Abstract
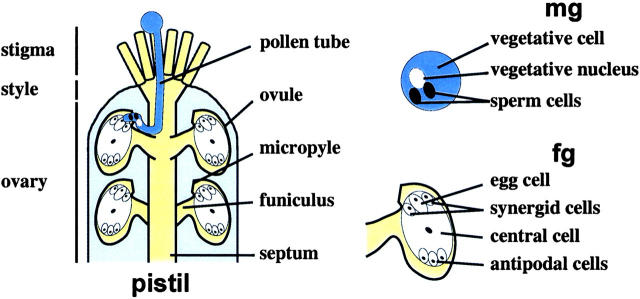
In flowering plants, the egg develops within a haploid embryo sac (female gametophyte) that is encased within the pistil. The haploid pollen grain (male gametophyte) extends a pollen tube that carries two sperm cells within its cytoplasm to the embryo sac. This feat requires rapid, precisely guided, and highly polarized growth through, between, and on the surface of the cells of the stigma, style, and ovary. Pollen tube migration depends on a series of long-range signals from diploid female cells as well as a short-range attractant emitted by the embryo sac that guides the final stage of tube growth. We developed a genetic screen in Arabidopsis thaliana that tags mutant pollen with a cell-autonomous marker carried on an insertion element. We found 32 haploid-disrupting (hapless) mutations that define genes required for pollen grain development, pollen tube growth in the stigma and style, or pollen tube growth and guidance in the ovary. We also identified genomic DNA flanking the insertion element for eleven hap mutants and showed that hap1 disrupts AtMago, a gene whose ortholog is important for Drosophila cell polarity.
IN flowering plants, haploid and diploid cells with distinct gene expression programs interact to produce a network of signals that guide pollen tube growth toward eggs (Figure 1). Mutations that eliminate the functions of the pollen [male gametophyte (MG)] or of the embryo sac [female gametophyte (FG)] cannot be transmitted through the defective gametes; consequently, these mutants can be carried only as heterozygotes. Here, we employed a novel strategy to identify heterozygous hapless (hap) mutants with alterations that impair the development or function of haploid gametophytes. This screen tagged mutant pollen tubes with an autonomous marker, yielding new mutant phenotypes that define key signaling events.
Figure 1.—
Arabidopsis reproductive structures. Key features of the pollen tube (MG) growth pathway through the stigma and style to the ovary are shown. At the septum, pollen tubes migrate into the ovary and approach an ovule containing an FG. The MG and FG comprise three and seven haploid cells, respectively.
Gametophytes are derived from the diploid sporophytic generation, the dominant stage of the angiosperm life cycle. The MG comprises three cells, two immobile sperm cells contained within a larger vegetative cell (reviewed in Twell 1994); the FG has seven cells, the egg, two synergids, the central cell, and three antipodals (Christensen et al. 1997). Upon pollination, MGs are partially desiccated and metabolically inactive; contact with a receptive stigma triggers hydration and germination of a pollen tube. The tip of this highly polarized cell travels across cell boundaries and through intracellular spaces and is guided by multiple discrete signals from the FG and the surrounding diploid cells (reviewed in Johnson and Preuss 2002). The pollen tube penetrates the stigma, grows through the transmitting tissue of the style and ovary, migrates along the septum to the funiculus of an ovule, and grows into the micropyle in response to signals emitted from the synergid cells and the ovule (Elleman et al. 1992; Hulskamp et al. 1995b; Ray et al. 1997; Higashiyama et al. 2001; Palanivelu et al. 2003). After entering the micropyle, the tube bursts to release two sperm, one of which fuses with the egg to produce a zygote, and the other merges with the central cell to generate the endosperm (reviewed in Faure and Dumas 2001).
Genetic screens for recessive loss-of-function mutations have identified several sporophytically expressed factors critical for gametogenesis (Schiefthaler et al. 1999; Hauser et al. 2000; Skinner et al. 2001; Wilson et al. 2001; Sorensen et al. 2003) and pollen-pistil interactions (Preuss et al. 1993; Hulskamp et al. 1995a; Wilhelmi and Preuss 1996; Palanivelu et al. 2003). Because these mutations affect genes expressed in diploid cells, plants heterozygous for a recessive sporophytic mutation can transmit the mutant allele efficiently through male and female gametophytes, forming homozygous progeny. In contrast, mutations that disrupt essential haploid-expressed functions show non-Mendelian inheritance: Because fully penetrant, unconditional mutations cannot be transmitted through the affected gamete, homozygotes cannot be obtained (reviewed in Drews and Yadegari 2002; Johnson and Preuss 2002).
Several groups have screened for gametophytic mutants by searching for aberrant transmission of an antibiotic resistance gene associated with a T-DNA or transposon insertion (Feldmann et al. 1997; Bonhomme et al. 1998; Christensen et al. 1998; Howden et al. 1998; Procissi et al. 2001; Huck et al. 2003; Oh et al. 2003; Lalanne et al. 2004). Coupling these screens with a visual assessment of seed formation has yielded mutants defective in FG development (Christensen et al. 1998, 2002) or in the ability of pollen tubes to release their sperm after entering the micropyle (Huck et al. 2003; Rotman et al. 2003). Distorted segregation screens have also identified mutations that affect pollen grain development and pollen tube growth (Howden et al. 1998; Procissi et al. 2001; Oh et al. 2003; Lalanne et al. 2004). Direct phenotypic screens have also been used to identify gametophytic mutations resulting in abnormal pollen grain development (Chen and McCormick 1996; Park et al. 1998; Johnson and McCormick 2001; Lalanne and Twell 2002). The large number of genetic resources available for reverse genetic studies in Arabidopsis are also leading to the discovery of new genes that are essential for pollen development (Gupta et al. 2002; Kang et al. 2003) and pollen tube growth (Sanderfoot et al. 2001; Mouline et al. 2002; Golovkin and Reddy 2003; Steinebrunner et al. 2003; Hicks et al. 2004). A limitation of screens for gametophytic mutants has been the difficulty in analyzing the phenotypes of mutations that disrupt late aspects of pollen tube growth or guidance. Gametophytic mutants must often be maintained as heterozygotes and the challenges of distinguishing mutant pollen tubes from wild type make it difficult to determine the role of genes involved in pollen tube guidance.
Here we describe an approach for identifying Arabidopsis gametophytic mutants that enables precise analysis of pollen mutant phenotypes. We mutagenized plants with a T-DNA that carried β-glucuronidase (GUS) under the control of the postmeiotic pollen-specific promoter, LAT52 (Twell et al. 1989); this cell-autonomous, MG-specific reporter is detectable in pollen grains and pollen tubes, making it possible to track mutant MGs throughout pollination. Phenotypes can be followed separately from the transmission of the selectable marker carried by the T-DNA, allowing cosegregation tests to unambiguously associate the mutant phenotype with a single T-DNA insertion. We also employed the quartet (qrt1) mutation, which causes pollen grains to be released as intact meiotic tetrads (Preuss et al. 1994) and makes the consequences of chromosomal rearrangements readily apparent. With qrt1 and the cell-autonomous GUS reporter, it is possible to classify plants as either homozygous or hemizygous for the T-DNA insertion by staining their pollen grains. We identified 30 MG mutants that can be grouped into three phenotypic classes: (1) pollen grain development, (2) pollen tube germination or growth within the stigma/style, and (3) pollen tube growth or guidance in the ovary. We identified T-DNA insertion sites for 11 hap mutations and showed that hap1, a mutation causing aberrant pollen tube growth, can be rescued with the tagged gene, Arabidopsis Mago nashi, the ortholog of a Drosophila gene required for oocyte polarity.
