Abstract
Multiple mating by females establishes the opportunity for postcopulatory sexual selection favoring males whose sperm is preferentially employed in fertilizations. Here we use natural variation in a wild population of Drosophila melanogaster to investigate the genetic basis of sperm competitive ability. Approximately 101 chromosome 2 substitution lines were scored for components of sperm competitive ability (P1′, P2′, fecundity, remating rate, and refractoriness), genotyped at 70 polymorphic markers in 10 male reproductive genes, and measured for transcript abundance of those genes. Permutation tests were applied to quantify the statistical significance of associations between genotype and phenotype. Nine significant associations were identified between polymorphisms in the male reproductive genes and sperm competitive ability and 13 were identified between genotype and transcript abundance, but no significant associations were found between transcript abundance and sperm competitive ability. Pleiotropy was evident in two genes: a polymorphism in Acp33A associated with both P1′ and P2′ and a polymorphism in CG17331 associated with both elevated P2′ and reduced refractoriness. The latter case is consistent with antagonistic pleiotropy and may serve as a mechanism maintaining genetic variation.
THERE are striking differences between strict monogamy and polygamy in the opportunity for sexual selection (Birkhead and Møller 1998). Parentage studies using highly polymorphic molecular markers indicate that at least 50% of wild-captured Drosophila melanogaster females had mated with more than a single male (Ochando et al. 1996; Harshman and Clark 1998). While the benefits to females are often debated (see Jennions and Petrie 2000; Birkhead and Pizzari 2002), polyandry provides the opportunity for postcopulatory sexual selection to influence patterns of reproductive success. In species with internal fertilization, reproductive success will likely be determined by complex interactions between sperm competition and cryptic female choice (Birkhead and Pizzari 2002). The ultimate outcome of these interactions will impact adaptive processes within populations and may influence higher-level processes such as speciation.
D. melanogaster males can increase their reproductive success not only by gaining additional mates (Bateman 1948), but also by influencing patterns of polyandry or affecting the use of sperm by females with whom they have mated. A male's reproductive success increases if he can prevent his mate from remating or if he can out-compete the sperm of other males to ensure that he fertilizes the majority of the female's eggs. Females, however, are unlikely to be passive recipients since natural selection is simultaneously maximizing their own reproductive success. If the reproductive interests of males and females differ, antagonistic coevolution between the sexes may influence postcopulatory sexual selection (Parker 1979; Rice and Holland 1997). Given the large effect that variation in reproductive success will have on overall fitness, traits affecting postcopulatory sexual selection are likely to be under strong selection in natural populations.
For selection to operate, variation must exist between individuals for traits affecting postcopulatory sexual selection, and empirical studies consistently demonstrate a genetic contribution to the observed phenotypic variance. For example, testis weight, ejaculate volume, and copulation duration have high heritabilities in dung beetles (Simmons and Kotiaho 2002). Both male investment in spermatogenesis (Pitnick et al. 2001b) and female remating rate (Pitnick et al. 2001a) also have strong genetic components. The effect of the male genotype (Civetta and Clark 2000; Nilsson et al. 2003), the female genotype (Clark and Begun 1998; Nilsson et al. 2003), and the interaction between the male and female genotype (Clark et al. 1999; Nilsson et al. 2003) on components of postcopulatory sexual selection have been shown using chromosome extraction lines. Quantitative genetic analysis reveals that most of the genetic variation in sperm precedence is nonadditive, and this finding is consistent with a form of balancing selection (Hughes 1997). Although these studies demonstrate a genetic component to phenotypic variation, they fail to provide a molecular basis for the observed variation.
Natural genetic variation in male reproductive fitness may be caused by variation in accessory gland proteins (Acps) or other male reproductive proteins that are transferred to the female as components of the seminal fluid (Clark et al. 1995). Acps affect a variety of processes that impact both male and female reproductive success (Wolfner 2002; Gillott 2003). For example, Acp26Aa increases egg-laying rate in mated females (Herndon and Wolfner 1995; Heifetz et al. 2000), providing a means for males to boost their reproductive success by manipulating female behavior. Acp70A influences oogenesis (Chen et al. 1988; Aigaki et al. 1991; Soller et al. 1999) and also decreases female receptivity to subsequent matings (Chen et al. 1988; Chapman et al. 2003; Liu and Kubli 2003). Variants in Acp70A could provide a key advantage if they allow a male to monopolize the reproductive efforts of his mates. Acp36DE is required for sperm storage (Neubaum and Wolfner 1999), and null mutants sire a smaller proportion of offspring when they are the second male to mate as compared to controls (Chapman et al. 2000).
If natural variation in male reproductive proteins affects reproductive success, selection may have left a molecular signature in current patterns of genetic diversity. Comparisons of sequences within and between closely related species suggest that male reproductive genes are under strong selective pressure (Civetta and Singh 1998; Swanson and Vacquier 2002). Adaptive evolution appears to be driving nonsynonymous substitutions in Acp26Aa (Aguadé 1998; Tsaur et al. 1998), Acp29Ab (Aguadé 1999; Begun et al. 2000), and Acp36DE (Begun et al. 2000) and this may be the result of antagonistic coevolution between the sexes (Parker 1979; Rice 1996). Strong selection, however, has not eliminated polymorphism in these genes. In fact, elevated levels of amino acid polymorphism compared to neutral expectations have been documented in several male reproductive proteins (Aguadé 1999; Begun et al. 2000; Tsaur et al. 2001). Several hypotheses, including nontransitivity among male genotypes (Clark et al. 2000), male-by-female interactions (Clark et al. 1999), and antagonistic pleiotropy (Prout and Clark 1996) have been proposed to explain the maintenance of genetic polymorphism. In addition to allelic variation in male reproductive genes, the quantity of seminal fluid proteins transferred to the female may play a key role in determining fertilization success. Differences in transcript production or stability may lead to such differences in protein content. Recent studies using microarrays (but not investigating Acps) have documented significant variation in transcript abundance at a variety of levels: between different individuals within a population (Oleksiak et al. 2002), between different strains of Drosophila (Meiklejohn et al. 2003), and among closely related mammalian species (Enard et al. 2002). Although it is not yet clear how variation in transcript abundance of male reproductive proteins might affect male fitness, studies of other phenotypes document strong links between transcript abundance and phenotype (see Purugganan 2000). Given the putative roles that male reproductive proteins play in postcopulatory sexual selection, variation in transcript abundance, in addition to variation at the nucleotide level, may be associated with differences in male reproductive success.
The goal of this study is to investigate whether natural variation in male reproductive proteins correlates with phenotypes likely to influence postcopulatory sexual selection. Across a set of lines derived from a natural population we have scored: natural polymorphisms in male reproductive genes (genotypes), variation in mRNA level of these genes (transcript abundance), and standard components of sperm competitive ability (phenotypes). Statistical tests were used to identify associations between genotype and transcript abundance, genotype and phenotype, and transcript abundance and phenotype. We report several associations between genotype and both transcript abundance and sperm competition phenotypes but no associations between transcript abundance and phenotype. One marker showed evidence for antagonistic pleiotropy between two phenotypes, suggesting a mechanism that maintains genetic variation.
MATERIALS AND METHODS
Drosophila lines:
A total of 101 chromosome 2 substitution lines derived from a natural population in State College, Pennsylvania (Lazzaro et al. 2004), were used as the experimental lines in this study. Lines are homozygous and identical for the third, fourth, and sex chromosomes, but each line contains a unique homozygous second chromosome. The tester males and the females used in this study were a standard lab cn bw strain (Civetta and Clark 2000). The experimental line carries the spapol mutation, giving those flies sparkling red eyes while the cn bw strain has recessive white eyes. Cultures were maintained on standard agar-dextrose-yeast media at 24° on a 12-hr light/dark cycle.
Scoring sperm competition phenotypes:
Both “defense” (experimental male is the first male to mate) and “offense” (experimental male is the second male to mate) components of sperm competition were measured in the experimental lines using a similar design as Clark et al. (1995). Six statistics were recorded for each line: P1′, P2′, fecundity from the offense and defense experiments, remating rate, and refractoriness (see below for details). P1′ and P2′ are the proportion of offspring sired by the experimental male when he is either the first or the second male to mate with a doubly mated female, respectively. The primes on P1 and P2 indicate that these metrics are based on females that are inferred to have mated with both males from the presence of both progeny types. Double matings that fail to produce progeny will be missed by this metric (Clark and Begun 1998). Fecundity was defined as the total number of offspring produced by each female across all vials. Remating rate is the proportion of experimental males to mate with an already mated female and refractoriness is the proportion of females that do not remate following mating to an experimental male. Strictly speaking, these phenotypes affect male reproductive fitness via differences in postcopulatory sexual selection, but we broadly refer to these statistics as components of sperm competitive ability or “sperm competition phenotypes.”
Virgin males and females were collected over CO2 and housed in single-sex vials at low density until 4–7 days old. Mass matings were conducted using 10 males and 10 females on the evening of day 0. On the morning of day 1, individual females were transferred, without anesthesia, to vial 1 and the males were discarded. On the evening of day 3, 2 virgin males were added to vial 1. On the morning of day 4, the females were transferred, without anesthesia, to vial 2 and the males were discarded. On day 7 the females were transferred, without anesthesia, to vial 3 and then discarded on day 14. Progeny were scored for eye color ∼15, 17, and 19 days after the initiation of egg laying for vials 1, 2, and 3, respectively. Only females that survived the entire experiment were used in analyses.
