Abstract
The invasive and filamentous growth forms of Saccharomyces cerevisiae are adaptations to specific environmental conditions, under particular conditions of limited nutrient availability. Both growth forms are dependent on the expression of the FLO11 gene, which encodes a cell-wall-associated glycoprotein involved in cellular adhesion. A complex regulatory network consisting of signaling pathways and transcription factors has been associated with the regulation of FLO11. Mss11p has been identified as a transcriptional activator of this gene, and here we present an extensive genetic analysis to identify functional relationships between Mss11p and other FLO11 regulators. The data show that Mss11p is absolutely required for the activation of FLO11 by most proteins that have previously been shown to affect FLO11 expression, including the signaling proteins Ras2p, Kss1p, and Tpk2p, the activators Tec1p, Flo8p, and Phd1p, and the repressors Nrg1p, Nrg2p, Sok2p, and Sfl1p. The genetic evidence furthermore suggests that Mss11p activity is not dependent on the presence of any of the above-mentioned factors and that the protein also regulates other genes involved in cellular adhesion phenotypes. Taken together, the data strongly suggest a central role for Mss11p in the regulatory network controlling FLO11 expression, invasive growth, and pseudohyphal differentiation.
IN Saccharomyces cerevisiae, the choice of a specific cellular growth form is frequently governed by nutrient availability. Nutrient-rich environments generally favor yeast-type unicellular multiplication, while nutrient-limited environments tend to support pseudohyphal and/or invasive growth. Nutrient depletion, on the other hand, leads to a different set of adaptations, either entry into the G0 phase of the cell cycle or meiosis and spore formation, depending on the cell type and on the nutritional composition of the growth substrate. The transition from unicellular to invasive and/or pseudohyphal growth is characterized by important morphological and physiological changes and referred to as a dimorphic switch or as pseudohyphal differentiation. This switch can be triggered by nitrogen source or amino acid limitation (Gimeno et al. 1992; Lambrechts et al. 1996a; Braus et al. 2003). It may also occur in response to carbon limitation, growth on alcohols (Cullen and Sprague 2000; Lorenz et al. 2000), or growth on other inefficiently used carbon sources, including starch (Lambrechts et al. 1996a). Invasive and pseudohyphal growth is characterized by directional unipolar budding and by cells that remain attached to each other after budding (Gimeno et al. 1992). Haploid cells growing as filaments display stronger adhesiveness than diploid cells and the filaments of haploid cells penetrate solid growth substrates more efficiently (Roberts and Fink 1994).
Complex regulatory networks govern the conversion of environmental signals into specific developmental outcomes. The nutrient-responsive regulatory network that controls haploid invasive growth consists of several signal transduction modules, including the nutrient-responsive mitogen-activated protein kinase (MAPK) cascade and the cyclic AMP-dependent protein kinase A (PKA) pathway (reviewed in Lengeler et al. 2000; Gancedo 2001; Gagiano et al. 2002). The MAP kinases Kss1p and Fus3p and the cAMP-dependent kinase Tpk1-3p activate or inactivate various transcription factors, which in turn control the expression of genes that are responsible for the cellular differentiation process (Cook et al. 1997; Madhani et al. 1997; Bardwell et al. 1998; Robertson and Fink 1998; Pan and Heitman 1999; Breitkreutz et al. 2003). Other signaling elements or regulators that have been associated with pseudohyphal differentiation and invasive growth include cell cycle regulators, in particular the G1 cyclins (Oehlen and Cross 1998; Loeb et al. 1999; Ahn et al. 2001), the amino-acid-specific response pathway (Braus et al. 2003), and the meiosis-specific regulator Rme1p (van Dyk et al. 2003).
FLO11/MUC1 and STA2 are two of the genes controlled by signaling pathways activated in response to the nutrient status of the growth substrate (Gagiano et al. 1999a,b; Rupp et al. 1999). FLO11 encodes a glycosyl-phosphatidylinositol-anchored cell surface protein, which is a member of the flocculin family, a group of proteins involved in cell-cell adhesion (Caro et al. 1997; Guo et al. 2000; Verstrepen et al. 2003). Apart from its previously reported role in flocculation (Lo and Dranginis 1996), the Flo11p glycoprotein was also shown to be required for invasive growth and pseudohyphal development (Lambrechts et al. 1996a; Lo and Dranginis 1998), cell-substrate adhesion (Guo et al. 2000), and biofilm formation (Reynolds and Fink 2001). The STA2 gene encodes a secreted α-glucoamylase, which enables yeast to degrade starch and to grow on starch-containing medium (Lambrechts et al. 1996a; Gagiano et al. 1999b). The promoters of FLO11 and STA2 are almost identical and as a consequence these genes are coregulated by a similar set of transcription factors (Gagiano et al. 1999a,b, 2003; van Dyk et al. 2003).
A large number of factors with complex functional relationships were shown to act on the ∼3-kb promoter of FLO11, one of the largest promoters identified in S. cerevisiae (reviewed in Gagiano et al. 2002; Palecek et al. 2002). On the basis of their roles, the regulators can be divided into different subgroups, which, apart from the general transcription machinery (RNA polymerase II and associated subcomplexes), include factors that act downstream of the cAMP and MAP kinase signaling modules. While these factors appear to be generally required for invasive growth, other factors appear to act only in response to the limitation of specific nutrients. Furthermore, FLO11 regulation has been linked to several chromatin-remodeling proteins, whose association with the above-mentioned signaling network has not yet been clearly established.
Signaling pathway-responsive regulators include the MAPK-controlled proteins Ste12p and Tec1p, which constitute a heterodimeric transcriptional activator that regulates genes involved in filamentous growth, including FLO11 (Madhani and Fink 1997; Rupp et al. 1999; Köhler et al. 2002; Zeitlinger et al. 2003). The cAMP-dependent PKA pathway regulates the activities of the Flo8p transcriptional activator and the Sfl1p transcriptional repressor (Liu et al. 1996; Robertson and Fink 1998; Pan and Heitman 1999, 2002). A 250-nucleotide stretch has been identified in the FLO11 promoter that is bound by both Flo8p and Sfl1p (Pan and Heitman 2002).
Nutrient-specific transcription factors include Gcn4p, Nrg1p, and Nrg2p. In response to amino acid starvation, Gcn4p upregulates FLO11 transcription, but does not appear to bind the FLO11 promoter (Braus et al. 2003). Snf1p, a highly conserved protein kinase required for the derepression of genes subject to glucose-repression (reviewed in Carlson 1999), activates FLO11 transcription by antagonizing the two repressors Nrg1p and Nrg2p (Kuchin et al. 2002).
Msn1p and Rme1p, two putative chromatin-remodeling factors (Covitz et al. 1994; Sidorova and Breeden 1999), were previously shown to activate FLO11 transcription when expressed from multi-copy plasmids (Gagiano et al. 1999a,b; van Dyk et al. 2003). MSN1 encodes a protein involved in the regulation of several genes, including HO and STA2 (Estruch and Carlson 1990; Lambrechts et al. 1996b; Sidorova and Breeden 1999). The regulator of meiosis, Rme1p, acts as a repressor of the early meiosis gene IME1 (Kassir et al. 1988; Covitz and Mitchell 1993) and as an activator of the G1-cyclin gene CLN2 (Toone et al. 1995) and of STA2 and FLO11 (van Dyk et al. 2003).
Finally, the activator-encoding gene PHD1 (Gimeno and Fink 1994) and the associated repressor Sok2p (Ward et al. 1995) are also involved in the regulation of FLO11 transcription. While their mode of action has not been established, activation by Phd1p was shown to be dependent on the presence of FLO8 (Pan and Heitman 2000).
Mss11p has previously been shown to play a role in invasive growth and starch degradation (Webber et al. 1997; Gagiano et al. 1999b, 2003). The data indicated that the protein is able to regulate the transcription of FLO11 and STA2 directly in response to nutritional signals (Gagiano et al. 2003). However, its relationship with other FLO11 regulators and its position within the FLO11 regulatory network have not been elucidated. Here we report on the genetic interactions between MSS11 and all of the genes encoding the aforementioned factors. The results indicate that the pronounced decrease in FLO11 expression observed in mss11Δ strains cannot be suppressed by most of the genes important for FLO11 regulation. The data show that the presence of MSS11 is absolutely required for the activation of FLO11 by hyperactive alleles or multiple copies of RAS2, KSS1, TPK2, TEC1, FLO8, and PHD1, as well as for derepression of the gene in strains deleted for SFL1, SOK2, NRG1, and NRG2. In our analysis, the only factors that appear to not depend on Mss11p are Msnp1 and Rme1p, which have been associated with chromatin-remodeling functions. Our data strongly suggest an essential and central role for Mss11p in the transcriptional regulation of FLO11 and that the protein acts in the very center of the regulatory network that specifically governs FLO11 transcription.
