Abstract
When the cytochrome-mediated mitochondrial electron transport chain of Neurospora crassa is disrupted, an alternative oxidase encoded by the nuclear aod-1 gene is induced. The alternative oxidase donates electrons directly to oxygen from the ubiquininol pool and is insensitive to chemicals such as antimycin A and KCN that affect the standard electron transport chain. To facilitate isolation of mutants affecting regulation of aod-1, a reporter system containing the region upstream of the aod-1 coding sequence fused to the coding sequence of the N. crassa tyrosinase gene (T) was transformed into a strain carrying a null allele of the endogenous T gene. In the resulting reporter strain, growth in the presence of chloramphenicol, an inhibitor of mitochondrial translation whose action decreases the level of mitochondrial translation products resulting in impaired cytochrome-mediated respiration, caused induction of both alternative oxidase and tyrosinase. Conidia from the reporter strain were mutagenized, plated on medium containing chloramphenicol, and colonies that did not express tyrosinase were identified as potential regulatory mutants. After further characterization, 15 strains were found that were unable to induce both the reporter and the alternative oxidase. Complementation analysis revealed that four novel loci involved in aod-1 regulation had been isolated. The discovery that several genes are required for regulation of aod-1 suggests the existence of a complex pathway for signaling from the mitochondria to the nucleus and/or for expression of the gene.
RESPIRATION in higher plants, apicomplexan parasites, some algal species, and many fungi can occur via the normal cytochrome-mediated pathway of electron transport or an alternative oxidase that accepts electrons from the ubiquinonol pool and donates them directly to oxygen. (Henry and Nyns 1975; Lambers 1982; McIntosh 1994; Vanlerberghe and McIntosh 1997; Siedow and Umbach 2000; Joseph-Horne et al. 2001; Roberts et al. 2004). Transfer of electrons via alternative oxidase bypasses two sites of proton pumping at respiratory complexes III and IV, which causes energy to be released as heat. The alternative oxidase is insensitive to classic inhibitors of the cytochrome-mediated pathway such as antimycin A and KCN, but is specifically inhibited by salicylhydroxamic acid (SHAM).
In plants, alternative oxidase can be induced by various stresses or developmental programming (Kearns et al. 1992; Whelan et al. 1996; Finnegan et al. 1997; Saisho et al. 1997; Vanlerberghe and McIntosh 1997; Considine et al. 2001; Djajanegara et al. 2002; Karpova et al. 2002). The enzyme has been shown to be constitutively present in the fungus Gaeumannomyces graminis (Joseph-Horne et al. 1998), and reporter expression directed by the promoter of the alternative oxidase encoding Aox1a gene in Candida albicans was also constitutive (Huh and Kang 2001). In N. crassa, alternative oxidase activity is not present under normal conditions but can be induced by mutations or chemicals that inhibit the cytochrome pathway (Lambowitz and Slayman 1971; Lambowitz et al. 1972, 1989; Li et al. 1996; Tanton et al. 2003). Nuclear run-on assays in Sauromatum guttatum (Rhoads and McIntosh 1992), Magnaporthe grisea (Yukioka et al. 1998), Trypanosoma brucei (Chaudhuri et al. 2002), and N. crassa (Tanton et al. 2003) revealed that alternative oxidase was constitutively transcribed at a low basal level even when the protein and enzyme activity were not detectable. Chemical induction caused an increase in transcription in M. grisea and N. crassa, but not in S. guttatum or T. brucei, suggesting that different organisms may regulate alternative oxidase expression by different means. Active degradation of alternative oxidase mRNA by a de novo synthesized degradation factor was shown to occur in M. grisea and T. brucei (Yukioka et al. 1998; Chaudhuri et al. 2002). The observation that uninduced cultures of N. crassa sometimes contain significant levels of aod-1 mRNA, but not protein (Tanton et al. 2003), suggests that translational control may also be involved in alternative oxidase expression.
Very little is known about the gene products responsible for alternative oxidase production and regulation in any organism. In N. crassa, two nuclear genes are known to be necessary for respiraton via the alternative pathway (Edwards et al. 1976; Bertrand et al. 1983). The alternative oxidase protein is encoded by the aod-1 gene, while the aod-2 gene product is believed to regulate alternative oxidase expression, although the identity and specific function of this gene have yet to be determined (Bertrand et al. 1983; Lambowitz et al. 1989; Li et al. 1996). A third gene, aod-3, encoding a second alternative oxidase has been identified, but no evidence of its expression was observed under any condition tested (Tanton et al. 2003). To identify other components involved in alternative oxidase regulation, we searched for new alternative oxidase regulatory mutants using a reporter system and have identified four new genes. In addition, our screen led to the isolation of a mutant that is resistant to chloramphenicol.
MATERIALS AND METHODS
Growth of N. crassa and induction of alternative oxidase:
Strains of N. crassa used are listed in Table 1 and were grown as described by Davis and De Serres (1970). To induce alternative oxidase, cultures were grown in the presence of chloramphenicol (2 mg/ml) or antimycin A (0.5 μg/ml). At the concentrations used, chloramphenicol reduces, but does not eliminate, mitochondrial translation. This results in deficiencies of oxidative phosphorylation complexes containing mitochondrially translated components and induction of alternative oxidase (Lambowitz and Slayman 1971; Li et al. 1996; Tanton et al. 2003). Antimycin A directly inhibits complex III and virtually eliminates cytochrome-mediated electron transport that leads to alternative oxidase induction (Lambowitz and Slayman 1971). Liquid cultures grown in the absence of these inhibitors (referred to as “noninduced”) were incubated with shaking at 30° for 12–18 hr, while cultures induced by growth in the presence of chloramphenicol or antimycin A (referred to as “induced”) were grown for 18–24 hr or 30–48 hr, respectively.
TABLE 1.
Strains used in this study
| Straina | Origin or source | Genotypeb | Original mutant isolation name |
|---|---|---|---|
| 2-195 | Mutagenesis of T11-76 | aod-4, T, al-2, a + pBATc | E1 |
| 4-294 | Mutagenesis of T11-76 | chl-1, T, al-2, a + pBAT | E2 |
| 5-34 | Mutagenesis of T11-76 | E4, T, al-2, a + pBAT | E4 |
| 5-14 | Mutagenesis of T11-76 | aod-2, T, al-2, a + pBAT | E3 |
| 6-280 | Mutagenesis of T11-76 | E7, T, al-2, a + pBAT | E7 |
| 7-64 | Mutagenesis of T11-76 | aod-7, T, al-2, a + pBAT | E15 |
| 763 | FGSC | nic-1, A | |
| 7064 | H. Bertrand | aod-2, nic-1, al-2, a | |
| 7207 | H. Bertrand | aod-1, pan-2, A | |
| 7263 | FGSC | helper + ad-2,am132, inl, inv, mei-2, A | |
| 7264 | FGSC | helper + trp-4, am132, inl, inv, mei-2, A | |
| EL62-2 | L2-62 × 7264 | aod-6, trp-4, A | E12 |
| EN195-109 | NCN233 × 2-195 | aod-4, pan-2, a | E1 |
| EN294-46 | NCN246 × 4-294 | chl-1, pyr-6, A | E2 |
| EN14-34 | 7263 × 5-14 | aod-2, ad-2, al-2, A + pBAT | E3 |
| L1-6 | Mutagenesis of T11-76 | E5, T, al-2, a + pBAT | E5 |
| L1-13 | Mutagenesis of T11-76 | E6, T, al-2, a + pBAT | E6 |
| L2-25 | Mutagenesis of T11-76 | aod-4, T, al-2, a + pBAT | E8 |
| L2-37 | Mutagenesis of T11-76 | aod-4, T, al-2, a + pBAT | E9 |
| L2-40 | Mutagenesis of T11-76 | aod-5, T, al-2, a + pBAT | E10 |
| L2-61 | Mutagenesis of T11-76 | aod-4, T, al-2, a + pBAT | E11 |
| L2-62 | Mutagenesis of T11-76 | aod-6, T, al-2, a + pBAT | E12 |
| L2-64 | Mutagenesis of T11-76 | aod-4, T, al-2, a + pBAT | E13 |
| L2-67 | Mutagenesis of T11-76 | aod-4, T, al-2, a + pBAT | E14 |
| NCN233 | Nargang lab | pan-2, A | |
| NCN246 | Nargang lab | pyr-6, A | |
| NL61-130 | 763 × L2-61 | aod-4, nic-1, A | E11 |
| PL40-23 | L2-40 × NCN233 | aod-5, pan-2, A | E10 |
| PN64-69 | 7-64 × NCN233 | aod-7, pan-2, a | E15 |
| T11-76 | This study | T, al-2, a + reporter | |
| T1P11 | S. Free | T, al-2, a |
FGSC, Fungal Genetics Stock Center.
