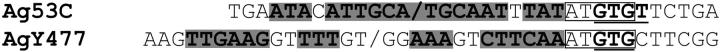Abstract
Satellite DNA is an enigmatic component of genomic DNA with unclear function that has been regarded as “junk.” Yet, persistence of these tandem highly repetitive sequences in heterochromatic regions of most eukaryotic chromosomes attests to their importance in the genome. We explored the Anopheles gambiae genome for the presence of satellite repeats and identified 12 novel satellite DNA families. Certain families were found in close juxtaposition within the genome. Six satellites, falling into two evolutionarily linked groups, were investigated in detail. Four of them were experimentally confirmed to be linked to the Y chromosome, whereas their relatives occupy centromeric regions of either the X chromosome or the autosomes. A complex evolutionary pattern was revealed among the AgY477-like satellites, suggesting their rapid turnover in the A. gambiae complex and, potentially, recombination between sex chromosomes. The substitution pattern suggested rolling circle replication as an array expansion mechanism in the Y-linked 53-bp satellite families. Despite residing in different portions of the genome, the 53-bp satellites share the same monomer lengths, apparently maintained by molecular drive or structural constraints. Potential functional centromeric DNA structures, consisting of twofold dyad symmetries flanked by a common sequence motif, have been identified in both satellite groups.
RECENT technological advances in genome research have led to the successful elucidation of genome sequence in a number of eukaryotic organisms. However, in each case only the euchromatic portion of the genome was completed to high quality. The often substantial heterochromatic portions remain poorly explored because repetitive DNA, a major constituent of heterochromatin, poses a considerable challenge for cloning, assembly, and annotation (Carvalho et al. 2003). Yet, heterochromatin has been demonstrated to be essential for cell and organismal viability. In addition to harboring vital genes, its proposed functions include centromere formation, meiotic pairing, and sister chromatid cohesion (McKee and Karpen 1990; Moore and Orr-Weaver 1998; Sullivan et al. 2001). Further studies of this biologically important sequence component are fundamental to a comprehensive understanding of eukaryotic genome structure, evolution, and function.
Heterochromatic sequences, often referred to as “junk DNA,” are composed primarily of transposable elements and satellite DNA (stDNA). The latter group is represented by tandemly repeated multicopy sequences, organized in long, often megabase-sized arrays. Such sequence organization dictates that stDNA is subject to different evolutionary forces compared to nontandem repetitive sequences. The salient characteristic of stDNA is concerted evolution, which maintains repeat unit homogeneity within arrays and among individuals within a population through unequal crossing over and gene conversion (Dover 1986). In sexually reproducing organisms the homogenization processes are significantly accelerated by meiotic recombination and redistribution of chromosomes to the next generation (Mantovani et al. 1997). In consequence, rapid divergence in satellite sequence, array size, or both, is usually observed between closely related species (Ugarkovic and Plohl 2002) and even between isolated populations (Elder and Turner 1994). However, examples from various taxa, including Drosophila, tenebrionid beetles, and fish demonstrate that stDNA can remain highly conserved for evolutionary periods exceeding 20–80 million years (Heikkinen et al. 1995; de la Herran et al. 2001; Mravinac et al. 2002).
The African malaria mosquito Anopheles gambiae, a major vector of malignant human malaria, has a karyotype consisting of two pairs of autosomes and a pair of sex chromosomes. From cytogenetic evidence, its Y chromosome is known to carry male determining gene(s), to be fully heterochromatic, and to vary in length and banding pattern in natural populations (Bonaccorsi et al. 1980; Clements 1992). Completion of the A. gambiae genome project did not result in the assembly of Y chromosome sequences, because of its peculiar structure, characterized by the massive accumulation of repetitive DNA (Holt et al. 2002; Krzywinski et al. 2004). However, by design, the genome project includes a large number of fragmented Y chromosome sequences that await identification, a goal we have been approaching with the use of bioinformatics tools. As part of this effort, we set out to characterize satellite sequences from the A. gambiae Y. Here, we have used a simple in silico strategy to identify four Y-linked satellite families and elucidate their spatial organization and evolution.
MATERIALS AND METHODS
Mosquitoes:
Specimens of A. gambiae were obtained from laboratory colonies maintained at the University of Notre Dame: PEST (Nigeria and Kenya; see Holt et al. 2002), SUA (Liberia), ZAN/U (Zanzibar), RSP (Kenya), GA-M-Mali (Mali), BKO (Mali), and GA-CAM (Cameroon). Specimens representing other species of the A. gambiae complex (A. arabiensis, A. melas, A. merus, A. quadriannulatus A) were F1 progeny of field-collected females. Genomic DNA was isolated from individual adult males and virgin females according to Collins et al. (1987). Where virgin females were unavailable, the distal part of the abdomen containing the spermatheca was removed prior to DNA extraction, to avoid contamination with male DNA transferred in sperm.
