Abstract
We have identified a novel gene named grappa (gpp) that is the Drosophila ortholog of the Saccharomyces cerevisiae gene Dot1, a histone methyltransferase that modifies the lysine (K)79 residue of histone H3. gpp is an essential gene identified in a genetic screen for dominant suppressors of pairing-dependent silencing, a Polycomb-group (Pc-G)-mediated silencing mechanism necessary for the maintenance phase of Bithorax complex (BX-C) expression. Surprisingly, gpp mutants not only exhibit Pc-G phenotypes, but also display phenotypes characteristic of trithorax-group mutants. Mutations in gpp also disrupt telomeric silencing but do not affect centric heterochromatin. These apparent contradictory phenotypes may result from loss of gpp activity in mutants at sites of both active and inactive chromatin domains. Unlike the early histone H3 K4 and K9 methylation patterns, the appearance of methylated K79 during embryogenesis coincides with the maintenance phase of BX-C expression, suggesting that there is a unique role for this chromatin modification in development.
THE homeotic genes of the Antennapedia (ANT-C) and Bithorax complexes (BX-C) are responsible for specifying parasegment identity in the fly. Early in development gap and pair-rule genes initiate parasegment-specific patterns of ANT-C and BX-C homeotic gene activity. The expression patterns established during the initiation phase are then sustained during the remainder of development by a maintenance system consisting of the trithorax-Group (trx-G) and the Polycomb-Group (Pc-G) genes, which have antagonistic functions (Kennison 1995; Mahmoudi and Verrijzer 2001; Simon and Tamkun 2002). trx-G proteins are required to maintain gene activity, and in trx-G mutants inactivation of one or more of the ANT-C and BX-C homeotic genes causes posterior-to-anterior transformations in parasegmental identity. Conversely, Pc-G proteins function as silencers, and in Pc-G mutants the inappropriate activation of homeotic genes causes anterior-to-posterior transformations in parasegmental identity.
The antagonistic activities and phenotypes associated with trx-G and Pc-G genes are a function of their distinct effects on chromatin structure. For example, the trx-G protein ASH1 is a histone methyltransferase that modifies histone H3 lysine (K) 4 and K9 residues as well as histone H4 K20 (Beisel et al. 2002). It is believed that this epigenetic modification functions in turn to recruit another TRX-G protein, Brahma, the fly homolog of the SNF2/SWI2 protein in yeast, which facilitates transcription via chromatin remodeling. Like ASH1, the PC-G protein E(Z) is also a histone methyltransferase, but has a different specificity, methylating K9 and K27 of histone H3 (Czermin et al. 2002). Nucleosomes possessing histone H3 methylated at these residues function to recruit Polycomb and other components of the PC-G silencing complex (Czermin et al. 2002).
In genetic screens designed to identify novel factors required for Pc-G-mediated silencing we recovered several alleles of a new Drosophila gene, grappa (gpp). gpp has the unusual property of exhibiting phenotypes and genetic interactions that are characteristic of both Pc-G and trx-G genes and is a member of the Enhancer of trithorax and Polycomb (ETP) class of genes (see Brock and van Lohuizen 2001). Moreover, gpp mutants dominantly suppress silencing by telomeric, but not centromeric heterochromatin. We show here that gpp is the fly ortholog of the Saccharomyces cerevisiae Dot1 gene, a gene that was originally identified in an overexpression screen for factors that perturb telomeric silencing (Singer et al. 1998). Analysis of gpp mutants indicates that, like Dot1 (Lacoste et al. 2002; Ng et al. 2002; van Leeuwen et al. 2002), GRAPPA functions as a methyltransferase and is required for the methylation of lysine 79 of histone H3 (H3meK79). We also describe novel features of the developmental profile and polytene chromosome distribution of the H3meK79 modification.
MATERIALS AND METHODS
gpp mutants:
gpp mutants were isolated by mating EMS-mutagenized w1 males containing Mcp or iab-7 PRE mini-white reporters (lines: Mcp, ff#10.5, Muller et al. 1999; iab-7 PRE 18.73.1, Hagstrom et al. 1997) to their sibs and screening F1 progeny for elevated mini-white expression. The mutant gene responsible for suppression of PRE-mediated silencing was named grappa because of the light yellow phenotype associated with the posterior-to-anterior transformation of the A5 and A6 male tergite plates. (This light color reminded one of the authors of a favorite drink from his not so distant youth and is thus the origin of the name.) X-ray alleles were isolated on the basis of their failure to complement gpp EMS mutants. Two of the X-ray alleles, gppX and gppXXV, have breakpoints in 83E4-83E8. Genomic DNA from these alleles was analyzed by probing with sequences amplified from BAC 32D5, which spans gpp (Hoskins et al. 2000). The primers used for probe 1 were Fwd, GGGCAGCGGCAGCAGATTTGCTGG; Rev, GATTGTCCGTATAGGAGGGG. Primers for probe 2 were Fwd, GCACTAGCTGAATGCCGCTTTGGC; Rev, CGCTTTAACTTTGAACTAAGTCGACTGC. The P-element insertion in gpp, gppIN1, was isolated by crossing pLac w males to Δ2-3 transposase females. Plasmid rescue was used to obtain clones from gpp.
Genetic interactions:
gpp interactions with the various Pc-G genes were carried out at 22°. All Pc-G lines were obtained from the Bloomington Stock Center. The data from these crosses were obtained by counting the male sex comb teeth present on all legs and averaging this number. The numbers of sex comb teeth present on the first tarsal leg were summed, averaged, and subtracted from the above number to generate the values depicted in Figure 2A. gpp alleles were reciprocally crossed against Abd-BM1, a null allele of the Abd-B gene at 22°. F1 trans-heterozygous males were scored for rotated genitalia. Chi-square analysis was used to determine if the number of rotated genitalia of a particular genotype was statistically different from control crosses.
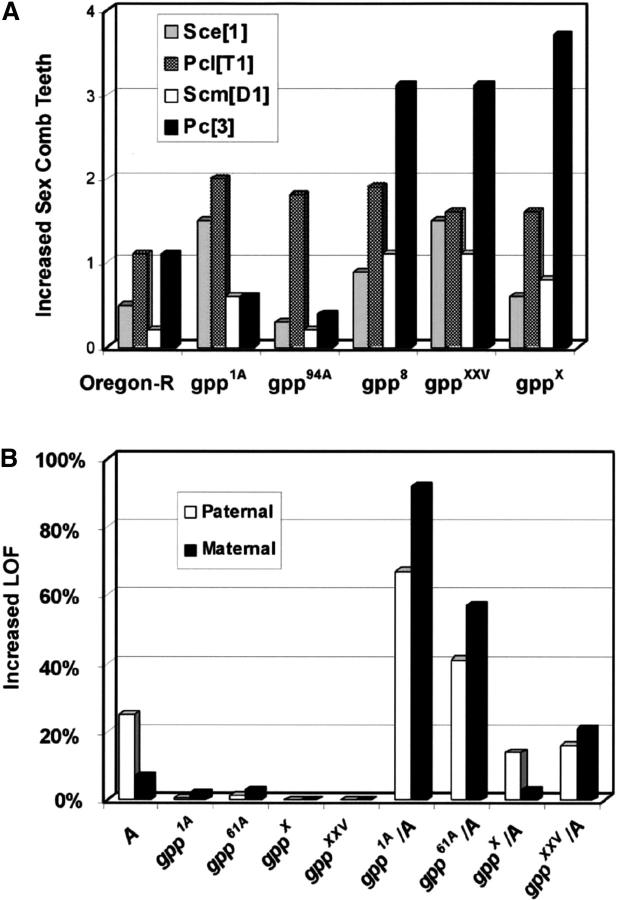
Figure 2.—
gpp alleles genetically enhance Pc-G and Abd-B mutant phenotypes. (A) gpp alleles were crossed against different Pc-G mutants. The y-axis shows the relative increase in the sex comb teeth associated with the double mutants. The number of sex comb teeth was calculated by counting the total number of teeth on all legs, averaging this number, and subtracting the averaged number of teeth present on the first tarsal leg. A minimum number of 54 flies were counted for each genotype. Sce1/TM6B, Sex combs extra; PclT1/Cyo, Polycomblike; ScmD1/TM6,Sb, Sex combs on the midleg; Pc3/TM3,Ser, Polycomb. (B) gpp alleles reciprocally crossed against w1;+;Abd-BM1/TM6B mutants enhance the male rotated genitalia phenotype. The increase in rotated genitalia of the trans-heterozygous combinations of gpp and Abd-BM1 is statistically significant. All P-values were <10−4 by the χ2 test. y-Axis, percentage change in genitalia phenotype; x-axis, trans-heterozygous combinations of gpp alleles and either w1 or w1;+;Abd-BM1/TM6,B,TB flies; LOF, loss of function; A, the w1;+;Abd-BM1 allele.