Analysis of variance (ANOVA, PROC MIXED) was used to test for differences in P1′, P2′, and fecundity across lines. The model is Pijk = μ + Li + B(L)ij + εijk, where Pijk is the observed statistic, μ is the overall mean, Li is the effect of the ith line, B(L)ij is the effect of the jth block (random factor) nested in the ith line, and εijk is the error term. P1′ and P2′ were arcsine square root transformed to improve the fit to normality (Clark et al. 1995). Least-square means for each line were calculated and used for association testing. Permutation tests based on chi-square statistics were performed in MATLAB and used to test for significant heterogeneity among lines in refractoriness and remating rate.
Measuring variation in mRNA level:
Total RNA was extracted from whole bodies of 15, 2-day-old virgin males using Trizol (Invitrogen, San Diego) according to the manufacturer's protocols and stored at −70°. Two-day-old virgins were chosen because all males in the sperm competition experiments were virgins; mating has been shown to upregulate expression of Acps (DiBenedetto et al. 1990; Herndon et al. 1997), and expression of Acps appears to be highest before day 4 posteclosion (Herndon et al. 1997). Two separate extractions were performed for each of the 101 experimental lines. Total RNA was resuspended in 80 μl of 0.1% DEPC-treated ddH2O and 8.0 μl of total RNA was treated with RNase-free DNase (Promega, Madison, WI) using twice the recommended concentration. First-strand cDNA synthesis using oligo(dT16) primers and M-MLV reverse transcriptase (Promega) was conducted according to manufacturer's protocols. The resulting cDNA was diluted to 1:16 for use in the quantitative real-time PCR reactions.
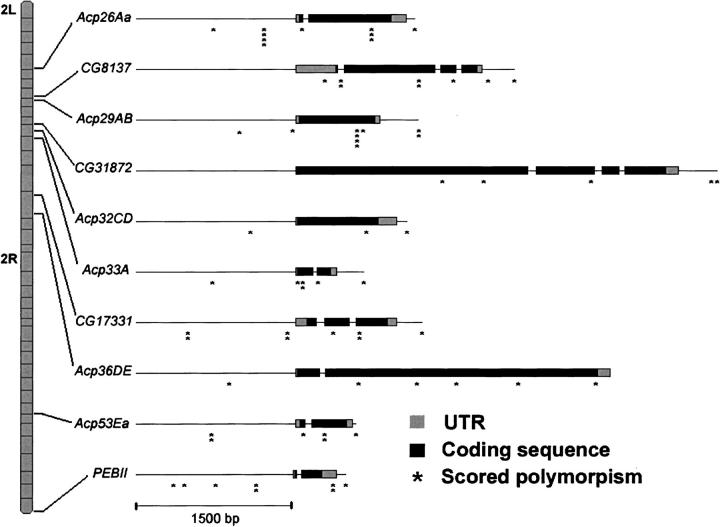
Standing mRNA levels of 10 male reproductive proteins (Acp26Aa, CG8137, Acp29AB, CG31872, Acp32CD, Acp33A, CG17331, Acp36DE, Acp53Ea, and PEBII) were measured using quantitative real-time PCR (Figure 1). Nine of these proteins are produced by the male accessory gland (Monsma and Wolfner 1988; Wolfner et al. 1997; Swanson et al. 2001) while PEBII is produced by the ejaculatory bulb (Dyanov and Dzitoeva 1995). Ribosomal protein L32 (RPL32) was used as a control gene to normalize the data (B. Bettencourt, unpublished primer sequences). Primer sequences are presented in supplemental Table 1 at http://www.genetics.org/supplemental/. Reactions were performed on an ABI 5700 using SYBR green I (Molecular Probes, Eugene, OR) as the fluorescent dye except for Acp29AB, which, due to a polymorphism under the original primer pair, was rerun on an ABI 7000 using a TaqMan probe (Applied Biosystems, Foster City, CA) and primers that avoided the known polymorphisms. We also verified that the qRT-PCR primers for the other assays were located in conserved regions. SYBR green reactions were performed in 50-μl volumes with final concentrations of 3 mm MgCl2, 1× Real-Time reaction buffer (Ambion, Austin, TX), 1/4000 SYBR green I (Molecular Probes), 60 nm ROX I (Synthegen), 0.2 mm each dNTP, 0.1 μm each primer, 1 unit Taq polymerase (Promega), and 5 μl of the diluted cDNA. Cycle conditions were 94° for 2 min followed by 40 cycles of 94° for 50 sec, 58° for 50 sec, and 72° for 60 sec with a final 7-min extension at 72°. A dissociation curve was run to ensure that only a single product was amplified and any reactions containing notable primer dimers were excluded from analysis. Acp29AB reactions were run according to manufacturer's recommendations for TaqMan probes. Standard curves were created using four PCR replicates from each of four different dilutions (1:8, 1:16, 1:32, 1:64) of a single RNA extraction and cDNA synthesis. Simple linear regression was used on log-transformed concentrations to calculate r2 and the slope of the standard curve was used to calculate assay efficiencies as: efficiency = [10(−1/slope)] − 1.
Figure 1.—
Schematic of location of genes and scored polymorphic sites on chromosome 2. The approximate location of each analyzed gene is shown relative to the cytological bands on the second chromosome. The approximate location of each scored polymorphic site is depicted on the gene structure running 5′–3′. Exons are depicted as boxes with the coding sequence solid and the untranslated regions shaded. The number of individual sites scored are indicated by the number of asterisks.
TABLE 1.
Summary of sperm competition phenotypes
| Phenotype | Mean across lines | Range of line means | d.f. | Test statistic |
|---|---|---|---|---|
| Defense | ||||
| P1′ | 0.41 | 0.004–0.79 | 94,1269 | F = 13.0*** |
| Fecundity-D | 79.6 | 48–107 | 94,1267 | F = 1.7*** |
| Refractoriness | 0.13 | 0–0.5 | 95 lines | χ2 = 16.5, permutation*** |
| Offense | ||||
| P2′ | 0.91 | 0.49–1.0 | 94,1142 | F = 4.7*** |
| Fecundity-O | 86.9 | 55–140 | 94,1142 | F = 1.7*** |
| Remating | 0.78 | 0.1–1.0 | 95 lines | χ2 = 47.2, permutation*** |
In defense tests, the experimental male was the first male to mate. P1′ is the proportion of offspring sired by the first male. Fecundity-D is the total fecundity of doubly mated females in the defense test. Refractoriness is the proportion of females that did not remate. In offense tests, the experimental male was the second male to mate. P2′ is the proportion of offspring sired by the second male. Fecundity-O is the total fecundity of doubly mated females in the offense test. Remating is the proportion of females that remated. *P < 0.05, **P < 0.01, ***P < 0.001 based on ANOVA or permutation tests (see text).
An incomplete block design was used to partition the individual lines across 66 different 96-well plates. Each plate contained both extractions for three randomly chosen lines. All 11 primer sets were run on each plate and one-third of the wells were randomly selected for PCR replication. Negative controls (no template) were run on each plate and all were free of contamination. Each line appeared on two different plates and replicate PCR reactions on the same plate were averaged prior to analysis. The default ΔRn analysis threshold (0.2) was used for each plate and the reciprocal of the resulting critical threshold (1/CT) was used in the following analyses (Applied Biosytems). For each extraction, 1/CT of the “gene of interest” was regressed against 1/CT of RPL32 and the residuals from the analysis were used. SAS procedure GLM was used to test for differences in transcript abundance among the lines. The model is Gijkl = μ + Pi + Lj + X(L)jk + εijkl, where Gijkl is the residual of transcript abundance from the above regression, μ is the overall mean, Pi is the effect of the ith plate, Lj is the effect of the jth line, X(L)jk is the effect of the kth extraction nested in the jth line, and εijkl is the error term. Least-squares line means were used to test for associations.
Preliminary experiments demonstrated that this approach accurately reconstructed relative transcript abundance. For these experiments, we varied the amount of male and female tissue prior to RNA extraction to produce samples that differed in the abundance of male reproductive transcripts but had the same overall amount of tissue (and therefore the same abundance of the control gene, RPL32). We analyzed “dilutions” of 100, 50, and 25% male tissue. Scaled relative to the samples with 100% male tissue, our method estimated transcript abundances of 0.49 ± 0.07 (SE) and 0.22 ± 0.03 (SE) for those samples containing 50 and 25% male tissue, respectively. Analyses using transcript abundance of RPL32 as a covariate produced nearly identical results (data not shown).
Identifying and genotyping polymorphisms:
Single nucleotide polymorphisms (SNPs) and insertion-deletion polymorphisms (indels) were identified from Genbank sequences and through additional sequencing of 10 of the 101 experimental lines. Surveyed regions included ∼1 kb upstream of the transcription start site to ∼500 bp downstream of the 3′-UTR. Attempts were made to genotype at least one polymorphism in each of the upstream, coding, and downstream regions from each gene (Figure 1). Seventy polymorphic sites were typed in the 10 male reproductive genes (see supplemental Table 2 at http://www.genetics.org/supplemental/ for descriptions). Four of these sites were indels that were scored on 4% agarose gels, while the remaining 66 sites were SNPs scored via Pyrosequencing (Ahmadian et al. 2000).
TABLE 2.
Correlations among line means for sperm competition phenotypes
| P1′ | Fecundity-D | Refractoriness | P2′ | Fecundity-O | |
|---|---|---|---|---|---|
| Fecundity-D | 0.205* | ||||
| Refractoriness | 0.217* | 0.083 | |||
| P2′ | 0.706** | 0.114 | 0.027 | ||
| Fecundity-O | 0.038 | −0.158 | −0.015 | 0.134 | |
| Remating | 0.139 | 0.079 | −0.092 | 0.305** | 0.100 |
In defense tests, the experimental male was the first male to mate. P1′ is the proportion of offspring sired by the first male. Fecundity-D is the total fecundity of doubly mated females in the defense tests. Refractoriness is the proportion of females that did not remate. In offense tests, the experimental male was the second male to mate. P2′ is the proportion of offspring sired by the second male. Fecundity-O is the total fecundity of doubly mated females in the offense tests. Remating is the proportion of females that remated. *P < 0.05, **P < 0.01 (significant after Bonferroni correction), ***P ≤ 0.001.