MATERIALS AND METHODS
Strains, media, and recombinant DNA techniques:
The yeast strains used in this study are listed in Table 1. Standard YEPD medium was used to cultivate yeast strains prior to transformation. Plasmids and disruption cassettes were introduced using the lithium acetate method described by Ausubel et al. (1994). Strains were cultivated at 30° and synthetic dropout medium containing 2% glucose, 0.67% yeast nitrogen base (Difco Laboratories, Detroit), and essential amino acids was used to propagate transformants (Sherman et al. 1991). Geneticin-resistant transformants were selected on YPED medium supplemented with 200 mg/liter geneticin (Sigma-Aldrich, St. Louis). Invasive growth was assessed on 0.2% glucose medium, and 2% potato starch (Sigma-Aldrich) was used as carbon source for starch (SCS) plates. SLAD (synthetic low ammonium dextrose) medium contained 2% glucose and SCGE (synthetic complete glycerol ethanol) medium was prepared using 3% glycerol and 3% ethanol. β-Galactosidase assays were performed on strains pregrown in liquid medium containing 2% glucose. All media were supplemented with 0.67% yeast nitrogen base (YNB) containing preadded ammonium sulfate (Difco), except for the SLAD medium, which contains 0.17% YNB to which 50 μm ammonium sulfate was separately added. In all cases 2% agar (Difco) was used as the solidifying agent for the plate media.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| YHUM272a | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG | H.-U. Mösch |
| ∑1278bflo8Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo8Δ::LEU2 | This study |
| ∑1278bkss1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG kss1Δ::kanMX4 | This study |
| ∑1278bmsn1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG msn1Δ::URA3 | This study |
| ∑1278bmss11Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG mss11Δ::LEU2 | This study |
| ∑1278bphd1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG phd1Δ::LEU2 | This study |
| ∑1278bsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG sfl1Δ::kanMX4 | This study |
| ∑1278bste12Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG ste12Δ::URA3 | This study |
| ∑1278btec1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG tec1Δ::LEU2 | This study |
| ∑1278btpk2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG tpk2Δ::kanMX4 | This study |
| ∑1278bflo8Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo8Δ::LEU2 sfl1Δ::kanMX4 | This study |
| ∑1278bmsn1Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG msn1Δ::URA3 sfl1Δ::kanMX4 | This study |
| ∑1278bmss11Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG mss11Δ::LEU2 sfl1Δ::kanMX4 | This study |
| ∑1278bste12Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG ste12Δ::URA3 sfl1Δ::kanMX4 | This study |
| ∑1278btec1Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG tec1Δ::LEU2 sfl1Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ flo8Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 flo8Δ::LEU2 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ gpa2Δras2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 gpa2Δ::LEU2 ras2Δ::kanMX4 | This study |
| ∑1278bflo11Δ:: lacZ kss1Δ |
MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 kss1Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ msn1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 msn1Δ::URA3 | van Dyk et al. (2003) |
| ∑1278bflo11Δ:: lacZ mss11Δ |
MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 mss11Δ::LEU2 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ nrg1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 nrg1Δ::kanMX4 | van Dyk et al. (2003) |
| ∑1278bflo11Δ:: lacZ nrg2Δ |
MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 nrg2Δ::kanMX4 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ phd1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 phd1Δ::LEU2 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ ras2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 ras2Δ::LEU2 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ sfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 sfl1Δ::kanMX4 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ sok2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 sok2Δ::kanMX4 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ Δste12 |
MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 ste12Δ::URA3 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ tec1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 tec1Δ::LEU2 | van Dyk et al. (2003) |
| ∑1278bflo11Δ::lacZ tpk2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 tpk2Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ flo8Δnrg1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 flo8Δ::LEU2 nrg1Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ flo8Δnrg2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 flo8Δ::LEU2 nrg2Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ flo8Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 flo8Δ::LEU2 sfl1Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ flo8Δsok2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 flo8Δ::LEU2 sok2Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ mss11Δnrg1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 mss11Δ::LEU2 nrg1Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ mss11Δnrg2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 mss11Δ::LEU2 nrg2Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ mss11Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 mss11Δ::LEU2 sfl1Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ mss11Δsok2Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 mss11Δ::LEU2 sok2Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ msn1Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 msn1Δ::URA3 sfl1Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ ste12Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 ste12Δ::URA3 sfl1Δ::kanMX4 | This study |
| ∑1278bflo11Δ::lacZ tec1Δsfl1Δ | MATα ura3-52 trp1Δ::hisG leu2Δ::hisG his3Δ::hisG flo11Δ::lacZ-HIS3 tec1Δ::LEU2 sfl1Δ::kanMX4 | This study |
| ISP15 | MATahis3 leu2 thr1 trp1 ura3 STA2 | This laboratory |
| ISP15msn1Δ ste12Δtec1Δ |
MATahis3 leu2 thr1 trp1 ura3 STA2 msn1Δ::ura3Δ::kanMX4 ste12Δ::URA3 tec1Δ::LEU2 | This study |
| ISP15flo8Δmsn1Δ ste12Δtec1Δ |
MATahis3 leu2 thr1 trp1 ura3 STA2 flo8Δ:: ura3Δ::loxP msn1Δ::URA3 ste12Δ::ura3Δ::kanMX4 tec1Δ::LEU2 | This study |
| BY4742b | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | EUROSCARF |
| BY4742kss1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 kss1Δ::kanMX4 | EUROSCARF |
| BY4742nrg1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nrg1Δ::kanMX4 | EUROSCARF |
| BY4742nrg2Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nrg2Δ::kanMX4 | EUROSCARF |
| BY4742ras2Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ras2Δ::kanMX4 | EUROSCARF |
| BY4742sfl1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sfl1Δ::kanMX4 | EUROSCARF |
| BY4742sok2Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sok1Δ::kanMX4 | EUROSCARF |
| BY4742tpk2Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tpk2Δ::kanMX4 | EUROSCARF |
YHUM272 (10560-6B) is from the ∑1278b background.
BY4742 is from the S288C genetic background (see Brachmann et al. 1998).
Escherichia coli strain DH5α was used for plasmid amplification (GIBCO BRL/Life Technologies, Rockville, MD). Plasmid-bearing bacterial strains were cultivated at 37° in Luria-Bertani broth. Standard bacterial transformations and plasmid isolation procedures were performed as described by Sambrook et al. (1989).
Plasmid construction and primers:
The constructs used in this study are listed in Table 2. KSS1 was excised from pXT1 (kindly provided by D. Engelberg, The Hebrew University of Jerusalem) as a 1647-bp EcoRI-SphI fragment and cloned into the corresponding sites of YEplac112 (Gietz and Sugino 1988) to generate YEplac112-KSS1. The TPK2 gene from the S288c genetic background was amplified with primers TPK2-Fp, 5′-ATATACGTACACACAATTCCATATCGAG-3′, and TPK2-Rp, 5′-GCAACGCTTGTTCTTCTCATCTCTT-3′. The resulting 1737-bp PCR product was cloned into pGEM-Teasy (Promega, Madison, WI), subsequently isolated as a SpeI-SnaBI fragment, and cloned into the XbaI-SmaI sites of YEplac112 to generate YEplac112-TPK2. The flo8Δ::LEU2 (pΔflo8-L) disruption cassette was constructed by replacing a 760-bp PstI-BglII fragment of YEplac112-FLO8 (Gagiano et al. 1999a) with a 2000-bp PstI-BamHI fragment containing the LEU2 marker of pJJ252 (Jones and Prakash 1990). MluNI and SnaBI restriction enzymes were used to remove the 4070-bp flo8Δ::leu2 disruption cassette from pΔflo8-L.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant genotype | Source or reference |
|---|---|---|
| YEplac112 | 2μ TRP1 | Gietz and Sugino (1988) |
| YEplac112-FLO8 | 2μ TRP1 FLO8 | Gagiano et al. (1999a) |
| YEplac112-KSS1 | 2μ TRP1 KSS1 | This study |
| YEplac112-MSN1 | 2μ TRP1 MSN1 | Gagiano et al. (1999b) |
| YEplac112-MSS11 | 2μ TRP1 MSS11 | Gagiano et al. (1999b) |
| YEplac112-PHD1 | 2μ TRP1 PHD1 | van Dyk et al. (2003) |
| YEplac112-STE12 | 2μ TRP1 STE12 | Gagiano et al. (1999b) |
| YEplac112-TEC1 | 2μ TRP1 TEC1 | van Dyk et al. (2003) |
| YEplac112-TPK2 | 2μ TRP1 TPK2 | This study |
| YCplac22-RAS2val19 | CEN4 TRP1 RAS2Val19 | Gagiano et al. (1999b) |
| pXT1a | 2μ LEU2 KSS1 | D. Engelberg |
| pGEM-Teasy | Promega | |
| pGEM-Teasy-TPK2 | PCR fragment containing TPK2 | This study |
| pUG6b | kanMXR | J. H. Hegemann |
| pSH47b | CEN6 URA3 | J. H. Hegemann |
| pΔura3::kan | ura3Δ::kanMX | Gagiano et al. (1999a) |
| pJJ252 | LEU2 | Jones and Prakash (1990) |
| pΔflo8-L | Flo8Δ::LEU2 | This study |
| pΔflo8 | Flo8Δ::URA3 | Gagiano et al. (1999a) |
| pΔgpa2 | gpa2Δ::LEU2 | van Dyk et al. (2003) |
| pΔmsn1 | msn1Δ::URA3 | Gagiano et al. (1999b) |
| pMSS11-Δ | mss11Δ::LEU2 | Webber et al. (1997) |
| pΔphd1 | phd1Δ::LEU2 | van Dyk et al. (2003) |
| pΔste12 | ste12Δ::URA3 | Gagiano et al. (1999b) |
| pΔtec1 | tec1Δ::LEU2 | van Dyk et al. (2003) |
pXT1 contains the original KSS1 isolate cloned in YEp13 (Courchesne et al. 1989).
Plasmids pUG6 and pSH47 are described in Güldener et al. (1996).
KSS1-Fp, 5′-GTACTTCCAATCTGTAGATATTGCACTTTATC-3′, and KSS1-Rp, 5′-CCGTTTAGGCAAAGCAGTGAAGA-3′, TPK2-Fp, and TPK2-Rp were used to PCR amplify kss1Δ::kanMX4 and tpk2Δ::kanMX4 from the genomic DNA of the corresponding BY4742 mutant strains.
Yeast strain construction:
Invasive growth and β-galactosidase assays were performed on strains derived from 1278b (10560-6B) (see Table 1 and van Dyk et al. 2003). The lacZ gene of ∑1278bflo11Δ::lacZ is under the control of the endogenous FLO11 promoter (van Dyk et al. 2003). The STA2-bearing strain ISP15 and isogenic mutants were used to assess starch degradation phenotypes (described in Gagiano et al. 1999b; van Dyk et al. 2003). The S288c-derived BY4742 strain collection (Brachmann et al. 1998) was purchased from the European Saccharomyces cerevisiae Archive for Functional Analysis (EUROSCARF). Disruption cassettes used to delete copies of FLO8, GPA2, MSN1, MSS11, NRG1, NRG2, PHD1, RAS2, SFL1, SOK2, STE12, and TEC1 in the ∑1278b wild-type and ∑1278bflo11Δ::lacZ reporter strains were obtained by means of PCR amplification using primers, genomic templates, and the disruption cassettes previously described in van Dyk et al. (2003). The genomic DNA of BY4742kss1Δ::kanMX4, BY4742ras2Δ::kanMX4, and BY4742tpk2Δ::kanMX4 (EUROSCARF) served as PCR templates for the amplification of the corresponding disruption cassettes.
To generate ISP15msn1Δste12Δtec1Δ and ISP15flo8Δmsn1Δste12Δtec1Δ, URA3 markers were recovered using the ura3Δ::kanMX disruption cassette of pΔura3::kan (Gagiano et al. 1999a). The kanMX gene was subsequently removed with the loxP-kanMX-loxP/Cre system as described by Güldener et al. (1996).
Invasive growth and starch degradation plate assays:
For the invasive growth, plate assay transformants were spotted onto 0.2% glucose medium and incubated for 5 days at 30°. Colonies were removed by vigorous rubbing under a constant stream of running water. The plates were subsequently allowed to dry on the bench before photographs were taken. Only cells of colonies that penetrated the growth medium were present after the washing procedure. The density of the residual cells observed under the surface of the growth medium reflects the ability of a strain to grow invasively.
Starch degradation was observed as transparent zones present around yeast colonies grown on solid starch-containing medium. The ability of strains to utilize starch was assessed by dropping 15 μl of synthetic complete dextrose (SCD) culture suspensions, grown to an optical density (OD600) of ∼1.0, onto starch (SCS) plates. The spotted cultures were incubated for 5 days at 30°. Starch precipitation was induced by adding 96% ethanol to the starch plates, after which the plates were incubated at 4°. Clear transparent zones around the colonies were observed within a few minutes of incubation.
