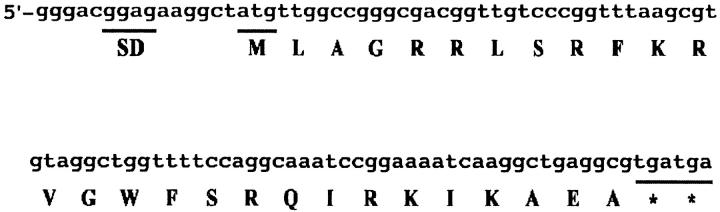Abstract
The dmd gene of bacteriophage T4 is required for the stability of late-gene mRNAs. When this gene is mutated, late genes are globally silenced because of rapid degradation of their mRNAs. Our previous work suggested that a novel Escherichia coli endonuclease, RNase LS, is responsible for the rapid degradation of mRNAs. In this study, we demonstrated that rnlA (formerly yfjN) is essential for RNase LS activity both in vivo and in vitro. In addition, we investigated a role of RNase LS in the RNA metabolism of E. coli cells under vegetative growth conditions. A mutation in rnlA reduced the decay rate of many E. coli mRNAs, although there are differences in the mutational effects on the stabilization of different mRNAs. In addition, we found that a 307-nucleotide fragment with an internal sequence of 23S rRNA accumulated to a high level in rnlA mutant cells. These results strongly suggest that RNase LS plays a role in the RNA metabolism of E. coli as well as phage T4.
THE control of mRNA stability is an important aspect of gene expression. mRNA degradation has been studied extensively in both prokaryotes and eukaryotes, and such studies have revealed that cis-acting RNA elements and trans-acting proteins influence mRNA stability (Schoenberg and Chernokalskaya 1997; Van Hoof and Green 1997; Coburn and Mackie 1999; Schwartz and Parker 2000). The most important factor among trans-acting proteins is an RNase that plays a central role in mRNA degradation. In Escherichia coli, the degradation of mRNA is usually initiated by endonucleolytic cleavage (Apirion 1973; Coburn and Mackie 1999; Kushner 2002). Under vegetative growth conditions, five endonucleases (RNases I*, III, E, G, P) are known or suggested to cleave mRNA (Portier et al. 1987; Arraiano et al. 1988; Bardwell et al. 1989; Cannistraro and Kennell 1991; Mackie 1991; Alifano et al. 1994; Umitsuki et al. 2001). Furthermore, two recently discovered endonucleases function under certain conditions. RelE cleaves mRNA positioned at the ribosomal A site when bacteria are starved for amino acids (Pedersen et al. 2003). MazF encoded by the mazEF addiction module (Aizenman et al. 1996) cleaves mRNAs when cells undergo programmed cell death (Zhang et al. 2003).
We have been investigating an mRNA cleavage activity that induces late gene silencing in bacteriophage T4 (Kai et al. 1996). The T4 dmd gene is required for the regulation of mRNA stability in a stage-dependent manner during infection (Ueno and Yonesaki 2001). When a T4 mutant defective in this gene infects E. coli cells at low temperatures, gene expression is globally silenced at late stages because of the rapid degradation of mRNA. The rapid degradation is caused by dmd mutant-specific cleavages of mRNA (Kai et al. 1996; Kai and Yonesaki 2002). Recently, we suggested that the host encodes an activity for performing such cleavage and we isolated E. coli std mutants that are defective in dmd mutant-specific cleavages. The loci of the std mutations as well as the effects of mutations in known RNase-encoding genes suggested that an RNA cleavage activity causing the dmd mutant-specific mRNA degradation could be attributed to a novel RNase (Otsuka et al. 2003). Hence, we propose calling this endonuclease RNase LS (late-gene silencing in T4).
RNase LS preferentially cleaves RNA 3′ to pyrimidines, but exceptions are observed. Some cleavages by RNase LS are introduced when the target RNA is translatable, while others are independent of translation (Kai et al. 1996; Kai and Yonesaki 2002). Thus, RNase LS seems to cleave RNA in a complex manner. The structural gene for RNase LS as well as its role in E. coli cells have not been described. The E. coli std-2 mutation abolishes RNase LS activity and allows T4 dmd to grow as well as wild-type phage (Otsuka et al. 2003). As an initial step to characterizing the RNase LS, we sought to identify the gene harboring the std-2 mutation and found that yfjN is the gene. Because yfjN is the name of a previously uncharacterized gene, we renamed it rnlA and its mutant alleles, std-2 and std-5, rnlA2 and rnlA5, respectively. Our results demonstrate that rnlA is essential for RNase LS activity in vitro as well as in vivo. To investigate the role of RNase LS in E. coli cells, we analyzed the phenotype of the rnlA2 mutant. The results strongly suggest that RNase LS plays a role in the RNA metabolism of E. coli during vegetative growth.
MATERIALS AND METHODS
Bacterial strains:
We used the E. coli K-12 strains MH1 (sup0 hsdR ΔlacX74 rpsL), TY0482 (MH1 rnlA2), TY1722 (MH1 std-3), TY1797 (MH1 std-4), and TY2133 (MH1 rnlA5; Otsuka et al. 2003). TY0324 (MH1 rnlA::kan) was constructed as described below. MG1655 was used for cloning bacterial genes.
Plasmids:
To clone rnlA-yfjO, the relevant DNA segment was amplified by PCR using strain MG1655 genomic DNA as a template and 5′-atgtttctatgggatccagg plus 5′-gctatttgatcatattggac as primers. The DNA segment was cloned into the EcoRV site in pBR322 to construct pBRNO and pBRON, in which rnlA-yfjO was cloned in both the same and the reverse directions as the tet promoter. To clone rnlA alone, pBRNO was digested with BanII and subsequently self-ligated to generate pBRN. pBRN(kan)O was constructed by inserting a HincII fragment containing a kanamycin-resistance marker derived from pUC4K (Pharmacia LKB) into the SphI site of pBRNO. The rnlA::kan DNA segment of pBRN(kan)O was amplified by PCR with the primer set used for constructing pBRNO and then cloned into the SmaI site in pKO3 (Link et al. 1997) to construct pKmyfjN.