For most Pyrosequencing assays, a universal biotinylated primer was used in combination with two locus-specific primers (e.g., Guo and Milewicz 2003), although direct-biotinylated locus-specific primers were used for some assays (primer sequences in supplemental Table 2 at http://www.genetics.org/supplemental/). PCR amplifications were carried out in 25- to 50-μl reactions with final concentrations of 1.5 mm MgCl2, 1× PCR buffer (Promega), 0.25 mm each dNTP, 0.3 μm primer lacking UB2 sequence, 0.03 μm primer with UB2 sequence, 0.26 μm 5′Bio/UB2 primer, and 0.5 units Taq Polymerase (Promega). When using direct-biotinylated locus-specific primers, each primer was at a final concentration of 0.3 μm. Cycle conditions were 95° for 2 min followed by 40 cycles of 95° for 15 sec, 55° for 30 sec, and 72° for 15 sec with a final 5-min extension at 72°. Single-strand PCR products were prepared according to manufacturer's protocols using a 96-pin vacuum preparation tool (Pyrosequencing) and combined with 0.3 μm sequencing primer in annealing buffer (20 mm Tris-Acetate; 2 mm MgAc2, pH 7.6). Sequences were analyzed using a PSQ 96MA (Pyrosequencing) instrument according to manufacturer's protocols, except all components of the 96 PSQ SNP reagent kit were diluted to 0.5×. Gametic disequilibrium was tested using Fisher's exact test as implemented in GENEPOP (Raymond and Rousset 1995).
Association analysis:
Associations between genotype, transcript abundance, and sperm competition phenotypes were conducted in MATLAB using simple linear regression and permutation tests (Churchill and Doerge 1994). For each phenotype, line means were randomly permuted across genotypes 5000 times and the maximum F-value for each predictor (i.e., individual marker or gene expression level) and the largest F-value across all predictors were recorded. The per-character experimentwise P-value for each phenotype was calculated by comparing the observed F-value to the maximum F-value distribution across the distribution of characters in the randomly permuted data, while the comparisonwise P-value was calculated according to the distribution of F-values for each respective predictor. The former test is overly conservative for a given phenotype because of differences in allele frequencies and number of scored lines across markers, while the latter fails to fully correct for the effect of testing multiple markers. Occasionally, lines were eliminated from individual analyses due to technical difficulties (e.g., failure to score a given marker) and thus sample sizes vary slightly (supplemental Table 2 at http://www.genetics.org/supplemental/). A total of 128 polymorphic markers in immunity genes were typed in these same lines (Lazzaro et al. 2004) and used as negative controls for this study.
RESULTS
Variation in sperm competitive ability:
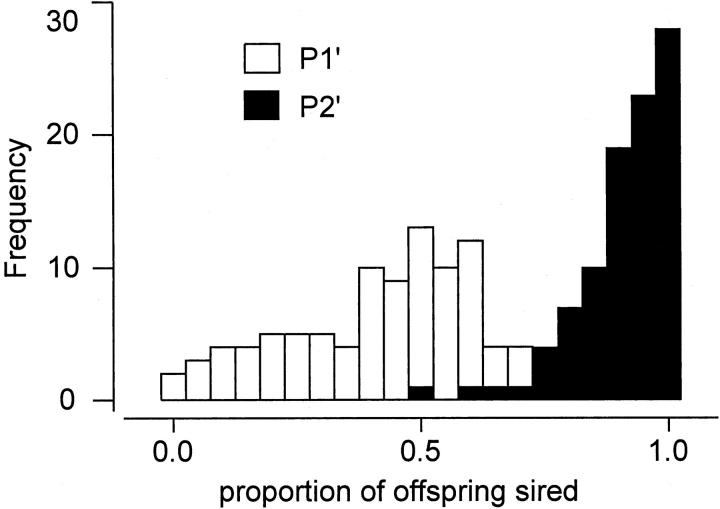
A total of 1547 females and 121,944 offspring from 95 lines were analyzed in the defense experiment while 1581 females and 132,578 offspring from 95 lines were analyzed in the offense experiment. Variation in development time prevented some lines from being included in the sperm competition experiments (if 4- to 7-day-old virgins were to be used) and accounts for having scored only 95 of the 101 lines. Significant line effects were detected using ANOVA for P1′, P2′, and fecundity in both offense and defense experiments and significant line effects were detected for remating rate and refractoriness using a permutation test (Table 1). These results indicate that significant genetic variation exists for components of male reproductive fitness. Overall, the second male to mate obtained the majority of the fertilizations; mean P1′ was 0.41 and mean P2′ was 0.91 (Table 1, Figure 2). P1′ was weakly positively correlated with the two other defense parameters, fecundity (defense), and refractoriness. P2′ was strongly positively correlated with P1′ and remating rate (Table 2). The majority of females mated with both males in the offense (78%) and defense (83%) experiments (Table 1) and doubly mated females had significantly higher fecundity than females mated to only a single male in both the offense (86.9 ± 1.1 and 53.9 ± 1.8 SE) and the defense experiments (79.6 ± 1.0 and 41.4 ± 2.9 SE).
Figure 2.—
Histograms of sperm competition phenotypes P1′ and P2′. The distribution of the fraction of offspring sired by the experimental males when they were the first males to mate with a doubly mated female, P1′, is indicated by open bars. The distribution of the fraction of offspring sired by the experimental males when they were the second males to mate with a doubly mated female, P2′, is indicated by solid bars. A total of 95 lines were analyzed.
Variation in mRNA levels:
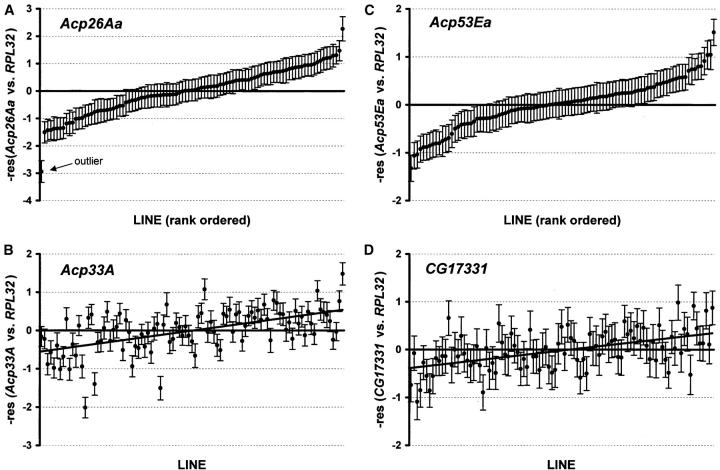
Transcript abundance of RPL32 was positively correlated with transcript abundance of each male reproductive protein (r2 = 0.57–0.85, Table 3), and thus the residuals from the regression have been used in all subsequent analyses. Even after normalization by RPL32, there were highly significant among-line effects for all the male reproductive genes investigated (Table 3, Figure 3), indicating extensive natural genetic variation in transcript level. Ranges of the residuals for the CT values varied from −5.9 to 2.8 for PEBII but only from −1.1 to 1.0 for CG17331 (Table 3). Taking into account the efficiencies of the real-time PCR assays (supplemental Table 1 at http://www.genetics.org/supplemental/), these values correspond to a 349-fold difference in transcript abundance between the highest and lowest lines for PEBII but only a 2.8-fold difference in gene expression of CG17331 (Table 3). Two genes, PEBII and Acp26Aa, each had a single line with dramatically reduced expression compared to the others. The standard error around the least-squares means for these two lines did not overlap with the standard error for any of the other lines (Table 3, Figure 3), but these were not the same line across the two genes. Removal of these outliers from the analysis lowered the observed differences between the highest and lowest lines from 37- to 14-fold at Acp26Aa and from 349- to 33-fold at PEBII. Transcript abundances of the 10 male reproductive genes were highly positively correlated across the experimental lines (Table 4, Figure 3): 40 of the 45 pairwise comparisons were significant at P < 0.05 and 34 of these pairwise comparisons remained significant after Bonferroni correction.
TABLE 3.
Summary of variation in transcript abundance
| Gene |
RPL32 abundance as predictor (r2)a |
Range of residuals (CTs) |
≈ Transcript fold differenceb |
d.f. | F-ratioc |
|---|---|---|---|---|---|
| Acp26Aa | 0.82*** | −2.9–2.3 | 37d | 96,242 | 5.3*** |
| CG8137 | 0.71*** | −1.7–2.4 | 13.3 | 95,227 | 6.5*** |
| Acp29AB | 0.67*** | −1.0–1.5 | 5.2 | 94,187 | 3.4*** |
| CG31872 | 0.80*** | −1.6–1.5 | 7.9 | 96,243 | 8.9*** |
| Acp32CD | 0.73*** | −1.5–1.3 | 4.4 | 96,263 | 7.4*** |
| Acp33A | 0.79*** | −2.0–1.5 | 10.7 | 96,237 | 5.3*** |
| CG17331 | 0.85*** | −1.1–1.0 | 2.8 | 96,251 | 2.5*** |
| Acp36DE | 0.76*** | −2.1–1.2 | 6.5 | 96,249 | 7.4*** |
| Acp53Ea | 0.83*** | −1.3–1.5 | 7.0 | 96,254 | 4.9*** |
| PEBII | 0.57*** | −5.9–2.8 | 349e | 96,262 | 2.8*** |
*P < 0.05, **P < 0.01, ***P ≤ 0.001 based on simple linear regression (see text).