Strains used only for complementation or preliminary mapping studies are not shown.
The aod-4, aod-5, aod-6, aod-7, and chl-1 gene designations have been assigned on the basis of the data described in this article.
The “pBAT” designation means that the strain carries the integrated reporter system as constructed on plasmid pBAT (see Figure 1).
Construction of the reporter strain:
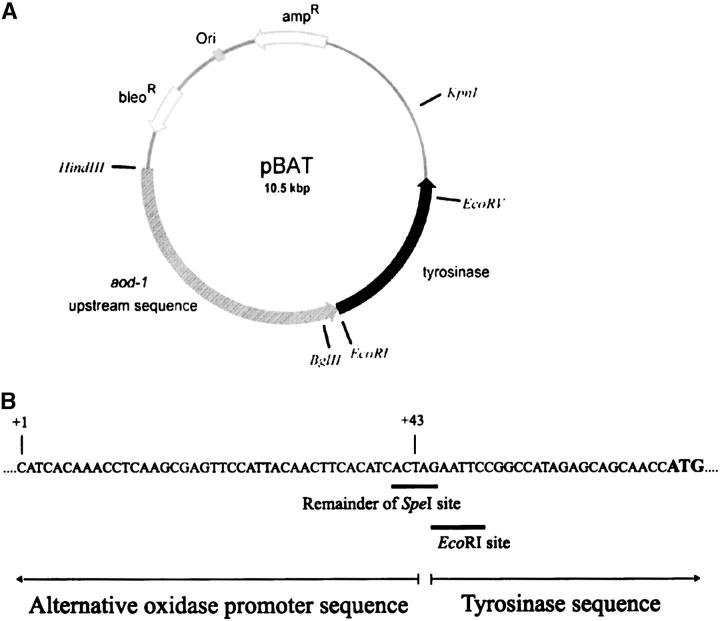
The use of the N. crassa tyrosinase enzyme as a general reporter was described previously (Kothe et al. 1993). Colonies expressing the enzyme turn brown when overlaid with a solution of tyrosine and Triton X-100. To use the system as a reporter for alternative oxidase, a 3347-bp HindIII/SpeI fragment spanning positions −3304 to +43 of the alternative oxidase upstream region [transcription start site defined as +1 (Li et al. 1996)] was isolated and the ends were blunted by end filling. The fragment was cloned into the filled EcoRI sites of the plasmid pTYR103 (Kothe et al. 1993), immediately upstream of the tyrosinase gene (gene symbol, T). The end filling and ligation restored a HindIII site at the EcoRI/HindIII junction and an EcoRI site at the EcoRI/SpeI junction. An XbaI fragment containing a bleomycin-resistance marker from plasmid pAB520 (Austin et al. 1990) was then cloned into the XbaI site of pTYR103, generating the reporter construct pBAT (Figure 1).
Figure 1.—
Alternative oxidase reporter construct. (A) Plasmid pBAT. The sequence upstream of the aod-1 structural gene (–3303 to +43) is indicated by the striped arrow and the T gene (tyrosinase) sequence is indicated by the solid arrow. The positions of the bleomycin-resistance cassette, the ampicillin-resistance cassette, and the bacterial ori sequence are also shown. Unique restriction sites on the plasmid are indicated. (B) Sequence of the fusion point of the 3′ end of the 3.3-kb aod-1 upstream region with the 5′ end of the tyrosinase sequence. The transcription initiation site (+1) of aod-1 (Li et al. 1996) up to nucleotide +43 is shown. The ATG start codon of the T gene sequence is in boldface type. The positions of the blunted SpeI site and the restored EcoRI site at the fusion point are shown by the solid bars below the sequence. In the endogenous aod-1 gene, the distance between the +1 transcription start site and the ATG start codon is 54 bp.
The tyrosinase null strain T1P11 (Fuentes et al. 1994) was confirmed to carry a tyrosinase gene inactivated by repeat induced point mutation (RIP; Selker 1990) by sequencing an appropriate PCR-amplified product from the strain (data not shown). T1P11 was transformed by electroporation (Tanton et al. 2003) with pBAT that had been linearized by digestion with KpnI. Transformants were grown for 3 days at 30° with 1 μg/ml bleomycin and 0.5 mg/ml caffeine present in both the top agar and the plates. Bleomycin-resistant colonies were picked to slants and allowed to conidiate. To promote the isolation of homokaryotic isolates, conidia were streaked onto plates containing bleomycin and caffeine, and single colonies were picked. Transformed strains were analyzed by Southern blot to determine the copy number of integrated pBAT sequences. Strain T11-76 contained one copy of pBAT (data not shown) and was selected as the reporter strain.
Tyrosinase plate assay:
The tyrosinase plate assay was as described previously (Kothe et al. 1993), with modifications specific for the alternative oxidase system. For testing individual strains, conidia were spread onto plates at a density of 50–100 colonies per plate. Uninduced controls were grown on minimal medium, plus appropriate supplements, at 30° for 2 days. Plates containing chloramphenicol (2 mg/ml) were used to induce reporter expression and required an additional day of growth at 30° since the inhibitor reduces the growth rate of N. crassa cells. The strongest formation of the brown pigment occurred when 5–10 ml of a freshly prepared solution of 10 mm tyrosine, 0.1% Triton X-100 was overlaid onto the plates containing colonies and allowed to incubate at 30° for several hours. The use of the assay to select mutants is described below.
EMS mutagenesis:
Conidia from the reporter strain T11-76 were treated with ethyl methanesulfonate (EMS) as described previously (Davis and De Serres 1970) with modifications. Conidia that were 7–10 days old were harvested in sterile distilled water and filtered through cheesecloth to remove hyphal fragments. The conidia were washed once in sterile distilled water and resuspended in 0.067 m phosphate buffer (pH 7.0) to a concentration of 2 × 107 conidia/ml. EMS was added to a final concentration of 0.1, 0.15, or 0.3 m. Conidia were incubated with EMS at 25° for 5 hr with shaking and then washed twice with sterile 1× Vogel's medium (Davis and De Serres 1970). In mutant isolation method 1, mutagenized conidia were diluted in 1× Vogel's and spread directly onto plates containing chloramphenicol. Plates were incubated at 30° for 4 days whereupon colonies were screened using the tyrosinase plate assay. In method 2, 1.8 × 109 EMS-treated conidia were placed in a 2-liter baffled flask containing 500 ml of medium plus antimycin A and incubated at 30° with shaking for 4 days. Every 24 hr the liquid cultures were filtered through sterile nylon stocking to remove any growing conidia. The rationale of this method was that mutants unable to produce alternative oxidase would not grow in the presence of antimycin A, which inhibits the cytochrome-mediated pathway, so that these cultures should become enriched for alternative oxidase mutants as the growing cells were removed. After 4 days, the remaining conidia were collected, plated on chloramphenicol-containing plates, and incubated at 30° for 4 days, and the colonies were screened by the tyrosinase plate assay. For both methods 1 and 2, colonies that remained white in a background of brown colonies in the tyrosinase plate assay were picked to slants and analyzed further.
