Abstract
During the development of the compound eye of Drosophila several signaling pathways exert both positive and inhibitory influences upon an array of nuclear transcription factors to produce a near-perfect lattice of unit eyes or ommatidia. Individual cells within the eye are exposed to many extracellular signals, express multiple surface receptors, and make use of a large complement of cell-subtype-specific DNA-binding transcription factors. Despite this enormous complexity, each cell will make the correct developmental choice and adopt the appropriate cell fate. How this process is managed remains a poorly understood paradigm. Members of the CREB binding protein (CBP)/p300 family have been shown to influence development by (1) acting as bridging molecules between the basal transcriptional machinery and specific DNA-binding transcription factors, (2) physically interacting with terminal members of signaling cascades, (3) acting as transcriptional coactivators of downstream target genes, and (4) playing a key role in chromatin remodeling. In a screen for new genes involved in eye development we have identified the Drosophila homolog of CBP as a key player in both eye specification and cell fate determination. We have used a variety of approaches to define the role of CBP in eye development on a cell-by-cell basis.
THE near-perfect ensemble of unit eyes or ommatidia composing the compound eye of Drosophila melanogaster is the result of a carefully choreographed series of morphogenetic movements, cell-specific gene expression patterns, and cell-cell communications (Ready et al. 1976; Dickson and Hafen 1993; Wolff and Ready 1993). These events begin early in the life of the fly when a small set of cells are set aside to form the eye anlagen during early embryogenesis (Cohen 1993). The earliest phase of eye development is characterized by rapid cell proliferation, the organization of several thousand cells into a single epithelial sheet called the eye imaginal disc, and the stepwise expression of a known set of eight nuclear factors collectively termed the “eye specification genes” (Baker 2001; Kumar and Moses 2001c; Mitashov and Koussulakos 2001). During the last larval instar a wave of differentiation begins at the posterior edge of the disc and sweeps across the eye field. The leading edge of this wave is visualized by a physical indentation within the epithelium, the morphogenetic furrow (Ready et al. 1976). As the furrow travels across the disc, the field of undifferentiated cells is transformed into a lattice of organized clusters of cells that self-assemble into ommatidia (Wolff and Ready 1991a). The cells within a developing unit eye undergo a precise order of recruitment starting with the R8 photoreceptor followed by the stereotyped addition of the R2/5, R3/4, and R1/6 cell pairs. The R7 neuron is the last photoreceptor to be recruited and is then followed by the addition of accessory cone and pigment cells (Ready et al. 1976; Tomlinson and Ready 1987; Cagan and Ready 1989a; Wolff and Ready 1993).
At least six signaling pathways, Ecdysteroids, Receptor Tyrosine Kinases (RTKs), Notch (N), Hedgehog (Hh), Decapentaplegic (Dpp), and Wingless (Wg), have been shown to exert positive and negative influences upon a plethora of downstream nuclear targets during successive stages of eye development (Cagan and Ready 1989b; Basler and Hafen 1990; Shilo 1992; Hafen et al. 1993; Heberlein et al. 1993; Ma et al. 1993; Heberlein and Moses 1995; Ma and Moses 1995; Treisman and Rubin 1995; Pignoni and Zipursky 1997; Royet and Finkelstein 1997; Brennan et al. 1998; Kurata et al. 2000; Kumar and Moses 2001a; Lee and Treisman 2001; Baonza and Freeman 2002; Cherbas et al. 2003). An individual cell within the developing eye will express many cell surface receptors and can expect to be presented simultaneously with several diffusible ligands (Voas and Rebay 2004). The expression patterns of specific DNA-binding factors that control eye development add an additional layer of complexity (Kumar and Moses 1997). Unlike very early predictions, each cell does not express “individualized” or mutually exclusive sets of transcription factors. Rather, cells within the eye express transcription factors in a complicated combinatorial pattern (Kumar and Moses 1997).
Thus, creating such a precise array of unit eyes reproducibly using multiple diffusible signals is an impressive feat. A key question is: How does an individual cell correctly relay the multiple bits of information received at the cell surface to the appropriate assortment of specific DNA-binding transcription factors and how is this information correctly used during cell fate decisions. A potential solution to this paradigm is to have a ubiquitously expressed protein act as a conduit for linking signaling pathways to nuclear transcription factors by interacting with (1) terminal members of the many signaling cascades and (2) the specific combination of transcription factors that are expressed in each different cell type. Such a system would also allow for several diffusible signals to ultimately generate a precise cellular pattern.
Drosophila CREB binding protein (CBP), which is encoded by the nejire (nej) locus, belongs to the CBP/p300 family of proteins (Akimaru et al. 1997b; Goodman and Smolik 2000). CBP was first identified on the basis of its physical interaction with the CREB transcription factor while p300 was identified on the basis of its ability to bind to adenoviral protein E1 (Chrivia et al. 1993; Kwok et al. 1994; Nordheim 1994). Since then CBP has been shown to bind to a large array of specific DNA-binding transcription factors as well as components of the basal transcriptional machinery, thereby acting as both a “bridging” molecule and a transcriptional coactivator (Arany et al. 1994; Kwok et al. 1994; Kee et al. 1996; Gu et al. 1997; Chan and La Thangue 2001; McManus and Hendzel 2001). An additional feature of this protein family is the presence of several protein interaction domains that have been shown to bind to nuclear hormone receptors, acetylated histones, and terminal components of several signal transduction pathways (Bannister and Kouzarides 1996; Chakravarti et al. 1996; Akimaru et al. 1997a; Avantaggiati et al. 1997; Goodman and Smolik 2000; Deng et al. 2003). Furthermore, CBP can modulate transcription and chromatin remodeling by acetylating histones (Bannister and Kouzarides 1996; Martinez-Balbas et al. 1998; Goodman and Smolik 2000). The ability to simultaneously bind so many diverse factors has led to the suggestion that CBP also functions as a “scaffolding” protein to link signaling cascades to transcriptional machinery and thereby influences developmental decisions (Goldman et al. 1997; Shi and Mello 1998; Goodman and Smolik 2000; Chan and La Thangue 2001). Further evidence has demonstrated that the ability of CBP to regulate signaling is itself a regulated process (through phosphorylation and protein-protein interactions), strengthening the argument for a role for CBP in patterning and development (Ait-Si-Ali et al. 1999; Shen et al. 2001). Consistent with such a role, human patients with lesions within the CBP gene suffer from Rubinstein-Taybi syndrome in which pattern formation proceeds incorrectly and is characterized by severe facial abnormalities, broad thumbs, broad big toes, and mental retardation (Petrij et al. 1995; Tanaka et al. 1997; Oike et al. 1999; Murata et al. 2001; Coupry et al. 2002; Kalkhoven et al. 2003). Strabismus, cataracts, juvenile glaucoma, and coloboma of the eyelid, iris, and lens are among the eye defects associated with this syndrome (Roy et al. 1968; Levy 1976; Ramakrishnan et al. 1990; Silengo et al. 1990; Guion-Almeida and Richieri-Costa 1992; van Genderen et al. 2000).
Similarly, mutations within the Drosophila CBP homolog have wide-ranging pleiotropic phenotypes. During embryogenesis alone, CBP (1) interacts with MAD protein to induce expression of Dpp pathway target genes in the dorsal ectoderm, (2) acts to regulate the function of the homeotic gene Deformed (Dfd) in the maxillary and mandibular head segments, (3) interacts with TRX protein to maintain the expression of another Hox gene Ultrabithorax (Ubx) in segments T3 and A1–A7, and (4) interacts with TCF to repress the Ubx enhancer in the embryonic midgut (Akimaru et al. 1997b; Florence and McGinnis 1998; Waltzer and Bienz 1998, 1999; Bantignies et al. 2000; Petruk et al. 2001; Takaesu et al. 2002; Lilja et al. 2003). The absence of CBP during embryogenesis leads to both early dorsoventral patterning abnormalities and later defects in segmentation of the head and trunk. Additional studies focusing on larval and pupal development have demonstrated roles for CBP in proper wing vein formation, dendritic and axonal morphogenesis, formation of the synapse, and the release of transmitters at the neuromuscular junction (Marek et al. 2000). Recently, the fly eye has been shown to be sensitive to the dosage of CBP, as expression of a full-length form of CBP during eye development results in a smooth external surface with a corresponding loss of ommatidia. Interestingly, these defects are not due to a failure of photoreceptor development but rather are the result of severe retinal degeneration (Ludlam et al. 2002). Consistent with these findings is the demonstration that the addition of CBP can reduce the severity of retinal degeneration and structural defects in the fly eye arising from polyglutamine disease (Taylor et al. 2003).
Here we report that Drosophila CBP is required at successive stages of compound eye development including photoreceptor cell fate determination. In a genetic screen designed to isolate new genes that could modify the no-eye phenotype of a dominant negative allele of the eye specification gene sine oculis (soD), we identified nej as a modifier. Loss-of-function nej mutations acted as enhancers of soD, while overexpression of CBP suppressed the no-eye phenotype. Using loss-of-function retinal mosaic clones, heteroallelic loss-of-function combinations, and RNA interference (RNAi) constructs we have extended our findings to demonstrate that CBP is necessary during eye determination and cell fate specification. Using a series of CBP variants we have used a “pathway interference” approach to determine that CBP activity is modular and functions during specific cell fate decisions. In particular, we show that CBP plays a role in the R3/4 cell fate choice and may also be a new member of the R7 pathway. Collectively, the data presented here represent a dissection of the role that CBP plays during the development of the Drosophila compound eye.
MATERIALS AND METHODS
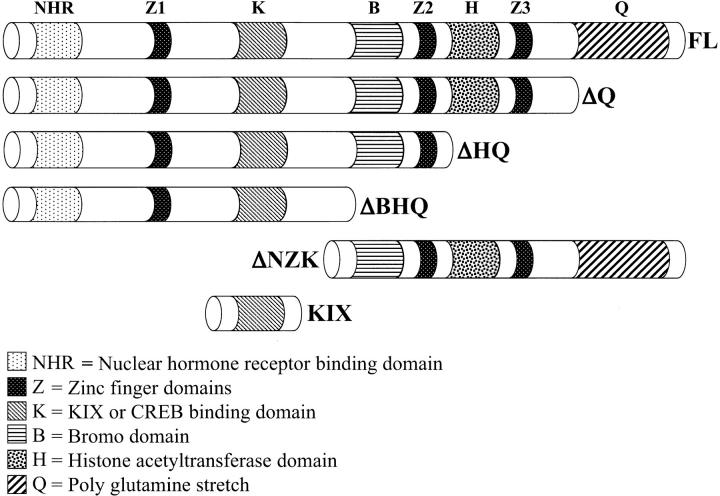
Fly stocks, deletion constructs, and plasmid construction:
The following fly alleles and insertions were obtained for the experiments described here: GMR-GAL4 (Lucy Cherbas), ey-GAL4 (Walter Gehring), soD, sev-GAL4, lz-GAL4, UAS-GFP, UAS-lacZ, ey-FLP, nej3, nejQ7, nejP (Bloomington Stock Center), nejTC41, nejS342, nejS103, nejTA57 (Bill McGinnis), nej131 (Norbert Perrimon), and UAS-CBP FL, UAS-CBP FL-AD (Sarah Smolik). CBP is a 3275-amino-acid protein and the coding regions for each of the CBP variants described in Figure 2 were subcloned into pUAST. Details of the generation of UAS-CBP FL (full-length wild-type version) and UAS-CBP FL-AD (acetylase dead version, F2161A) plasmids can be found within Ludlam et al. (2002) and the cloning strategies for the following deletion constructs are available on request. UAS-CBP ΔNZK contains an ATG followed by amino acids 1030–3275; UAS-CBP ΔQ contains amino acids 1–2677; UAS-CBP ΔHQ contains amino acids 1–1998; UAS-CBP ΔBHQ contains amino acids 1-1501; and UAS-CBP KIX contains an ATG followed by amino acids 800–1090. UAS-CBP RNAi was generated by sequentially cloning a 697-bp fragment (269 bp of 5′-UTR + 428 bp of 5′ coding sequence) into pUAST in the antisense and sense orientations. Expression experiments using the UAS-CBP KIX and UAS-CBP RNAi responders were conducted at 27°, while all remaining experiments using other UAS responders were conducted at 25°.
