Abstract
Transcription of the Saccharomyces MAL structural genes is induced 40-fold by maltose and requires the MAL-activator and maltose permease. To identify additional players involved in regulating MAL gene expression, we carried out a genetic selection for MAL constitutive mutants. Strain CMY4000 containing MAL1 and integrated copies of MAL61promoter-HIS3 and MAL61promoter-lacZ reporter genes was used to select constitutive mutants. The 29 recessive mutants fall into at least three complementation groups. Group 1 and group 2 mutants exhibit pleiotropic phenotypes and represent alleles of Mediator component genes RGR1 and SIN4, respectively. The rgr1 and sin4 constitutive phenotype does not require either the MAL-activator or maltose permease, indicating that Mediator represses MAL basal expression. Further genetic analysis demonstrates that RGR1 and SIN4 work in a common pathway and each component of the Mediator Sin4 module plays a distinct role in regulating MAL gene expression. Additionally, the Swi/Snf chromatin-remodeling complex is required for full induction, suggesting a role for chromatin remodeling in the regulation of MAL gene expression. A sin4Δ mutation is unable to suppress the defects in MAL gene expression resulting from loss of the Swi/Snf complex component Snf2p. The role of the Mediator in MAL gene regulation is discussed.
SACCHAROMYCES maintains a variety of nutrient-sensing mechanisms that enable it to respond to different nutrients and monitor nutrient levels. These include sensing mechanisms for carbon sources, particularly glucose but also other fermentable carbon sources (reviewed in Ozcan and Johnston 1999); nitrogen sources, including ammonia, urea, and amino acids in general (reviewed in Forsberg and Ljungdahl 2001; ter Schure et al. 2000); and other requirements such as phosphate (Wykoff and O'Shea 2001). At least three sensing mechanisms are utilized to monitor glucose levels alone: the Snf1 protein kinase pathway, the Rgt2/Snf3 receptor pathway, and the Gpr1/Gpa2 signaling pathway (reviewed in Johnston 1999; Thevelein and de Winde 1999; Versele et al. 2001). Systems for sensing specific sugars, such as galactose or maltose, or specific amino acids, such as histidine or proline, also are present. Both the specific systems and the more global regulatory systems are integrated via multiple mechanisms.
A major interest of our laboratory is the sensing mechanism for maltose and other α-glucosides. Studies of maltose fermentation undertaken during the last 50 years, including work from our laboratory, demonstrate that maltose induction of MAL gene expression depends on the MAL-activator and maltose permease (Charron et al. 1986, 1989). Deletion of the gene encoding maltase causes a nonfermentable phenotype but maltose induction of maltose permease is unaffected, indicating that this enzyme is not required for induction but only for utilization of maltose (Charron et al. 1986). Our previous work reported that the role of maltose permease in induction is the accumulation of intracellular maltose but the means of sensing the presence of intracellular maltose remain undetermined (Wang et al. 2002). It is possible that the MAL-activator itself is the maltose-binding sensor. Alternatively, other positive or negative regulators may be involved but may not have been identified as yet because they are encoded by repeated or essential genes.
To identify possible additional players involved in regulating MAL gene expression, we designed a sensitive genetic selection for MAL constitutive (Malc) mutants using a MAL61promoter-HIS3 reporter. This approach should allow us to identify dominant constitutive alterations in positive regulators or recessive constitutive alterations in previously unidentified negative regulators. Here we report the identification of two genes, SIN4 and RGR1, in which recessive mutations cause constitutive MAL gene expression. Our results indicate that Rgr1p and Sin4p are negative regulators of basal, but not induced, expression of the MAL structural genes. Because Sin4p and Rgr1p are both components of the Sin4 module of the yeast Mediator complex, we compared the roles of the other components of the Sin4 module in MAL gene regulation and found that each plays a distinct role in basal and induced expression. The interplay between the Mediator and the Swi/Snf complex as it relates to MAL gene regulation is explored.
MATERIALS AND METHODS
Yeast strains and plasmids:
The strains used in this study are listed in Table 1. CMY1001 is described in Medintz et al. (1996). It contains a single MAL1 locus at which the MAL11 maltose permease gene is replaced by the HA-tagged MAL61, referred to as mal11Δ::MAL61/HA. No other MAL genes are present in this strain. Strain CMY4000 was constructed by inserting two YIp365-based plasmids (Myers et al. 1986) carrying MAL61promoter-lacZ and MAL61promoter-HIS3 reporter genes into the leu2 gene of CMY1001 by targeted integration. Strain CMY4001 was created by changing mating type of CMY4000 from mating type a to α, using plasmid pGHOT obtained from R. Rothstein (Columbia University). The URA3 plasmid YIp355 was integrated into the ura3-52 gene of CMY4001 to create a strain CMY4002. Strains CM-31 and CM-33 were isolated by UV mutagenesis as described below.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference and source |
|---|---|---|
| CMY1001 | MATaMAL61/HA MAL12 MAL13 GAL leu2 ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-200 | Medintz et al. (1996) |
| CMY4000 | MATamal11Δ::MAL61/HA MAL12 MAL13 GAL ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-200 leu2::MAL61pro-LacZ::MAL61pro-HIS3 | This study |
| CMY4001 | MATα mal11Δ::MAL61/HA MAL12 MAL13 GAL leu2 ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-200 leu2::MAL61pro-LacZ::MAL61pro-HIS3 | This study |
| CMY4002 | MATα mal11Δ::MAL61/HA MAL12 MAL13 GAL leu2 ura3-52: YIp355 lys2-801 ade2-101 trp1-Δ63 his3-200 leu2:MAL61pro-LacZ:MAL61pro-HIS3 | This study |
| CM-31 | rgr1-31 (isogenic to CMY4000) | This study |
| CM-33 | sin4-33 (isogenic to CMY4000) | This study |
| CMY5003 | mal13Δ::G418 (isogenic to CMY4000) | This study |
| CMY5004 | mal11Δ::G418 (isogenic to CMY4000) | This study |
| CMY5005 | rgr1-31 mal13Δ::G418 (isogenic to CMY4000) | This study |
| CMY5006 | sin4-33 mal13Δ::G418 (isogenic to CMY4000) | This study |
| CMY5007 | rgr1-31 mal11Δ::G418 (isogenic to CMY4000) | This study |
| CMY5008 | sin4-33 mal11Δ::G418 (isogenic to CMY4000) | This study |
| CMY5009 | sin4Δ::G418 (isogenic to CMY4000) | This study |
| CMY5010 | pgd1Δ::G418 (isogenic to CMY4000) | This study |
| CMY5011 | med2Δ::G418 (isogenic to CMY4000) | This study |
| CMY5012 | gal11Δ::G418 (isogenic to CMY4000) | This study |
| CMY5013 | mig1Δ::G418 (isogenic to CMY4000) | This study |
| CMY5014 | mig2Δ::HygB (isogenic to CMY4000) | This study |
| CMY5015 | mig1Δ::G418 mig2Δ::HygB (isogenic to CMY4000) | This study |
| CMY5016 | rgr1-31 mig1Δ::G418 (isogenic to CMY4000) | This study |
| CMY5017 | rgr1-31 mig2Δ::HygB (isogenic to CMY4000) | This study |
| CMY5018 | rgr1-31 mig1Δ::G418 mig2Δ::HygB (isogenic to CMY4000) | This study |
| CMY5019 | sin4-33 mig1Δ::G418 (isogenic to CMY4000) | This study |
| CMY5020 | sin4-33 mig2Δ::HygB (isogenic to CMY4000) | This study |