Transcript fold difference calculated as: difference = (1 + PCR efficiency)(largest residual CT−smallest residual CT).
*P < 0.05, **P < 0.01, ***P ≤ 0.001 based on ANOVA test for heterogeneity among lines (see text).
Excluding outlier results in a 14-fold difference.
Excluding outlier results in a 33-fold difference. This outlier was not the same line as the outlier in Acp26Aa.
Figure 3.—
Natural variation in transcript abundance across four male reproductive genes. Line means and standard errors for the residuals of the regression of 1/CT for “gene of interest” vs. 1/CT for RPL32 are shown. Higher residuals indicate higher relative mRNA levels. (A) Rank-ordered lines for expression of Acp26Aa with an outlier showing reduced gene expression. (B) Expression of Acp33A is positively correlated with expression of Acp26Aa. Lines ranked according to expression of Acp26Aa from A (r2 = 0.67, P < 0.001). (C) Rank-ordered lines for expression of Acp53Ea. (D) Expression of CG17331 is positively correlated with expression of Acp53Ea. Lines ranked according to expression of Acp53Ea from C (r2 = 0.56, P < 0.001).
TABLE 4.
Correlations among line means for transcript abundance
| Acp26Aa | CG8137 | Acp29AB | CG31872 | Acp32CD | Acp33A | CG17331 | Acp36DE | Acp53Ea | |
|---|---|---|---|---|---|---|---|---|---|
| CG8137 | 0.539*** | ||||||||
| Acp29AB | 0.529*** | 0.468*** | |||||||
| CG31872 | 0.579*** | 0.576*** | 0.461*** | ||||||
| Acp32CD | 0.153 | 0.343*** | 0.170 | 0.362*** | |||||
| Acp33A | 0.667*** | 0.606*** | 0.391*** | 0.657*** | 0.446*** | ||||
| CG17331 | 0.518*** | 0.193 | 0.261* | 0.403*** | 0.290** | 0.410*** | |||
| Acp36DE | 0.492*** | 0.513*** | 0.438*** | 0.470*** | 0.325*** | 0.591*** | 0.174 | ||
| Acp53Ea | 0.609*** | 0.369*** | 0.526*** | 0.561*** | 0.478*** | 0.629*** | 0.561*** | 0.478*** | |
| PEBII | 0.309*** | 0.167 | 0.226 | 0.275** | 0.392*** | 0.284** | 0.366*** | 0.211* | 0.489*** |
P < 0.05,
P < 0.01,
P ≤ 0.001 (significant after Bonferroni correction).
Genotype data:
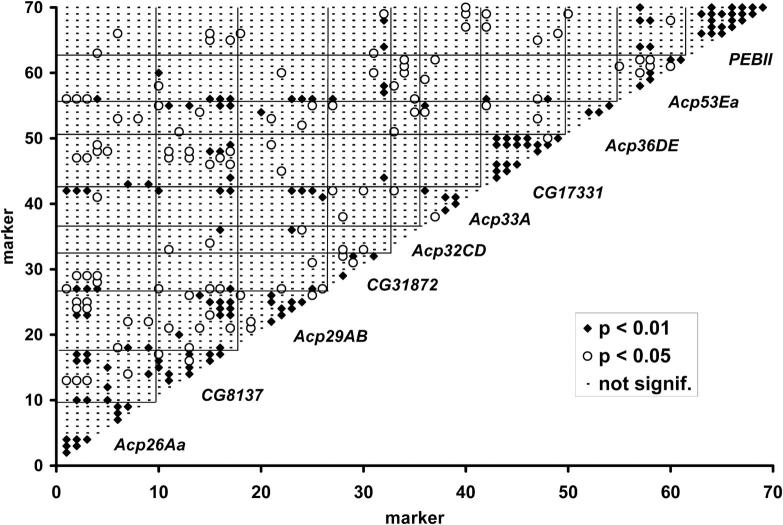
Most of the 70 scored polymorphisms had relatively complete data with 53 successfully genotyped in at least 90 of the lines (supplemental Table 2 at http://www.genetics.org/supplemental/). The common allele was at a frequency of <80% in 46 of the markers and only 10 markers had the common allele at a frequency of >90% (supplemental Table 2 at http://www.genetics.org/supplemental/). Independence of sites across genes is important for testing associations between SNPs and sperm competitive phenotypes. In general, linkage disequilibrium was most common within genes, but occasional long-distance associations were observed (Figure 4).
Figure 4.—
Linkage disequilibrium across the 70 markers scored in the chromosome 2 substitution lines. Highly significant disequilibrium (P < 0.01, Fisher's exact test) is depicted by solid diamonds, and significant disequilibrium (P < 0.05, Fisher's exact test) is depicted by open circles.
Associations:
Nine significant associations (comparisonwise P < 0.01 or experimentwise P < 0.05) were identified between genotype and the measured sperm competition phenotypes (3 offense and 6 defense, Table 5), and an additional 24 associations were suggestive (comparisonwise P < 0.05, supplemental Table 3 at http://www.genetics.org/supplemental/). Of the 9 significant associations, 6 were in noncoding regions (3 upstream, 2 downstream, and 1 in an intron), two markers were synonymous changes, and one was a nonsynonymous change. P1′ was affected by a marker in the intron of CG8137 (CG8137s1910) and also by a marker downstream of Acp33A (Acp33s2125). Refractoriness was associated with a marker upstream of CG17331 (CG17331s1421). Two markers upstream of CG17331 (CG17331s1411 and CG17331s1421) affected P2′; these were adjacent markers in linkage disequilibrium and may represent only a single true association. A synonymous change in Acp29AB (Acp29s2072), a marker downstream of Acp33A (Acp33s2125), and a radical amino acid change from serine to isoleucine at position 207 of Acp26Aa (Acp26s2201) were also associated with the proportion of offspring sired by the second male, P2′. Remating rate was affected by a synonymous change in CG31872 (CG31872s4328). No significant associations were identified for fecundity in either offense or defense experiments.
TABLE 5.
| Phenotype | Marker | Frequency of “fit” allele | Marker typea | Adjusted r2b | Other associationsc |
|---|---|---|---|---|---|
| Defense | |||||
| P1′ | CG8137s1910** | 0.84 | Intron | 0.08 | 33,17331 (+) |
| Acp33s2125** | 0.78 | Downstream | 0.08 | P2, Rem (+) | |
| Fecundity-D | None | NA | NA | NA | NA |
| Refractoriness | CG17331s1421** | 0.18 | Upstream | 0.06 | P2 (−): FecO (+) |
| Offense | |||||
| P2′ | Acp26s2201**** | 0.72 | AGC(S):ATA(I) | 0.13 | 53 (−) |
| Acp29s2072** | 0.73 | ACA(un.):ACC(pref.) | 0.07 | P1 (+) | |
| Acp33s2125**** | 0.78 | Downstream | 0.16 | P1, Rem (+) | |
| CG17331s1411** | 0.91 | Upstream | 0.08 | FecO (+) | |
| CG17331s1421** | 0.82 | Upstream | 0.08 | Refr (−): FecO (+) | |
| Fecundity-O | None | NA | NA | NA | NA |
| Remating | CG31872s4328** | 0.34 | CTT(un.):CTC(un.) | 0.05 | None |
In defense tests the experimental male was the first male to mate. P1′ is the proportion of offspring sired by the first male. Fecundity-D is the total fecundity of doubly mated females in the defense tests. Refractoriness (Refr) is the proportion of females that did not remate after mating to experimental males. In offense tests the experimental male was the second male to mate. P2′ is the proportion of offspring sired by the second male. Fecundity-O (FecO) is the total fecundity of doubly mated females in the offense tests. Remating (Rem) is the proportion of females that remated in the offense tests.
Comparisonwise P < 0.05,
P < 0.01,
P ≤ 0.001,
Experimentwise P < 0.05. NA, not applicable.
High-fitness codon shown on the left with its amino acid for nonsynonymous changes or usage; preferred (pref.) and unpreferred (un.) for synonymous changes indicated in parentheses.
Calculated using a simple linear regression considering only the individually significant marker (see text).
“Other associations” indicates other phenotypes that were associated with that marker. Significant associations are underlined. Antagonistic associations indicated by (−) and protagonistic associations indicated by (+). Markers associated with both phenotype and expression were considered protagonistic if the same allele produced higher fitness and higher transcript abundance. Genes are listed excluding the “CG” or the “Acp” prefix.