Conidial DNA preparation and PCR:
A “small pea-sized” clump of conidia was mixed with 100 μl of cracking buffer [1 m sorbitol, 20 mm EDTA, 3 mg/ml lysing enzyme (Sigma, Oakville, ON)]. The mixture was incubated at 37° for 10 min and then spun in a microcentrifuge at 14,000 rpm for 10 min at room temperature. The pellet was washed with 500 μl 1 m sorbitol, 20 mm EDTA, resuspended in 100 μl sterile distilled water, subjected to a standard glassmilk purification protocol (Geneclean II kit, Q Biogene, Carlsbad, CA), and eluted in 50 μl of sterile distilled water. For a standard PCR reaction, 10 μl of the preparation was used. A 1.3-kb region of the endogenous alternative oxidase upstream sequence [from −1216 to +111 of the aod-1 gene (Li et al. 1996)] was amplified with primers FNA88 (5′ CCTTCCCTCCAGAAGGCTTTCTGCG 3′) and ao5 (5′ TTAGTTGGGCCGCTTGTCC 3′). The ectopically integrated tyrosinase reporter construct was amplified using primers FNA88 and a tyrosinase gene-specific primer, ADE19 (5′ GGAGGTAGAGATTGAACTGCTCCGG 3′), which generated a 1.6-kb PCR product (from −1216 of the aod-1 upstream region to +402 of the T gene) if the reporter construct was present.
RNA isolation:
Mycelium was harvested by vacuum filtration and a portion of the pad was immediately wrapped in aluminum foil, frozen in liquid nitrogen, and stored at −80° until needed. Pieces of ∼100 mg were ground in liquid nitrogen using a mortar and pestle. RNA was isolated using either an RNeasy Plant Mini Kit (QIAGEN, Mississauga, ON) or hot phenol extraction as described previously (Verwoerd et al. 1989) with the following modifications. After grinding in liquid nitrogen, the mycelial powder was mixed with 500 μl of fresh extraction mix (0.1 m LiCl, 100 mm Tris Cl, pH 8.0, 10 mm EDTA, 1% SDS, 50% phenol) at 65°. This solution was mixed with 250 μl of 24:1 chloroform:isoamylalcohol and then centrifuged in a microcentrifuge for 5 min at 4°. The aqueous phase was taken to a clean tube and mixed with an equal volume of 4 m LiCl. This mixture was cooled at −20° for 1 hr and then centrifuged for 15 min at 4°. The pellet was dissolved in 250 μl diethylpyrocarbonate-treated distilled water and mixed with 25 μl 3 m sodium acetate, pH 5.2, and 550 μl cold 95% ethanol. RNA was allowed to precipitate at −20° for 15 min whereupon the sample was spun at 14,000 rpm for 15 min at 4°. The pellet was washed with 50 μl 70% ethanol and then dissolved in 50 μl diethylpyrocarbonate-treated water.
DNA sequencing and analysis:
DNA sequence was obtained by the Molecular Biology Service Unit, Department of Biological Sciences, University of Alberta. Labeled products obtained using a DyeNamic Sequencing Kit system (Amersham Pharmacia Biotech, San Francisco) were analyzed with a model 373 Stretch Sequencer Separation system (Applied Biosystems, Foster City, CA), and sequence profiles were generated by Applied Biosystems sequence analysis software (version 3.4.1). Sequences were analyzed using DNAMAN (version 4.13) software (Lynnon Biosoft, Vaudreuil, PQ).
Antisera:
The generation of antiserum to N. crassa AOD-1 was described previously (Tanton et al. 2003). Antiserum to Tom70 was a generous gift from W. Neupert.
Other techniques:
Standard procedures were used for growth and transformation of Escherichia coli cells, DNA modification and isolation, Southern analysis, labeling of DNA with random primers, PCR, Northern analysis, SDS-PAGE, Western blotting, and immunodetection (Sambrook and Russell 2001). Previously described protocols were used for isolation of mitochondria (Pfanner and Neupert 1985) and generation of cytochrome spectra (Bertrand and Pittenger 1969). Assay of the induction of alternative oxidase activity in N. crassa liquid cultures was performed as described (Tanton et al. 2003) using a Yellow Springs Instruments (Yellow Springs, OH) oxygen monitor. Oxygen utilization that was insensitive to KCN and sensitive to SHAM was indicative of the presence of the alternative pathway. Manufacturer's instructions were followed for the isolation of plasmid or cosmid DNA from E. coli cells using QIAGEN Miniprep or Midi Kits and determination of protein concentration with Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA).
RESULTS
Creation of the tyrosinase reporter system:
Previous attempts to isolate mutations affecting alternative respiration used the inability of strains lacking alternative oxidase to grow in the presence of antimycin A as the sole basis of selection. This approach resulted in the isolation of 20 mutations in the aod-1 structural gene and 4 mutations in the aod-2 regulatory gene (Edwards et al. 1976; Bertrand et al. 1983). To avoid the apparent bias toward isolation of mutants in the structural gene and enhance the isolation of regulatory mutations, we developed a reporter gene system (Figure 1) consisting of 3.3 kb of sequence upstream of the aod-1 structural gene fused to the coding sequence of tyrosinase, which has previously been used as a reporter in N. crassa (Kothe et al. 1993). The fusion was carried on the plasmid pBAT, which also contained a bleomycin-resistance gene for selection of transformants in N. crassa (Austin et al. 1990). The construct was transformed into the tyrosinase null strain T1P11 (Fuentes et al. 1994). Transformant T11-76, which contained a single inserted copy of the reporter (data not shown), was chosen for further work.
To test the efficacy of the reporter system, T1P11 and T11-76 were examined by the tyrosinase plate assay (see materials and methods). Conidia from both strains were spread onto plates containing chloramphenicol to induce alternative oxidase and also on plates without chloramphenicol as a control. Colonies of strain T1P11 remained white under both growth conditions. Colonies of the T11-76 reporter strain, grown on noninducing medium also remained white. However, T11-76 colonies grown on chloramphenicol-containing medium began to turn brown within 2–4 hr after application of the tyrosinase solution (Figure 2A). Thus, the reporter tyrosinase gene in T11-76 was expressed in a fashion comparable to the aod-1 gene, which is induced by growth in the presence of chloramphenicol.
Figure 2.—
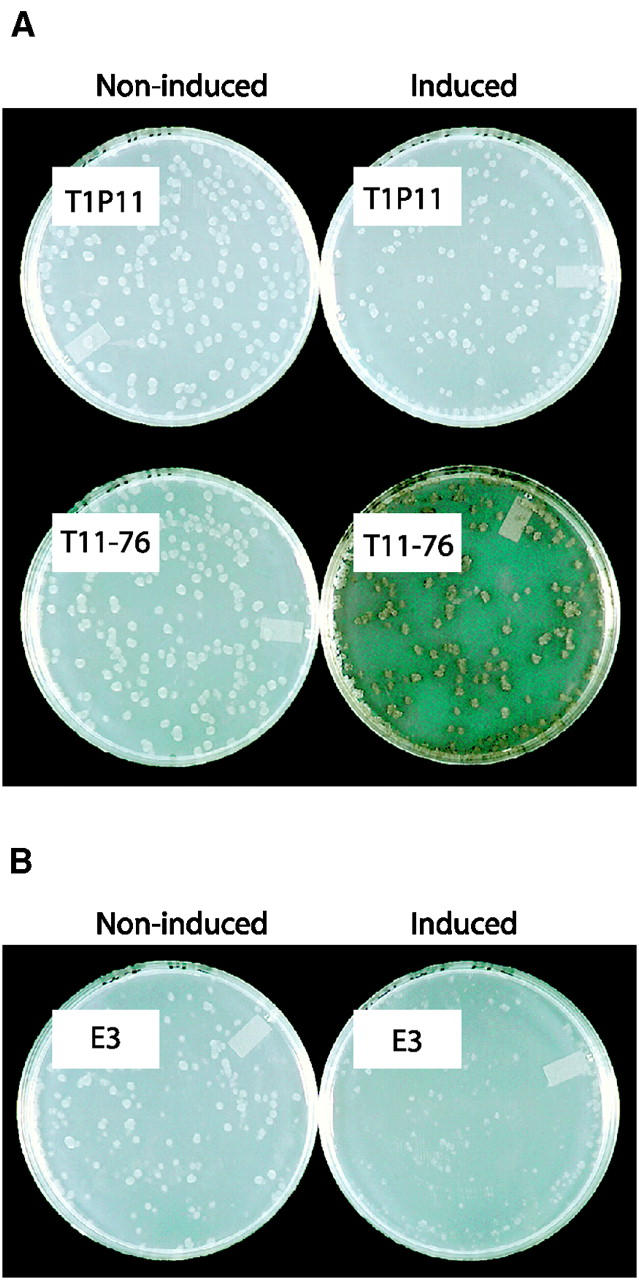
Tyrosinase plate assay. (A) Parent strain T1P11 and the reporter strain T11-76. Conidia from each strain were harvested and spread on both noninducing and chloramphenicol-containing plates. Conidia on noninducing plates were allowed to grow for 2 days at 30° while conidia on chloramphenicol-containing plates were allowed to grow for 3 days. Tyrosine solution was then added and pictures were taken after incubation with the solution for 24 hr. (B) As for A, but showing mutant isolate E3.
