Abstract
Species-level genetic diversity and recombination in bacterial pathogens of wild plant populations have been nearly unexplored. Pseudomonas viridiflava is a common natural bacterial pathogen of Arabidopsis thaliana, for which pathogen defense genes and mechanisms are becoming increasing well known. The genetic variation contained within a worldwide sample of P. viridiflava collected from wild populations of A. thaliana was investigated using five genomic sequence fragments totaling 2.3 kb. Two distinct and deeply diverged clades were found within the P. viridiflava sample and in close proximity in multiple populations, each genetically diverse with synonymous variation as high as 9.3% in one of these clades. Within clades, there is evidence of frequent recombination within and between each sequenced locus and little geographic differentiation. Isolates from both clades were also found in a small sample of other herbaceous species in Midwest populations, indicating a possibly broad host range for P. viridiflava. The high levels of genetic variation and recombination together with a lack of geographic differentiation in this pathogen distinguish it from other bacterial plant pathogens for which intraspecific variation has been examined.
MUCH of the nature of bacterial species and their diversity remains a mystery due to the very large numbers of taxa, the potential difficulty in quantifying them, and the fact that they are relatively cryptic in nature. Of the many bacterial species that live on the leaves of plants, only those associated with agricultural crop plants tend to be isolated, identified, and characterized. These species are under an unusual selection regime in that their hosts are often isogenic and planted in monoculture. This may be biasing our understanding of the population structure and diversity of plant pathogenic bacteria.
Natural plant communities vary dramatically in their chemistry, their leaf surface environment, and their ability to mount specific defenses against bacterial pathogens, and thus phytopathogenic bacteria must be able to survive under a potentially wide range of conditions (Beattie and Lindow 1995; Andrews and Harris 2000). Yet, with the exception of a few Pseudomonas and Xanthomonas species that attack economically important crops (e.g., Denny et al. 1988; Ardales et al. 1996; Little et al. 1998; Restrepo et al. 2000; Sarkar and Guttman 2004), there are limited data available on the population genetic variation in pathogenic bacteria of plants, particularly wild plants. Meanwhile, there is increasing interest in studies of the population structure and genetic variation in many other types of common bacteria, such as clinical species (e.g., Souza et al. 1999; Spratt and Maiden 1999), Bacillus (e.g., Istock et al. 1992; Duncan et al. 1994; Roberts and Cohan 1995) and other environmental bacteria (Wise et al. 1995, 1996), rhizobia (e.g., Souza et al. 1992; Hagen and Hamrick 1996), and rhizosphere-associated bacteria (Di Cello et al. 1997; Dalmastri et al. 1999; Sikorski et al. 2001).
Pseudomonas viridiflava is a common pathogen of the annual weed Arabidopsis thaliana and is one of only a few known natural pathogens of A. thaliana (Jakob et al. 2002). Extensive knowledge of the patterns of variation in A. thaliana makes its natural pathogens especially attractive study systems. A. thaliana has a broad distribution (Hoffmann 2002) and shows phenotypic and genetic diversity across populations (e.g., Miyashita et al. 1999; Alonso-Blanco and Koornneef 2000; Erschadi et al. 2000). Genetic variation in A. thaliana is widely distributed on a worldwide scale, although there can be limited genetic variation within local populations (Bergelson et al. 1998). Of the loci for which variation has been characterized in A. thaliana, the nucleotide-binding site and leucine-rich repeat encoding genes, whose primary role is the recognition of pathogens, appear to be some of the most dynamic. The A. thaliana genome is known to contain at least 149 of these genes (Arabidopsis Genome Initiative 2000). Some of these resistance genes (R-genes) are known to have widely distributed variation, to show patterns of positive or balancing selection at the molecular level, and to segregate for resistance and susceptibility alleles within populations (Caicedo et al. 1999; Stahl et al. 1999; Bergelson et al. 2001; Tian et al. 2002; Mauricio et al. 2003).
The presence of long-lived balanced polymorphisms for pathogen recognition in this host plant suggests that its pathogens will likewise be polymorphic for virulence factors, a dynamical polymorphism termed “trench warfare” (Stahl et al. 1999). Alternatively, selective sweeps will predominate under an evolutionary arms race, in which adaptive mutants are selected for in both host and pathogen. The impact that balanced polymorphism and directional selection has on linked sites depends on the extent of recombination, and bacterial species exhibit a wide range of recombination rates (Feil and Spratt 2001). Clonal species are expected to be especially vulnerable to selective sweeps (periodic selection; Levin 1981) and therefore to have lower genetic variation. In contrast, more recombinogenic bacteria can potentially maintain higher levels of variation even as selection acts on an adaptive allele. Recombination rates are therefore expected to result in very different population structures among bacterial plant pathogens evolving in response to host resistance.
In this study, we examine genetic variation in P. viridiflava by sequencing five genomic DNA fragments using a worldwide sample of 93 isolates from >15 A. thaliana populations. Our goal is to characterize levels of genetic variation and recombination contained within local and global samples of this plant pathogen. In particular, Is there evidence for strong clonal structure with little polymorphism or is polymorphism high and recombination frequent?
MATERIALS AND METHODS
Collection and Identification:
The P. viridiflava isolates used in this study were collected from a number of A. thaliana populations in the Midwest United States; from three sites in North Carolina; from two adjacent sites in Spain; and from one site each in England, Sweden, and Japan. Most isolates were from diseased A. thaliana leaves (Table 1A) but we also included isolates collected from plant species co-occurring with A. thaliana from a subset of the Midwestern sites (Table 1B). The Midwestern populations range from 500 m apart (RM and PT) to ∼100 km apart. Collections of multiple isolates from Midwestern A. thaliana populations in 2001 were from plants located within a single 10-m diameter circle at each site.
TABLE 1.
Summary of sequencedP. viridiflava isolates
| No. in clade |
Polymorphisma (π)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Country | Location | Site | Year | No. of isolates |
A | B | A | B |
| A. P. viridiflava isolates from A. thaliana | ||||||||
| United States | St. Joseph, Michigan | RMX | 2001 | 3 | 3 | 0 | 0.013 | — |
| 2000 | 1 | 1 | 0 | — | — | |||
| 1998 | 1 | 0 | 1 | — | — | |||
| Knox, Indiana | KNOX | 2001 | 6 | 0 | 6 | — | 0.005 | |
| 1998 | 1 | 1 | 0 | — | — | |||
| Berrien County, Michigan | LH | 2001 | 6 | 5 | 1 | 0.008 | — | |
| Michigan City, Indiana | LP | 2000 | 1 | 1 | 0 | — | — | |
| Benton Harbor, Michigan | ME | 2001 | 6 | 6 | 0 | 0.024 | — | |
| 1998 | 1 | 1 | 0 | — | — | |||
| Laporte County, Indiana | PT | 2001 | 6 | 3 | 3 | 0.029 | 0.008 | |
| Spinks Corners, Michigan | PNA | 1998 | 1 | 1 | 0 | — | — | |
| Laporte County, Indiana | RM | 2001 | 6 | 4 | 2 | 0.002 | 0.003 | |
| La Porte, Indiana | RT | 2001 | 6 | 3 | 3 | 0.015 | 0.014 | |
| North Liberty, Indiana | SL | 2001 | 6 | 6 | 0 | 0.015 | — | |
| Durham, North Carolina | DUS | 2002 | 1 | 0 | 1 | — | 0.006b | |
| Durham, North Carolina | DUB | 2002 | 1 | 1 | 0 | |||
| Durham, North Carolina | DUD | 2002 | 5 | 0 | 5 | |||
| England | Silwood Park | SP | 2002 | 6 | 6 | 0 | 0.014 | — |
| Japan | Kyoto | KY | 2002 | 5 | 3 | 2 | 0.022 | 0 |
| Spain | Boadilla, site 1 | BOR | 2002 | 3 | 3 | 0 | 0.018c | — |
| Boadilla, site 2 | BOG | 2002 | 4 | 4 | 0 | |||
| Sweden | Lund | LU | 2002 | 4 | 4 | 0 | 0.001 | — |
| Total | 0.022 | 0.009 | ||||||
| Site | Plant Species | Clade | Isolate | |||||
| B. Other hosts from which one P. viridiflava isolate was sequenced (collected in 2002) | ||||||||
| ME | Cerastium vulgatum (Mouse-ear Chickweed) | A | ME751.1a | |||||
| Draba verna (Whitlow Grass) | A | ME753.1a | ||||||
| Stellaria media (Common Chickweed) | A | ME754.1a | ||||||
| Veronica sp. (Speedwell sp.) | A | ME755.1a | ||||||
| Cardamine parviflora (Small-flowered Bitter Cress) | A | ME756.1a | ||||||
| Lamium purpureum (Purple Dead Nettle) | A | ME758.1a | ||||||
| RM | Unidentified sp. | B | RM752.1a | |||||
| Stellaria media (Common Chickweed) | A | RM754.1a | ||||||
| Lepidium campestre (Field Peppergrass) | A | RM755.1a | ||||||
| Veronica sp. (Speedwell sp.) | A | RM757.1a | ||||||
| KN | Unidentified sp. | A | KNOX752.1a | |||||
| Unidentified sp. | B | KNOX753.1a | ||||||
| RT | Unidentified sp. | A | RT751.1a | |||||
Polymorphism was calculated using all five sequence fragments.