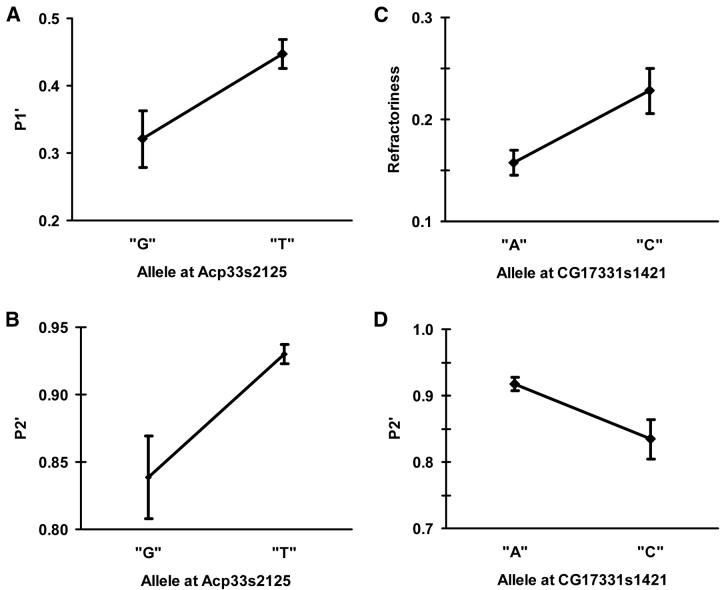
Two different markers had pleiotropic effects on multiple sperm competition phenotypes (Figure 5). A marker downstream of Acp33A (Acp33s2125) influenced P1′ and P2′ with the same allele conferring an advantage for both phenotypes. A marker upstream of CG17331 (CG17331s1421) was associated with both P2′ and refractoriness, but in this case the alleles acted antagonistically. Males carrying the more common allele sired a greater proportion of offspring when they were the second to mate, but females were more likely to remate after copulating with these males. Excluding the marker with antagonistic effects, the allele with higher fitness was at a higher frequency in six of the seven associations (Table 5).
Figure 5.—
Pleiotropy at male reproductive genes. Protagonistic pleiotropy at marker Acp33s2125 for P1′ and P2′ (A and B). (A) Mean P1′ and standard errors for lines with alternative alleles at Acp33s2125. (B) Mean P2′ and standard errors for lines with alternative alleles at Acp33s2125. Antagonistic pleiotropy at marker CG17331s1421 for P2′ and refractoriness (C and D). (C) Mean refractoriness and standard errors for lines with alternative alleles at CG17331s1421. (D) Mean P2′ and standard errors for lines with alternative alleles at CG17331s1421.
Thirteen markers were associated with transcript levels of eight different male reproductive genes (Table 6), and an additional 38 associations were suggestive (supplemental Table 4 at http://www.genetics.org/supplemental/). Of the 13 significant associations, 4 were between markers in and transcript level of the same gene: a synonymous substitution in CG8137 (CG8137s2957), a marker upstream of CG17331 (CG17331s486), and two markers in the coding sequence of Acp36DE (a synonymous difference at Acp36s2084 and an 8-amino-acid insertion deletion at Acp36Indel2623). The two markers in Acp36DE are adjacent markers in linkage disequilibrium and thus may represent only a single true association. In the remaining 9 cases, a marker in one gene was associated with transcript abundance of another gene (Table 6). One marker, CG8137s2957, was associated with the transcript level of two different genes (CG8137 and Acp36DE) and the same allele was associated with higher transcript abundance for both. In no case was transcript level of a male reproductive gene associated with any of the sperm competition phenotypes.
TABLE 6.
Genotype-to-transcript-level associations*
| Phenotype | Marker | Frequency of “fit” allele | Marker typea | Adjusted r2b | Other associationsc |
|---|---|---|---|---|---|
| Acp26Aa | Acp33s1559** | 0.91 | GAT(pref.):GAC(un.) | 0.05 | None |
| CG8137 | CG8137s2957** | 0.50 | GGA(un.):GGT(un.) | 0.08 | 29, 33 (−): 36 (+) |
| Acp29AB | CG17331s2065**** | 0.70 | AAC(pref.):AAT(un.) | 0.15 | P1 (+) |
| CG31872 | None | NA | NA | NA | NA |
| Acp32CD | None | NA | NA | NA | NA |
| Acp33A | Acp29s2633*** | 0.95 | Downstream | 0.10 | 32, 8137 (+) |
| CG17331 | CG17331s486*** | 0.33 | Upstream | 0.08 | 53 (+) |
| Acp36DE | CG8137s2957** | 0.50 | GGA(un.):GGT(un.) | 0.07 | 29, 33 (−): FecO, 8137 (+) |
| Acp36s2084** | 0.52 | CUA(un.):GUC(un.) | 0.09 | None | |
| Acp36Indel2623**** | 0.47 | LLREAQQK pos. 253 | 0.12 | None | |
| PEBs1155*** | 0.44 | Upstream | 0.11 | None | |
| Acp53Ea | Acp26s2193*** | 0.29 | AGC(pref.):AGT(un.) | 0.11 | 26, 33, 36, 31872 (+) |
| Acp26s2201** | 0.28 | ATA(I):AGC(S) | 0.07 | P2 (−) | |
| PEBII | Acp53s1760** | 0.91 | ACC(pref.):ACT(un.) | 0.10 | None |
| Acp53s1766** | 0.70 | CGT(un.):CGC(pref.) | 0.08 | 26, 29, 33, 31872 (+) |
Comparisonwise P < 0.05,
P < 0.01,
P ≤ 0.001,
Experimentwise P < 0.05. NA, not applicable.
High-fitness codon shown on the left with its amino acid for nonsynonymous changes or usage; preferred (pref.) and unpreferred (un.) for synonymous changes indicated in parentheses.
Calculated using a simple linear regression considering only the individually significant marker (see text).
“Other associations” indicates other phenotypes that were associated with that marker. Significant associations are underlined. Antagonistic associations indicated by (−) and protagonistic associations indicated by (+). Markers associated with both phenotype and expression were considered protagonistic if the same allele produced higher fitness and higher transcript abundance. Genes are listed excluding the “CG” or “Acp” prefix.
A simple linear regression was run on each of the individually significant associations and the adjusted r2 was recorded. Most markers explained only a small proportion of the variance with the adjusted r2 ranging from 0.05 to 0.16, and many markers explain ∼8% of the variance (Tables 5 and 6). Only a single significant association (comparisonwise P < 0.01) was identified between the negative control immunity genotypes and a sperm competition phenotype, and overall there was a significant deficit of associations as compared to the male reproductive genes (χ2 = 28.2, d.f. = 1, P ≪ 0.001). The false discovery rate (Storey and Tibshirani 2003 and references within) was also calculated, using all of the measured phenotypes. Among the 22 associations identified at P < 0.01, 8 may represent false positives. Only 1 false positive is expected among the 8 associations identified at P < 0.001 and none of the associations at the experimentwise P < 0.05 are expected to be false positives.
DISCUSSION
In this study we use natural variation in a population of D. melanogaster to investigate the genetic basis of sperm competitive ability. We document significant genetic effects for both offense and defense components of sperm competitive ability and provide one of the most comprehensive studies of genetic variation affecting transcript level of male reproductive genes. We also identify several associations among genotypic variation in male reproductive genes (SNPs and indels) and both transcript abundance and sperm competition phenotypes. Furthermore, our results indicate that antagonistic pleiotropy is operating among alleles at CG17331 that are associated with the proportion of offspring sired by the second male to mate (P2′) and with refractoriness. These associations provide important insights into the molecular basis of sperm competition, postcopulatory sexual selection, and the evolution of male reproductive proteins.
Correlations among components of sperm competitive ability:
Several of the components of sperm competitive ability were positively correlated across lines in this study (Table 2) and in the investigation by Clark et al. (1995). Both studies identified positive correlations between P1′ and refractoriness; individuals that sire more offspring when they are the first male to mate are also better at preventing the female from mating to the second male. Both studies also identified positive correlations between P2′ and remating rate; male genotypes that are better at gaining access to previously mated females are also more effective at gaining fertilizations when they are the second male to mate. At least three explanations exist for these observations: females could be mating more than once in a given vial, females could be using indicators of male quality to influence mate choice, or phenotypes share a common genetic basis.
Triple matings, rather than the expected double matings, could lead to the observed positive correlations among sperm competition phenotypes. If males from high-remating lines copulated with a given female more than once, this could elevate P2′ and lead to the observed positive correlation between remating rate and P2′. Similarly, if the males that induced higher refractoriness in the defense experiment did so by mating twice with the females, then P1′ could have been elevated, leading to the positive correlation between refractoriness and P1′. Although possible, we find this scenario unlikely for several reasons. First, previous experiments have shown that the time the males and females were together yields relatively few triple matings (Civetta and Clark 2000). Second, this hypothesis predicts that remating rate and refractoriness would be positively correlated and they are not in this experiment and have been shown to be negatively correlated in other experiments (Clark et al. 1995). Finally, courtship rates of virgin males from these same lines were not correlated with any of the components of sperm competitive ability (K. A. McKean, unpublished results).
Alternative hypotheses for the observed correlations predict that females are active participants in postcopulatory sexual selection and are using indicators of male quality in mate choice (see Birkhead and Pizzari 2002). Females could thus be gaining indirect genetic benefits from being polyandrous (see Jennions and Petrie 2000) in addition to the direct benefits of higher fecundity. The “phenotype-linked fertility” hypothesis predicts that males with higher precopulatory success will also have higher fertilization success (Sheldon 1994). If females are choosing a trait in males that predicts fertilization success, then P2′ and remating rate should be positively correlated and so would P1′ and refractoriness, as we observe. Furthermore, the “trade-up” hypothesis predicts that if females are basing current mate choice decisions on the perceived quality of their previous mate (Halliday 1983), then they will be more likely to mate with males that are of higher quality than their previous mate. Under these conditions, P2′ and remating rate, as well as P1′ and refractoriness, could be positively correlated.
The observed correlations may also be due to a common genetic basis among the phenotypes. We have identified cases where single markers have pleiotropic effects on male reproductive fitness (see below). In fact, variation at Acp33s2125 was associated with both P2′ and remating rate (although only suggestive for remating rate) and the same allele was favored in both phenotypes. These protagonistic associations could result in the observed positive correlations among sperm competition phenotypes. Although these alternative hypotheses—female choice or pleiotropy—are attractive and our results are consistent with such explanations, more research is required to tease these hypotheses apart.