| CMY5021 | sin4-33 mig1Δ::G418 mig2Δ::HygB (isogenic to CMY4000) | This study |
| CMY5022 | snf2Δ::HygB (isogenic to CMY4000) | This study |
| CMY5023 | sin4Δ::G418 snf2Δ::HygB (isogenic to CMY4000) | This study |
| CMY5030 | rgr1-31 sin4Δ::G418 (isogenic to CMY4000) | This study |
| BLY1 | MATα lys2-801 his3-Δ200 ura3-52 | Brehon C. Laurent |
| BLY3 | MATα snf5-Δ2 his3-Δ200 ura3-52 ade2-101 | Brehon C. Laurent |
| BLY4 | MATasnf2-141oc his3-Δ200 ura3-52 suc2 | Brehon C. Laurent |
| BLY5 | MATα snf6-Δ2 his3-Δ200 ura3-52 suc2 | Brehon C. Laurent |
| BLY13 | MATaswi1Δ::LEU2 his3-Δ200 lys2-801 ura3-52 ade2-101 trp1Δ1 leu2-Δ1 | Brehon C. Laurent |
| BLY14 | MATaswi3Δ::TRP1 his3-Δ200 lys2-801 ura3-52 ade2-101 trp1Δ1 leu2-Δ1 | Brehon C. Laurent |
| BLY16 | MATα snf2Δ1::HIS3 his3-Δ200 lys2-801 ura3-52 | Brehon C. Laurent |
Gene disruptions were done by the PCR-based one-step gene replacement method in appropriate strains. The primer pairs used for each different gene disruption were determined on the basis of the sequence of S288C available at the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/). The appropriate upstream and downstream primers were used to amplify the G418 resistance marker gene using pFA2-kanMX2 as template or the hygromycin resistance marker gene using pAG32 as template (Wach et al. 1994). Candidate disruptants were confirmed by PCR analysis. Plasmid pLN1384 containing SNF2 (obtained from Brehon Laurent) was introduced into strain CMY5022 (snf2Δ::HygB) prior to the disruption of the chromosomal copy of SIN4 to avoid difficulties resulting from the very slow growth rate of sin4 snf2 double-mutant strains. Following the successful disruption of sin4, the plasmid was cured from the strain to create CMY5023 (sin4Δ::G418 snf2Δ::HygB).
Plasmid pUN30-MAL61promoter-ADE2 carrying the ADE2 open reading frame under the control of the MAL61 promoter was constructed as follows. Plasmid YIp365-I61 (Danzi et al. 2000) carrying the MAL61 promoter was digested with EcoRI and SalI to liberate a 0.9-kb fragment containing base pairs −874 to −1 of the MAL61 promoter. This was subcloned into vector pUN30 (Elledge and Davis 1988), forming pUN30-MAL61pro. The ADE2 open reading frame (∼1.7 kb) was amplified by PCR from plasmid pRS402 (ATCC87477) with primers 5′-GGGGGTCGACATGGATTCTAGAACAGTTGG-3′ and 5′-GGGGGCATGCAGATCTTATGTATGAAATTC-3′. This amplified PCR product was digested with SalI and SphI and inserted downstream of the MAL61 promoter in pUN30-MAL61pro to create pUN30-MAL61pro-ADE2.
Mutagenesis and isolation of MAL constitutive mutants:
Strain CMY4000 was grown in YPD to midlog phase. Cells were collected by centrifugation, washed, and resuspended in sterile water. The cell suspension was mutagenized by exposure to UV light of wavelength 254 nm to ∼15% survival. The mutagenized cells were immediately plated onto minimal medium containing 2% galactose, 3% glycerol, and 2% lactate (SGalG/L) lacking histidine, and the plates were incubated in the dark for 5 days at 25° until His+ colonies appeared. The potential Malc mutants were screened by assaying expression of the MAL61promoter-lacZ reporter using the standard β-galactosidase plate assay and MAL12 expression by assaying maltase activity levels in galactose-grown cells.
Cloning of wild-type alleles of a mutant gene in strains CM-31 and CM-33:
Constitutive mutants CM-31 and CM-33 were chosen as representatives of complementation groups 1 and 2, respectively. CM-31 and CM-33 are pink because of the presence of the ade2-101 mutant allele. Each strain was transformed with pUN30-MAL61promoter-ADE2. The resulting transformants form white colonies on SGalG/L media because of the constitutive expression of the MAL61promoter-ADE2 reporter. These were then transformed with a centromere-based YCp50 genomic library prepared from strain S288C. Pink Ura+ transformants on SGalG/L media were isolated as potential carriers of the dominant wild-type allele of the mutation present in CM-31 or CM-33. Dependence of pink color on the presence of the library plasmid and complementation of the constitutive maltase expression phenotype was determined for each transformant. The library plasmid was isolated from each transformant and reintroduced into the CM-31 [pUN30-MAL61promoter-ADE2] or CM-33 [pUN30-MAL61promoter-ADE2] mutant strains to confirm the complementation. The yeast insert in each library plasmid was identified by sequencing the YCp50-insert junction.
Sequencing of rgr1 mutant alleles:
The genomic copy of each of the rgr1 mutant alleles was amplified by PCR using primers 5′-GTAGAGGTCTGTTGTAAAGATCATC-3′ (53–77 bases before the start codon) and 5′-TTCAGGAGAGGGGTTACAATCTCC-3′ (complementary to sequence 36–59 bases after the stop codon) and high-fidelity platinum Taq DNA polymerase (Invitrogen, San Diego) to ensure the fidelity of amplification product. Seven sequencing primers were designed, each annealing to sites about every 500 bp along the RGR1 ORF, on the basis of sequence of this gene in S288C (http://genome-www.stanford.edu/Saccharomyces/). The site of the alteration was sequenced using DNA from independent amplifications to ensure that the detected alteration does not result from a PCR error.
Maltase assay:
Maltase activity was determined in total cell extracts as described by Dubin et al. (1985). Activity is expressed as nanomoles of ρ-nitrophenyl β-d-glucopyranoside (PNPG) hydrolyzed per milligram of total protein per minute. The values reported are the average of duplicate assays obtained with extracts from at least duplicate cultures of the same strain. The values from different cultures varied ∼15%.
β-Galactosidase plate assay:
Cells were patched onto a plate containing the appropriate selective medium and grown for 2 days. A substrate-agarose mixture was prepared by mixing melted agarose with 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-Gal) solution and maintained as a liquid at 55°–60°. The final mixture contains 0.5% agarose, 0.5 m Na2HPO4-NaH2PO4 pH 7 buffer, 0.1% SDS, 2% dimethylformamide, and 0.05% X-Gal. Approximately 10 ml of this mixture was poured over the surface of the culture plate and photographs were taken following 6–12 hr of blue color development.
Flocculation assay:
Cells were grown overnight in 5 ml liquid minimal medium to approximately midlog phase. The culture was vortexed briefly to separate and suspend the cells in the medium as best as possible, and the culture tubes were photographed immediately and after 15 and 30 min of standing in a test tube rack.
RESULTS
Isolation and genetic analysis of MAL constitutive mutants:
To select for Malc mutants we constructed strain CMY4000, which carries the complete MAL1 locus encoding maltose permease (mal11Δ::MAL61/HA), maltase (MAL12), and the MAL-activator (MAL13) as well as integrated copies of both MAL61promoter-HIS3 and MAL61promoter-lacZ reporter genes (see materials and methods). The ability of CMY4000 to grow in the absence of histidine is dependent on the presence of maltose in the growth medium. Mutations causing constitutive MAL gene expression should provide the ability to grow in the absence of histidine even under uninduced growth conditions (galactose or glycerol/lactate medium) and should allow for the constitutive expression of β-galactosidase, maltose permease, and maltase.