Calculated for combined DUS, DUB, and DUD isolates.
Calculated for combined BOR and BOG isolates.
Leaves appearing to be diseased were collected into sterile 1.6-ml microcentrifuge tubes, surface sterilized in 70% ethanol, and ground with a pestle in 200 μl buffered water (10 mm MgSO4). Leaves collected from outside the Midwest were partially processed on site to reduce plant nutrients available for growth of saprophytic bacteria and fungi during transport. Specifically, leaves were surface sterilized, ground in buffered water, and centrifuged at low speed; the supernatant was removed and the pellet was resuspended prior to transport. Leaf homogenate (50 μl) was plated on King's Medium B and incubated at 28° for 48 hr. Colonies from these plates were screened using the standard LOPAT determinative tests for fluorescent Pseudomonads (Lelliott et al. 1966). P. viridiflava was distinguished from other related species in our samples by a hypersensitive response in tobacco when infiltrated with a concentrated suspension of bacteria, a lack of oxidase activity, and an ability to rot potato (Schaad 1988). A PCR assay based on a diagnostic restriction site in the 16S ribosomal RNA gene and 16S gene sequence for a subsample of isolates (Jakob et al. 2002) confirmed our identification of isolates as P. viridiflava. In addition, four isolates were independently identified as P. viridiflava by Biolog analysis (Biolog, Hayward, CA), which characterizes bacterial species on the basis of the ability to metabolize 95 different carbon sources. The LOPAT protocol may have selected against nonpathogenic P. viridiflava, as only pathogenic strains are thought to cause a hypersensitive response in tobacco. It is similarly possible that we excluded P. viridiflava isolates that were atypical in their response to the other tests.
Isolates were named according to site abbreviation (Table 1A), followed by plant number, a decimal point, leaf number, and isolate as represented by a lowercase letter. Isolate numbers in the 700s are all collected from other host species, not A. thaliana (Table 1B).
Sequencing:
A total sample of 93 P. viridiflava isolates was assembled (Table 1, A and B). Specific isolates from each site from A. thaliana (80) and from non-A. thaliana hosts (13) were randomly selected. Total genomic DNA was extracted from each isolate using the CTAB method of Ausubel et al. (1996).
Because few gene sequences were available for P. viridiflava, four genomic fragments were obtained by shotgun sequencing total genomic DNA from the isolate KNOX3.4a. Genomic DNA was digested with MboI and fragments of 500–1000 bp were gel purified, incubated with T4 DNA polymerase, and blunt ligated into the positive selection cloning vector pZero2.0 (Invitrogen, Carlsbad, CA). Inserts were then sequenced using universal M13 primers. Four shotgun fragments (7, 17, 20, and 26) were chosen for resequencing on the basis of length and primer efficiency. The primers used for both PCR and sequencing (5′–3′), with annealing temperature, are as follows: fragment 7, GTTCCTTGAAGTGCCTGA and GTTTTCGTAGCGGTTGCG, 50°; fragment 17, GATTTGACGAAGTGACCT and GTAAAGCAACTTGTCCAC, 56°; fragment 20, CGCCGTTTTCGTTCTTGT and GCATGGAATACGCCGACA, 58°; and fragment 26, GTTTACGCTGACCTGACC and CACGATGCTCAGAAACAG, 58°. We were not able to amplify fragment 17 for isolate LU9.1e from Sweden. This isolate and one other from the Midwest (PNA3.3a) did not amplify with the original fragment 20 primers and an alternative set of primers was used to obtain a shorter sequence: CGAGCACGTCCAGCTTGG and GCGACGGCAAGGACATCA, 58°.
We also sequenced the gyrase subunit B gene (gyrB), which has recently been used as a higher-resolution alternative to the 16S rRNA gene for phylogenetic studies of Pseudomonas species (Yamamoto et al. 2000). Two sets of primers were designed on the basis of the partial gyrB sequence of strain ICMP/PDDCC 2848 (GenBank AB039427), TGGGCGTCTCGGTAGTAAAC and AGACCAGCGATGTCCAATGC, 50°; and strain HRI 2673C (AB039489), CGTGGGTGTCTCGGTAGTAA and CAGACCTGCGATGTCCAATG, 50°. These sequences are P. viridiflava (Yamamoto et al. 2000), although they have been mislabeled as P. syringae in GenBank. Again, isolate LU9.1e did not amplify with these primer sets. Sequencing was performed for both strands with CEQ DTCS Quick Start Mix (Beckman Coulter) and run on Beckman Coulter CEQ 8000 capillary sequencers.
Analysis:
Sequences were edited and aligned using Sequencher v. 4.1.2 (Gene Codes, Ann Arbor, MI). Coding status and homology of the sequenced P. viridiflava fragments were determined by BLAST searches against the sequenced genomes of P. syringae pv. tomato (Pto) DC3000 (Buell et al. 2003), P. syringae pv. syringae (Psy) B728a (DOE-JGI), and P. syringae pv. phaseolicola (Pph) 1448a (TIGR/Cornell University).
Phylogenetic trees for each sequence fragment were inferred using parsimony, maximum likelihood, and neighbor-joining methods using PAUP 4.0b10 (Swofford 2003) and Bayesian analysis as implemented in MrBayes v3.0b4 (Huelsenbeck and Ronquist 2001). A general time-reversible model of evolution with gamma rate variation across sites was used for all sequence fragments. Heuristic searches were used for maximum-parsimony and maximum-likelihood analyses. In MrBayes, Markov chain Monte Carlo analysis was run for 300,000 generations, generating 3000 trees. The first 1000 trees were discarded and the remaining 2000 trees were summarized in a consensus tree. Results were compared in multiple independent runs to ensure parameter convergence.
We concatenated the sequence of all five loci and used the same evolutionary model as above with the addition of the invariable sites parameter to generate a total evidence tree. Bayesian analysis generated ∼100,000 trees, of which the last 7000 were retained in two independent runs. A consensus tree was produced using the 14,000 trees from these two runs.
Tree topologies were compared using the Shimodaira-Hasegawa test (Shimodaira and Hasegawa 1999; Goldman et al. 2000) as implemented in PAUP 4.0b10 using the RELL method and 1000 bootstrap replicates.
Population genetic analyses were conducted with DNAsp v. 3.53 (Rozas and Rozas 1999). Analysis of molecular variance (AMOVA; Excoffier et al. 1992) was calculated using Arlequin ver. 2.000 (Schneider et al. 2000), using Jukes and Cantor distances.
Linkage disequilibrium among loci was tested using both the method of Maynard Smith et al. (1993), using the program available at www.mlst.net, and the method of Haubold et al. (1998), using LIAN 3.1 (Haubold and Hudson 2000).
Split decomposition was used to build a phylogenetic network for each locus. This is a method that takes into account support for different or conflicting phylogenies within a single data set by producing a tree-like network (Bandelt and Dress 1992). Reticulations in this tree may indicate past recombination events. Split decomposition networks were constructed in SplitsTree v4b6 (Huson 1998; D. H. Huson and D. Bryant, unpublished data) using Hamming distances, equal angles without branch weights, and 1000 bootstrap replicates.