To construct plasmids pBSrpsA and pBRbla, DNA fragments containing either rpsA or bla were amplified by PCR using MG1655 DNA as a template with each set of primers and ligating into the EcoRV site of pBluescript II KS+ or pBR322. The primers were 5′-ggatccggcagccgatgctttagtg-3′ plus 5′-ggatcctgcaatctgtcaagtaaac-3′ for pBSrpsA and 5′-ggagcaggcgcataaatg-3′ plus 5′-cctgttcctgatgatcgttc-3′ for pBRbla.
Construction of strain TY0324:
Following transformation of MH1 cells with pKmyfjN, the chromosomal rnlA gene was replaced with rnlA::kan as described by Link et al. (1997). Briefly, transformants were plated at 42°, a temperature nonpermissive to the pKO3 replicon, to select cells with chromosomal integrants of the plasmid. Subsequently, these cells were appropriately diluted and plated at 30° on LB plates containing 5% sucrose to select cells that had lost the plasmid. Finally, sucrose-resistant, kanamycin-resistant, chloramphenicol-sensitive colonies were screened. The candidates were examined by PCR for an insertion of kan in the target gene.
Primer extension:
Total RNA from a 1.5-ml culture of infected cells was isolated as described (Kai et al. 1996). Primer-extension analysis of soc RNA was performed using soc primer 2 as described (Kai and Yonesaki 2002). The reaction products were denatured by boiling for 2 min and analyzed by electrophoresis through a 5% polyacrylamide gel containing 7 m urea.
Functional decay rate:
MH1 or TY0482 cells were grown at 30° in M9 minimal medium supplemented with 0.3% casamino acids, 1 μg/ml thiamine, and 20 μg/ml tryptophan. When the cell density reached 4 × 108 cells/ml, rifampicin was added to 500 μg/ml (zero time). [35S]Methionine/cysteine (American Radiolabeled Chemicals, St. Louis; >37 TBq/mmol) was added to a 100-μl culture to 3.7 MBq/ml at various times to label newly synthesized protein for 1 min. Labeled proteins from an equal portion of cell culture were analyzed by electrophoresis through a 12.5% polyacrylamide gel containing 0.1% SDS. An autoradiograph was taken with a Bio-Image analyzer (Fuji BAS-1800). To calculate the functional half-life of net mRNA, all the signal intensities of bands in each lane were summed and plotted to measure the time required for a 50% reduction of total intensity.
Northern blot analysis:
Cells were grown to 4 × 108 cells/ml as described above and rifampicin was added to 500 μg/ml (zero time). At various times thereafter, a 1.5-ml culture was quickly chilled on ice and the cells were harvested by centrifugation. Total RNA extraction and Northern blotting were carried out according to Kai et al. (1996). A radioactive probe for each gene transcript was prepared by PCR with either T4 DNA or E. coli DNA (MG1655) as a template. One of the PCR primers was 5′ end-labeled by incubation with T4 polynucleotide kinase (Toyobo) and [γ-32P]ATP (Institute of Isotopes of the Hungarian Academy of Sciences, Hungary; 259 TBq/mmol). The sequences of primers are as follows: for the soc gene, 5′-32P-gttattaaccagttactttc and 5′-cctgcagtaacaagttcggctc; for the rpsO gene, 5′-32P-ttgcttcagtacttagagac and 5′-gcttaacgtcgcgtaaattg; for the rpsA gene, 5′-32P-ggatcctgcaatctgtcaagtaaac and 5′-ggatccggcagccgatgctttagtg; for the bla gene, 5′-32P-agcggggtaattgtgcgatg and 5′-ggatcctgttcctgatgatcgttc; for the ompA gene, 5′-32P-ttggatttagtgtctgcacg and 5′-atgaaaaagacagctatcgc. After autoradiography with a Bio-Image analyzer, the signal intensity was quantified with the National Institutes of Health (NIH) image program. To calculate the half-life of each mRNA, the amount of the full-length transcript was quantified by NIH image and its relative amount was plotted to measure the time required for a 50% reduction. Except for rpsA, we performed two experiments for ompA mRNA and three for the other mRNAs.
Analysis of RNA accumulated in an rnlA2 mutant:
Total RNA was extracted from cells according to Kai et al. (1996). Total RNA from TY0482 cells was electrophoresed through a 5% polyacrylamide gel and stained with ethidium bromide. Comparing RNA bands between MH1 and TY0482, the RNA species specific to TY0482 cells was eluted from the gel slice, precipitated with ethanol, and resuspended in water. To further purify this RNA species, the same manipulation was carried out using a 5% polyacrylamide gel containing 7 m urea. The purified RNA was used as a radioactive probe for dot-blot hybridization. The 5′ end of this RNA was dephosphorylated by incubating with calf intestine alkaline phosphatase (Toyobo) and phosphorylated by incubating with T4 polynucleotide kinase and [γ-32P]ATP. Dot-blot hybridization for the cosmid library of the K-12 W3110 chromosome (Tabata et al. 1989), which was kindly provided by A. Nishimura at the National Institute for Genetics, was performed as follows. A solution containing each cosmid plasmid was spotted onto a nylon membrane. After denaturing the DNAs by exposing the membrane to 0.5 n sodium hydroxide, the DNAs were crosslinked by ultraviolet irradiation and were hybridized with a 32P-labeled probe at 45° overnight in 50% formamide, 0.25 m sodium chloride, and 3.5% SDS. The membrane was washed at 45° with 2× SSC containing 0.1% SDS. A cosmid clone, E3107, was cleaved with BamHI and EcoRI and ligated into a plasmid pBluescript II (Stratagene, La Jolla, CA) previously cut with the same enzymes, yielding pBS109. pBS109 was digested with BamHI and NruI, blunted with T4 DNA polymerase, and self-ligated with T4 DNA ligase to construct pBS109BN. The DNA fragment in pBS109BN was sequenced using two primers (KS primer 5′-tcgaggtcgacggtatc and T3 primer 5′-aattaaccctcactaaaggg). The nucleotide sequences at the 5′ and 3′ termini of the purified RNA fragment were determined by RNA ligation, cDNA sequencing, and primer extension analysis as described previously (Yonesaki 2002).