RNA isolation and Northern blotting:
Total RNA was isolated from adult flies using an acid phenol extraction protocol (Samuels et al. 1991). Poly(A)+ RNA was isolated from 24-hr-old embryos and third instar larvae using the μmACS mRNA isolation kit (Miltenyi Biotec). RNA (10 μg per lane) was loaded and the blot was probed with gpp cDNAs.
Western analysis:
Six imaginal discs or central nervous systems (CNSs) were isolated from third instar larvae and immediately homogenized in gel loading buffer (125 mm Tris, pH 6.9, 6% SDS, 50% glycerol, 10% β-mercaptoethanol). The samples were electrophoresed on a 20% SDS PAGE gel and transferred to membranes. A 1:6000 dilution of histone H3 dimethyl K79 (H3dmeK79) antibody was incubated with the blots [a generous gift from Yi Zhang (Ng et al. 2002) and Michael Grunstein]. The same results were observed using anti-H3dmeK79 commercially available from Upstate Biotechnology (Lake Placid, NY). Blots were rehybridized with anti-snf monoclonal antibody 4G3 serum at a 1:10 dilution as a control. This antibody recognizes the SNF protein involved in nuclear mRNA splicing and is present in all cells.
Immunocytochemistry:
Imaginal wing discs isolated from wild-type and gppX third instar larvae were fixed in a 4% solution of formaldehyde in PBS and washed and blocked in PBTX (1× PBS, 0.3% Triton X-100, and 0.3% BSA). Staged embryos were fixed and stained following the protocol of Deshpande et al. (1995). Discs and embryos were incubated with K79 antibody (1:6000) and then stained with secondary antibodies (Alexa fluor 546 and 488; Molecular Probes, Eugene, OR).
RESULTS
gpp mutants suppress pairing-dependent silencing:
PC-G silencing complexes are targeted to cis-acting targets known as Polycomb response elements (PREs) that are located in the ANT-C and BX-C regions. When mini-white transgenes containing PREs are paired in vivo, a phenomenon known as pairing-sensitive silencing (PSS) represses reporter gene expression. To identify genes involved in PSS we screened for second-site mutations that dominantly suppress silencing induced by the BX-C PREs Mcp and iab-7. While many of the mutations recovered corresponded to known Pc-G genes (Vazquez et al. 1993), several appeared to represent novel loci. One of the novel genes, called grappa (gpp), was defined by four independent alleles. Three of the alleles, gpp1A, gpp72A, and gpp94A are viable, while the fourth, gpp61A, is lethal (Table 1).
TABLE 1.
Heteroallelic combinations ofgpp alleles result in an enhancedAbd-B LOF phenotype
| gpp1A | gpp61A | gpp72A | gpp94A | gpp8 | gppIN1 | gppX | gppXXV | Df(3R) WIN11 | |
|---|---|---|---|---|---|---|---|---|---|
| gpp1A | E | ||||||||
| gpp61A | E | L | |||||||
| gpp72A | E | E | E | ||||||
| gpp94A | E | E | E | E | |||||
| gpp8 | E | L | E | E | L | ||||
| gppIN1 | E | L | E | E | L | L | |||
| gppIN2 | ND | L | ND | ND | ND | L | L | ND | ND |
| gppX | E | L | E | E | L | L | L | ||
| gppXXV | E | L | E | E | L | L | L | L | |
| Df(3R) WIN11 | E | L | E | E | L | L | L | L | L |
E, genetic combination of these gpp alleles enhances the Abd-B LOF phenotype; L, genetic combination of these gpp alleles results in lethality; ND, not done.
As illustrated for gpp1A in Figure 1A, all four gpp alleles dominantly suppress Mcp silencing of mini-white. gpp mutants also suppress silencing by the iab-7 and bxd PREs (data not shown). On the basis of the effects of these mutations on PRE activity, gpp would be classified as a Pc-G gene. This classification is supported by the finding that gpp61A pharate adults (as well as two alleles isolated in other screens, gpp8 and gppIN-1) exhibit aristae-to-leg transformations (see Figure 1B; Duncan et al. 1998). On the other hand, a number of additional phenotypes are observed that suggest that gpp is not a typical Pc-G gene. First, gpp mutants exhibit a rough eye phenotype that differs in severity depending on the allele. Second, as described further below, trx-G-like, not Pc-G-like transformations are observed in abdominal segments and legs of gpp mutants. Third, an additional EMS allele of gpp, gpp8, was identified in a screen for suppressors of a dominant gain-of-function hedgehog (hh) mutation, Moonrat (hhMrt; Felsenfeld and Kennison 1995). gpp8 is embryonic lethal when homozygous and is pharate adult lethal when combined in trans with gpp61A (see Table 1). Since hhMrt activates hh transcription in inappropriate tissues and cell types, suppression by a gpp mutation also points to a role in transcriptional activation rather than silencing. Given this unusual combination of phenotypes, it was of interest to further characterize gpp.
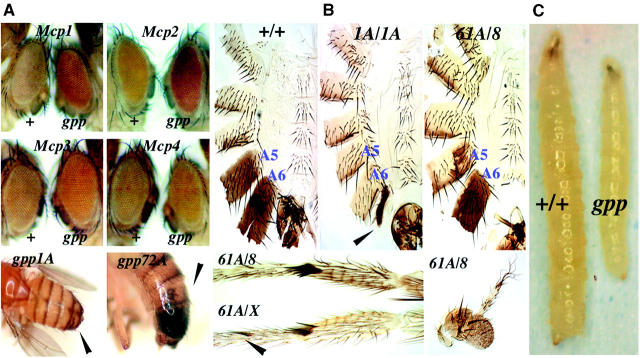
Figure 1.—
gpp mutants suppress PRE-mediated chromatin silencing and exhibit numerous phenotypes. (A) gpp1A mutants suppress both Mcp cis- and trans-silencing. The Mcp1 panel depicts flies containing homozygously paired Mcp mini-white reporter transgenes. The fly on the right contains paired Mcp transgenes recombined with the gpp1A mutation. In this fly, the gpp1A mutation suppresses trans-silencing of the Mcp constructs as evidenced by the increase in mini-white expression. Mcp2, -3, and -4 display fly lines containing cis-Mcp constructs (see Table 2). The flies on the right-hand side of each of these contain different Mcp cis lines that are trans-heterozygous with the gpp1A mutant. In each case, the cis-silencing associated with these lines is disrupted by the gpp1A mutation. The bottom displays prominent trx-G phenotypes associated with two different gpp alleles, gpp1A and gpp72A. The arrowheads identify the posterior-to-anterior transformation of adult male tergite plates associated with these two alleles of gpp. (B) A number of additional phenotypes associated with gpp alleles. The top depicts tergite filets dissected from wild-type, gpp1A, and gpp61A/gpp8 males. Note that in gpp1A there is a complete loss of tergite coloration relative to wild type. This is indicative of the transformation of adult segment A5 and A6 into an A4 anterior segmental identity. Also note that in these flies the ventral sternite hairs are transformed into a more anterior identity. The arrowhead points to a vestigial A7 tergite not normally present in males. In gpp61A/gpp8 there is a loss of A5 tergite coloration and additional sternite hairs characteristic of a trx-G phenotype. The bottom left shows male legs from gpp61A/gpp8 and gpp61A/gppX combinations that possess a sex comb reduced phenotype. However, the gpp61A/gppX animals exhibit an extra sex comb on the first leg as indicated by the arrowhead. The bottom right displays an aristae-to-leg transformation associated with gpp61A/gpp8 mutant flies. (C) The slow growth phenotype associated with homozygous gppX larvae.