The Burst algorithm on www.mlst.net was used to search for isolates that vary in sequence at only one of the five loci (single-locus variants, SLVs), which were used to investigate recent recombination events within loci.
The two isolates, PNA3.3a and LU9.1e, for which we had only 334 bp of fragment 20, were excluded from some analyses, but were identical to the isolates LU5.1a and ME210.1b over the sequenced nucleotides at fragment 20.
RESULTS
P. viridiflava homology to other Pseudomonas species:
We sequenced four genomic fragments, hereafter referred to as fragments 7, 17, 20, and 26, and 741 bp of gyrB in 93 P. viridiflava isolates (Table 1, A and B). Fragments 7, 20, and 26 are largely coding sequence based on homology to the annotated genome sequence of P. syringae pv. tomato DC3000; however, we do not have any direct evidence that they are actually expressed. Fragment 7 (395 bp) is homologous to a putative radical sterile alpha motif domain protein (PSPTO3969); fragment 20 (372 bp) is homologous to the adenylosuccinate synthetase gene purA (PSPTO4937); and fragment 26 (442 bp) includes partial coding sequences of two different putative genes, an amino acid ABC transporter, periplasmic amino acid-binding protein (PSPTO4136) in the first half of the fragment and an amino acid ABC transporter, permease protein (PSPTO4137) at the end of the fragment. Fragment 17 (429 bp) is homologous to a 130-bp hypothetical protein (PSPTO1938) in P. syringae from positions 57 to 186, but due to a stop codon in this region in P. viridiflava the entire fragment is considered noncoding in the following analyses. The five sequence locations are well distributed across the P. syringae pv. syringae DC3000 chromosome, but we do not know their locations on the P. viridiflava chromosome.
Multilocus sequence typing (MLST) studies have recently become a popular way to characterize the population genetic structure of large samples of clinical bacterial isolates and have started to be extended to other bacterial species. These studies typically use 400- to 500-bp fragments from seven housekeeping genes. Both gyrB and purA (fragment 20) are genes that have been used in MLST, specifically for P. syringae (gyrB; Sarkar and Guttman 2004) and Escherichia coli (gyrB and purA; http://web.mpiib-berlin.mpg.de/mlst/). We also have partial sequence from three putative housekeeping genes (fragment 7 and two different coding regions in fragment 26). Thus, 1840 bp from four different genomic locations are likely to represent housekeeping gene sequence in this study. Furthermore, the sequenced noncoding regions were found in all isolates, suggesting that these sequences are all “core” genome, i.e., found in most or all of the P. viridiflava isolates in the groups represented.
Nucleotide identity of P. viridiflava sequences to homologous sequences in P. syringae pv. phaseolicola 1448A, P. syringae pv. syringae B728a, and P. syringae pv. tomato DC3000 ranged from 74 to 92% (Table 2). Average synonymous site divergence, Ks, in coding regions between P. viridiflava and P. syringae is high, ranging from ∼0.38 to almost 0.89 after correcting for multiple substitutions (Table 2).
TABLE 2.
Nucleotide identity (whole sequence) and synonymous site divergence (in coding regions) betweenP. viridiflava and threeP. syringae pathovars averaged over allP. viridiflava isolates
| % average pairwise identity |
% average synonymous site divergence (JC-corrected) |
||||||
|---|---|---|---|---|---|---|---|
| Fragment | Coding sites in P. virdiflava fragment |
Pph | Psy | Pto | Pph | Psy | Pto |
| 7 | 98% (9–395) | 85.0 | 84.7 | 86.1 | 52.1 (89.0) | 52.1 (89.0) | 46.6 (72.7) |
| 17 | 0 | 79.5 | 76.7 | 78.9 | — | — | — |
| 20 | 100% (1–372) | 92.1 | 91.5 | 91.0 | 30.1 (38.4) | 36.1 (49.3) | 36.1 (49.2) |
| 26 | 77% (1–234; 337–442) | 88.2 | 85.7 | 88.4 | 39.4 (55.8) | 43.9 (66.0) | 38.4 (53.9) |
| gyrB | 100% (1–741) | 89.7 | 90.1 | 89.9 | 42.3 (62.3) | 39.8 (56.7) | 40.7 (58.6) |
P. viridiflava from alternative hosts:
The P. viridiflava isolates found on herbaceous species that co-occur with A. thaliana at Midwest sites do not appear to be genetically distinct from the population infecting A. thaliana (Figures 1 and 2). There is little differentiation at the sequence level and there are no fixed differences between these groups, even though these isolates were collected in a different year from those from A. thaliana. Analysis of AMOVA confirms the lack of significant variation between A. thaliana and other hosts and indicates that nearly 100% of the sequence variation is contained within rather than among hosts (not shown). Isolates from both A. thaliana and other hosts are therefore pooled in all of the following analyses.
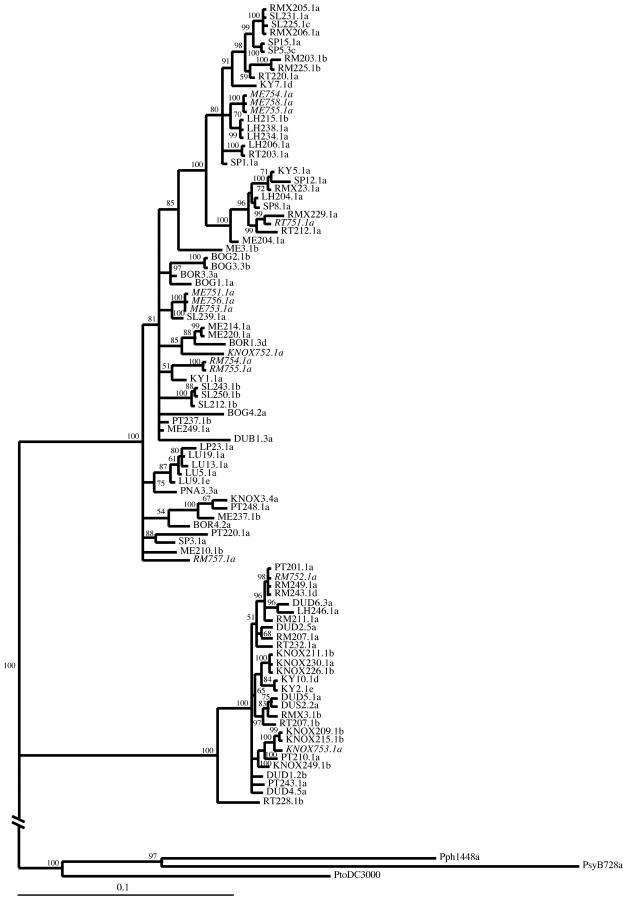
Figure 1.—
Consensus of 14,000 trees generated by Bayesian inference for the concatenated data set. Branch length values represent only trees on which those branches were present. Posterior probabilities (×100) of clades are given. The tree was rooted with three P. syringae sequences (Pph 1448a, Psy B728a, and Pto DC3000). Isolates collected from other host species in A. thaliana populations are shown in italics.