Figure 2.—
Schematic of CBP variants. Each CBP variant is expressed within subdomains of the developing eye using the UAS/GAL4 misexpression system. See materials and methods for cloning strategies. Individual protein domains are coded.
Genetic screen:
We crossed the 235 stocks that constitute the Drosophila deficiency kit, which provides nearly 90% coverage of the genome, to soD/CyO flies and assayed the ability of each deficiency to modify the no-eye phenotype of soD/+ heterozygotes. Seven deficiencies scored positive in our assay: six suppressed and one enhanced the soD/+ retinal phenotype. We mapped the suppression and enhancing activities by crossing single-gene mutations that were uncovered by the seven deficiencies to soD/+ and scored the ability of each single-gene mutation to mimic that of the overlying deficiency. We identified six complementation groups that suppressed (to be described elsewhere) and the nejire locus that enhanced (this report) the soD/+ retinal phenotype.
Generation of mosaic clones:
Loss-of-function nejire alleles were recombined onto an X chromosome containing the FRT101 element. FRT101 nej[LOF]/FM7; eyFLP females were crossed to either FRT101 Ub-GFP or FRT101 P[w+] males to induced retinal clones that could be analyzed within either the eye imaginal disc or adult retinas, respectively. Clones within the developing and adult eyes were negatively marked and identified in the imaginal disc by the absence of a GFP reporter and in the adult by the absence of red pigment.
Antibodies, immunohistochemistry, and light/confocal microscopy:
The following primary antibodies were used: rat anti-Elav (1:100, gift of Gerald Rubin), rabbit anti-Atonal (1:2000, gift of Andrew Jarman), mouse anti-Dachshund (1:100, gift of Graeme Mardon), mouse anti-Eyes Absent (1:10, gift of Seymour Benzer), chicken anti-CBP (1:500, gift of Sarah Smolik), and rabbit anti-β-galactosidase (1:100, Cortex Biochemical). The following secondary antibodies were obtained from Jackson Laboratories: goat anti-mouse-FITC (1:100), goat anti-mouse-HRP (1:100), goat anti-rabbit-HRP (1:100), goat anti-rabbit-TRITC (1:100), and rabbit anti-chicken-FITC (1:100). F-actin was visualized using phalloidin-TRITC (1:500, Molecular Probes, Eugene, OR). Imaginal discs were dissected and treated as described in Kumar et al. (1998). Pupal retinas were dissected and treated as described in Wolff and Ready (1991b). Adult eyes were prepared for scanning electron microscopy as described in Kumar et al. (1998). Adult eyes were sectioned for light microscopy as described in Kumar et al. (2001). Embryos were stained with antibodies as described in Kumar and Moses (2001b) with the exception that secondary antibodies were conjugated to HRP and detected with the horseradish peroxidase conjugate substrate kit (Bio-Rad, Richmond, CA).
RESULTS
soD encodes a putative dominant negative allele:
The early development of the compound eye is regulated in part by a regulatory network of genes that include the Pax genes twin of eyeless (toy), eyeless (ey), twin of eyegone (toe), and eyegone (eyg); the founding members of the Dach and Eya gene families dachshund (dac) and eyes absent (eya); and the Six class genes optix and sine oculis (Treisman and Heberlein 1998; Kumar 2001; Kumar and Moses 2001c). Extracellular instructions from the Hh, Dpp, Egfr, Notch, and Wg signaling cascades are integrated into this network at several levels creating additional layers of complexity (Halder et al. 1995; Callaerts et al. 1997; Treisman and Heberlein 1998; Kumar 2001; Voas and Rebay 2004). We used a dominant allele of sine oculis, soD, as the starting material for a genetic screen to isolate new genes involved in eye specification. The soD allele appears to function as a dominant negative mutant. First, soD heterozygotes lack compound eyes while heterozygotes of the so3 null allele have wild-type eyes; thus soD is a dominant mutant. Second, embryonic lethality results if the soD mutation is placed in trans to the so3 allele (soD/so3). Third, compound eye development is restored in soD mutants by the addition of wild-type SO protein via UAS-so transgenes; thus soD has an inhibitory function. We sequenced the open reading frame of the soD mutant and found a single valine-to-aspartic acid substitution at amino acid 200 (V200D). This mutation occurs within the Six domain, which is implicated in both DNA-binding and protein-protein interactions with EYA (Pignoni et al. 1997). Mutations within this domain of SO could negatively affect eye development by either altering its interactions with potential binding partners or causing inappropriate transcriptional regulation of downstream target genes.
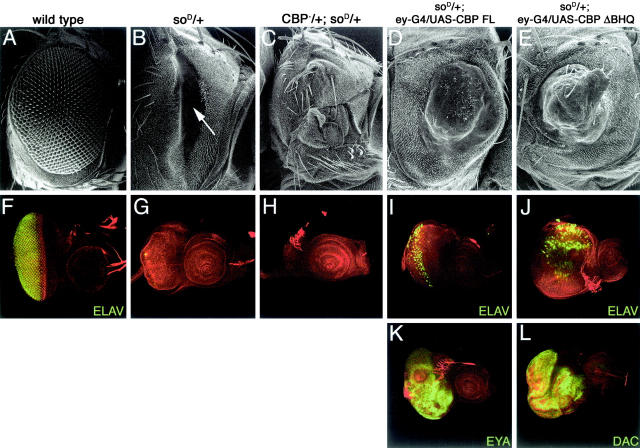
The retinal phenotypes of the eye-specific so1 loss-of-function mutant and the soD dominant negative allele differ slightly from one another. SO protein levels are below detection in so1 mutant eye discs (Pignoni et al. 1997; Halder et al. 1998) while remaining at wild-type levels in soD discs (data not shown). Similarly, the expression of several other genes that are required for eye development, such as dpp and dac, are not reduced in soD mutants while being disrupted in so1 mutants (to be described elsewhere). Furthermore, in so1 adults the region normally occupied by the compound eyes is replaced by surrounding head tissue. In contrast, soD flies have a large nonpigmented and nondifferentiated field (Figure 1, A and B, arrow). The lack of retinal tissue in soD adults can be traced back to a complete lack of photoreceptor differentiation during larval eye imaginal disc development as assayed by the absence of ELAV, a pan-neural protein (Figure 1, F and G). The presence of this nondifferentiated field in soD adults allows for the isolation of both suppressor and enhancer mutations. We recovered six complementation groups that suppressed (to be described elsewhere) and one complementation group (this report) that enhanced the soD no-eye phenotype. The enhancing locus is nej, the gene that encodes CBP in Drosophila (Akimaru et al. 1997b).
Figure 1.—
CBP interacts genetically with soD. Scanning electron micrographs of adult eyes are shown in A–E. Confocal images of third instar imaginal discs are shown in F–L. All genotypes are at top of each column. Red is F-actin; green is identified in each panel. Anterior is to the right. G4, GAL4.
CBP interacts with soD during eye development:
Removal of one copy of nej in a soD background results in an eye phenotype that is now indistinguishable from so1 loss-of-function mutants (Figure 1, C and H). Similar to so1 mutants, eye imaginal discs from nej3/+; soD/+ heterozygotes (nej3 is a null allele) are small and undergo increased levels of cell death (Figure 1H), while adults lack the nondifferentiated field and instead contain only head tissue (Figure 1C). Conversely, expression of CBP throughout the soD retinal field suppresses the no-eye phenotype (Figure 1, D and I). Eye imaginal discs are near normal in size and contain large numbers of photoreceptor cell clusters (Figure 1I), and adult eyes are fully pigmented although not normally patterned (Figure 1D).
Functional dissection of CBP during early eye development:
CBP is a large protein containing >3200 amino acids and features several different functional domains (Goodman and Smolik 2000). The N terminus contains several protein interaction domains including a region that binds hormone receptors and a domain (KIX) that binds the CREB transcription factor (Chrivia et al. 1993; Kwok et al. 1994). Subsequent work has demonstrated that this domain binds several other transcription factors as well (Frangioni et al. 2000). The C-terminal half of the protein contains three major regions: (1) a BROMO domain that binds to acetylated lysine residues, (2) a HAT domain that acetylates lysine 8 of histone H4, and (3) a glutamine-rich stretch that is implicated in transcriptional activation (Kraus et al. 1999; Goodman and Smolik 2000; Manning et al. 2001). To functionally dissect the CBP during early eye development, we made several CBP variants lacking either single or multiple protein domains (Figure 2) and expressed them throughout the eye in soD mutants. A truncated protein lacking the N-terminal half of CBP was insufficient to rescue soD (ΔNZK, data not shown) while several proteins that retained the N-terminal portion of CBP (ΔHQ and ΔBHQ) rescued the eye phenotype of soD flies to the same degree as the full-length protein (Figure 1, E and J). The CBP variant lacking just the long glutamine-rich stretch (ΔQ) functions as a potent dominant negative protein (see below) and was unable to rescue the soD phenotype. It is interesting to note that both the ΔHQ and the ΔBHQ variant proteins retain the KIX or CREB binding domain (Figure 2). In addition to binding to the transcription factor CREB, the KIX domain binds the transcription factor Cubitus Interruptus (CI) the terminal member of the Hh signaling cascade (Akimaru et al. 1997a), which functions during successive stages of eye development and is required for eye specification (Kumar 2001; Pappu et al. 2003; Voas and Rebay 2004). Consistent with the above data, expression of Hh and CI proteins in soD mutant eyes also rescues the soD phenotype to the levels of full-length CBP (data not shown). It has been recently reported that mammalian CBP may function within the context of a SIX-EYA-DAC transcriptional complex by mediating physical interactions between the mouse DACH1 and EYA1 proteins (Ikeda et al. 2002). Expression of the full-length, ΔHQ or ΔBHQ CBPs is sufficient not only to support photoreceptor development but also to promote the expression of the eye specification genes dac and eya (Figure 1, K and L).
CBP is expressed in the developing eye:
The genetic interactions observed between nej and soD suggested that CBP is expressed in the eye. We confirmed the expression profile of nej within the developing eye imaginal disc by using an antibody directed against CBP (gift of Sarah Smolik; Figures 3 and 4). CBP is detected in all cells ahead of the morphogenetic furrow as determined by either individual staining (Figure 3, A and B) or costaining with antibodies against EYA, DAC, and SO proteins (Figure 4, A–C and G–I; data not shown). The coexpression of all four proteins ahead of the morphogenetic furrow is supportive of a role for CBP in mediating SO-EYA-DAC interactions as suggested by studies in mammalian systems (Ikeda et al. 2002). Posterior to the furrow, CBP can be found in the eight photoreceptors and four cone cells of each ommatidium (Figure 3, D–F). The identity of each cell was determined by their stereotyped position within the developing ommatidial cluster and by their costaining with an antibody against the pan-neuronal protein ELAV (Figure 4, D–F; data not shown). It remains unclear if CBP is present within each pigment cell subtype. Within the eye disc CBP is also present within the undifferentiated cells as determined by coexpression of CBP and a transcriptional reporter that faithfully reflects the expression pattern of the transcription factor lozenge (Figure 3, G–I, arrows; Flores et al. 1998). The presence of CBP within each cell of the developing eye is consistent with its proposed role as both a “bridging” protein during transcription and a “scaffolding” protein during signaling.