To carry out the selection for Malc mutants, ∼107 cells of CMY4000 were mutagenized with UV-light and plated directly onto minimal medium containing galactose plus glycerol/lactate and lacking histidine. A total of 33 potential Malc mutant colonies were isolated. Of these, 31 also exhibited constitutive expression of the MAL61promoter-lacZ reporter gene on the basis of plate assay and constitutive maltase expression ranging from 10 to 70% of fully induced levels (data not shown).
The 31 Malc mutant strains were mated to strain CMY4002 and the phenotype of the diploids was determined. Two mutants, CM-24 and CM-32, were found to be dominant. Complementation analysis was performed on the remaining 29 recessive mutants. Due to very poor sporulation and spore viability, we were able to obtain sufficient four-spore tetrads to follow segregation of the Malc phenotype only for mutants CM-31 and CM-33. For the crosses involving CM-31 and CM-33, the Malc phenotype segregated 2:2, indicating that both carried a single-mutant alteration. Haploid MATα segregants carrying the Malc mutation obtained from the cross of CM-31 with CMY4002 or CM-33 with CMY4002 were mated to each of the 29 recessive Malc mutants. The results indicate that these 29 Malc mutants fall into at least three complementation groups. Group 1 includes 10 Malc mutants, CM-7, -9, -10, -13, -17, -18, -19, -20, -30, and -31; and group 2 includes 5 Malc mutants, CM-5, -21, -28, -29, and -33. The remaining 14 recessive mutants lie in at least one additional complementation group but have not been studied further.
Group 1 and group 2 mutations are pleiotropic:
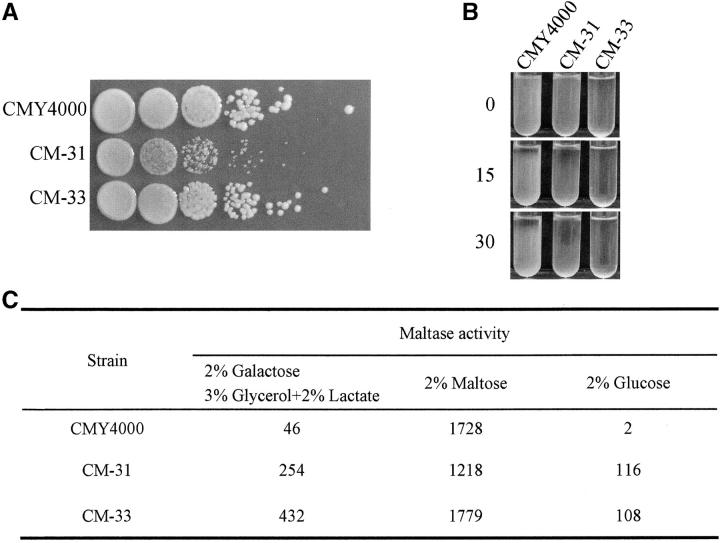
During the genetic analysis of the Malc mutants, we observed that some grew significantly more slowly than the parental strain and also that some were flocculent; that is, when grown in liquid media the cells quickly settled to the bottom of the culture tube. We compared growth rates on rich media (YPD) and flocculation rates of all mutant strains. Figure 1 shows the results obtained with mutants CM-31 and CM-33, representatives from group 1 and group 2, respectively, but in all cases members of the same complementation group exhibited similar phenotypes (data not shown).
Figure 1.—
Phenotypic consequences of mutations in CM-31 (group 1) and CM-33 (group 2). (A) Growth rates of CM-31, CM-33, and CMY4000 on YPD plates. (B) Flocculation phenotype. Cultures of CMY4000, CM-31, and CM-33 were grown overnight in liquid minimal medium, vortexed briefly to separate and suspend the cells in the culture medium, and allowed to stand without further agitation. The culture tubes were photographed immediately and after 15 and 30 min. (C) Effects of mutations on glucose repression. CMY4000, CM-31, and CM-33 were grown in minimal media under uninduced (2% galactose, 3% glycerol, and 2% lactate), induced (2% maltose), and repressed (2% glucose) conditions and maltase activity was assayed as described in materials and methods.
CM-31 exhibits a slow growth phenotype but CM-33 grows normally, as can be seen from the dilution assay shown in Figure 1A. To measure the extent of flocculation, CM-31, CM-33, and CMY4000 were grown overnight in liquid minimal medium, vortexed briefly to separate and suspend the cells in the culture medium, and the culture tubes were photographed immediately and after 15 and 30 min. The results presented in Figure 1B demonstrate that CM-31 is modestly flocculent compared to nonflocculent CMY4000. In contrast, CM-33 is so flocculent that the cells grow in clumps, cannot be adequately resuspended even after vigorous vortexing, and largely remain at the bottom of the tube.
Glucose as the preferred carbon source inhibits transcription of the MAL genes, a phenomenon referred to as glucose repression. We found that both group 1 and group 2 mutants relieve glucose repression. Maltase expression was assayed following growth under maltose-induced, glucose-repressed, and uninduced growth conditions. The results for strains carrying the mutant allele from CM-31 and CM-33 are presented in Figure 1C and clearly demonstrate that mutations in both genes partially relieve glucose repression. Similar results were found for other group 1 and group 2 mutants (data not shown).
Taken together, these results indicate that the group 1 and group 2 mutations are pleiotropic, suggesting that the genes encode global regulators controlling the expression of diverse genes, not specifically the MAL genes.
Group 1 and group 2 Malc mutations represent alleles of RGR1 and SIN4, respectively:
The wild-type alleles of the group 1 and group 2 Malc mutations were cloned by complementation from a low-copy yeast genomic library (YCp50-based), using the following strategy. A reporter plasmid carrying the ADE2 gene under the control of the MAL61 promoter was introduced into the Malc mutant strain, either CM-31 or CM-33, which also carries the ade2-101 mutation. As a result of the constitutive expression of the plasmid-borne MAL61promoter-ADE2 gene the transformant strains form white colonies on galactose-containing medium. Library plasmids carrying the dominant wild-type allele should restore the maltose-inducible phenotype, thereby blocking expression of the MAL61promoter-ADE2 reporter, and thus should produce pink colonies on galactose-containing medium. Transformant colonies carrying a library plasmid were selected on minimal medium lacking uracil with 2% galactose and screened for pink colonies. Dependence on the library plasmid of the pink colony color was confirmed by plasmid loss. The pink transformants were screened to identify those that also exhibited low uninduced levels of maltase expression and normal flocculation rates. The library plasmid was recovered from each independent transformant and the sequence of the ends of the yeast insert determined.
