Abstract
We report the discovery, mapping, and characterization of a meiotic drive locus (D) exhibiting nearly 100% nonrandom transmission in hybrids between two species of yellow monkeyflowers, outcrossing Mimulus guttatus and selfing M. nasutus. Only 1% of F2 hybrids were M. nasutus homozygotes at the marker most tightly linked to D. We used a set of reciprocal backcrosses to distinguish among male-specific, female-specific, and zygote-specific sources of transmission ratio distortion. Transmission was severely distorted only when the heterozygous F1 acted as the female parent in crosses to either parental species, ruling out pollen competition and zygote mortality as potential sources of drive. After four generations of backcrossing to M. nasutus, nearly isogenic lines were still >90% heterozygous at markers linked to D, suggesting that heterozygosity at the drive locus alone is sufficient for nonrandom transmission. A lack of dramatic female fitness costs in these lines rules out alternatives involving ovule or seed mortality and points to a truly meiotic mechanism of drive. The strength and direction of drive in this system is consistent with population genetic theory of selfish element evolution under different mating systems. These results are the first empirical demonstration of the strong female-specific drive predicted by new models of selfish centromere turnover.
OBSERVATIONS of non-Mendelian inheritance date back almost to the rediscovery of Mendel's work (Gershenson 1928) and have become increasingly common as molecular markers have replaced phenotypic variants as the primary tool of quantitative and molecular genetics (Jenczewski et al. 1997; Rieseberg et al. 2000). Unequal representation of parental alleles in a progeny population can result from differential inclusion in the products of meiosis, differential survival or fertilization success of haploid gametes, or differential survival of the diploid zygotes. Determining which of these processes contributes to the observed transmission ratio distortion (TRD; Pardo-Manuel de Villena and Sapienza 2001b) is a first step in analyzing any non-Mendelian pattern of genetic transmission.
Only the first cause of TRD, the nonrandom inclusion of chromosomes or alleles in the products of meiosis, is meiotic drive as originally defined (Sandler and Novitski 1957). Historically, classic cases of TRD (e.g., Spore-killer in Neurospora, Segregation Distorter in Drosophila, and t-haplotypes in mouse; see Lyttle 1991 for review) have also been termed meiotic drive, but these well-known systems all involve postmeiotic dysfunction of spores or gametes. Evolutionary analyses of such distorters have provided tremendous insight into the dynamics of selfish elements within populations (e.g., Hartl 1970; Feldman and Otto 1991), as well as evidence for a role in species barriers (e.g., Dermitzakis et al. 2000; Tao et al. 2001) and even phenotypic evolution (Wilkinson et al. 1998). Despite similar selfishness, however, true meiotic drive fundamentally differs from postmeiotic transmission distortion systems, and we still know little of its incidence, mechanisms, and consequences.
As recently formalized by Pardo-Manuel de Villena and Sapienza (2001b), the necessary and sufficient conditions for nonrandom segregation of chromosomes or chromatids during meiosis are remarkably unrestrictive. In most plants and animals, female meiosis is intrinsically asymmetric: three of the four products of meiosis degenerate and only one goes on to form a gamete. Thus, any chromosomal variant that can preferentially segregate to the functional meiotic product gains an inherent transmission advantage; this asymmetry of cell division and cell fate continually selects for selfish chromosomal elements. Cytogenetic analyses demonstrate that female meiosis can act as a predictable arena for chromosomal competition and selfish evolution. For example, heterochromatic knobs in maize (Rhoades 1952; Dawe and Cande 1996; Buckler et al. 1999) and some B chromosomes experience preferential transmission through female meiosis. Similarly biased transmission of certain chromosomal rearrangements has been reported (Foster and Whitten 1991; Pardo-Manuel de Villena and Sapienza 2001c) and has been proposed as an explanation for large-scale patterns of karyotypic evolution in mammals (Pardo-Manuel de Villena and Sapienza 2001a).
Recent work suggests that centromeres, the primary sites of kinetochore assembly and spindle attachment (Dawe 1998), may also routinely compete during asymmetric meiosis (Henikoff et al. 2001a; Henikoff and Malik 2002; Malik and Henikoff 2002). However, because female drive by centromeres does not involve obvious chromosomal polymorphism and does not necessarily cause the biases in sex ratio or reductions in reproductive fitness that characterize male segregation distorter systems (Taylor and Ingvarsson 2003), it may be impossible to detect without detailed genotypic analyses. Furthermore, population genetic theory of outbred species predicts the rapid spread and fixation of competitively superior drive elements that do not entail fitness costs (Burt and Trivers 1998; Hurst and Werren 2001). Alternatively, suppression of drivers may rapidly evolve and restore equal segregation (Sandler and Novitski 1957). In either case, selfish chromosomal elements may be only transiently polymorphic within populations and the female meiotic drive causing their turnover would be rarely observed in within-species analyses.