MATERIALS AND METHODS
Genetic screening:
Arabidopsis thaliana qrt1 lines (Columbia ecotype) carrying T-DNA insertions were generated by Agrobacterium-mediated transformation with the pCSA110 T-DNA (McElver et al. 2001). pCSA110 encodes GUS under the control of the postmeiotic, pollen-specific LAT52 promoter (Twell et al. 1989), as well as resistance to the herbicide Basta (BastaR). Individual BastaR transgenic plants (primary transformants, T1) were self-fertilized to yield T2 seed stocks. T2 stocks were plated on Murashige and Skoog (MS) medium [MS salts (4.33 g/liter; Carolina Biological Supply), 10% sucrose, pH 5.7 (KOH), 7% Bacto Agar] containing 50 mg/liter Basta (glufosinate ammonium; Crescent Chemical) and the percentage of BastaR seedlings was determined. One stage 14 flower (Smyth et al. 1990) from each BastaR plant retained was fixed (80% acetone, 30 min, 22°) and stained in X-Gluc (5 mm potassium ferrocyanide, 5 mm potassium ferricyanide, 50 mm NaPO4, pH 7, 0.5 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) overnight at 37°.
Pollen tetrads were assayed for meiotic segregation of the LAT52:GUS gene using an inverted Zeiss Telaval 31 microscope. Transmission of the T-DNA through the MG was tested by pollinating stage 14 (Smyth et al. 1990) ms1 flowers (male-sterile, Landsberg erecta ecotype) with mature pollen from hap+/− plants; transmission through the FG was monitored by emasculating stage 12 hap+/− flowers and pollinating them 24–40 hr later with qrt1 (Columbia ecotype) pollen. F1 seed was plated on Basta-containing MS media and seedlings were scored for resistance or sensitivity. A frequency of ≤30% BastaR progeny in the F1 of either cross differs significantly from the expected frequency of 50% (χ2, P = 0.01) when ≥50 progeny are scored.
Phenotypic analysis:
Pollen behavior was examined after crossing three plants from each hap line to three or more ms1 pistils and allowing 12 hr for pollen tube growth. Pistils were excised and mounted on double-sided tape; ovary walls were then removed under a dissecting scope using a 27.5-gauge needle (Becton Dickinson, Franklin Lakes, NJ). Pistils were immediately placed in a microtiter dish containing 100 μl 80% acetone for 30 min to fix cells and remove chlorophyll. Pistils were then incubated overnight at 37° in X-Gluc at high humidity. Pistils were mounted on microscope slides in 50% glycerol and imaged using DIC optics on a Zeiss Axioskop. For closer examination of pollen tube behavior on the ovule, pistils were stained in 0.1% Congo red following incubation in X-Gluc. Congo red is a fluorescent dye that stains pollen tubes along with other cells, allowing analysis by confocal laser scanning microscopy (Palanivelu et al. 2003; Zeiss LSM 510 microscope).
Identification of HAP genes:
Genomic sequences flanking the right and left T-DNA borders were amplified with thermal asymmetric interlaced (TAIL) PCR (McElver et al. 2001). The HAP1 gene was amplified by PCR (primer sequences 5′-TGCACAAACACAAGCCAGTCC-3′ and 5′-GCGAAATTCAACAGCCCTCCTTAC-3′), sequenced, cloned into pCAMBIA2200 (GenBank no. AF234313), and introduced into Agrobacterium (GV3101); hap1 plants were transformed with the floral dip method (Clough and Bent 1998).
RESULTS
A novel screen for mutations that distort Mendelian inheritance:
Our screening procedure utilized two features that facilitated the identification of gametophytic mutants by distorted segregation. First, in addition to an herbicide resistance marker (Basta), the T-DNA element used for mutagenesis, pCSA110 (McElver et al. 2001), contained a LAT52:GUS pollen-specific reporter gene, providing a cell-autonomous tag for pollen grains that carry an insertion. Second, the screen was carried out in the qrt1 mutant, which sheds pollen tetrads at dehiscence, allowing rapid monitoring of transgene inheritance in each strain. We screened 10,074 families derived from the self-fertilization of primary transformants and identified 32 hap mutants.
The screen was conducted in three stages (Figure 2):
Between 50 and 100 seeds descended from each T1 plant were plated on media containing Basta. Mendelian inheritance predicts 75% of the seeds derived from self-fertilization of a hemizygous T1 plant will be BastaR. However, if the T-DNA insertion impairs the development or function of either gametophyte, the percentage of BastaR plants will be reduced because the mutant allele is not transmitted as frequently as the wild-type allele. We retained lines whose T1 progeny yielded <70% BastaR as candidate hap mutants (1391 lines).
Twelve BastaR seedlings from each candidate line were transferred to soil and flowers were stained to reveal GUS expression. Observing GUS segregation in qrt1 pollen tetrads makes it possible to distinguish homozygous (4 GUS+:0 GUS−) from hemizygous (2 GUS+:2 GUS−) plants. An insertion with no effect on the gametophytes is expected to segregate one homozygous plant (4 GUS+:0 GUS−) for every two hemizygotes (2 GUS+:2 GUS−), when only BastaR plants are analyzed. We retained lines as candidate hap mutants if all 12 BastaR plants were hemizygotes (81 lines). Some plants produced pollen tetrads with a mixture of patterns (2:2, 3:1, and 4:0) or lacked GUS expression entirely. Lines with these expression patterns were discarded as they could have resulted from multiple insertions, incomplete insertions, or silencing of the T-DNA.
The 81 hemizygous lines that were retained after the first two assays were reciprocally crossed to wild type, and the percentage of BastaR in the F1 progeny from this cross was determined. If mutant and wild-type gametophytes are equally functional, this cross is expected to yield 50% hemizygous (Basta resistant) and 50% wild-type (Basta sensitive) progeny. Lines that showed ≤30% BastaR F1 progeny, whether the hap plant was crossed as a male or female, were considered to have a gametophytic mutation; 32 hap mutants met these criteria. These reciprocal crosses also allowed us to eliminate recessive embryo-lethal mutations, which were expected to pass the first two stages of the screen. Although these mutations yield 66% BastaR T2 progeny and no homozygous mutant plants, they show no transmission defect when hemizygotes are crossed to wild type.
Figure 2.—

The hapless screen. (A) 32 hap mutant lines were identified by screening >10,000 T1 plants for distorted segregation of BastaR and LAT52:GUS. (B) Segregation of LAT52:GUS in pollen tetrads was used to assign genotypes to the descendants of transformed plants; homozygous wild type (0 GUS+:4 GUS−, left), hemizygotes (2 GUS+:2 GUS−, middle), and homozygotes carrying a GUS transgene (4 GUS+:0 GUS−, right) are shown. A control line exhibited Mendelian (1:2:1) segregation of wild type, GUS+/−, and GUS+/+ plants. An anther locule (top; bar, 50 μm) and individual pollen tetrads (bottom; bar, 25 μm) are shown.
Further tests of BastaR segregation confirmed reduced transmission of hap mutations (Table 1). A hap mutant with one completely nonfunctional gametophyte is expected to yield 50% BastaR offspring when self-fertilized; ∼21 lines had this phenotype. Nine lines, however, produced significantly <50% BastaR progeny, indicating both male and female gametophytes are likely affected. Two others showed >50% BastaR (but <75%), indicating milder gametophytic defects.