Isolation of alternative oxidase regulatory mutants:
We predicted that a mutation in any gene involved in regulating alternative oxidase via transcription at the aod-1 promoter or via an effect on a signal transduction component would also affect expression of the reporter gene. Two general types of mutations were possible. The first would inactivate negative control components and result in constitutive reporter and alternative oxidase expression so that affected colonies would turn brown when grown under both inducing and noninducing conditions. However, it would be impossible to discern such mutations against a large background of mutations affecting the biogenesis of the cytochrome-mediated electron transport chain, which would also result in constitutive aod-1 and reporter gene expression. The second type of expected mutations would inactivate components necessary for both alternative oxidase and reporter gene expression and would cause colonies to remain white even when grown under aod-1 inducing conditions. We decided to focus on the latter mutants to increase the likelihood of specifically obtaining alternative oxidase regulatory mutants.
Two schemes were used to screen for mutants unable to induce expression of tyrosinase in the reporter strain T11-76 as described in materials and methods. Approximately 130,000 colonies from method 1 were screened directly by the tyrosinase plate assay and 1583 colonies that remained white during the assay were picked to slants (Table 2). Conidia from these isolates were spread onto chloramphenicol-containing plates for individual rescreening using the tyrosinase plate assay. Of the original strains picked, 146 remained white when rescreened as shown for one such mutant (E3) in Figure 2B. For the filtration enrichment approach (method 2), 1.8 × 109 mutagenized conidia were inoculated into liquid medium containing antimycin A (Table 2). Following 4 days of growth with daily filtration, the remaining cells were plated to chloramphenicol-containing plates, and ∼7500 colonies were formed. The tyrosinase plate assay revealed 116 colonies that remained white. When picked to slants and retested using the plate assay, 42 strains were chosen as potential alternative oxidase regulatory mutants.
TABLE 2.
Mutants isolated by EMS mutagenesis
| Method | No. of colonies screened |
No. picked |
No. characterized by respiration | Mutations isolated |
|---|---|---|---|---|
| 1 | 130,000 | 1583 | 146 | E1, E2, E3, E4, E7, E15 |
| 2 | 1.8 × 109, 7500a | 116 | 42 | E5, E6, E8, E9, E10, E11, E12, E13, E14 |
The first number indicates the total number of conidia treated with EMS and inoculated into antimycin A-containing liquid medium. The second number indicates the number of colonies that grew when the conidia remaining after filtration enrichment were plated.
To rule out the possibility that colonies chosen from either method remained white for undesired reasons, we took advantage of the fact that the endogenous aod-1+ gene was intact in the reporter strain. Thus, the mutant strains were examined for their ability to induce bona fide alternative oxidase activity by assaying for the presence of KCN-insensitive respiration following growth in liquid medium containing chloramphenicol. This screen revealed 15 mutant strains (named E1–E15) that were unable to induce both tyrosinase and alternative oxidase activities; 6 from method 1 and 9 from method 2. We chose 11 of these isolates for further analysis (E1–E3 and E8–E15). The remaining 4 strains were found to be leaky or inconsistent with respect to the inability to induce alternative oxidase when grown in the presence of chloramphenicol and have not been studied further.
Complementation analysis and gene assignment:
To determine if the regulatory mutations carried by the strains obtained were allelic to each other or to the previously described regulatory gene aod-2, we wished to perform complementation analysis in forced heterokaryons. To develop strains with nutritional requirements, the mutants were crossed to various strains carrying auxotrophic markers. Progeny from the crosses were screened and isolates were identified containing both the regulatory mutation of interest and a suitable auxotrophic marker. A complication in the development of these strains was that the original mutant isolates contained the integrated reporter construct so that the endogenous and ectopic copies of the aod-1 upstream region could serve as substrates for RIP during sexual crosses (Selker 1990). Therefore, the region upstream of the endogenous aod-1 gene in all progeny to be used for further work was amplified by PCR and sequenced to demonstrate that no RIPs were present following the cross (data not shown). This ensured that lack of induction of alternative oxidase in the progeny was due to the regulatory mutation of interest and not inactivation of the endogenous aod-1 promoter. To prevent the need for continual sequencing of aod-1 in progeny from subsequent crosses, isolates were chosen that had lost the reporter construct through random segregation as judged by inability to obtain a fusion-specific PCR product from isolated genomic DNA.
Once mutant strains with appropriate nutritional markers were isolated, forced heterokaryons were established and tested for their ability to induce alternative oxidase activity when grown in minimal medium in the presence of chloramphenicol. This analysis showed that we had isolated four new complementation groups involved in regulating alternative oxidase activity (Table 3). These complementation groups have been assigned the gene names aod-4, aod-5, aod-6, and aod-7 (Table 4). (An additional complementation group was assigned the gene name chl-1; see below.) Mutations in the aod-4 complementation group were the most frequently isolated, with mutants obtained from both the direct plating method (E1) and the filter enrichment technique (E8, E9, E11, E13, and E14). Each of the other complementation groups contained a single mutant allele. Strains carrying mutation E3 failed to complement the previously identified regulatory mutant, aod-2 (Bertrand et al. 1983). Furthermore, preliminary mapping studies revealed that the E3 mutation was found on linkage group II, near the arg-5 marker as described previously for aod-2. Thus, E3 is likely a new allele of aod-2. All of our mutant strains complemented aod-1 (Table 3), confirming that in each strain the endogenous aod-1 gene is functional. The fact that each mutation was able to complement mutations from other groups suggested that none of the mutations was dominant. In crosses of the mutant strains to various auxotrophic and mapping strains, the regulatory mutations were all inherited in a Mendelian fashion, demonstrating that the mutations are in nuclear encoded genes.
TABLE 3.
Complementation analysis using heterokaryotic strains
| Mutant strains |
E1 | E2 | E3 | E8 | E9 | E10 | E11 | E12 | E13 | E14 | E15 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| aod-1 | +a | + | + | + | + | + | + | + | + | + | + |
| aod-2 | + | + | − | + | + | + | + | + | + | + | + |
| E1 | + | + | − | − | ND | − | + | − | − | + | |
| E2 | + | + | + | + | + | + | + | + | + | ||
| E3 | + | + | ND | + | + | + | + | + | |||
| E8 | − | ND | − | + | − | − | + | ||||
| E9 | + | − | + | − | − | + | |||||
| E10 | + | + | ND | ND | + | ||||||
| E11 | + | − | − | + | |||||||
| E12 | + | + | + | ||||||||
| E13 | − | + | |||||||||
| E14 | ND |
Complementation is indicated by a plus, lack of complementation is indicated by a minus, and ND indicates no data. (Once it was discovered that mutations E1, E8, E9, E11, and E14 were allelic and that mutations aod-2 and E3 were allelic, not all combinations with each of these mutations were analyzed.)
TABLE 4.
Assigned gene names and strains used in analysis of alternative oxidase mutants
| Original mutation | Assigned gene name |
Strains used for further analysis |
|---|---|---|
| E1,a E8, E9, E11, E13, E14 | aod-4 | EN195-109b NL61-130c |
| E10 | aod-5 | PL40-23 |
| E12 | aod-6 | EL62-2, EL62-25 |
| E15a | aod-7 | PN64-69 |
| E3a | Allelic to aod-2 | EN14-34 |
| E2a | chl-1 | EN294-46 |
| E4,a E5, E6, E7a | Not characterized |
Isolated using selection method 1. All other mutants were obtained by method 2.
Contains aod-4 allele E1.