Figure 3.—
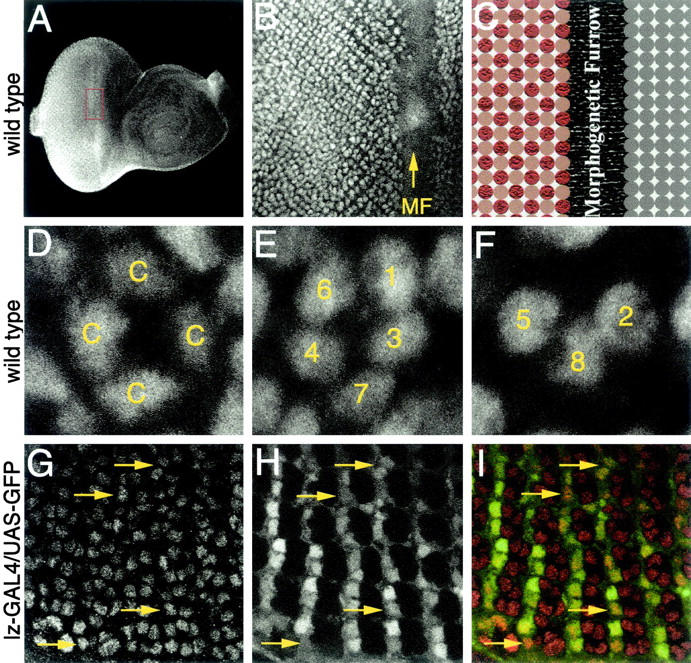
CBP is expressed in all cells of the developing eye imaginal disc. Confocal images of third instar imaginal discs are shown. Genotypes are listed at the left of each row. (A and B) Low and high magnification view of CBP expression ahead of and behind the morphogenetic furrow (MF). (C) Schematic of cells within the eye disc. Gray circles represent cells ahead of the furrow. Red circles represent ommatidial clusters. Brown circles represent intervening cells. (D–F) CBP is present in cone (c) and photoreceptor cells (1–8). (G–I) CBP is present in the intervening cells. (G) CBP; (H) lz-GAL4/UAS-GFP; and (I) merge of G and H. Yellow arrows indicate intervening cells. Anterior is to the right.
Figure 4.—
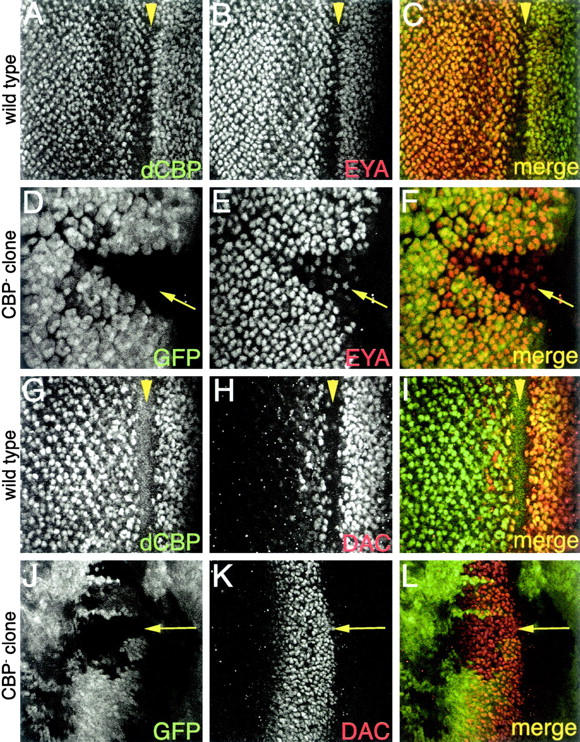
CBP regulates eyes absent but not dachshund expression during eye development. Confocal images of third instar imaginal discs are shown. All genotypes are to the left of each row. (A–C) CBP and Eya are coexpressed. (D–F) Eya protein levels are reduced in CBP loss-of-function retinal clones. (G–I) CBP and Dac are coexpressed. (J–L) Dac protein levels are not regulated by CBP. Molecules visualized are listed in each panel. Arrows mark CBP loss-of-function clones. Arrowheads mark the morphogenetic furrow. Anterior is to the right.
nejire mutants affect the eye specification genes in the eye and embryo:
The expression profile of CBP within the developing eye field (Figures 3 and 4) led us to determine if the expression of the eye specification genes are dependent upon nej function (Figure 4). We focused on the expression of the eya, dac, and so genes since their mammalian counterparts appear to interact with mouse CBP (Ikeda et al. 2002). Due to the embryonic lethality associated with nej loss-of-function mutations we attempted to generate large retinal clones of seven nej loss-of-function alleles, including the molecularly characterized null allele nej3. Consistent with null alleles being cell lethal, only clones of the strong hypomorphic alleles nejTC41 and nejS342 survived to be analyzed. Clones of either allele gave identical results (see below) and therefore only nejTC41 clones are shown. In nej clones the level of so-lacZ expression (data not shown) and EYA protein was dramatically reduced (Figure 4, D–F). Note that within the clone EYA protein levels are lower compared to the adjacent wild-type tissue. We did not observe an elevated level of cell death in these clones. These results are suggestive of a role for CBP in the regulation of both so and eya during eye specification. In contrast, the level of DAC protein was not affected by the loss of CBP (Figure 4, J–L). Note that DAC protein levels remain the same within the wildtype and clonal tissue. This result is consistent with reports that some DAC protein remains in so and eya single and double loss-of-function mutants (data not shown).
Expression of the “eye specification” genes is not confined to the developing eye but is detected in dynamic spatial and temporal patterns within several other tissues (Cheyette et al. 1994; Mardon et al. 1994; Quiring et al. 1994; Serikaku and O'Tousa 1994; Jones et al. 1998; Leiserson et al. 1998; Czerny et al. 1999; Seimiya and Gehring 2000; Kumar and Moses 2001b). For example, during the extended germband stages of embryogenesis EYA protein is present within the clypeolabrum, the protocerebrum, and the visual primordium (Figure 5, A and C). At these same stages DAC protein is detected within a subdomain of the protocerebrum and within cells of the maxillary and mandibular head segments (Figure 5, E and G), while transcription of so is detected within the visual primordium and within small groups of unidentified cells at the segmental grooves (Figure 5, I and K). While the expression of so, eya, and dac is restricted to specific embryonic domains, CBP appears to be ubiquitously expressed. Although CBP is maternally contributed, homozygous nej mutants die as embryos and have a characteristic twisted phenotype (Akimaru et al. 1997b). In nej mutants, EYA protein remains within the clypeolabrum and the visual primordium. However, the protein is lost from the protocerebrum and the mesoderm (Figure 5, B and D). In contrast, the expression of so is nearly completely abolished from the visual primordium (Figure 5, J and L). Furthermore, the level of DAC protein is also drastically reduced in nej homozygous embryos (Figure 5, F and H), whereas it remained unaffected in nej retinal clones (Figure 4, J–L). These results suggest that the regulatory relationships between CBP and SO, DAC, and EYA proteins are likely to be tissue and even cell subtype dependent. This would be consistent with a role for CBP in acting as a scaffolding protein during development to link signaling cascades to specific DNA-binding transcription factors.
Figure 5.—
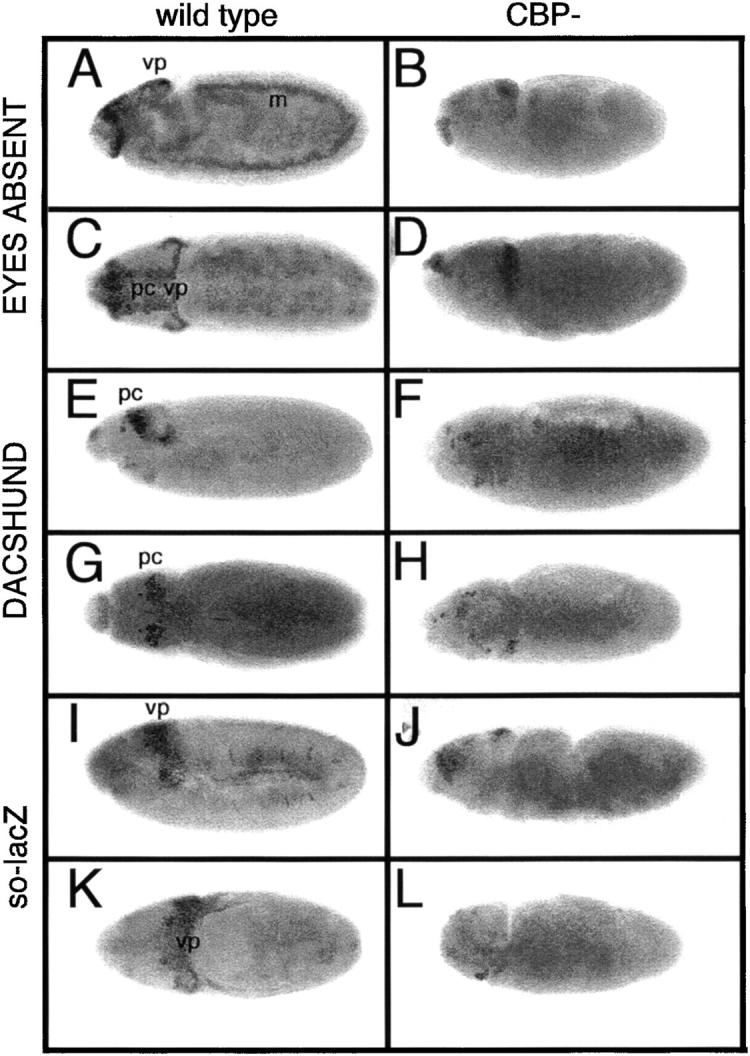
Mutations within CBP alter dac, eya, and so expression in the embryonic visual system. Light microscope images of wild type (A, C, E, G, I, and K) and nejTC41 mutant embryos (B, D, F, H, J, and L) are shown. Genotypes are at the top of each column. Molecules visualized are listed to the left of each panel. Lateral views of embryos are shown in A, B, E, F, I, and J and dorsal views are shown in C, D, G, H, K, and L. Eyes Absent and Dachshund proteins are detected with antibodies. β-Gal antibodies were used to detect the pattern of so-lacZ. vp, visual primordium; pc, protocerebrum; m, mesoderm. Anterior is to the left.
nejire is required for photoreceptor specification:
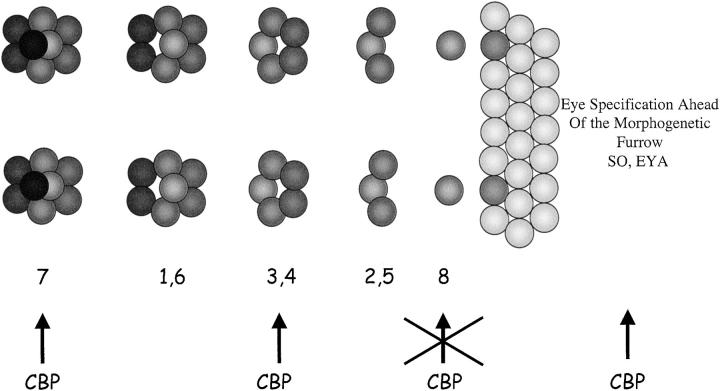
The expression of CBP within all photoreceptors suggests a role for CBP in their recruitment and/or maintenance. We sought to determine the requirement for nej in photoreceptor specification by using loss-of-function retinal clones, heteroallelic combinations, and RNAi. Retinal clones of the strong loss-of-function alleles nejTC41 and nejS342 were generated and analyzed in developing eye imaginal discs and adult eyes using confocal, light, and scanning electron microscopy (Figure 6). Adult compound eyes containing clones of nej mutant tissue have a disorganized external surface (Figure 6, A and C). Retinal sections through mutant eyes show variable numbers of ommatidia and photoreceptor cells (Figure 6, B and D). The central domains of large clones are completely devoid of photoreceptor cells and appear to be replaced by pigment cells, suggesting a cell fate switch has occurred (asterisk in Figure 6D). The outer edges of the clones contain ommatidia with variable numbers of photoreceptors (arrows in Figure 6D) and we did not observe any ommatidia that were genetically mutant for nej and were also morphologically wild type. This suggests that CBP is required for the formation of all photoreceptor cell subtypes.
