Abstract
The cornerstone of population genetics is a probabilistic understanding of the ultimate fate—survival or extinction—of rare mutations. If a mutation is beneficial, it enables its carrier to reproduce faster than native wild-type individuals. In classic derivations and in the considerable body of research that has followed, “faster” has been defined mathematically to mean “able to produce more surviving offspring per generation.” Many organisms, however, may increase their reproductive rate by producing the same number of offspring in a shorter generation time: a mutant bacterium, for example, may complete the cell cycle and produce two offspring more quickly than the wild type. We find that the ultimate fixation probability of a mutation conferring a shorter generation time differs from that of a mutation conferring more offspring by a factor of 2 ln(2)—nearly 40%. This predicts a reduction in the overall substitution rate for any mutation that decreases the generation time: fixation probability is biased toward increased offspring number.
THE selective advantage, as classically defined by Fisher (1930) and Wright (1931), is realized in terms of fecundity: if the wild type has on average one offspring per generation, a rare beneficial mutant has on average (1 + s) offspring per generation. This model of the selective advantage is fundamental to the classic publications in population genetics (Haldane 1927, 1932; Kimura 1957, 1962) and has been assumed, explicitly or implicitly, in the considerable body of literature that has followed. Using increased fecundity in this way to model the selective advantage is not only mathematically convenient, but also appropriate from a biological point of view. Specifically, the (1 + s) model captures any mechanism that changes the mean number of offspring that survive to reproductive maturity.
For many organisms, however, a mutant may have the same number of offspring as the wild type on average, but may produce these offspring in a reduced generation time. A simple example here is bacterial fission. In an environment that favors growth, each bacterium will produce, on average, close to two surviving offspring; the enhanced growth of a beneficial mutant can then be realized as a reduced cell cycle time. This may be particularly true in the presence of antibiotics, which reduce the rate of cell growth in drug-sensitive strains. Similarly, the number of virions released during lysis may be limited by the size of the host cell; mutant strains that have a shorter replication time will realize their selective advantage by reaching this limit more quickly. To our knowledge, the effect of this alternative mechanism of the selective advantage has not been explored.
In the sections that follow, the fate of a rare beneficial mutation is determined, assuming that the selective advantage is realized in terms of decreased generation time. Perhaps surprisingly, the fixation probability of such a mutant differs significantly from the fixation probability for a mutant with an equivalent advantage in fecundity. Before describing the analytical work, however, we present an illustrative example to build some intuition about this effect.
ILLUSTRATIVE EXAMPLE
For our example, we consider a population of lytic viruses. As is typical for fixation probabilities, the exact size of the population is unimportant, as long as the wild-type population is sufficiently “large.” We also allow unmitigated exponential growth, at least over a few generations, in our sample population. While ignoring the need to keep growth in check greatly simplifies the example, we do not allow unbounded growth in the analysis that follows.
We assume that the viruses are not perfectly adapted to their environment; that is, beneficial mutations are possible. In particular, we consider two different mutations that might theoretically be available to our population. Mutation F, for fecundity, increases the number of infectious virions produced per infected cell. Mutation G, for generation time, produces the same number of virions as the wild type, but produces them in a shorter time, decreasing the time until cell lysis.
To make our example concrete, we suppose that each cell infected by the wild-type virus will release, on average, two infectious virions at cell lysis, which occurs 1 time unit after the cell was infected. Mutant F, in contrast, is able to produce 2.02 infectious virions on average. Mutant G, like the wild type, produces two infectious virions on average, but produces these in a slightly reduced time, 0.986 time units.
This value of the generation time was chosen such that the long-term growth rate of the two mutants is equivalent. In particular, the expected number of infectious virions produced in t time units by virus F is 2.02t. Likewise, the number of copies of virus G after t time units is 2t/0.986. Manipulating the latter expression we find that
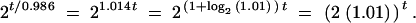 |
Thus the long-term growth rates of mutant F and mutant G are exactly the same. Typically, if we were introducing either mutant into a mathematical model we would say that the growth rate is given by (2(1 + s))t and that both mutants F and G have the same selective advantage, s = 0.01. Likewise, if we were experimentally measuring the growth rate of F or G in a fitness assay, we would measure the same value for s. Finally, if either F or G occurred de novo in a population, the expected time to fixation would be precisely the same for the two mutant strains.