Interpreting associations:
Several factors were considered prior to interpreting the results of the association analyses. First, the lack of a significant association does not exclude a gene from affecting a given phenotype and could simply imply the lack of segregating variation in the investigated population or reduced statistical power due to highly skewed allele frequencies. On the flip side, a significant association between genotype and phenotype does not indicate that the typed marker is the causative polymorphism. The actual causative site may be physically linked or simply in linkage disequilibrium with the typed marker. Population admixture can result in spurious genotype-to-phenotype associations (e.g., Remington et al. 2001). We did observe long-distance disequilibrium, as would be expected if admixture were present, but we did not observe more than the expected number of random associations with the immunity genes scored by Lazzaro et al. (2004). In fact, we observed significantly fewer associations than expected by chance (P < 0.001), a result that is incompatible with the idea that the sample is seriously admixed. Caveats aside, radical amino acid substitutions, such as the serine-to-isoleucine change identified in Acp26Aa, are promising candidates to be the causal agent and provide testable hypotheses to begin formal molecular and/or biochemical characterizations of the different alleles.
As with any association study, determining the appropriate significance threshold can be difficult, and interpretation of the rate of false positives can be a challenge in the context of multiple testing. Several factors impact the ability to detect significant associations. Sample sizes of the lines that were successfully genotyped vary across the markers as do the allele frequencies at each marker. Markers with fewer genotyped lines or highly skewed allele frequencies will not have the same power as those with complete genotypes or equal allele frequencies. Bonferroni correction also assumes that all of the sites are independent, which is not always the case due to linkage disequilibrium. These difficulties can confound a simple Bonferroni correction and lead to highly conservative conclusions and missed associations (see McIntyre et al. 2000). Experimentwise permutation tests (e.g., Doerge and Churchill 1996) can also be affected when either allele frequencies or the number of scored genotypes differs across markers and also if more than a single marker is expected to associate with the phenotype. Sperm competition phenotypes are quantitative traits likely to be influenced by multiple genes. On the other hand, per-character experimentwise P-values could be liberal as they fail to correct for multiple comparisons. Given these considerations, we have focused on associations with comparisonwise P < 0.01 or per-character experimentwise P < 0.05, but have presented other suggestive associations (comparisonwise P < 0.05) in the supplemental material (supplemental Tables 3 and 4 at http://www.genetics.org/supplemental/). Among the 62 suggestive associations, calculation of the false discovery rate shows that ∼31 may represent false positives (Storey and Tibshirani 2003 and references within). A 50% false positive rate may seem very high, but if the subsequent test is not costly, then testing 62 genes may reveal 31 with a real effect and the effort would be well rewarded. By making the test more conservative, the false positive rate declines, but this comes at the expense of losing true positives as well.
There are some limitations to the study of homozygous chromosomal extraction lines for evolutionary inference. Inbred lines cannot be used to estimate the additive genetic variance and inbreeding has the potential to confound interpretations. It seems unlikely that our conclusions are compromised due to inbreeding depression, however. First, our sperm competition phenotypes were not correlated with male courtship rate (K. A. McKean, unpublished results) or with the immunocompetence phenotype scored by Lazzaro et al. (2004). Furthermore, sperm competition phenotypes tended to be associated with variation in male reproductive genes (this study) while immune phenotypes tested in the same lines were associated with variation in immunity genes (Lazzaro et al. 2004), suggesting that these studies were not mapping variation conferring overall line health. Finally, it seems unlikely that other studies would identify male-by-female interactions (Clark et al. 1999) or nontransitivity among male genotypes (Clark et al. 2000) if inbreeding depression were the major factor contributing to the observed genetic effects. Although our study did not estimate additive genetic variation, Hughes (1997) concluded that most of the variation in sperm precedence was nonadditive and consistent with a form of balancing selection. Our observation of antagonistic pleiotropy (see below) is consistent with induced overdominance leading to a form of balancing selection and could theoretically explain Hughes (1997) observation of nonadditivity.
Regulation of male reproductive genes:
Transcript levels of the 10 male reproductive genes were highly positively correlated across the experimental lines (Table 4). Normalization by a control gene, RPL32, is expected to control for differences in overall mRNA levels or differences in body size among the lines. Thus, the positive correlations were likely due to either coregulation of the male reproductive genes or differences between lines in the size of their gonads relative to their total body size (i.e., gonadosomatic index).
Although the molecular mechanisms regulating expression of Acps and other male reproductive genes are not yet understood, coregulation is consistent with evidence that transcription of Acps declines as virgin males age, as well as the observation that mating increases both rates of transcription and translation (Schmidt et al. 1985; DiBenedetto et al. 1990; Monsma et al. 1990; Bertram et al. 1992; Herndon et al. 1997; Cho et al. 2000). Proteins such as hormones (Herndon et al. 1997) or the transcription factor paired (Xue and Noll 2002) also affect Acp expression. Furthermore, we have identified genetic polymorphisms associated with variation in transcript abundance. In several cases, a marker associated with transcript level was identified within or near the gene itself. Such cases likely represent evidence for allelic differences in cis regulation. In other cases, markers in one gene were associated with transcript abundance of other male reproductive genes. While this is consistent with linked markers affecting trans regulation of these genes, we are unable as yet to provide any molecular mechanism by which this regulation is operating. Several markers were associated with transcript abundance of multiple genes (e.g., CG8137s2957 is associated with both CG8137 and Acp53Ea), further supporting the hypothesis that Acps and perhaps other male reproductive genes are coregulated.
Although coregulation of Acps seems likely, it is possible that the positive correlations we observed were due to differences between the lines in their gonadosomatic index. Theory predicts that under some conditions males should invest more heavily in traits related to the production of sperm when the risk of sperm competition is high (Parker 1990a,b) and many empirical studies have found correlations between sperm competition risk and male investment in gametogenesis (Harcourt et al. 1981; Gage 1994; Stockley et al. 1997; Morrow and Gage 2000). Similar findings in the experimental lines would be interesting from a life history perspective and could provide important insights into mechanisms controlling alternative male reproductive tactics.
Genetic basis of sperm competitive ability:
On the basis of observed associations, natural polymorphisms in the investigated male reproductive genes appear to be important in determining male fitness. Six of the 10 genes had significant associations with at least one of the measured sperm competition phenotypes and all 10 genes had associations that were suggestive (comparisonwise P < 0.05). Given the molecular and biochemical data indicating that Acps influence female behavior and physiology postmating (reviewed in Wolfner 2002; Gillott 2003), it is not surprising that natural variants of these genes are important determinants of male reproductive success. On the basis of our current understanding of the functions of these genes some of the identified associations, however, were not predicted. Thus, our findings indicate that these genes may play additional roles in determining male reproductive success. For example, Acp26Aa and Acp29AB previously have been shown to correlate with P1′ (Clark et al. 1995), yet both were strongly associated with P2′ in this study. Given the role Acp26Aa has in egg laying (Herndon and Wolfner 1995; Heifetz et al. 2000; Chapman et al. 2001), one might expect it to influence P2′ through differences in fecundity, although no association between Acp26Aa and fecundity was observed in this study. Since Acp26Aa increases egg-laying rate for only 1 day (Herndon and Wolfner 1995), our measures, which summed fecundity over multiple days, may have been insensitive to these effects.
Potential roles for previously uncharacterized genes have also been identified. A polymorphism near Acp33A was associated with both P1′ and P2′. Little is known about the function of Acp33A, but these results suggest it to be important in male reproductive fitness. CG8137, a predicted serine protease inhibitor, was associated with the defense component P1′ and previous researchers have speculated that protease inhibitors could function to prevent premature processing of Acps or to protect sperm from proteolysis (see Lung et al. 2002; J. L. Mueller, D. R. Ripoll, C. F. Aquadro and M. F. Wolfner, unpublished results) or may even be involved with the coagulation of the mating plug (Coleman et al. 1995). Variation in CG17331, an endopeptidase, was associated with both refractoriness and P2′. It is unlikely that CG17331 is transferred to the female during mating since it does not appear to have a signal sequence (Swanson et al. 2001) and currently it is unclear how CG17331 could affect these phenotypes.
In addition to the associations that we did identify, information is present in associations that were not detected. For example, two variants of Acp36DE failed to associate with sperm competition phenotypes. The first was an 8-amino-acid insertion deletion (at amino acid 253) and the other was a premature stop codon that shortens the protein by 440 amino acids. Given this gene's important role in sperm storage (Bertram et al. 1996; Neubaum and Wolfner 1999; Qazi and Wolfner 2003), a priori one might expect these dramatic changes to have important effects on male fitness (Chapman et al. 2000). It is unlikely that low statistical power could account for the lack of associations as the rare allele was at a frequency of at least 20% and both markers were significantly associated with transcript abundance of Acp36DE. Variation observed at the transcript level could have compensated for the differences in amino acid composition, but this hypothesis would require further testing. The lack of any association with the premature stop codon is also consistent with the observation that males carrying a truncated version of Acp36DE show normal fertility (see Neubaum and Wolfner 1999; M. C. Bloch Qazi and M. F. Wolfner, unpublished results).