Contains aod-4 allele E11.
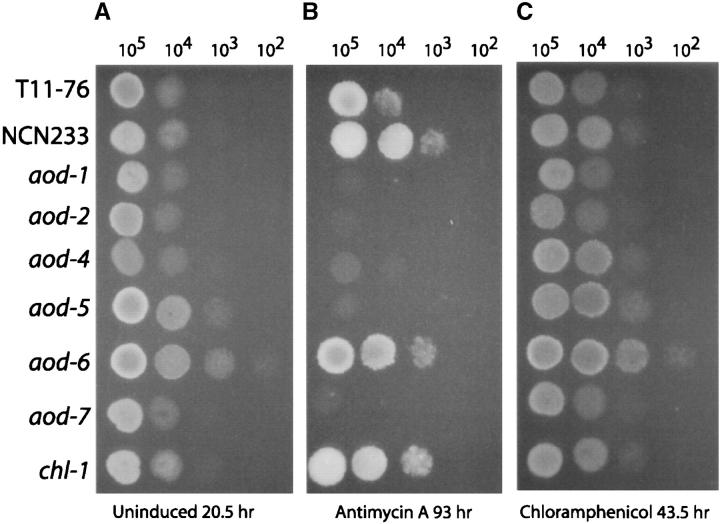
To address the possibility that the mutant strains did not induce alternative oxidase under our selection schemes because they had become resistant to chloramphenicol, each mutant strain was grown in the presence of antimycin A, which blocks electron transport through complex III, resulting in induction of alternative oxidase. Wild-type strains grow at slightly reduced rates in antimycin A but known alternative oxidase mutants have an extremely slow growth rate or fail to grow at all in the presence of the drug. Of the mutants isolated here, aod-4, aod-5, and aod-7 failed to grow in the presence of antimycin A, as did the previously isolated aod-1 and aod-2 mutants (Figure 3, A and B). Although the aod-6 mutant did grow in the presence of antimycin A it was still considered an alternative oxidase mutant because it contained very low levels of KCN-insensitive respiration under this condition as compared to an aod+ control strain (T11-76, Figure 4). Mutant E2 was also able to grow in the presence of antimycin A, but contained high levels of KCN-insensitive respiration when grown in this inhibitor. Thus, the inability of this mutant to induce alternative oxidase during growth in chloramphenicol probably reflects a resistance to the drug so that this strain was considered to be a chloramphenicol-resistant mutant, rather than an alternative oxidase regulatory mutant. E2 was the sole chloramphenicol-resistant mutant isolated and has been named chl-1 (Table 4; Figure 4). Taken together, these data demonstrate that we have identified four new genes that encode factors necessary for efficient expression of alternative oxidase. The respiratory behavior of a mutant from each of the complementation groups is shown in Figure 4.
Figure 3.—
Plate growth assays. Plates containing media with appropriate nutritional supplements (uninduced, A), antimycin A (B), or chloramphenicol (C) were spotted with conidia from the indicated strains (number spotted indicated above each column) in 10 μl sterile water and incubated at 30°.
Figure 4.—
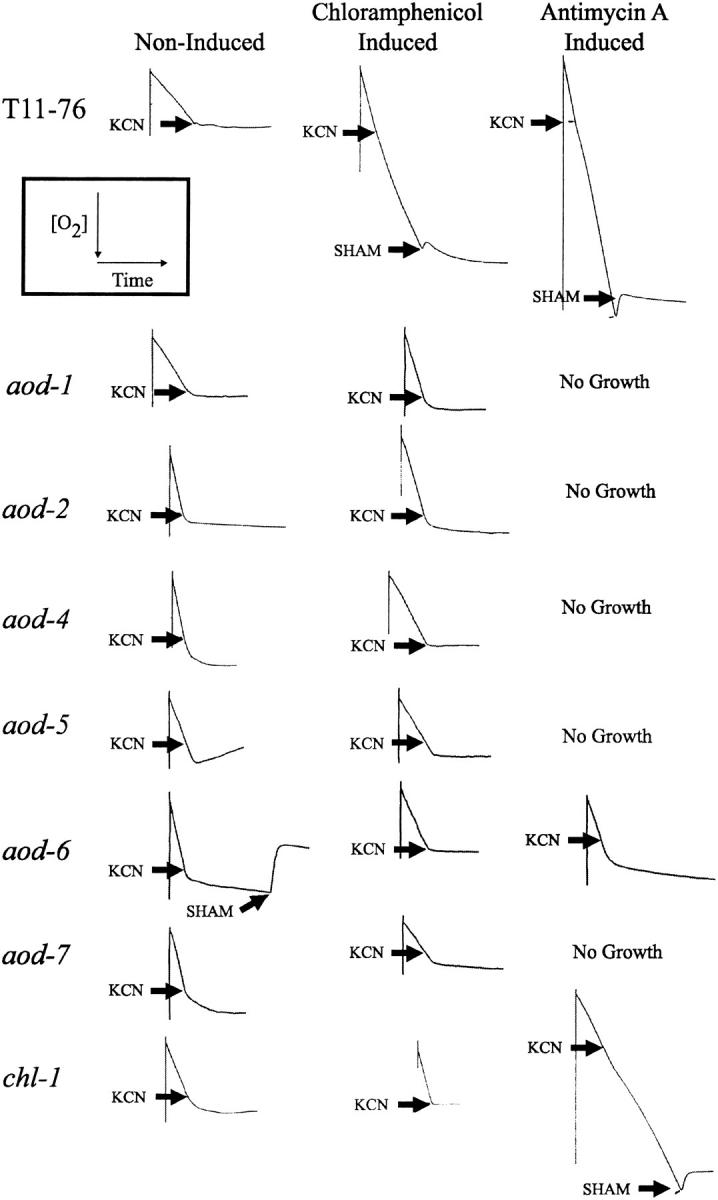
Respiratory behavior of regulatory mutants. Respiration tracings of cultures grown under noninducing conditions, in the presence of chloramphenicol, or in the presence of antimycin A show the consumption of oxygen over time (inset, upper left). The mutants isolated in this study were compared with the existing aod-1 and aod-2 alternative oxidase mutants, as well as the reporter strain T11-76 as a control. KCN and SHAM were added at the points indicated by the arrows. Strains aod-1, aod-2, aod-4, aod-5, and aod-7 did not grow in the presence of antimycin A.
When strains were grown on noninducing medium, no gross differences in growth rate were observed among strains containing the aod mutations or the chl-1 mutation and wild type (Figure 3A), although the aod-4 strain appears to have a slightly reduced growth rate. The growth of aod-6 on plates containing antimycin A is difficult to explain considering its low level of KCN-insensitive respiration when grown in liquid medium containing this inhibitor (Figure 4). In fact, in liquid cultures containing antimycin A, aod-6 did grow significantly slower than wild type. From similar inocula, a wild-type control strain produced 3 gm fresh weight of mycelium in 25 hr whereas 51 hr were required to produce a similar amount from aod-6. Little difference in growth on chloramphenicol-containing medium was observed for any strain (Figure 3C). This was surprising for the chl-1 strain since it was expected to grow somewhat faster than other strains in the presence of the drug. The lack of a differential effect of chloramphenicol on growth rates of the strains could be due to the fact that the drug only partially inhibits mitochondrial translation at the concentration used and/or because mitochondrially encoded components of complex I, which precedes the branch point for alternative oxidase and cytochrome-mediated respiration, are also affected by the action of the drug.