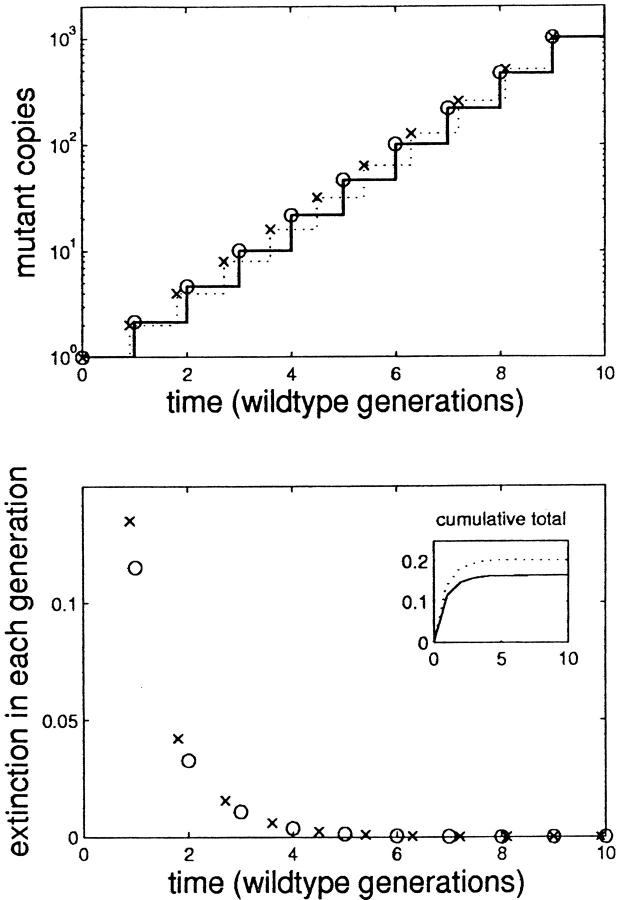
This effect is illustrated in the top of Figure 1. The solid line shows the growth of mutant F (fecundity advantage) over 10 generations of unimpeded growth; in each generation, the number of copies of F increases by a factor of 2(1 + s). The dotted line, in contrast, shows the growth of mutant G (reduced generation time). The number of copies of G increases by only a factor of 2 in each generation, but the generation times are slightly shorter. Circles and crosses show the number of copies after each lysis event and illustrate that after 9 time units, both F and G have exactly the same number of copies at exactly the same instant. The overall growth rates of the two mutant strains are identical.
Figure 1.—
Growth and extinction probability of two mutant strains. In the top, the solid line shows the growth of a viral mutant that has a fecundity advantage, mutant F. In each generation, the number of copies of F is Poisson distributed and increases by a factor of 2(1 + s). The dotted line shows the growth of a different mutant that has a reduced generation time, mutant G. The number of copies of G increases by only a factor of 2 in each generation, but the generation times are slightly shorter. Circles and crosses show the number of copies after each lysis event and illustrate that after 9 time units, both F and G have exactly the same number of copies at exactly the same instant. The overall growth rates of the two mutant strains are identical. The bottom shows the probability that either F (circles) or G (crosses) have zero offspring in each successive generation. During the first few generations, mutant G has a slightly higher extinction probability because of its lower fecundity. The inset plots the cumulative extinction probability after 10 generations for mutants G (dotted line) and F (solid line).
The bottom part of Figure 1, however, illustrates that extinction probabilities for F (circles) and G (crosses) are not the same. First, note that once either mutant has survived four or five generations, the chance of extinction in future generations becomes negligible. During the first few critical generations, however, mutant G has a slightly higher extinction probability because of its lower fecundity. We have assumed a Poisson distribution of offspring, but the effect we describe depends only on the reasonable assumption that lower fecundity entails a higher probability of having zero surviving offspring in a single generation.
This difference in extinction probability between the two mutant strains accrues such that the cumulative extinction probability, shown in the inset, differs substantially depending on the mechanism of the selective advantage. Put another way, we see in the top of Figure 1 that strain G gets an extra generation of growth and “catches up” with strain F, such that over 9 time units their growth rate is precisely equivalent. Unfortunately, strain G survives long enough to realize this advantage only 80% of the time. We find that a mutation is more likely to spread through the population if it confers an increase in fecundity, rather than what would be measured in a fitness assay as an equivalent reduction in generation time. This is because even a modest increase in fecundity reduces the chance of extinction in these first, critical generations.
In the sections that follow, we derive the fixation probability for mutations that confer either a fecundity or generation time advantage. We compare these values when the population size is constant and then extend our results to consider populations in environments where growth is favored. In the latter case we assume that growth is ultimately kept in check by periodic population bottlenecks.
METHODS: ANALYTICAL DERIVATION
Our analytical work simply extends the classic derivation of the fixation probability described by Haldane (1927). We outline the basic techniques and assumptions for the general reader below; a detailed derivation is presented in the appendix.
Constant population, fecundity advantage:
In brief, we assume that a wild-type individual produces on average r offspring per generation and thus produces rt offspring in t generations. For the classic model of an increase in fecundity, we consider a mutant lineage in which the generation time remains unchanged from the wild-type value, but more offspring are produced per generation by the mutant. Thus a mutant with selective advantage s would produce (r(1 + s))t offspring in the same amount of time.