TABLE 1.
hap mutations display distorted segregation, affecting the male, female, or both gametophytes
| Self-fertilized
|
Male
|
Female
|
||||
|---|---|---|---|---|---|---|
| % BastaR | n | % BastaR | n | % BastaR | n | |
| Male onlya | ||||||
| hap1 | 43.2 | 1308 | 1.8 | 328 | 54.0 | 174 |
| hap2 | 52.8 | 1581 | 0.7 | 403 | 47.1 | 467 |
| hap3 | 50.0 | 1987 | 1.3 | 151 | 52.3 | 155 |
| hap6 | 48.4 | 153 | 0.0 | 411 | 39.0 | 118 |
| hap9 | 46.6 | 1518 | 8.7 | 585 | 42.9 | 196 |
| hap10 | 51.1 | 2051 | 28.9 | 166 | 48.5 | 233 |
| hap11 | 71.5b | 899 | 18.0 | 133 | 53.5 | 198 |
| hap13 | 52.4 | 1356 | 0.0 | 135 | 61.1 | 113 |
| hap14 | 61.5 | 1582 | 14.7 | 109 | 45.8 | 144 |
| hap15 | 55.2 | 1343 | 0.9 | 109 | 50.1 | 363 |
| hap16 | 49.4 | 875 | 0.0 | 115 | 54.0 | 139 |
| hap21 | 48.1 | 828 | 5.1 | 551 | 46.5 | 258 |
| hap24 | 54.9 | 886 | 20.1 | 413 | 48.2 | 110 |
| hap26 | 55.8 | 265 | 15.9 | 453 | 50.1 | 407 |
| hap28 | 55.2 | 1915 | 4.4 | 1196 | 50.0 | 252 |
| hap31 | 57.1 | 303 | 18.0 | 523 | 48.1 | 135 |
| Female onlyc | ||||||
| hap19 | 55.2 | 647 | 40.4 | 109 | 22.4 | 147 |
| hap29 | 55.3 | 485 | 48.1 | 341 | 4.2 | 72 |
| Affect bothd | ||||||
| hap4 | 42.7 | 1814 | 16.9 | 498 | 40.7 | 236 |
| hap5 | 50.4 | 1275 | 19.3 | 316 | 29.7 | 148 |
| hap7 | 10.6 | 1222 | 0.0 | 124 | 5.9 | 271 |
| hap8 | 52.2 | 1037 | 5.2 | 461 | 32.0 | 172 |
| hap12 | 41.6 | 1282 | 0.0 | 126 | 25.2 | 206 |
| hap17 | 35.9 | 395 | 1.2 | 244 | 32.1 | 156 |
| hap18 | 38.8 | 201 | 8.9 | 621 | 37.0 | 319 |
| hap20 | 24.1 | 826 | 0.5 | 622 | 32.0 | 275 |
| hap22 | 50.5 | 105 | 14.7 | 693 | 28.2 | 117 |
| hap23 | 13.7 | 497 | 4.8 | 832 | 9.1 | 187 |
| hap25 | 36.6 | 123 | 27.6 | 174 | 31.0 | 174 |
| hap27 | 38.9 | 610 | 20.2 | 391 | 0.0 | 70 |
| hap30 | 40.4 | 178 | 18.9 | 386 | 0.0 | 146 |
| hap32 | 29.1 | 2245 | 1.3 | 237 | 35.5 | 169 |
| Controle | 75.9b | 686 | 53.4 | 706 | 49.5 | 188 |
Self fertilized: hap+/− × hap+/−, data are shown for the T3 or T4 generation. Male: ms1 × hap+/−, ms1 females were hand pollinated using hap+/− anthers. Female: hap+/− × qrt1, hap+/− females were emasculated and hand pollinated using qrt1 anthers. % BastaR, the percentage of BastaR F1 progeny from the indicated cross.
Significantly different from 1:1 in male cross but not in female cross (χ2, P < 0.01).
Not significantly different from 3:1 (χ2, P > 0.01).
Significantly different from 1:1 in female cross but not in male cross (χ2, P < 0.01).
Significantly different from 1:1 in both the male and female crosses (χ2, P < 0.01).
Not significantly different from 1:1 in male or female crosses (χ2, P > 0.01).
To determine the extent to which a given hap mutation affected the MG, FG, or both gametophytes, we collected large sets of F1 progeny from reciprocal crosses of hap+/− to HAP+/+ (Table 1). The number of BastaR F1 progeny was monitored, allowing a direct comparison of the reproductive success of each hap gametophyte relative to HAP counterparts produced by the same meiosis. When a hemizygous control line was used as either the male or the female parent in crosses to wild type, 50% BastaR progeny were obtained, indicating that expression of LAT52:GUS does not impair transmission (Table 1). Sixteen mutants showed reduced transmission through the MG but did not appreciably affect the FG; 14 hap defects were general, decreasing, but not eliminating, the function of both gametophytes, and two hap mutations disrupted only the FG (Table 1). Three of the male-specific mutants (hap6, hap13, and hap16) completely disrupted male functions, siring no BastaR progeny when crossed to wild type, and 6 had a strong, but not complete impact on pollen function (≤5% BastaR F1). Of the two female-specific hap mutations, hap29 was nearly completely penetrant (4.2% BastaR F1), and hap19 had a milder impact (22.4% BastaR F1). Two other mutations completely eliminated the function of the FG while having a mild impact on MG function (hap27 and hap30). Last, of the mutations that affected both sexes, some were severe in both males and females (hap7 and hap23), whereas others severely affected one gametophyte while only modestly disrupting the other (hap12, hap27, and hap30).
Assaying pollen phenotypes:
We used the LAT52:GUS reporter to track the behavior of pollen grains and tubes carrying the T-DNA insertion. Tetrads from hemizygous controls produced two GUS+ pollen grains and two GUS− pollen grains (Figure 3A), all of which germinated efficiently. Assays for later stages of growth were performed 12 hr after pollination; in control lines, GUS+ pollen tubes germinated, penetrated the stigmatic papillae, grew through the style, entered the ovary through the transmitting tract, and migrated to the ovule (Figures 4, A and B, and 5, A and B). After entering the micropyle, GUS+ pollen tubes burst, releasing an aggregate of GUS activity (Figure 5B, arrowhead) that marks the final stage of pollen tube function. We pollinated wild type with a hemizygous GUS+ control and after 12 hr of pollen tube growth found 45% (n = 422) of the ovules showed GUS staining; the slight decrease from the expected rate of 50% suggests that 12 hr of pollen growth was too brief for the fertilization of every ovule.
Figure 3.—
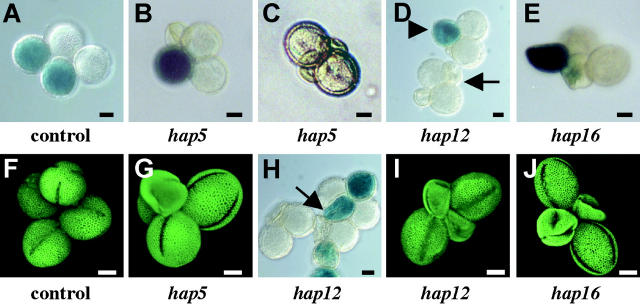
Class 1 hap mutations disrupt pollen grain development. (A and F) A control line hemizygous for a LAT52:GUS insertion produced tetrads with two morphologically normal GUS+ grains. Lines carrying hap5 (B, C, and G) and hap12 (D, H, and I) mutations produced tetrads that had two normal GUS− pollen grains and two grains with varying degrees of abnormality; development of these tetrads was sometimes arrested, inhibiting their ability to express GUS (D, arrow), while in other cases GUS expression was seen in apparently normal (D, arrowhead) or misshapen (H, arrow) grains. Lines carrying hap16 (E and J) produced two aberrant GUS+ pollen grains in each tetrad. Light micrographs (A–E and H, differential interference contrast optics) and fluorescent micrographs (F, G, I, and J, auramine-O stained to highlight pollen surface, confocal microscopy) are shown; bar, 10 μm.
Figure 4.—
Class 2 hap mutations alter early stages of pollen tube growth. (A and B) Pollinating wild type with a LAT52:GUS control revealed GUS+ pollen tubes that germinated, penetrated the stigmatic papillae, grew through the style, entered the ovary via the transmitting tract, and targeted ovules for fertilization. Only GUS+ pollen tubes are visible. (C) Pollen grains carrying a hap6 defect germinated, but failed to exit the style. (D) hap7 pollen tubes were less likely to germinate and rarely entered the style. Bars, 100 μm.