Figure 6.—
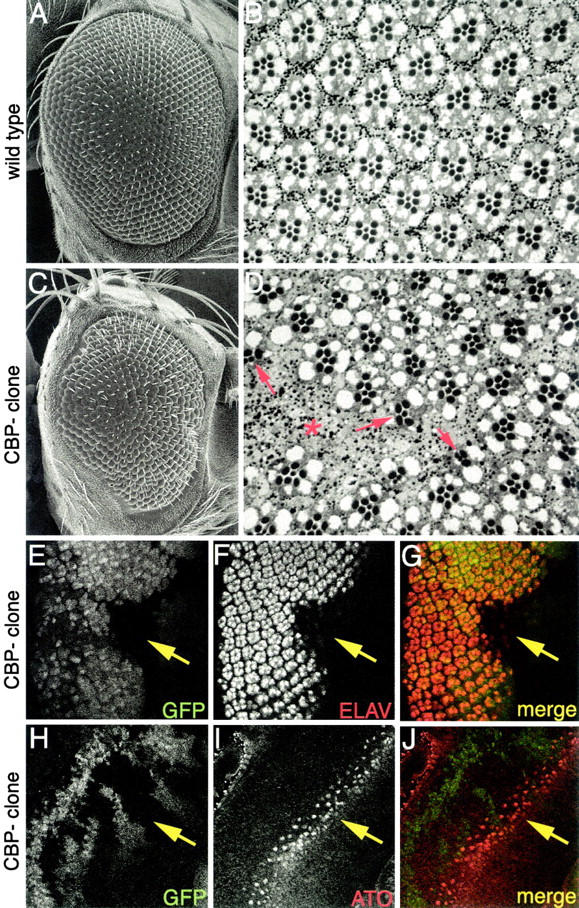
CBP mutants inhibit eye development. (A and C) Scanning electron micrographs of adult eyes. (B and D) Light microscope sections of adult retinas. (E–J) Confocal images of third instar imaginal discs. Genotypes are listed to the left of each row. Wild-type eyes are shown in A and B. CBP clones are shown in C–J. Molecules visualized are listed within each panel. Red asterisk in D marks center of large clone that is devoid of photoreceptors. Red arrows in D mark ommatidia at the borders of clones. Yellow arrows in E–J mark position of CBP retinal mosaic clone. Note the reduction of Elav staining within the clone. Also note that Atonal expression within the clone remains normal. Anterior is to the right.
Eye imaginal discs were stained with an antibody that recognizes the ELAV protein. nej mutant clones showed a significant loss of ELAV positive cells (Figure 6, E–G), which is consistent with the loss of photoreceptor cells seen in adult sections. Although an analysis of adult clones suggested that nej function was necessary for the specification of all photoreceptor cells, mutant nejTC41 retinal clones still maintained normal expression of the proneural transcription factor Atonal (Figure 6, H–J), which is the primary determinant for selection of the R8 photoreceptor. These results suggest that nej is not required for R8 selection. However, clonal analysis in eye discs and adult retinal sections indicates that all other photoreceptor cell types appear to be affected by the loss of nej function.
We looked for heteroallelic combinations of the known nej mutants that would generate adult flies. Two combinations, nej131/nejP and nejTC41/nejP, give viable adults (nej131/nejP ∼15% and nejTC41/nejP ∼5%) and have moderately rough eyes, further suggesting that CBP plays a role in eye development (Figure 7A). Sections of nej131/nejP adult retinas reveal that ommatidia within these mutant eyes have variable numbers of photoreceptors (Figure 7B). Furthermore, it appears that both the R7 and the outer photoreceptors (R1–R6) are affected by loss of nej. This is consistent with our clonal analysis indicating that photoreceptor development requires CBP (Figure 6). No survivors were recovered for any other heteroallelic combination (nejS342/nejP, nejS103/nejP, nejTA57/nejP, nej3/nejP, and nejQ/nejP).
Figure 7.—
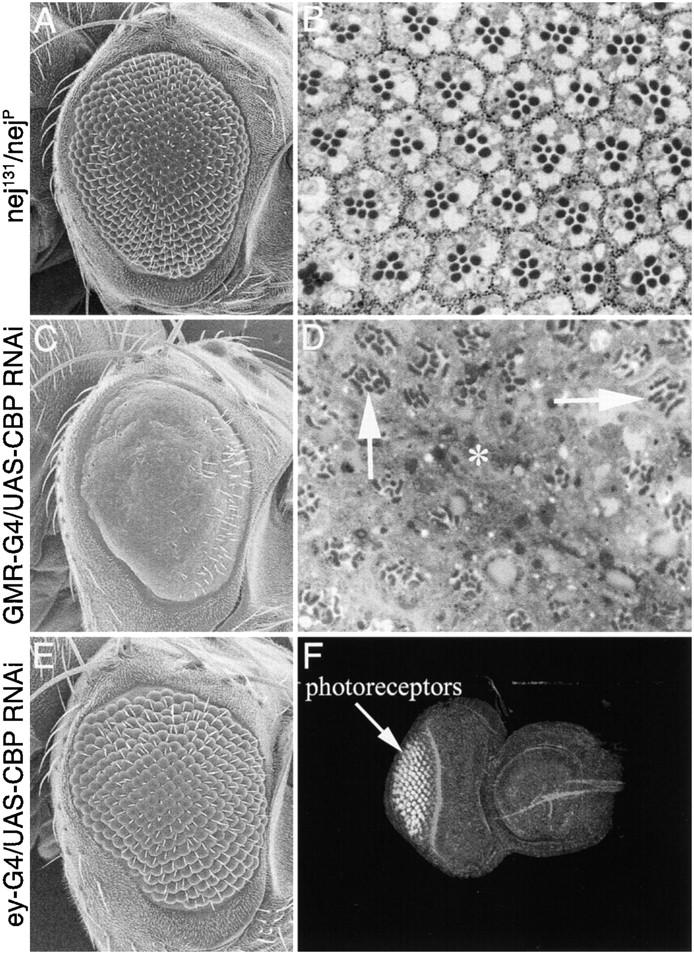
CBP mutants inhibit eye development. (A, C, and E) Scanning electron micrographs of adult eyes. (B and D) Light microscope sections of adult retinas. (F) Confocal image of third instar imaginal disc. Genotypes are listed to the left of each row. nej heteroallelic (nej131/nejP) eyes are shown in A and B. GMR-GAL4/UAS-dCBP RNAi is shown in C and D. ey-GAL4/UAS-dCBP RNAi is shown in E and F. Asterisk in D marks region of eye that is devoid of photoreceptors. Arrows in D mark ommatidia with malformed rhabdomeres. Anterior is to the right.
In addition, we have used RNAi interference to knock down the levels of CBP within the developing eye. We generated a CBP RNAi snapback construct (see materials and methods) and expressed it both ahead of and posterior to the morphogenetic furrow using ey-GAL4 and GMR-GAL4 insertions (Figure 7). The GMR element (Hay et al. 1995) directs expression to all cells posterior to the morphogenetic furrow. The external surface of GMR-GAL4/UAS-CBP RNAi eyes is relatively smooth, individual facets cannot be distinguished, and the number of interommatidial bristles is dramatically reduced (Figure 7C). Although retinal sections confirm the loss of many ommatidia (asterisk in Figure 7D), a majority of the retina has photoreceptor clusters. Each surviving ommatidial cluster appears to have fewer than the normal number of photoreceptors. The number of neurons appears to be somewhat variable, suggesting that each cell is equally susceptible to the loss of CBP. The remaining photoreceptors appear to have defectively formed rhabdomeres (arrows in Figure 7D). Although the effects of our RNAi snapback construct on photoreceptor development are somewhat weaker than those of the loss-of-function mutant phenotypes, the data are consistent between both experiments. It is likely that the amount of CBP RNA in photoreceptor cells is at a very high level and our CBP RNAi construct is not efficiently reducing the levels of CBP in these cells. The smooth external surface does suggest, however, that the level of CBP RNAi is sufficient to affect the cone, pigment, and mechanosensory bristle cells.
The ey enhancer element (Hauck et al. 1999) directed expression of the CBP RNAi snapback construct to cells ahead of the furrow and resulted in the inhibition of eye development (Figure 7, E and F). Note that the eye disc is smaller than wild-type discs and there are fewer ommatidial clusters (compare to Figure 1F). The continued presence of substantial retinal tissue may reflect either insufficient knockdown of endogenous CBP RNA levels or a partial requirement for CBP in eye specification. A closer examination indicates that the surviving ommatidia are constructed properly and have the normal numbers of photoreceptors and accessory cells. This is consistent with the expression pattern of the eyeless enhancer, which directs expression ahead of the morphogenetic furrow. Thus the reduction in the overall number of unit eyes is likely due to the elimination of precursor cells ahead of the furrow.
Distinct roles for CBP in ommatidial assembly:
The list of proteins that physically interact with members of the CBP family of proteins has grown to >100 factors (Goodman and Smolik 2000). While nej loss-of-function mutants have revealed a general role for CBP in fly retinal cell fate specification, loss-of-function experiments are complicated by the effects of such large-scale disrupted interactions. We used a “pathway interference” approach to determine if CBP played a more defined role in ommatidial assembly than that revealed by the simple analysis of loss-of-function phenotypes. We expressed CBP variants (Figure 2) in the developing eye under the control of the GMR enhancer element, which drives expression in all cells posterior to the morphogenetic furrow. We then analyzed photoreceptor, cone, and pigment cell development in eye imaginal discs, pupal discs, and adult retinas (Figure 8). The different truncated proteins are expected to act as “protein sinks” by soaking up sets of transcription factors, thus depleting cells of crucial proteins required during cell fate specification. Since each CBP truncated protein retains a different set of protein domains, the expression of each variant protein is expected to yield a different phenotype. Collectively, the phenotypes obtained by this set of deletion proteins should provide a deeper insight into the role played by CBP in cell fate decisions in the fly eye.