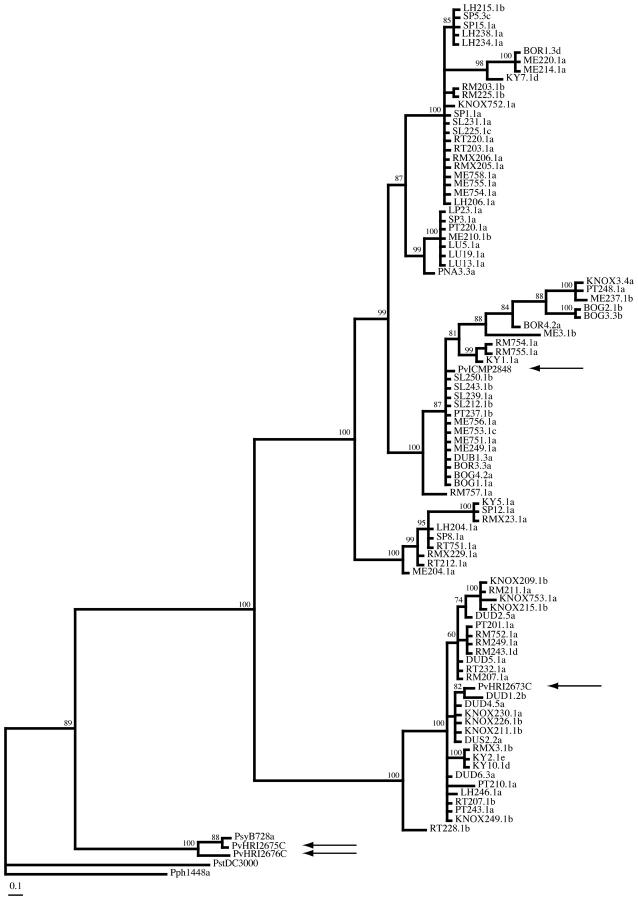
Figure 2.—
Consensus of 2000 trees generated by Bayesian inference for gyrB. Branch length values represent only trees on which those branches were present. Posterior probabilities (×100) of clades are given. The tree was rooted with P. syringae sequences as in Figure 1. Arrows indicate the P. viridiflava isolates sequenced by Yamamoto et al. (2000).
Genetic variation in P. viridiflava:
We found high levels of genetic variation within our sample of 93 P. viridiflava isolates: 376 segregating sites in 2379 bp of sequence and 15.8% nucleotide diversity (π) at synonymous sites (17.8% after Jukes-Cantor correction). Phylogenetic trees constructed with the concatenated sequence reveal that isolates segregate into two distinct clades with strong support (Figure 1). The larger of the two clades, hereafter referred to as clade A, contains 65 isolates and the smaller clade B contains the remaining 28 isolates. Substantial divergence between clades A and B can be seen in each of the five loci examined (Table 3). In particular, synonymous site nucleotide divergence, Ks [with Jukes-Cantor (JC) correction], averages 33.4% between clades compared to 9.3 and 2.3% within clade A and clade B, respectively. The average nucleotide divergence, Dxy, between P. viridiflava clades is less than but comparable to the nucleotide divergence over the same sequence between three P. syringae pathovars (Table 3), which represent three different major clades within the P. syringae species complex (Sawada et al. 1999; Sarkar and Guttman 2004). Since we set out to examine genetic variation in what we thought was a single taxon, we chose primers that amplified most or all of the isolates in the sample. The LOPAT test used for screening isolates also did not distinguish isolates in the A and B clades. It is therefore unlikely that we missed any isolates that may have been hybrids of the two clades.
TABLE 3.
Divergence within and betweenP. viridiflava clades and comparison toP. syringae pathovars
| Whole-sequence % nucleotide differences, Dxy(JC-corrected)a |
% synonymous site divergence, Ks (JC-corrected)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fragment | Pv A vs. B | Pph vs. Psy | Pph vs. Pto | Psy vs. Pto | Pv A | Pv B | Pv A vs. B | Pph vs. Psy | Pph vs. Pto | Psy vs. Pto |
| 7 | 7.9 (8.3) | 11.4 (12.4) | 10.8 (11.6) | 10.1 (10.9) | 7.2 (7.7) | 3.2 (3.3) | 28.8 (36.4) | 41.0 (59.3) | 38.2 (53.3) | 35.2 (47.6) |
| 17 | 8.2 (8.7) | 12.8 (14.0) | 10.7 (11.5) | 14.9 (16.6) | — | — | — | — | — | — |
| 20 | 5.8 (6.0) | 6.7 (7.0) | 6.2 (6.5) | 7.8 (8.2) | 7.2 (7.7) | 2.4 (2.4) | 22.5 (26.7) | 25.6 (31.3) | 23.5 (28.3) | 28.6 (36.0) |
| 26 | 3.6 (3.7) | 7.6 (8.0) | 6.0 (6.2) | 6.4 (6.7) | 6.3 (6.6) | 2.2 (2.2) | 16.6 (18.7) | 24.2 (29.3) | 19.1 (22.1) | 21.6 (25.4) |
| gyrB | 7.9 (8.4) | 7.4 (7.8) | 8.3 (8.7) | 8.5 (9.0) | 10.3 (11.6) | 4.6 (5.5) | 33.4 (44.2) | 31.4 (40.6) | 33.4 (44.2) | 36.1 (49.3) |
Values for P. viridiflava clades are averages over all pairs.
Sequence from the gyrB locus confirms that the A and B clades form a monophyletic group in the Pseudomonas genus when compared to the available gyrB sequence for other Pseudomonas (Yamamoto et al. 2000). Of the P. viridiflava isolates sequenced for the Yamamoto et al. (2000) study, the pathotype strain ICMP 2848 (isolated from bean in Switzerland) falls within clade A and HRI 2673C falls within clade B (Figure 2). The two other P. viridiflava isolates included in Yamamoto et al. (2000), HRI 2675C and HRI 2676C, fall outside these clades and appear to more closely resemble P. syringae (Figure 2). 16S rRNA gene sequence for 10 isolates from clade A and 5 isolates from clade B (GenBank nos. AY574907, AY574908, AY574909, AY574910, AY574911, AY574912 and AY604840, AY604841, AY604842, AY604843, AY604844, AY604845, AY604846, AY604847, AY604848) contained no fixed differences between clades in 1481 bp of sequence.
Much of the variation seen in the total sample is due to differences between the clades because of the extensive divergence between them. Yet, there is considerable variability within both clades as well. A total of 254 and 102 segregating sites in clades A and B, respectively, produce 52 different haplotypes in the 65 clade A isolates and 21 haplotypes in 28 clade B isolates. Nucleotide diversity averaged over all loci and sites is 2.2% for clade A and <0.9% for clade B. The variation within clades is predominantly silent, with few amino acid substitutions in the identified coding regions and low Ka/Ks values (Table 4). Tajima's D is not significantly different from zero for any locus in either clade (not shown). Comparing variation within and between the two clades, the McDonald-Kreitman test for selection is not significant for any locus (not shown), indicating that mutations at these loci do not deviate from those expected under neutrality. This further suggests that these coding regions could likely be considered housekeeping genes, since it appears that they are not under positive selection.
TABLE 4.
Segregating sites by locus, clade, and coding status
| Clade A
|
Clade B
|
Both clades
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fragment | Length | No. sites | Seg. sites | Ka/Ks | No. sites | Seg. sites | Ka/Ks | Seg. sites | Ka/Ks | Fixed differences between clades |
| 7 | ||||||||||
| Total | 395 | |||||||||
| Coding | 387 | 42 | 0.033 | 19 | 0.015 | 66 | 0.020 | 13 | ||
| Synonymous | 102.05 | 37 | 101.82 | 17 | 59 | 13 | ||||
| Replacement | 284.95 | 5 | 285.18 | 2 | 7 | 0 | ||||
| Noncoding | 8 | 2 | 0 | 2 | 0 | |||||
| 17 | ||||||||||
| Total (noncoding) | 429 | 429 | 92 | — | 429 | 32 | — | 114 | — | 8 |
| 20 | ||||||||||
| Total (coding)a | 372 | 27 | 0 | 10 | 0 | 43 | 0 | 9 | ||
| Synonymous | 97.22 | 27 | 98.0 | 10 | 43 | 9 | ||||
| Replacement | 271.78 | 0 | 271.0 | 0 | 0 | 0 | ||||
| 26 | ||||||||||
| Total | 442 | |||||||||
| Coding | 339 | 23 | 0.004 | 11 | 0.025 | 42 | 0.075 | 10 | ||
| Synonymous | 83.80 | 21 | 83.69 | 9 | 36 | 8 | ||||
| Replacement | 255.10 | 2 | 255.31 | 2 | 6 | 2 | ||||
| Noncoding | 103b | 1b | 1 | 2 | 0 | |||||
| gyrB | ||||||||||
| Total (coding) | 741 | 66 | 0.002 | 29 | 0.003 | 110 | 0.001 | 28 | ||
| Synonymous | 169.8 | 65 | 170.54 | 28 | 108 | 28 | ||||
| Replacement | 568.2 | 1 | 567.46 | 1 | 2 | 0 | ||||
Seg., segregating.