To determine the fixation probability in the simplest case with a constant population size, we use Haldane's method. If N0 denotes the total, constant size of the population, it is clear that no matter how big r is, only N0 of all newly created offspring can survive each generation. Haldane used probability-generating functions (see appendix) to describe the processes of creating new offspring and randomly determining which offspring survive to form the next generation. The effects of these two processes—growth and sampling—are combined to determine the probability that a rare mutant ultimately leaves no descendants. If fn(0) gives the probability that zero offspring have survived after n generations of growth and sampling, we find that the extinction probability v is given by
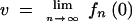 |
1 |
(Haldane 1927).
Following Haldane and others, we define the extinction probability, v, as the probability of ultimately leaving no descendants, and therefore the fixation probability u is simply given by u = 1 − v. Since we do not allow our populations to grow without bound, this is strictly true. Another implicit assumption, however, is that a beneficial mutation can be lost only by producing zero offspring in either the growth or the sampling processes. This is not strictly true, since the lineage may also be eliminated by other factors such as clonal interference (see Gerrish and Lenski 1998). The effects we describe in the sections that follow are therefore mitigated if clonal interference accounts for a substantial component of the extinction probability.
Population bottlenecks, fecundity advantage:
A population bottleneck is a severe reduction in population size, typically caused by intense competition, adverse environmental conditions, or parasitism. In many natural populations, seasonality imposes regular bottlenecks. Natural populations of parasites, similarly, are subject to severe bottlenecks in their transmission from host to host. In laboratory populations, population bottlenecks are often an inherent feature of the experimental protocol; bacterial populations, for example, may be sampled at ratios of 1:100 or more during serial passaging, usually on a daily basis (Lenski et al. 1991; Lenski and Travisano 1994). Experimental evolution in viruses likewise involves serial passaging (Kichler Holder and Bull 2001) or periodic sampling when chemostat tubes sustaining populations of phage and host are changed (Bull et al. 1997).
Haldane's approach can naturally be extended to include populations that experience growth followed by bottlenecks. We assume that these bottlenecks occur at fixed times, every τ generations. (Our analysis does not yet include situations in which population bottlenecks occur randomly, at variable times.) Once again, the appendix describes the derivation in detail. We find that the extinction probability is again given by Equation 1, but fn(x) in this case includes τ growth processes followed by one sampling process, the population bottleneck.
An important point to note is that for the typically assumed case of Poisson-distributed offspring and random sampling of offspring to obtain the next generation, a constant population size is formally equivalent to a population that undergoes bottlenecks at rate τ = 1. When deriving the fixation probability for a mutation with a reduced generation time, we therefore investigate the general case of a bottlenecked population for any integer τ. To address a constant population size, we can then consider the case τ = 1.
Reduced generation time, constant or bottlenecked population:
To model the selective advantage in terms of a reduced generation time, we find a generation time for the mutant that would produce the same overall growth rate as a (1 + s) fecundity advantage. Since the wild-type generation time is by definition 1 time unit, we use td to denote the generation time for the mutant, with td < 1. The average number of offspring for the wild type in t time units (wild-type generations) is rt. As illustrated in the example, for a mutation with a fecundity advantage, the mean number of offspring is (r(1 + s))t; for a generation time advantage, this number is rt/td.
The overall growth rate of the mutant will then be the same in both cases when the mutant generation time is
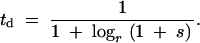 |
2 |
Once again, this equivalence is illustrated in the example.
For a small selective advantage (td ≈ 1), it is clear that after the favorable mutation first appears, both mutant and wild type will likely experience the same number of generations before the next bottleneck. The replication times of the mutant will occur progressively earlier in the growth phase, however, as the generation time advantage of the mutant accrues. Eventually, the mutant population will experience τ + 1 replications between two bottlenecks. It is fairly straightforward to include this effect in Haldane's probability-generating functions, as described in the appendix. The fixation probability can then be determined for a mutation conferring a reduced generation time but no increased fecundity.
RESULTS
Useful approximations:
For a population of constant size, it is well known that the fixation probability can be approximated by 2s, for a mutation conferring a fecundity advantage (Haldane 1927). When population bottlenecks are included in the analysis, this approximation changes to τs for moderate values of τ (Heffernan and Wahl 2002). Our overall goal was to find similar approximations for mutations conferring reduced generation times.
For an analytically tractable approximation, we imposed the condition that the mutant with a reduced generation time experiences an extra generation regularly every b bottlenecks. (Thus, in a constant population, an extra generation occurs every b wild-type generations.) This allows us to write td as the fraction τb/(τb + 1), where both τ and b are integers. For example, in Figure 1 the mutant doubles 10 times in the time it takes the wild type to double 9 times, and td would therefore be given by 18/19.
We can then use the Kolmogorov forward equation (Crow and Kimura 1970), in a derivation analogous to that described in Wahl and Gerrish (2001). In brief, the approach is an extension of classic work and allows us to approximate the fixation probability as u ≈ 1 − e−2M/V, where M is the mean number of offspring after some time step, and V is the variance of this number. In our case, we take a “time step” to be b bottlenecks. The appendix describes the calculation of M and V in more detail.