Two plasmids were isolated that fully restored inducible expression of the MAL61promoter-ADE2 reporter in CM-31. The overlapping region of the insert fragments is derived from the right arm of chromosome XII and contains six intact ORFs (YLR071C–YLR076C). Subcloning identified a BglII-EcoRI fragment capable of partially complementing the Malc phenotype of CM-31 and fully complementing the flocculation phenotype. This fragment contains only one complete ORF, RGR1. The basis of the partial complementation by this shorter insert fragment is unexplained but is observed with both multicopy and CEN vectors carrying only the RGR1 gene. In heterozygous diploids all of the rgr1 mutant alleles are purely recessive to RGR1 for all phenotypes. Thus, the partial complementation is unlikely to be of functional significance. Nonetheless, it concerned us so the complete sequence of the open reading frame of all 10 presumed rgr1 mutant alleles obtained by our selection scheme was determined to confirm the presence of a mutation. All 10 rgr1 alleles contain a single alteration, either a nonsense or a frameshift mutation, located in the region of codons 710–910 of this 1082-codon ORF (listed in Table 2). Thus, group 1 mutants are alleles of RGR1, an essential gene encoding a scaffold-like component in the middle and tail regions of the RNA polymerase II (RNAPII) mediator complex (Asturias et al. 1999; Dotson et al. 2000). It is interesting to note that the rgr1 alleles isolated here differed in the level of constitutive maltase expression and the severity of the flocculation phenotype but no clear correlation between their phenotype and the position of the mutant alteration is evident (data not shown).
TABLE 2.
Mutant alterations ofrgr1 alleles
| Allele | Mutation | Amino acid replacements |
|---|---|---|
| rgr1-7 | C2689T | Q897* |
| rgr1-9 | C2128T | Q710* |
| rgr1-10 | C2689T | Q897* |
| rgr1-13 | A2674T | K892* |
| rgr1-17 | 2547: insertion of A | N849KF* |
| rgr1-18 | T2193A | Y731* |
| rgr1-19 | A2674T | K892* |
| rgr1-20 | 2561: deletion of A | N854TSR* |
| rgr1-30 | A2674T | K892* |
| rgr1-31 | 2730: insertion of T | F910FLR* |
, stop codon.
Six plasmids were isolated from the CEN genomic library that restored inducible expression of the MAL61promoter-ADE2 reporter in CM-33. These plasmids complemented both the Malc and the flocculation phenotypes. The overlap of these insert fragments contains two intact ORFs, SIN4 and YNL235C. SIN4 was amplified by PCR from CMY4000, using primers that exclude the promoter region of YNL235C. Subsequent cloning of this fragment in pUN70 enabled us to demonstrate that the complementing region is the SIN4 gene, suggesting that the group 2 mutations are alleles of SIN4. Sin4p is a component in the tail region of the RNAPII mediator complex (Asturias et al. 1999; Myers et al. 1999; Dotson et al. 2000). We deleted the nonessential SIN4 in strain CMY4000 to create CMY5009 and observed similar levels of constitutive maltase expression in the sin4Δ null allele as in all five of the sin4 mutant strains (data not shown). Thus, loss of Sin4p results in constitutive maltase expression.
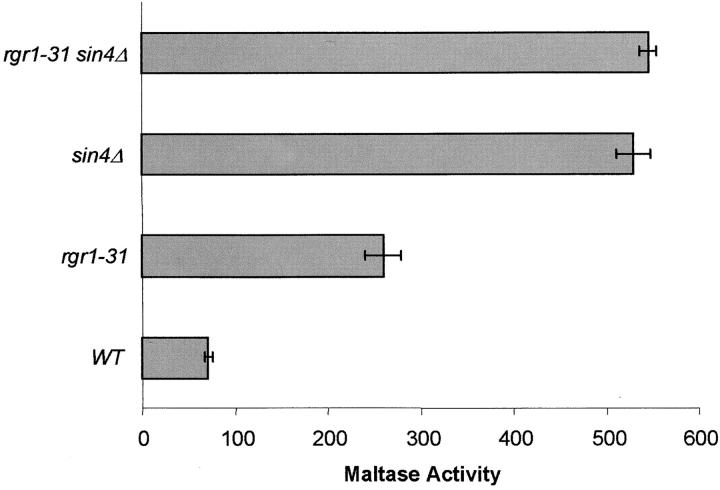
SIN4 and RGR1 regulate MAL gene expression in a common pathway:
Although Sin4p and Rgr1p are both components of the mediator complex, it is possible that each could act independently to regulate MAL gene expression. To test this, SIN4 was deleted in CMY4000 and the isogenic rgr1-31 strain (CM-31) to create CMY5009 and CMY5030, respectively. Maltase activity was assayed in cells grown under uninduced conditions. As shown in Figure 2, the double mutant rgr1-31 sin4Δ displays a similar level of constitutive maltase expression compared to the single mutant sin4Δ, indicating that RGR1 and SIN4 function in a common pathway to regulate MAL gene expression. This conclusion is not in conflict with the fact that lower levels of maltase activity were observed in rgr1-31 (CM-31) than in sin4Δ (CMY5009) because RGR1 is an essential gene and rgr1-31 is only a partial loss-of-function allele.
Figure 2.—
RGR1 and SIN4 are in a common pathway for MAL gene regulation. CMY4000 (wild type), CM-31 (rgr1-31), CMY5009 (sin4Δ::G418), and CMY5030 (rgr1-31 sin4Δ::G418) were grown under uninduced conditions in minimal media with 2% galactose, 3% glycerol, and 2% lactate. Maltase activity was assayed as described in materials and methods.
The constitutive phenotype of rgr1 and sin4 is not dependent on either the MAL-activator or maltose permease:
The MAL-activator gene (MAL13) or the maltose permease gene (mal11Δ::MAL61) were deleted from CMY4000 (RGR1 SIN4), CM-31 (rgr1-31 SIN4), and CM-33 (RGR1 sin4-33), all of which carry an integrated reporter MAL61promoter-HIS3 gene. The resulting strains were tested for their ability to grow on maltose medium lacking histidine and galactose medium lacking histidine, and maltase expression levels were assayed in galactose-grown cells. As was found previously (Charron et al. 1986, 1989), strains lacking the MAL-activator (mal13Δ) or maltose permease (mal11Δ) are not maltose inducible (Table 3). The double-mutant strains, rgr1-31 mal13Δ and rgr1-31 mal11Δ, are able to grow in galactose medium lacking histidine and express similar levels of maltase as the strains carrying the rgr1-31 mutation alone. Similarly, loss of the MAL-activator or maltose permease has no effect on MAL gene expression in the sin4-33 mutant strain (Table 3). These results indicate that constitutive expression of the MAL genes in rgr1 or sin4 mutant strains is not dependent on either the MAL-activator or maltose permease. It should be noted that the constitutive level of MAL gene expression in the rgr1 or sin4 mutant strains lacking a MAL-activator is not sufficient to allow for growth on maltose medium lacking histidine.
TABLE 3.
The constitutive phenotype ofrgr1 andsin4 is not dependent on eitherMAL-activator or maltose peamease
| Growth
|
||||
|---|---|---|---|---|
| Strain | Relative genotype | Mal − His | Gal − His | Maltase activity (SGalG/L) |
| CMY4000 | RGR1 SIN4 mal11Δ::MAL61 MAL13 | + | − | 46 |
| CMY5004 | RGR1 SIN4 mal11Δ MAL13 | − | − | 27 |
| CMY5003 | RGR1 SIN4 mal11Δ::MAL61 mal13Δ | − | − | 21 |
| CMY5001 | rgr1-31 SIN4 mal11Δ::MAL61 MAL13 | + | + | 254 |
| CMY5007 | rgr1-31 SIN4 mal11Δ MAL13 | − | + | 280 |
| CMY5005 | rgr1-31 SIN4 mal11Δ::MAL61 mal13Δ | − | + | 285 |
| CMY5002 | RGR1 sin4-33 mal11Δ::MAL61 MAL13 | + | + | 432 |
| CMY5008 | RGR1 sin4-33 mal11Δ MAL13 | − | + | 326 |
| CMY5006 | RGR1 sin4-33 mal11Δ::MAL61 mal13Δ | − | + | 347 |
The MAL-activator gene (MAL13) or the maltose permease gene (mal11Δ::MAL61) was disrupted from CMY4000 (RGR1 SIN4 mal11Δ::MAL61 MAL13), CMY5001 (rgr1-31 SIN4 mal11Δ::MAL61 MAL13), and CMY5002 (RGR1 sin4-33 mal11Δ::MAL61 MAL13). The resulting strains were streaked for single colonies on minimal media lacking histidine with either 2% maltose or 2% galactose and growth was monitored for 3 days. Maltase activity was assayed in cells grown in minimal media with 2% galactose, 3% glycerol, and 2% lactate (SGalG/L) as described in materials and methods.