PCR, cloning, and sequencing:
PCR mixtures contained 1 μl template DNA (1/100 of the DNA extracted from a single mosquito), 1.5 mm MgCl2, 20 mm Tris-HCl (pH 8.4), 50 mm KCl, 0.2 mm each dNTP, 25 pmol of each primer (supplementary Table 1 at http://www.genetics.org/supplemental/), and 2.5 units Taq polymerase in a total volume of 50 μl. PCR reactions were performed in a Perkin-Elmer 9600 thermocycler with an initial denaturation at 94° for 3 min, followed by 35 cycles of 94° for 30 sec, 51°–55° for 30 sec, and 72° for 30–60 sec, followed by final elongation step at 72° for 10 min. PCR products were gel purified using a QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA) and cloned into the pGEM-T Easy vector (Promega, Madison, WI). Cloned PCR templates were PCR amplified and gel purified prior to sequencing. Sequencing was performed using ABI BigDye terminator chemistry (Perkin-Elmer Applied Biosystems) on an ABI 377 or 3700 sequencer. Sequences were assembled and verified by inspection of both strands using ABI Sequence Navigator software (Perkin-Elmer Applied Biosystems).
TABLE 1.
A. gambiae satellite families identifiedin silico
| Hits to database
|
||||
|---|---|---|---|---|
| Satellite familya | Exemplary scaffold | % GC | Male only | Whole genome |
| AgY53A | AAAB01005036 | 31 | 25 | 25 |
| AgY53B | AAAB01004290 | 34 | 23 | 24 |
| Ag53C | AAAB01007101 | 33 | 5 | 24 |
| Ag72 | AAAB01003201 | 43 | 1 | 90 |
| Ag93 | AAAB01003426 | 36 | 30 | 436 |
| Ag113 | AAAB01003510 | 43 | 3 | 119 |
| Ag404 | AAAB01001799 | 44 | 4 | 38 |
| AgY477 | AAAB01004782 | 41 | 14 | 15 |
| AgComplex-Ab | AAAB01004063 | 60 | 1 | 6 |
| AgComplex-Bc | AAAB01002959 | 45 | 8 | 108 |
Satellites from the Y chromosome are in italics.
Family names indicate monomer size in base pairs.
Composed of irregularly alternating short runs of 66- and 102-bp monomers, which differ by a 36-bp indel.
Composed of a 54-bp unit repeat, with short irregular runs of 64-bp units that differ by a 10-bp tandem duplication.
Southern and dot-blot analysis:
Southern analysis was conducted at high stringency as previously described (Krzywinski et al. 2004). For dot blots, a parallel series of dilutions of known quantities of A. gambiae male, female, and plasmid DNA containing a monomer sequence of AgY477 or AgY53A were transferred to Hybond-N+ membrane using Bio-Dot (Bio-Rad, Richmond, CA). The membrane was hybridized with cloned satellite monomer, washed three times at high stringency (allowing detection of sequences at least 95% identical to the probes), and scanned using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Estimation of the satellite DNA copy number was based on a densitometric analysis of pixel intensity using the ImageQuant program (Molecular Dynamics). Calculations assumed 0.27 pg DNA per haploid mosquito genome (Besansky and Powell 1992).
Fluorescent in situ hybridization:
Polytene chromosome spreads were obtained from females of the A. gambiae RSP or SUA strains as described previously (Kumar and Collins 1994). Using ∼1 μg of cloned satellite DNA, probes were labeled with Cy3-AP3-dUTP (Amersham, Arlington Heights, IL) using the Nick Translation kit (Amersham). Fluorescent in situ hybridization (FISH) was performed as described by Sharakhov et al. (2002).
Sequence analysis:
Tandem repeated sequences were identified using Tandem Repeats Finder (http://tandem.biomath.mssm.edu/trf/trf.submit.options.html). Pairs of sequences were aligned using BLAST2 (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html). Multiple sequence alignment was performed using ClustalX (Thompson et al. 1997) and subsequently refined manually using YAMSAT (http://bioinformatics.picr.man.ac.uk/bioinf/download_yam.jsp). Restriction sites in the DNA sequences were identified using Webcutter 2.0 (http://rna.lundberg.gu.se/cutter2/). Phylogenetic analyses were conducted under a maximum parsimony criterion (with and without inclusion of gap information) using PAUP* 4.0b10 (Swofford 2002) and the neighbor-joining algorithm under Kimura's two-parameter model as implemented in MEGA 2.1 (Kumar et al. 2001).