The lack of associations between steady-state transcript level of male reproductive genes and sperm competition phenotypes also provides important information for understanding the genetic basis of sperm competitive ability. It seems unlikely that deficiencies in statistical power explain the lack of associations, because multiple genotype-to-transcript abundance and genotype-to-phenotype associations were successfully identified using the same lines. Therefore, it is probable that a biological explanation is responsible for the lack of associations between transcript abundance and sperm competition phenotypes. Although many have argued that transcriptional regulation plays an important role in phenotypic evolution (reviewed in Wray et al. 2003), it is possible that the extent of variation in transcript abundance among our experimental lines does not affect sperm competitive ability. Transcript abundance in the experimental lines may exceed a required threshold value, and any variation above this level is selectively neutral. It is also possible that protein abundance, not transcript abundance, determines the ultimate phenotype. Steady-state mRNA levels are often, but not always, positively correlated with protein abundance (Futcher et al. 1999; Griffin et al. 2002), and protein turnover rates (Pratt et al. 2002) may be the important determinants for phenotypes affecting sperm competition. It is also possible that the lack of association was the result of the mRNA levels and sperm competition phenotypes being measured nearly 8 months apart, suggesting that transcript abundance may be strongly affected by the environment. Hopefully, future microarray studies will yield a more complete understanding of how interactions between genotype and the environment mediate gene expression levels and perhaps will provide additional insights into the role that transcriptional regulation plays in phenotypic evolution.
Maintaining genetic polymorphism:
Extensive genetic variation exists for phenotypes associated with male reproductive fitness (Clark et al. 1995; Service and Vossbrink 1996; Civetta and Clark 2000; Simmons and Kotiaho 2002; Nilsson et al. 2003; this study) and high levels of polymorphism have been observed in accessory gland proteins (Aguadé 1999; Begun et al. 2000; Tsaur et al. 2001). Mutation-selection balance may be responsible for some of the extant variation with low-frequency deleterious alleles segregating in this population. At two of the polymorphic sites that were associated with sperm competition phenotypes (CG8137s1910 and Acp33s2125) the high-fitness allele was the more common allele and was also ancestral, as indicated by its presence in Drosophila simulans. Background selection, however, cannot explain the elevated levels of polymorphism observed in some Acps (Aguadé 1999; Begun et al. 2000; Tsaur et al. 2001) and several other hypotheses have been proposed (see Clark 2002).
Many of the alternative mechanisms to maintain genetic variation at genes affecting sperm competition phenotypes invoke complex fitness relationships between alternative alleles. Antagonistic pleiotropy has been proposed as a mechanism maintaining genetic variation at genes affecting sperm competitive ability (Prout and Clark 1996), but empirical evidence has been lacking and researchers have formulated alternative explanations. Nontransitivity among males and male-by-female interactions (i.e., reproductive “rock-paper-scissor”-type competitions) have gained both theoretical and empirical support (Prout and Bundgaard 1977; Clark et al. 1999; 2000; Zamudio and Sinervo 2000) and focus has turned away from antagonistic pleiotropy. In this study, however, we have collected strong evidence that antagonistic pleiotropy is operating on a gene affecting two different sperm competition phenotypes. A polymorphism in CG17331 was associated with both P2′ and refractoriness (Figure 5). Males that carried the “A” allele at position CG17331s1421 sired a larger proportion of the offspring when they were the second males to mate (i.e., had a higher P2′ value) but at the cost of reduced refractoriness in the females when these males were the first to mate. Phenotypic tradeoffs between different life history characteristics are not unexpected (see Roff 2002) but the observation of antagonistic pleiotropy was surprising, given the trend toward positive correlations among many sperm competition phenotypes. In this study P2′ and refractoriness were uncorrelated, suggesting that negative genetic correlations are not required to detect antagonistic pleiotropy operating at the genotypic level. Our results support the hypothesis that antagonistic pleiotropy may still be an important mechanism maintaining genetic variation for sperm competition phenotypes and is consistent with a form of balancing selection producing the nonadditive genetic variance documented by Hughes (1997). The observation of antagonistic pleiotropy is particularly exciting as it suggests that association studies might also be able to identify polymorphisms in genes that are responsible for the observed nontransitivity and perhaps even to understand the molecular basis of male-by-female interactions.
Acknowledgments
We thank B. Haerum (Hughes Undergraduate Scholar), S. Galasinski, and B. Sceurman for their assistance with DNA sequencing and sperm competition scoring. B. Bettencourt kindly provided sequences for the RPL32 real-time PCR primers. M. Aguadé, C. Aquadro, B. Lazzaro, L. McGraw, K. McKean, K. Montooth, T. Schlenke, K. Thornton, T. Wittkopp, J. Walters, M. Wolfner, A. Wong, and two anonymous reviewers provided comments on early versions of this manuscript or suggestions during the course of this study. This work was supported by a National Science Foundation grant (DEB-0242987 to A.G.C.) and a National Institutes of Health Ruth L. Kirschstein Postdoctoral Fellowship (NGA 1 F32 GM70300-01 to A.C.F.).
References
- Aguadé, M., 1998. Different forces drive the evolution of the Acp26Aa and Acp26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics 150: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian, A., B. Gharizadeh, A. C. Gustafsson, F. Sterky, P. Nyren et al., 2000. Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem. 280: 103–110. [DOI] [PubMed] [Google Scholar]
- Aigaki, T., I. Fleischmann, P. S. Chen and E. Kubli, 1991. Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron 7: 557–563. [DOI] [PubMed] [Google Scholar]
- Bateman, A. J., 1948. Intra-sexual selection in Drosophila. Heredity 2: 349–368. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., P. Whitley, B. L. Todd, H. M. Waldrip-Dail and A. G. Clark, 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156: 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, M. J., G. A. Akerkar, R. L. Ard, C. Gonzalez and M. F. Wolfner, 1992. Cell type-specific gene expression in the Drosophila melanogaster male accessory gland. Mech. Dev. 38: 33–40. [DOI] [PubMed] [Google Scholar]
- Bertram, M. J., D. M. Neubaum and M. F. Wolfner, 1996. Localization of the Drosophila male accessory gland protein Acp36DE in the mated female suggests a role in sperm storage. Insect Biochem. Mol. Biol. 26: 971–980. [DOI] [PubMed] [Google Scholar]
- Birkhead, T. R., and A. P. Møller, 1998 Sperm Competition and Sexual Selection. Academic Press, San Diego/New York/London.
- Birkhead, T. R., and T. Pizzari, 2002. Postcopulatory sexual selection. Nat. Rev. Genet. 3: 262–273. [DOI] [PubMed] [Google Scholar]
- Chapman, T., D. M. Neubaum, M. F. Wolfner and L. Partridge, 2000. The role of male accessory gland protein Acp36DE in sperm competition in Drosophila melanogaster. Proc. R. Soc. Lond. B Biol. Sci. 267: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T., L. A. Herndon, Y. Heifetz, L. Partridge and M. F. Wolfner, 2001. The Acp26Aa seminal fluid protein is a modulator of early egg hatchability in Drosophila melanogaster. Proc. R. Soc. Lond. B Biol. Sci. 268: 1647–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T., J. Bangham, G. Vinti, B. Seifried, O. Lung et al., 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100: 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P. S., E. Stumm-Zollinger, T. Aigaki, J. Balmer, M. Bienz et al., 1988. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell 54: 291–298. [DOI] [PubMed] [Google Scholar]
- Cho, K. S., D. H. Won, G. H. Cha and C. C. Lee, 2000. Regulation of Mst57Dc expression in male accessory glands of Drosophila melanogaster. Mol. Cell 10: 180–185. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., and A. G. Clark, 2000. Chromosomal effects on male and female components of sperm precedence in Drosophila. Genet. Res. 75: 143–151. [DOI] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1998. Sex-related genes, directional sexual selection, and speciation. Mol. Biol. Evol. 15: 901–909. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., 2002. Sperm competition and the maintenance of polymorphism. Heredity 88: 148–153. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., and D. J. Begun, 1998. Female genotypes affect sperm displacement in Drosophila. Genetics 149: 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., M. Aguadé, T. Prout, L. G. Harshman and C. H. Langley, 1995. Variation in sperm displacement and its association with accessory-gland protein loci in Drosophila melanogaster. Genetics 139: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., D. J. Begun and T. Prout, 1999. Female × male interactions in Drosophila sperm competition. Science 283: 217–220. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., E. T. Dermitzakis and A. Civetta, 2000. Nontransitivity of sperm precedence in Drosophila. Evolution 54: 1030–1035. [DOI] [PubMed] [Google Scholar]
- Coleman, S., B. Drahn, G. Petersen, J. Stolorov and K. Kraus, 1995. A Drosophila male accessory-gland protein that is a member of the serpin superfamily of proteinase-inhibitors is transferred to females during mating. Insect Biochem. Mol. Biol. 25: 203–207. [DOI] [PubMed] [Google Scholar]