We must also decide how to model the growth and sampling of the population. If we assume that each individual in the population has exactly r offspring in each generation, and that individuals are chosen at random to survive the bottleneck, it is straightforward to show that M = r and V = r(1 + r(b − 1))(1 − r−τ). Thus
 |
in this case.
For a Poisson distribution of offspring with mean r, the derivation of V is quite difficult. In this situation, we restrict our attention to a population in which each individual produces an average of two offspring per replication, that is, r = 2. This is the natural case to consider for bacterial fission, for example. This allows us to compute M = 2 and V = 4(2b − 1)(1 − 2−τ), yielding
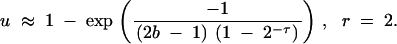 |
Although this equation is still fairly unwieldy, it allows us to make a number of further approximations. For a constant population, τ = 1. If b is moderately large (such that 2b − 1 ≈ 2b), we find u ≈ 1 − exp(−1/b). The condition that b is large implies that s is small, and thus we have the further approximation
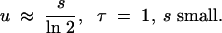 |
If we had incorrectly assumed that the mutation of interest conferred a fecundity advantage, the fixation probability would be given by 2s (Haldane 1927). Thus the error made in assuming a fecundity advantage, if in fact the mutation confers a generation time advantage, is 2 ln 2 − 1, or 39%.
Similarly, when τ is moderately large (more than about five generations between bottlenecks) we have 1 − 22−τ ≈ 1, which yields u ≈ 1 − exp(−1/2b) when s is small. Again, this yields
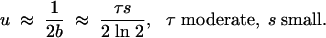 |
Assuming a fecundity advantage, the fixation probability for population doublings under bottlenecks is approximately τs for moderate values of τ (Heffernan and Wahl 2002). Thus the error in assuming a fecundity advantage is again 39%.
Numerical and simulation results:
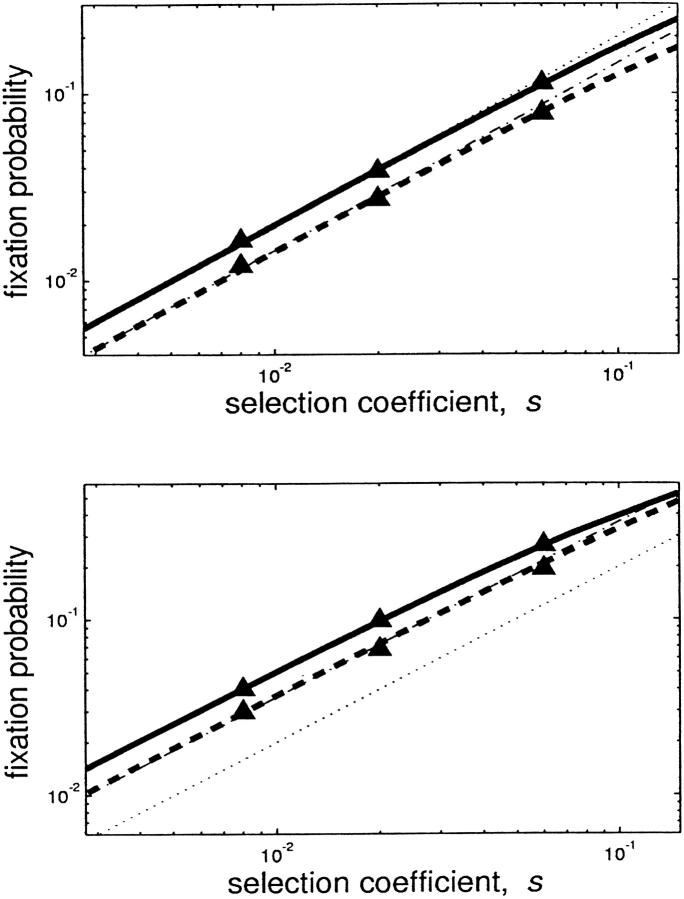
The fixation probability can also be determined numerically, that is, by taking the limit described in Equation 1 using standard computational methods. Over a large range of s, this technique verifies that the fixation probability of the mutant is significantly affected by the mechanism of the selective advantage. The top of Figure 2 shows the fixation probability when fecundity is increased (solid line) or when generation time is decreased (dashed line) as functions of s for a constant population (τ = 1). The analytical approximations 2s (dotted line) and s/ln 2 (dotted-dashed line) are also shown. The logarithmic plot shown here illustrates the power-law relation between fixation probability and s, but obscures the true magnitude of the differences involved; see Figure 3 for clarification. The bottom of Figure 2 shows the same results for τ = 5; in this case the appropriate analytical approximation is τs/(2 ln 2) (dotted-dashed line). For comparison, the classic fixation probability, 2s, is also shown (dotted line). Note that when τ ≠ 1, 2s is a very poor approximation to the fixation probability because the effects of population growth between bottlenecks have been neglected (Wahl and Gerrish 2001). These numerical results were verified using Monte Carlo simulation, in which large populations were modeled at the level of the individual bacterium (triangles).
