 1989. Pathophysiology of the Lyme disease spirochete, Borrelia burgdorferi, in ixodid ticks. Rev. Infect. Dis. 11(Suppl 6): S1442–S1450. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., M. Nordborg and D. Charlesworth, 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70: 155–174. [DOI] [PubMed] [Google Scholar]
- Czochor, R. J., and K. J. Leonard, 1982. Multiple-niche polymorphism in haploid microorganisms. Am. Nat. 119: 293–296. [Google Scholar]
- de Silva, A. M., and E. Fikrig, 1997. Borrelia burgdoferi genes selectively expressed in ticks and mammals. Parasitol. Today 13: 267–270. [DOI] [PubMed] [Google Scholar]
- Dressler, F., J. A. Whalen, B. N. Reinhardt and A. C. Steere, 1993. Western blotting in the serodiagnosis of Lyme disease. J. Infect. Dis. 167: 392–400. [DOI] [PubMed] [Google Scholar]
- Dykhuizen, D. E., and G. Baranton, 2001. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9: 344–350. [DOI] [PubMed] [Google Scholar]
- Dykhuizen, D. E., D. S. Polin, J. J. Dunn, B. Wilske, V. Preacmursic et al., 1993. Borrelia burgdorferi is clonal—implications for taxonomy and vaccine development. Proc. Natl. Acad. Sci. USA 90: 10163–10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers, C. H., and D. S. Samuels, 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181: 7308–7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers, C. H., S. Casjens, S. F. Hayes, C. F. Garon, C. J. Damman et al., 2000. Bacteriophages of spirochetes. J. Mol. Microbiol. Biotechnol. 2: 365–373. [PubMed] [Google Scholar]
- Engstrom, S. M., E. Shoop and R. C. Johnson, 1995. Immunoblot interpretation criteria for serodiagnosis of early Lyme disease. J. Clin. Microbiol. 33: 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerle, V., U. Hauser, G. Liegl, B. Petko, V. Preac-Mursic et al., 1995. Expression of outer surface proteins A and C of Borrelia burgdorferi in Ixodes ricinus. J. Clin. Microbiol. 33: 1867–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, B. P., G. L. McHugh, J. M. Leong and A. C. Steere, 1994. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of the immunoglobulin M response in the serodiagnosis of early infection. Infect. Immun. 62: 3213–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, R. D., K. J. Kappel, M. C. Dolan, T. R. Burkot and B. J. B. Johnson, 1996. Outer surface protein C (OspC), but not P39, is a protective immunogen against a tick-transmitted Borrelia burgdorferi challenge: evidence for a conformational protective epitope in OspC. Infect. Immun. 64: 2234–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliddon, C., and C. Strobeck, 1975. Necessary and sufficient conditions for multiple-niche polymorphism in haploids. Am. Nat. 109: 233–235. [Google Scholar]
- Goodwin, B. J., R. S. Ostfeld and E. M. Schauber, 2001. Spatiotemporal variation in a Lyme disease host and vector: black-legged ticks on white-footed mice. Vector Borne Zoonotic Dis. 1: 129–138. [DOI] [PubMed] [Google Scholar]
- Gromko, M. H., 1977. What is frequency-dependent selection. Evolution 31: 438–442. [DOI] [PubMed] [Google Scholar]
- Hedrick, P. W., 1986. Genetic-polymorphism in heterogeneous environments—a decade later. Annu. Rev. Ecol. Syst. 17: 535–566. [Google Scholar]
- Jaurisheipke, S., R. Fuchs, M. Motz, V. Preacmursic, E. Schwab et al., 1993. Genetic heterogeneity of the genes coding for the outer surface protein C (Ospc) and the flagellin of Borrelia burgdorferi. Med. Microbiol. Immunol. 182: 37–50. [DOI] [PubMed] [Google Scholar]
- Johns, R., J. Ohnishi, A. Broadwater, D. E. Sonenshine, A. M. De Silva et al., 2001. Contrasts in tick innate immune responses to Borrelia burgdorferi challenge: immunotolerance in Ixodes scapularis versus immunocompetence in Dermacentor variabilis (Acari: Ixodidae). J. Med. Entomol. 38: 99–107. [DOI] [PubMed] [Google Scholar]
- Kimura, M., and T. Ohta, 1971. Protein polymorphism as a phase of molecular evolution. Nature 229: 467. [DOI] [PubMed] [Google Scholar]
- Lane, R. S., J. Piesman and W. Burgdorfer, 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36: 587–609. [DOI] [PubMed] [Google Scholar]
- Levene, H., 1953. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87: 331–333. [Google Scholar]