Non-Mendelian inheritance of molecular markers in hybrid linkage mapping populations, which is commonly observed but largely unexplained (Jenczewski et al. 1997; Harushima et al. 2001), provides a novel opportunity to detect and study chromosomal elements that may have diverged via female drive. Here, we investigate the mechanism of segregation distortion in hybrids between two species of the flowering plant Mimulus (Scrophulariaceae sensu lato). Mimulus guttatus, a predominantly outcrossing species with showy, insect-pollinated flowers, and M. nasutus, a highly self-fertilizing species with reduced, often cleistogamous flowers, are closely related members of the broadly interfertile yellow monkeyflower species complex (Vickery 1964). Pre- and postzygotic barriers partially isolate M. guttatus and M. nasutus (Vickery 1964; Fishman and Willis 2001), but hybrids are often observed at sympatric sites (Ritland and Ritland 1996) and there is evidence of ongoing introgression at nuclear loci in some parts of the shared range (Sweigart and Willis 2003). Thus, artificial hybrids between these taxa provide insight into the genetics of divergence in incipient species and also predict the consequences of natural hybridization.
Previous linkage mapping revealed a striking pattern of transmission ratio distortion in M. nasutus × guttatus F2 hybrids (Fishman et al. 2001). M. guttatus parental alleles were overrepresented in at least nine distinct genomic regions on the 14 linkage groups; only two regions showed significant, but relatively slight, excesses of M. nasutus alleles. In this report, we focus on the mechanism of non-Mendelian inheritance in the most severely distorted region of the hybrid genome, a portion of linkage group 11 (Fishman et al. 2001). Interactions between the heterospecific genomes must underlie this severe distortion; because the parental lines were highly homozygous inbred lines and a single F1 was selfed to form the entire F2 mapping population, single-locus recessive lethals (inbreeding depression) cannot contribute to non-Mendelian inheritance in this cross.
The observed transmission bias on linkage group 11 (LG11) could be caused by preferential segregation of M. guttatus alleles/chromosomes during F1 female meiosis, by postmeiotic differences in the fitness of haploid F1 gametophytes or gametes, or by differential survival of diploid zygote genotypes. Because of the mating system difference between the parental species, selection at quantitative trait loci underlying pollen performance was a particularly attractive hypothesis; the outcrosser M. guttatus outcompetes M. nasutus in mixed pollinations (Diaz and Macnair 1999) and the mapping cross created the opportunity for competition among recombinant F1 pollen grains. Given that genic interactions underlie partial hybrid sterility in this cross (Fishman and Willis 2001), postfertilization loss of particular multilocus hybrid genotypes (zygote inviability) was also a reasonable alternative for some or all of the distortion. To discriminate among these hypotheses, we use reciprocal backcrosses to isolate the effects of F1 male function (meiosis and gamete performance), F1 female function (meiosis and gamete performance), and progeny class (nuclear genotype) on the transmission of LG11 markers. We further characterize the female-specific distortion in a set of nearly isogenic lines (NILs). We propose a single meiotic drive locus on LG11 that experiences nearly 100% preferential transmission of the M. guttatus allele in interspecific heterozygotes by biasing segregation at meiosis I and argue that extreme drive in hybrids reflects different histories of selfish evolution in the two species.
MATERIALS AND METHODS
Linkage and distortion mapping:
The initial F2 mapping population (N = 526) was generated by selfing a single F1 formed by crossing an inbred line of M. guttatus (IM62; Iron Mountain, OR; pollen parent) with an inbred line of M. nasutus (SF5; Sherar's Falls, OR; seed parent). The populations from which these lines were drawn are highly divergent in floral morphology (Fishman et al. 2002) and mating system (F = 1 for Sherar's Falls M. nasutus, Kelly and Willis 1998; F = 0.19 for Iron Mountain M. guttatus, Sweigart et al. 1999). The framework linkage map consisted of 174 markers spanning 1780 cM Kosambi on 14 linkage groups (Fishman et al. 2001). Additional codominant markers, including MgSTS87, MgSTS19, MgSTS20, and MgSTS26 on linkage group 11, were subsequently placed by genotyping a subset of the original F2 hybrids (N = 288) and constructing linkage maps using similar protocols in Mapmaker (Lander and Botstein 1989). The MgSTS markers are length polymorphisms in intronic regions of single-copy nuclear genes. These gene regions were discovered as part of a collaborative project to develop genomic tools for Mimulus by partially sequencing random cDNA clones generated from M. guttatus floral bud RNA. The resulting sequences were assembled into contigs and primers were designed to flank putative intron locations (J. H. Willis, L. Fishman, T. J. Vision and F. Deitrich, unpublished data). The primers for MgSTS markers used in this project are as follows: MgSTS19 (F-primer, 5′-ATTTGCCGTTCCACAATCTC; R-primer, 5′-AGTTCCATTCGACCGATACG), MgSTS-20 (F-primer, 5′-ACTGGTGCCAAGACGAGAAT; R-primer, 5′-ATCCCCTCGTAAAAGGCTGT), MgSTS26 (F-primer, 5′-GATGGAAAGCTCTCGCAACT; R-primer, 5′-CGCTTAAACATGGGAGCATT), and MgSTS87 (F-primer, 5′-CTTCGACGATGCAGAGAGT; R-primer, 5′-ACATAAGCCCTCCTCGTGAA). The marker bc374 on LG11 is a dominant amplified fragment length polymorphism (AFLP), which was not informative for use in the later experiments. Because of the large interval between bc374 and the next nearest marker and its lower information content, its map position is relatively uncertain.