β-Galactosidase assays:
The composition of the media, culture preparation, and growth conditions were previously described (van Dyk et al. 2003). Assays for β-galactosidase activity were performed exactly according to the instructions of Ausubel et al. (1994). At least three independent transformants were used for each experiment, and experiments were performed in triplicate. Every data point represents the average of three transformants with a standard deviation of <15%. Activity of β-galactosidase is expressed in Miller units (Ausubel et al. 1994).
RNA extraction and Northern analysis:
Test tubes containing 5 ml of SCD (2% glucose) selective media were inoculated to an OD600 of ∼0.05 from overnight precultures. The cultures were grown to an OD600 of ∼1.0 and cells were harvested from 4 ml of cell suspension. Total RNA was extracted by the glass bead mechanical disruption method (Ausubel et al. 1994). Approximately 10 μg of total RNA was separated on 1.2% formaldehyde agarose gels. The RNA was transferred and crosslinked to BioBond-Plus nylon membranes (Sigma-Aldrich).
Probes to detect the mRNA of the FLO11 and of the actin-encoding ACT1 genes were generated through PCR with primers FLO11-Fprobe, 5′-TCACGACGGCTATTCCAACC-3′; FLO11-Rprobe, 5′-TTAGAATACAACTGGAAGAGCGAG-3′; ACT1-F probe, 5′-GACGCTCCTCGTGCTGTCTT-3′; and ACT1-Rprobe, 5′-GGAAGATGGAGCCAAAGCGG-3′; they were then labeled using the PCR DIG probe synthesis kit (Roche Diagnostics). The labeled PCR products correspond to the nucleotides +3700–+4104 and +73 and +972 of the FLO11 and ACT1 open reading frames (ORFs), respectively. After hybridization, the probe-target hybrids were visualized as described in the digoxigenin (DIG) application manual (Roche Diagnostics).
RESULTS
MSS11 is required for the regulation of invasive growth and FLO11 expression by FLO8:
The effect of MSS11 on invasive growth phenotypes and on FLO11 transcription was investigated in a ∑1278b strain and compared to the effect of FLO8 (Figure 1A). Previous studies showed that multiple copies of MSS11 were able to suppress the phenotypic effect of a FLO8 deletion in a starch-degrading ISP20 strain (Gagiano et al. 1999a). The data presented in Figure 1A show that the same observations can be made in the ∑1278b genetic background. Deletions of either FLO8 or MSS11 resulted in loss of ability of ∑1278b to grow invasively. However, while high-copy expression of MSS11 restored the agar-invasion defect of the flo8Δ mutant, the 2μ-FLO8 plasmid was unable to suppress the invasive growth defect displayed by mss11Δ.
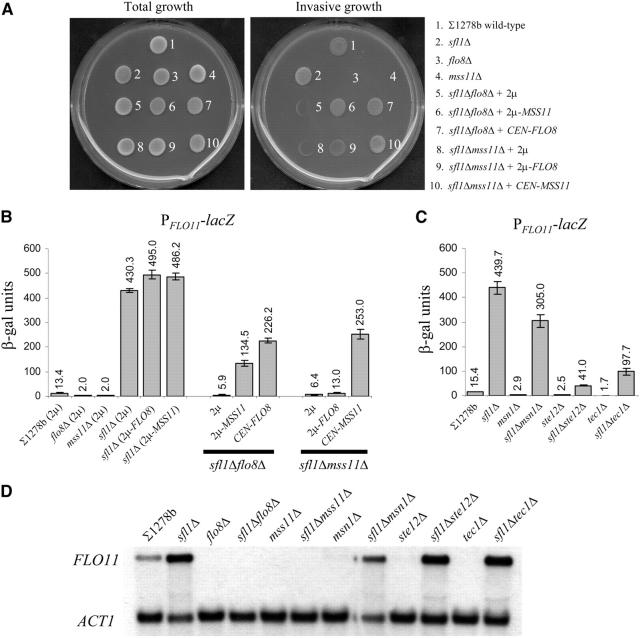
Figure 1.—
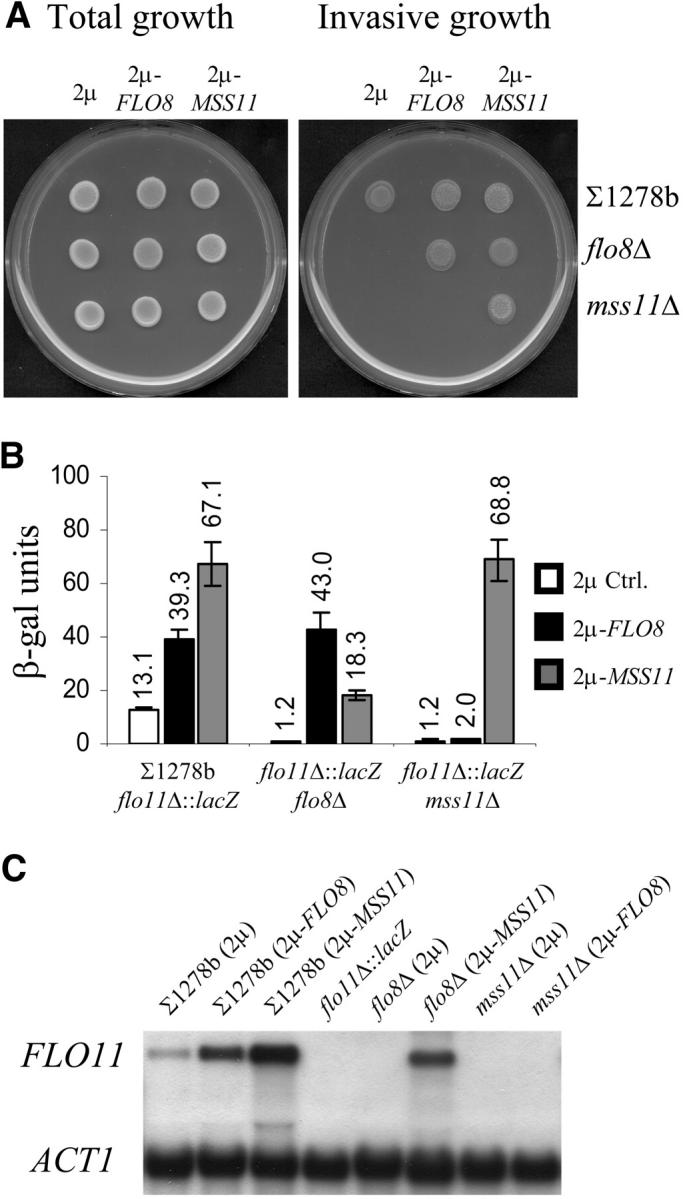
FLO8 requires MSS11 for the regulation of invasive growth and PFLO11-lacZ expression. (A) Total growth (before washing) and invasive growth (cells observed after the plate surface was rubbed under running water) phenotypes of isogenic strains ∑1278b wild type, flo8Δ, and mss11Δ transformed with YEplac112 (2μ), YEplac112-FLO8 (2μ-FLO8), or YEplac112-MSS11 (2μ-MSS11) on low-glucose (0.2%) medium. Multiple copies of FLO8 were insufficient to suppress the invasion defect of mss11Δ, while 2μ-MSS11 restored the phenotype of flo8Δ. (B) β-Galactosidase activity of isogenic ∑1278bflo11Δ::lacZ strains grown in SCD (2% glucose) liquid medium. The genomic ORF of FLO11 was replaced with lacZ in the wild-type ∑1278b strain (described in van Dyk et al. 2003), and FLO8 and MSS11 deletions were subsequently created in the ∑1278bflo11Δ::lacZ strain. β-Galactosidase activity is expressed in Miller units (Ausubel et al. 1994). (C) Northern blot showing the mRNA transcript levels of FLO11 and ACT1 in the ∑1278b strains used for the invasive growth plate assay (A). The ∑1278bflo11Δ::lacZ reporter strain served as a negative control and ACT1 was used as the internal standard.
To assess whether this effect was directly linked to the transcriptional regulation of FLO11, strains in which the endogenous FLO11 open reading frame had been replaced with a lacZ ORF were used (van Dyk et al. 2003). The PFLO11-lacZ expression data presented in Figure 1B are consistent with the invasive growth data (Figure 1A). Compared to the wild type (which shows 13.1 units of β-galactosidase activity), both flo8Δ and mss11Δ exhibited an 11-fold reduction (1.2 units) in β-galactosidase activity. Introduction of the 2μ-FLO8 or 2μ-MSS11 plasmids into the ∑1278bflo11Δ::lacZ strain led to a 3-fold (from 13.1 to 39.3 units) and a five-fold (from 13.1 to 67.1 units) increase, respectively, and restored lacZ expression in the corresponding flo8Δ and mss11Δ strains. However, when multiple copies of FLO8 were assessed in the mss11Δ strain, the low lacZ expression levels remained unaffected, while 2μ-MSS11 in the flo8Δ mutant resulted in a 15-fold induction and a higher FLO11 expression level than that in the wild-type control strain. These results were further verified by Northern analysis (Figure 1C). The data show an excellent correlation between the concentrations of FLO11 mRNA and the lacZ expression data for all the investigated strains.
sfl1Δ-dependent derepression of FLO11 requires MSS11 and FLO8:
Previous investigations indicated that the activity of the Flo8p transcriptional activator is controlled by the cAMP-dependent PKA pathway, which appears to control FLO11 transcription by regulating the antagonistic activities of Flo8p and Sfl1p (Pan and Heitman 2002). The authors' data suggested that FLO8 was required for Sfl1p-dependent derepression of FLO11. Since our results indicated that Flo8p function depends on the presence of MSS11, we assessed whether FLO11 derepression, caused by a SFL1 deletion, was also dependent on MSS11 through phenotype analysis (Figure 2A), PFLO11-lacZ expression (Figure 2, B and C), and Northern blot analysis (Figure 2D).
Figure 2.—
Assessment of MSS11 and FLO8 function in strains lacking SFL1. The strains listed in A are from a ∑1278b background. Low-copy (CEN) and high-copy (2μ) plasmids were used to either serve as positive controls (the low-copy YCplac22-FLO8 and YCplac22-MSS11 plasmids) or determine the effect of multiple copies (2μ-plasmid, YEplac112) of FLO8 and MSS11 on invasive growthin the sfl1Δflo8Δ and sfl1Δmss11Δ double mutants. MSS11 (2μ) significantly enhanced agar invasion in sfl1Δflo8Δ, whereas FLO8 (2μ) only slightly increased invasion of sfl1Δmss11Δ. (B) β-Galactosidase activities of the corresponding PFLO11-lacZ strains as well as a sfl1Δ mutant carrying YEplac112-FLO8 or YEplac112-MSS11. The quantitative data are consistent with the phenotypes observed in A. (C) Deletions of MSN1, STE12, or TEC1 in the ∑1278bflo11Δ::lacZsfl1Δ strain do not abolish derepression of FLO11 in a sfl1Δ background. (D) Northern blot analysis of the transcript levels of FLO11 and ACT1 in the corresponding ∑1278b (FLO11) strains.