Figure 8.—
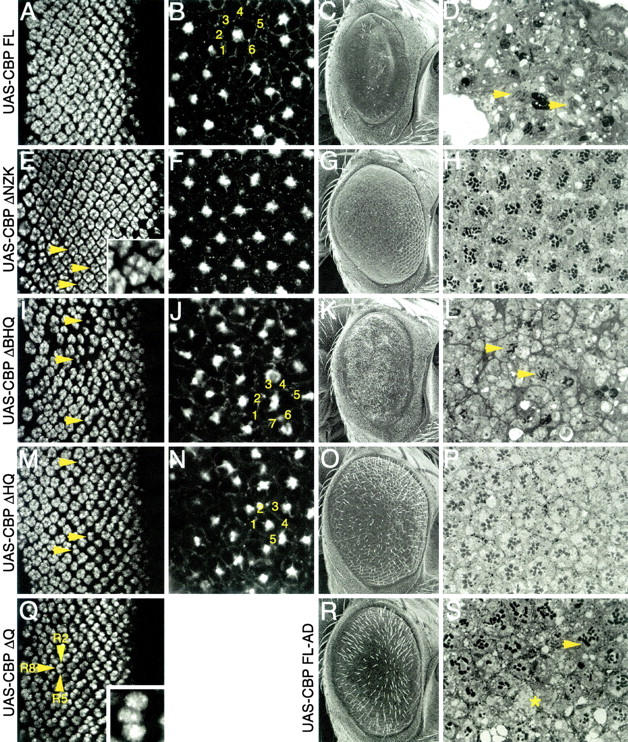
Expression of individual CBP variants results in specific effects on photoreceptor and cone cell development. (A, E, I, M, and Q) Confocal images of third instar eye discs. (B, F, J, and N) Confocal images of pupal retinas. (C, G, K, O, and R) Scanning electron micrographs of adult eyes. (D, H, L, P, and S) Light microscope sections of adult retinas. Genotypes are listed to the left of each row. In all cases the CBP variant is expressed from a UAS construct driven by GMR-GAL4. Eye discs are stained with an antibody against Elav. Pupal retinas are stained for F-actin. Yellow numbers mark the number of cone cells within an ommatidium. Yellow asterisk in S marks area that is devoid of photoreceptors. Yellow arrowheads mark photoreceptor clusters. Anterior is to the right.
Normal adult eyes are characterized by their crystalline-like external appearance and an underlying perfect arrangement of ommatidia (Figure 6, A and B). The developing photoreceptor clusters are best visualized within the eye imaginal disc while the accessory cone and pigment cells are arranged in a near-perfect array midway through pupal development. As a control we first expressed a full-length version of wild-type CBP in the eye. This resulted in a severe rough eye with very few, if any, surviving photoreceptors in freshly eclosed adults (Figure 8, C and D). Consistent with an earlier report, the photoreceptor cells appear to be recruited and specified correctly within the eye imaginal disc (Figure 8A). The defects in eye development appeared to be the result of two independent events: (1) degeneration of photoreceptor cells after they are initially specified and (2) faulty cone cell formation during pupal development (Figure 8B). During midpupal development the accessory ommatidial cells form a near-perfect lattice structure. Overexpression of wild-type CBP appears to increase the number of cone cells per ommatidium from four in wild type to five or six cells (yellow numbers in Figure 8B). A mutation that inactivates the acetyltransferase activity of CBP alleviates this phenotype moderately (Figure 8R). However, many ommatidia in the adult retina either are still lacking photoreceptor cells (asterisk, Figure 8S) or have gross defects in rhabdomere development (arrow, Figure 8S).
The expression of mutant CBPs was surprisingly useful in teasing out potential roles for CBP in eye development and the roles of individual protein domains. For example, expression of CBP ΔNZK (Figure 2) resulted in a slight roughening of the external retinal surface and a loss of ommatidial bristles (Figure 8G). Optical sections of pupal retinas indicated a normal complement of accessory cone and pigment cells (Figure 8F), while adult retinal sections revealed the recruitment of one to two extra photoreceptors per ommatidium (Figure 8H), which can also be seen in eye imaginal disc preparations (Figure 8E). These two extra photoreceptors were in the right anatomical position to be the so-called “mystery cells” being recruited into the ommatidium. Normally, the mystery cells leave the assembling ommatidia (Tomlinson and Ready 1987). Thus it is likely that CBP is blocking an inhibitory signal, therefore committing the mystery cells to a photoreceptor cell fate.
An equally striking phenotype is observed in eye discs expressing the CBP ΔQ protein, in which the poly(Q) trans-activation domain has been deleted (Figure 2). Flies expressing this construct died shortly after cessation of larval development, precluding any study of pupal and adult eye development. During larval eye disc development each ommatidium contained only the R8, R2, and R5 photoreceptors (Figure 8Q). It appeared that this mutant protein blocked ommatidial assembly specifically at the recruitment of the R3/R4 photoreceptor pair. A likely explanation is that CBP ΔQ is binding to factors that are required for R3/R4 specification but cannot activate transcription of downstream targets due to the absence of the poly(Q) domain. Such a scenario is consistent with the dominant negative activity of the CBP ΔQ protein. This phenotype has been observed in retinas that are mutant for both the glass and the rough loci, two genes previously shown to be involved in photoreceptor cell determination (Tomlinson et al. 1988; Moses et al. 1989; Treisman and Rubin 1996).
The HAT activity of CBP has also been implicated in regulating transcription during development by acetylating histones at lysine residues (Goodman and Smolik 2000; Ludlam et al. 2002). The expression of CBP ΔHQ mutant protein, in which both poly(Q) and HAT domains are deleted (Figure 2), within cells posterior to the furrow caused adult flies to have a moderate roughening of the external surface of the compound eye (Figure 8O). Each ommatidium contained a variable number of photoreceptors (PFigure 8, M and P) while often recruiting additional numbers of accessory cone cells (Figure 8N). Since the activity of the BROMO and HAT domains appears to be functionally linked, it is unsurprising that the retinal phenotypes associated with expression of CBP ΔBHQ, which removes the BROMO, HAT, and poly(Q) domains (Figure 2), are significantly more severe than those observed by expression of the CBP ΔHQ protein. The external surface of the adult eye was flattened, reduced in size, and covered with small bristles (Figure 8K). Many ommatidial clusters showed a severe reduction in the number of photoreceptor cells (Figure 8, I and L) while containing an increased number of accessory cone cells (Figure 8N). It has been suggested that the photoreceptors play a role in recruiting the accessory cone cells. Thus the simultaneous loss of photoreceptors and gain of cone cells that results from expression of CBP FL, ΔHQ, and ΔBHQ proteins is unusual and may suggest that a cell fate switch has taken place.
CBP functions in the recruitment of the R3/R4 and R7 photoreceptors:
We sought to further define the role that CBP plays in ommatidial assembly by using the sevenless (sev) enhancer to drive expression of CBP variants in a more restricted pattern (Figure 9). The sev enhancer directs expression within the R3, R4, R1, R6, and R7 photoreceptors and cone cells (Basler et al. 1989; Bowtell et al. 1991). Expression of the CBP ΔNZK protein appeared to have no effect within the SEV expressing cells (Figure 9, A and B), while expression of the CBP ΔQ protein variant resulted in the near deletion of the compound eye (data not shown).
Figure 9.—
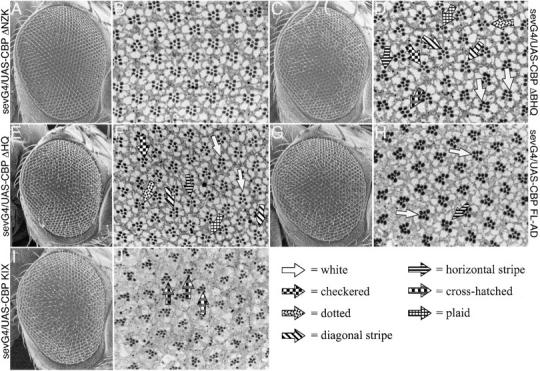
CBP functions during R3, R4, and R7 photoreceptor cell specification. (A, C, E, G, and I) Scanning electron micrographs of adult eyes. (B, D, F, H, and J) Light microscope sections of adult retinas. Genotypes are listed at the sides of each row; G4 stands for GAL4. In all cases the CBP variant is expressed from a UAS construct driven by sev-GAL4. (A–H) White arrows mark ommatidia with two R3 cells. Diagonal stripe arrows mark ommatidia that have opposite chirality. Dotted arrows mark ommatidia in which the R3 cell has transformed into an R7. Horizontal stripe arrows mark ommatidia in which R4 has transformed into R7. Checkered arrows mark ommatidia in which R4 has not been specified. Orange arrow marks an ommatidium in which R7 has been deleted. Crosshatched arrows mark ommatidia in which both R3 and R4 have been transformed into R7, resulting in three R7 cells per cluster. (J) Plaid arrows mark the large outer photoreceptor that occupies the R7 cell position. Arrow key is at bottom right of figure.
In contrast, expression of the CBP ΔBHQ protein variant led to several surprising and interesting assembly phenotypes (Figure 9, C and D). The external surface of the compound eyes was near wild type with the only overt defect being the frequent loss or mispositioning of the interommatidial bristles (Figure 9C). A careful analysis of adult retinal sections revealed several defects in ommatidial assembly. First, many individual putative R4 photoreceptors failed to make the correct choice and adopted the R3 cell fate (white arrows, Figure 9D). Second, at a lower frequency the R3 and R4 cells adopted the opposite cell fates and the ommatidium rotated in the wrong direction (diagonal stripe arrows, Figure 9D). These phenotypes are similar to those observed in alleles of the WNT receptor frizzled (fz; Zheng et al. 1995). A possible conclusion is that CBP cooperates with Wnt signaling to establish R3/R4 cell identities. Third, in some ommatidia the R4 failed to be specified (checkered arrow, Figure 9D). Fourth, in many ommatidia either the R3 or the R4 cell adopted an R7 cell fate (dotted and horizontal stripe arrows, Figure 9D). However, in some cases both cells adopted the R7 fate, resulting in an ommatidial cluster containing three R7 neurons (cross hatched arrow, Figure 9D). The transformation of R3/R4 cells into R7 photoreceptors implicates CBP as a possible member of the Sevenless signaling cascade. Finally, in rare cases the presumptive R7 cell failed to differentiate (plaid arrow, Figure 9D). All of these phenotypes were also observed when CBP ΔHQ expression was directed by the sev enhancer element (Figure 9, E and F). However, some of these phenotypes, such as loss of R7 cells and the presence of three R7 cells within an ommatidum, were reduced in frequency. In addition, expression of a full-length CBP with dramatically reduced HAT activity (CBP FL-AD) could also redirect the presumptive R4 cell into an R3 cell fate and could occasionally transform either the R3 or the R4 into an R7 photoreceptor (Figure 9, G and H). It should be noted that the ability to respecify the R3/R4 fate into R7 is very rare in this situation and, unlike expression of CBP ΔBHQ and ΔHQ, expression of CBP FL-AD never leads to the loss of the R4 or the R7 cells. Together, these results suggest that the N-terminal half of CBP is acting as a protein sink that sequesters factors required for correct R3/R4 specification, and the C-terminal half of the protein is actively involved in R7 development. It is possible that CBP promotes R7 development by regulating downstream genes through either the zinc finger domain or the transcriptional activation domain located at the C terminus. The role that CBP plays in R7 development may be even more complicated since expression of just the KIX domain is able to transform the small inner R7 cell into a large outer photoreceptor (Figure 9, I and J). The KIX domain is known to bind the transcription factor CREB. It would be interesting to determine if the Drosophila homolog of the CREB transcription factor functions during R7 photoreceptor specification.