Figure 2.—
Generation time vs. fecundity advantage. The fixation probability for a rare beneficial mutation with selective advantage s was computed as described in the text for mutants with a fecundity advantage (solid line) or an equivalent generation time advantage (dashed line). The analytical approximations 2s (dotted line) and s/ln 2 (dotted-dashed line) are also shown for comparison. In the top, the population has a constant size (τ = 1). In the bottom, the population grows for five wild-type generations, after which a bottleneck reduces the population to its initial size; this process repeats indefinitely. For both top and bottom r = 2. Triangles show the same results calculated by simulating the fate of a single beneficial mutation 100,000 times. The standard deviation of the simulated data was calculated as (u(1 − u)/105)1/2, but the resulting error bars are too small to discern on the scale of the figure.
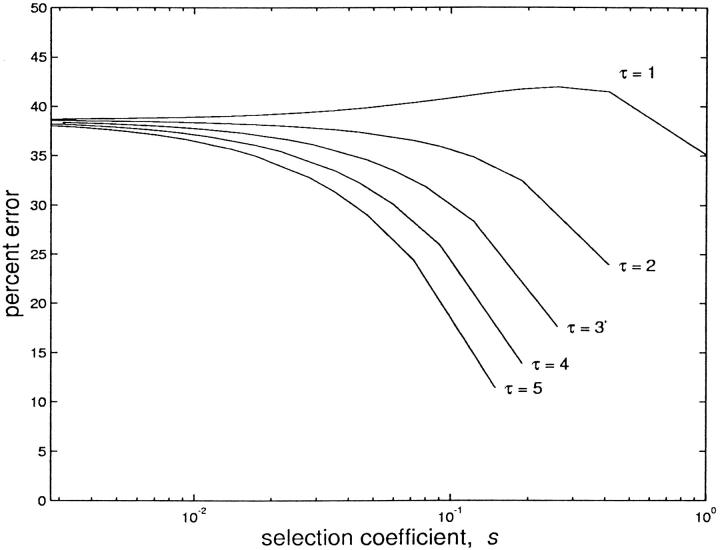
Figure 3.—
Percentage difference in fixation probability. The difference between the fixation probabilities determined with a fecundity or generation time advantage is plotted as a percentage of the fixation probability with a generation time advantage. Populations grow for τ wild-type generations between population bottlenecks, which reduce the population to its initial size; the case τ = 1 corresponds to a constant population size. In all cases r = 2. Because we consider only a limited set of s values [such that td = τb/(τb + 1)], the maximum value of s varies with τ.
The overall importance of the mechanism of the selective advantage is more clearly illustrated in Figure 3, which shows the percentage error introduced by assuming a fecundity advantage when a mutation actually confers a generation time advantage. We see that for a constant population, or for a population experiencing regular bottlenecks, this error approaches 40% for typical values of s.
Our analytical work assumes that the bottleneck occurs after an integer number of wild-type generations. We relaxed this assumption using the Monte Carlo approach, investigating bottlenecks that occur at various times between the first and second generations during the growth phase. As shown in Figure 4, the fixation probabilities for mutations conferring either a fecundity or generation time advantage are affected in complex ways by changes in the bottleneck time. Briefly, this is because the pattern of the number of generations between bottlenecks can be vastly different for noninteger bottleneck times and can in some cases greatly reduce the fixation probability conferred by a fecundity advantage. Thus, although the difference between the two cases is clearly mitigated, fixation probabilities are still significantly higher, on balance, for a fecundity advantage.
Figure 4.—
Fixation probability vs. bottleneck time. The fixation probability, u, is plotted for a fecundity advantage (circles) or a generation time advantage (squares), for a bottleneck that occurs at τ wild-type generations. Values are calculated through Monte Carlo simulation of 100,000 replications with r = 2 and s = 0.0093; error bars show the standard deviation of the computed values, given by (u(1 − u)/105)1/2.