Preparation of cell extract:
MH1 or TY0482 cells were grown to 3 × 108 cells/ml in 300 ml of Luria broth (LB) medium, harvested by centrifugation, and washed twice with cold TMCK buffer [10 mm Tris-HCl (pH 7.5), 10 mm magnesium acetate, 30 mm KCl, 0.5 mm DTT]. The cells were frozen and stored at −80° until used in the following procedures, which were carried out at 4°: The frozen cells were thawed and ground with 0.75 g of aluminium oxide. Subsequently, 150 μl of TMCK buffer containing 1.5 units of RNase-free DNase (Nippon Gene) was added to the cell paste. The suspension was kept on ice for 10 min, and the aluminum oxide was removed by centrifugation at 15,000 × g for 20 min to obtain a clear lysate. This lysate was centrifuged at 30,000 × g for 30 min to prepare the S30 fraction.
Assay for RNA cleavage in vitro:
soc RNA was synthesized by T7 RNA polymerase with pTK40 (Kai and Yonesaki 2002) as a template and subjected to cleavage in a buffer supplied for the in vitro translation kit (Ambion, Austin, TX). The standard 10-μl reaction mixture contained 2 μl of the master mixture, 0.4 μl of T7 RNA polymerase, 0.3 μg pTK40 DNA, and 100 μg protein from the S30 fraction of MH1 or TY0482 cell extract. The reactions were incubated for 30 min at 30°. RNAs were extracted twice with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) previously equilibrated with 0.2 m sodium acetate (pH 5.2), collected by ethanol precipitation, and suspended in water. A total of 7.5 μg RNA recovered from each reaction mixture was used for a primer-extension assay.
RESULTS
Involvement of rnlA in RNase LS activity:
Previously, we isolated five E. coli std mutants that impair RNase LS activity (Otsuka et al. 2003). The std-1, std-3, and std-4 mutants weakly decrease RNase LS activity and the growth defect of T4 dmd phages. Their effects require iscR (Otsuka et al. 2003), which encodes a transcriptional repressor of the iscRSUA operon (Schwartz et al. 2001). When cloned in a multicopy plasmid, iscR can suppress the growth defect of the dmd mutant (T. Yonesaki, unpublished results), suggesting that an increase of intracellular IscR protein counteracts RNase LS activity. Thus, the mutational effects of std-1, -3, and -4 on RNase LS seem to be indirect.
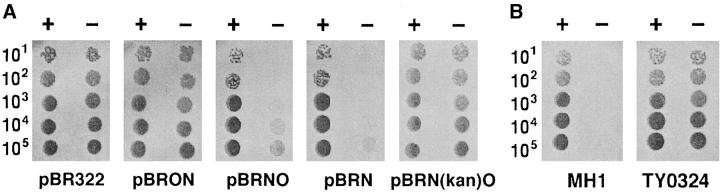
In contrast, rnlA2 and rnlA5 strongly reduce RNase LS activity and the growth defect of the T4 dmd mutant. In addition, their effects did not require iscR. Accordingly, these mutations may directly alter RNase LS. P1-mediated transduction showed that the rnlA2 and rnlA5 mutations were in the vicinity of yfjK (Otsuka et al. 2003). To identify genes harboring these mutations, we cloned a number of genes located in the vicinity of yfjK and looked for a clone that rendered rnlA2 or rnlA5 mutants unable to support the growth of a T4 dmd mutant. A clone, pBRNO, reduced the efficiency of plating of the dmd mutant on rnlA2 cells <10−2 and sharply reduced the plaque size (Figure 1A). Thus, this clone could complement the rnlA2 mutation. pBRNO contains two genes of previously unknown function, rnlA and yfjO. pBRON, in which rnlA-yfjO is cloned in the reverse direction to the vector tet promoter, could not complement the rnlA2 mutation.
Figure 1.—
Growth capacity of the T4 dmd mutant. (A) A solution containing T4 wild-type (+) or dmd mutant (−) phage was serially diluted in 10-fold steps, and 2 μl containing the number of phages indicated in the left margin was spotted onto a plate seeded with TY0482 cells harboring the plasmid indicated below the figure. See detail in text. (B) Spot tests were performed as in A on a plate seeded with MH1 or TY0324. Photographs were taken after overnight incubation at 30°.
Next, we determined which gene was effective in complementation. When rnlA (pBRN) or yfjO [pBRN(kan)O] alone was introduced into rnlA2 cells, a T4 dmd mutant was able to grow in the presence of pBRN(kan)O but not in the presence of pBRN. Similar results were obtained with the rnlA5 mutation (data not shown). These results strongly suggested that rnlA2 and rnlA5 were alleles of rnlA. Therefore, we sequenced rnlA from the rnlA2 and rnlA5 mutants. rnlA from the rnlA2 mutant had one base substitution that converted codon 32 from GAA (gln) to TAA (ochre). rnlA from the rnlA5 mutant had two amino acid substitutions: from glu (GAG) to lys (AAG) at codon 188 and from asp (GAT) to asn (AAT) at codon 196.
To confirm that rnlA is responsible for the RNase LS activity, we constructed the rnlA-disrupted strain TY0324 (see materials and methods). A T4 dmd mutant exhibited an efficiency of plating of <10−5 on parental MH1 cells (Figure 1B). In contrast, the T4 dmd mutant grew well on TY0324 cells with an efficiency of plating of ∼1. The burst size of a T4 dmd mutant on TY0324 cells was the same as that of wild-type phage (data not shown). Therefore, like the rnlA2 mutation, disruption of rnlA completely restored the growth of a T4 dmd mutant.
To clarify that rnlA disruption eliminates RNase LS activity, the mRNA of T4 late-gene soc was examined by Northern blot and primer-extension analyses. The half-life of soc mRNA was 2.2 min when the T4 dmd mutant infected wild-type cells, while it was 40 min in rnlA-disrupted cells infected with the same phage (data not shown). Primer-extension analysis was performed using total RNA from T4-infected cells at 21 min after infection (Figure 2). The cleavages of soc mRNA by RNase LS were represented by cDNA bands indicated by arrowheads in Figure 2 (compare lanes 1 and 2). These cleavage sites are 3′ of nucleotide positions 135, 144, 145, 153, 172, 185, and 207, respectively (Kai and Yonesaki 2002). Although the cleavage at 135 was previously uncharacterized, we sometimes observed this cleavage associated with a dmd mutant in repeated experiments. The cleavage at 153 was previously suggested to be translation termination dependent because it was particularly frequent when a premature termination codon was placed just upstream of this cleavage site (Kai and Yonesaki 2002). However, in later experiments, we noted that this cleavage was also detectable, although weakly, even when the premature termination codon was absent or when translation was blocked (Figure 2; data not shown). Therefore, we marked all of these cleavages as dmd mutant-specific cleavages. As expected, these cleavages were not detected with RNA from T4 dmd mutant-infected TY0324 cells (lane 4). This result clearly indicated that disruption of rnlA entirely eliminated RNase LS activity. Putting all these results together, we conclude that rnlA is essential for RNase LS activity in vivo.