- Dibenedetto, A. J., H. A. Harada and M. F. Wolfner, 1990. Structure, cell-specific expression, and mating-induced regulation of a Drosophila melanogaster male accessory-gland gene. Dev. Biol. 139: 134–148. [DOI] [PubMed] [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyanov, H. M., and S. G. Dzitoeva, 1995. Method for attachment of microscopic preparations on glass for in situ hybridization, PRINS and in situ PCR studies. Biotechniques 18: 822–826. [PubMed] [Google Scholar]
- Enard, W., P. Khaitovich, J. Klose, S. Zollner, F. Heissig et al., 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296: 340–343. [DOI] [PubMed] [Google Scholar]
- Futcher, B., G. I. Latter, P. Monardo, C. S. Mclaughlin and J. I. Garrels, 1999. A sampling of the yeast proteome. Mol. Cell. Biol. 19: 7357–7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage, M. J. G., 1994. Associations between body-size, mating pattern, testis size and sperm lengths across butterflies. Proc. R. Soc. Lond. B Biol. Sci. 258: 247–254. [Google Scholar]
- Gillott, C., 2003. Male accessory gland secretions: modulators of female reproductive physiology and behavior. Annu. Rev. Entomol. 48: 163–184. [DOI] [PubMed] [Google Scholar]
- Griffin, T. J., S. P. Gygi, T. Ideker, B. Rist, J. Eng et al., 2002. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 1: 323–333. [DOI] [PubMed] [Google Scholar]
- Guo, D. C., and D. M. Milewicz, 2003. Methodology for using a universal primer to label amplified DNA segments for molecular analysis. Biotechnol. Lett. 25: 2079–2083. [DOI] [PubMed] [Google Scholar]
- Halliday, T., 1983. Behavioral ecology—Do frogs and toads choose their mates? Nature 306: 226–227. [Google Scholar]
- Harcourt, A. H., P. H. Harvey, S. G. Larson and R. V. Short, 1981. Testis weight, body-weight and breeding system in primates. Nature 293: 55–57. [DOI] [PubMed] [Google Scholar]
- Harshman, L. G., and A. G. Clark, 1998. Inference of sperm competition from broods of field-caught Drosophila. Evolution 52: 1334–1341. [DOI] [PubMed] [Google Scholar]
- Heifetz, Y., O. Lung, E. A. Frongillo and M. F. Wolfner, 2000. The Drosophila seminal fluid protein Acp26Aa stimulates release of oocytes by the ovary. Curr. Biol. 10: 99–102. [DOI] [PubMed] [Google Scholar]
- Herndon, L. A., and M. F. Wolfner, 1995. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg laying in females for 1 day after mating. Proc. Natl. Acad. Sci. USA 92: 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon, L. A., T. Chapman, J. M. Kalb, S. Lewin, L. Partridge et al., 1997. Mating and hormonal triggers regulate accessory gland gene expression in male Drosophila. J. Insect Physiol. 43: 1117–1123. [DOI] [PubMed] [Google Scholar]
- Hughes, K. A., 1997. Quantitative genetics of sperm precedence in Drosophila melanogaster. Genetics 145: 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennions, M. D., and M. Petrie, 2000. Why do females mate multiply? A review of the genetic benefits. Biol. Rev. Camb. Philos. Soc. 75: 21–64. [DOI] [PubMed] [Google Scholar]
- Lazzaro, B. P., B. K. Sceurman and A. G. Clark, 2004. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303: 1873–1876. [DOI] [PubMed] [Google Scholar]
- Liu, H. F., and E. Kubli, 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, O., U. Tram, C. M. Finnerty, M. A. Eipper-Mains, J. M. Kalb et al., 2002. The Drosophila melanogaster seminal fluid protein Acp62F is a protease inhibitor that is toxic upon ectopic expression. Genetics 160: 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre, L. M., E. R. Martin, K. L. Simonsen and N. L. Kaplan, 2000. Circumventing multiple testing: a multilocus Monte Carlo approach to testing for association. Genet. Epidemiol. 19: 18–29. [DOI] [PubMed] [Google Scholar]
- Meiklejohn, C. D., J. Parsch, J. M. Ranz and D. L. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100: 9894–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsma, S. A., and M. F. Wolfner, 1988. Structure and expression of a Drosophila male accessory gland gene whose product resembles a peptide pheromone precursor. Genes Dev. 2: 1063–1073. [DOI] [PubMed] [Google Scholar]
- Monsma, S. A., H. A. Harada and M. F. Wolfner, 1990. Synthesis of two Drosophila male accessory gland proteins and their fate after transfer to the female during mating. Dev. Biol. 142: 465–475. [DOI] [PubMed] [Google Scholar]
- Morrow, E. H., and M. J. G. Gage, 2000. The evolution of sperm length in moths. Proc. R. Soc. Lond. B Biol. Sci. 267: 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubaum, D. M., and M. F. Wolfner, 1999. Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics 153: 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, T., C. Fricke and G. Arnqvist, 2003. The effects of male and female genotype on variance in male fertilization success in the red flour beetle (Tribolium castaneum). Behav. Ecol. Sociobiol. 53: 227–233. [Google Scholar]
- Ochando, M. D., A. Reyes and F. J. Ayala, 1996. Multiple paternity in two natural populations (orchard and vineyard) of Drosophila. Proc. Natl. Acad. Sci. USA 93: 11769–11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiak, M. F., G. A. Churchill and D. L. Crawford, 2002. Variation in gene expression within and among natural populations. Nat. Genet. 32: 261–266. [DOI] [PubMed] [Google Scholar]
- Parker, G. A., 1979 Sexual selection and sexual conflict, pp. 123–166 in Sexual Selection and Reproductive Competition in Insects, edited by M. S. Blum and N. A. Blum. Academic Press, New York.
- Parker, G. A., 1990. a Sperm competition games—raffles and roles. Proc. R. Soc. Lond. B Biol. Sci. 242: 120–126. [Google Scholar]
- Parker, G. A., 1990. b Sperm competition games—sneaks and extra-pair copulations. Proc. R. Soc. Lond. B Biol. Sci. 242: 127–133. [Google Scholar]
- Pitnick, S., W. D. Brown and G. T. Miller, 2001. a Evolution of female remating behaviour following experimental removal of sexual selection. Proc. R. Soc. Lond. B Biol. Sci. 268: 557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick, S., G. T. Miller, J. Reagan and B. Holland, 2001. b Males' evolutionary responses to experimental removal of sexual selection. Proc. R. Soc. Lond. B Biol. Sci. 268: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, J. M., J. Petty, I. Riba-Garcia, D. H. L. Robertson, S. J. Gaskell et al., 2002. Dynamics of protein turnover, a missing dimension in proteomics. Mol. Cell. Proteomics 1: 579–591. [DOI] [PubMed] [Google Scholar]
- Prout, T., and J. Bundgaard, 1977. The population genetics of sperm displacement. Genetics 85: 95–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout, T., and A. G. Clark, 1996. Polymorphism in genes that influence sperm displacement. Genetics 144: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan, M. D., 2000. The molecular population genetics of regulatory genes. Mol. Ecol. 9: 1451–1461. [DOI] [PubMed] [Google Scholar]
- Qazi, M. C. B., and M. F. Wolfner, 2003. An early role for the Drosophila melanogaster male seminal protein Acp36DE in female sperm storage. J. Exp. Biol. 206: 3521–3528. [DOI] [PubMed] [Google Scholar]
- Raymond, M., and F. Rousset, 1995. Genepop (Version-1.2)—population-genetics software for exact tests and ecumenicism. J. Hered. 86: 248–249. [Google Scholar]
- Remington, D. L., J. M. Thornsberry, Y. Matsuoka, L. M. Wilson, S. R. Whitt et al., 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W. R., 1996. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381: 232–234. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., and B. Holland, 1997. The enemies within: intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav. Ecol. Sociobiol. 41: 1–10. [Google Scholar]
- Roff, D. A., 2002 Life History Evolution. Sinauer Associates, Sunderland, MA.
- Schmidt, T., E. Stummzollinger and P. S. Chen, 1985. Protein-metabolism of Drosophila melanogaster male accessory-glands 3. Stimulation of protein-synthesis following copulation. Insect Biochem. 15: 391–401. [Google Scholar]
- Service, P. M., and R. E. Vossbrink, 1996. Genetic variation in “first” male effects on egg laying and remating by female Drosophila melanogaster. Behav. Genet. 26: 39–48. [DOI] [PubMed] [Google Scholar]
- Sheldon, B. C., 1994. Male phenotype, fertility, and the pursuit of extra-pair copulations by female birds. Proc. R. Soc. Lond. B Biol. Sci. 257: 25–30. [Google Scholar]
- Simmons, L. W., and J. S. Kotiaho, 2002. Evolution of ejaculates: patterns of phenotypic and genotypic variation and condition dependence in sperm competition traits. Evolution 56: 1622–1631. [DOI] [PubMed] [Google Scholar]
- Soller, M., M. Bownes and E. Kubli, 1999. Control of oocyte maturation in sexually mature Drosophila females. Dev. Biol. 208: 337–351. [DOI] [PubMed] [Google Scholar]
- Stockley, P., M. J. G. Gage, G. A. Parker and A. P. Møller, 1997. Sperm competition in fishes: the evolution of testis size and ejaculate characteristics. Am. Nat. 149: 933–954. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaur, S. C., C. T. Ting and C.-I Wu, 1998. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila: II. Divergence versus polymorphism. Mol. Biol. Evol. 15: 1040–1046. [DOI] [PubMed] [Google Scholar]
- Tsaur, S. C., C. T. Ting and C.-I Wu, 2001. Sex in Drosophila mauritiana: a very high level of amino acid polymorphism in a male reproductive protein gene, Acp26Aa. Mol. Biol. Evol. 18: 22–26. [DOI] [PubMed] [Google Scholar]
- Wolfner, M. F., 2002. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88: 85–93. [DOI] [PubMed] [Google Scholar]
- Wolfner, M. F., H. A. Harada, M. J. Bertram, T. J. Stelick, K. W. Kraus et al., 1997. New genes for male accessory gland proteins in Drosophila melanogaster. Insect Biochem. Mol. Biol. 27: 825–834. [DOI] [PubMed] [Google Scholar]
- Wray, G. A., M. W. Hahn, E. Abouheif, J. P. Balhoff, M. Pizer et al., 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377–1419. [DOI] [PubMed] [Google Scholar]
- Xue, L., and M. Noll, 2002. Dual role of the Pax gene paired in accessory gland development of Drosophila. Development 129: 339–346. [DOI] [PubMed] [Google Scholar]
- Zamudio, K. R., and E. Sinervo, 2000. Polygyny, mate-guarding, and posthumous fertilization as alternative male mating strategies. Proc. Natl. Acad. Sci. USA 97: 14427–14432. [DOI] [PMC free article] [PubMed] [Google Scholar]