The two shorter sequences (PNA3.3a and LU9.1e) in clade A were excluded from this analysis. Including these two sequences and limiting the analysis to 334 bp results in 26 segregating sites in clade A and 40 segregating sites when both clades are combined.
In clade A, 26 isolates have a 1-bp deletion in the noncoding region and a single isolate has a different 1-bp deletion in the noncoding region; all 27 of these isolates have a true sequence length of 441 bp. These deletions are not included in the number of segregating sites.
Recombination in P. viridiflava:
In conducting phylogenetic analyses for each sequence fragment, changing associations among isolates in trees for the different loci were evident. Furthermore, phylogenetic trees inferred from the concatenated sequence (Figure 1) show a substantial loss in substructure compared to trees for the individual loci (e.g., gyrB, Figure 2). These observations suggest recombination between loci. The observation of changing tree topologies among loci was examined with the Shimodaira-Hasegawa (SH) test of phylogenetic congruence, using the Bayesian consensus tree for each sequenced fragment. The differences between the likelihoods of each tree and the “best” tree for each sequence (in all cases the tree inferred from that sequence) were always statistically significant (Table 5). All comparisons were similarly statistically significant when neighbor-joining trees were used. The SH test thus indicates changing tree topologies among the five sequence fragments.
TABLE 5.
Shimodaira-Hasegawa test of tree topologies
| Consensus tree
|
|||||
|---|---|---|---|---|---|
| Sequence fragment |
Fragment 7 | Fragment 17 | Fragment 20 | Fragment 26 | gyrB |
| 7 | 1278* | 427* | 580* | 1039* | |
| 17 | 672* | 359* | 453* | 1097* | |
| 20 | 704* | 1171* | 485* | 1171* | |
| 26 | 730* | 1254* | 438* | 1284* | |
| gyrB | 635* | 1155* | 362* | 531* | |
Values are differences in −ln likelihoods between the Bayesian tree for each sequence fragment and trees for each of the other sequenced loci. Statistically significant differences are indicated by asterisks. *P < 0.001.
A commonly used test of recombination among loci in bacteria is that of Maynard Smith et al. (1993), which is a significance test for linkage disequilibrium using the index of association among loci. The variance in the index of association (IA) is not significantly different from that expected under linkage equilibrium for clade A isolates (IA = 0.72). There is significant linkage disequilibrium at the P = 0.01 level among clade B isolates (IA = 0.69), but using only a single example of each five-locus haplotype removes any significant pattern of linkage disequilibrium (IA = 0.11). The index of association for the entire sample is 0.80. An alternative test, proposed by Haubold et al. (1998), produces “standardized IA” values of 0.22, 0.24, and 0.25 for clade A, clade B, and the entire sample, respectively, and indicates significant linkage disequilibrium for all three of these groups. Nevertheless, there are at least 22 examples (20 in clade A and 2 in clade B) of pairs of loci in which all four possible combinations of haplotypes are found in our sample of isolates, which is strong evidence for recombination between loci.
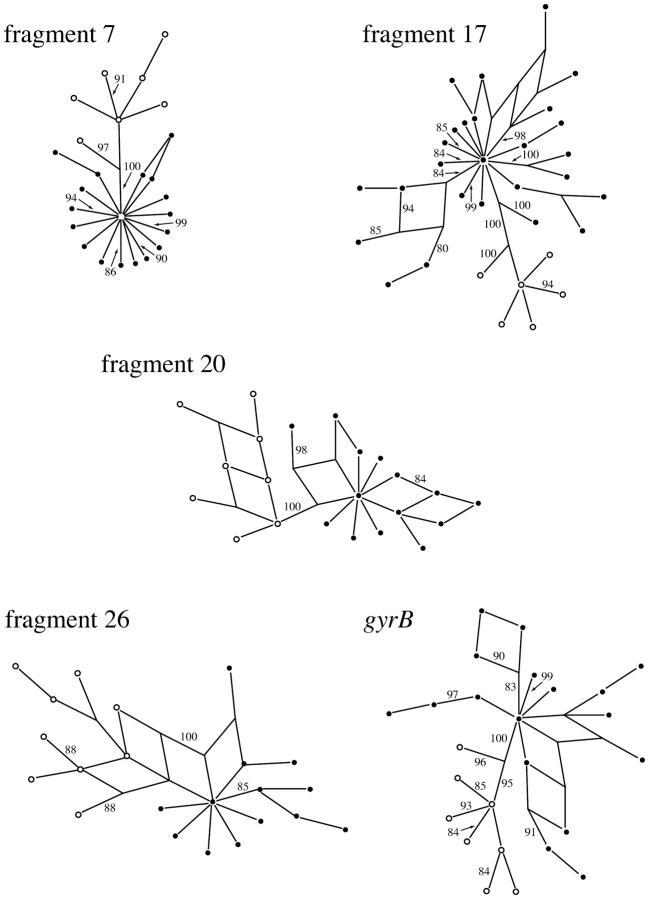
We also investigated recombination within each of the sequenced loci. Split decomposition analyses suggest the possibility of recombination within sequenced fragments, indicated by reticulations in the gene trees (Figure 3). In fact, the predicted minimum number of recombination events within a fragment (Hudson and Kaplan 1985) reaches as high as 10 (Table 6). In addition, the recombination parameter C = 2Nc (Hudson 1987) for the sequenced fragments is often greater than the mutation parameter θ = 2Nμ within clades (Table 6). In contrast, mutation rates exceed recombination rates when clades are combined, indicating that the vast majority of recombination is occurring within rather than between clades.
Figure 3.—
Split decomposition networks for the five sequence fragments. Solid circles are clade A haplotypes, and open circles are clade B haplotypes. Bootstrap scores >80% are shown.
TABLE 6.
Minimum number of recombination events within each locus and ratio ofC, the recombination parameter, to θ
| A only
|
B only
|
All isolates
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fragment | Min. events | C | θ | C/θ | Min. events | C | θ | C/θ | Min. events | C | θ | C/θ |
| 7 | 10 | 24.0 | 8.3 | 2.9 | 2 | 1.5 | 3.4 | 0.4 | 11 | 4.4 | 17.6 | 0.3 |
| 17 | 10 | 8.3 | 13.8 | 0.6 | 4 | 14.7 | 6.5 | 2.3 | 14 | 10.2 | 22.3 | 0.5 |
| 20 | 2 | 16.7 | 6.2 | 2.7 | 1 | 58.3 | 2.3 | 35.4 | 6 | 6.6 | 11.8 | 0.6 |
| 26 | 6 | 24.9 | 5.3 | 4.7 | 0 | 23.8 | 2.0 | 11.9 | 6 | 8.5 | 9.6 | 0.9 |
| gyrB | 9 | 16.9 | 16.7 | 1.0 | 2 | 3.4 | 4.1 | 0.8 | 20 | 4.1 | 33.9 | 0.1 |
| All | 41 | 68.1 | 50.4 | 1.4 | 13 | 22.5 | 18.4 | 1.2 | 60 | 6.4 | 94.5 | 0.7 |
The relative contribution of recombination vs. point mutation to genetic variation has recently been estimated for bacterial populations by comparing single-locus variants (SLVs) from MLST (Feil et al. 1999, 2000). The rationale for this approach is that strains that differ at only a single locus (out of five to seven sequenced loci) have a relatively recent common ancestor compared to strains that differ at multiple loci, since recombination in bacteria occurs over short stretches of sequence. The number of nucleotide differences between SLVs at the variable locus should indicate whether the variation at this locus is due to recombination or point mutation. If the sequences differ by multiple nucleotide substitutions, this variation is more likely to have originated by a recent recombination event. If the sequences differ by a single substitution that is unique or at very low frequency within the sample population, then it is more likely to be due to point mutation. The P. viridiflava sample had only nine sets of isolates that were SLVs (Table 7). Sequence differences between SLVs range from 4 to 36 nucleotides with only three sets of SLVs that differ by a single nucleotide. Of these three, two were unique substitutions, not seen in any other of the 92 isolates included in the analysis. The third single base-pair difference distinguishes two common haplotypes for fragment 26 and therefore is unlikely to have been a recent mutation. Of the nine sets of SLVs, seven could be attributed to recombination and two to point mutation. On a per site basis, 75 nucleotides were affected by recombination and only 2 by point mutation, yielding a per site relative contribution of mutation vs. recombination to genetic variation of 1:38. However, the number of isolates used in this study is much less than that used in the MLST studies for which this method was designed.