Reciprocal backcross experiment:
A single F1 individual (M. nasutus SF5 × M. guttatus IM62) was used to generate four reciprocal backcross populations (BC) and a replicate F2 generation for the test of gender-specific transmission ratio distortion. For all crosses, the female parent was emasculated in the bud 1–2 days prior to hand-pollination to prevent autogamous selfing. Three of the BC classes (M. nasutus × F1, F1 × M. nasutus, and F1 × M. guttatus) and the F2 hybrids have the M. nasutus cytoplasmic genomes, whereas the M. guttatus × F1 BC progeny have the M. guttatus cytoplasmic genomes. The five progeny classes were grown in a common garden at the Duke University Research greenhouse. Tissue for DNA extraction and genotyping was collected from young rosettes to minimize opportunities for postgermination selection. Genotypic sample sizes ranged from 130 to 185 individuals. Because the replicate F2 showed the same pattern as the F2 mapping population, those data are not included in Figure 2.
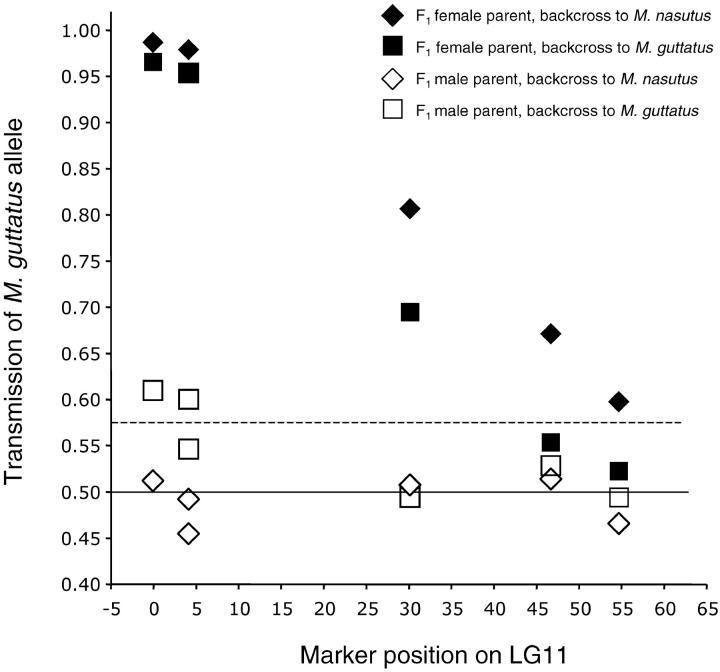
Figure 2.—
Transmission ratio distortion at LG11 markers in backcross populations. Data for codominant markers MgSTS87-MgSTS26 are shown. The solid line is the Mendelian expectation of 0.50. Values above the dotted line deviated significantly (P < 0.05) from the Mendelian expectation by one-tailed χ2 with 1 d.f., but each value was tested against a slightly different threshold because of differences in sample size. Because linked markers are not independent and the pattern in F1 female backcrosses is clear, we did not correct for multiple tests. The most distorted marker in the F1 male × M. guttatus backcross (MgSTS87; 61% M. guttatus transmission through F1) deviates from the Mendelian expectation with P = 0.0027, suggesting that milder distortion in this cross also has a real biological basis.
Test for cytoplasmic effects on drive:
As part of a separate project on nucleo-cytoplasmic male sterility in hybirds, we formed a single M. guttatus-cytoplasm F1 hybrid of the same parental lines and then backcrossed it (as the seed parent) to the M. nasutus SF5 line. The backcross progeny were grown in the Duke University greenhouse. A subset (N = 90) were genotyped at MgSTS87, the marker most tightly linked to the LG11 distortion locus.
Construction and genotyping of NILs:
The NILs descend from a replicate BC1 population [(SF5 × IM62) × SF5]; N > 500 initially) derived from the same F1 cross. NIL construction was designed to minimize opportunities for fecundity/fertility selection; each BC2–BC4 line descended from a different BC1 individual, was used as the seed parent in the backcrosses to the recurrent parent (M. nasutus SF5), and was maintained by random single-seed descent. Seed parents were emasculated in the bud to prevent self-pollination. The BC4 NILs (N = 187) were grown in a common garden under the same conditions as previous generations. To estimate the NIL genomic background, we genotyped 48 additional codominant markers. These control markers are unlinked to LG11, but were otherwise chosen haphazardly from a pool of markers polymorphic in these lines.