Levels of aod-1 mRNA and AOD-1 protein in the regulatory mutants:
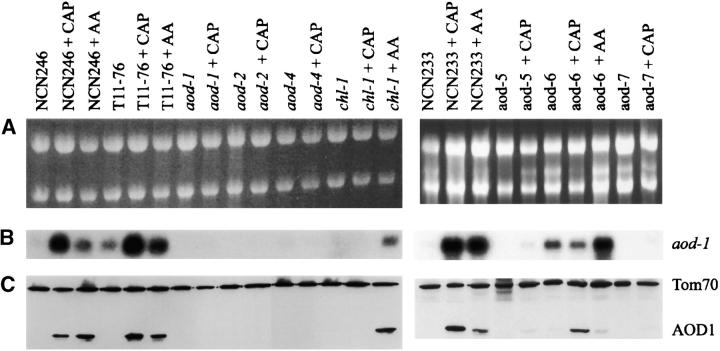
Mutant strains were examined for expression of aod-1 mRNA and protein and compared to the previously isolated aod-1 and aod-2 mutants as well as wild-type controls (Figure 5). As expected from previous studies, the amount of aod-1 mRNA present in wild-type cultures increased upon growth in medium containing chloramphenicol or antimycin A (Li et al. 1996; Tanton et al. 2003). This increase correlated with the appearance of AOD1 protein. Unexpectedly, we found that the tyrosinase mutant strain T1P11 (data not shown) and the reporter strain T11-76, which was derived from T1P11, differed from other strains that are wild type with respect to respiration, because they contain significant levels of aod-1 transcript under noninducing conditions (Figure 5). However, even though the mRNA was present, no AOD-1 protein could be detected in these noninduced cultures (Figure 5), nor was there KCN-insensitive respiration (Figure 4), in agreement with our previous suggestion that alternative oxidase may also be regulated at the level of translation in N. crassa (Tanton et al. 2003). We analyzed eight random ascospore progeny containing a nonfunctional T gene that were derived from a cross between T1P11 and a strain (NCN233) with a wild-type respiratory phenotype. Five of these progeny contained aod-1 mRNA under noninducing conditions and three did not. Thus, the presence of aod-1 mRNA in noninduced cultures did not segregate with the mutant T gene and an unknown factor is responsible for the accumulation of aod-1 mRNA in strains T1P11 and T1176. It is unlikely that this factor had any effect on our results since our screen was designed for the isolation of mutants with signaling or transcriptional defects. These should be epistatic to any mechanism of translational control.
Figure 5.—
Northern and Western blot analysis of alternative oxidase expression. (A) Ethidium bromide-stained gels of total RNA (5 μg per lane) isolated from the strains indicated and electrophoresed on a 1% gel. Cultures were either uninduced or induced for alternative oxidase by growth in the presence of chloramphenicol (CAP) or antimycin A (AA). Strains NCN247, T11-76, and NCN233 served as controls. (B) The gels in A were blotted to nylon membranes and probed. The probe was a 32P-labeled 1.3-kb cDNA copy of aod-1. The band recognized is 1.3 kb. (C) Mitochondria were isolated from the same cultures from which the RNA was derived, and 25 μg of mitochondrial protein for each sample was subjected to SDS-PAGE. The gels were blotted to nitrocellulose and immunodecorated with antiserum to Tom70 (a 70-kD mitochondrial outer membrane protein) or antiserum against the AOD1 protein (36 kD).
Strains carrying mutations in aod-4, aod-5, and aod-7, as well as the previously isolated aod-1 and aod-2 strains, contained severely reduced levels of transcript and no AOD1 protein when grown under either inducing or noninducing conditions. The aod-1 allele analyzed (aod-1-7) was previously shown to be deficient in aod-1 mRNA under inducing conditions (Li et al. 1996; Tanton et al. 2003). The strain contains a frameshift mutation (Li et al. 1996) and the mRNA produced from the strain is likely subject to nonsense-mediated decay. Since the mutations in the regulatory mutants do not affect the aod-1 structural gene, the inability of the aod-2, aod-4, aod-5, and aod-7 mutants to properly induce alternative oxidase activity must be due to either defects in their ability to produce aod-1 mRNA under inducing conditions or an inability to prevent its rapid degradation prior to translation, although it seems unlikely that the latter type would be isolated via our reporter-based selection scheme. The aod-6 mutant strain contained aod-1 transcript under all conditions and AOD1 protein under alternative oxidase inducing conditions (Figure 5), but exhibited little to no KCN-insensitive respiration when grown in the presence of either chloramphenicol or antimycin A (Figure 4). The aod-6 mutation might affect alternative oxidase maturation or assembly rather than the regulation of expression of the aod-1 transcript, although it is difficult to explain such a mutation being selected by the reporter-based screen. In agreement with the finding that the aod-1 and aod-6 (E12) mutants complemented each other (Table 3), sequence analysis of the aod-1 coding region in the aod-6 mutant (data not shown) revealed no mutations, thus eliminating the possibility of a missense mutation inactivating the enzyme in aod-6. The chl-1 mutant strain contained both aod-1 transcript and protein when grown in the presence of antimycin A, but not when grown in chloramphenicol. Thus, chl-1 does not affect alternative oxidase regulation directly, but rather confers resistance to chloramphenicol.
Cytochrome spectra:
To determine if the isolated mutations had any affect on the cytochrome components of the standard electron transport chain, mitochondrial cytochrome spectra were obtained from representative strains (Figure 6). When grown under noninducing conditions, the aod-4 strain had moderate deficiencies of cytochromes aa3 and b, suggesting that the effects of mutations in this gene may not be limited to alternative oxidase regulation, but may have more global effects on the regulation of respiratory system biogenesis. The cytochrome deficiency of aod-4 may explain its slightly reduced growth rate (Figure 3A). All of the strains except the chl-1 strain, showed reduced levels of cytochrome aa3 and b when grown in the presence of chloramphenicol, since these cytochromes contain components synthesized on mitochondrial ribosomes (Attardi and Schatz 1988). The levels of cytochromes aa3 and b in the chl-1 mutant strain did not change appreciably when the strain was grown in chloramphenicol. Therefore, it seems likely that respiration through the cytochrome-mediated pathway in chl-1 was unaffected by growth in chloramphenicol and alternative oxidase was not induced, in agreement with the observation that the strain contains only cyanide-sensitive respiration when grown in the presence of chloramphenicol (Figure 4). The chl-1 and aod-6 strains were the only mutant strains able to grow in antimycin A, and the cytochrome spectra following growth in this inhibitor are similar to those in the control strain.
Figure 6.—
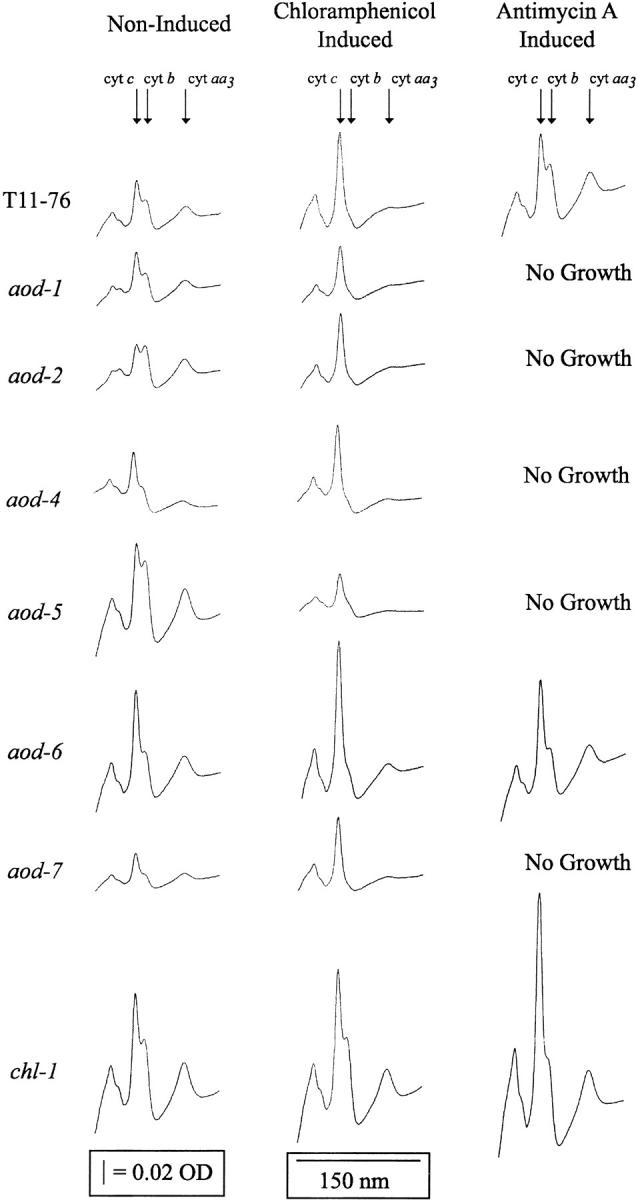
Qualitative cytochrome spectra of the reporter and mutant strains. The cytochrome content of cultures grown under noninducing conditions, in the presence of chloramphenicol, or in the presence of antimycin A was determined by scanning a reduced sample vs. an oxidized reference from 650 to 500 nm. The peaks corresponding to cytochromes aa3 (605 nm), b (560 nm), and c (550 nm) are indicated above the spectrum shown for the T11-76 tracings. Strains aod-1, aod-2, aod-4, aod-5, and aod-7 did not grow in the presence of antimycin A.