Figure 5.—
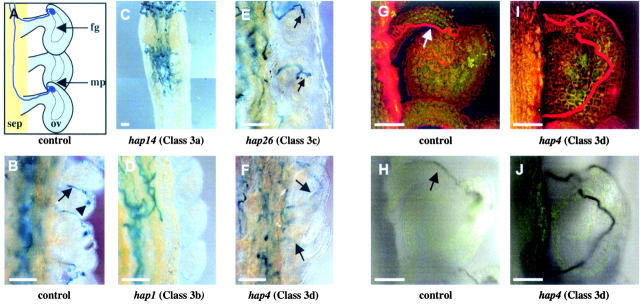
Class 3 hap mutations alter late stages of pollen tube growth or guidance. (A) Diagram of wild-type pollen tube paths (blue); sep, septum; ov, ovule; mp, micropyle; fg, female gametophyte. (B–F) LAT52:GUS staining of pollen tubes growing in wild-type pistils (pollen tube, arrow; GUS activity in ovule, arrowhead). (B) HAP pollen tubes carrying LAT52:GUS grow up the funiculus, penetrate the micropyle, and deposit an aggregate of GUS activity in the FG. (C) hap14 mutants (class 3a) grow short pollen tubes that target the ovules they reach. (D) hap1 pollen tubes (class 3b) fail to exit the septum. (E) hap26 pollen tubes (class 3c) grow up the funiculus, but fail to enter the micropyle. (F) hap4 pollen tubes (class 3d) exhibit random growth on the surface of ovules. Confocal laser scanning micrographs of control (G) and hap4 (I) pollen tubes stained with Congo red show that hap4 pollen tubes grow past the micropyle without entering. Transmitted light micrographs of the same samples (H and J) show GUS activity in pollen tubes. The ovule shown in G and H has two pollen tubes growing on it: a GUS− pollen tube (arrow) is visible in G, while a GUS+ pollen tube (arrow) is visible in H. Collecting both images allows GUS− and GUS+ pollen tubes to be compared. The pollen tube visible in I is GUS+ (J) and therefore carries the hap4 mutation. Bars: A–F, 100 μm; G–J, 50 μm.
We monitored pollen development and pollen tube growth behavior for each hap mutant, examining self-pollinated flowers and crosses of hap pollen onto wild type. This analysis placed each hap mutant into one of four major phenotypic classes (Table 2), having alterations in pollen grain development (class 1; 3 mutants), defective pollen tube growth through the style (class 2; 12 mutants), alterations in pollen tube growth or guidance in the ovary (class 3; 14 mutants), or no obvious defect (class 4; 3 mutants). In instances where some mutant pollen arrested at an earlier stage and other pollen progressed to a later stage, the mutant was assigned to the class representing the majority of mutant pollen.
TABLE 2.
hap mutants fall into four pollen phenotypic classes
| Class | Description | n | Mutants |
|---|---|---|---|
| 1 | Disrupted pollen grain development | 3 | hap5, hap12, hap16 |
| 2 | Short pollen tube growth—failure to exit style | 12 |
hap3, hap6, hap7, hap8, hap9, hap13, hap15, hap17, hap20, hap21, hap28, hap32 |
| 3 | Pollen tube growth in the ovary and/or guidance is disrupted | ||
| 3a | Growth in ovary is short, but pollen tubes target the ovules they reach |
4 | hap10, hap14, hap23, hap31 |
| 3b | Pollen tubes fail to leave the septum | 3 | hap1, hap22, hap18 |
| 3c | Pollen tube growth path appears normal, yet tubes fail to enter the micropyle |
3 | hap11, hap26, hap30 |
| 3d | Pollen tube growth is chaotic in the ovary | 4 | hap2, hap4, hap24, hap27 |
| 4 | No obvious defect | 3 | hap19, hap25, hap29 |
Class 1, alterations in pollen grain development:
Three hap mutants (hap5, hap12, and hap16) affected pollen grain development (Table 2, Figure 3). Neither hap12 nor hap16 pollen grains sired progeny, while hap5 was less extreme, showing a 2.6-fold decrease from wild-type transmission levels (Table 1). The morphology of hap16 pollen grains was consistently aberrant, with tetrads typically forming two almond-shaped GUS+ (hap16) grains that were slightly collapsed and two normal GUS− (HAP16) pollen grains. In some cases hap16 pollen did not express GUS (Figure 3, E and J), and mutant grains never formed pollen tubes; these phenotypes are consistent with an essential function expressed early in pollen development.
The morphological defects of hap5 and hap12 were less severe and showed variable expressivity. By monitoring GUS staining and pollen grain morphology, we scored tetrads as follows: (i) four grains with normal morphology (2 GUS+:2 GUS−); (ii) three normal and one morphologically aberrant grain (2GUS−:1 GUS+:1 aberrant); and (iii) two normal and two morphologically aberrant grains (2 GUS−:2 aberrant). For hap 5, categories i, ii, and iii contained, respectively, 12, 75, and 13% of the tetrads (n = 187; Figure 3, A–C, F, and G); and for hap 12, 0, 47, and 53% of the tetrads (n = 127; Figure 3H, arrow; 3D, arrowhead; 3D, arrow; and 3I). Despite these morphological defects, both hap5 and hap12 produced some pollen tubes that germinated; those from hap12 were arrested in the style (as in class 2 below), while hap5 pollen grains that developed normally were able to fertilize FGs.
Class 2, defective pollen tube growth at the stigma or style:
After pollen tubes emerge from the grain, they establish polarized growth, grow within the cell wall of the stigmatic papillae cells, and then enter the style where they grow through the nutrient-rich extracellular matrix of the transmitting tract. We identified 12 hap mutations that were defective in these stages of pollen tube growth (Figure 4, Table 2). These mutations affected pollen tube germination and growth and when they were reciprocally crossed to wild type, they all showed extreme defects in male reproductive success; five exhibited a 100-fold or greater decrease in transmission and the remainder showed a reduction between 6- and 50-fold (Table 1). One group was male specific (hap3, -6, -9, -13, -15, -21, and -28), and the other (hap7, -8, -17, -20, and -32) affected both sexes.
Class 3, alterations in pollen tube growth or guidance in the ovary:
Despite intensive screening for sporophytic sterile mutations, few genes affecting pollen tube growth or guidance from the style to the ovule have been identified, presumably because such genes are gametophytic. In this screen we identified 14 mutants with alterations in these later stages of pollination (Table 2), two of which (hap1 and hap2) are male specific and virtually 100% penetrant (Table 1). Class 3 mutants fell into four phenotypic groups that genetically define critical steps in the pollen tube growth and guidance process: (a) short tubes that target the ovules they reach (hap10, -14, -23, and -31); (b) pollen tubes that remain on the septum (hap1, -18, and -22); (c) pollen tubes that grow normally to the ovules, but fail to enter the micropyle (hap11, -26, and -30); and (d) pollen tubes that grow chaotically in the ovary (hap2, 4, 24, and 27).
Class 3a mutants demonstrate that pollen tube growth can be genetically separated from guidance; for example, hap14 pollen tubes never grew more than one-third of the way down the septum (Figure 5C), but nonetheless sired 14.7% of F1 progeny when crossed to wild type (Table 1). In contrast, pollen tubes from class 3b (hap1) grew the length of the pistil, but did not migrate toward the ovules and sired only 1.8% of F1 progeny when crossed to wild type (Figure 5D, Table 2); hap18 and hap22 had a similar phenotype. Upon close inspection of class 3b pollen tubes, we found they remained on the septum or in the transmitting tract (Figure 5D).
Class 3c mutants also exhibited a novel phenotype, with the growth of the pollen tubes indistinguishable from wild type except for the final step—targeting the micropyle. For example, hap26 pollen tubes often grew up the funiculus but then stopped and failed to enter the micropyle (Figure 5E); hap11 and hap30 pollen tubes were similarly arrested. We did not identify any hap mutants with pollen tubes that entered the micropyle but continued to grow without bursting as has been observed in the FG mutants fer and srn (Huck et al. 2003; Rotman et al. 2003). The class 3c defects were all subtle with relatively modest (two- to threefold) decreases in transmission through the MG.