Examination of NIL seed development:
To explore the hypothesis that severely biased transmission at D was caused by the loss of female gametophytes or developing seeds with the disfavored genotype, we compared seed development in D-heterozygous NILs and the D-homozygous SF5 recurrent parent. To produce the observed female-specific transmission bias, all ovules (or their descendant seeds) carrying the M. nasutus allele at D would have to be eliminated prior to germination. This differential fate should translate into a bimodal distribution of ovule/seed sizes at one or more stages of ovary development, with at least 50% of ovules not developing into mature seeds. Because of the possibility for reproductive compensation (i.e., maximum seed set despite high ovule or seed mortality), we chose to examine the size distributions of developing ovules/seeds rather than count absolute numbers of mature seeds. Ovaries were collected at pre- and postpollination (by SF5) stages from a sample of NILs and SF5 controls and preserved in ethanol. Each ovary was macerated with forceps to free the ovules/developing seeds, which were suspended in H2O on microscope slides and examined under a Leica MZ6 dissecting scope equipped with a video camera and a transmitted-light base producing a high-contrast dark-field image. Images were captured with frame-grabbing software and analyzed with Scion (Frederick, MD) Image 1.62c. For each ovary, we measured the major and minor elliptical axes of a random sample of 60–200 ovules/seeds. We then examined the distributions of the major elliptical axis (length) for each ovary sample. For the bimodal postfertilization distributions, we counted the number of objects in the large (seed) and small size classes (ovule/aborted seeds) and calculated the proportion in the small class to the total.
Genotyping:
Genomic DNA was extracted from leaf tissue using a CTAB/chloroform protocol (Doyle and Doyle 1990) modified for use in 96-well format. The MgSTS markers were amplified using standard touchdown PCR conditions (annealing temperatures incremented from 62° to 52° for the first 10 cycles and then an additional 30 cycles at 52°). All marker genotyping was performed by sizing PCR-amplified DNA fragments with an incorporated 5′ fluorescent-labeled primer on an ABI 3700 automated capillary sequencer (Applied Biosystems, Foster City, CA). Marker genotypes were assigned automatically using the program Genotyper (Applied Biosystems) and then verified by eye.
RESULTS
Linkage and distortion mapping:
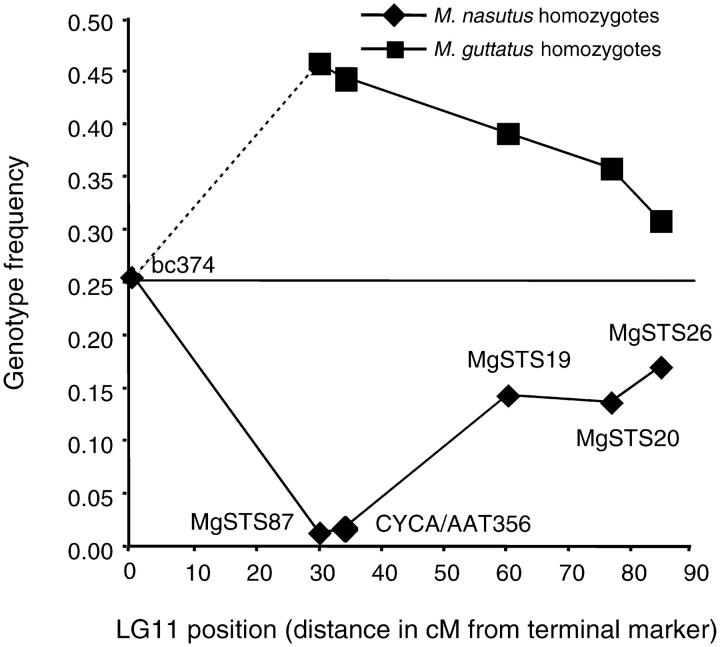
The new map of LG11 confirms the pattern of severe distortion previously reported, with greater resolution from additional codominant makers. At the most distorted marker in this region (MgSTS87), only 1% of individuals in the F2 mapping population are M. nasutus homozygotes, whereas the proportions of heterozygotes and M. guttatus homozygotes are both near 50% (Figure 1). The magnitude of transmission ratio distortion diminishes proportionately at increasingly distant markers, suggesting a single distorter locus (hereafter D) that is tightly linked to MgSTS87 and causes a 98–100% bias against the M. nasutus homozygote.
Figure 1.—
Map position and transmission ratio distortion of markers on LG11 in the M. nasutus × M. guttatus F2 mapping population (N = 287). The horizontal line indicates the Mendelian expectation for either parental homozygote class (0.25). The dotted segment between bc374 and MgSTS87 continues the symmetry of homozygote frequencies shown by the other markers, but we do not have data on the frequency of M. guttatus homozygotes at bc374 because it is a dominant AFLP. Distances are in cM Kosambi.
Reciprocal backcross experiment:
To determine whether extreme transmission bias at D in the F2 generation originated in F1 female function, F1 male function, or postfertilization selection among zygotes, we compared genotypic ratios in pairs of reciprocal backcrosses to each parental line. Strikingly, M. nasutus alleles were severely underrepresented at markers for the D locus only when the F1 was the female parent (Figure 2). In backcrosses to both parental species, only 1–4% of progeny inherited the M. nasutus allele at MgSTS87 from their F1 mothers, whereas transmission through an F1 male parent was Mendelian or only slightly skewed (Figure 2).