Deletion of SFL1 significantly enhanced invasive growth (Figure 2A) and led to very high levels of PFLO11-lacZ expression (430.3 units; Figure 2B). The data show that MSS11 is required to the same degree as FLO8 for the hyperinvasive phenotype and the high FLO11 expression levels displayed by sfl1Δ mutant. PFLO11-lacZ-dependent β-galactosidase activity in the sfl1Δmss11Δ strain (6.4 units) is indeed similar to the activity found in the sfl1Δflo8Δ strain (5.9 units).
However, multiple copies of either FLO8 or MSS11 were not able to significantly enhance the high β-galactosidase expression levels in the sfl1Δ genetic background. This may suggest that these genes are dependent on the presence of Sfl1p to activate FLO11 and that Mss11p might act within the cAMP-dependent transcription complex. However, when the 2μ-MSS11 was transformed into the flo8Δsfl1Δ double-mutant strain, an invasive growth phenotype similar to the wild-type strain was observed (Figure 2A). The strain also displayed PFLO11-lacZ-dependent β-galactosidase expression levels that were higher (134.5 units) than those observed in the wild-type strain transformed with the same plasmid (67.1 units; Figures 2B and 1B). Multiple copies of FLO8, on the other hand, only slightly enhanced the invasive growth phenotype of the sfl1Δmss11Δ strain (Figure 2A) and resulted in a twofold increase in PFLO11-lacZ expression (Figure 2B). Taken together, the data therefore suggest that Mss11p does not require either FLO8 or SFL1 to activate FLO11.
Since derepression of PFLO11-lacZ in a sfl1Δ strain requires both MSS11 and FLO8, other transcriptional activators of FLO11 were also assessed for their ability to support Sfl1p-dependent derepression. The data show that MSN1, STE12, and, as previously reported by Pan and Heitman (2002), TEC1 appear to not be required for this purpose, since their absence in a sfl1Δ background continued to result in PFLO11-lacz expression levels that are significantly above those found in the wild type, with 305, 41, and 97 units, respectively (Figure 2C).
All of the PFLO11-lacZ data were further verified through Northern blot analysis (Figure 2D). Again, the mRNA levels confirmed the data generated through measurement of β-galactosidase activity, indicating that these data accurately reflected variations in mRNA concentration.
Deletion of MSS11 blocks Ras2p- and Tpk2p-dependent regulation of FLO11:
TPK2 and RAS2Val19, respectively, encode one of the catalytic subunits of the PKA complex and a hyperactive protein that activates the Kss1p-MAPK and PKA pathways (Mösch et al. 1996, 1999; Kübler et al. 1997; Robertson and Fink 1998; Pan and Heitman 1999). These genes were included in the genetic analysis to further assess the relationship between the PKA pathway and MSS11.
As seen in Figure 3, A–C, multiple copies of TPK2 enhanced the invasive growth phenotype and increased FLO11 mRNA levels in the wild-type strain and PFLO11-lacZ expression three-fold (from 13.1 to 41.5 units) in the PFLO11-lacZ strain. The 2μ-TPK2 plasmid was able to partially suppress the invasive growth defect of the mss11Δ and flo8Δ strains. However, this suppression was not due to increased FLO11 transcription since both the PFLO11-lacZ-driven β-galactosidase activity (Figure 3B) and the Northern blot (Figure 3C) indicated that FLO11 transcription remained unaffected by TPK2 expression levels in these strains. The data therefore suggest that Tpk2p can regulate other genes that lead to invasive growth independently of FLO11 and that this regulation does not require FLO8 or MSS11. Similarly, the introduction of RAS2Val19 resulted in a 3.5-fold increase of FLO11 transcription in the wild-type strain, but failed to induce the β-galactosidase activity of the PFLO11-lacZ mss11Δ strain (Figure 3B) or to increase mRNA levels in the mss11Δ strain (Figure 3C). To assess whether MSS11 acted as a general suppressor of RAS2Val19-dependent phenotypes, other phenotypes associated with the expression of RAS2Val19, including very slow growth on nonfermentable carbon sources and reduced survival in stationary phase, were also assessed. These phenotypes, however, were unaffected by the deletion of MSS11 (data not shown).
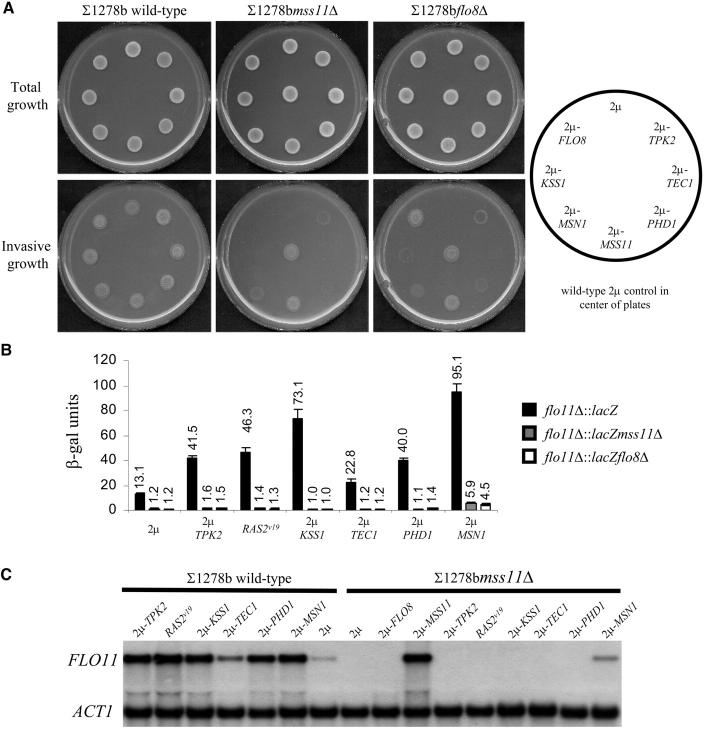
Figure 3.—
MSS11 and FLO8 are required by the Kss1p-MAPK and PKA pathways and PHD1, but not by MSN1. (A) Invasive growth on 0.2% glucose medium of ∑1278b wild type and ∑1278bmss11Δ and ∑1278bflo8Δ strains, transformed with YEplac112 (2μ) without insert or with the same plasmid carrying a single copy of FLO8, KSS1, MSN1, MSS11, PHD1, TEC1, and TPK2. The central colony in the panels showing the phenotypes of ∑1278bmss11Δ and ∑1278bflo8Δ is the wild-type strain transformed with YEplac112. (B) β-Galactosidase activity of the corresponding PFLO11-lacZ strains, grown in 2% glucose liquid medium. (C) FLO11 and ACT1 transcript levels of ∑1278b wild type and ∑1278bmss11Δ carrying the aforementioned genes in multiple copies or a single copy of the hyperactive RAS2Val19 allele.
While these data show that the PKA pathway requires MSS11 and FLO8 to activate FLO11 transcription, they do not exclude that a functional PKA pathway may be required for MSS11-dependent invasive growth and PFLO11-lacZ expression. Deletion of RAS2 and GPA2 negatively affects filamentous growth presumably by diminishing the intracellular cAMP levels of the cell (Kübler et al. 1997; Lorenz and Heitman 1997). We therefore generated a gpa2Δras2Δ double mutant and a tpk2Δ single mutant. Deletion of GPA2 and RAS2 resulted in a fivefold decrease in PFLO11 lacZ-driven β-galactosidase activity (Table 3), but introduction of 2μ-MSS11 increased the activity to 67.1 units, which is comparable to the 70.7 units observed in the wild-type strain transformed with the same plasmid. Deletion of TPK2 also resulted in a fivefold decrease in β-galactosidase activity of the PFLO11-lacZ strain, but 22.3 units of activity were obtained in the presence of the 2μ-MSS11 plasmid. In all cases, multiple copies of MSS11 induced invasive growth significantly (Figure 4).
TABLE 3.
Expression of PFLO11-lacZ in ∑1278b mutant strains
| Mean β-galactosidase activity (Miller units ±SD) |
|||
|---|---|---|---|
| Relevant genotype | 2μ | 2μ-MSS11 | 2μ-FLO8 |
| ∑1278bflo11Δ::lacZ | 12.9 ± 0.8 | 70.7 ± 4.3 | 37.2 ± 3.7 |
| gpa2Δras2Δ | 2.8 ± 0.2 | 67.1 ± 0.9 | 15.8 ± 1.3 |
| tpk2Δ | 2.8 ± 0.1 | 22.3 ± 1.6 | 7.7 ± 0.4 |
| kss1Δ | 4.3 ± 0.4 | 50.8 ± 4.5 | 22.1 ± 3.3 |
| ste12Δ | 2.5 ± 0.1 | 16.0 ± 0.2 | 5.5 ± 0.4 |
| tec1Δ | 1.7 ± 0.2 | 26.2 ± 0.4 | 12.2 ± 1.6 |
| phd1Δ | 5.0 ± 0.6 | 63.2 ± 1.2 | 35.1 ± 4.0 |
| msn1Δ | 2.2 ± 0.2 | 28.4 ± 3.3 | 7.1 ± 0.5 |
The listed mutants are isogenic to the ∑1278bflo11Δ::lacZ reporter strain. Strains were transformed with YEplac112, YEplac112-MSS11, and YEplac112-FLO8 and assayed following growth in SCD (2% glucose) liquid medium (see materials and methods). At least three transformants were assayed and the average β-galactosidase activity is presented. The error margins for the presented average data do not exceed 15%. The experiment was performed in triplicate and similar tendencies were observed.