TABLE 7.
Sequence differences between all pairs of single-locus variants and possible source of recombinant sites
| Single-locus variants | Clade | Variable locus | Sequence differences (bp) |
Source |
|---|---|---|---|---|
| LH204.1a and SP8.1a | A | 17 | 1 | Unique |
| KY5.1a and SP12.1a | A | 17 | 10 | Multiple?a |
| LP23.1a and PT220.1a | A | 17 | 36 | Multiple?b |
| RMX205.1a and (RMX206.1a, SL231.1a, SL225.1c) | A | 26 | 1 | Unique |
| KY5.1a and RMX23.1a | A | 26 | 1 | Clade Ac |
| RM225.1b and RM203.1b | A | 26 | 4 | Clade A |
| LH204.1a and RT751.1a | A | 26 | 11 | Clade A |
| LH204.1a and RMX23.1a | A | gyrB | 9 | Multiple?d |
| RM211.1a and (PT201.1a, RM243.1d, RM249.1a, RM752.1a) | B | gyrB | 4 | Multiple?e |
Five sites are polymorphic in both clades and five sites are polymorphic only in clade A, with four of these at approximately equal frequency.
One unique site, 27 sites polymorphic only in A at low frequency and 8 polymorphic only in A but the lower frequency state is fixed in B.
Both are common haplotypes for fragment 26.
One site is polymorphic in both clades, five sites are polymorphic in A at low frequency, and three sites are polymorphic in A with the lower-frequency state fixed in B.
One site is polymorphic in both clades, one site is polymorphic only in B at low frequency, and two sites with lower-frequency state are fixed in clade A.
The source of the observed recombination is variable and unclear (Table 7). In fragment 26, the differences between the SLVs are most likely due to recombination between clade A isolates. Yet, there are several sets of SLVs for fragment 17 and gyrB in which the source of the polymorphic sites could be either clade A or clade B. Therefore, we cannot exclude the possibility that some recombination has occurred between clades. The only other indication of recombination between clades that we have observed is a reticulation between the clades in the split decomposition tree of fragment 26 (Figure 3). However, given the high degree of synonymous divergence between clades (Table 3), the numerous fixed differences between clades (Table 4), and the lower ratios of recombination to mutation when clades are considered together (Table 6), the rate of recombination between clades is clearly much less than that within clades.
Population-level and geographic variation in P. viridiflava:
Isolates from both A and B clades can often be found together in populations (Table 1). The three major European sites, SP, BOG/BOR, and LU, contained only clade A isolates, but this could be due to the limited size and number of samples. A general lack of geographic differentiation within clades is suggested by minimal grouping by geographic location in the gene trees (Figure 1). Nucleotide divergence (Dxy) among regions averages 0.025, ranging from 0.017 to 0.031 (Table 8) and AMOVA of these geographic regions (nested within clade) shows no significant genetic variation among regions (not shown). Nucleotide polymorphism within individual populations in a particular year varied from 0.001 to 0.029 in clade A and 0 to 0.014 in clade B (Table 1), but an AMOVA of populations within clades shows no significant variation among these populations (Table 9).
TABLE 8.
Dxy among regional samples, calculated separately for clade A isolates (top right) and clade B isolates (bottom left)
| Spain | Sweden | England | Japan | North Carolina | Midwestern United States |
|
|---|---|---|---|---|---|---|
| Spain | 0.025 | 0.026 | 0.024 | 0.030 | 0.024 | |
| Sweden | — | 0.021 | 0.026 | 0.031 | 0.023 | |
| England | — | — | 0.017 | 0.028 | 0.018 | |
| Japan | — | — | — | 0.028 | 0.020 | |
| North Carolina | — | — | — | 0.007 | 0.029 | |
| Midwestern United States | — | — | — | 0.008 | 0.007 | 0.021/0.008a |
Nucleotide diversity (π) for combined Midwestern populations (clade A/clade B). See Table 1 for other values of π.
TABLE 9.
Analysis of molecular variance
| Source of variation | d.f. | Sum of squares | Variance component | % variation |
|---|---|---|---|---|
| Clade | 1 | 2713 | 67.95 | 39.77a |
| Among populations, within clades | 21 | 1638 | −8.73b | −5.11b |
| Within populations, within clades | 69 | 7701 | 111.62 | 65.34 |
| Total | 91 | 12052 | 170.84 |
P < 0.0001.
Value is negative due to greater variation within populations than among populations.
DISCUSSION
We set out to examine the patterns of genetic variation in the bacterial phytopathogen P. viridiflava collected from A. thaliana populations in Spain, England, Sweden, Japan, North Carolina, and the Midwest United States. In our 93-isolate sample we found two cryptic clades that are present in close proximity within several A. thaliana populations, frequent recombination and high levels of polymorphism within each clade, a general lack of geographic differentiation in both clades, and the apparent ability of isolates in both clades to infect other herbaceous species that commonly co-occur with A. thaliana.
P. viridiflava, like its host A. thaliana, has a broad distribution with little geographic structure. However, variation in A. thaliana tends to be differentiated by local population (Bergelson et al. 1998), whereas for P. viridiflava variation within populations appears to be about equivalent to variation between populations. This could suggest that P. viridiflava is not adapted to A. thaliana at the local level. That the genetic variation observed within P. viridiflava clades is unstructured across a global sample of populations stands in contrast to previous studies of related plant pathogenic bacteria, which have generally shown either little variation or high levels of geographically structured variation. For example, a study of genetic variation in a worldwide collection of 17 P. syringae pv. tomato isolates revealed very little genetic variation on the basis of multilocus enzyme electrophoresis profiles using 26 enzyme loci; 13 of the 17 isolates were identical (Denny et al. 1988). Similarly, of 89 P. syringae pv. syringae isolates collected from stone fruit orchards in northern California, 81 of them had one of four enterobacterial repetitive intergenic consensus PCR patterns (Little et al. 1998). However, another study of a worldwide sample of P. syringae pv. tomato and P. syringae pv. maculicola showed unique fingerprints for all but 4 of 30 isolates (Clerc et al. 1998). The best examples of genetically diverse plant pathogenic bacteria come from Xanthomonas species, but these species also show geographic differentiation between regions (Ardales et al. 1996; Gagnevin et al. 1997; Restrepo and Verdier 1997; Restrepo et al. 2000).
P. viridiflava may be distinguished from many of the above pathogens in that it appears to be a generalist, able to attack a variety of host species in the sampled A. thaliana populations. These species, such as common chickweed (Stellaria media), purple dead nettle (Lamium purpureum), and other weedy mustards, are some of the most common herbaceous species in Midwestern A. thaliana populations. P. viridiflava is also frequently characterized as a “weak” or opportunistic pathogen and thus could experience selection pressures during the epiphytic phase of its life history that are less prevalent in pathogenic species that depend on a single host.
We found high rates of recombination in this worldwide sample of P. viridiflava within each of two genetic clades, while finding little evidence for recombination across clades. Fluorescent pseudomonads and other plant pathogens are generally not known to be naturally transformable. However, P. fluorescens, which is not known as a naturally competent species in vitro, seems to be able to naturally transform in soil microcosms (Demaneche et al. 2001). Many more bacteria strains may be able to transform only under narrow, but natural conditions; such species may not yet have been identified as transformable. Furthermore, plant pathogenic Pseudomonads have rarely been studied at the population level necessary to detect recombination in nature, yet several studies suggest genetic stability in P. syringae. In one study, two different P. syringae races (i.e., isolates with different virulence profiles) that were harvested from a single field pea crop in Australia in 1992–1993 were found to be genetically similar to isolates collected from field pea in 1967 and 1980 (Hollaway et al. 1997). Similarly, P. syringae strains from stone fruit orchards in California had similar genomic fingerprints to strains that had been in culture for >30 years (Little et al. 1998). In fact, this pattern has recently been observed in several P. syringae pathovars in a multilocus sequence typing study of P. syringae (Sarkar and Guttman 2004).