Test for cytoplasmic effects on drive:
The female-specific distortion on LG11 is not restricted to the M. nasutus cytoplasmic background. We observed similarly strong distortion at MgSTS87 when an M. guttatus-cytoplasm F1 was used as the female parent in a backcross to the M. nasutus parental line [6% SF homozygotes, N = 90; not significantly different (P > 0.05) by χ2 test from comparable M. nasutus-cytoplasm BC1 in the reciprocal backcross experiment].
Persistence of drive in nearly isogenic lines:
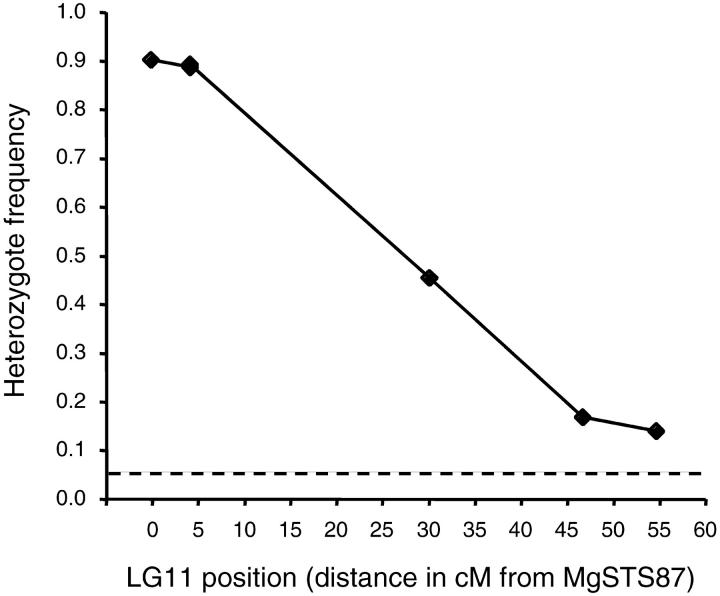
To characterize female-specific distortion in a more uniform genetic background, we examined the genotypes and phenotypes of NILs formed by four generations of backcrossing to the M. nasutus parent. In the absence of selection, average heterozygosity should be reduced by 50% with each backcross generation, such that 93.75% of these NILs are expected to be M. nasutus homozygotes at any given locus. However, because our breeding design always used M. nasutus as the pollen donor, each successive backcross also recreated conditions for female-specific drive. In contrast to Mendelian expectations, markers on LG11 maintain extremely high heterozygosity (Figure 3). About 90% (160/178) of the NILs were heterozygous at MgSTS87, whereas more distant markers were near the Mendelian expectation (6.25%) and the experimental mean for unlinked markers [7.75% ± 2.87% (SD) heterozygotes, N = 48 markers].
Figure 3.—
LG11 heterozygosity in NILs formed by four generations of backcrossing to the M. nasutus parental line (BN4, N = 187). Data for codominant markers MgSTS87-MgSTS26 are shown. The dashed line indicates the expected (Mendelian) heterozygote frequency (0.0625). The F2, BC1, and NIL data suggest that the D locus is within 2 cM of MgSTS87.
Seed development in D-heterozygous NILs:
As a screen for the postmeiotic elimination of female gametophytes with the M. nasutus allele as ovules or developing seeds, we dissected and examined NIL ovaries (pre- and postpollination) for evidence of the 50% mortality necessary to nearly completely bias allelic transmission at D. Prior to fertilization, the distribution of ovule sizes was unimodal in both the NILs and the SF5 parental controls (N = 10 and 9, respectively). In fruits collected several days after pollination, differential seed development produced clear bimodality, with a discrete set of ovule-sized objects as well as larger developing seeds observed in both NIL and M. nasutus ovaries. In both types of plants, the aborted/unfertilized ovule size class accounted for ∼30% of the total distribution (NIL range, 0.24–0.47; mean, 0.34 ± 0.08 SD; N = 7; SF range, 0.18–0.56; mean, 0.29 ± 0.10 SD, N = 14). Because unfilled ovules appear to be a normal part of fruit development in the parental line, any postmeiotic mechanism of female drive would need to shift the proportion of undeveloped seeds to 50–80% of total ovule production in the drive-heterozygous NILs. There is no evidence of such a dramatic increase in ovule or seed mortality.
DISCUSSION
We report the discovery of a unique segregation distorter locus (D) in Mimulus hybrids. This locus is remarkable in its strength: in heterozygotes, the M. guttatus (outcrosser) allele at D experiences close to 100% preferential transmission over its M. nasutus (selfer) homolog. There are a surprisingly large number of possible causes of such transmission ratio distortion in plant hybrids: nonrandom segregation of chromosomes in female meiosis (true meiotic drive), differential male gamete survival or performance (including interactions with each other and with paternal sporophyte and maternal stylar genotypes), differential female gamete survival or performance (including interactions with maternal nuclear and cytoplasmic genotypes), and differential seed survival (including direct lethality of nuclear zygote genotypes, competitive interactions between endosperm and embryo genotypes, and interactions between seed and maternal genotypes). Because the extreme distortion was initially identified in an F2 mapping population in which all of these are possibilities, we undertook a series of additional crossing experiments to dissect the mechanism of D drive.