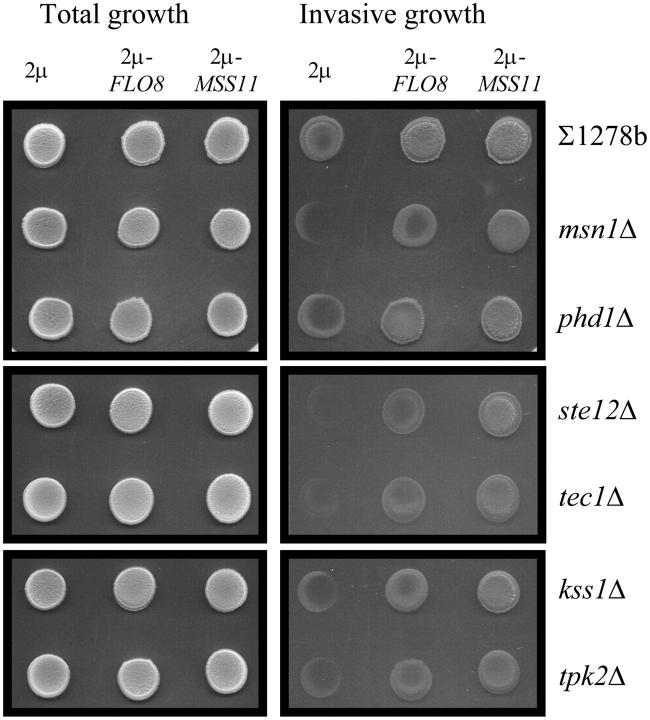
Figure 4.—
FLO8 and MSS11 regulate invasive growth independently of MSN1, PHD1, and genes of the Kss1p-MAPK and cAMP-PKA pathways. ∑1278b wild type and the isogenic mutant strains msn1Δ, phd1Δ, ste12Δ, tec1Δ, kss1Δ, and tpk2Δ were transformed with YEplac112 (2μ), YEplac112-FLO8 (2μ-FLO8), and YEplac112-MSS11 (2μ-MSS11) and were subsequently spotted onto 0.2% glucose-containing medium. High-copy expression of FLO8 and MSS11 enhanced the invasiveness of all the tested strains.
Introduction of 2μ-FLO8 into the wild-type PFLO11-lacZ strain resulted in 37.2 units of β-galactosidase activity, while the lacZ expression levels of the gpa2Δras2Δ and tpk2Δ mutants were increased to only 15.8 and 7.7 units, respectively (Table 3). These significantly reduced expression levels are consistent with the direct regulation of Flo8p by the PKA pathway. Mss11p, on the other hand, appears to be much less affected by this pathway. Indeed, the reduced activation observed in the TPK2 deletion strain can probably be explained by the effect of Tpk2p on Flo8p, since induction by 2μ-MSS11 is similar in the tpk2Δ and flo8Δ deletion strain (Figure 1B and Table 3).
Deletion of TPK2 did not prevent the 2μ-FLO8 plasmid from enhancing invasive growth (Figure 4), suggesting either that other Tpks can phosphorylate Flo8p in the absence of Tpk2p or that gene dosage and not only PKA-mediated protein modification of Flo8p contributes to FLO11 expression.
MSS11 is required by the Kss1p-MAPK pathway:
Figure 3, A–C, shows that the agar-invasion phenotypes, PFLO11-lacZ expression levels, and mRNA concentrations of the wild-type strains increased in the presence of multiple copies of KSS1 and TEC1. The PFLO11-lacZ expression levels increased five- and twofold in strains transformed with the 2μ-KSS1 and 2μ-TEC1 plasmids, respectively. The data show that these increases are entirely dependent on MSS11 and FLO8, except for some invasion observed for the flo8Δ strain that carries multiple copies of KSS1. This invasive phenotype appears to be clearly independent of FLO11, since no increase in expression levels was observed for this gene. High-copy expression of TEC1 did not suppress the FLO8 mutation as had previously been reported (Pan and Heitman 1999). This discrepancy may be accounted for by differences in TEC1 expression levels, since in the previous study TEC1 expression was controlled by the TDH1 promoter (Pan and Heitman 1999), while the episomal plasmid used in this study carries a copy of TEC1 with its native promoter.
KSS1, STE12, or TEC1 were deleted to ascertain whether the disruption of elements of the MAPK pathway would affect the effects of 2μ-MSS11 and 2μ-FLO8. Compared to the control strain, ∑1278bflo11Δ::lacZ, the gene deletions decreased lacZ expression between three- and sevenfold (Table 3), which is reflected in decreased invasive growth phenotypes of the corresponding FLO11-carrying strains (Figure 4). With the introduction of 2μ-MSS11, the lacZ expression levels increased in all three mutants, with the level of induction being at least similar to that observed for 2μ-MSS11 in the wild-type strain. In the case of the 2μ-FLO8 plasmid, similar results were observed. In all cases, the FLO11 expression data and the level of invasiveness paralleled each other.
The dependency of the MAPK pathway elements on the presence of Mss11p appears specific to invasive growth and FLO11 transcription. Indeed, the mss11Δ strain did not display any mating-associated defects, as does, for example, the ste12Δ strain (data not shown).
The hyperactive STE11-4 allele and 2μ-STE12 were also transformed into the ∑1278b and ∑1278bflo11Δ::lacZ strains, but severe growth defects were observed. Invasive growth and β-galactosidase activity did not increase in the transformed strains. To verify previous reports (Mösch et al. 1999; Köhler et al. 2002) and the functionality of the two constructs, their effect on FRE(Ty)::lacZ expression was assessed. Both plasmids significantly induced this reporter system, and 2μ-STE12 also restored the ability of a sterile ste12Δ mutant to mate (results not shown). The data indicate that the regulation of the FRE element does not reflect FLO11 transcription.
Relationships among MSS11, FLO8, and other genes encoding regulators of FLO11:
It has been reported that the Phd1p-Sok2p activator and repressor module requires FLO8 to regulate FLO11 transcription (Pan and Heitman 2000). It is therefore not surprising that our data indicate that the same applies to the relation between this module and MSS11. As shown in Figure 3, A–C, introduction of 2μ-PHD1 led to enhanced agar invasion, induced lacZ expression, and increased FLO11 mRNA levels in the ∑1278b and ∑1278bflo11Δ::lacZ control strains, but not in the mss11Δ and flo8Δ mutants. Introduction of 2μ-MSS11 and 2μ-FLO8 in the phd1Δ strains, on the other hand, effectively suppressed the defects in agar invasion (Figure 4) and lacZ expression (Table 3). The data obtained with sok2Δ strains confirm the observations made for PHD1 since no derepression is observed in these strains in the absence of MSS11 or FLO8 (Figure 5), while both genes activate FLO11 efficiently in this strain (Table 4). Flo8p and Mss11p function does not depend on Phd1p and Sok2p, while these factors clearly require the presence of both.
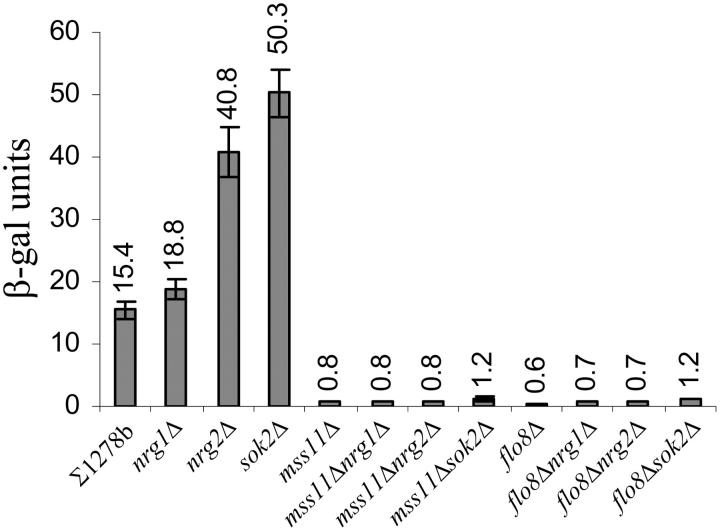
Figure 5.—
PFLO11-lacZ derepression is dependent on MSS11 and FLO8. Deletion of NRG1, NRG2, or SOK2 in the ∑1278bflo11Δ::lacZ strain resulted in elevated β-galactosidase activity. Increases in activity were abolished when MSS11 or FLO8 was deleted in the corresponding repressor mutants.
TABLE 4.
Expression of PFLO11-lacZ in repressor mutants
| Mean β-galactosidase activity (Miller units ±SD)
|
|||||
|---|---|---|---|---|---|
| Plasmids | ∑1278b | nrg1Δ | nrg2Δ | sok2Δ | sfl1Δ |
| YEplac112 | 13.1 ± 0.6 | 16.8 ± 0.4 | 35.9 ± 4.5 | 44.7 ± 5.7 | 434.3 ± 3.9 |
| YEplac112-MSS11 | 70.7 ± 4.3 | 96.4 ± 3.9 | 147.4 ± 4.8 | 185.5 ± 1.7 | 486.2 ± 14.1 |
| YEplac112-FLO8 | 37.2 ± 3.7 | 36.4 ± 4.2 | 102.1 ± 7.4 | 133.5 ± 8.8 | 495.0 ± 17.7 |
To test whether Mss11p and Flo8p might activate gene transcription by antagonizing other identified transcriptional repressors of FLO11, the effect of multiple copies of MSS11 and FLO8 was assessed in the corresponding mutants. As can be seen in Table 4 and Figure 5, deletion of NRG2 led to a significant increase in the activity of β-galactosidase, while deletion of NRG1 resulted in a less severe increase. Expression of both 2μ-MSS11 and 2μ-FLO8 in these strains induced PFLO11-lacZ expression, with a level of induction similar to that observed in the wild-type strain (Table 4). The absence of these repressors therefore did not prevent multiple copies of FLO8 and MSS11 from further enhancing the expression of the lacZ reporter gene.
To assess whether derepression in the absence of the repressors is dependent on functional FLO8 and MSS11 alleles, deletions of either MSS11 or FLO8 were combined with deletions of each of the repressor genes. The data in Figure 5 show that the deletion of MSS11 and FLO8 in the nrg1Δ and nrg2Δ strains abolished the increases in β-galactosidase activities observed in the single mutants. Since similar observations were made when MSS11 or FLO8 were deleted in the sfl1Δ background (Figure 2A), the data show that mss11Δ and flo8Δ block FLO11 derepression in all the repressor mutants that were assessed.
Msn1p does not require MSS11 or FLO8 to activate FLO11:
Lambrechts et al. (1996a) previously reported on the ability of multiple copies of MSN1 to activate FLO11. Our data show that 2μ-MSN1 continued to lead to notable agar-invasion phenotypes in both the mss11Δ and the flo8Δ mutant strain (Figure 3A), as well as increased lacZ expression in the corresponding PFLO11-lacZ strains (Figure 3B). Most importantly, the fold induction observed in the presence of multiple copies of MSN1 is similar in the wild-type strain and the two mutant strains. 2μ-MSS11 and 2μ-FLO8, on the other hand, also suppressed the defects of the ∑1278bmsn1Δ and ∑1278bflo11Δ::lacZmsn1Δ strains (Figure 4 and Table 3), indicating that the regulatory role of Msn1p is independent from that of Mss11p and Flo8p. These observations are similar to those made for RME1 overexpression and deletion (van Dyk et al. 2003).