Figure 2.—
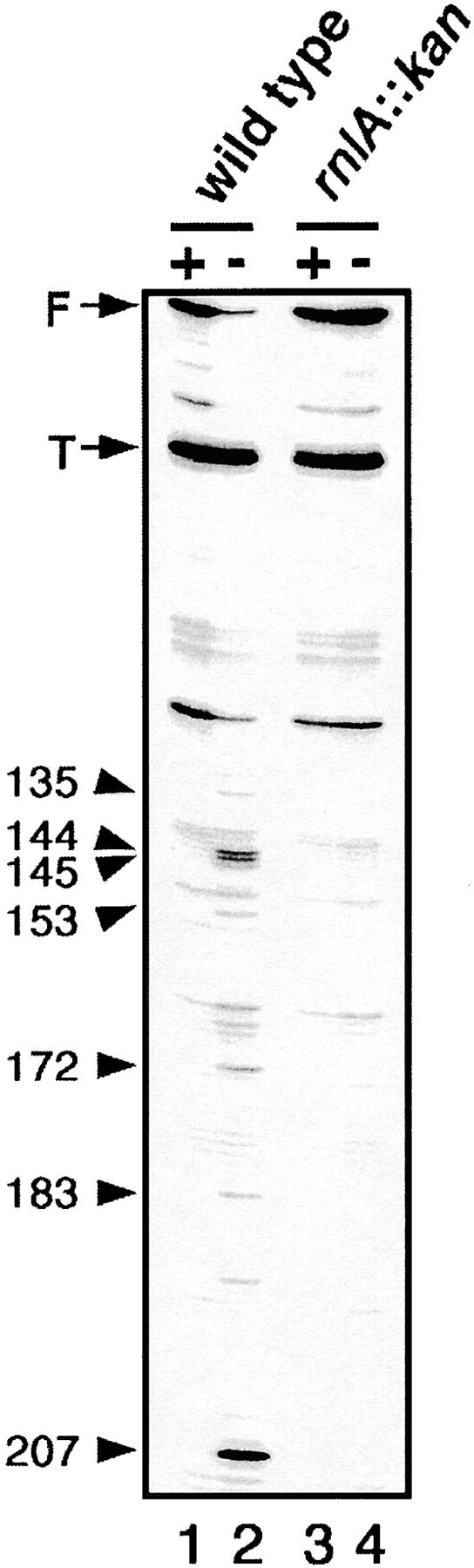
Effects of rnlA on dmd mutant-specific cleavages. MH1 and TY0324 cells were grown to 4 × 108 cells/ml and infected with T4 wild-type (+) or dmd mutant (−) phage. Total RNAs were extracted at 21 min after infection and used as templates for primer extension as described in materials and methods. Band F corresponds to full-length soc RNA. Band T corresponds to an RNase E-cleaved product (Otsuka et al. 2003). Arrowheads marked with the position of nucleotide 3′ of the cleavage site indicate bands corresponding to the dmd mutant-specific cleavages (see detail in text).
Detection of rnlA-dependent endoribonuclease activity in a cell extract:
To investigate the requirement of RnlA for RNase LS activity at the molecular level, we attempted to detect the activity of RNase LS in vitro. For this purpose, we prepared cell extract and incubated it with soc RNA synthesized by T7 RNA polymerase. After the incubation, RNAs were extracted and utilized for primer-extension analysis of soc RNA. Using the S30 fraction of the wild-type cell extract, we detected cDNA bands corresponding to soc RNA cleaved at various sites (Figure 3). Some of the cDNA bands, marked with the position of nucleotide 3′ of the cleavage site, 135, 144, 145, 153, and 207, were identical to those specific to the in vivo cleavages by RNase LS (lane 1 in Figure 3 and refer to Figure 2; note that the bands corresponding to the cleavages at 144 and 145 were between two close bands). As expected, all of these bands were not detected when the S30 fraction of the rnlA2 cell extract was used instead (lane 2 in Figure 3). This result suggests that these cleavages are caused by RNase LS and that RnlA is required for the RNase LS activity in vitro.
Figure 3.—
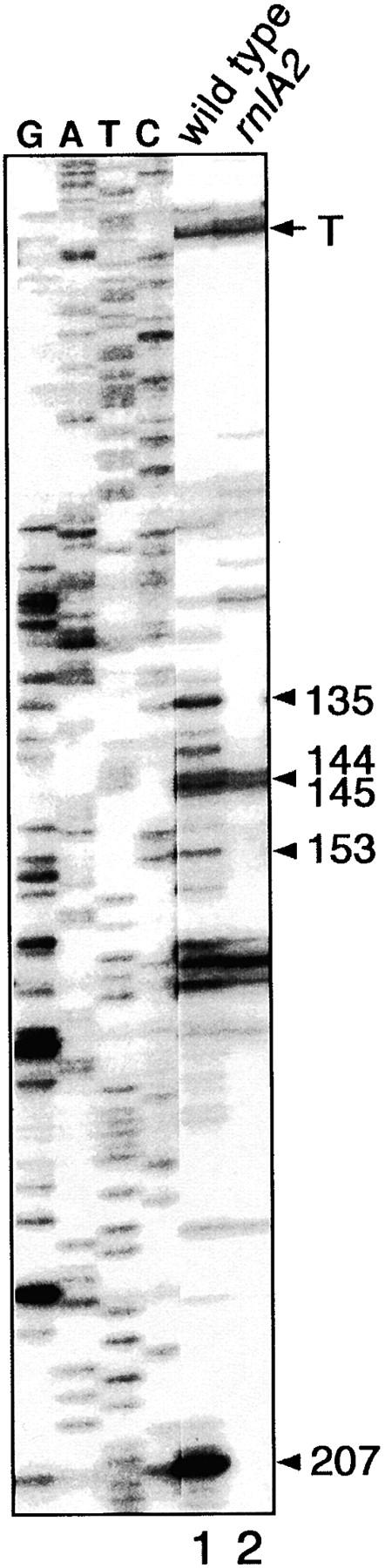
In vitro cleavage of soc RNA. After soc RNA was cleaved in the presence of the S30 fraction of MH1 or TY0482 cell extract, the RNA was used as a template for primer extension as described in materials and methods. A set of sequence ladders for wild-type soc obtained by the dideoxy-sequencing method were presented. Bands marked with the position of nucleotide 3′ of the cleavage site correspond to RNase LS-cleaved products (see detail in text), while band T corresponds to an RNase E-cleaved product (Otsuka et al. 2003).