CBP variants display context and dose-dependent effects:
The complex phenotypes that we observed with the CBP variants led us to determine the functional nature of each variant. We expressed each of the CBP variants listed in Figure 2 ahead of the morphogenetic furrow in an otherwise wild-type background (Figure 10) using an ey-GAL4 driver that faithfully reflects the expression pattern of the endogenous ey gene. In contrast to the rescue of the soD no-eye phenotype, the expression of each variant protein in an otherwise wild-type genetic background inhibited eye development (Figure 10). This suggests that the effects of these variant proteins are both context dependent and dose sensitive. Interestingly, the relative strength of that inhibition varies according to the construct (Figure 10), which is not surprising since each variant retains a differing subset of protein interactions domains. We were able to rule out insertion-specific effects by testing five independent insertions for each variant type and obtaining similar if not identical results from each insertion. Expression of the CBP variant lacking the N-terminal half (CBP ΔNZK; see Figure 2) ahead of the advancing furrow appeared to strongly inhibit eye development within the dorsal half of the eye while pattern formation proceeds in the ventral domain (Figure 10, A and D). The CBP variant lacking just the C-terminal glutamine-rich region (CBP ΔQ; see Figure 1) was the strongest inhibitor of eye development. Expression of this construct blocked initiation of pattern formation within the eye disc, thus completely deleting the compound eyes in the adult (Figure 10, B and E). The deleted segment of the CBP ΔQ variant contains several polyglutamine and polyalanine stretches. Both glutamine- and alanine-rich domains have been implicated in the activation of transcription in several systems. The severe effects of the CBP ΔQ variant on eye development may be the result of this mutant protein retaining the ability to bind to and deplete cells of dozens of transcription factors while lacking the ability to activate transcription.
Figure 10.—
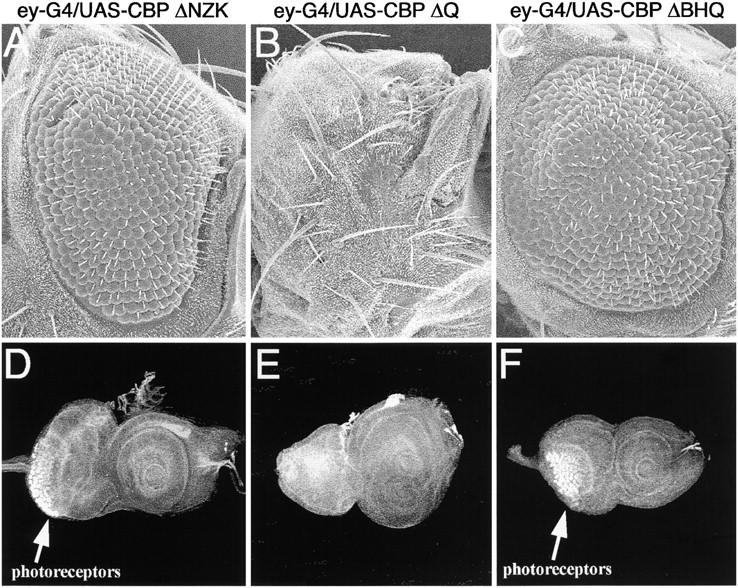
Expression of CBP variant proteins ahead of the furrow inhibits eye development. Scanning electron micrographs of adult eyes are in shown A–C. Confocal images of third instar imaginal discs are shown in D–F. All genotypes are at the top of each column. In all cases the CBP variant is expressed from a UAS construct driven by ey-GAL4. Anterior is to the right.
The expression of variant proteins that retain the N-terminal half of the protein but lack varying amounts of the C-terminal half allowed for the initiation of the morphogenetic furrow at the posterior margin of the disc but inhibited its continuous reinitiation along the posterior-lateral domains (CBP ΔBHQ; Figure 10, C and F; CBP ΔHQ data not shown). This phenotype is similar to situations in which Notch and Egfr signaling is inhibited along the margins of the eye disc. It is noteworthy that although the CBP ΔHQ and ΔBHQ protein variants also lack the trans-activation domain, their overexpression phenotypes were different and significantly less severe than the overexpression phenotype of the ΔQ variant. The increased severity of the ΔQ variant may be due to the inability of the mutant protein to activate transcription while maintaining protein-protein interactions between signaling molecules and the HAT and BROMO domains.
The inhibition of eye development that results from the expression of CBP variants prompted us to determine the activity of these molecules—i.e., are they functioning as dominant gain-of-function or dominant negative proteins (Table 1). We expressed each variant listed in Figure 2 in a subset of cells posterior to the morphogenetic furrow using the GMR-GAL4 insertion. Expression of each construct altered the structure of the compound eye (Table 1). We repeated this experiment in a nej3 null mutant heterozygote background. As expected, the severe rough eye that resulted from the expression of full-length CBP was moderately suppressed by the loss of one copy of nej (Table 1). Removal of one copy of nej led to suppression of the rough eye phenotype that is associated with the expression of the KIX domain, suggesting a gain-of-function role for the KIX domain (Table 1). The CBP ΔQ and CBP ΔNZK rough eye overexpression phenotypes were enhanced by the loss of one copy of nej, suggesting that these are functioning as dominant negative proteins (Table 1). Surprisingly, the rough eye phenotypes associated with the expression of the ΔHQ and ΔBHQ variants were unaffected by the removal of one copy of nej (Table 1). This result might suggest that these variants have neomorphic activities. It will be very informative to determine the exact molecular and biochemical role that each CBP domain plays in retinal development. Of particular interest is the identification of binding partners that also play a role in eye formation. The construction of the described CBP variants has been a good first step toward dissecting the role that CBP plays in eye specification and photoreceptor cell determination. The identification of interacting partners in this process will certainly move our understanding of eye development considerably further.
TABLE 1.
Activity of CBP variants
| Driver | Responder | nej+ | nej3/+ | Activity | |
|---|---|---|---|---|---|
| GMR-GAL4 | UAS-CBP FL | Severe rough eye | Suppress | Gain of function | |
| GMR-GAL4 | UAS-CBP ΔNZK | Moderate rough eye | Enhance | Dominant negative | |
| GMR-GAL4 | UAS-CBP ΔBHQ | Severe rough eye | Enhance | Dominant negative | |
| GMR-GAL4 | UAS-CBP ΔHQ | Mild rough eye | No effect | ? | |
| sev-GAL4 | UAS-CBP ΔQ | Severe rough eye | No effect | ? | |
| GMR-GAL4 | UAS-CBP KIX | Very mild rough eye | Suppress | Gain of function | |
| Driver | Responder | Phenotype | UAS-ci | UAS-Mad | UAS-panTCF |
| GMR-GAL4 | UAS-CBP FL | Severe rough eye | No effect | Enhance | No effect |
| GMR-GAL4 | UAS-CBP ΔNZK | Moderate rough eye | NA | Enhance | Enhance |
| GMR-GAL4 | UAS-CBP ΔBHQ | Severe rough eye | No effect | NA | NA |
| GMR-GAL4 | UAS-CBP ΔHQ | Mild rough eye | No effect | NA | NA |
| GMR-GAL4 | UAS-CBP ΔQ | Severe rough eye | No effect | Enhance | No effect |
NA, not applicable.
DISCUSSION
The optical constraints of the adult Drosophila compound eye require that during development every cell must make the appropriate cell fate choice and position itself correctly within the growing retinal lattice. Early models predicted that each cell would express an “individualized” set of membrane-bound receptors and specific DNA-binding transcription factors, which would then be linked to the basal transcriptional machinery by yet another set of “personalized” bridging molecules. However, experimental evidence points to a much more complicated mechanism for producing the fly eye. It is clear that a cell within the developing eye will be presented with many extracellular signals and will express several receptors along with overlapping sets of transcription factors. How a cell sorts through this information and ultimately makes the correct choice is a problem that is not restricted to the insect eye but rather is a common theme in metazoan development. The fly eye has proven to be a tractable model system for unraveling this paradigm because it has a relatively small number of different cell types, its stereotyped development has been extensively studied, and it is amenable to a wide range of genetic and molecular manipulations.
In a screen for new genes involved in eye development we identified Drosophila CBP as a modifier of a dominant negative allele of the eye specification gene sine oculis. CBP is encoded by the nejire locus and belongs to the CBP/p300 family of proteins (Akimaru et al. 1997b; Goodman and Smolik 2000). Mutations within human CBP are the underlying cause of Rubinstein-Taybi syndrome and as such CBP/p300 has been implicated in regulating key events in development including the formation of the eye (Roy et al. 1968; Levy 1976; Ramakrishnan et al. 1990; Silengo et al. 1990; Guion-Almeida and Richieri-Costa 1992; Petrij et al. 1995; Tanaka et al. 1997; Oike et al. 1999; van Genderen et al. 2000; Murata et al. 2001; Coupry et al. 2002; Kalkhoven et al. 2003). Of particular interest are the demonstrated roles of CBP in (1) bridging specific DNA-binding transcription factors to the basal transcriptional machinery; (2) regulating transcription on a global scale by acetylating histones; (3) serving as a molecular scaffold by interacting simultaneously with a myriad of proteins whose numbers to date have swelled past 100; and (4) activating transcription through its alanine- and glutamine-rich domains. Furthermore, CBP is known to bind to terminal members of several signaling cascades that are known to function during retinal development and is suggested to interact with the mammalian homologs of the eye specification genes sine oculis, eyes absent, and dachshund (Goldman et al. 1997; Goodman and Smolik 2000; Ikeda et al. 2002).
In this report we have demonstrated that CBP plays a crucial role in eye development at successive stages. First, we have shown that CBP is expressed in all cells within the developing eye imaginal disc. Second, we have demonstrated that CBP interacts genetically with a member of the eye specification cascade and that eye development is sensitive to the levels of CBP. Third, loss-of-function CBP mutations affect the expression of several eye specification genes within the embryonic visual system, protocerebrum, mesoderm, and the developing eye imaginal disc. Fourth, using viable loss-of-function allelic combinations, loss-of-function retinal clones, and RNAi interference, we have demonstrated that each cell type in the developing eye, with the exception of the founder R8 photoreceptor, requires CBP for its specification. Finally, using a “pathway interference” approach we have shown that CBP likely functions in the R3/R4 cell fate choice and in the specification of the R7 photoreceptor.
The results presented here indicate a role for CBP in a myriad of developmental decisions within the developing fly retina. It remains to be determined if these effects are through repeated interactions with a small set of master regulatory proteins or with a larger set of signaling molecules and cell-subtype-specific transcription factors. It is more likely that the latter scenario will be correct. This is based on the large body of biochemical data that suggest CBP interacts with >100 proteins that are members of many diverse signaling cascades. Furthermore, to our knowledge no single gene has been shown to affect all of the processes that require the activity of CBP. Thus our hypothesis is that CBP functions as a connecting point for signaling, transcription, and chromatin remodeling during all phases of fly eye development.
The sheer number of potential interactions mediated by CBP makes an analysis of this protein inherently difficult. To circumvent this potential problem, we used a pathway interference approach to dissect CBP function by expressing a series of truncated CBPs within the developing eye. The underlying idea behind this approach is that each protein variant will act as a protein sink and soak up a unique set of endogenous factors, thus providing insight into the processes that are affected by CBP. It also provides a first step toward understanding the role that each domain of CBP plays in the developmental process and lays the groundwork for identifying critical components using more biochemical methods. The target proteins are likely to interact with CBP at stoichiometric levels during normal development. However, by increasing the dosage of CBP, the amount of these proteins within a cell becomes limiting and loss-of-function phenotypes can be observed. This approach successfully revealed roles for CBP in the R3/R4 cell fate choice and in R7 fate specification.
How CBP functions in any of these processes is still an unanswered question. Our attempts to identify additional components of the regulatory network disrupted by expression of variant CBPs through the restoration of putative interacting and downstream factors were unsuccessful. The addition of any one single factor was insufficient to rescue the effects of any of the CBP variants (Table 1). Although it is possible that none of the correct factors were tested, it is more likely that the observed phenotypes result from the loss of several proteins and adding just one is insufficient to restore normal eye development.