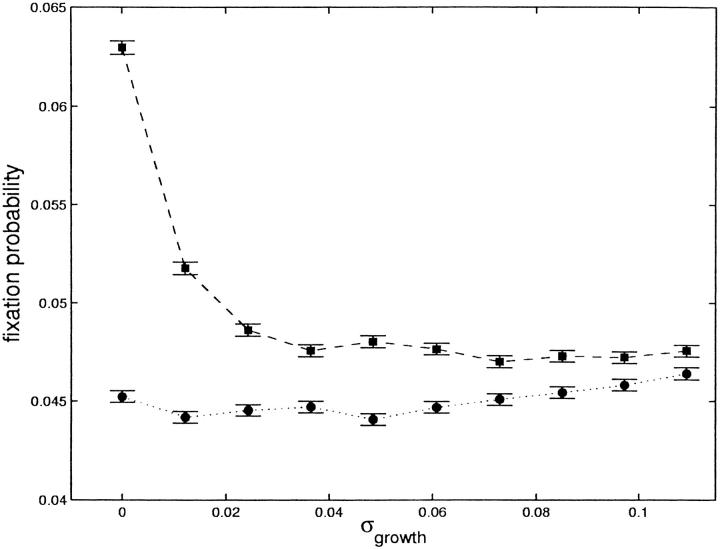
Our analytical work also assumes that the generation time for individuals carrying a beneficial mutation is constant, such that the descendants of a rare mutant all undergo fission, for example, at the same time for all future generations. Again, we investigated the effect of relaxing this assumption using the Monte Carlo approach. Here we test a case in which each offspring of the mutant, although genetically identical, has a small variation in fission time; bottlenecks occur regularly between the sixth and seventh generations of the growth phase. Figure 5 shows the fixation probability for a fecundity advantage (circles) or generation time advantage (squares) in this case. We find that fixation probability for a fecundity advantage decreases when the lifetimes of genetically identical mutants have some variation. This effect gradually erodes the difference between the two types of advantage when lifetimes are sufficiently variable. Because even small biases become relevant on evolutionary timescales, however, we predict that the bias in favor of a fecundity advantage will persist in populations with variable generation times, but will be most prevalent when there is minimal variability in the length of the reproductive cycle.
Figure 5.—
Fixation probability with stochastic generation times. The fixation probability, u, is plotted for a fecundity advantage (circles) or a generation time advantage (squares), for a bottleneck that occurs at τ = 6.91 wild-type generations; r = 2 and s = 0.0093. Values are calculated through Monte Carlo simulation with 500,000 replications; error bars show the standard deviation (u(1 − u)/5 × 105)1/2. Each generation time for each individual in the simulation is drawn from a normal distribution with standard deviation σ; the x-axis plots the total standard deviation for the descendants of the mutant at the end of the growth phase. Thus, on the right side, the fission times of the mutant offspring after the growth phase occur within a 20% window around the mean ∼95% of the time.
DISCUSSION
Theoretically, our results demonstrate that the mechanism of the selective advantage is of critical importance. In particular, the classic “2s” approximation for fixation probability does not hold for any mutation that reduces the generation time: in this case, the fixation probability should instead be approximated by s/ln 2 in a population of constant size. Figure 1 demonstrates the reasons underlying this difference: although two beneficial mutations might have the same selective advantage s, the same growth rate against the wild type, and the same expected time to fixation, the mutation that increases fecundity is more likely to survive its first few generations of growth.
In describing this process mathematically, we were forced to explicitly define the processes underlying growth in our model populations, entailing a number of assumptions. We emphasize, however, that the general results we describe depend only on one critical (but reasonable) assumption: that a slight increase in fecundity implies a slight decrease in the probability of having zero offspring in each generation.
Practically, our results will be important in estimating the substitution rate for beneficial mutations in natural and experimental populations, since the overall fixation rate will be lower than classically estimated if reductions in generation time are possible. For example, since mutations conferring antibiotic resistance typically allow bacteria to complete the cell cycle more quickly than their drug-sensitive counterparts, we might model such mutations as having a reduced generation time. Thus when predicting how often these mutations will arise de novo and survive the first few generations of growth and sampling, their reduced fixation probability should be taken into account. Our results indicate that such mutations will survive less frequently than has been previously predicted under the assumption that they confer a fecundity advantage (Levin et al. 2000).
The effect we describe is not conditional upon successive generations reproducing in lock-step. Variance in the generation times of individuals carrying the mutation of interest does, however, significantly mitigate this “fecundity bias.” Similarly, variation in the time at which the bottleneck occurs reduces this effect. Since the bias is of the order of 40% without these mitigating factors, however, reduction due to either source of variation is unlikely to eliminate the effect, especially when evolutionary timescales come into play.
Although related to the classic evolutionary trade-off between increased fecundity and reduced development time (Stearns 1992), the fecundity bias we report does not directly pertain to the optimization of these life-history parameters. Theories of “rK-selection” assume that decreased development time implies reduced fecundity (and vice versa); that is, you cannot have it both ways. In contrast, our results pertain to populations in which both strategies are available and potentially advantageous to the organism. When either reduced generation times or increased fecundity is possible, and even if they confer an equivalent growth advantage, we find that fixation is biased in favor of fecundity. This predicts, for example, that if mutations of both types are available and occur at roughly equal rates, increases in fecundity are a more likely adaptive response to a changing environment. Studies of the adaptation of fruit flies to bottle culture support our predictions, exhibiting an increase in fecundity that was not matched by reductions in generation time (Sgro and Partridge 2000). Similarly, increases in cell size are one of the clearest phenotypic changes in the adaptation of bacterial populations to serial passaging environments (Lenski et al. 1991). An increase in cell size is consistent with increased offspring survival, but is not likely to accompany decreased generation time.