Gagiano et al. (1999a)(b) presented phenotype-based data suggesting that deletion of MSS11 may block activation by Msn1p. The data here indicate that this apparent suppression was probably due to the very low basal FLO11 expression levels in the MSS11 deletion strain, which did not allow Msn1p-dependent induction of invasive growth. Indeed, our data show that the basal FLO11 expression levels of the ISP strains are much lower than those in ∑1278b. It is therefore probable that in the ISP mss11Δ strains, 2μ-MSN1 does not raise FLO11 expression levels efficiently enough to result in invasive growth phenotypes, and hence the misinterpretation.
SFL1 deletion induces invasive growth independently of FLO11:
Previous reports suggest that invasive growth is not solely dependent on FLO11 (Palecek et al. 2000), but can also be induced by overexpression of FIG2 and FLO10 (Guo et al. 2000; Halme et al. 2004). Deletion of SFL1 in a flo11Δ strain also results in agar invasion (Robertson and Fink 1998), which is attributed to the upregulation of FLO10 (Guo et al. 2000; Halme et al. 2004). We similarly showed that 2μ-MSS11 is able to induce invasive growth in a flo11Δ background (Gagiano et al. 1999b). These observations prompted us to assess the genetic relationships among FLO8, SFL1, and MSS11 in FLO11-deleted strains.
As can be seen in Figure 6A, the flo11Δ and the double-deleted strains flo11Δflo8Δ and flo11Δmss11Δ, as expected, were unable to grow invasively, unlike the flo11Δsfl1Δ strain, confirming the previous report of Guo et al. (2000). The residual invasive growth phenotype of flo11Δsfl1Δ was abolished by deletions of FLO8 and MSS11 (Figure 6B). Multiple copies of MSS11 significantly enhanced agar invasion in the flo11Δsfl1Δflo8Δ strain, but 2μ-FLO8 did not suppress the defect of the flo11Δsfl1Δmss11Δ triple mutant. The genetic relationships among FLO8, SFL1, and MSS11 are therefore not merely relevant to FLO11 expression, but also to invasive growth in general, which implies that other target genes are also regulated by a mechanism that involves Flo8p, Sfl1p, and Mss11p.
Figure 6.—
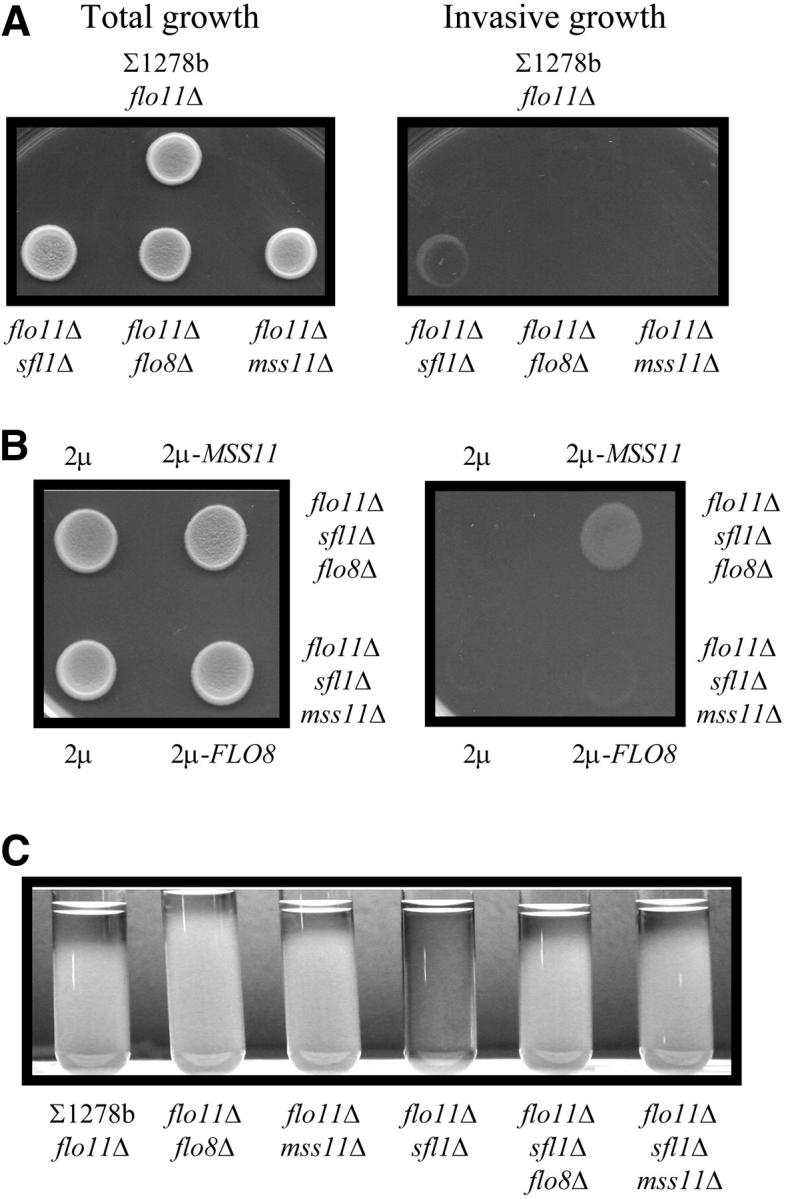
MSS11 and FLO8 controls invasive growth and flocculation independently of FLO11. (A and B) Invasive growth phenotypes on 0.2% glucose medium of FLO11 mutant strains from the ∑1278b genetic background. Deletion of SFL1 in ∑1278bflo11Δ restored agar invasion (A). This phenotype of ∑1278bflo11Δsfl1Δ was abolished when FLO8 or MSS11 was deleted (B). YEplac112-MSS11 suppressed the invasion defect of ∑1278bflo11Δsfl1Δflo8Δ, but YEplac112-FLO8 was unable to complement the ∑1278bflo11Δsfl1Δmss11Δ defect. (C) Flocculation phenotypes of the strains used in A and B. Deletion of SFL1 in ∑1278bflo11Δ resulted in flocculation that is dependent on FLO8 and MSS11. Single colonies were inoculated in 5 ml YPED liquid media and allowed to grow for 48 hr at 30° on a rotating wheel. The cultures were vortexed vigorously for 1 min to ensure that flocculating cells are resuspended and were subsequently placed on the bench to allow for cell aggregation and sedimentation. The cultures were photographed after 20 min.
The hyperflocculation phenotype of a sfl1Δ mutant depends on MSS11 and FLO8, even in the absence of FLO11:
The introduction of multiple copies of FLO8 and MSS11 in the S288c genetic background leads to flocculation (results not shown). This, however, is not the case in the ∑1278b strain, in which all the FLO genes, with the exception of FLO11, are repressed (Halme et al. 2004). In this strain, the role of FLO8 and MSS11 in flocculation became apparent only when SFL1 was deleted. To further establish the involvement of a general regulatory mechanism/complex (Flo8p, Sfl1p, and Mss11p) in the regulation of FLO11-unrelated target genes, we assessed the flocculation phenotypes of flo11Δ strains.
The flo11Δ, flo11Δflo8Δ, and flo11Δmss11Δ mutants shown in Figure 6C exhibited similar sedimentation phenotypes as can be deduced from the transparent sections of the corresponding cell cultures. The flo11Δsfl1Δ strain, on the other hand, was highly flocculent and the cells settled immediately after agitation. Deletions of FLO8 and MSS11 abrogated the hyperflocculation phenotype of the flo11Δsfl1Δ strain and the phenotypes of these strains were comparable to those of flo11Δflo8Δ and flo11Δmss11Δ strains. Mss11p and Flo8p therefore appeared to regulate flocculation independently of FLO11 when SFL1 is disrupted. The transcription of other FLO genes was shown to be under the control of Sfl1p and Flo8p (Kobayashi et al. 1999; Guo et al. 2000; Halme et al. 2004), but our genetic data strongly suggest that Mss11p is also required for this regulation.
Interaction between the Kss1p-MAPK and PKA pathways:
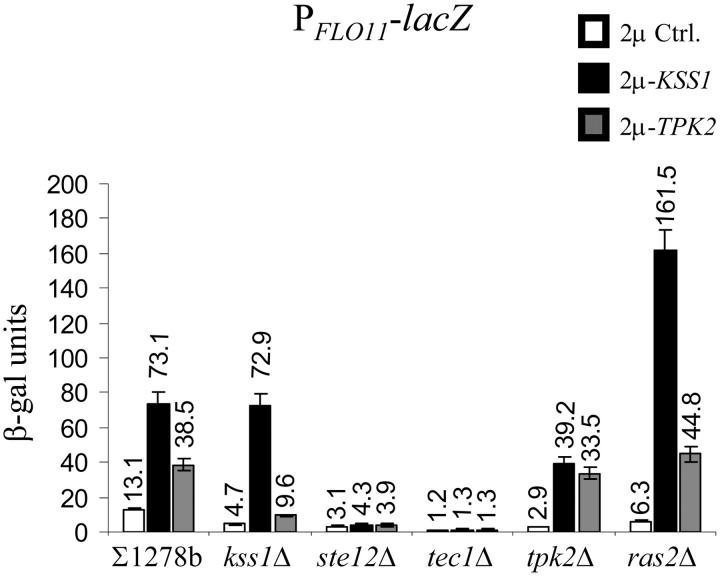
High-copy expression of TPK1, TPK2, and TPK3 was previously shown to stimulate FRE(Ty)::lacZ expression in a ras2Δ mutant (Mösch et al. 1999). The elevated expression was dependent on STE12 and TEC1, and it was therefore suggested that the Tpk subunits act on the Ste12p-Tec1p transcription factor to induce downstream target genes. This result implies that the Kss1p-MAPK and PKA pathways are interconnected at the level of specific transcription factors. In agreement with this report, we observed that multiple copies of TPK2 induce PFLO11-lacZ expression threefold (Figure 7) and that this induction was dependent on the presence of STE12, TEC1, and to a lesser extent KSS1, but not on RAS2. High-copy expression of KSS1 in a ras2Δ strain led to increased PFLO11-lacZ-dependent β-galactosidase activity (161.5 units), when compared to 73.1 units observed for the wild type.
Figure 7.—
Interaction between the Kss1p-MAPK and cAMP-dependent PKA pathways. YEplac112, YEplac112-KSS1, and YEplac112-TPK2 were introduced to ∑1278bflo11Δ::lacZ and to five isogenic mutants with deleted copies of KSS1, STE12, TEC1, TPK2, and RAS2, respectively.
The 5.6-fold increase in reporter gene activity conferred by 2μ-KSS1 in the wild type was dependent on STE12 and TEC1, a result that was anticipated since Kss1p is an upstream regulator of the Ste12p-Tec1p transcription factor. Reporter gene activity in a strain carrying 2μ-KSS1 was also dependent on FLO8 for the induction of PFLO11-lacZ (Figure 3B), suggesting that the Kss1p-MAPK and PKA pathways are interdependent. On the other hand, Kss1p activity was not dependent on a functional copy of TPK2, since a 13.5-fold increase in β-galactosidase activity was obtained when 2μ-KSS1 was introduced into the PFLO11-lacZ tpk2Δ strain.