Genetic mapping:
The aod-1 and aod-2 genes were previously mapped near the centromeres of linkage groups IV and II, respectively (Bertrand et al. 1983). Preliminary mapping data place the aod-4 gene on the right arm of linkage group V, the aod-5 gene on linkage group VI, and the aod-7 gene on the left arm of linkage group IV. The chl-1 gene was mapped to the right arm of linkage group II between arg-5 and arg-12, near nuc-2 (data not shown). Although our mapping data are preliminary, they demonstrate that three of the newly isolated aod mutants map to different chromosomes and thus affect different genes, in agreement with the complementation analysis. These data also rule out the possibility that intraallelic complementation caused an overestimation of the number of new alternative oxidase regulatory genes identified.
DISCUSSION
Prior to this study, only one gene had been identified as a regulatory gene for alternative oxidase induction in any system—the aod-2 gene of N. crassa. In an effort to find additional genes controlling alternative oxidase production, we developed a scheme for isolation of regulatory mutants. Previous attempts to identify such mutants, using the inability of alternative oxidase-deficient strains to grow in the presence of antimycin A, had shown a bias toward isolation of mutations in the alternative oxidase structural gene (Edwards et al. 1976; Bertrand et al. 1983). Therefore, we developed a reporter strain carrying the upstream region of aod-1 fused to the coding sequence for the enzyme tyrosinase. Growth of this strain in the presence or absence of chloramphenicol clearly demonstrated that expression of the tyrosinase reporter was controlled in a manner similar to aod-1 gene expression.
Two methods were used to generate mutant strains that did not properly induce alternative oxidase. Both methods produced a number of false positive colonies that remained white when rescreened on chloramphenicol-containing plates, but were able to produce KCN-insensitive respiration when grown in the presence of the inhibitor. These false positives could be the result of mutations to the integrated reporter construct itself, mutations affecting factors involved in protyrosinase maturation (Lerch et al. 1982; Kupper et al. 1989), or mutations affecting tyrosine uptake. Four novel regulatory genes have been isolated using our mutagenesis and selection schemes. Multiple isolates were found only for the aod-4 complementation group, suggesting that our screen has not been saturated. Characterization of the gene products of the loci that were mutagenized may provide insight into why the aod-4 complementation group was isolated with a relatively high frequency. The existence of five different regulatory mutants (aod-2, aod-4, aod-5, aod-6, and aod-7) and an unsaturated mutant screen suggest the existence of a relatively complex pathway of alternative oxidase regulation. The most obvious possibilities for the function of the components in such a pathway include products that recognize changes in efficiency of mitochondrial function, signal transduction components, and/or transcription factors. In support of these suggestions, an effect on alternative oxidase expression has been observed in a Candida albicans histidine kinase mutant (Huh and Kang 2001). Studies on Saccharomyces cerevisiae identified a system whereby defects in mitochondrial function are communicated to the nucleus via the retrograde regulation system (Butow and Avadhani 2004). This system utilizes regulatory proteins that include transcription factors, a sensor of mitochondrial dysfunction, and a number of other interacting proteins. Although this system controls carbon and nitrogen metabolism in yeast, related systems may facilitate the regulation of other nuclear genes that respond to mitochondrial dysfunction.
Previous Northern blot analysis had revealed that induction of alternative oxidase activity in wild-type cells by growth in the presence of chloramphenicol or antimycin A caused an increase in the steady-state levels of aod-1 mRNA (Li et al. 1996; Tanton et al. 2003). Nuclear run-on assays showed that noninduced wild-type cultures constitutively expressed aod-1 at a low level, and treatment with antimycin A caused an increase in the amount of transcription (Tanton et al. 2003). Thus, regulation of aod-1 expression in response to inducing treatments occurs through increased transcription of the gene and possibly increased stability of the mRNA. Northern blots of noninduced cultures usually show little to no aod-1 mRNA accumulation, and Western blots of these noninduced cultures show no AOD1 protein (Li et al. 1996; Tanton et al. 2003). The lack of AOD1 protein in noninduced cultures suggests that aod-1 regulation also occurs post-transcriptionally, by reducing the stability of the small amount of constitutively expressed mRNA and/or by preventing its translation. Unexpectedly, we found that noninduced cultures of the T mutant strain T1P11 and the reporter strain T11-76 consistently contained readily detectable steady-state levels of aod-1 mRNA. Accumulation of aod-1 mRNA has been seen occasionally in noninduced cultures of other alternative oxidase wild-type strains (Tanton et al. 2003) but never as consistently as in strains T1P11 and T11-76. Strains T1P11 and T11-76 could have a higher level of constitutive aod-1 transcription than wild type or a reduced rate of degradation that allows higher accumulation of aod-1 mRNA. The possibility that the increased aod-1 mRNA in the noninduced cultures of these strains is due to the inactive tyrosinase gene product was ruled out by demonstrating that the mutant T allele does not cosegregate with the ability to accumulate aod-1 mRNA under noninducing conditions. In all cases where aod-1 mRNA has been found in noninduced cultures, no alternative oxidase activity or AOD1 protein has been detected. This observation directly implies regulation at the level of translation.
Growth in the presence of chloramphenicol did not affect the levels of cytochromes aa3 and b in the strain carrying the chl-1 mutation. In addition, the strain did not induce alternative oxidase in the presence chloramphenicol, but did contain KCN-insensitive respiration when grown in the presence of antimycin A. These observations demonstrate that this mutation results in chloramphenicol resistance. Since the levels of cytochromes aa3 and b were not reduced when this strain was grown in chloramphenicol, there was likely no respiratory deficiency under this condition. This would explain why alternative oxidase was not induced by this treatment. To date, there are no known nuclear or mitochondrial encoded chloramphenicol-resistance markers in N. crassa (Perkins et al. 2001). Mutations causing chloramphenicol resistance are known in other fungi, but these affect mtDNA (Gunatilleke et al. 1975; Belcour and Begel 1977; Waxman et al. 1979). Chloramphenicol inhibits bacterial and mitochondrial translation by binding to the ribosome and preventing the association of tRNA with the A site, thereby blocking peptidyl transfer (Schlunzen et al. 2001). The nuclear chl-1 mutation might affect a mitochondrial ribosomal protein, prevent the uptake of the inhibitor, or cause its inactivation.
Further characterization of the alternative oxidase regulatory mutant strains is focused on identifying the genes affected. This may lead to an increased understanding of the regulatory mechanisms affecting alternative oxidase production and, by extension, to more general mechanisms of nuclear-mitochondrial communication.
Acknowledgments
We are grateful for excellent technical assistance from Tehnia Aziz and Kim Lam who were supported by summer studentships from the Natural Sciences and Engineering Research Council of Canada (NSERC). We thank Steve Free for supplying strain T1P11 and plasmid pTYR103. A.T.D. was supported by scholarships from NSERC and the Alberta Heritage Foundation for Medical Research. This work was supported by a Discovery grant from NSERC to F.E.N.