- Lin, T., J. H. Oliver, Jr. and L. Gao, 2002. Genetic diversity of the outer surface protein C gene of southern Borrelia isolates and its possible epidemiological, clinical, and pathogenetic implications. J. Clin. Microbiol. 40: 2572–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGiudice, K., R. S. Ostfeld, K. A. Schmidt and F. Keesing, 2003. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc. Natl. Acad. Sci. USA 100: 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnarelli, L. A., J. F. Anderson and D. Fish, 1987. Transovarial transmission of Borrelia burgdorferi in Ixodes dammini (Acari:Ixodidae). J. Infect. Dis. 156: 234–236. [DOI] [PubMed] [Google Scholar]
- Margolis, N., D. Hogan, W. Cieplak, T. G. Schwan and P. A. Rosa, 1994. Homology between Borrelia burgdorferi OspC and members of the family of Borrelia hermsii variable major proteins. Gene 143: 105–110. [DOI] [PubMed] [Google Scholar]
- Masuzawa, T., T. Komikado and Y. Yanagihara, 1997. PCR-restriction fragment length polymorphism analysis of the ospC gene for detection of mixed culture and for epidemiological typing of Borrelia burgdorferi sensu stricto. Clin. Diagn. Lab. Immunol. 4: 60–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, G., F. Shaw, H. Badrane and X. Vekemans, 1999. The signature of balancing selection: fungal mating compatibility gene evolution. Proc. Natl. Acad. Sci. USA 96: 9172–9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, R. R., S. E. Malawista, K. J. M. Feen and L. K. Bockenstedt, 1996. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 183: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nupp, T. E., and R. K. Swihart, 1996. Effect of forest patch area on population attributes of white-footed mice (Peromyscus leucopus) in fragmented landscapes. Can. J. Zool. 74: 467–472. [Google Scholar]
- Ohta, T., 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23: 263–286. [Google Scholar]
- Ostfeld, R., and F. Keesing, 2000. a The function of biodiversity in the ecology of vector-borne zoonotic diseases. Can. J. Zool. 78: 2061–2078. [Google Scholar]
- Ostfeld, R. S., and F. Keesing, 2000. b Biodiversity and disease risk: the case of Lyme disease. Conserv. Biol. 14: 722–728. [Google Scholar]
- Ostfeld, R. S., M. C. Miller and K. R. Hazler, 1996. Causes and consequences of tick (Ixodes scapularis) burdens on white-footed mice (Peromyscus leucopus). J. Mammal. 77: 266–273. [Google Scholar]
- Ostfeld, R. S., E. M. Schauber, C. D. Canham, K. Keesing, C. G. Jones et al., 2001. Effects of acorn production and mouse abundance on abundance and Borrelia burgdorferi infection prevalence of nymphal Ixodes scapularis ticks. Vector Borne Zoonotic Dis. 1: 55–64. [DOI] [PubMed] [Google Scholar]
- Patrican, L. A., 1997. Acquisition of Lyme disease spirochetes by cofeeding Ixodes scapularis ticks. Am. J. Trop. Med. Hyg. 57: 589–593. [DOI] [PubMed] [Google Scholar]
- Piesman, J., and C. M. Happ, 2001. The efficacy of co-feeding as a means of maintaining Borrelia burgdorferi: a North American model system. J. Vector Ecol. 26: 216–220. [PubMed] [Google Scholar]
- Preac-Mursic, V., B. Wilske, E. Patsouris, S. Jauris, G. Will et al., 1992. Active immunization with pC protein of Borrelia burgdorferi protects gerbils against B. burgdorferi infection. Infection 20: 342–349. [DOI] [PubMed] [Google Scholar]
- Probert, W. S., M. Crawford, R. B. Cadiz and R. B. LeFebvre, 1997. Immunization with outer surface protein (Osp) A, but not OspC, provides cross-protection of mice challenged with North American isolates of Borrelia burgdorferi. J. Infect. Dis. 175: 400–405. [DOI] [PubMed] [Google Scholar]
- Qiu, W. G., E. M. Bosler, J. R. Campbell, G. D. Ugine, I. N. Wang et al., 1997. A population genetic study of Borrelia burgdorferi sensu stricto from eastern Long Island, New York, suggested frequency-dependent selection, gene flow and host adaptation. Hereditas 127: 203–216. [DOI] [PubMed] [Google Scholar]