Mss11p is still functional in mutants deleted for several activators:
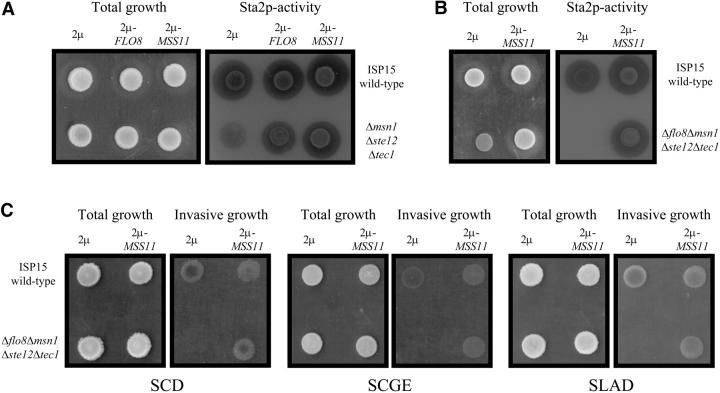
Yeast strains carrying one of the three glucoamylase-encoding STA genes are able to utilize starch as a sole carbon source. The STA1-3 genes and FLO11 have almost identical promoters and are therefore controlled by a similar set of transcription factors, which include Mss11p, Flo8p, Msn1p, Ste12p, Tec1p, Nrg1p, Sfl1p, and Sok2p (Gagiano et al. 1999a,b; Park et al. 1999; van Dyk et al. 2003; Kim et al. 2004). Multiple deletion mutants, including strains msn1Δste12Δtec1Δ and flo8Δmsn1Δste12Δtec1Δ, were generated in the STA2-bearing ISP15 genetic background to assess whether our data regarding the ability of Mss11p to activate transcription in mutants deleted for individual activator genes may be the result of multiple interactions of the protein with several positive regulators.
As can be seen in Figure 8, A and B, both triple and quadruple mutants were able to grow on starch-containing SCS medium, but displayed significantly reduced glucoamylase activity. The quadruple mutant in particular displays a severe growth defect on this medium, whereas growth on glucose-containing medium was unaffected. High-copy expression of FLO8 and MSS11 suppressed the starch degradation defect of the triple mutant (Figure 8C), while introduction of 2μ-MSS11 in the quadruple mutant restored the strain's ability to degrade starch and also suppressed the severe growth defect (Figure 8B).
Figure 8.—
Mss11p regulates starch degradation and invasive growth under different nutritional conditions. Total growth and starch degradation phenotypes of (A) ISP15 wild type and ISP15msn1Δste12Δtec1Δ and (B) ISP15flo8Δmsn1Δste12Δtec1Δ, grown on starch-containing medium at 30° for 6 days. Transparent zones are indicative of Sta2p-glucoamylase activity. YEplac112-FLO8 and YEplac112-MSS11 restored Sta2p-glucoamylase activity in the triple mutant (A), while the 2μ-plasmid carrying MSS11 also restored growth and starch degradation phenotypes in the quadruple mutant (B). (C) Invasive growth phenotypes of ISP15 wild type and the quadruple mutant on SCD (glucose repressed), SCGE (glucose derepressed), and SLAD (limited nitrogen) media.
We also assessed whether Mss11p activity is linked to glucose repression in the same genetic background. The quadruple mutant was unable to grow invasively on media containing either 2% glucose or 3% glycerol and ethanol (Figure 8C). However, agar invasion was restored under both conditions with the introduction of 2μ-MSS11, and similar data were obtained when invasive growth phenotypes were assessed for the quadruple mutant on medium containing limited amounts of nitrogen (Figure 8C).
DISCUSSION
Mss11p is a central element in the regulation of invasive growth:
The results presented here strongly suggest a central role for Mss11p in the regulation of FLO11 expression and invasive growth. At least four arguments can be made in support of this hypothesis:
The deletion of MSS11 completely suppressed the activation of FLO11 by hyperactive alleles and multiple copies of genes encoding components of the nutrient-responsive MAP kinase cascade and of the cAMP-signaling pathway that activate FLO11 in the wild type.
Similarly, multiple copies of genes encoding transcriptional activators, including FLO8, TEC1, and PHD1, were no longer able to increase FLO11 expression in the mss11Δ background.
In the same mss11Δ background, no derepression of FLO11 was observed in strains with deletions of the previously identified FLO11 repressor-encoding genes NRG1, NRG2, SOK2, and SFL1.
Multiple copies of MSS11 activated FLO11 expression, even in the absence of the above-mentioned individual activators.
The same was true in strains with combinations of deletions in activator-encoding genes. In many regards, the genetic interactions displayed by MSS11 are very similar to those observed for FLO8. However, MSS11 was clearly epistatic to FLO8, since deletion of MSS11 was not suppressed by multiple copies of FLO8, whereas multiple copies of MSS11 suppressed the effects of a FLO8 deletion efficiently. Multiple copies of MSS11 were unable to further enhance FLO11 transcription in a sfl1Δ strain. However, these data probably reflect saturation of the transcription capacity of the promoter in this strain. Indeed, Mss11p is clearly not dependent on the presence of SFL1, since it efficiently activates transcription in a flo8Δsfl1Δ double mutant.
MSS11 also affects other genes involved in flocculation:
Our data show that Mss11p and Flo8p are required for Sfl1p-dependent flocculation, even in the absence of FLO11. Mss11p, similar to Flo8p and Sfl1p, is required for the expression of genes involved in flocculation. Microarray analysis using mss11Δ and MSS11 multiple copy strains suggests that the dominant flocculation gene FLO1 might be the specific target gene responsible for these phenotypes (our unpublished data), but additional confirmation of these data is required.
MSS11 specifically affects flocculation and invasive growth-related phenotypes:
While MSS11 is essential for FLO11 expression and affects transcription of other flocculation genes, it does not appear to affect other cellular functions. Indeed, the mss11Δ strain displayed only FLO11 and flocculation-related phenotypes. As reported previously, this strain does not present any morphological or growth defects in a range of conditions, including growth on various carbon or nitrogen sources (Gagiano et al. 1999b). When compared to the wild-type strain, the mss11Δ strain did not display significant differences in its viability upon nutrient depletion and in its response to heat stress, osmotic shock, or salt toxicity (Gagiano et al. 1999b and data not shown). Furthermore, while suppressing the effect of the cAMP/PKA pathway and of the MAP kinase cascade on FLO11 expression, it does not affect any of the other phenotypes that are associated with these pathways. Indeed, a RAS2Val19 mss11Δ double mutant displays RAS2Val19 phenotypes with regard to viability under starvation conditions, growth on nonfermentable carbon sources, and glycogen accumulation (data not shown). Mating efficiency of mss11Δ strains is also similar to wild-type strains, suggesting that the gene does not affect the mating signaling pathway, which shares many elements with the nutrient-responsive MAP kinase cascade. Finally, the microarray analysis of strains deleted for MSS11 or carrying MSS11 on a multiple copy plasmid reveal only very few genes whose expression was significantly affected by modified MSS11 expression levels. Of these genes, FLO11 is consistently identified as the most significantly affected gene (our unpublished data).
Role of Mss11p in FLO11 expression:
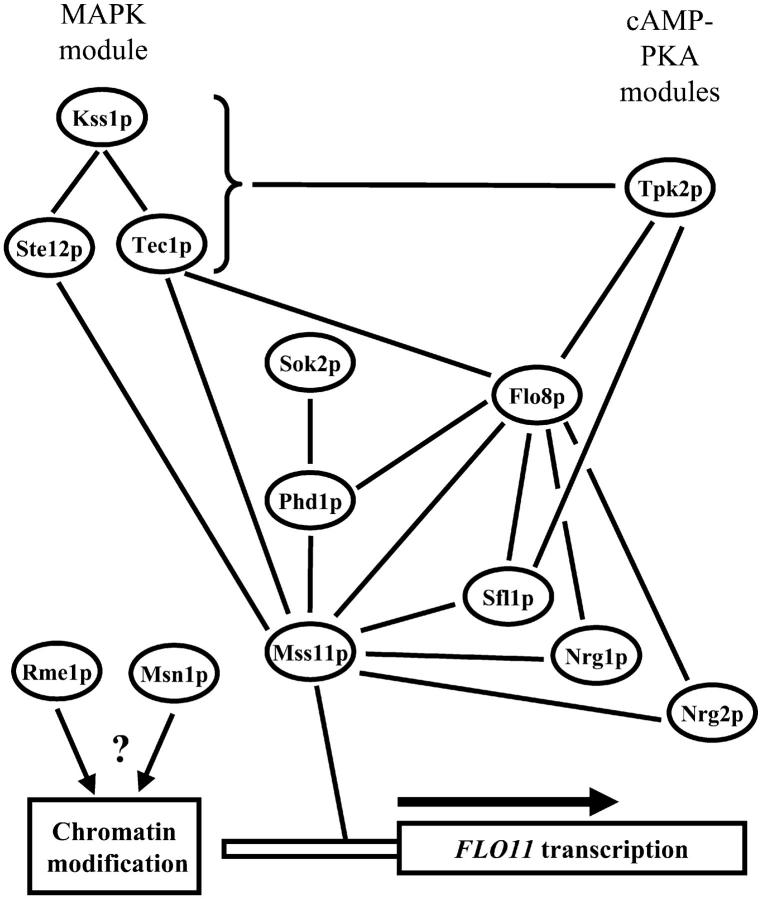
The results of the genetic analysis presented in this article are summarized in Figure 9. The central position of Mss11p and the ability of Mss11p to activate transcription on its own (Gagiano et al. 2003) strongly suggest a direct link between this protein and essential elements of the RNA polymerase II transcription machinery. However, various attempts through immunoprecipitation and several two-hybrid screens using Mss11p or nonactivating domains of Mss11p as bait have failed to yield any information regarding its interaction with other proteins. The genetic analysis suggests a close link of Mss11p with Flo8p and Sfl1p. This is supported by the fact that the only significant homology between Mss11p and any other protein consists of two small domains that are shared by Mss11p and Flo8p (Gagiano et al. 2003). Furthermore, Mss11p appears to have the same range of target genes as Flo8p. However, direct attempts to show interactions between Flo8p and Mss11p in the two-hybrid system have failed (data not shown).
Figure 9.—
A model summarizing the genetic interactions among Mss11p and other factors implicated in the transcriptional control of FLO11. The lines do not imply physical or functional interactions.
Mss11p does not appear to have any ortholog in other species, including in closely related organisms. No protein with significant homology to Mss11p was identified in the genomes of Candida albicans, Cryptococcus neoformans, and Neurospora crassa. This suggests that MSS11 might be of relatively recent evolutionary origin. It also raises the question of the nature of interactions between Mss11p and other proteins. It has recently been suggested that proteins with strong and multiple protein interactions are better conserved evolutionarily and therefore have a higher probability of having well-conserved orthologs in other species (Pagel et al. 2004).