The RNase LS cleavages at 172 and 185 detected in vivo for soc RNA (Figure 2) were not detected in this experiment. The failure of detecting these cleavages seemed rather reasonable, because the cleavages at 172 and 185 depend on translation (Kai and Yonesaki 2002) and the factors required for translation likely were not sufficiently active under the present experimental conditions.
Stabilization of transcripts by the rnlA2 mutation:
Our experiments using T4 phage were performed with actively dividing E. coli cells. RNase LS degrades T4 mRNAs even though T4 infection shuts off the expression of host genes within a few minutes. Accordingly, it is probable that RNase LS is constitutively expressed in growing cells. To investigate the function of RNase LS in E. coli cells, we examined the phenotype of the rnlA2 mutant during growth. The rnlA2 cells formed slightly smaller colonies than the wild-type control on a minimal agar plate at 30°, although no such difference was detected on a rich medium plate or on a minimal agar plate at 37° or 42° (data not shown). Therefore, a mutation in rnlA hardly affects cell growth.
To investigate the role of RNase LS in mRNA degradation, we examined the effect of the rnlA2 mutation on functional decay rates of mRNAs. The functional decay rate is estimated from the ability of mRNA to direct protein synthesis after transcription has been blocked. Rifampicin was added to a log-phase culture of bacteria and proteins newly synthesized at various times in wild-type or rnlA2 cells were pulse labeled with [35S]methionine/cysteine and analyzed with SDS polyacrylamide gel electrophoresis (Figure 4). Overall, the synthesis of most proteins was significantly lowered by 7 min after adding rifampicin to wild-type cells, while it was still prominent at 7 min and lowered by 9 min in rnlA2 cells. Indeed, the functional half-life of net mRNA was 5 min in wild-type cells vs. 6.5 min in the rnlA2 cells in one experiment and 6 min vs. 8 min in another experiment (data not shown). Comparison of each protein band between the wild type and the mutant revealed that the half-lives of some proteins were not changed, as indicated by the arrows in Figure 4. In contrast, the half-lives of others were remarkably extended (more than twofold) by the rnlA2 mutation, as shown by the arrowheads.
Figure 4.—
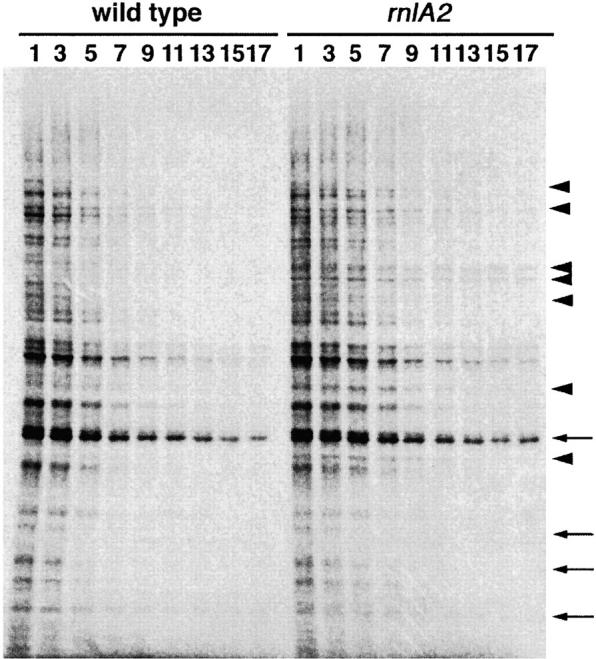
Functional decay of E. coli mRNAs. Rifampicin was added to 500 μg/ml to MH1 or TY0482 cells grown to 4 × 108 cells/ml. Newly synthesized proteins were pulse labeled for 1 min with [35S]methionine/cysteine at various times as indicated in the figure and analyzed by 12.5% polyacrylamide gel electrophoresis as described in materials and methods. Arrowheads indicate the proteins, whose mRNAs functionally decayed at a lower rate in TY0482 than in MH1. Arrows indicate the proteins, whose mRNAs functionally decayed at a similar rate in two E. coli strains.
Chemical decay of individual transcripts was also examined. Tested transcripts were from four E. coli genes (ompA, rpsA, rpsO, and bla) and the T4 soc gene. Because bla was expressed at a very low level, its stability was examined with a transcript from pBRbla. This plasmid has a single promoter for bla, while the chromosomal bla is transcribed from two different promoters (Deana et al. 1996). For the analysis of the T4 soc transcript, we used the plasmid pTK44 carrying the whole soc gene, including its cognate promoter. The soc gene could be transcribed from pTK44 in uninfected cells because an E. coli promoter-like sequence overlaps the soc promoter (Otsuka et al. 2003). To determine the stability of each transcript, rifampicin was added to bacterial cell cultures and total RNAs were extracted at various times to analyze the decay rate of each transcript by Northern blotting (Figure 5). For the ompA and rpsA transcripts, there was virtually no difference in decay rates between wild-type and rnlA2 cells (t1/2 = 13 and 5 min for ompA and rpsA, respectively). For the rpsO and soc transcripts, a modest decrease in the decay rate was observed for rnlA2 cells. The half-life (± the standard deviation) for rpsO was 1.9 ± 0.4 min in the wild-type cells and 3.8 ± 0.4 min in the rnlA2 cells. As for soc, it was 11 ± 1.4 min in the wild-type cells and 17 ± 2.8 min in the rnlA2 cells. The stability of the bla transcript was increased more in rnlA2 mutant cells (t1/2 = 4.9 ± 1.4 min) compared with wild-type cells (t1/2 = 1.5 ± 0.3 min). Thus, consistent with the result of functional decay, the degree of stabilization by the rnlA2 mutation was different with individual mRNAs.