The pattern of distortion in reciprocal backcross populations revealed that extreme drive at D is female specific, occurring when the heterozygous F1 acts as the female in crosses to itself or to either parental species (Figure 2). Female-specific drive clearly eliminates the possibility that D is a pollen performance QTL or a segregation distorter mediating interactions between male gametes. This immediately places D in a class distinct from the well-studied transmission ratio distortion systems caused by male-gamete competition (Lyttle 1991; Taylor and Ingvarrson 2003). The female specificity of drive also rules out direct lethal effects of diploid nuclear genotype as a source of transmission bias. Each pair of reciprocal backcrosses should contain identical pools of zygotic genotypes, but only those crosses with F1 mothers were severely distorted. The relatively mild transmission ratio distortion at some markers in the M. guttatus female × F1 male backcross is most likely due to a second linked locus (such as a pollen performance QTL) with male-specific effects. However, it could conceivably be a pleiotropic effect of the D locus.
The backcross data are consistent with drive through female meiosis, but there are other potential sources of female-specific distortion. In hermaphroditic flowering plants such as Mimulus, the female parent transmits the cytoplasmic genomes, surrounds the developing gametophytes and seeds, and makes the majority contribution to the embryo-fostering triploid endosperm. Interactions involving these genomes multiply the opportunities for differential female gametophyte or zygote survival in hybrids. In this experiment, the F1 parent and all but one of the experimental backcross populations carry M. nasutus cytoplasmic DNA, so we can immediately rule out negative heterospecific interactions with the maternal or zygotic cytoplasmic genotype in the strong bias against M. nasutus alleles at D. In addition, the M. nasutus cytoplasmic background does not appear to be necessary for biased transmission; the backcross progeny of an F1 female with M. guttatus cytoplasm showed a similar pattern of distortion at D-linked markers.
The female specificity of drive allowed us to assess drive in a set of NILs constructed by repeated backcrossing of the F1 (and later hybrids) to the M. nasutus parental line. Drive persists in this nearly isogenic M. nasutus background, with >90% of BN4 NILs remaining heterozygous at marker MgSTS87 (Figure 3) compared to >8% at unlinked markers. This is precisely the expectation if the M. guttatus allele at D invariably outcompetes the alternative allele each generation, and recombination rarely disassociates it from the M. guttatus allele of the closely linked marker. These data suggest that only heterozygosity at D itself (or tightly linked modifiers) is required for biased transmission; any unlinked M. guttatus modifiers of drive would be retained in only a minority of NILs. Because the NILs are essentially M. nasutus plants except for their heterozygosity at D, the persistence of drive in these lines also rules out lethal interactions between developing ovules and the maternal nuclear genotype (e.g., a dominant M. guttatus allele at another locus) as the source of female-specific distortion. Whatever the mechanism, it almost certainly involves only direct interactions between alternative genotypes at D. Although it is possible that interactions with an unlinked locus with similarly extreme transmission bias could be necessary for female drive at D, individual unlinked markers (N = 48; L. Fishman and J. H. Willis, unpublished data) were either not distorted or only mildly distorted in the BN4 NIL population. Thus, it is unlikely that the genome contains another region with extreme female drive.
The backcross and NIL genotypic data rule out several postmeiotic sources of female-specific distortion, but some alternatives to meiotic drive remained. In particular, these data alone cannot rule out lethal ovule-ovule or endosperm-endosperm interactions analogous to ascomycete spore-killer systems (Nauta and Hoekstra 1993). It is difficult to envision a physiological mechanism for consistently lethal competition between developing gametophytes or seeds, which are encased in maternal tissue, but such interactions are theoretically possible given the divergent histories of the parental species. Unlike true drive through female meiosis, however, these and any other postmeiotic processes must reduce female fecundity—the observed genotypic bias could result only from the loss of all developing ovules or seeds with the disfavored genotype (i.e., 50% mortality). The NILs, which are almost entirely homozygous M. nasutus elsewhere in the genome, allow us to isolate such fitness effects of D from other hybrid incompatibilities expressed in the F1 and F2 generations (Fishman and Willis 2001).
To explore the hypothesis that nearly completely biased transmission at D was caused by the loss of female gametophytes or developing seeds with the disfavored genotype, we compared seed development in D-heterozygous NILs and the D-homozygous SF5 recurrent parent. We saw no evidence of an additional 50% female gamete death due to drive in D-heterozygotes; the NILs and the M. nasutus parental line both displayed ∼30% unfertilized or aborted ovules after fertilization, on average, and no NILs had >50% unfilled seeds. Additional experiments will be necessary to fully characterize any subtler fitness effects of the D genotype, such as increased rates of chromosomal nondisjunction (Zwick et al. 1999), but mass loss of female gametophytes or developing zygotes does not appear to account for nearly completely biased allelic transmission at D by heterozygous individuals.