In our analysis, of the previously identified genes that encode factors that positively affect FLO11 transcription, only two were shown to be able to activate FLO11 in the absence of MSS11, MSN1, and RME1. Indeed, multiple copies of these two genes result in similar fold increases of PFLO11-lacZ expression in the wild-type strain and in the mss11Δ strain, although the expression levels in the latter strain remain low since they increase from a very low base. Both these factors have been linked to chromatin-related modes of action (Covitz et al. 1994; Sidorova and Breeden 1999), while the factors whose role in FLO11 expression is suppressed by MSS11 have all been associated more directly with the RNA polymerase II transcriptional machinery. It is therefore possible that Msn1p and Rme1p activate FLO11 transcription through more nonspecific means, for example, by modifying nucleosome positioning and by rendering a general promoter element such as the TATA box more accessible. It is a well-established fact that such elements can be recognized in a nonspecific way by the transcription machinery and can lead to transcriptional activation without the requirement for a specific activator (Roeder 1996).
The regulation of FLO11 recently has been shown to be subjected to epigenetic regulation (Halme et al. 2004). Sfl1p was identified as one of the essential elements within the epigenetic regulatory network. The suppression of Sfl1p- and Flo8p-dependent regulation of FLO11 by Mss11p strongly suggests that Mss11p also plays a role in epigenetic regulation.
Acknowledgments
The authors thank H.-U. Mösch, J. H. Hegemann, D. Engelberg, and E. Blachinsky for strains and constructs, and P. Young for technical support with Northern blotting. This work was supported by grants from the South African Wine Industry (Winetech) and the National Research Foundation of South Africa.
References
- Ahn, S. H., B. T. Tobe, J. N. Fitz-Gerald, S. L. Anderson, A. Acurio et al., 2001. Enhanced cell polarity in mutants of the budding yeast cyclin-dependent kinase Cdc28p. Mol. Biol. Cell 12: 3589–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1994 Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Bardwell, L., J. G. Cook, D. Voora, D. M. Baggott, A. R. Martinez et al., 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12: 2887–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Braus, G. H., O. Grundmann, S. Bruckner and H. U. Mösch, 2003. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 4272–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz, A., L. Boucher, B.-J. Breitkreutz, M. Sultan, I. Jurisca et al., 2003. Phenotypic and transcriptional plasticity directed by yeast mitogen-activated protein kinase network. Genetics 165: 997–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M., 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2: 202–207. [DOI] [PubMed] [Google Scholar]
- Caro, L. H., H. Tettelin, J. H. Vossen, A. F. Ram, H. van den Ende et al., 1997. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13: 1477–1489. [DOI] [PubMed] [Google Scholar]
- Cook, J. G., L. Bardwell and J. Thorner, 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390: 85–88. [DOI] [PubMed] [Google Scholar]
- Courchesne, W. E., R. Kunisawa and J. Thorner, 1989. A putative protein kinase overcomes pheromone-induced arrest of cell cycling in S. cerevisiae. Cell 58: 1107–1119. [DOI] [PubMed] [Google Scholar]
- Covitz, P. A., and A. P. Mitchell, 1993. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 7: 1598–1608. [DOI] [PubMed] [Google Scholar]
- Covitz, P. A., W. Song and A. P. Mitchell, 1994. Requirement for RGR1 and SIN4 in RME1-dependent repression in Saccharomyces cerevisiae. Genetics 138: 557–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P. J., and G. F. Sprague, Jr., 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97: 13619–13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch, F., and M. Carlson, 1990. Increased dosage of the MSN1 gene restores invertase expression in yeast mutants defective in the SNF1 protein kinase. Nucleic Acids Res. 18: 6959–6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagiano, M., D. van Dyk, F. F. Bauer, M. G. Lambrechts and I. S. Pretorius, 1999. a Divergent regulation of the evolutionarily closely related promoters of the Saccharomyces cerevisiae STA2 and FLO11 genes. J. Bacteriol. 181: 6497–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagiano, M., D. van Dyk, F. F. Bauer, M. G. Lambrechts and I. S. Pretorius, 1999. b Msn1p/Mss10p, Mss11p and Flo11p/Flo11p are part of a signal transduction pathway downstream of Mep2p regulating invasive growth and pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Microbiol. 31: 103–116. [DOI] [PubMed] [Google Scholar]
- Gagiano, M., F. F. Bauer and I. S. Pretorius, 2002. The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res. 2: 433–470. [DOI] [PubMed] [Google Scholar]
- Gagiano, M., M. Bester, D. van Dyk, J. Franken, F. F. Bauer et al., 2003. Mss11p is a transcription factor regulating pseudohyphal differentiation, invasive growth and starch metabolism in Saccharomyces cerevisiae in response to nutrient availability. Mol. Microbiol. 47: 119–134. [DOI] [PubMed] [Google Scholar]
- Gancedo, J. M., 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25: 107–123. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and A. Sugino, 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Gimeno, C. J., and G. R. Fink, 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 14: 2100–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno, C. J., P. O. Ljungdahl, C. A. Styles and G. R. Fink, 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090. [DOI] [PubMed] [Google Scholar]
- Güldener, U., S. Heck, T. Fiedler, J. Beinhauer and J. H. Hegemann, 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 14: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B., C. A. Styles, Q. Feng and G. R. Fink, 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97: 12158–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme, A., S. Bumgarner, C. Styles and G. R. Fink, 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415. [DOI] [PubMed] [Google Scholar]
- Jones, J. S., and L. Prakash, 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6: 363–366. [DOI] [PubMed] [Google Scholar]
- Kassir, Y., D. Granot and G. Simchem, 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52: 853–862. [DOI] [PubMed] [Google Scholar]
- Kim, T. S., J. Y. Ahn, J. H. Yoon and H. S. Kang, 2004. STA10 repression of STA gene expression is caused by a defective activator, flo8, in Saccharomyces cerevisiae. Curr. Genet. 44: 261–267. [DOI] [PubMed] [Google Scholar]
- Kobayashi, O., H. Yoshimoto and H. Sone, 1999. Analysis of the genes activated by the FLO8 gene in Saccharomyces cerevisiae. Curr. Genet. 36: 256–261. [DOI] [PubMed] [Google Scholar]
- Köhler, T., S. Wesche, N. Taheri, G. H. Braus and H.-U. Mösch, 2002. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryot. Cell 1: 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübler, E., H.-U. Mösch, S. Rupp and M. P. Lisanti, 1997. Gpa2p, a G protein α-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272: 20321–20323. [DOI] [PubMed] [Google Scholar]
- Kuchin, S., V. K. Vyas and M. Carlson, 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22: 3994–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts, M. G., F. F. Bauer, J. Marmur and I. S. Pretorius, 1996. a Flo11, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 93: 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts, M. G., P. Sollitti, J. Marmur and I. S. Pretorius, 1996. b A multicopy suppressor gene, MSS10, restores STA2 expression in Saccharomyces cerevisiae strains containing the STA10 repressor gene. Curr. Genet. 29: 523–529. [PubMed] [Google Scholar]
- Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen et al., 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64: 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., C. A. Styles and G. R. Fink, 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144: 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, W. S., and A. M. Dranginis, 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 178: 7144–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, W. S., and A. M. Dranginis, 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, J. D., T. A. Kerentseva, T. Pan, M. Sepulveda-Becerra and H. Liu, 1999. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153: 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., and J. Heitman, 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16: 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M. C., N. S. Cutler and J. Heitman, 2000. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 183–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani, H. D., and G. R. Fink, 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275: 1314–1317. [DOI] [PubMed] [Google Scholar]
- Madhani, H. D., C. A. Styles and G. R. Fink, 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 28: 673–684. [DOI] [PubMed] [Google Scholar]
- Mösch, H.-U., R. L. Roberts and G. R. Fink, 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 28: 5352–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch, H.-U., E. Kubler, S. Krappmann, G. R. Fink and G. H. Braus, 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10: 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen, L. J., and F. R. Cross, 1998. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J. Biol. Chem. 25: 25089–25097. [DOI] [PubMed] [Google Scholar]
- Pagel, P., H.-W. Mewes and D. Frishman, 2004. Conservation of protein-protein interactions–lessons from ascomycota. Trends Genet. 20: 72–76. [DOI] [PubMed] [Google Scholar]
- Palecek, S. P., A. S. Parikh and S. J. Kron, 2000. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics 156: 1005–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek, S. P., A. S. Parikh and S. J. Kron, 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148: 893–907. [DOI] [PubMed] [Google Scholar]
- Pan, X., and J. Heitman, 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., and J. Heitman, 2000. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol. Cell. Biol. 20: 8364–8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., and J. Heitman, 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22: 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S. H., S. S. Koh, J. H. Chun, H. J. Hwang and H. S. Kang, 1999. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 2044–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, T. B., and G. R. Fink, 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291: 878–881. [DOI] [PubMed] [Google Scholar]
- Roberts, R. L., and G. R. Fink, 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8: 2974–2985. [DOI] [PubMed] [Google Scholar]
- Robertson, L. S., and G. R. Fink, 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95: 13783–13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, R. G., 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21: 327–335. [PubMed] [Google Scholar]
- Rupp, S., E. Summers, H. J. Lo, H. Madhani and G. R. Fink, 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18: 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sherman, F., G. R. Fink and J. Hicks, 1991 Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sidorova, J., and L. Breeden, 1999. The MSN1 and NHP6A genes suppress SWI6 defects in Saccharomyces cerevisiae. Genetics 151: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone, W. M., A. L. Johnson, G. R. Banks, J. H. Toyn, D. Stuart et al., 1995. Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J. 14: 5824–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dyk, D., G. Hansson, I. S. Pretorius and F. F. Bauer, 2003. Cellular differentiation in response to nutrient availability: the repressor of meiosis, Rme1p, positively regulates invasive growth in Saccharomyces cerevisiae. Genetics 165: 1045–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen, K. J., G. Derdelinckx, H. Verachtert and F. R. Delvaux, 2003. Yeast flocculation: what brewers should know. Appl. Microbiol. Biotechnol. 61: 197–205. [DOI] [PubMed] [Google Scholar]
- Ward, M. P., C. J. Gimeno, G. R. Fink and S. Garrett, 1995. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol. Cell. Biol. 15: 6854–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, A. L., M. G. Lambrechts and I. S. Pretorius, 1997. MSS11, a novel yeast gene involved in the regulation of starch metabolism. Curr. Genet. 32: 260–266. [DOI] [PubMed] [Google Scholar]
- Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert et al., 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404. [DOI] [PubMed] [Google Scholar]