Figure 5.—
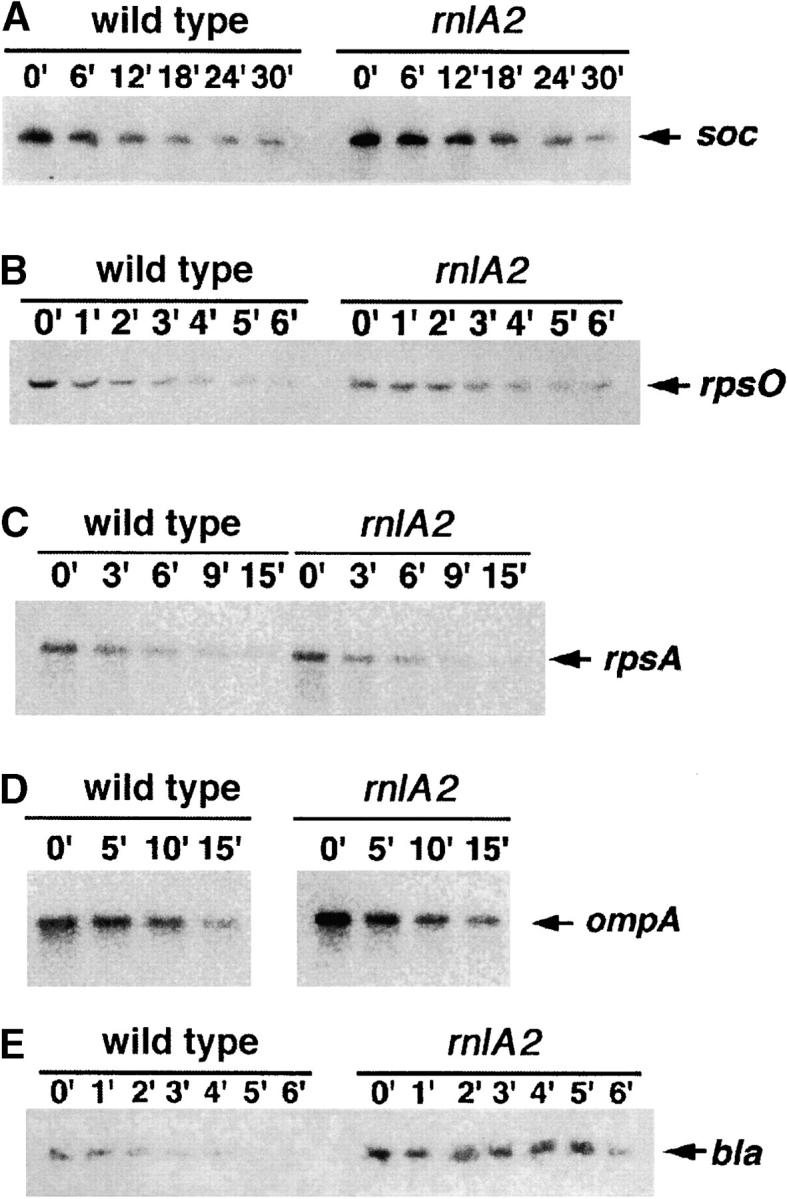
Northern blots of specific transcripts. Rifampicin was added to 500 μg/ml to MH1 or TY0482 cells grown to 4 × 108 cells/ml. Total RNAs were extracted at the indicated times and 5 μg of each total RNA were analyzed by Northern blot assay with probes for specific E. coli or T4 transcripts. The arrow indicates a transcript of each gene.
Accumulation of a fragment with an internal sequence of 23S rRNA:
During the course of Northern blot analyses, we found a unique property associated with the rnlA2 mutation. An extra band of RNA species was reproducibly observed with total RNAs from rnlA2 cells after polyacrylamide gel electrophoresis and ethidium bromide staining. This species was not visible with total RNAs from wild-type cells. The accumulation of this RNA was also observed with rnlA5, std-3, and std-4 mutations (Figure 6A). Because these mutations were isolated as RNase LS hypomorphs (Otsuka et al. 2003), the accumulation of specific RNA could be attributable to reduced RNase LS activity.
Figure 6.—
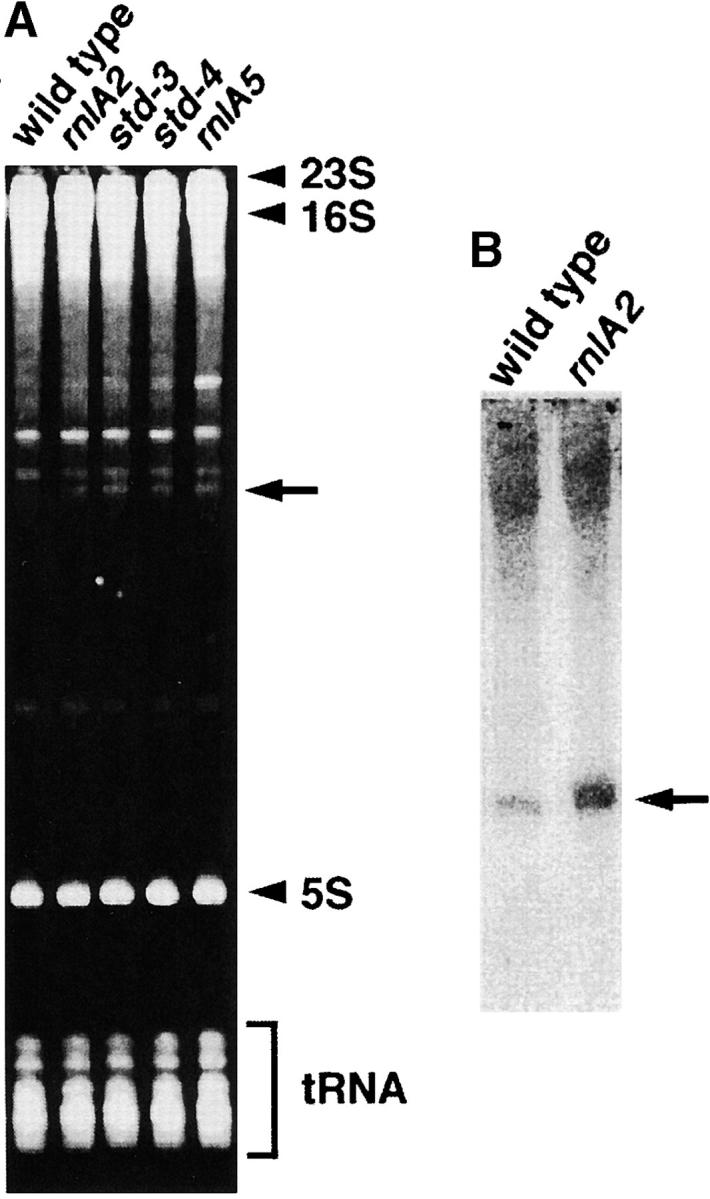
Accumulation of an internal fragment of 23S rRNA. (A) Ten micrograms of each total RNA extracted from MH1, TY0482, TY1722, TY1797, and TY2133 cells was electrophoresed through a 5% polyacrylamide gel containing 7.5 m urea and the gel was stained with ethidium bromide. Bands corresponding to 23S rRNA, 16S rRNA, 5S rRNA, and tRNAs are indicated. The arrow indicates the 307-nt RNA specific to the std mutants. (B) Northern blotting of the 23S-rRNA fragment. Total RNAs were extracted from MH1 or TY0482 cells grown to midlog phase and 5 μg of each total RNA was analyzed by Northern blotting with a probe complementary to the 307-nt fragment.