Removal or impairment of the FG has been shown to result in chaotic pollen tube growth on affected ovules (Ray et al. 1997; Shimizu and Okada 2000), similar to that observed in the sporophytic mutant, pop2 (Palanivelu et al. 2003). Here we have defined a set of pollen mutants with this phenotype (class 3d); the pollen tubes of hap2, -4, -24, and -27 exit the transmitting tract, yet take unorthodox paths within the ovary and meander along the surface of the ovules (Figure 5, F, I, and J). Two of these (hap2 and -24) are male specific, while one (hap27) also completely impairs FG function. In all of these cases, the principle defect does not appear to be one of pollen tube extension; instead, it is more likely that the gametophytes are defective in responding to signals or, alternatively, in forming appropriate surface contacts necessary for guidance.
T-DNA insertion sites:
Genomic DNA was amplified from 15 of the hap mutants by TAIL PCR and putative insertion sites were successfully identified for 11 mutants (Liu et al. 1995). The junction between the T-DNA and the genome was identified on both sides of the insertion for four of these (hap2, -4, -6, and -15; Table 3). Comparison with the sequenced, annotated Arabidopsis genome localized the TAIL-PCR products to specific loci (Table 3). Other than hap3 and hap11, which fell between two genes, all of the amplified insertion junctions were within a single gene or in the immediately adjacent 5′ or 3′ DNA (Table 3). With the exception of hap2, an expressed sequence tag or a full-length cDNA sequence supported the annotation of each of these genes (Table 3). Current annotation of the Arabidopsis genome describes the role of most of these genes as “unclassified”; the exceptions are hap4 and hap12, which have been assigned roles in protein synthesis and transcription, respectively (Arabidopsis Genome Initiative 2000).
TABLE 3.
Putative insertion sites forhapless mutations
| Mutant | Genea | cDNAb | Description or reference genec | Insertion sitef |
|---|---|---|---|---|
| hap1d | At1g02140 | FL | Mago nashi (Drosophila melanogaster, e = 3 × 10−61; Boswell et al. 1991) | 93 bp upstream |
| hap2e | At4g11720 | None | Unknown | Exon 12 of 14 |
| hap3 | At1g66570 | EST | Sucrose transporter, SUC1 (A. thaliana, e = 0.0; Stadler et al. 1999) | 502 bp upstream |
| At1g66580 | FL | 60S ribosomal protein L10 | 1814 bp upstream | |
| hap4e | At3g52590 | FL | Ubiquitin extension protein 1 (UBQ1)/60S ribosomal protein L40 (Callis et al. 1990) |
Intron 4 of 4 |
| hap5d | At1g30450 | EST | Cation-chloride cotransporter (Nicotiana tabacum, e = 0.0; Harling et al. 1997) |
Exon 13 of 13 |
| hap6e | At4g21150 | FL | Ribophorin II (Homo sapiens, e = 8 × 10−19; Crimaudo et al. 1987) |
186 bp upstreamg |
| hap8 | At5g56250 | FL | Unknown | Exon 3 of 4 |
| hap11d | At5g47020 | EST | Unknown | 495 bp downstream |
| At5g47030 | FL | Mitochondrial ATP synthase δ chain (Ipomoea batatas, e = 6 × 10−72; Morikami et al. 1992) | 89 bp upstream | |
| hap12 | At4g36900 | FL | Contains AP2 domain (RAP2.10) | 62 bp downstreamh |
| hap13 | At1g60780 | FL | Clathrin adapter medium chain, MU1B (Mus musculs, e = 10−154; Ohno et al. 1999) | Exon 8 of 11 |
| hap15e | At1g20200 | FL | 26s proteasome regulatory subunit S3 | Exon 3 of 9 |
Arabidopsis gene names; two genes are listed when insertions were found between two genes.
FL, annotation supported by full-length cDNA; EST, expressed sequence tag in GenBank; none, no EST or full-length cDNA in databases.
Protein sequences were compared with GenBank's nonredundant database (Altschul et al. 1990). Genes with significant similarity for which functional data have been published are noted; e-value from Blast is given.
In addition to TAIL-PCR, one T-DNA border was confirmed by a secondary PCR.
Both T-DNA borders were recovered by PCR.
Based on full-length cDNA if available, or most recent annotation; for insertions between genes, positions are relative to the translational start codon (upstream) or the translational stop codon (downstream).
Insertion in 5′-untranslated region.
Insertion in 3′-untranslated region.
We used the predicted amino acid sequence of each gene to search for similar sequences that might provide insight into their biochemical functions and found similarity to proteins involved in gene expression (hap1, hap3, hap4, hap12, and hap15), secretion (hap6 and hap13), molecular transport (hap3 and hap5), and cellular energy production (hap11; Table 3). The hap8 insertion disrupts a gene with no matches in databases and therefore appears to be unique to the Arabidopsis genome sequence; the hap2 insertion and one of the genes potentially disrupted by hap11 are found only in Arabidopsis and rice and may, therefore, be plant specific.
Molecular complementation of hap1:
The hap1 insertion was found just upstream of the Arabidopsis ortholog of Mago nashi, a Drosophila gene required for localization of oskar mRNA and, consequently, for differentiation of the oocyte posterior and germ-line formation (Boswell et al. 1991; Mohr et al. 2001). To verify the identity of HAP1, we introduced a wild-type copy (HAP1tr), including 730 bp upstream and 700 bp downstream of the open reading frame, into hap1 mutants. A T-DNA carrying HAP1tr and a kanamycin resistance gene was introduced into heterozygous hap1 plants; these plants were self-pollinated and kanamycin-resistant (KanR) progeny were selected. As expected, half of these plants produced pollen tetrads with 2 GUS+ and 2 GUS− grains and thus carried the original hap1 insertion. These plants were self-fertilized, and the segregation of the Basta and Kan markers was analyzed by plating on selective MS medium. Lines with multiple unlinked insertions of the KanR gene were discarded (>90% KanR). A self cross of a line heterozygous for hap1+/− (BastaR+/−) and HAP1tr+/− (KanR+/−) is expected to produce F2 progeny that segregate ∼67% BastaR (8/12) and ∼83% KanR (10/12); this segregation pattern results because hap1 MGs function only when they carry the HAP1tr construct. Two independent transgenic lines were thoroughly analyzed: F2 progeny were 70% (n = 380) and 69% (n = 377) BastaR and 79% (n = 482) and 82% (n = 355) KanR, respectively. Untransformed hap1 lines showed 48% BastaR progeny (see also Table 1). These results indicate that HAP1tr restores function to hap1 MGs. Further confirmation was obtained when pollen tetrads were analyzed in KanR F2 plants: plants with pollen tetrads that were 4 GUS+ to 0 GUS− were found at expected frequencies (∼25%) among KanR plants (24/112 plants); these were never found in hap1 lines that were not transformed with HAP1tr (n > 200). In addition, HAP1tr restored the ability of hap1 pollen to migrate from the transmitting tract and to fertilize ovules (data not shown).
DISCUSSION
Haploid-specific genes required for pollen tube growth and guidance:
Here, we describe 32 Arabidopsis mutations that affect genes expressed in haploid gametophytes. Thirty hap mutants altered MG functions, significantly expanding the set of previously described MG mutations. Mutant pollen cells were tagged with an autonomous marker that facilitated precise phenotypic analysis, allowing the placement of each mutant into one of three phenotypic categories: (1) altered pollen grain development, (2) failure of pollen tube growth within the stigma and/or style, or (3) failure of pollen tube growth within the ovary (Table 2).