Isolation of gpp rearrangements:
Recombination mapping relative to P-element insertions along the third chromosome placed gpp in the centromere proximal region of 3R. It was localized to the 83-84 interval by the failure of gpp61A (and several other lethal alleles) to complement Df(3R) WIN11 and Df (3R)Dfd13. To further analyze the genetic properties of gpp and obtain DNA rearrangements that would facilitate molecular isolation, P-element- and X-ray-induced gpp alleles were isolated. The P-element allele, gppIN-1, is homozygous lethal at the pharate adult stage. It is also lethal in combination with Df(3R)WIN11, gpp61A, or gpp8. In situ hybridization localized the P-element insertion to 83E on 3R. The gppX X-ray allele exhibits no obvious patterning defects and is lethal during late larval stages. The growth rate of gppX larvae is reduced relative to similarly aged wild-type larvae (Figure 1C). Cytological examination of gppX reveals a small inversion with breakpoints in 83C8-D1, 2 and 83 E4-8. The other X-ray allele, gppXXV, is embryonic lethal, and it has both an inversion (75 C1, 2; 83 E4-8) and a small deletion (of 75C1, 2-E). The small deletion rather than the lesion in gpp seems to be responsible for the embryonic lethality of gppXXV.
Phenotypic effects of gpp mutants:
gpp suppresses Pc-G-mediated silencing:
To compare the effects of different gpp alleles on PSS of mini-white by PREs, we took advantage of the unusual ability of Mcp to establish silencing interactions in cis between transgenes inserted at distant sites on the same chromosome (Muller et al. 1999). We tested all of the gpp alleles (except for the P transposon induced) with three different Mcp-mini-white transgene cis combinations. Most of the alleles suppress long distance Mcp silencing; however, the strength of suppression varies with the gpp allele and the Mcp cis combination (Table 2). gpp1A shows the most dramatic effects on Mcp silencing, and it strongly suppresses all three Mcp cis combinations. gpp72A also suppress the three cis combinations; however, the increase in mini-white expression is not as pronounced for two of the cis combinations as it is with gpp1A. While none of the remaining EMS and X-ray alleles suppress the Mcp4 combination, all suppress one or both of the other combinations. For example, gpp8 and gppXXV suppress Mcp silencing in both the Mcp2 and the Mcp3 combinations, while gppX suppresses silencing only in the Mcp3 combination.
TABLE 2.
Effect ofgpp mutations onMcp-mediated long-range silencing
|
gpp alleles
|
|||||||
|---|---|---|---|---|---|---|---|
| Mcp cis-silencing linesa | gpp1A | gpp72A | gpp94A | gpp61A | gpp8 | gppX | gppXXV |
| Mcp2 (ff#13.101 w#14.29) | S | S | ws | ws | S | N | ws |
| Mcp3 (w11.16, 11.102) | S | ws | ws | N | ws | S | ws |
| Mcp4 (wy2.63, ff#15.30) | S | ws | N | N | N | N | N |
The tabulated results represent trans-heterozygous combinations of Mcp lines with different gpp alleles. S, suppression of silencing; N, no effect; ws, weak suppression of silencing.
See Muller et al. 1999.
In addition to testing the cis Mcp lines, we examined the effect of the gpp1A allele on various hemizygous and homozygous PRE-containing transgene lines. We found that gpp1A increases the eye color of both hemizygous and homozygous Mcp, iab-7 PRE and bxd PRE lines but has no effect on the endogenous white locus (data not shown).
gpp interacts with Pc-G genes:
Males heterozygous for Pc-G mutations often exhibit ectopic sex comb teeth or complete sex combs on the second and third tarsal legs. These transformations result from a loss of Pc-G silencing and can be enhanced by double-mutant combinations with other Pc-G genes. To explore the role of gpp in Pc-G-dependent silencing, we tested whether gpp dominantly enhances the phenotypic effects of mutations in Sex combs extra (Sce), Polycomblike (Pcl), Sex combs on the midleg (Scm), and Polycomb (Pc).
The simplest interactions are observed for gpp8 and the two X-ray alleles, gppX and gppXXV. As shown in Figure 2 these alleles increase the number of sex comb teeth in males heterozygous for all four Pc-G mutations. Moreover, for all three alleles, the strongest interactions are observed for Scm and Pc3. As the two X-ray alleles are presumed to be simple loss-of-function, rather than antimorphic or neomorphic mutations, these findings would be consistent with the classification of gpp as a Pc-G gene. More complex interactions are observed for gpp1A and gpp94A. While gpp1A enhances the sex combs phenotype of Sce, Pcl, and Scm, it has the opposite effect on Pc3, suppressing the sex combs phenotype. Similarly, gpp94A acts as both an enhancer and a suppressor of the sex comb phenotype, depending upon the particular Pc-G mutation. Why these two gpp alleles do not interact in a consistent pattern with the Pc-G mutations is unclear; it is possible that this reflects some unusual properties of the mutant GRAPPA (GPP) protein.
gpp affects telomeric silencing but not centromeric silencing:
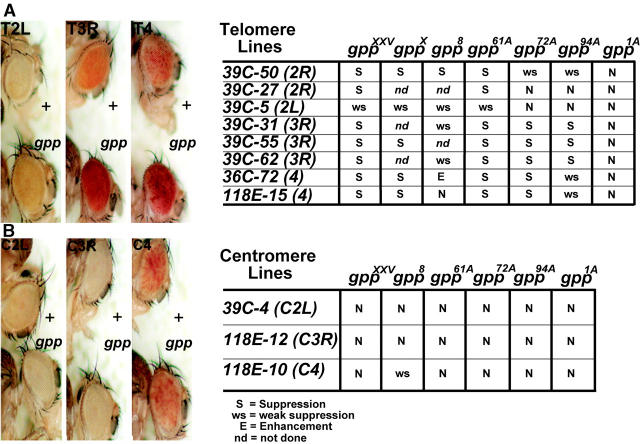
The suppression of Pc-G-mediated silencing of mini-white as well as the genetic interactions with mutations in several Pc-G genes would be consistent with the idea that gpp is a member of the Pc-G family. While the silencing activity of many Pc-G family members is restricted to euchromatic genes, a subset of the PC-G proteins function in telomeric silencing [telomeric position effect (TPE); Cryderman et al. 1999; Boivin et al. 2003]. To determine whether gpp is required for TPE, we asked whether gpp mutations dominantly suppress the silencing of mini-white transgenes inserted into telomeric heterochromatin of the second, third, and fourth chromosomes (Cryderman et al. 1999; Figure 3A). gppXXV, gppX, and gpp61A mutants suppressed TPE of all mini-white transgene inserts. As illustrated for gppX in Figure 3A, suppression by these three gpp alleles is quite strong relative to eye color controls. The one exception is the telomeric insert in 2L, which is suppressed only weakly by gppX and the two other gpp alleles; however, this insertion is also only weakly suppressed by mutations in other Pc-G genes required for TPE, such as Su(z)25 (our unpublished data; Cryderman et al. 1999). Three other alleles, gpp8, gpp72A and gpp94A, also suppressed TPE at most but not all telomeric sites.