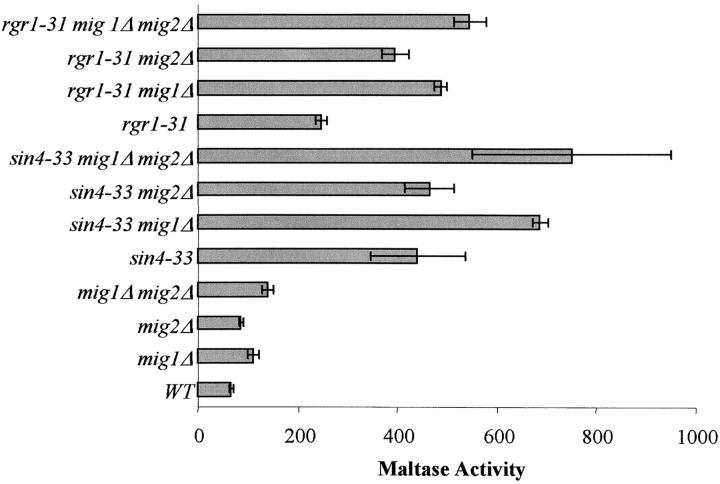
SIN4 and RGR1 act synergistically with MIG1 and MIG2 in the repression of MAL gene expression:
The Sin4 module of the Saccharomyces Mediator is thought to sense signals from gene-specific activators and repressors. Other than the MAL-activator the only gene-specific regulators of the MAL structural genes identified are the Mig1,2 repressors and thus these are possible candidates for interaction with the Mediator. We tested the possibility that Mig1p and/or Mig2p repress the MAL gene expression through direct or indirect interaction with the Sin4 module. MIG1, MIG2, and both were disrupted in strain CMY4000, the rgr1-31 mutant (CM-31), and the sin4-33 mutant (CM-33). Maltase expression levels were determined under uninduced growth conditions (SGalG/L; Figure 3). Consistent with previous studies (Hu et al. 2000), deletion of MIG1, MIG2, or both modestly increases the basal level of maltase expression to levels about four times lower than the maltase levels observed in sin4 and rgr1 mutants, indicating that MIG1 and MIG2 play only minor roles in repressing basal-level MAL gene expression. Disruption of either MIG1 or MIG2 in either rgr1-31 or sin4-33 mutant strains causes further increases in maltase expression levels and disruption of both MIG1 and MIG2 in the rgr1-31 or sin4-33 strains leads to even higher levels of maltase activity than in either strain containing the single mig1Δ or mig2Δ alone. These findings indicate that Mig1p and Mig2p repress the expression of MAL genes independently of the Mediator Sin4 module and act to repress MAL gene expression via a separate pathway.
Figure 3.—
SIN4 and RGR1 act synergistically with MIG1 and MIG2 in the repression of MAL gene expression. MIG1, MIG2, and both were deleted from strain CMY4000, the rgr1-31 mutant strain CMY5001, and the sin4-33 mutant strain CMY5002. Maltase activity was assayed in cells grown under uninduced conditions in minimal media with 2% galactose, 3% glycerol, and 2% lactate (SGalG/L) as described in materials and methods.
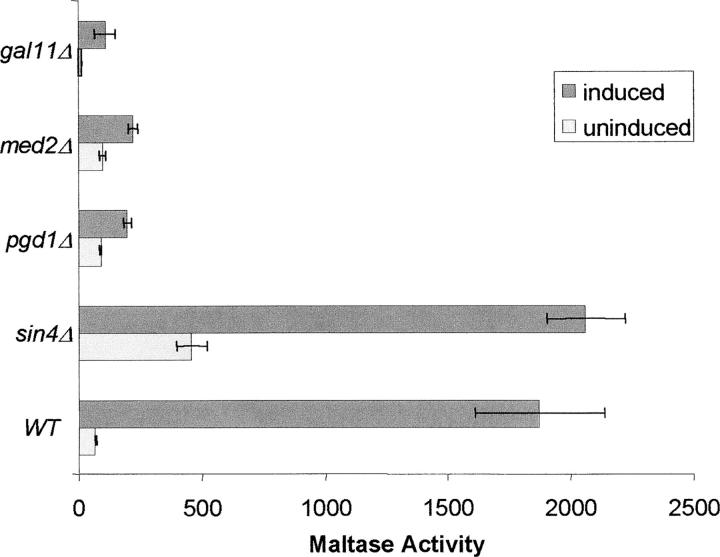
Each component of the Sin4 module of the Mediator plays a distinct role in regulation of MAL gene expression:
Biochemical analysis indicates that the Saccharomyces Sin4 module of the Mediator complex contains Sin4p, Gal11p, Med2p, and Pgd1p. These bind to Rgr1p that reportedly serves as a bridge connecting the Sin4 module to the Med9/Med10 module (Li et al. 1995; Myers et al. 1999). Genetic evidence reveals negative as well as positive regulatory roles of the Sin4 module and Rgr1p, depending on the promoters (Myers and Kornberg 2000).
To elucidate the role of the various Sin4 module subunits in MAL gene regulation, the effects of deletion of the nonessential SIN4, GAL11, MED2, and PGD1 genes on maltase expression were tested. Strains CMY5009 (sin4Δ), CMY5010 (pgd1Δ), CMY5011 (med2Δ), and CMY5012 (gal11Δ) were constructed by one-step gene replacement and maltase expression levels assayed under induced (SMal) and uninduced (SGalG/L) growth conditions. As shown in Figure 4, the sin4Δ strain displays a significant increase in basal maltase expression with no significant effect on induced expression levels. In contrast, the med2Δ and pgd1Δ strains exhibit no significant impact on basal expression but maltose-induced expression of maltase is dramatically decreased. Deletion of GAL11 significantly decreases both basal and induced expression of maltase. Basal expression of maltase is extremely low (11 units) in the gal11Δ strain but ∼10-fold induction is observed. This is significant but lower than the 30-fold induction observed in strain CMY4000. Thus, the components of the Sin4 module have distinct effects on basal and induced MAL gene expression. Moreover, Sin4p is a negative regulator of basal expression of the MAL structural genes, Med2p and Pgd1p are positive regulators of induced expression, and Gal11p is required for both basal and induced expression.
Figure 4.—
Effects of deletion of the Sin4 module components on basal maltase expression and maltase induction. The SIN4, GAL11, MED2, or PGD gene was deleted from strain CMY4000. Maltase activity was assayed in cells grown in minimal media with 2% galactose, 3% glycerol, and 2% lactate (SGalG/L, uninduced conditions) and with 2% maltose (SMal, induced conditions) as described in materials and methods.