This study identified new mutations that cause novel pollen tube growth phenotypes, as well as mutations that are phenotypically similar to previously characterized sporophytic or gametophytic mutants. The 12 class 2 hap mutations disrupted pollen tube germination or growth through the stigma and style similar to the previously identified mad4, syp21-1, npg1, AtAPY1/AtAPY2, kip, seth1, and seth2 gametophytic mutations (Grini et al. 1999; Sanderfoot et al. 2001; Golovkin and Reddy 2003; Procissi et al. 2003; Steinebrunner et al. 2003; Lalanne et al. 2004). Four class 3a hap mutations arrested tube growth in the upper ovary and, like the gametophytic tip1 defect (Ryan et al. 1998), did not impair the tube's ability to target ovules. Three class 3b hap mutations produced tubes that grew along the septum but failed to exit onto the placenta surface, reminiscent of the major defect caused by the sporophytic pop2 mutation (Palanivelu et al. 2003). Three class 3c hap mutations yielded pollen tubes that had normal growth to the micropyle yet failed to penetrate ovules; this phenotype has not been previously described. Finally, like the secondary pop2 defect (Palanivelu et al. 2003), the four class 3d hap mutations generated tubes that grew toward ovules, but failed to adhere to the funicular surface or target the micropyle; no MG mutants with this defect were previously known.
Beyond defining functions required for assembling pollen grains and extending tubes, each hap mutant class could include MG genes that mediate responses to female signals directing pollen tube growth. Class 2 hap mutations might disrupt the initial interactions between pollen and stigma cells, including the pollen's ability to hydrate, establish polarity, germinate, and penetrate stigmatic papillae (Johnson and Preuss 2002). Sporophytically expressed pollen coat components, including lipids and proteins, are critical for pollen hydration (Preuss et al. 1993; Lush et al. 1998), yet MG-expressed factors likely mediate pistil interactions soon after germination. A lily in vitro system recently led to the identification of a complex of pectin and a small cysteine-rich protein (SCA) that forms on the stigma and transmitting tract to promote pollen tube attachment and growth (Lord 2003). Pollen tube factors that bind the pectin/SCA matrix are not known, and hap mutants that fail to grow in the transmitting tract may reveal components of a matrix-driven translocation system. Additionally, chemocyanin, a stigma protein that redirects pollen tube growth in vitro, was also recently purified from lily (Kim et al. 2003). Whether a similar factor directs Arabidopsis pollen tube growth remains to be determined; however, Arabidopsis does encode a protein that is 60% identical to chemocyanin (Kim et al. 2003). Class 2 hap mutants may be an important resource to identify new components of an Arabidopsis analog to this lily signaling system.
Class 3 hap defects could disrupt MG responses to FG signals that mediate transmitting tract exit, ovule choice, funicular growth, or micropylar targeting. Genetic ablation of the FG causes pollen tubes to bypass the affected ovules (Ray et al. 1997), and more subtle defects in FG development implicate signals that regulate adhesion to the funiculus and micropyle targeting (Shimizu and Okada 2000). Class 3c hap mutants, which arrest tube growth just before the micropyle (Figure 5E), may be defective in their response to these late guidance cues, representing male counterparts of the maa1 and maa3 FG mutants (Shimizu and Okada 2000). Class 3d hap pollen tubes resemble pop2 pollen tubes that exit the transmitting tract, but fail to target the micropyle. In pop2, a 100-fold increase in gamma amino butyric acid (GABA) concentration causes aberrant tube migration; therefore, class 3d mutants may define components of a pollen-expressed GABA response pathway.
The hap screen complements previous screens based on distorted segregation:
Many gametophytic mutants have been identified by monitoring the segregation of antibiotic resistance markers in lines generated by insertion mutagenesis (Feldmann et al. 1997; Bonhomme et al. 1998; Christensen et al. 1998; Howden et al. 1998; Procissi et al. 2001; Huck et al. 2003; Oh et al. 2003; Lalanne et al. 2004). Non-Mendelian, distorted segregation ratios identified numerous FG mutations, as well as MG mutations that alter pollen grain development, pollen tube germination, and tube growth. The inability to differentiate mutant from wild-type pollen tubes in heterozygous plants hampered characterization of MG mutants affecting the final stages of pollen tube growth (Procissi et al. 2001). Here, we assayed distorted segregation of an herbicide resistance marker and a pollen-specific reporter gene (LAT52:GUS) that tagged mutant pollen in qrt1 plants, enabling a focus on single insertions that potentially affect any stage of MG growth.
Gametophytic mutations can be pleiotropic, incompletely penetrant, and can display variable expressivity (Feldmann et al. 1997; Bonhomme et al. 1998; Howden et al. 1998; Grini et al. 1999; Drews and Yadegari 2002). The inclusion of a cell-autonomous tag in the hap screen makes analysis of these characteristics more efficient. For example, the pollen grain defects in hap16, hap5, and hap12 varied from strong (no GUS expression, completely collapsed) to weak (normal GUS expression, normal pollen grain), and this phenotypic variation was evident because the pollen tetrads were marked by LAT52:GUS (Figure 3). By tagging the mutant pollen tubes, it was clear that rare hap12 pollen grains germinated pollen tubes that failed to leave the stigma, and that hap3 pollen tubes occasionally grew down the length of the ovary but failed to target ovules. These variable phenotypes could result from unequal inheritance of gene products expressed in the diploid meiocyte or from variable expression of gametophytically expressed genes.
The method of Agrobacterium-mediated transformation used here targets the FG, potentially limiting our ability to recover insertions that result in FG lethality (Clough and Bent 1998; McElver et al. 2001; Drews and Yadegari 2002). Nevertheless, we did identify two hap defects that completely blocked FG function (Table 1, hap27 and hap30), suggesting that these genes function at a developmental stage that occurs prior to the Agrobacterium targeting event. Consistent with previous observations (Feldmann et al. 1997; Bonhomme et al. 1998; Grini et al. 1999; Christensen et al. 2002), approximately half (14) of the hap mutations affected both the MG and FG to some degree, suggesting they alter basic functions required by male and female haploid cells. Interestingly, some hap mutants with MG and FG defects extend full-length pollen tubes with a reduced capacity to target ovules (hap4, hap18, hap22, hap27, and hap30), raising the possibility that they define factors required for signaling between male and female gametophytes. On the other hand, a greater number (16) were pollen specific, identifying a set of genes required by the MG that have no essential role in MG development or function.
By performing the hap screen in the qrt1 background, we were able to efficiently discard numerous lines with unwanted alterations that are byproducts of T-DNA mutagenesis, including multiple unlinked insertions, secondary untagged mutations, complex or incomplete insertions, and translocations (Castle et al. 1993; Feldmann et al. 1997; Nacry et al. 1998). We retained BastaR plants that produced only 2 GUS+ to 2 GUS− tetrads; plants that carry multiple T-DNA insertions are readily differentiated by their altered patterns of GUS segregation. By monitoring the inheritance of BastaR, GUS, and the gametophytic phenotype, we were assured that the hap mutation is caused by a single insertion. Furthermore, while translocations and other genomic rearrangements have contaminated previous screens for gametophytic genes (Feldmann et al. 1997; Bonhomme et al. 1998), tetrad analysis readily identifies these events. Translocations produce two types of tetrads in equal proportion: (1) four viable pollen grains with balanced chromosomes and (2) four inviable (shriveled) pollen grains that carry duplications and deficiencies (Kindiger et al. 1991). In QRT1+/+ plants, translocations yield 50% aborted pollen and are indistinguishable from a heterozygous gametophytic lethal mutation; however, translocations are obvious in qrt1, where two types of tetrads are produced as described above.
Functional genomics of the male gametophyte:
A complete understanding of the ∼26,000 Arabidopsis genes requires a thorough analysis of the gametophytic generation. Between 13 and 20% of Arabidopsis genes are expressed in pollen and as many as 5% are pollen specific (Becker et al. 2003; Honys and Twell 2003); functional analysis of only a small fraction of these genes has been performed. None of the genes identified in this study were previously identified by mutations in plants (Table 3). Screens for gametophytic mutations are efficient, rapid, and can be performed on a genome-wide scale. Approximately 180,000 T-DNA insertions are required to achieve a 95% probability of identifying at least one insertion in every Arabidopsis gene, with an average of three insertions per gene (Krysan et al. 1999). Here, we identified 30 MG mutants in a screen of 10,074 transgenic lines (0.3%); this number underestimates the mutant/insertion rate because we discarded ∼47% of the lines because of multiple insertions (Budziszewski et al. 2001). Correcting for multiple inserts, we calculate that MG hap mutations represent 1/180 insertions. A saturation screen would consequently yield ∼1000 hap mutations representing ∼330 MG genes (assuming three alleles per gene). Traditional tests of allelism cannot be performed with gametophytic mutants because the affected cells are haploid; therefore, determining the allelic relationship between hap mutations requires identifying the responsible gene. The identity of HAP1 has been confirmed by molecular complementation, and provisional assignments of several other HAP genes have been made by PCR. Because each hap strain has a single T-DNA insertion, it is likely that many of these assignments will prove correct.