Figure 3.—
gpp mutants suppress TPE but not PEV. (A) Flies heterozygous for P-element reporters inserted into the subtelomeric chromatin of chromosomes 2L, 3R, and 4. The three bottom flies are trans-heterozygous combinations of the telomere inserts and the gppX allele. TPE is suppressed in these flies as evidenced by the increase in mini-white expression of the telomeric inserts relative to the heterozygous controls. The telomere lines y1w1;39C-5;+;+, y1w1; +;39C-55;+, and y1w1;+;+;118E-15 were used (Cryderman et al. 1999). The table to the right documents the effect on TPE of trans-heterozygous combinations of various gpp alleles with different second, third, and fourth chromosome telomere P-element inserts. The genetic effect on suppression or enhancement of TPE by gpp mutations was measured by comparing trans-heterozygous eye colors relative to heterozygous controls. S, strong suppression of TPE; ws, weak suppression of TPE; N, no effect on TPE; E, enhancement of TPE; nd, not done. (B) Flies heterozygous for centromeric reporter P elements. Bottom flies are trans-heterozygous combinations of the different centromeric inserts and the gppX allele. PEV silencing is not affected by gpp mutations. The centromeric P-element lines y1w1;39C-4;+, y1w1;+;118E-12, and y1w1;+;+;118E-10 were used in this figure (from Cryderman et al. 1998). The table to the right documents the effect of various gpp alleles on the centromeric P-element inserts. The genetic effect on suppression or enhancement of PEV by gpp mutations was measured as above against controls. N, no effect on PEV; ws, weak suppression of PEV.
We also tested whether gpp has any dominant effects on silencing by centromeric heterochromatin [position effect variegation (PEV)]. None of the gpp alleles had any effect on PEV silencing associated with mini-white insertions into second or third chromosome centromeric heterochromatin (Figure 3B). Assuming that the unusual effect of gpp8 on the fourth chromosome insert is unique to this particular allele, these findings suggest that gpp does not function in centromeric silencing.
gpp is also a member of the trx-G gene family:
While the effects of gpp mutations on Pc-G and telomeric silencing suggest that it functions in the establishment and/or maintenance of repressive chromatin structures, gpp mutants also exhibit phenotypes and genetic interactions that are characteristic of mutations in trx-G genes. trx-G-like phenotypes were first observed in gpp61A/+ males as a transformation of the A5 tergite into a tergite that resembles A4. This phenotype is characteristic of mutations that compromise Abd-B activity in parasegment 10 and would be expected for a mutation in a trx-G, not a Pc-G gene. While gpp61A/gpp61A flies don't survive to the adult stage, males homozygous for gpp1A, gpp72A, and gpp94A can be obtained. In these males A5 and also A6 is transformed toward an A4 identity. The severity of this Abd-B-like loss-of-function phenotype ranges from the moderate transformation seen in gpp72A and gpp94A to near complete loss of male pigmentation in both A5 and A6 and the appearance of an “A7” tergite in homozygous gpp1A (see Figure 1, A and B). Also indicative of A6 to A5/A4 transformation, gpp1A, gpp72A, and gpp94A males have hairs on the sixth sternite. Similar transformations of A5 and A6 toward A4 are observed when viable gpp alleles are combined with some of the lethal alleles (see gpp1A/gpp8 in Figure 1B). The severity of the transformations in segment identity seen in homozygous mutant males is influenced by the maternal genotype. For example, the phenotype in the progeny is more severe when the mothers are homozygous for the gpp mutation than when the mothers are heterozygous.
While the Abd-B loss-of-function (LOF) phenotype is not observed in flies heterozygous for gpp8, for either X-ray allele or the P-element insertion gppIN-1, dissection of homozygous male gppIN-1 pupae reveals that A5 and A6 are transformed toward an A4 identity. The gppIN-1 males also have a reduced number of sex combs, indicating that expression of homeotic genes in the ANT-C is altered. Similar loss-of-function transformations of A5 and A6 and a reduction in the number of sex combs are observed in dissected gpp61A/gpp61A pupae (not shown). A reduction in the number of sex comb teeth (and sex combs on the fourth tarsus) is also observed in dissected gpp61A/gpp8 and gpp61A/gppX pupae (see Figure 1B). In addition, as shown in Table 1, gppX, gppXXV, gppIN-1, and gpp8 all enhance the transformation of A5 and A6 when trans to a viable gpp allele.
Since these phenotypes suggest that gpp functions in promoting gene expression, we tested whether gpp mutations dominantly enhance the weak haplo-insufficiency of the Abd-B gene (Figure 2B). While neither X-ray allele shows strong interactions, both gpp1A and gpp61A are potent enhancers of the Abd-B haplo-insufficiency. Moreover, this interaction is consistently stronger when the gpp mutation is inherited from the mother. These findings, taken together with the trx-G-like phenotypes seen in different gpp mutant backgrounds would be consistent with the idea that gpp has some type of function in gene activation or the maintenance of gene activity.
Molecular isolation of the gpp gene:
Clones containing DNA flanking the gppIN-1 P-element were isolated by plasmid rescue and the insertion site was determined by comparison with the genomic sequence. On the centromere proximal side, the transposon is 1.5 kb from the 5′ end of the Celera predicted gene CG1021, while on the centromere distal side it is ∼30 kb from the predicted 5′ end of CG10272. An additional P-element line, gppIN-2 [l(3)0334203342; Spradling et al. 1999] is inserted into the first intron of CG10272 and is ∼17 kb distal to the gppIN-1 P-element insertion site (see Figure 4A). The sequence insertion site for the gppIN-2 P-element was determined by the Berkeley gene disruption project (Spradling et al. 1999). The gppIN-2 P-element insertion does not complement lethal gpp alleles (Table 1). In addition to the P-element insertions, the position of the X-ray breakpoint alleles was determined. We found that the gppX breakpoint maps to a restriction fragment located ∼13 kb distal to the gppIN-1 P-element and 4 kb proximal to gppIN-2 (Figure 4A). However, the gppXXV breakpoint is located ∼35 kb from gppIN-1 inside the predicted coding region for the CG10272 gene. This finding suggested that CG10272 might correspond to gpp.
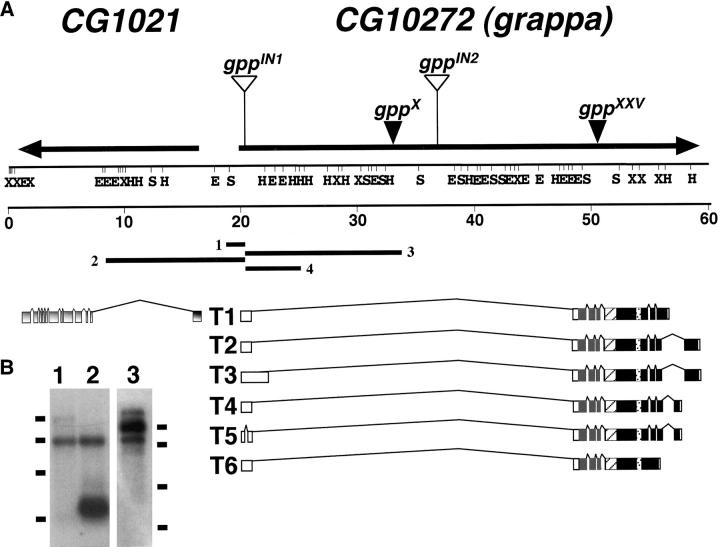
Figure 4.—
The molecular organization of the gpp locus and the developmental expression of gpp RNAs. (A) Genomic organization and restriction map of the gpp locus along the 83 E4-8 region on the third chromosome. The location and the direction of transcription (solid arrows) of the CG1021 and CG10272 (gpp) genes are depicted at the top. The location of the P-element alleles gppIN-1 and gppIN-2[l(3)03342; see Spradling et al. 1999] and the X-ray breakpoint alleles (gppX and gppXXV) are depicted above the gpp transcription unit. A restriction enzyme map and a scale bar (from 0 to 60 kb) are presented below the transcription units. Plasmid rescue clones (1–4) isolated from gppIN-1 flies are positioned around the proximal P-element insert. A putative CG1021 transcript is pictured on the left side of the figure (in gray). Six alternatively spliced gpp cDNAs are shown below the map. The white boxes on the cDNAs represent 5′- and 3′-UTRs. Solid boxes represent gene exons. Transcripts T1–T6 represent different alternatively spliced gpp transcripts. The size (in kilobases) of the transcripts is as follows: T1, 7.6; T2, 8.7; T3, 10.1; T4, 7.6; T5, 7.3; and T6, 6.6. Protein domains overlaying respective encoding exons are depicted on the cDNAs: gray box, the domain homologous to DOT1; hatched box, a potential coiled-coil domain; stippled box, a potential ATP/GTP binding domain. Restriction enzyme sites: S, SalI; H, HindIII; E, EcoRI; X, XhoI. (B) Northern analysis of gpp transcripts. Lanes 1 and 2 contain total RNA isolated from adult w1; TM3,Ser/Sb1 and gppXXV/TM3, Ser flies, respectively. Lane 3 contains mRNA isolated from 24-hr-old w1 embryos. Size markers (in kilobases) are represented as bars on either side of the blots, representing molecular sizes of 9.5, 7.46, 4.4, and 2.37 kb.