- Qiu, W. G., D. E. Dykhuizen, M. S. Acosta and B. J. Luft, 2002. Geographic uniformity of the Lyme disease spirochete (Borrelia burgdorferi) and its shared history with tick vector (Ixodes scapularis) in the northeastern United States. Genetics 160: 833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, W. G., S. E. Schutzer, J. F. Bruno, O. Attie, Y. Xu et al., 2004. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc. Natl. Acad. Sci. USA 101: 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph, S., and L. Gern, 2003. Co-feeding transmission and its contribution to the perpetuation of the Lyme disease spirochete Borrelia afzelii. Emerg. Infect. Dis. 9: 893–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph, S. E., L. Gern and P. A. Nuttall, 1996. Co-feeding ticks: epidemiological significance for tick-borne pathogen transmission. Parasitol. Today 12: 472–479. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., X. Vekemans and D. Charlesworth, 2000. The effect of subdivision on variation at multi-allelic loci under balancing selection. Genet. Res. 76: 51–62. [DOI] [PubMed] [Google Scholar]
- Schmidt, K. A., and R. S. Ostfeld, 2001. Biodiversity and the dilution effect in disease ecology. Ecology 82: 609–619. [Google Scholar]
- Schmidt, K. A., R. S. Ostfeld and E. M. Schauber, 1999. Infestation of Peromyscus leucopus and Tamias striatus by Ixodes scapularis (Acari: Ixodidae) in relation to the abundance of hosts and parasites. J. Med. Entomol. 36: 749–757. [DOI] [PubMed] [Google Scholar]
- Schwan, T. G., and J. Piesman, 2000. Temporal changes in outer surface proteins A and C of the lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38: 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan and P. A. Rosa, 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92: 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seinost, G., D. E. Dykhuizen, R. J. Dattwyler, W. T. Golde, J. J. Dunn et al., 1999. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun. 67: 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, C. M., and L. L. Chao, 2002. Genetic analysis of the outer surface protein C gene of Lyme disease spirochaetes (Borrelia burgdorferi sensu lato) isolated from rodents in Taiwan. J. Med. Microbiol. 51: 318–325. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995 Biometry. W. H. Freeman, New York.
- Stevenson, B., and S. W. Barthold, 1994. Expression and sequence of outer surface protein C among North American isolates of Borrelia burgdorferi. FEMS Microbiol. Lett. 124: 367–372. [DOI] [PubMed] [Google Scholar]
- Stevenson, B., and J. C. Miller, 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57: 309–324. [DOI] [PubMed] [Google Scholar]
- Stevenson, B., S. Casjens and P. Rosa, 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144(Pt. 7): 1869–1879. [DOI] [PubMed] [Google Scholar]
- Strobeck, C., 1979. Haploid selection with N-alleles in M-niches. Am. Nat. 113: 439–444. [Google Scholar]
- Thrall, P. H., A. Biere and M. K. Uyenoyama, 1995. Frequency dependent disease transmission and the dynamics of the Silene ustilago host-pathogen system. Am. Nat. 145: 43–62. [Google Scholar]
- Uyenoyama, M. K., 2000. Evolutionary dynamics of self-incompatibility alleles in Brassica. Genetics 156: 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama, M. K., Y. Zhang and E. Newbigin, 2001. On the origin of self-incompatibility haplotypes: transition through self-compatible intermediates. Genetics 157: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans, X., and M. Slatkin, 1994. Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics 137: 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I. N., D. E. Dykhuizen, W. G. Qin, J. J. Dunn, E. M. Bosler et al., 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilske, B., V. Preac-Mursic, G. Schierz and K. V. Busch, 1986. Immunochemical and immunological analysis of European Borrelia burgdorferi strains. Zentralbl. Bakteriol. Mikrobiol. Hyg. [A] 263: 92–102. [DOI] [PubMed] [Google Scholar]
- Wilske, B., V. Preacmursic, S. Jauris, A. Hofmann, I. Pradel et al., 1993. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect. Immun. 61: 2182–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M. L., and A. Spielman, 1985. Seasonal activity of immature Ixodes dammini (Acari: Ixodidae). J. Med. Entomol. 22: 408–414. [DOI] [PubMed] [Google Scholar]