To characterize this RNA, it was purified and used as a probe to screen the E. coli K-12 W3110 cosmid library (see materials and methods). One positive clone, E3107, was identified. Because E3107 carries a sequence of ∼30 kbp, the region corresponding to the RNA was narrowed and subcloned. The subcloned fragment in plasmid pBS109BN was ∼2.4 kbp long and subsequent sequencing revealed that this fragment contained the sequence of an internal region of 23S ribosomal RNA (data not shown). To establish a fine structure of the RNA species, its 5′ and 3′ termini were ligated by T4 RNA ligase and a region containing the joint was amplified by RT-PCR. Then the nucleotide sequences at the 5′ and 3′ termini were deduced by sequencing the cDNA. This RNA was found to be the 307 nt corresponding to nt 1304–1610 of 23S rRNA. The accumulation of this RNA in rnlA2 mutant cells was further confirmed by Northern blotting (Figure 6B). The result showed that the short RNA was detected in wild-type cells as well as in rnlA2 cells and that the accumulation was fivefold higher in rnlA2 cells than in wild-type cells.
DISCUSSION
The present study demonstrates that rnlA is essential for RNase LS activity in vitro as well as in vivo (Figures 2 and 3). On the basis of the deduced amino acid sequence, rnlA encodes a 40-kD protein. By computer search, RnlA was found to have weak homology with proteins encoded by plasmids residing in E. coli O157 and Salmonella typhimurium. No significant homolog other than E. coli K-12 was found among proteins encoded by chromosomal genes in prokaryotes. In addition, RnlA appears to lack a known RNA-binding motif. As described elsewhere, we found that upon fractionation of the RNase LS activity in cell extract, at least two different fractions, one of which contained RnlA, were required for the activity. In addition, RnlA involved in the fraction sedimented as a large complex through a sucrose gradient. Accordingly, RNase LS may consist of multiple components for its function. Identification of rnlA would give a clue to identifying other components of RNase LS.
In E. coli cells, the initiation of mRNA decay is triggered by an endonucleolytic cleavage (Apirion 1973; Coburn and Mackie 1999; Kushner 2002). Among previously identified endonucleases, only RNase E is known to affect the stability of many mRNAs during growth (Ono and Kuwano 1979; Lopez et al. 1999; Kushner 2002). However, even in cells deficient in RNase E, some mRNAs (such as trxA mRNA) are still degraded rapidly (Ow et al. 2000). The rnlA2 mutation, which completely eliminates RNase LS activity (Otsuka et al. 2003), stabilizes numerous mRNAs modestly and a subset of mRNAs considerably in growing E. coli cells (Figures 4 and 5). Thus, RNase E and RNase LS may have different preferences for different mRNAs and may cooperate in initiating mRNA degradation. The discovery of RNase LS provides a clue to understanding the mechanism for initiating mRNA decay in E. coli.
bla mRNA was significantly stabilized by the rnlA2 mutation, while ompA and rpsA mRNAs were not (Figure 5). Because these mRNAs were almost the same length, other sequence features must be responsible for the diverse stabilization effects of rnlA2. The frequency of rare codons in rpsA and ompA mRNAs is ∼5% of total codons, whereas bla mRNA contains ∼28% of rare codons. Indeed, frequency use of rare codons is reported to affect the stability of mRNAs (Deana et al. 1996). The replacement of seven consecutive frequently used codons with synonymous infrequently used codons reduced the stability of ompA mRNA. On the other hand, the exchange of 24 rare codons for most frequently used synonymous codons extended bla mRNA longevity (Deana et al. 1996). If the differential effects of rnlA2 on mRNA stability reflect the frequency of rare codons, then RNase LS would preferably cleave mRNAs containing rare codons. This idea may be consistent with the previous notion that RNase LS can cleave mRNA at sites when a region containing cleavage sites is translatable, suggesting intimate linkage of cleavages with peptide chain elongation (Kai and Yonesaki 2002). Rare codons lead to a decrease in the rate of peptide chain elongation, which may stimulate cleavage by RNase LS.
RNase LS rapidly degrades the T4 soc transcript (t1/2 = 2–3 min) when a dmd mutant infects E. coli (Otsuka et al. 2003; data not shown). However, the half-life of the soc transcript was 11 min in uninfected cells and increased only 1.5-fold in rnlA2 cells (t1/2 = 17 min; Figure 5). Therefore, the contribution of RNase LS to soc mRNA degradation is much lower in uninfected cells than in T4 dmd mutant-infected cells. This fact adds another line of evidence in support of the notion that RNase LS activity is activated after middle stages of T4 infection (Kai et al. 1998; Ueno and Yonesaki 2001).