MAL gene induction is defective in strains carrying Swi/Snf complex mutations:
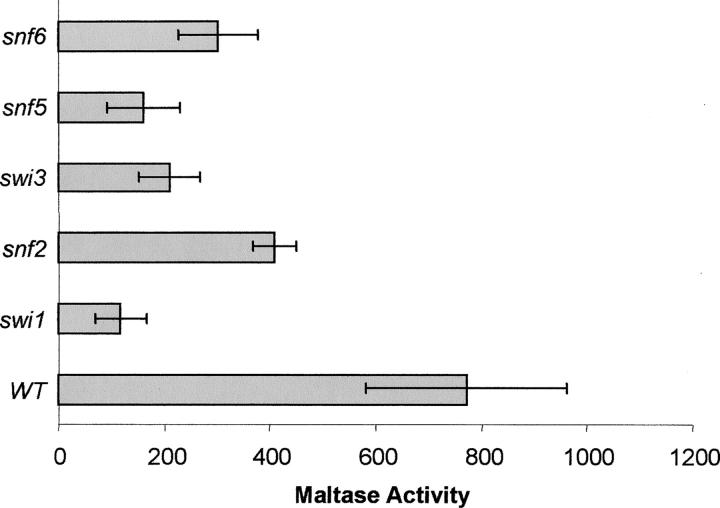
Swi/Snf is a 2-MD multisubunit complex that plays key roles in the regulation of eukaryotic gene expression (Peterson and Workman 2000). Swi/Snf is required for changes in chromatin structure that accompany transcriptional induction of SUC2 and PHO8 and other yeast promoters (Wu and Winston 1997; Gregory et al. 1998, 1999). To examine whether the Swi/Snf complex is required for MAL gene induction, we measured maltase induction in an isogenic series of strains containing mutations in SWI1, SNF2, SWI3, SNF5, or SNF6 encoding components of the Swi/Snf complex. As shown in Figure 5, loss of any one of these functions causes a dramatic decrease in induced maltase expression compared to that of the isogenic wild-type strain. Thus, the Swi/Snf complex is required for the full induction of MAL gene expression. In contrast, mutations in components of the SAGA complex, including Ada2p, Ada3p, and Gcn5p, have no significant effect on MAL gene expression, suggesting the acetylation of the chromatin template by Gcn5p is not required for maltose induction (B. Zhang and C. A. Michels, unpublished results).
Figure 5.—
The Swi/Snf complex is involved in maltose induction. Plasmid YCp50-MAL63 carrying the MAL63 MAL-activator gene (Danzi et al. 2000) was transformed into strain BLY1 and the isogenic strain series, BLY13 (swi1Δ), BLY16 (snf2Δ), BLY14 (swi3Δ), BLY3 (snf5Δ), and BLY5 (snf6Δ). Cells were grown under uninduced conditions in selective media containing 0.2% glucose to early-log phase. Cells were then collected and transferred to selective induced media containing 2% maltose. After 6 hr, maltase activity was assayed as described in materials and methods.
Constitutive MAL gene expression caused by loss of SIN4 is dependent on the Swi/Snf complex:
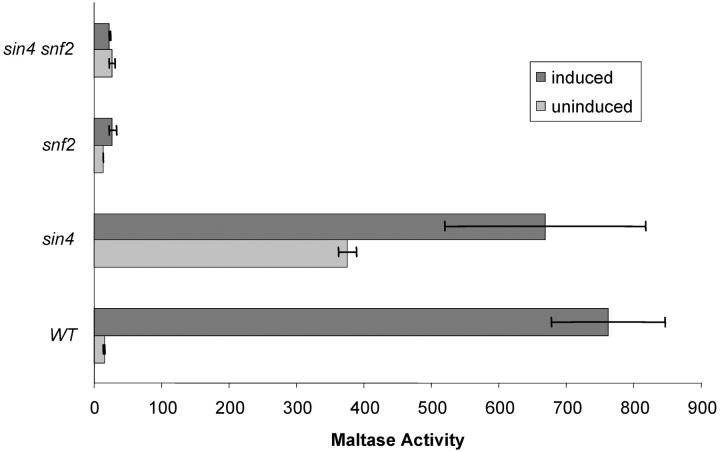
Previous studies have suggested that Sin4p negatively regulates gene transcription by acting to inhibit chromatin reorganization and/or maintain the inactive chromatin structure (Jiang and Stillman 1992; Jiang et al. 1995; Macatee et al. 1997; Moss and Laybourn 2000). To investigate the relationship between these two complexes in regulation of MAL gene transcription SIN4 and SNF2 were individually and together deleted in strain CMY1001. These strains were grown under uninduced and induced conditions and maltase activity was determined. As shown in Figure 6, and consistent with the findings shown above in Figure 5, the snf2Δ mutation causes a severe defect in maltase expression under induced conditions that is not suppressed by sin4Δ. In contrast, no effect is observed on uninduced maltase expression levels in snf2Δ but there is an approximately twofold increase in maltase activity in the sin4Δ snf2Δ double mutant (Figure 6). It is interesting to note that the effects of snf2Δ are significantly greater in this strain series compared to those in the strains shown in Figure 5 and probably are a function of differences in the strain backgrounds and/or the MAL loci of these strains. The results in Figure 6 indicate that the constitutive expression of MAL genes caused by loss of Sin4p is largely dependent on the Swi/Snf complex, and that Sin4p acts upstream of the Swi/Snf in controlling basal MAL gene expression.
Figure 6.—
Effects of sin4Δ and snf2Δ on MAL gene expression. Maltase expression was determined in the isogenic strain series CMY4000 (SIN4 SNF2), CMY5009 (sin4Δ SNF2), CMY5022 (SIN4 snf2Δ), and CMY5023 (sin4Δ snf2Δ). Strains were grown to midlog in synthetic medium containing 0.1% glucose plus either 2% maltose (induced) or 2% galactose (uninduced) and harvested, and maltase activity was assayed as described in materials and methods. The low concentration of glucose is not repressing but is required because of the very slow growth rate of CMY5023, in particular.
DISCUSSION
The Mediator complex plays an essential role in both activation and repression of RNA polymerase II-mediated transcription and the Sin4 module to act in regulatory signal transduction. Our results suggest that the components of the Sin4p module of the yeast Mediator function differentially and in distinct combinations to modulate MAL gene transcription and regulate basal expression via as yet unidentified MAL promoter-binding repressor(s) and activator(s).
Sin4p and the C-terminal region of Rgr1p repress basal level expression of MAL genes, but have little or no effect on maltose induction:
Mutations in RGR1 that truncate up to 270 residues of the C-terminal region and SIN4 null mutations increase maltase expression to ∼20–30% of the fully induced levels under uninduced growth conditions and partially relieve glucose repression independent of the MAL-activator and maltose permease. Thus, the C terminus of Rgr1p and Sin4p repress basal expression of the MAL genes.
Mig1,2 repressor regulates glucose repression of MAL gene transcription (Hu et al. 1995) but actions of this repressor are distinct from the Sin4p effects observed here. Mig1p exerts its repressive effects by recruiting the Ssn6-Tup1 corepressor complex, which reportedly interacts with the Mediator (Treitel and Carlson 1995; Papamichos-Chronakis et al. 2000). Nonetheless, deletion of MIG1, MIG2, or both in strains carrying rgr1-31 or sin4-33 mutations shows that Mig1,2 repressor further increases maltase expression and thus acts synergistically with the sin4 and rgr1 mutations (Figure 3), indicating that Sin4p and the C terminus of Rgr1p are not involved in transmission of the Mig1,2p repression signal but instead function via an independent repression pathway.