Structure of the gpp transcription unit:
We further characterized the gpp transcription unit by analyzing cDNAs isolated from various ovarian, embryonic, and larval libraries, by RT-PCR and by Northern blotting. As shown in Figure 4A, gpp is 42 kb in length and encodes mRNAs that range in size from 6.5 to 10 kb. The 5′ end of the gene is just upstream of the gppIN-1 transposon, and because of alternative splicing several different 5′-UTRs can be produced. The alternative exon sequences at the 5′ end of the transcription unit are separated from the main body of the gene by an ∼30-kb intron and they are spliced to a common exon containing additional 5′-UTR sequences plus the translation start site for all known gpp mRNAs. The main body of the mRNA has 5 exons that are common to all gpp mRNAs and encode the bulk of the protein. There are multiple 3′ exon combinations that differ from each other by the use of alternative 3′ and 5′ splice sites and poly(A) addition sites. gpp could potentially code for 12 different mRNAs; however, only six variants were detected in our analysis.
In 0-24 embryos the major species is ∼9 kb and there are less abundant RNAs of 7.5 and 10 kb (Figure 4B). This profile changes as development proceeds. In larvae (not shown), the 9- and 7.5-kb species are equally abundant while a smaller 6.5-kb species appears. In adults (see Figure 4B) the predominant mRNA is 7.5 kb. On the basis of our analysis of the different mRNAs, the 9-kb species seen in embryos and larvae is likely to correspond to T2, while the 7.5-kb species seen in embryos, larvae, and adults could correspond to T1, T4, and T5 as all three of these mRNAs are between 7.3 and 7.7 kb. T3 is 10 kb, about the size of the very large mRNA in embryos, while T6 is 6.6 kb and could correspond to the small mRNA seen at the larval stages.
Inspection of the gpp transcription unit reveals that the gppIN-1 P-element is inserted in the first exon, the gppIN-2 P-element is inserted midway through the first intron, the gppX breakpoint is located in the large intron, and the gppXXV breakpoint is in the protein coding sequence. Since these mutations disrupt the gpp gene, they might be expected to produce aberrant transcripts. To determine if this is the case, Northern blots of RNA from heterozygous flies were probed with gpp cDNAs. While we did not detect any unusual transcripts for gppIN-1/TM3 or gppX/TM6, an ∼4-kb RNA species is present in RNA prepared from gppXXV/TM3 flies (Figure 3B). Moreover, consistent with the disruption of the gpp transcription unit in gppXXV, this species is detected with probes upstream of the breakpoint, but not downstream.
The predicted GPP protein:
The gpp mRNAs code for proteins with a predicted mass ranging from 171 to 232 kD. Homology searches revealed that the common N-terminal domain of GPP shares significant sequence similarity to the S. cerevisiae DOT1 protein. DOT1 is a novel histone H3 methyltransferase that modifies the K79 residue inside the first globular domain of H3 (Lacoste et al. 2002; Ng et al. 2002; van Leeuwen et al. 2002). The sequence conservation between the GPP N terminus and DOT1 is ∼42%. However, the GPP sequence shows 100% homology to the DOT1 MT methyltransferase fold, which is required for methylation of histone H3 (Feng et al. 2002). This suggests that GPP is likely to be a Drosophila H3 K79 methyltransferase. GPP has additional protein domains that are not present in DOT1. These include a putative coiled-coil domain and an ATP/GTP binding domain. The presence of these domains suggests that GPP may dimerize or interact with other proteins. The GPP protein also contains several regions rich in alanine, proline, histidine, and glutamine. While yeast DOT1 does not contain these additional domains, the coiled-coil motif and the proline-rich region are found in human, Caenorhabditis elegans, Drosophila pseudoobscura, and Anopheles gambia DOT1-like proteins.
gpp functions as a histone H3 K79 methyltransferase:
The sequence similarity between gpp and Dot1 suggested that GPP protein might function as an H3 K79 methyltransferase. If this is correct, one might expect to find that H3meK79 is depleted in gpp mutants. Since our genetic experiments indicated that the gpp gene product is supplied maternally and the phenotypic effects of gpp mutants in the zygote are not observed until the larval stage, we examined K79 methylation in larval tissues. Imaginal discs from wild-type and gppX mutant third instar larvae were probed with antibodies raised against H3dmeK79 (Ng et al. 2002). Figure 5A shows that H3dmeK79 can be detected in all nuclei of wild-type wing discs. In contrast, the level of H3dmeK79 antibody staining is substantially reduced in wing discs from homozygous gppX larvae. In the second experiment, Western blots of proteins from wing discs, eye discs, and the CNS of wild-type and gppX mutant larvae were probed with antibodies against either monomethyl (not shown) or dimethyl K79 (Figure 5B). While H3 mono- and dimethyl K79 could be detected in proteins extracted from wild-type larval tissue, we found little or no mono- or dimethyl K79 in tissues from gppX mutants. This would suggest that like its counterparts in other organisms, GPP functions as a histone H3 K79 methyltransferase.
Figure 5.—
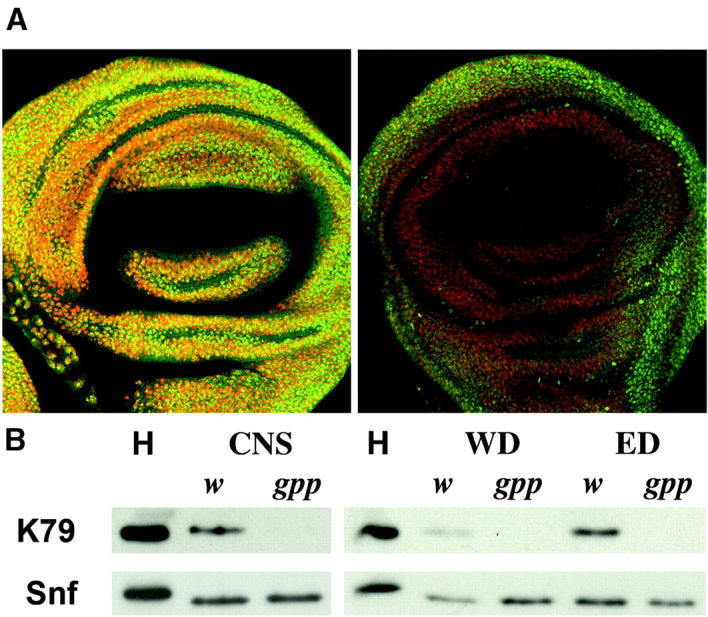
gppX mutant flies are deficient in methylation of H3dmeK79 relative to wild type. (A) Fluorescent images of imaginal wing discs from w1 (left) and gppX/gppX (right) third instar larvae stained with anti-H3dmeK79 antibody identified with fluorescent secondary antibody (green) and DNA dye (red). Both wing discs were imaged at the same fluorescent intensity using confocal microscopy. (B) Western blot analysis of different imaginal tissue removed from w1 and gppX third instar larvae. K79 levels are absent in the gppX lanes relative to wild type. H, acid-extracted histone proteins used as a control; central nervous systems (CNS), wing discs (WD), and eye discs (ED) were removed from third instar larvae and prepared as described in materials and methods. Snf antibody was used to verify equal loading of protein in the lanes.