The 307-nt fragment derived from 23S rRNA accumulated in rnlA2 cells to a level visible with ethidium bromide staining after electrophoresis of total RNAs (Figure 6). This result indicates that RNase LS plays a role in the metabolism of this small RNA. Recently, Cheng and Deutscher (2003) described quality control of rRNA by cooperation between polynucleotide phosphorylase and RNase R to abolish rRNA fragments presumably produced by misprocessing; when both RNases are deficient, rRNA fragments accumulate. RNase LS might also function in the quality control of rRNA. However, the 307-nt fragment was also detected by Northern blotting in wild-type cells. Therefore, this RNA might not be refuse and may have its own function. It is worthy of note that the short RNA contains a putative Shine-Dalgarno (SD) sequence and an open reading frame encoding a polypeptide of 27 amino acids (Figure 7). Because the SD-like sequence is properly located upstream of the initiation codon, this RNA could encode the polypeptide. It may not be surprising that ribosomal RNA functions as an mRNA because a functional peptide comprising five amino acids may be encoded in the 23S rRNA, overexpression of the pentapeptide rendering cells resistant to erythromycin (Tenson et al. 1996).
Figure 7.—
Partial 23S rRNA nucleotide sequence. The sequence is from nt 1424 to 1525 of E. coli 23S rRNA. The putative SD sequence, initiation codon (M), and two termination codons (*) are underlined. Amino acids deduced from codons are also shown below the nucleotide sequence.
Acknowledgments
We cordially thank John W. Drake at the National Institute of Environmental Health Sciences for invaluable help with the manuscript. We are grateful to Mary Berlyn at E. coli Genetic Stock Center for her professional advice on naming the E. coli gene. We thank the staff of the Radioisotope Research Center at Toyonaka, Osaka University, for facilitating our research because all of our experiments using radioisotopes were carried out there. This work was supported in part by a grant from the program Grants-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Aizenman, E., H. Engelberg-Kulka and G. Glaser, 1996. An Escherichia coli chromosomal “addiction module” regulated by 3′, 5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93: 6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifano, P., F. Rivellini, C. Piscitelli, C. M. Arraiano, C. B. Bruni et al., 1994. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 8: 3021–3031. [DOI] [PubMed] [Google Scholar]
- Apirion, D., 1973. Degradation of RNA in Escherichia coli. A hypothesis. Mol. Gen. Genet. 122: 313–322. [DOI] [PubMed] [Google Scholar]
- Arraiano, C. M., S. D. Yancey and S. R. Kushner, 1988. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 170: 4625–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell, J. C. A., P. Regnier, S. M. Chen, Y. Nakamura, M. Grunberg-Manago et al., 1989. Autoregulation of RNase III operon by mRNA processing. EMBO J. 8: 3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro, V. J., and D. Kennell, 1991. RNase I*, a form of RNase I, and mRNA degradation in Escherichia coli. J. Bacteriol. 173: 4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z. F., and M. P. Deutscher, 2003. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. USA 100: 6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn, G. A., and G. A. Mackie, 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62: 55–108. [DOI] [PubMed] [Google Scholar]
- Deana, A., R. Ehrlich and C. Reiss, 1996. Synonymous codon selection controls in vivo turnover and amount for mRNA in Escherichia coli bla and ompA genes. J. Bacteriol. 178: 2718–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, T., and T. Yonesaki, 2002. Multiple mechanisms for degradation of bacteriophage T4 soc mRNA. Genetics 160: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, T., E. H. Selick and T. Yonesaki, 1996. Destabilization of bacteriophage T4 mRNAs by a mutation of gene 61.5. Genetics 144: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, T., H. Ueno and T. Yonesaki, 1998. Involvement of other bacteriophage T4 genes in the blockade of protein synthesis and mRNA destabilization by a mutation of gene 61.5. Virology 248: 148–155. [DOI] [PubMed] [Google Scholar]
- Kushner, S. R., 2002. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184: 4658–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, A. J., D. Phillips and G. M. Church, 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179: 6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, P. J., I. Marchand, S. A. Joyce and M. Dreyfus, 1999. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33: 188–199. [DOI] [PubMed] [Google Scholar]
- Mackie, G. A., 1991. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J. Bacteriol. 173: 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, M., and M. Kuwano, 1979. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 129: 343–357. [DOI] [PubMed] [Google Scholar]
- Otsuka, Y., H. Ueno and T. Yonesaki, 2003. Escherichia coli endoribonucleases involved in the cleavage of bacteriophage T4 mRNAs. J. Bacteriol. 185: 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow, M. C., Q. Liu and S. R. Kushner, 2000. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 38: 854–866. [DOI] [PubMed] [Google Scholar]
- Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes et al., 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112: 131–140. [DOI] [PubMed] [Google Scholar]
- Portier, C., L. Dondon, M. Grunberg-Manago and P. Regnier, 1987. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is ribonuclease III processing at the 5′ end. EMBO J. 6: 2165–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg, D. R., and E. Chernokalskaya, 1997 Ribonucleases involved in eukaryotic mRNA turnover, pp. 217–240 in mRNA Metabolism and Post-transcriptional Gene Regulation, edited by J. B. Harford and D. R. Morris. Wiley-Liss, New York.
- Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka et al., 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98: 14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D. C., and R. Parker, 2000 Interaction of mRNA translation and mRNA degradation in Saccharomyces cerevisiae, pp. 807–825 in Translational Control of Gene Expression, edited by N. Sonenberg, J. W. B. Hershey and M. B. Mathews. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Tabata, S., A. Higashitani, M. Takanami, K. Akiyama, Y. Kohara et al., 1989. Construction of an ordered cosmid collection of the Escherichia coli K-12 W3110 chromosome. J. Bacteriol. 171: 1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson, T., A. Deblasio and A. Mankin, 1996. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc. Natl. Acad. Sci. USA 93: 5641–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, H., and T. Yonesaki, 2001. Recognition and specific degradation of bacteriophage T4 mRNAs. Genetics 158: 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umitsuki, G., M. Wachi, A. Takada, T. Hikichi and K. Nagai, 2001. Involvement of RNase G in in vivo mRNA metabolism in Escherichia coli. Genes Cells 6: 403–410. [DOI] [PubMed] [Google Scholar]
- Van Hoof, A., and P. J. Green, 1997 Control of mRNA decay in plants, pp. 201–216 in mRNA Metabolism and Post-transcriptional Gene Regulation, edited by J. B. Harford and D. R. Morris. Wiley-Liss, New York.
- Yonesaki, T., 2002. Scarce adenylation in bacteriophage T4 mRNAs. Genes Genet. Syst. 77: 219–225. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing et al., 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12: 913–923. [DOI] [PubMed] [Google Scholar]