How are so many developmental decisions in the developing eye regulated by CBP? On the basis of reported roles for CBP/p300 in mammalian development, CBP would appear to be the perfect candidate to act as a “network manager” during eye development. A scenario can be envisioned in which every cell within the eye disc expresses CBP and a specific combination of transcription factors; some are present in restricted expression patterns while other are more promiscuously expressed. As signals are interpreted at the cell surface and transmitted into the nucleus, the CBP-transcription factor scaffold would interact with terminal members of signaling cascades and execute these instructions by modulating transcription of downstream target genes. Late in development this would translate into the differentiation of specific cell types—photoreceptors, cone cells, pigment cells, and mechanosensory bristles. This is an attractive model for several reasons. First, the uncommonly high number of described biochemical interactions suggests that CBP may act as a link between signaling pathways, specific DNA-binding proteins, and the basal transcriptional machinery. These qualities have been shown to be true in vitro. Second, it allows for individual cells to receive several common-use signals but then personalize the output. Third, the ability to interact with members of signaling pathways as well as remodel chromatin allows for very efficient transduction of extracellular instructions. This may be important for the recruitment of photoreceptors into the ommatidial cluster, a process that occurs over a relatively short period of time. This model can be extended to early events in eye specification. CBP is expressed in all cells of the eye and antennal tissues during early development (data not shown), while expression of selector genes is restricted to the individual tissues (Kumar and Moses 2001a; Kenyon et al. 2003). Signaling pathways that include Notch, Egfr, Hh, Dpp, and Wg are known to influence both eye and antennal development (Heberlein et al. 1993; Ma et al. 1993; Ma and Moses 1995; Treisman and Rubin 1995; Hsiao et al. 2001; Kumar 2001; Kumar and Moses 2001a; Baonza and Freeman 2002; Voas and Rebay 2004). CBP may mediate the interactions between signaling pathways and these selector genes, thereby participating in the process of subdividing the eye-antennal disc into the eye and antenna proper.
Previous reports of CBP in the eye have focused on the role of CBP in the modulation of polyglutamine diseases and retinal degeneration (Ludlam et al. 2002; Taylor et al. 2003). The work presented here extends these results and points to a role for CBP both in early eye determination and later in cell fate specification (Figure 11). Our results that pertain to early eye determination are supported by the synergistic interactions between CBP and SIX, EYA, and DACH proteins observed in mammals (Ikeda et al. 2002). Furthermore, we have demonstrated a role for CBP in the development of several photoreceptor cell subtypes including the R7 neuron (Figure 11). In recent years it has become increasingly clear that the molecules and mechanisms that control eye development have been preserved in both mammalian and invertebrate retinal systems. It will be interesting to elucidate the molecular and biochemical mechanisms by which CBP influences early eye specification and later photoreceptor cell fate decisions in both invertebrate and mammalian retinal systems.
Figure 11.—
Schematic of steps in eye development regulated by CBP. CBP has been shown to interact with the eye specification gene sine oculis and regulates the expression of eyes absent. These interactions happen ahead of the advancing morphogenetic furrow (purple text). Behind the morphogenetic furrow CBP functions during many stages of ommatidial assembly. Interestingly, the development of the R8 founder cell is not dependent upon CBP. We have demonstrated a strong requirement for CBP in the R3, R4, and R7 photoreceptors. We have yet to determine a requirement for CBP in the R2/R5 and R1/R6 pairs.
Acknowledgments
We thank Kathy Matthews, Susan Strome, and Peter Cherbas for comments and suggestions on this manuscript. We also thank Andrew Jarman for providing the Atonal antibody; Sarah Smolik for providing the CBP antibody and both UAS-CBP FL and UAS-CBP FL-AD fly stocks; Lucy Cherbas for providing the GMR-GAL4 fly stock; Walter Gehring for providing the ey-GAL4 fly stock; Nancy Thompson for generating the UAS-CBP ΔHQ and UAS-CBP RNAi transformants; the Bloomington Stock Center for the lz-GAL4, sev-GAL4, soD/CyO, eyFLP, and FRT101 fly stocks; and the Developmental Studies Hybridoma Bank for the Dachshund and Eyes Absent antibodies. This work has been supported by a grant from the National Eye Institute (R01 EY014863) to J.P.K.
References
- Ait-Si-Ali, S., D. Carlisi, S. Ramirez, L. C. Upegui-Gonzalez, A. Duquet et al., 1999. Phosphorylation by p44 MAP kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun. 262: 157–162. [DOI] [PubMed] [Google Scholar]
- Akimaru, H., Y. Chen, P. Dai, D. X. Hou, M. Nonaka et al., 1997. a Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386: 735–738. [DOI] [PubMed] [Google Scholar]
- Akimaru, H., D. X. Hou and S. Ishii, 1997. b Drosophila CBP is required for dorsal-dependent twist gene expression. Nat. Genet. 17: 211–214. [DOI] [PubMed] [Google Scholar]
- Arany, Z., W. R. Sellers, D. M. Livingston and R. Eckner, 1994. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 77: 799–800. [DOI] [PubMed] [Google Scholar]
- Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine et al., 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89: 1175–1184. [DOI] [PubMed] [Google Scholar]
- Baker, N. E., 2001. Cell proliferation, survival, and death in the Drosophila eye. Semin. Cell Dev. Biol. 12: 499–507. [DOI] [PubMed] [Google Scholar]
- Bannister, A. J., and T. Kouzarides, 1996. The CBP co-activator is a histone acetyltransferase. Nature 384: 641–643. [DOI] [PubMed] [Google Scholar]
- Bantignies, F., R. H. Goodman and S. M. Smolik, 2000. Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers. Mol. Cell. Biol. 20: 9317–9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baonza, A., and M. Freeman, 2002. Control of Drosophila eye specification by Wingless signalling. Development 129: 5313–5322. [DOI] [PubMed] [Google Scholar]
- Basler, K., and E. Hafen, 1990. Receptor tyrosine kinases mediate cell-cell interactions during Drosophila development. Prog. Growth Factor Res. 2: 15–27. [DOI] [PubMed] [Google Scholar]
- Basler, K., P. Siegrist and E. Hafen, 1989. The spatial and temporal expression pattern of sevenless is exclusively controlled by gene-internal elements. EMBO J. 8: 2381–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowtell, D. D., T. Lila, W. M. Michael, D. Hackett and G. M. Rubin, 1991. Analysis of the enhancer element that controls expression of sevenless in the developing Drosophila eye. Proc. Natl. Acad. Sci. USA 88: 6853–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, C. A., M. Ashburner and K. Moses, 1998. Ecdysone pathway is required for furrow progression in the developing Drosophila eye. Development 125: 2653–2664. [DOI] [PubMed] [Google Scholar]
- Cagan, R. L., and D. F. Ready, 1989. a The emergence of order in the Drosophila pupal retina. Dev. Biol. 136: 346–362. [DOI] [PubMed] [Google Scholar]
- Cagan, R. L., and D. F. Ready, 1989. b Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 3: 1099–1112. [DOI] [PubMed] [Google Scholar]
- Callaerts, P., G. Halder and W. J. Gehring, 1997. PAX-6 in development and evolution. Annu. Rev. Neurosci. 20: 483–532. [DOI] [PubMed] [Google Scholar]
- Chakravarti, D., V. J. LaMorte, M. C. Nelson, T. Nakajima, I. G. Schulman et al., 1996. Role of CBP/P300 in nuclear receptor signalling. Nature 383: 99–103. [DOI] [PubMed] [Google Scholar]
- Chan, H. M., and N. B. La Thangue, 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114: 2363–2373. [DOI] [PubMed] [Google Scholar]
- Cherbas, L., X. Hu, I. Zhimulev, E. Belyaeva and P. Cherbas, 2003. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development 130: 271–284. [DOI] [PubMed] [Google Scholar]
- Cheyette, B. N., P. J. Green, K. Martin, H. Garren, V. Hartenstein et al., 1994. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12: 977–996. [DOI] [PubMed] [Google Scholar]
- Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy et al., 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859. [DOI] [PubMed] [Google Scholar]
- Cohen, S. M., 1993 Imaginal disc development, pp. 747–841 in The Development of Drosophila melanogater, edited by M. Bate and A. Martinez Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Coupry, I., C. Roudaut, M. Stef, M. A. Delrue, M. Marche et al., 2002. Molecular analysis of the CBP gene in 60 patients with Rubinstein-Taybi syndrome. J. Med. Genet. 39: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny, T., G. Halder, U. Kloter, A. Souabni, W. J. Gehring et al., 1999. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell 3: 297–307. [DOI] [PubMed] [Google Scholar]
- Deng, Z., C. J. Chen, M. Chamberlin, F. Lu, G. A. Blobel et al., 2003. The CBP bromodomain and nucleosome targeting are required for Zta-directed nucleosome acetylation and transcription activation. Mol. Cell. Biol. 23: 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, B., and E. Hafen, 1993 Genetic dissection of eye development in Drosophila, pp 1327–1362 in The Development of Drosophila melanogater, edited by M. Bate and A. Martinez Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Florence, B., and W. McGinnis, 1998. A genetic screen of the Drosophila X chromosome for mutations that modify Deformed function. Genetics 150: 1497–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, G. V., A. Daga, H. R. Kalhor and U. Banerjee, 1998. Lozenge is expressed in pluripotent precursor cells and patterns multiple cell types in the Drosophila eye through the control of cell-specific transcription factors. Development 125: 3681–3687. [DOI] [PubMed] [Google Scholar]
- Frangioni, J. V., L. M. LaRiccia, L. C. Cantley and M. R. Montminy, 2000. Minimal activators that bind to the KIX domain of p300/CBP identified by phage display screening. Nat. Biotechnol. 18: 1080–1085. [DOI] [PubMed] [Google Scholar]
- Goldman, P. S., V. K. Tran and R. H. Goodman, 1997. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog. Horm. Res. 52: 103–119. [PubMed] [Google Scholar]
- Goodman, R. H., and S. Smolik, 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14: 1553–1577. [PubMed] [Google Scholar]
- Gu, W., X. L. Shi and R. G. Roeder, 1997. Synergistic activation of transcription by CBP and p53. Nature 387: 819–823. [DOI] [PubMed] [Google Scholar]
- Guion-Almeida, M. L., and A. Richieri-Costa, 1992. Callosal agenesis, iris coloboma, and megacolon in a Brazilian boy with Rubinstein-Taybi syndrome. Am. J. Med. Genet. 43: 929–931. [DOI] [PubMed] [Google Scholar]
- Hafen, E., B. Dickson, T. Raabe, D. Brunner, N. Oellers et al., 1993 Genetic analysis of the sevenless signal transduction pathway of Drosophila. Dev. Suppl., 41–46. [PubMed]
- Halder, G., P. Callaerts and W. J. Gehring, 1995. New perspectives on eye evolution. Curr. Opin. Genet. Dev. 5: 602–609. [DOI] [PubMed] [Google Scholar]
- Halder, G., P. Callaerts, S. Flister, U. Walldorf, U. Kloter et al., 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125: 2181–2191. [DOI] [PubMed] [Google Scholar]
- Hauck, B., W. J. Gehring and U. Walldorf, 1999. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc. Natl. Acad. Sci. USA 96: 564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, B. A., D. A. Wassarman and G. M. Rubin, 1995. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83: 1253–1262. [DOI] [PubMed] [Google Scholar]
- Heberlein, U., and K. Moses, 1995. Mechanisms of Drosophila retinal morphogenesis: the virtues of being progressive. Cell 81: 987–990. [DOI] [PubMed] [Google Scholar]