Developmental regulation of H3 K79 methylation:
We have previously examined the pattern of histone H3 methylated on K4 and K9 during embryogenesis (Schaner et al. 2003). The developmental profile of these two H3 modifications is consistent with findings in other organisms indicating that histone H3 methylated K4 (H3meK4) is generally associated with active chromatin, while histone H3 methylated K9 (H3meK9) is usually a marker for inactive or silenced chromatin (Schaner et al. 2003). During the rapid nuclear cleavage stages there is little if any H3meK4, while H3meK9 can be detected. H3meK4 remains low even after the nuclei migrate to the periphery of the embryo at nuclear cycle 9–10 and only around cycle 12 does the level of H3meK4 begin to increase in somatic cells. Concomitant with the increase in H3meK4, transcription is upregulated in somatic nuclei at about this stage. At cellularization all somatic nuclei have high levels of H3meK4. In contrast, there is little or no K4 methylation in the pole cells, while there are high levels of K9. This difference reflects the fact that germ cells are transcriptionally quiescent until much later in development. The correlation between transcriptional activity and methylation of K4 or K9 suggested that it would be of interest to examine the developmental profile of K79 methylation.
Like K4, little if any H3 mono- or dimethyl K79 could be detected in early nuclear cleavage stage embryos. However unlike K4, K79 methylation does not appear to be activated when transcription commences in syncytial blastoderm embryos. As shown for a precellular blastoderm embryo in Figure 6, H3meK79 seems to be absent not only from the somatic nuclei, which are still dividing in these embryos, but also from the pole cell nuclei, which are arrested in the cell cycle in G2 (Su et al. 1998). H3 mono- (not shown) and dimethyl K79 (Figure 6) is first readily detected by antibody staining much later in development in germband extended embryos. At this stage H3meK79 accumulation appears to be coupled to the cell cycle. The highest levels of K79 methylation are found in cells undergoing mitosis and as shown by the stage 8/9 embryo in Figure 6 these cells are often clustered in small mitotic domains. In addition to the cells that are in mitosis, some nuclei have high levels of dimethyl K79 histone H3 but do not appear to be in the process of dividing. Usually in these nuclei, antibody staining is concentrated in one or two spots, while the distribution of chromosomal DNA appears to be more diffuse. The remaining nuclei in the germband extended embryos have either low levels of antibody staining or apparently none at all. This general pattern persists through stages 10–11 until just before germband retraction. At this point the level of mono- (not shown) and dimethyl K79 histone H3 (Figure 6) begins to increase substantially, particularly in epidermal nuclei. As illustrated by the stage 15–16 embryo, high levels of dmeK79 histone H3 are observed in virtually all epidermal nuclei at this stage and in subsequent stages. A different pattern is seen in the CNS: in some CNS nuclei there seems to be a high level of dmeK79, while in many other nuclei there is only a low level of this modification. The CNS differs from epidermis at this stage of development in that the neuroblasts and ganglion mother cells (GMCs) in the CNS are still dividing, while only the progeny of the GMCs, the neurons, have stopped dividing (Lee and Orr-Weaver 2003).
Figure 6.—
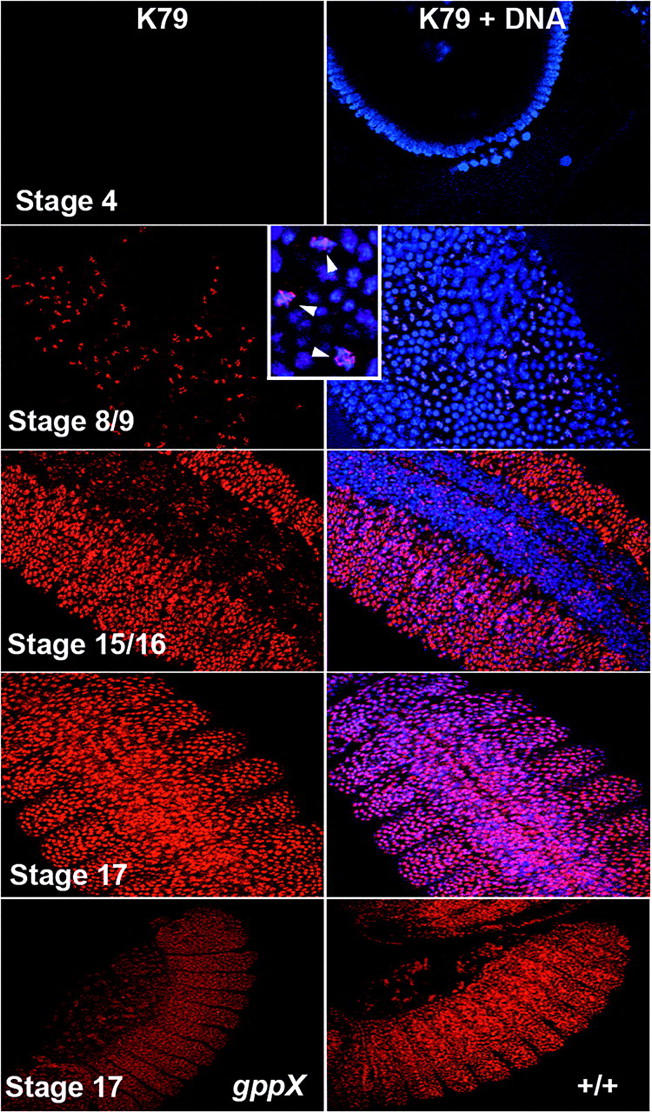
Methylation of H3 K79 in embryos is coincident with the developmental stage of homeotic gene expression. w1 embryos from different developmental stages were fixed and stained with anti-H3dmeK79. Inset shows a close-up view of mitotic cells stained with K79. Note the difference in the stage 17 embryo CNS K79 staining vs. stage 15/16. Bottom compares K79 staining intensity in late-stage gppX homozygotes and w1 embryos. H3 K79 is in red and DNA is blue.
On the basis of our genetic analysis of gpp mutants, we anticipated that there would be a substantial maternal contribution of gpp gene products and that dmeK79 would be detected in gpp homozygous mutant embryos. This expectation was correct since the level of dmeK79 in both gppX mutants and wild type is similar in pattern through the blastoderm to stage 11–12 embryos. However, in older embryos the level of H3dmeK79 appears to be reduced compare to wild-type embryos at a similar stage. The difference in the level of antibody staining between wild-type and gppX embryos in two stage 17 embryos is shown Figure 6. At this stage, the level of H3dmeK79 in the mutant is about one-half that in wild type when imaged at the same intensity.
Distribution of H3dmeK79 in polytene chromosomes:
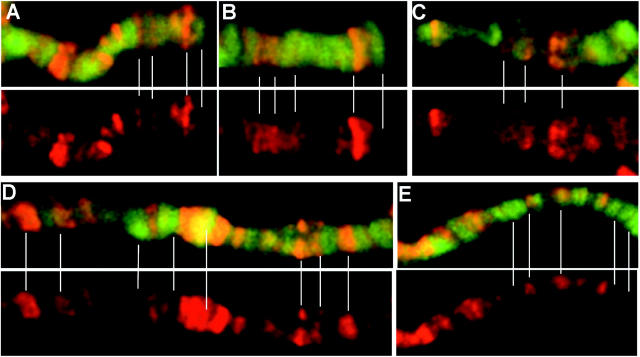
The phenotypic effects of gpp mutants point to a role in both gene activation and silencing. Consequently, we examined the distribution of H3dmeK79 in salivary polytene chromosomes to determine if this modification was preferentially localized to active or inactive chromatin. As depicted in Figure 7 several findings are of interest. First, H3dmeK79 appears to be underrepresented at the telomeres compared to other nearby chromosomal segments (Figure 7, A and B). Second, H3dmeK79 is often enriched in puffs and interbands (Figure 7, C, D, and E). Since puffs and interbands are thought to correspond to active chromatin domains, this suggests that there may be a connection between methylation of histone H3 K79 and transcription. However, this connection must be domain specific in that H3dmeK79 is not enriched at all puffs and interbands. Third, H3dmeK79 can also be found localized in bands. As observed for puffs and interbands, H3dmeK79 enrichment in bands is domain specific. Fourth, as evident from the green-to-yellow to orange-to-red staining in different bands, interbands, and puffs, the relative level of H3dmeK79 per “unit” of DNA seems to vary substantially from one chromosomal domain to the next.