Structural organization of the Sin4 module at the MAL promoter:
The Sin4 module of the Mediator complex contains Sin4p, Gal11p, Med2p, and Pgd1p (Myers et al. 1999). Biochemical analyses and electron microscopy imaging propose that Sin4p anchors the other module components to the Mediator complex through its interaction with the C terminus of Rgr1p. Mediator complexes purified from sin4Δ and rgr1-Δ2 strains lack all of the components of the Sin4 module (Jiang et al. 1995; Li et al. 1995; Asturias et al. 1999; Myers et al. 1999; Dotson et al. 2000). Pgd1p, Med2p, and Gal11p are dependent on each other in regard to their ability to form a stable association within the Mediator but their loss has little effect on the association of Sin4p with Rgr1p (Lee et al. 1999; Myers et al. 1999).
Our results conflict with this proposed anchoring function of Sin4p. Given the essential requirement of Gal11p, Pgd1p, and Med2p in MAL gene induction demonstrated here, loss of Sin4p should block MAL gene induction. This is clearly not the observed result (Table 3 and Figure 4). Both sin4-33 and sin4Δ mutant strains exhibit wild-type levels of induced maltase expression. Thus, Pgd2p, Med2p, and Gal11p binding to Sin4p could not be required for their interaction with the Mediator. We suggest instead that Sin4p interacts with the C-terminal 270 residues of Rgr1p and that Gal11p, Pgd1p, and Med2p bind elsewhere on Rgr1p. Loss of Sin4p may destabilize the complex in vitro but not in vivo, at least not enough to alter induced expression at the MAL promoter.
Swi/Snf chromatin reorganization is required for MAL gene expression.
The results reported in Figures 5 and 6 indicate that the Swi/Snf chromatin-reorganizing complex is required for induced MAL gene expression as well as for the elevated basal rate of MAL gene expression exhibited in sin4 mutants. We suggest that Sin4p negative regulation of basal MAL gene transcription is achieved by blocking Swi/Snf-dependent chromatin reorganization. Previous studies have suggested that Sin4p and Rgr1p affect transcription by altering chromatin structure. Mutations in SIN4 and RGR1 increase the linking number of plasmid DNA (Jiang and Stillman 1992; Jiang et al. 1995). Loss of SIN4 results in an increase in chromatin accessibility as measured by increased sensitivity to micrococcal nuclease digestion but does not appear to alter nucleosome positioning, histone expression, or histone modification (Macatee et al. 1997). Rgr1p is required for nucleosomal repression of transcription in a plasmid-chromosome transcriptional system (Moss and Laybourn 2000). Moreover, defects in components of both the Mediator and the Swi/Snf complex are suppressed by similar mutations in chromatin components, the so-called sin mutations in histones and associated factors (Prelich and Winston 1993; Kruger et al. 1995).
We report little or no suppression of the snf2Δ defect in basal and induced maltase expression by sin4Δ (Figure 6). Several swi/snf mutations had been reported to be partially suppressed by a sin4 null mutation (Sternberg et al. 1987; Stillman et al. 1994), which would suggest a role for Sin4p downstream of this chromatin-reorganizing complex. These finding may be an artifact since suppression is observed only with LacZ reporter genes and not with the genomic copy of the same genes (Yu et al. 2000). Thus, our results, similar to results for the HO gene, indicate that Sin4p acts upstream of the Swi/Snf complex to maintain the low basal level of MAL gene expression probably by blocking chromatin reorganization and/or maintaining an inactive chromatin structure.
Role of the Sin4 module in MAL gene regulation:
Sin4p and Gal11p play opposing roles in regulating basal expression from the MAL promoter. Sin4p is a negative regulator of basal MAL gene expression. Gal11p is a positive regulator of both basal and MAL-activator-dependent maltose-induced MAL gene expression. We propose that Sin4p represses basal expression of MAL gene promoters by blocking Gal11p-mediated chromatin reorganization. Our findings are consistent with the reported roles of these Mediator components in yeast transcription regulation. Gal11p is considered a positive regulator of transcription, although modest negative effects have been reported (Yu and Fassler 1993; Nishizawa et al. 1994; Nishizawa 2001). Artificial tethering of a Gal11p fusion protein to the promoter region of a reporter construct produces strong activation of reporter gene expression (Jiang and Stillman 1992; Barberis et al. 1995). Alternately, with the exception of CTS1, MATα2, Ty1, and HIS4, Sin4p is regarded as a negative regulator of transcription (Jiang and Stillman 1992; Jiang et al. 1995). We propose that Sin4p-bound Mediator complex interacts with a DNA-bound repressor and blocks the functions of Gal11p. In the absence of Sin4p, Gal11p interacts with a DNA-bound activator, recruits Mediator to the activator, and activates basal transcription in a Swi/Snf-dependent manner. It should be noted that interactions between DNA-bound Mig1,2 repressor and Mediator do not similarly interfere with this Gal11p-dependent activation of basal expression.
Sin4p is not involved in regulation of maltose-induced MAL gene expression (Figure 1). For maltose-induced expression, we propose that a Mediator complex containing Gal11p, Med2p, and Pgd1p interacts with the MAL-activator and activates transcription in a Swi/Snf-dependent manner. Thus, the Mediator plays different roles in MAL gene expression and acts as both an antagonist and protagonist of MAL gene expression under different growth conditions by utilizing different components of the Sin4p module. Whether these results suggest different structural conformations of the Mediator Sin4p module or that the composition the Mediator Sin4 module is heterogeneous and varies from promoter to promoter, as a function of the growth medium, or when bound to different DNA-binding transcription factors remains to be determined.
Studies support the proposal that the Mediator complex is recruited to promoters by means of interactions between the Sin4p module and DNA-bound transcription regulators (Lee et al. 1999; Bhoite et al. 2001). Evidence of direct interaction between Gal11p and several transcription activators including Gal4p, Gcn4p, and the VP16 activation domain has been reported by Park et al. (2000). Bhoite et al. (2001) show that Swi5 activator recruits Mediator to the HO promoter via direct interaction. Sin4p has been shown to immunoprecipitate with Sfl1p, a repressor of SUC2, FLO11, and HSP26 (Conlan and Tzamarias 2001). What DNA-binding transcription factors are involved in regulating basal MAL gene expression? Sequence analysis of the bidirectional MAL61 and MAL62 promoter (Krull et al. 2003; Matys et al. 2003) and multiple-alignment analysis of MAL promoters from S. mikatae, S. kudriavzevii, and S. bayanus were largely uninformative. Deletion analysis carried out by Levine et al. (1992) points to a possible repressor-binding site located in base pairs −348 to −514 upstream of the MAL61 ORF, a region that exhibits sequence similarities to the proposed Sfl1p-binding site (Conlan and Tzamarias 2001). We are currently involved in studies to identify novel regulators of basal MAL gene transcription with particular attention to Sfl1 repressor.
Acknowledgments
We thank Brehon C. Laurent for Swi/Snf mutant strains and plasmid pLN1384, Michael Hampsay for SAGA mutant strains, Lucy Robinson for theYCp50 genomic library, and David Stillman for pCEN-RGR1. We are also grateful to Paul Cliften and Mark Johnston for doing the ClustalW analysis and David Stillman, Judith Jaehning, and Zhen Hu for valuable discussions. This work was supported by grants from the National Institutes of Health (GM28216 and GM49280) to C.A.M. and is in partial fulfillment of the requirements of the Ph.D. degree of the Graduate School of CUNY (X.W.).