- Heberlein, U., T. Wolff and G. M. Rubin, 1993. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75: 913–926. [DOI] [PubMed] [Google Scholar]
- Hsiao, F. C., A. Williams, E. L. Davies and I. Rebay, 2001. Eyes Absent mediates cross-talk between retinal determination genes and the receptor tyrosine kinase signaling pathway. Dev. Cell 1: 51–61. [DOI] [PubMed] [Google Scholar]
- Ikeda, K., Y. Watanabe, H. Ohto and K. Kawakami, 2002. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol. Cell. Biol. 22: 6759–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, N. A., Y. M. Kuo, Y. H. Sun and S. K. Beckendorf, 1998. The Drosophila Pax gene eye gone is required for embryonic salivary duct development. Development 125: 4163–4174. [DOI] [PubMed] [Google Scholar]
- Kalkhoven, E., J. H. Roelfsema, H. Teunissen, A. den Boer, Y. Ariyurek et al., 2003. Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 12: 441–450. [DOI] [PubMed] [Google Scholar]
- Kee, B. L., J. Arias and M. R. Montminy, 1996. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J. Biol. Chem. 271: 2373–2375. [DOI] [PubMed] [Google Scholar]
- Kenyon, K. L., S. S. Ranade, J. Curtiss, M. Mlodzik and F. Pignoni, 2003. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell 5: 403–414. [DOI] [PubMed] [Google Scholar]
- Kraus, W. L., E. T. Manning and J. T. Kadonaga, 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19: 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, J. P., 2001. Signalling pathways in Drosophila and vertebrate retinal development. Nat. Rev. Genet. 2: 846–857. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., and K. Moses, 1997. Transcription factors in eye development: A georgeous mosaic? Genes Dev. 11: 2023–2028. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., and K. Moses, 2001. a EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell 104: 687–697. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., and K. Moses, 2001. b Expression of evolutionarily conserved eye specification genes during Drosophila embryogenesis. Dev. Genes Evol. 211: 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, J. P., and K. Moses, 2001. c Eye specification in Drosophila: perspectives and implications. Semin. Cell Dev. Biol. 12: 469–474. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., M. Tio, F. Hsiung, S. Akopyan, L. Gabay et al., 1998. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development 125: 3875–3885. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., G. S. Wilkie, H. Tekotte, K. Moses and I. Davis, 2001. Perturbing nuclear transport in Drosophila eye imaginal discs causes specific cell adhesion and axon guidance defects. Dev. Biol. 240: 315–325. [DOI] [PubMed] [Google Scholar]
- Kurata, S., M. J. Go, S. Artavanis-Tsakonas and W. J. Gehring, 2000. Notch signaling and the determination of appendage identity. Proc. Natl. Acad. Sci. USA 97: 2117–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger et al., 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370: 223–226. [DOI] [PubMed] [Google Scholar]
- Lee, J. D., and J. E. Treisman, 2001. The role of Wingless signaling in establishing the anteroposterior and dorsoventral axes of the eye disc. Development 128: 1519–1529. [DOI] [PubMed] [Google Scholar]
- Leiserson, W. M., S. Benzer and N. M. Bonini, 1998. Dual functions of the Drosophila eyes absent gene in the eye and embryo. Mech. Dev. 73: 193–202. [DOI] [PubMed] [Google Scholar]
- Levy, N. S., 1976. Juvenile glaucoma in the Rubinstein-Taybi syndrome. J. Pediatr. Ophthalmol. 13: 141–143. [PubMed] [Google Scholar]
- Lilja, T., D. Qi, M. Stabell and M. Mannervik, 2003. The CBP coactivator functions both upstream and downstream of Dpp/Screw signaling in the early Drosophila embryo. Dev. Biol. 262: 294–302. [DOI] [PubMed] [Google Scholar]
- Ludlam, W. H., M. H. Taylor, K. G. Tanner, J. M. Denu, R. H. Goodman et al., 2002. The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosophila melanogaster. Mol. Cell. Biol. 22: 3832–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C., and K. Moses, 1995. Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development 121: 2279–2289. [DOI] [PubMed] [Google Scholar]
- Ma, C., Y. Zhou, P. A. Beachy and K. Moses, 1993. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75: 927–938. [DOI] [PubMed] [Google Scholar]
- Manning, E. T., T. Ikehara, T. Ito, J. T. Kadonaga and W. L. Kraus, 2001. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21: 3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon, G., N. M. Solomon and G. M. Rubin, 1994. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120: 3473–3486. [DOI] [PubMed] [Google Scholar]
- Marek, K. W., N. Ng, R. Fetter, S. Smolik, C. S. Goodman et al., 2000. A genetic analysis of synaptic development: pre- and postsynaptic dCBP control transmitter release at the Drosophila NMJ. Neuron 25: 537–547. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas, M. A., A. J. Bannister, K. Martin, P. Haus-Seuffert, M. Meisterernst et al., 1998. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 17: 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus, K. J., and M. J. Hendzel, 2001. CBP, a transcriptional coactivator and acetyltransferase. Biochem. Cell Biol. 79: 253–266. [PubMed] [Google Scholar]
- Mitashov, V. I., and S. Koussulakos, 2001. Molecular mechanisms of development and differentiation of eye structures in Drosophila and vertebrates. Ontogenez 32: 14–28 (in Russian). [PubMed] [Google Scholar]
- Moses, K., M. C. Ellis and G. M. Rubin, 1989. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature 340: 531–536. [DOI] [PubMed] [Google Scholar]
- Murata, T., R. Kurokawa, A. Krones, K. Tatsumi, M. Ishii et al., 2001. Defect of histone acetyltransferase activity of the nuclear transcriptional coactivator CBP in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 10: 1071–1076. [DOI] [PubMed] [Google Scholar]
- Nordheim, A., 1994. Transcription factors. CREB takes CBP to tango. Nature 370: 177–178. [DOI] [PubMed] [Google Scholar]
- Oike, Y., A. Hata, T. Mamiya, T. Kaname, Y. Noda et al., 1999. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum. Mol. Genet. 8: 387–396. [DOI] [PubMed] [Google Scholar]
- Pappu, K. S., R. Chen, B. W. Middlebrooks, C. Woo, U. Heberlein et al., 2003. Mechanism of hedgehog signaling during Drosophila eye development. Development 130: 3053–3062. [DOI] [PubMed] [Google Scholar]
- Petrij, F., R. H. Giles, H. G. Dauwerse, J. J. Saris, R. C. Hennekam et al., 1995. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376: 348–351. [DOI] [PubMed] [Google Scholar]
- Petruk, S., Y. Sedkov, S. Smith, S. Tillib, V. Kraevski et al., 2001. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294: 1331–1334. [DOI] [PubMed] [Google Scholar]
- Pignoni, F., and S. L. Zipursky, 1997. Induction of Drosophila eye development by decapentaplegic. Development 124: 271–278. [DOI] [PubMed] [Google Scholar]
- Pignoni, F., B. Hu, H. Z. Kenton, J. Xiao, P. A. Garrity et al., 1997. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91: 881–891. [DOI] [PubMed] [Google Scholar]
- Quiring, R., U. Walldorf, U. Kloter and W. J. Gehring, 1994. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 265: 785–789. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, S., D. C. Sharma, V. Ramakrishnan, P. S. Parihar, S. Sharma et al., 1990. Rubinstein-Taybi syndrome. Indian Pediatr. 27: 404–405. [PubMed] [Google Scholar]
- Ready, D. F., T. E. Hanson and S. Benzer, 1976. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53: 217–240. [DOI] [PubMed] [Google Scholar]
- Roy, F. H., R. L. Summitt, R. L. Hiatt and J. G. Hughes, 1968. Ocular manifestations of the Rubinstein-Taybi syndrome. Case report and review of the literature. Arch. Ophthalmol. 79: 272–278. [DOI] [PubMed] [Google Scholar]
- Royet, J., and R. Finkelstein, 1997. Establishing primordia in the Drosophila eye-antennal imaginal disc: the roles of decapentaplegic, wingless and hedgehog. Development 124: 4793–4800. [DOI] [PubMed] [Google Scholar]
- Seimiya, M., and W. J. Gehring, 2000. The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development 127: 1879–1886. [DOI] [PubMed] [Google Scholar]
- Serikaku, M. A., and J. E. O'Tousa, 1994. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics 138: 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W. F., K. Krishnan, H. J. Lawrence and C. Largman, 2001. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell. Biol. 21: 7509–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., and C. Mello, 1998. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12: 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo, B. Z., 1992. Roles of receptor tyrosine kinases in Drosophila development. FASEB J. 6: 2915–2922. [DOI] [PubMed] [Google Scholar]
- Silengo, M. C., M. Lerone, A. Pelizza, R. Gatti, A. Barabino et al., 1990. A new syndrome with cerebro-oculo-skeletal-renal involvement. Pediatr. Radiol. 20: 612–614. [DOI] [PubMed] [Google Scholar]
- Takaesu, N. T., A. N. Johnson, O. H. Sultani and S. J. Newfeld, 2002. Combinatorial signaling by an unconventional Wg pathway and the Dpp pathway requires Nejire (CBP/p300) to regulate dpp expression in posterior tracheal branches. Dev. Biol. 247: 225–236. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., I. Naruse, T. Maekawa, H. Masuya, T. Shiroishi et al., 1997. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. USA 94: 10215–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J. P., A. A. Taye, C. Campbell, P. Kazemi-Esfarjani, K. H. Fischbeck et al., 2003. Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev. 17: 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson, A., and D. F. Ready, 1987. Cell fate in the Drosophila ommatidium. Dev. Biol. 123: 264–275. [DOI] [PubMed] [Google Scholar]
- Tomlinson, A., B. E. Kimmel and G. M. Rubin, 1988. rough, a Drosophila homeobox gene required in photoreceptors R2 and R5 for inductive interaction in the developing eye. Cell 55: 771–784. [DOI] [PubMed] [Google Scholar]
- Treisman, J. E., and U. Heberlein, 1998. Eye development in Drosophila: formation of the eye field and control of differentiation. Curr. Top. Dev. Biol. 39: 119–157. [DOI] [PubMed] [Google Scholar]
- Treisman, J. E., and G. M. Rubin, 1995. wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development 121: 3519–3527. [DOI] [PubMed] [Google Scholar]
- Treisman, J. E., and G. M. Rubin, 1996. Targets of glass regulation in the Drosophila eye disc. Mech. Dev. 56: 17–24. [DOI] [PubMed] [Google Scholar]
- van Genderen, M. M., G. F. Kinds, F. C. Riemslag and R. C. Hennekam, 2000. Ocular features in Rubinstein-Taybi syndrome: investigation of 24 patients and review of the literature. Br. J. Ophthalmol. 84: 1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voas, M. G., and I. Rebay, 2004. Signal integration during development: insights from the Drosophila eye. Dev. Dyn. 229: 162–175. [DOI] [PubMed] [Google Scholar]
- Waltzer, L., and M. Bienz, 1998. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature 395: 521–525. [DOI] [PubMed] [Google Scholar]
- Waltzer, L., and M. Bienz, 1999. A function of CBP as a transcriptional co-activator during Dpp signalling. EMBO J. 18: 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1991. a The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113: 841–850. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1991. b Cell death in normal and rough eye mutants of Drosophila. Development 113: 825–839. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1993 Pattern formation in the Drosophila retina, pp. 1277–1326 in The Development of Drosophila melanogaster, edited by M. Bate and A. Martinez Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Zheng, L., J. Zhang and R. W. Carthew, 1995. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development 121: 3045–3055. [DOI] [PubMed] [Google Scholar]