Figure 7.—
Anti-H3dmeK79 staining of w1 polytene chromosomes reveals that dmeK79 is found in both band and interband regions, but is not present at the telomeres. (A–D) Top, merged image of the H3dmeK79 (red) and DNA (green) stained chromosomes; bottom, polytenes stained only with K79. White lines connecting the bottom and top identify regions of interest. (A and B) K79 staining is missing from telomeres. (C) K79 staining is present in a puff. (D and E) The distribution of K79 is diverse in its localization to band and interband regions.
DISCUSSION
Recent studies on telomeric silencing in S. cerevisiae have led to the identification of a histone methlylase, DOT1, which has a number of unusual properties (Singer et al. 1998; Feng et al. 2002; Lacoste et al. 2002; van Leeuwen et al. 2002). First, unlike the previously identified histone methylases, DOT1 does not have a canonical SET domain. Instead, the DOT1 protein resembles a family of S-adenosyl methione methyltransferases that modify arginine residues (Feng et al. 2002; Lacoste et al. 2002; van Leeuwen et al. 2002). DOT1 methylates histone H3 at lysine 79 only when it is assembled into nucleosomes and methylation strongly depends upon prior Rad6 dependent ubiquitination of histone H2B at K123 (Briggs et al. 2002). Second, in yeast, deletion or overexpression of Dot1 disrupts TPE and also silencing of the mating-type loci (Singer et al. 1998). In contrast, silencing in the yeast ribosomal gene cluster is disrupted only when DOT1 is overexpressed (Singer et al. 1998). Third, both telomeric and mating-type silencing are disrupted by mutations in the lysine 79 residue of histone H3. Fourth, methylation of K79 appears to influence the recruitment of the SIR silencing proteins to the telomeres (van Leeuwen et al. 2002; Ng et al. 2003a). The SIR silencing proteins appear to preferentially associate with chromatin that is deficient in K79 methylation, while the proteins are generally not associated with chromatin in which there is an enrichment for K79 methylated H3 (van Leeuwen et al. 2002; Ng et al. 2003a). Fifth, there is evidence that K79 methylation is coordinated with polymerase transcription via the COMPASS complex (Krogan et al. 2003). Consistent with the idea that K79 methylation might be coordinated with transcription, H3meK79 is enriched in transcribed sequences in yeast and mammals (Im et al. 2003; Ng et al. 2003b). Interestingly, the distribution of H3meK79 in the β-globin locus differs from H3meK4 in that it is not found at the locus control region (Im et al. 2003). These findings have led to a model in which H3meK79 serves as a marker for transcribed sequences where it functions to block the association of chromatin proteins that mediate transcriptional silencing.
While Dot1 homologs have been identified in higher eukaryotes, little is known about their biological functions (Feng et al. 2002). In this report we have characterized the Drosophila Dot1 ortholog gpp. The gpp transcription unit is >40 kb in length and it encodes a complex array of alternatively spliced transcripts that range in size from 6.5 to >9 kb and are expressed at different developmental stages. Consistent with our assignment of the gpp gene, P-element and X-ray mutations disrupt this large transcription unit and in at least one case lead to the production of truncated mRNAs. The gpp transcripts are predicted to encode 170- to 232-kD polypeptides that share a common N-terminal domain that corresponds to about two-thirds of the protein but have different C-terminal domains. The common N-terminal domain contains the Dot1 homology region including the MT methyltransferase fold required for methylation of histone H3 (Feng et al. 2002). Mutation of conserved glycine residues in the active site of both yeast and human DOT1 protein inactivates the enzyme (Feng et al. 2002; van Leeuwen et al. 2002). GPP also contains domains that are not present in DOT1 including a coiled-coil motif also found in the human, C. elegans, D. pseudoobscura, and A. gambia DOT1-like proteins. In yeast, K79 is mono-, di-, and trimethylated and Dot1 is responsible for all three modifications (Feng et al. 2002; van Leeuwen et al. 2002). The different methylated states of H3 at K79 suggest that multiple regulatory activities are conferred on these modified nucleosomes (Ng et al. 2002; van Leeuwen et al. 2002). However, in fly tissue culture cells, the mono- and di- but not the trimethylated form is observed (Mckittrick et al. 2004). Since database searches indicate that gpp is the only fly Dot1 homolog, it should also be the sole fly protein in this class that methylates histone H3 on K79. Consistent with this suggestion, discs and other tissues isolated from gpp mutant larvae have little if any H3 mono- or dimethyl K79.
Like its yeast counterpart, gpp is required for the silencing of reporter transgenes inserted into telomeric heterochromatin. However suppression of silencing associated with pericentric heterochromatin is unaffected by mutations in gpp. While these observations point to a role of gpp in silencing specific for telomeric heterochromatin, our antibody staining experiments indicate that there is a paucity of H3dmeK79 at telomeres in polytene chromosomes compared to many other chromosomal DNA segments. In this respect it is interesting that both telomeric and mating-type chromatin in yeast are hypomethylated on K79 compared to “bulk” chromatin even though DOT1 is required for SIR silencing in each case (Ng et al. 2003a). It has been suggested that the meK79 modification in euchromatic nucleosomes blocks SIR protein association and that silencing is lost in the absence of DOT1 because the SIR proteins spread into euchromatin (van Leeuwen et al. 2002). On the other hand, in flies, since many euchromatic domains in wild-type polytene chromosomes have only little H3meK79, it is difficult to see how telomeric silencing proteins would be restricted to telomeres by this modification even when gpp is fully active.
gpp also has functions in flies besides telomeric silencing. Unlike Dot1, gpp is essential for viability. Although the underlying cause of lethality remains to be established, gpp mutant larvae grow more slowly than wild type and this potentially implicates gpp in pathways that control growth rates and size in flies. In addition, gpp mutants display defects that are characteristic of both Pc-G and trx-G genes. The first gpp alleles were recovered as dominant suppressors of mini-white silencing by two BX-C PREs. Consistent with a role in Pc-G silencing, gpp mutants enhance the segmentation defects of several Pc-G genes. In this context, it is interesting to note that several Pc-G genes have recently been shown to play a role not only in the repression of genes in the homeotic complexes but also in telomeric silencing (Boivin et al. 2003). Thus, it is possible that gpp activity in telomeric silencing may be linked in some manner to its role in Pc-G silencing.
gpp mutants also exhibit transformations in segment identity and genetic interactions with Abd-B that are characteristic of trx-G mutations. This would point to a role in promoting rather than repressing gene expression. Some function in transcription would be consistent with studies in other systems as well as with the enrichment of meK79 seen in many polytene interbands and puffs. However, this correlation is not complete. Thus, there are many puffs and interbands that have only little H3dmeK79. Conversely, H3dmeK79 is sometimes enriched in bands. These findings would argue that in Drosophila, meK79 is not a ubiquitous marker for transcriptionally active chromatin, but rather may have functions that are specific to particular chromatin domains. In this case, the disruptions in homeotic gene expression seen in gpp mutants could reflect a special requirement for H3meK79 in the transcription of these particular genes. Domain-specific requirements for gpp activity in transcription could also potentially account for the effects of gpp mutations on Pc-G and telomeric silencing. In this model, Pc-G and telomeric silencing would be disrupted in gpp mutants because the expression of one or more Pc-G (and/or telomeric heterochromatin) genes is downregulated when gpp activity is compromised.