In aggregrate, our data strongly suggest that severe distortion at D results from biased chromosomal segregation during female meiosis. What functional difference between chromosomal homologs could cause nearly completely asymmetric meiotic transmission? The best-understood case of true meiotic drive, the Ab10/knob system in maize, involves genic activation of neocentromeric knob regions that competitively bind microtubules and orient the carrier chromatids toward the outer spindle poles at meiosis II (Rhoades 1952; Dawe and Cande 1996; Yu et al. 1997). Like proposed telomeric drive elements (Zwick et al. 1999), knobs must assort independently from the centromere to drive and thus have a maximum transmission advantage of 83.3% (Buckler et al. 1999). Because we observe >98% distortion at linked markers, D is almost certainly not a knob or telomeric element driving at meiosis II. The strength of distortion instead argues that direct competition between homologous centromeres at meiosis I produces the observed deviation from Mendelian segregation. If true, the functionally heterozygous driving element at D must be either the LG11 centromere itself or one or more tightly linked genes that modify the behavior of the LG11 centromere at meiosis. Experimental confirmation of this inference will require mapping of driven LG11 markers relative to the LG11 centromere. At this point, we have no information on centromere positions in Mimulus; however, the ongoing development of bacterial artificial chromosome libraries and a physical map of M. guttatus will facilitate the joint localization of the D locus and the LG11 centromere using fluorescence in situ hybridization.
Why extreme functional divergence at D? Recent molecular work argues that chromosomal competition during female meiosis drives rapid turnover of centromeres and associated histone proteins within populations, potentially accelerating the development of reproductive incompatibilities between species (Henikoff et al. 2001b). This constant centromeric arms race is limited to outcrossing species, however; population genetic theory shows that selfish chromosomal elements cannot spread in highly selfing populations (Burt and Trivers 1998; Hurst and Werren 2001). Therefore, extreme drive at D, as well as milder bias against M. nasutus alleles in other regions of the hybrid genome (Fishman et al. 2001), may reflect ongoing selection for selfish centromeres in outcrossing M. guttatus and its relaxation in strictly selfing M. nasutus. Asymmetric divergence in these species generates a remarkable new system for the study of true meiotic drive, including its functional mechanisms, dynamics within populations, and role in the development of species barriers. By demonstrating the selective power of female meiosis, our results further suggest that chromosomal drive may be a hidden contributor to divergence in many eukaryotic systems.
Acknowledgments
We thank A. Sweigart, J. Aagaard, and A. Kelly for assistance with marker development; D. Pedersen, L. Bukovnik, and A. Sweigart for assistance with genotyping; D. Emlen and S. Miller for sharing equipment for the image analysis of developing seeds; and B. Calhoun for plant care. We are grateful to N. Martin, M. Hall, A. Sweigart, Y.-W. Lee, A. Cooley, A. Case, L. Moyle, S. Otto, and two anonymous reviewers for helpful comments on earlier versions of the manuscript. The manuscript also benefited from illuminating discussions with B. Charlesworth, D. Charlesworth, R. K. Dawe, C. Langley, B. Nicklas, M. Uyenoyama, and M. Zwick. This work was supported by National Science Foundation awards DEB-0075704 to J.W. and L.F., DEB-0316786 and DBI-0321329 to L.F., and FIBR-0328636 to J.W.
References
- Buckler, E. S., IV, T. L. Phelps-Durr, C. S. K. Buckler, R. K. Dawe, J. F. Doebley et al., 1999. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt, A., and R. Trivers, 1998. Selfish DNA and breeding system in flowering plants. Proc. R. Soc. Lond. Ser. B 265: 141–146. [Google Scholar]
- Dawe, R. K., 1998. Meiotic chromosome organization and segregation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 371–395. [DOI] [PubMed] [Google Scholar]
- Dawe, R. K., and W. Z. Cande, 1996. Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc. Natl. Acad. Sci. USA 93: 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E. T., J. P. Masly, H. M. Waldrip and A. G. Clark, 2000. Non-Mendelian segregation of sex chromosomes in heterospecific Drosophila males. Genetics 154: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, A., and M. R. Macnair, 1999. Pollen tube competition as a mechanism of prezygotic reproductive isolation between Mimulus nasutus and its presumed progenitor M. guttatus. New Phytol. 144: 471–478. [DOI] [PubMed] [Google Scholar]
- Doyle, J. J., and J. L. Doyle, 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Feldman, M. W., and S. P. Otto, 1991. A comparative approach to the population-genetics theory of segregation distortion. Am. Nat. 137: 433–456. [Google Scholar]