Many candidate MG genes have predicted functions that could meet the unusual demands of pollen tube growth. The pollen tube extends at an astounding rate, growing >100-fold in length by absorbing metabolites from floral tissues, converting them into energy, and delivering newly synthesized membrane and cell wall components to the tube tip (Hepler et al. 2001). Therefore, it is not surprising that hap mutants with short pollen tubes implicate genes with predicted roles in sucrose transport (hap3; Stadler et al. 1999) or membrane trafficking (hap6 and hap13; Crimaudo et al. 1987; Ohno et al. 1999). hap5 acts much earlier, causing defects in pollen grain development (Figure 3, B, C, and G); this phenotype could result from alterations in a predicted cation-chloride cotransporter (Harling et al. 1997). Intriguingly, hap11 acts at a very late stage, affecting pollen tube entry into the micropyle; this mutant has an insertion just upstream of a predicted mitochondrial ATP synthase δ chain (Morikami et al. 1992), suggesting an unexpected late-stage energy requirement. Microarray analysis of mature pollen (Becker et al. 2003; Honys and Twell 2003) and profiles of pollen gene expression (Mascarenhas 1990) suggest that the mature pollen grain is packed with transcripts that are translated upon tube germination. A large set of hap genes may regulate the expression or stability of these MG gene products (Table 3), affecting mRNA metabolism/localization (hap1), transcription (hap12), protein synthesis (hap3 and hap4), and protein degradation (hap15). hap1 alters late stages of tube growth, and its correspondence to Mago nashi, a highly conserved protein associated with mRNA processing and translocation in animals (Palacios 2002), is particularly intriguing. In Drosophila, Mago is predominantly nuclear, but shuttles to the cytoplasm as part of a complex that is required for proper localization of Oskar mRNA (Micklem et al. 1997; Hachet and Ephrussi 2001; Mohr et al. 2001); consequently, hap1 could point to an important requirement for subcellular mRNA localization in pollen tube growth and guidance.
Considerable effort will be required to uncover all of the MG functions necessary for pollen cell maturation, growth, and communication with female cells. This work will also be valuable for understanding the functions of genes that are critical for later stages in plant development because many MG mutations identify genes that also have important sporophytic functions. Dissecting these roles will require comparing the outcomes of gametophyte screens with those of saturation screens for embryo-lethal mutations, analyzing the phenotype of rare homozygotes recovered from gametophytic mutants, or examining the sporophytic development of gametophytic mutants rescued by a gametophytically expressed transgene. Such efforts will considerably advance the goal of defining the function of all Arabidopsis genes within this decade.
Acknowledgments
We thank Janine Hill, Jason Wilcox, Eric McClure, Geoffrey Schultz, Jacob Shreckengost, and Benjamin Maher for technical assistance and the staff of The University of Chicago Greenhouse for excellent plant growth facilities. We thank Gregory Copenhaver, Jean Greenberg, Jocelyn Malamy, and Ravishankar Palanivelu for helpful discussions and comments on the manuscript. This work was supported in part by the U.S. Department of Energy, the National Science Foundation, the Searle Scholars Program, and the Howard Hughes Medical Institute. M.A.J. was supported by a National Institutes of Health, Ruth L. Kirschstein National Research Service Award; K.v.B. was supported by the University of Chicago Medical Scientist Training Program.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Becker, J. D., L. C. Boavida, J. Carneiro, M. Haury and J. A. Feijo, 2003. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 133: 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme, S., C. Horlow, D. Vezon, S. de Laissardiere, A. Guyon et al., 1998. T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distortion alone. Mol. Gen. Genet. 260: 444–452. [DOI] [PubMed] [Google Scholar]
- Boswell, R. E., M. E. Prout and J. C. Steichen, 1991. Mutations in a newly identified Drosophila melanogaster gene, Mago nashi, disrupt germ-cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development 113: 373–384. [DOI] [PubMed] [Google Scholar]
- Budziszewski, G. J., S. P. Lewis, L. W. Glover, J. Reineke, G. Jones et al., 2001. Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics 159: 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis, J., J. A. Raasch and R. D. Vierstra, 1990. Ubiquitin extension proteins of Arabidopsis thaliana. Structure, localization, and expression of their promoters in transgenic tobacco. J. Biol. Chem. 265: 12486–12493. [PubMed] [Google Scholar]
- Castle, L. A., D. Errampalli, T. L. Atherton, L. H. Franzmann, E. S. Yoon et al., 1993. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol. Gen. Genet. 241: 504–514. [DOI] [PubMed] [Google Scholar]
- Chen, Y. C., and S. McCormick, 1996. Sidecar pollen, an Arabidopsis thaliana male gametophytic mutant with aberrant cell divisions during pollen development. Development 122: 3243–3253. [DOI] [PubMed] [Google Scholar]
- Christensen, C. A., E. J. King, J. R. Jordan and G. N. Drews, 1997. Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10: 49–64. [Google Scholar]
- Christensen, C. A., S. Subramanian and G. N. Drews, 1998. Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202: 136–151. [DOI] [PubMed] [Google Scholar]
- Christensen, C. A., S. W. Gorsich, R. H. Brown, L. G. Jones, J. Brown et al., 2002. Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14: 2215–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crimaudo, C., M. Hortsch, H. Gausepohl and D. I. Meyer, 1987. Human ribophorins I and II: the primary structure and membrane topology of two highly conserved rough endoplasmic reticulum-specific glycoproteins. EMBO J. 6: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G. N., and R. Yadegari, 2002. Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36: 99–124. [DOI] [PubMed] [Google Scholar]
- Elleman, C. J., V. Franklin Tong and H. G. Dickinson, 1992. Pollination in species with dry stigmas: the nature of the early stigmatic response and the pathway taken by pollen tubes. New Phytol. 121: 413–424. [DOI] [PubMed] [Google Scholar]
- Faure, J. E., and C. Dumas, 2001. Fertilization in flowering plants. New approaches for an old story. Plant Physiol. 125: 102–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, K. A., D. A. Coury and M. L. Christianson, 1997. Exceptional segregation of a selectable marker (KanR) in Arabidopsis identifies genes important for gametophytic growth and development. Genetics 147: 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin, M., and A. S. Reddy, 2003. A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc. Natl. Acad. Sci. USA 100: 10558–10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini, P. E., A. Schnittger, H. Schwarz, I. Zimmermann, B. Schwab et al., 1999. Isolation of ethyl methanesulfonate-induced gametophytic mutants in Arabidopsis thaliana by a segregation distortion assay using the multimarker chromosome 1. Genetics 151: 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R., J. T. Ting, L. N. Sokolov, S. A. Johnson and S. Luan, 2002. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14: 2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet, O., and A. Ephrussi, 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11: 1666–1674. [DOI] [PubMed] [Google Scholar]
- Harling, H., I. Czaja, J. Schell and R. Walden, 1997. A plant cation-chloride co-transporter promoting auxin-independent tobacco protoplast division. EMBO J. 16: 5855–5866. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hauser, B. A., J. Q. He, S. O. Park and C. S. Gasser, 2000. TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development 127: 2219–2226. [DOI] [PubMed] [Google Scholar]
- Hepler, P. K., L. Vidali and A. Y. Cheung, 2001. Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187. [DOI] [PubMed] [Google Scholar]