Our results do not address the underlying rates at which mutations that either increase fecundity or decrease generation time might spontaneously occur. For example, if mutations that reduce the generation time occur 40% more frequently than mutations that increase fecundity, the difference in fixation probability that we describe would be entirely offset. It is perhaps impossible to predict which of these alternate mechanisms will be more “accessible,” by mutation, to an evolving population. Nonetheless we are able to conclude that the fixation probability for any mutation that decreases the generation time will be significantly less than we would previously have predicted on the basis of the selective advantage alone. Thus, observed substitution rates will not be an accurate reflection of the underlying mutation rates unless the mechanism of the fitness increase for each mutation is known. The interesting point here is that even if the population size, structure, and mating behavior can be well estimated, the fitness of a beneficial mutation is not enough to predict its fixation probability, as we had previously assumed. The mechanism by which the selective advantage is conferred is also of critical importance.
Our model assumes that the survival probability to reproductive age is the same for both fecundity and generation-time mutants. This assumption is clearly invalid when the generation times in the two cases differ significantly, suggesting a direction for future work. In particular, consider the possibility of a mutation that delays reproduction, but has a corresponding increase in fecundity, such that the overall growth rate is still (r(1 + s))t. Using the current model, we might predict that such a mutation would have an even higher fixation probability than the classic 2s approximation, implying that delaying reproduction indefinitely would maximize fixation probability. This describes a scenario in which the assumption of equal survival probabilities is clearly invalid and further assumes that an unlimited number of offspring are possible in one reproduction event.
We frame our analysis in terms that include both populations of constant size and populations that sustain periods of growth followed by population bottlenecks. Bottlenecks are ubiquitous in natural systems and may in fact be more the rule than the exception for natural populations in the face of constant seasonality and rapidly evolving parasitism. In experimental studies of evolution, serial passaging imposes severe bottlenecks, often on a daily basis; these bottlenecks have profound effects on evolutionary dynamics (Wahl et al. 2002). The results of this study may have particular relevance for these experimental systems, many of which involve microbial populations in an environment that favors growth. In this situation a decrease in generation time is clearly a possible mechanism for the selective advantage; for lytic viruses and bacterial populations in favorable environments, a shorter replication time is often a more natural assumption than an increase in offspring number. Now that detailed experimental work has begun to elucidate the molecular mechanisms underlying increased fitness in these systems (see, for example, Bull et al. 2000), it will be fascinating to determine whether the predictions of this study are realized in the laboratory.
Acknowledgments
We thank two anonymous referees for their insightful comments. This work was supported by the Natural Sciences and Engineering Research Council of Canada, the Ontario Ministry of Science, Technology, and Industry, and the SHARCNET parallel computing facility.
APPENDIX
Fixation probability with a fecundity advantage:
We assume that a wild-type individual produces on average r offspring per generation and thus produces rt offspring in t generations. A mutant with a fecundity advantage s produces (r(1 + s))t offspring in the same amount of time.
We first introduce a general method of determining the fixation probability in the classic case of a fecundity advantage and constant population size. Following Haldane (1927), we imagine that a large number of offspring are produced (the “growth” phase), of which a fraction survive (or are “sampled”) to form the next generation. Let g(x) be the probability-generating function (pgf) that describes the number of offspring produced in a single mutant lineage during one generation. A pgf is a mathematically convenient way to express a discrete probability distribution and is defined in the following way: if the probability of producing i offspring is pi, g(x) is simply formed by writing g(x) = p0 + p1x + p2x2 + … (Harris 1963). As described above, g(x) will reflect the selective advantage of the mutant and will have a larger mean than the analogous growth distribution for the wild type. For Poisson growth with mean 2(1 + s), we note that g(x) can also be written in the short-hand notation g(x) = e2(1+s)(x−1).
Let N0 denote the total size of the population, including both mutant and wild type. To maintain a constant population size, only N0 of the newly created offspring will survive each generation. Thus, if each wild-type individual produces r offspring per generation on average, the chance that each offspring survives is 1/r. We can then write the pgf for this sampling process, h(x), in exactly the same way as we formed g(x). In particular, for the sampling process we know that the probability that an individual survives is p1 = 1/r, while the probability that the individual does not survive sampling is p0 = 1 − 1/r. This allows us to write h(x) = 1 − 1/r + (1/r)x.
One of the reasons why pgfs are convenient is that to determine the overall pgf of process g followed by process h, we can simply take the composition g(h(x)), also written as g ∘ h(x). The overall probability-generating function for the mutant lineage after one generation is then f(x) = g ∘ h(x). After n generations, the probability-generating function for the total number of offspring in the mutant lineage will be
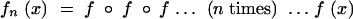 |
For any pgf f(x) = p0 + p1x + p2x2 … evaluating at x = 0 eliminates the higher terms and leaves us with p0. Haldane therefore computed the extinction probability, the probability that the lineage is ultimately eliminated, as v = limn→∞fn(0) (Haldane 1927).