- Fishman, L., and J. H. Willis, 2001. Evidence for Dobzhansky-Muller incompatibilities contributing to the sterility of hybrids between Mimulus guttatus and M. nasutus. Evolution 55: 1932–1942. [DOI] [PubMed] [Google Scholar]
- Fishman, L., A. Kelly, E. Morgan and J. H. Willis, 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman, L., A. J. Kelly and J. H. Willis, 2002. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution 56: 2138–2155. [DOI] [PubMed] [Google Scholar]
- Foster, G. G., and W. J. Whitten, 1991. Meiotic drive in Lucilia cuprina and chromosomal evolution. Am. Nat. 137: 403–415. [Google Scholar]
- Gershenson, S., 1928. A new sex ratio abnormality in Drosophila obscura. Genetics 13: 488–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, D. L., 1970. Analysis of a general population genetic model of meiotic drive. Evolution 24: 538–545. [DOI] [PubMed] [Google Scholar]
- Harushima, Y., M. Nakagahra, M. Yano, T. Sasaki and N. Kurata, 2001. A genome-wide survey of reproductive barriers in an intraspecific hybrid. Genetics 159: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and H. S. Malik, 2002. Selfish drivers. Nature 417: 227. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. a The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. b Speciation and centromere evolution. Science 294: 2478–2480. [DOI] [PubMed] [Google Scholar]
- Hurst, G. D. D., and J. H. Werren, 2001. The role of selfish genetic elements in eukaryotic evolution. Nat. Rev. Genet. 2: 597–606. [DOI] [PubMed] [Google Scholar]
- Jenczewski, E., M. Gherardi, I. Bonnin, J. M. Prosperi, I. Olivieri et al., 1997. Insight on segregation distortions in two intraspecific crosses between annual species of Medicago (Leguminosae). Theor. Appl. Genet. 94: 682–691. [Google Scholar]
- Kelly, A. J., and J. H. Willis, 1998. Polymorphic microsatellite loci in Mimulus guttatus and related species. Mol. Ecol. 7: 769–774. [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyttle, T. W., 1991. Segregation distorters. Annu. Rev. Genet. 25: 511–557. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2002. Conflict gets complexity: the evolution of centromeres. Curr. Opin. Genet. Dev. 12: 711–718. [DOI] [PubMed] [Google Scholar]
- Nauta, M. J., and R. F. Hoekstra, 1993. Evolutionary dynamics of spore killers. Genetics 135: 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., and C. Sapienza, 2001. a Female meiosis drives karyotypic evolution in mammals. Genetics 159: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., and C. Sapienza, 2001. b Nonrandom segregation during meiosis: the unfairness of females. Mamm. Genome 12: 331–339. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., and C. Sapienza, 2001. c Transmission ratio distortion in offspring of heterozygous female carriers of Robertsonian translocations. Hum. Genet. 108: 31–36. [DOI] [PubMed] [Google Scholar]
- Rhoades, M. M., 1952 Preferential segregation in maize, pp. 66–80 in Heterosis, edited by J. W. Gowen. Iowa State Press, Ames, IA.
- Rieseberg, L. H., S. J. E. Baird and K. A. Gardner, 2000. Hybridization, introgression and linkage evolution. Plant Mol. Biol. 42: 205–224. [PubMed] [Google Scholar]
- Ritland, K., and C. Ritland, 1996. Inferences about quantitative inheritance based on natural population structure in the yellow monkeyflower, Mimulus guttatus. Evolution 50: 1074–1082. [DOI] [PubMed] [Google Scholar]
- Sandler, L., and E. Novitski, 1957. Meiotic drive as an evolutionary force. Am. Nat. 91: 105–110. [Google Scholar]
- Sweigart, A. L., and J. H. Willis, 2003. Patterns of nucleotide diversity are affected by mating system and asymmetric introgression in two species of Mimulus. Evolution 57: 2490–2506. [DOI] [PubMed] [Google Scholar]
- Sweigart, A. L., K. Karoly, A. Jones and J. H. Willis, 1999. The distribution of individual inbreeding coefficients and pairwise relatedness in a population of Mimulus guttatus. Heredity 83: 625–632. [DOI] [PubMed] [Google Scholar]
- Tao, Y., D. L. Hartl and C. C. Laurie, 2001. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc. Natl. Acad. Sci. USA 98: 13183–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D. R., and P. K. Ingvarsson, 2003. Common features of segregation distortion in plants and animals. Genetica 117: 27–35. [DOI] [PubMed] [Google Scholar]
- Vickery, R. K., Jr., 1964. Barriers to gene exchange between members of the Mimulus guttatus complex (Scrophulariaceae). Evolution 18: 52–69. [Google Scholar]
- Wilkinson, G. S., D. C. Presgraves and L. Crymes, 1998. Male eye span in stalk-eyed flies indicates genetic quality by meiotic drive suppression. Nature 391: 276–279. [Google Scholar]
- Yu, H.-G., E. N. Hiatt, A. Chan, M. Sweeney and R. K. Dawe, 1997. Neocentromere-mediated chromosome movement in maize. J. Cell Biol. 139: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick, M. E., J. L. Salstrom and C. H. Langley, 1999. Genetic variation in rates of nondisjunction: associations of two naturally occurring polymorphisms in the chromokinesin nod with increased rates of nondisjunction in Drosophila melanogaster. Genetics 152: 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]