References
- Attardi, G., and G. Schatz, 1988. The biogenesis of mitochondria. Annu. Rev. Cell Biol. 4: 289–333. [DOI] [PubMed] [Google Scholar]
- Austin, B., R. M. Hall and B. M. Tyler, 1990. Optimized vectors and selection for transformation of Neurospora crassa and Aspergillus nidulans to bleomycin and phleomycin resistance. Gene 93: 157–162. [DOI] [PubMed] [Google Scholar]
- Belcour, L., and O. Begel, 1977. Mitochondrial genes in Podospora anserina: recombination and linkage. Mol. Gen. Genet. 153: 11–21. [DOI] [PubMed] [Google Scholar]
- Bertrand, H., and T. H. Pittenger, 1969. Isolation and classification of extranuclear mutants of Neurospora crassa. Genetics 61: 643–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, H., A. Argan and N. A. Szakacs, 1983 Genetic control of the biogenesis of cyanide insensitive respiration in Neurospora crassa, pp. 495–507 in Mitochondria, edited by R. J. Schweyen, K. Wolf and F. Kaudewitz. Walter de Gruyter, Berlin.
- Butow, R. A., and N. G. Avadhani, 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14: 1–15. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, M., R. Sharan and G. C. Hill, 2002. Trypanosome alternative oxidase is regulated post-transcriptionally at the level of RNA stability. J. Eukaryot. Microbiol. 49: 263–269. [DOI] [PubMed] [Google Scholar]
- Considine, M. J., D. O. Daley and J. Whelan, 2001. The expression of alternative oxidase and uncoupling protein during fruit ripening in mango. Plant Physiol. 126: 1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R. H., and F. J. De Serres, 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17A: 70–143. [Google Scholar]
- Djajanegara, I., P. M. Finnegan, C. Mathieu, T. McCabe, J. Whelan et al., 2002. Regulation of alternative oxidase gene expression in soybean. Plant Mol. Biol. 50: 735–742. [DOI] [PubMed] [Google Scholar]
- Edwards, D. L., J. H. Chalmers, Jr., H. J. Guzik and J. T. Warden, 1976 Assembly of the cyanide-insensitive respiratory pathway in Neurospora crassa, pp. 865–872 in Genetics and Biogenesis of Chloroplasts and Mitochondria, edited by T. H. Bucher, W. Neupert, W. Sebald and S. Werner. Elsevier/North-Holland Biomedical Press, Amsterdam.
- Finnegan, P. M., J. Whelan, A. H. Millar, Q. Zhang, M. K. Smith et al., 1997. Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol. 114: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes, A. M., I. Connerton and S. J. Free, 1994. Production of tyrosinase defective mutants of Neurospora crassa. Fungal Genet. Newsl. 41: 38–39. [Google Scholar]
- Gunatilleke, I. A., C. Scazzocchio and H. N. Arst, Jr., 1975. Cytoplasmic and nuclear mutations to chloramphenicol resistance in Aspergillus nidulans. Mol. Gen. Genet. 137: 269–276. [DOI] [PubMed] [Google Scholar]
- Henry, M. F., and E. J. Nyns, 1975. Cyanide insensitive respiration: an alternative mitochondrial pathway. Subcell. Biochem. 4: 1–65. [PubMed] [Google Scholar]
- Huh, W., and S. Kang, 2001. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 356: 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Horne, T., P. M. Wood, C. K. Wood, A. L. Moore, J. Headrick et al., 1998. Characterization of a split respiratory pathway in the wheat “take-all” fungus, Gaeumannomyces graminis var. tritici. J. Biol. Chem. 273: 11127–11133. [DOI] [PubMed] [Google Scholar]
- Joseph-Horne, T., D. W. Holloman and P. M. Wood, 2001. Fungal respiration: a fusion of standard and alternative components. Biochim. Biophys. Acta 1504: 179–195. [DOI] [PubMed] [Google Scholar]
- Karpova, O. V., E. V. Kuzmin, T. E. Elthon and K. J. Newton, 2002. Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14: 3271–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, A., J. Whelan, S. Young, T. E. Elthon and D. A. Day, 1992. Tissue-specific expression of the alternative oxidase in soybean and siratro. Plant Physiol. 99: 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe, G., M. Deak and S. J. Free, 1993. Use of the Neurospora tyrosinase gene as a reporter gene in transformation experiments. Fungal Genet. Newsl. 40: 43–45. [Google Scholar]
- Kupper, U., D. M. Niedermann, G. Travaglini and K. Lerch, 1989. Isolation and characterization of the tyrosinase gene from Neurospora crassa. J. Biol. Chem. 264: 17250–17258. [PubMed] [Google Scholar]
- Lambers, H., 1982. Cyanide-resistant respiration: a non-phosphorylating electron transport pathway acting as an energy overflow. Physiol. Plant. 55: 478–485. [Google Scholar]
- Lambowitz, A. M., and C. W. Slayman, 1971. Cyanide-resistant respiration in Neurospora crassa. J. Bacteriol. 108: 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz, A. M., E. W. Smith and C. W. Slayman, 1972. Oxidative phosphorylation in Neurospora mitochondria. Studies on wild type, poky, and chloramphenicol-induced wild type. J. Biol. Chem. 247: 4859–4865. [PubMed] [Google Scholar]
- Lambowitz, A. M., J. R. Sabourin, H. Bertrand, R. Nickels and L. McIntosh, 1989. Immunological identification of the alternative oxidase of Neurospora crassa mitochondria. Mol. Cell. Biol. 9: 1362–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch, K., C. Longoni and E. Jordi, 1982. Primary structure of tyrosinase from Neurospora crassa. I. Purification and amino acid sequence of the cyanogen bromide fragments. J. Biol. Chem. 257: 6408–6413. [PubMed] [Google Scholar]
- Li, Q., R. G. Ritzel, L. T. T. McLean, L. McIntosh, T. Ko et al., 1996. Cloning and analysis of the alternative oxidase gene of Neurospora crassa. Genetics 142: 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, L., 1994. Molecular biology of the alternative oxidase. Plant Physiol. 105: 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D., A. Radford and M. S. Sachs, 2001 The Neurospora Compendium Chromosomal Loci. Academic Press, San Diego.
- Pfanner, N., and W. Neupert, 1985. Transport of proteins into mitochondria: a potassium diffusion potential is able to drive the import of ADP/ATP carrier. EMBO J. 4: 2819–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads, D. M., and L. McIntosh, 1992. Salicylic acid regulation of respiration in higher plants: alternative oxidase expression. Plant Cell 4: 1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C. W., F. Roberts, F. L. Henriquez, D. Akiyoshi, B. U. Samuel et al., 2004. Evidence for mitochondrial-derived alternative oxidase in the apicomplexan parasite Cryptosporidium parvum: a potential anti-microbial agent target. Int. J. Parasitol. 34: 297–308. [DOI] [PubMed] [Google Scholar]
- Saisho, D., E. Nambara, S. Naito, N. Tsutsumi, A. Hirai et al., 1997. Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol. Biol. 35: 585–596. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj et al., 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413: 814–821. [DOI] [PubMed] [Google Scholar]
- Selker, E. U., 1990. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 24: 579–613. [DOI] [PubMed] [Google Scholar]
- Siedow, J. N., and A. L. Umbach, 2000. The mitochondrial cyanide-resistant oxidase: structural conservation amid regulatory diversity. Biochim. Biophys. Acta 1459: 432–439. [DOI] [PubMed] [Google Scholar]
- Tanton, L. L., C. E. Nargang, K. E. Kessler, Q. Li and F. E. Nargang, 2003. Alternative oxidase expression in Neurospora crassa. Fungal Genet. Biol. 39: 176–190. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe, G. C., and L. McIntosh, 1997. Alternative oxidase: from gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 703–734. [DOI] [PubMed] [Google Scholar]
- Verwoerd, T. C., B. M. M. Dekker and A. Hoekema, 1989. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman, M. F., J. A. Knight and P. S. Perlman, 1979. Suppression of mitochondrially-determined resistance to chloramphenicol and paromomycin by nuclear genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 167: 243–250. [DOI] [PubMed] [Google Scholar]
- Whelan, J., A. H. Millar and D. A. Day, 1996. The alternative oxidase is encoded in a multigene family in soybean. Planta 198: 197–201. [DOI] [PubMed] [Google Scholar]
- Yukioka, H., S. Inagaki, R. Tanaka, K. Katoh, N. Miki et al., 1998. Transcriptional activation of the alternative oxidase gene of the fungus Magnaporthe grisea by a respiratory-inhibiting fungicide and hydrogen peroxide. Biochim. Biophys. Acta 1442: 161–169. [DOI] [PubMed] [Google Scholar]