Population bottlenecks:
The derivation above can naturally be extended to include populations that experience growth followed by bottlenecks. We assume that these bottlenecks occur at fixed times, every τ generations. In this case the overall generating function for the mutant lineage after one growth phase (τ successive generations of growth) and one bottleneck is
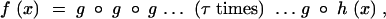 |
which we also denote gτ ∘ h(x).
An important point to note is that for the typical case of Poisson-distributed offspring and binomial sampling, a constant population size is formally equivalent to a population that undergoes bottlenecks at rate τ = 1. This is because for g(x) = er(1+s)(x−1) and h(x) = 1 − 1/r + (1/r)x, we find that g ∘ h(x) = e(1+s)(x−1).
Fixation probability with a reduced generation time:
We let td denote the generation time for a mutant with a reduced generation time (but no fecundity advantage), such that td < 1. The average number of offspring for the wild type in t time units (wild-type generations) is rt. For a mutation with a fecundity advantage, the mean number of offspring is (r(1 + s))t; for a generation time advantage, this number is rt/td.
The overall growth rate of the mutant will then be the same in both cases when the mutant generation time is td = 1/(1 + logr(1 + s)) = 1/(1 + s′). Here for convenience the notation s′ has been introduced, where s′ = logr(1 + s); note that when s is small, s′ ≈ s/log(r).
For the mutation with a generation time advantage, the mutant population will eventually experience τ + 1 generations between two bottlenecks. For a mutation that first arises at the beginning of a growth phase, this will first occur before bottleneck n1, where
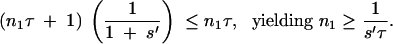 |
Extending this logic, it can be seen that the successive bottlenecks ni before which the mutant strain experiences an extra generation are given by the smallest integer that is greater than i/(s′τ).
The probability-generating function for the mutant lineage is therefore a composition of f+(x) = gτ+1 ∘ h(x) for growth and sampling phases where an extra generation occurs, and f(x) = gτ ∘ h(x) for all other growth and sampling phases. For example, if τ = 2 and s′ = 1/6, the generating function for the mutant strain after six bottlenecks would be f6(x) = f ∘ f ∘ f+ ∘ f ∘ f ∘ f+.
Once again, the extinction probability can then be determined by evaluating Equation 1.
Finally, we make use of the known mean and variance of a pgf to evaluate M and V as described in the Useful approximations section. For any pgf f(x), the mean of the underlying distribution is given by f′(1) and the variance by f′′(1) + f′(1) − [f′(1)]2 (Harris 1963). We can therefore determine M and V for the approximations described in the text by simply taking the first and second derivatives of fb(x) = f ∘ f ∘ … ∘ f+(x).
References
- Bull, J. J., M. R. Badgett, H. A. Wichman, J. P. Huelsenbeck, D. M. Hillis et al., 1997. Exceptional convergent evolution in a virus. Genetics 147: 1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J. J., M. R. Badgett and H. A. Wichman, 2000. Big-benefit mutations in a bacteriophage inhibited with heat. Mol. Biol. Evol. 17: 942–950. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970 An Introduction to Population Genetics Theory. Harper & Row, New York.
- Fisher, R. A., 1930 The Genetical Theory of Natural Selection. Oxford University Press, Oxford.
- Gerrish, P. J., and R. E. Lenski, 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102/103: 127–144. [PubMed] [Google Scholar]
- Haldane, J. B. S., 1927. The mathematical theory of natural and artificial selection. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Haldane, J. B. S., 1932 The Causes of Evolution. Harper Brothers, New York.
- Harris, T. E., 1963 The Theory of Branching Processes. Springer-Verlag, Berlin/New York.
- Heffernan, J. M., and L. M. Wahl, 2002. The effects of genetic drift in experimental evolution. Theor. Popul. Biol. 62: 349–356. [DOI] [PubMed] [Google Scholar]
- Kichler Holder, K., and J. J. Bull, 2001. Profiles of adaptation in two similar viruses. Genetics 159: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1957. Some problems of stochastic processes in genetics. Ann. Math. Stat. 28: 882–901. [Google Scholar]
- Kimura, M., 1962. On the probability of fixation of mutant genes in a population. Biometrics 19: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski, R. E., and M. Travisano, 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA 91: 6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski, R. E., M. R. Rose, S. C. Simpson and S. C. Tadler, 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- Levin, B. R., V. Perrot and N. Walker, 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgro, C. M., and L. Partridge, 2000. Evolutionary responses of the life history of wild-caught Drosophila melanogaster to two standard methods of laboratory culture. Am. Nat. 156: 341–353. [Google Scholar]
- Stearns, S. C., 1992 The Evolution of Life Histories. Oxford University Press, Oxford.
- Wahl, L. M., and P. J. Gerrish, 2001. Fixation probability in populations with periodic bottlenecks. Evolution 55(12): 2606–2610. [DOI] [PubMed] [Google Scholar]
- Wahl, L. M., I. Saika-Voivod and P. J. Gerrish, 2002. Evaluating the impact of population bottlenecks in experimental evolution. Genetics 162: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]