The developmental profile of H3dmeK79 also suggests that this modification cannot be a ubiquitous marker for either transcriptionally active or silenced chromatin. High levels of Pol II transcription in somatic nuclei begin in the precellular blastoderm stage around nuclear cycle 11/12. Concomitant with the activation of transcription, H3meK4 can be first be detected at this stage, and the level of meK4 then increases through cellularization (Schaner et al. 2003). By contrast, little if any H3 mono- or dimethyl K79 is in either the transcriptionally active somatic nuclei or the transcriptionally quiescent pole cell nuclei. H3meK79 can first be readily detected only later in development in germband extended embryos. However, at this stage accumulation is restricted primarily to a subset of cells in the embryo, most of which seem to be in the process of cell division. High levels of H3meK79 are not observed until stages 13–15, long after the initial upregulation of transcription in the early zygote. This result also suggests that the homeotic transformations seen in gpp mutants are unlikely to be due to defects in the initial establishment of parasegment-specific patterns of homeotic gene expression by the gap and pair-rule genes. Rather, these transformations probably reflect a requirement for gpp activity later in development during the maintenance phase of homeotic gene regulation—a phase that is dependent upon Pc-G and trx-G genes. In this respect it is curious that homeotic transformations are not observed in gpp embryos when they hatch as first instar larvae. Maternally derived gpp activity in homozygous mutant embryos maybe sufficient to maintain specific parasegmental patterns of homeotic gene expression through the end of embryogenesis. Alternatively, there may not be absolute requirement for H3meK79 in maintaining appropriate parasegmental patterns of homeotic expression during embryogenesis.
The developmental profile of H3meK79 indicates that this modification is present at low levels in specific developmental stages and tissues (CNS) undergoing active cell division. In contrast, the highest levels of H3meK79 are observed in epidermal cells that have exited the cell cycle and are undergoing differentiation. Thus, it seems possible that this modification may be activated when specific chromatin configurations, active or inactive, need to be maintained for extended periods of time in the absence of de novo DNA synthesis/chromatin assembly. In this respect it is interesting that Mckittrick et al. (2004) have reported that the highest levels of meK79 are found in a histone H3 variant, H3.3, which is assembled into chromatin by a replication-independent mechanism. Further studies of gpp in Drosophila will be required to understand the mechanisms governing the temporal and tissue-specific regulation of the K79 modification and how this relates to the functions of this particular histone modification during development. Understanding this aspect of the histone code in a multicellular organism such as Drosophila will lead to a better understanding of chromatin regulatory mechanisms during development.
Acknowledgments
We thank the Bloomington Stock Center for mailing stocks throughout the course of this work; the Princeton, Szeged, Geneva, and Basel fly food facilities for reliable supply of quality fly food; Girish Deshpande, Joseph Goodhouse, Tony Greenberg, Ruth Steward, Lori Wallrath, the Schedl lab, Jill Kohler, Radhika Mohan, Yi Zhang, Qin Feng, Michael Grunstein, Maria Vogelauer, Kirsten Hagstrom, Julio Vazquez, Judy Kassis, Francois Karch, Daniel Pauli, Rakesh Mishra, Ivan Dellino, Christian Sigrist, Frank Hirth, Lars Kammermeier, and Heinrich Reichert for discussions and help. We are indebted to Jim Kennison for sharing information on the hhMrt suppressor screen and the gpp8 allele. This work was supported by a National Institutes of Health grant to P.S. G.S. was supported by an NIH postdoctoral fellowship. While working in P.S.'s lab, M.M. was supported by a European Molecular Biology Organization long-term fellowship and the Swiss National Science Foundation. M.M. is grateful to Vincenzo Pirrotta in whose lab part of this work was done.
References
- Beisel, C., A. Imhof, J. Greene, E. Kremmer and F. Sauer, 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857–862. [DOI] [PubMed] [Google Scholar]
- Boivin, A., C. Gally, S. Netter, D. Anxolabehere and S. Ronsseray, 2003. Telomeric associated sequences of Drosophila recruit Polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164: 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz et al., 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Brock, H. W., and M. van Lohuizen, 2001. The Polycomb group—no longer an exclusive club. Curr. Opin. Genet. Dev. 11: 175–181. [DOI] [PubMed] [Google Scholar]
- Cryderman, D. E., M. H. Cuaycong, S. C. Elgin and L. L. Wallrath, 1998. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107: 277–285. [DOI] [PubMed] [Google Scholar]
- Cryderman, D. E., E. J. Morris, H. Biessmann, S. C. Elgin and L. L. Wallrath, 1999. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. EMBO J. 18: 3724–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin, B., R. Melfi, D. Mccabe, V. Seitz, A. Imhof et al., 2002. Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196. [DOI] [PubMed] [Google Scholar]
- Deshpande, G., J. Stukey and P. Schedl, 1995. scute (sis-b) function in Drosophila sex determination. Mol. Cell. Biol. 15: 4430–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, D. M., E. A. Burgess and I. Duncan, 1998. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 12: 1290–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld, A. L., and J. A. Kennison, 1995. Positional signaling by hedgehog in Drosophila imaginal disc development. Development 121: 1–10. [DOI] [PubMed] [Google Scholar]
- Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst et al., 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12: 1052–1058. [DOI] [PubMed] [Google Scholar]
- Hagstrom, K., M. Muller and P. Schedl, 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila Bithorax Complex. Genetics 146: 1365–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., C. R. Nelson, B. P. Berman, T. R. Laverty and R. A. George, 2000. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287: 2271–2274. [DOI] [PubMed] [Google Scholar]
- Im, H., C. Park, Q. Feng, K. D. Johnson, C. M. Kiekhaefer et al., 2003. Dynamic regulation of histone H3 methylated at lysine 79 within a tissue-specific chromatin domain. J. Biol. Chem. 278: 18346–18352. [DOI] [PubMed] [Google Scholar]
- Kennison, J. A., 1995. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29: 289–303. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney et al., 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23: 4207–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier and J. Cote, 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem 277: 30421–30424. [DOI] [PubMed] [Google Scholar]
- Lee, L. A., and T. L. Orr-Weaver, 2003. Regulation of cell cycles in Drosophila development: intrinsic and extrinsic cues. Annu. Rev. Genet. 37: 545–578. [DOI] [PubMed] [Google Scholar]
- Mahmoudi, T., and C. P. Verrijzer, 2001. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene 20: 3055–3066. [DOI] [PubMed] [Google Scholar]
- Mckittrick, E., P. Gafken, K. Ahmad and S. Henikoff, 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101: 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, M., K. Hagstrom, H. Gyurkovics, V. Pirrotta and P. Schedl, 1999. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153: 1333–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst et al., 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16: 1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger and K. Struhl, 2003. a Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100: 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H. H., S. Dole and K. Struhl, 2003. b The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278: 33625–33628. [DOI] [PubMed] [Google Scholar]
- Samuels, M. E., P. Schedl and T. W. Cline, 1991. The complex set of late transcripts from the Drosophila sex determination gene sex-lethal encodes multiple related polypeptides. Mol. Cell. Biol. 11: 3584–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner, C. E., G. Deshpande, P. D. Schedl and W. G. Kelly, 2003. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J. A., and J. W. Tamkun, 2002. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 12: 210–218. [DOI] [PubMed] [Google Scholar]
- Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson et al., 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, T. T., Campbell, S. D., and O'Farrell, P. H., 1998. The cell cycle program in germ cells of the Drosophila embryo. Dev. Biol. 196: 160–170. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F., P. R. Gafken and D. E. Gottschling, 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756. [DOI] [PubMed] [Google Scholar]
- Vazquez, J., G. Farkas, M. Gaszner, A. Udvardy, M. Muller et al., 1993. Genetic and molecular analysis of chromatin domains. Cold Spring Harbor Symp. Quant. Biol. 58: 45–54. [DOI] [PubMed] [Google Scholar]