References
- Asturias, F. J., Y. W. Jiang, L. C. Myers, C. M. Gustafsson and R. D. Kornberg, 1999. Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283: 985–987. [DOI] [PubMed] [Google Scholar]
- Barberis, A., J. Pearlberg, N. Simkovich, S. Farrell, P. Reinagel et al., 1995. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell 81: 359–368. [DOI] [PubMed] [Google Scholar]
- Bhoite, L. T., Y. Yu and D. J. Stillman, 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15: 2457–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron, M. J., R. A. Dubin and C. A. Michels, 1986. Structural and functional analysis of the MAL1 locus of Saccharomyces cerevisiae. Mol. Cell. Biol. 6: 3891–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron, M. J., E. Read, S. R. Haut and C. A. Michels, 1989. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics 122: 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan, R. S., and D. Tzamarias, 2001. Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. J. Mol. Biol. 309: 1007–1015. [DOI] [PubMed] [Google Scholar]
- Danzi, S. E., B. Zhang and C. A. Michels, 2000. Alterations in the Saccharomyces MAL-activator cause constitutivity but can be suppressed by intragenic mutations. Curr. Genet. 38: 233–240. [DOI] [PubMed] [Google Scholar]
- Dotson, M. R., C. X. Yuan, R. G. Roeder, L. C. Myers, C. M. Gustafsson et al., 2000. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA 97: 14307–14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin, R. A., R. B. Needleman, D. Gossett and C. A. Michels, 1985. Identification of the structural gene encoding maltase within the MAL6 locus of Saccharomyces carlsbergensis. J. Bacteriol. 164: 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge, S. J., and R. W. Davis, 1988. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene 70: 303–312. [DOI] [PubMed] [Google Scholar]
- Forsberg, H., and P. O. Ljungdahl, 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40: 91–109. [DOI] [PubMed] [Google Scholar]
- Gregory, P. D., A. Schmid, M. Zavari, L. Lui, S. L. Berger et al., 1998. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell 1: 495–505. [DOI] [PubMed] [Google Scholar]
- Gregory, P. D., A. Schmid, M. Zavari, M. Munsterkotter and W. Horz, 1999. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18: 6407–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z., J. O. Nehlin, H. Ronne and C. A. Michels, 1995. MIG1-dependent and MIG1-independent glucose regulation of MAL gene expression in Saccharomyces cerevisiae. Curr. Genet. 28: 258–266. [DOI] [PubMed] [Google Scholar]
- Hu, Z., Y. Yue, H. Jiang, B. Zhang, P. W. Sherwood et al., 2000. Analysis of the mechanism by which glucose inhibits maltose induction of MAL gene expression in Saccharomyces. Genetics 154: 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. W., and D. J. Stillman, 1992. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 4503–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. W., P. R. Dohrmann and D. J. Stillman, 1995. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics 140: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, M., 1999. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 15: 29–33. [DOI] [PubMed] [Google Scholar]
- Kruger, W., C. L. Peterson, A. Sil, C. Coburn, G. Arents et al., 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 9: 2770–2779. [DOI] [PubMed] [Google Scholar]
- Krull, M., N. Voss, C. Choi, S. Pistor, A. Potapov et al., 2003. TRANSPATH((R)): an integrated database on signal transduction and a tool for array analysis. Nucleic Acids Res. 31: 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. C., J. M. Park, S. Min, S. J. Han and Y. J. Kim, 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19: 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, J., L. Tanouye and C. A. Michels, 1992. The UASMAL is a bidirectional promotor element required for the expression of both the MAL61 and MAL62 genes of the Saccharomyces MAL6 locus. Curr. Genet. 22: 181–189. [DOI] [PubMed] [Google Scholar]
- Li, Y., S. Bjorklund, Y. W. Jiang, Y. J. Kim, W. S. Lane et al., 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92: 10864–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macatee, T., Y. W. Jiang, D. J. Stillman and S. Y. Roth, 1997. Global alterations in chromatin accessibility associated with loss of SIN4 function. Nucleic Acids Res. 25: 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys, V., E. Fricke, R. Geffers, E. Gossling, M. Haubrock et al., 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31: 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz, I., H. Jiang, E. K. Han, W. Cui and C. A. Michels, 1996. Characterization of the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. J. Bacteriol. 178: 2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, D. R., and P. J. Laybourn, 2000. Upstream nucleosomes and Rgr1p are required for nucleosomal repression of transcription. Mol. Microbiol. 36: 1293–1305. [DOI] [PubMed] [Google Scholar]
- Myers, L. C., and R. D. Kornberg, 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69: 729–749. [DOI] [PubMed] [Google Scholar]
- Myers, A. M., A. Tzagoloff, D. M. Kinney and C. J. Lusty, 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45: 299–310. [DOI] [PubMed] [Google Scholar]
- Myers, L. C., C. M. Gustafsson, K. C. Hayashibara, P. O. Brown and R. D. Kornberg, 1999. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl. Acad. Sci. USA 96: 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa, M., 2001. Negative regulation of transcription by the yeast global transcription factors, Gal11 and Sin4. Yeast 18: 1099–1110. [DOI] [PubMed] [Google Scholar]
- Nishizawa, M., S. Taga and A. Matsubara, 1994. Positive and negative transcriptional regulation by the yeast GAL11 protein depends on the structure of the promoter and a combination of cis elements. Mol. Gen. Genet. 245: 301–312. [DOI] [PubMed] [Google Scholar]
- Ozcan, S., and M. Johnston, 1999. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63: 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis, M., R. S. Conlan, N. Gounalaki, T. Copf and D. Tzamarias, 2000. Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase II holoenzyme. J. Biol. Chem. 275: 8397–8403. [DOI] [PubMed] [Google Scholar]
- Park, J. M., H. S. Kim, S. J. Han, M. S. Hwang, Y. C. Lee et al., 2000. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 20: 8709–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C. L., and J. L. Workman, 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10: 187–192. [DOI] [PubMed] [Google Scholar]
- Prelich, G., and F. Winston, 1993. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, P. W., M. J. Stern, I. Clark and I. Herskowitz, 1987. Activation of the yeast HO gene by release from multiple negative controls. Cell 48: 567–577. [DOI] [PubMed] [Google Scholar]
- Stillman, D. J., S. Dorland and Y. Yu, 1994. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SW15 transcriptional activator. Genetics 136: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Schure, E. G., N. A. van Riel and C. T. Verrips, 2000. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 24: 67–83. [DOI] [PubMed] [Google Scholar]
- Thevelein, J. M., and J. H. de Winde, 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33: 904–918. [DOI] [PubMed] [Google Scholar]
- Treitel, M. A., and M. Carlson, 1995. Repression by SSN6–TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. USA 92: 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele, M., K. Lemaire and J. M. Thevelein, 2001. Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2: 574–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang, X., M. Bali, I. Medintz and C. A. Michels, 2002. Intracellular maltose is sufficient to induce MAL gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 1: 696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., and F. Winston, 1997. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 25: 4230–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff, D. D., and E. K. O'Shea, 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159: 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., and J. S. Fassler, 1993. SPT13 (GAL11) of Saccharomyces cerevisiae negatively regulates activity of the MCM1 transcription factor in Ty1 elements. Mol. Cell. Biol. 13: 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y., P. Eriksson and D. J. Stillman, 2000. Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol. 20: 2350–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]