- Hicks, G. R., E. Rojo, S. Hong, D. G. Carter and N. V. Raikhel, 2004. Germinating pollen has tubular vacuoles, displays highly dynamic vacuole biogenesis, and requires VACUOLESS1 for proper function. Plant Physiol. 134: 1227–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama, T., S. Yabe, N. Sasaki, Y. Nishimura, S. Miyagishima et al., 2001. Pollen tube attraction by the synergid cell. Science 293: 1480–1483. [DOI] [PubMed] [Google Scholar]
- Honys, D., and D. Twell, 2003. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132: 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden, R., S. K. Park, J. M. Moore, J. Orme, U. Grossniklaus et al., 1998. Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck, N., J. M. Moore, M. Federer and U. Grossniklaus, 2003. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., S. D. Kopczak, T. F. Horejsi, B. K. Kihl and R. E. Pruitt, 1995. a Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J. 8: 703–714. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., K. Schneitz and R. E. Pruitt, 1995. b Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M. A., and D. Preuss, 2002. Plotting a course: multiple signals guide pollen tubes to their targets. Dev. Cell 2: 273–281. [DOI] [PubMed] [Google Scholar]
- Johnson, S. A., and S. McCormick, 2001. Pollen germinates precociously in the anthers of raring-to-go, an Arabidopsis gametophytic mutant. Plant Physiol. 126: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B. H., D. M. Rancour and S. Y. Bednarek, 2003. The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J. 35: 1–15. [DOI] [PubMed] [Google Scholar]
- Kim, S., J. C. Mollet, J. Dong, K. Zhang, S. Y. Park et al., 2003. Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl. Acad. Sci. USA 100: 16125–16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindiger, B., J. B. Beckett and E. H. Coe, 1991. Differential effects of specific chromosomal deficiencies on the development of the maize pollen grain. Genome 34: 579–594. [Google Scholar]
- Krysan, P. J., J. C. Young and M. R. Sussman, 1999. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne, E., and D. Twell, 2002. Genetic control of male germ unit organization in Arabidopsis. Plant Physiol. 129: 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne, E., D. Honys, A. Johnson, G. H. Borner, K. S. Lilley et al., 2004. SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. G., N. Mitsukawa, T. Oosumi and R. F. Whittier, 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463. [DOI] [PubMed] [Google Scholar]
- Lord, E. M., 2003. Adhesion and guidance in compatible pollination. J. Exp. Bot. 54: 47–54. [DOI] [PubMed] [Google Scholar]
- Lush, W. M., F. Grieser and M. Wolters-Arts, 1998. Directional guidance of Nicotiana alata pollen tubes in vitro and on the stigma. Plant Physiol. 118: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas, J. P., 1990. Gene activity during pollen development. Annu. Rev. Plant Physiol. Mol. Biol. 41: 317–338. [Google Scholar]
- McElver, J., I. Tzafrir, G. Aux, R. Rogers, C. Ashby et al., 2001. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem, D. R., R. Dasgupta, H. Elliott, F. Gergely, C. Davidson et al., 1997. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol. 7: 468–478. [DOI] [PubMed] [Google Scholar]
- Mohr, S. E., S. T. Dillon and R. E. Boswell, 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15: 2886–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikami, A., K. Aiso, T. Asahi and K. Nakamura, 1992. The delta′-subunit of higher-plant 6-subunit mitochondrial F1-Atpase is homologous to the delta-subunit of animal mitochondrial F1-Atpase. J. Biol. Chem. 267: 72–76. [PubMed] [Google Scholar]
- Mouline, K., A. A. Very, F. Gaymard, J. Boucherez, G. Pilot et al., 2002. Pollen tube development and competitive ability are impaired by disruption of a Shaker K(+) channel in Arabidopsis. Genes Dev. 16: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry, P., C. Camilleri, B. Courtial, M. Caboche and D. Bouchez, 1998. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S. A., S. K. Park, I. Jang, R. Howden, J. M. Moore et al., 2003. halfman, an Arabidopsis male gametophytic mutant associated with a 150 kb chromosomal deletion adjacent to an introduced Ds transposable element. Sex. Plant Reprod. 16: 99–102. [Google Scholar]
- Ohno, H., T. Tomemori, F. Nakatsu, Y. Okazaki, R. C. Aguilar et al., 1999. mu 1B, a novel adaptor medium chain expressed in polarized epithelial cells. FEBS Lett. 449: 215–220. [DOI] [PubMed] [Google Scholar]
- Palacios, I. M., 2002. RNA processing: splicing and the cytoplasmic localisation of mRNA. Curr. Biol. 12: R50–R52. [DOI] [PubMed] [Google Scholar]
- Palanivelu, R., L. Brass, A. F. Edlund and D. Preuss, 2003. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59. [DOI] [PubMed] [Google Scholar]
- Park, S. K., R. Howden and D. Twell, 1998. The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799. [DOI] [PubMed] [Google Scholar]
- Preuss, D., B. Lemieux, G. Yen and R. W. Davis, 1993. A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 7: 974–985. [DOI] [PubMed] [Google Scholar]
- Preuss, D., S. Y. Rhee and R. W. Davis, 1994. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460. [DOI] [PubMed] [Google Scholar]
- Procissi, A., S. de Laissardiere, M. Ferault, D. Vezon, G. Pelletier et al., 2001. Five gametophytic mutations affecting pollen development and pollen tube growth in Arabidopsis thaliana. Genetics 158: 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procissi, A., A. Guyon, E. S. Pierson, A. Giritch, B. Knuiman et al., 2003. KINKY POLLEN encodes a SABRE-like protein required for tip growth in Arabidopsis and conserved among eukaryotes. Plant J. 36: 894–904. [DOI] [PubMed] [Google Scholar]
- Ray, S. M., S. S. Park and A. Ray, 1997. Pollen tube guidance by the female gametophyte. Development 124: 2489–2498. [DOI] [PubMed] [Google Scholar]
- Rotman, N., F. Rozier, L. Boavida, C. Dumas, F. Berger et al., 2003. Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 13: 432–436. [DOI] [PubMed] [Google Scholar]
- Ryan, E., C. Grierson, A. Cavell, M. Steer and L. Dolan, 1998. TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol. 138: 49–58. [Google Scholar]
- Sanderfoot, A. A., M. Pilgrim, L. Adam and N. V. Raikhel, 2001. Disruption of individual members of Arabidopsis syntaxin gene families indicates each has essential functions. Plant Cell 13: 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefthaler, U., S. Balasubramanian, P. Sieber, D. Chevalier, E. Wisman et al., 1999. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K. K., and K. Okada, 2000. Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127: 4511–4518. [DOI] [PubMed] [Google Scholar]
- Skinner, D. J., S. C. Baker, R. J. Meister, J. Broadhvest, K. Schneitz et al., 2001. The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 13: 2719–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D. R., J. L. Bowman and E. M. Meyerowitz, 1990. Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, A. M., S. Krober, U. S. Unte, P. Huijser, K. Dekker et al., 2003. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33: 413–423. [DOI] [PubMed] [Google Scholar]
- Stadler, R., E. Truernit, M. Gahrtz and N. Sauer, 1999. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 19: 269–278. [DOI] [PubMed] [Google Scholar]
- Steinebrunner, I., J. Wu, Y. Sun, A. Corbett and S. J. Roux, 2003. Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiol. 131: 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D., 1994 The Diversity and Regulation of Gene Expression in the Pathway of Male Gametophyte Development. Cambridge University Press, Cambridge/London/New York.
- Twell, D., R. Wing, J. Yamaguchi and S. McCormick, 1989. Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 217: 240–245. [DOI] [PubMed] [Google Scholar]
- Wilhelmi, L. K., and D. Preuss, 1996. Self-sterility in Arabidopsis due to defective pollen tube guidance. Science 274: 1535–1537. [DOI] [PubMed] [Google Scholar]
- Wilson, Z. A., S. M. Morroll, J. Dawson, R. Swarup and P. J. Tighe, 2001. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28: 27–39. [DOI] [PubMed] [Google Scholar]