The highest levels of nucleotide variation in bacteria have been found in highly recombinogenic species. Synonymous site variation in the naturally competent Neisseria meningitidis and Helicobacter pylori ranges from 5.9 to 26.8% across 11 housekeeping genes (averaging 13.4%) and from ∼15 to 23% across 3 genes, respectively (Suerbaum et al. 1998). In comparison, synonymous variation in E. coli is highly variable, ranging from 0.99% for gapA to 28.8% for gnd, which is linked to the highly polymorphic rfb locus involved in O antigen synthesis (Guttman and Dykhuizen 1994). Synonymous variation in clade A of our P. viridiflava sample ranged from 6.6 to 11.6% over five loci and overall nucleotide variation observed in clade A averaged 2.2%. Nucleotide variation in protein-coding genes in ecologically distinct taxa of bacteria is typically ≤1% (Palys et al. 1997).
An obvious comparison to P. viridiflava is the closely related P. syringae. The P. syringae species complex contains at least four major clades (Sarkar and Guttman 2004) and nine genomic species (Gardan et al. 1997). It has further been divided into >50 pathovars (Clerc et al. 1998) on the basis of host range and symptom development (Young et al. 1992), and strains of several pathovars have been found that do not fall in the same genomic species as other strains of the same pathovar (Gardan et al. 1997; Clerc et al. 1998; Sarkar and Guttman 2004). The type strain of P. viridiflava, which is a member of clade A as described here, generally appears as an outgroup of the P. syringae complex. The P. viridiflava isolates sequenced here form a monophyletic cluster, but as at least two P. viridiflava isolates fall within the P. syringae cluster (Yamamoto et al. 2000), it appears that P. viridiflava as a whole may also be polyphyletic.
This study is a focused examination of the population genetic structure of a pathogen collected primarily from a single host. In contrast, phylogenetic and MLST studies of P. syringae have generally set out to capture the whole extent of the variation contained within the P. syringae species complex. For example, the sample of Sarkar and Guttman (2004) contained 21 pathovars from 30 host plant species, which resulted in four major clades of isolates. Since the divergence between the A and B clades of P. viridiflava is equal to or less than the divergence between P. syringae pathovars representing three of the P. syringae clades (Table 3), the genetic variation that we observe within P. viridiflava clades may be on a scale equivalent to that contained within a single pathovar or subset of pathovars within one clade of the P. syringae complex. Thus, the observation by Sarkar and Guttman (2004) that there may be recombination at the tips of their gene trees seems to be consistent with our observation of frequent recombination within but not between clades.
The clonality observed within the P. syringae species complex as a whole and the consistent divergence between clade A and B isolates in P. viridiflava across genomic fragments are not unexpected under the recently proposed “core genome hypothesis” (Lan and Reeves 2000, 2001; Hacker and Carniel 2001). The general idea is that core (i.e., housekeeping) genes can be used to define species boundaries because they tend to represent clonal descent rather than lateral gene transfer, which is common among genes involved in adaptation (e.g., genes for pathogenicity, antibiotic resistance, symbiosis, etc.). The core genome hypothesis may therefore serve as a logical, although imperfect, extension of the biological species concept to prokaryotes (Wertz et al. 2003).
The evidence for genome-wide divergence between the P. viridiflava clades of the magnitude we observed (∼30%), coupled with frequent recombination within clades, unambiguously shows that these two clades have been evolving as distinct groups for many millions of generations. Given the currently accepted criteria for species definition in bacteria, these clades might be considered subspecies of P. viridiflava, as there are other examples of ecologically distinct groups of bacteria that are indistinguishable by 16S rRNA sequence variation (Palys et al. 1997). However, by extension of the biological species concept using the idea of the core genome, they should likely be considered independent species.
Within clades, frequent recombination across broad geographic scales appears to protect P. viridiflava against reductions in variation due to periodic selection. Recombination and considerable haplotype diversity in P. viridiflava raise the prospect of using population genetic approaches, such as linkage disequilibrium or association mapping, in investigations of genetic variation for virulence. The level of recombination in P. viridiflava, together with the fact that it is a pathogen of the plant genetic model system A. thaliana, makes it a good candidate for investigation of the genetic basis of virulence traits.
It is unclear at this time whether the genetic diversity observed within P. viridiflava has been maintained as a result of an interaction with variable resistance mechanisms in a single plant host species, or if this pathogen's apparent interaction with numerous plant hosts can better explain this genetic variation. Population genetic and molecular evolutionary studies of virulence and pathogenicity genes in P. viridiflava may shed further light on this. Additional studies of bacterial pathogens of plants will be necessary to determine whether the levels of genetic variation and recombination observed in P. viridiflava are common among generalist pathogens and/or bacterial species primarily associated with wild plant populations.
Acknowledgments
We thank David Guttman, Fred Cohan, Hitoshi Araki, and two anonymous reviewers for improving the manuscript; Katrin Jakob, Hitoshi Araki, Brian Traw, Carlos Alonzo-Blanco, Jenny Hagenblad, Diana Wolf, and Jed Kim for their indispensable assistance with the collections; and Gale Wichmann, Jean Gladstone, and Dacheng Tian for technical assistance. This work was supported by a National Science Foundation Doctoral Dissertation Improvement Grant (DEB-0309028) to E.M.G., a National Institutes of Health Grant (GM57994) to J.B., and a Department of Education Graduate Assistance in Areas of National Need Training Grant in Ecology (P200A040070).
References
- Alonso-Blanco, C., and M. Koornneef, 2000. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci. 5: 22–29. [DOI] [PubMed] [Google Scholar]
- Andrews, J. H., and R. F. Harris, 2000. The ecology and biogeography of microorganisms on plant surfaces. Annu. Rev. Phytopathol. 38: 145–180. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Ardales, E. Y., H. Leung, C. M. Vera Cruz, T. W. Mew, J. E. Leach et al., 1996. Hierarchical analysis of spatial variation of the rice bacterial blight pathogen across diverse agroecosystems in the Philippines. Phytopathology 86: 241–252. [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann et al., 1996 Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Bandelt, H.-J., and A. W. M. Dress, 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1: 242–252. [DOI] [PubMed] [Google Scholar]
- Beattie, G. A., and S. E. Lindow, 1995. The secret life of foliar bacterial pathogens on leaves. Annu. Rev. Phytopathol. 33: 145–172. [DOI] [PubMed] [Google Scholar]
- Bergelson, J., E. A. Stahl, S. Dudek and M. Kreitman, 1998. Genetic variation within and among populations of Arabidopsis thaliana. Genetics 148: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J., M. Kreitman, E. A. Stahl and D. Tian, 2001. Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen et al., 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100: 10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A. L., B. A. Schaal and B. N. Kunkel, 1999. Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc, A., C. Manceau and X. Nesme, 1998. Comparison of randomly amplified polymorphic DNA with amplified fragment length polymorphism to assess genetic diversity and genetic relatedness within genospecies III of Pseudomonas syringae. Appl. Environ. Microbiol. 64: 1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmastri, C., L. Chiarini, C. Cantale, A. Bevivino and S. Tabacchioni, 1999. Soil type and maize cultivar affect the genetic diversity of maize root-associated Burkholderia cepacia populations. Microb. Ecol. 38: 273–284. [DOI] [PubMed] [Google Scholar]
- Demaneche, S., E. Kay, F. Gourbiere and P. Simonet, 2001. Natural transformation of Pseudomonas fluorescens and Agrobacterium tumefaciens in soil. Appl. Environ. Microbiol. 67: 2617–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny, T. P., M. N. Gilmour and R. K. Selander, 1988. Genetic diversity and relationships of two pathovars of Pseudomonas syringae. J. Gen. Microbiol. 134: 1949–1960. [DOI] [PubMed] [Google Scholar]
- Di Cello, F., A. Bevivino, L. Chiarini, R. Fani, D. Paffetti et al., 1997. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 63: 4485–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, K. E., N. Ferguson, K. Kimura, X. Zhou and C. A. Istock, 1994. Fine-scale genetic and phenotypic structure in natural populations of Bacillus subtilis and Bacillus licheniformis: implications for bacterial evolution and speciation. Evolution 48: 2002–2025. [DOI] [PubMed] [Google Scholar]
- Erschadi, S., G. Haberer, M. Schoniger and R. A. Torres-Ruiz, 2000. Estimating genetic diversity of Arabidopsis thaliana ecotypes with amplified fragment length polymorphisms (AFLP). Theor. Appl. Genet. 100: 633–640. [Google Scholar]
- Excoffier, L., P. Smouse and J. Quattro, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil, E. J., and B. G. Spratt, 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55: 561–590. [DOI] [PubMed] [Google Scholar]
- Feil, E. J., M. C. J. Maiden, M. Achtman and B. G. Spratt, 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16: 1496–1502. [DOI] [PubMed] [Google Scholar]
- Feil, E. J., J. Maynard Smith, M. C. Enright and B. G. Spratt, 2000. Estimating recombinational parameter in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnevin, L., J. E. Leach and O. Pruvost, 1997. Genomic variability of the Xanthomonas pathovar mangiferaeindicae, agent of mango bacterial black spot. Appl. Environ. Microbiol. 63: 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardan, L., H. L. Shafik and P. A. D. Grimont, 1997 DNA relatedness among pathovars of P. syringae and related bacteria, pp. 445–448 in Pseudomonas syringae Pathovars and Related Pathogens, edited by K. Rudolph, T. J. Burr, J. W. Mansfield, D. E. Stead, A. Vivian et al. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Goldman, N., J. P. Anderson and A. G. Rodrigo, 2000. Likelihood-based tests of topologies in phylogenetics. Syst. Biol. 49: 652–670. [DOI] [PubMed] [Google Scholar]
- Guttman, D. S., and D. E. Dykhuizen, 1994. Detecting selective sweeps in naturally occurring Escherichia coli. Genetics 138: 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker, J., and E. Carniel, 2001. Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep. 2: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, M. J., and J. L. Hamrick, 1996. A hierarchical analysis of population genetic structure in Rhizobium leguminosarum bv. trifolii. Mol. Ecol. 5: 177–186. [DOI] [PubMed] [Google Scholar]
- Haubold, B., and R. R. Hudson, 2000. Lian 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16: 847–848. [DOI] [PubMed] [Google Scholar]
- Haubold, B., M. Travisano, P. B. Rainey and R. R. Hudson, 1998. Detecting linkage disequilibrium in bacterial populations. Genetics 150: 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. H., 2002. Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J. Biogeogr. 29: 125–134. [Google Scholar]
- Hollaway, G. J., M. R. Gillings and P. C. Fahy, 1997. Use of fatty acid profiles and repetitive element polymerase chain reaction (PCR) to assess the genetic diversity of Pseudomonas syringae pv. pisi and Pseudomonas syringae pv. syringae isolated from field peas in Australia. Australas. Plant Pathol. 26: 98–108. [Google Scholar]
- Hudson, R. R., 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50: 245–250. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Huson, D. H., 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14: 68–73. [DOI] [PubMed] [Google Scholar]
- Istock, C. A., K. E. Duncan, N. Ferguson and X. Zhou, 1992. Sexuality in a natural population of bacteria—Bacillus subtilis challenges the clonal paradigm. Mol. Ecol. 1: 95–103. [DOI] [PubMed] [Google Scholar]
- Jakob, K., E. M. Goss, H. Araki, T. Van, M. Kreitman et al., 2002. Pseudomonas viridiflava and P. syringae—natural pathogens of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 15: 1195–1203. [DOI] [PubMed] [Google Scholar]
- Lan, R., and P. R. Reeves, 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8: 396–401. [DOI] [PubMed] [Google Scholar]
- Lan, R., and P. R. Reeves, 2001. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 9: 419–424. [DOI] [PubMed] [Google Scholar]
- Lelliott, R. A., E. Billing and A. C. Hayward, 1966. A determinative scheme for the fluorescent plant pathogenic pseudomonads. J. Appl. Bacteriol. 29: 470–489. [DOI] [PubMed] [Google Scholar]
- Levin, B. R., 1981. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics 99: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, E. L., R. M. Bostock and B. C. Kirkpatrick, 1998. Genetic characterization of Pseudomonas syringae pv. syringae strains from stone fruits in California. Appl. Environ. Microbiol. 64: 3818–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio, R., E. A. Stahl, T. Korves, D. Tian, M. Kreitman et al., 2003. Natural selection for polymorphism in the disease resistance gene Rps2 of Arabidopsis thaliana. Genetics 163: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., N. H. Smith, M. O'Rourke and B. G. Spratt, 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita, N. T., A. Kawabe and H. Innan, 1999. DNA variation in the wild plant Arabidopsis thaliana revealed by amplified fragment length polymorphism analysis. Genetics 152: 1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palys, T., L. K. Nakamura and F. M. Cohan, 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47: 1145–1156. [DOI] [PubMed] [Google Scholar]
- Restrepo, S., and V. Verdier, 1997. Geographical differentiation of the population of Xanthomonas axonopodis pv. manihotis in Colombia. Appl. Environ. Microbiol. 63: 4427–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, S., C. M. Velez and V. Verdier, 2000. Measuring the genetic diversity of Xanthomonas axonopodis pv. manihotis within different fields in Colombia. Phytopathology 90: 683–690. [DOI] [PubMed] [Google Scholar]
- Roberts, M. S., and F. M. Cohan, 1995. Recombination and migration rates in natural populations of Bacillus subtilis and Bacillus mojavensis. Evolution 49: 1081–1094. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Sarkar, S. F., and D. S. Guttman, 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, H., F. Suzuki, I. Matsuda and N. Saitou, 1999. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J. Mol. Evol. 49: 627–644. [DOI] [PubMed] [Google Scholar]
- Schaad, N. W., 1988 Laboratory Guide for the Identification of Plant Pathogenic Bacteria. American Phytopathological Society, St. Paul.
- Schneider, S., D. Roessli and L. Excoffier, 2000 Arlequin Ver. 2.000: A Software for Population Genetics Data Analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva.
- Shimodaira, H., and M. Hasegawa, 1999. Multiple comparisions of log-likelihoods with application to phylogenetic inference. Mol. Biol. Evol. 16: 1114–1116. [Google Scholar]
- Sikorski, J., H. Jahr and W. Wackernagel, 2001. The structure of a local population of phytopathogenic Pseudomonas brassicacearum from agricultural soil indicates development under purifying selection pressure. Environ. Microbiol. 3: 176–186. [DOI] [PubMed] [Google Scholar]
- Souza, V., T. T. Nguyen, R. R. Hudson, D. Pinero and R. E. Lenski, 1992. Hierarchical analysis of linkage disequilibrium in Rhizobium populations: Evidence for sex? Proc. Natl. Acad. Sci. USA 89: 8389–8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, V., M. Rocha, A. Valera and L. E. Eguiarte, 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65: 3373–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt, B. G., and M. C. J. Maiden, 1999. Bacterial population genetics, evolution and epidemiology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354: 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, E. A., G. Dwyer, R. Mauricio, M. Kreitman and J. Bergelson, 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671. [DOI] [PubMed] [Google Scholar]
- Suerbaum, S., J. Maynard Smith, K. Bapumia, G. Morelli, N. H. Smith et al., 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95: 12619–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 2003 PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods), Version 4.0b10. Sinauer Associates, Sunderland, MA.
- Tian, D., H. Araki, E. A. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz, J. E., C. Goldstone, D. M. Gordon and M. A. Riley, 2003. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J. Evol. Biol. 16: 1236–1248. [DOI] [PubMed] [Google Scholar]
- Wise, M. G., L. J. Shimkets and J. V. McArthur, 1995. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl. Environ. Microbiol. 61: 1791–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, M. G., J. V. McArthur, C. Wheat and L. J. Shimkets, 1996. Temporal variation in genetic diversity and structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl. Environ. Microbiol. 62: 1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S., H. Kasai, D. L. Arnold, R. W. Jackson, A. Vivian et al., 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146: 2385–2394. [DOI] [PubMed] [Google Scholar]
- Young, J. M., Y. Takikawa, L. Gardan and D. E. Stead, 1992. Changing concepts in the taxonomy of plant pathogenic bacteria. Annu. Rev. Phytopathol. 30: 67–105. [Google Scholar]