RESULTS
In silico screen for Y chromosome-specific satellite DNA:
Whole-genome shotgun sequencing of A. gambiae employed libraries prepared from males and females separately, to facilitate identification of male Y chromosome sequences (Holt et al. 2002). After genome assembly, sequences originating from the Y were expected to be included among the unmapped scaffolds, as polytene chromosomes used for physical mapping in A. gambiae derive only from female tissue. Of 8845 unmapped scaffolds, 975 derived exclusively from male libraries were identified and downloaded to a “male-only” database (Krzywinski et al. 2004). Each such scaffold was screened for the possible presence of stDNA using Tandem Repeats Finder software, which revealed complex and simple tandem repeats that could be grouped into families according to period size and sequence similarity (Table 1). As a preliminary indication of Y specificity, we used the consensus sequence of each tandem repeat family as a query in local BLASTN searches against two databases: the entire A. gambiae genome database consisting of all 8987 scaffolds and the male-only subset of 975 scaffolds. We reasoned that Y-specific stDNA queries should return the same number of hits in both databases, whereas X-linked and autosomal stDNA should return many more hits from the genome database. The three candidate Y-specific stDNA families (AgY53A, AgY53B, and AgY477) that gave a comparable number of hits to both databases (Table 1) were analyzed further, together with Ag53C for reference.
AgY477 satellite:
PCR:
To test for Y-linkage, PCR primers (supplementary Table 1 at http://www.genetics.org/supplemental/) were designed within a consensus monomer of the AgY477 array. Male-specific amplification of the 477-bp product was achieved in different A. gambiae strains (data not shown). This product did not amplify in either sex of four other tested species in the A. gambiae complex. However, an unexpected 367-bp product amplified in both sexes of all tested species. Subsequent sequence analysis showed the 367- and 477-bp products to differ by an insertion-deletion (indel), but they are otherwise very closely related (see below).
Southern analysis:
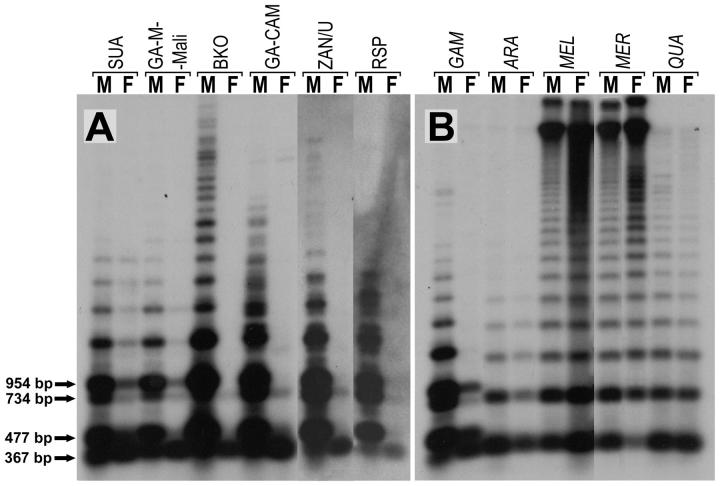
The 477-bp monomer PCR product from male A. gambiae was cloned and used to probe Southern blots prepared from DNA digested with NsiI, an enzyme that cleaves once within consensus monomers. In males of all A. gambiae strains, the AgY477 probe hybridized to the 477-bp monomer and a ladder of its multimeric variants, produced by loss(es) of the NsiI site, the pattern expected for long tandem arrays of imperfectly homogenized repeats. Some interstrain variation also was detected (Figure 1). These results were consistent across multiple specimens of each A. gambiae strain (data not shown).
Figure 1.—
Southern blot of genomic DNA from different geographic isolates of A. gambiae (A) and other species of the A. gambiae complex (B) probed with the AgY477 satellite monomer. GAM, A. gambiae; ARA, A. arabiensis; MEL, A. melas; MER, A. merus; QUA, A. quadriannulatus. Bands corresponding to monomers and dimers of the AgX367 and AgY477 satellite DNAs are marked with arrows and their respective sizes are given.
Unexpectedly, analysis of overexposed films showed that females of all A. gambiae strains except BKO also revealed a faint 477-bp ladder pattern (Figure 1). We believe that this is due to an independent (but related to the AgY477) satellite family. See discussion for details.
In addition to the 477-bp ladder, the AgY477 probe hybridized to a 367-bp ladder apparent in both sexes and all strains of A. gambiae on overexposed films. This 367-bp ladder also was present in four other species of the A. gambiae complex (Figure 1). Subsequent analysis showed that the 367-bp monomer corresponds to the 367-bp PCR product generated with the AgY477 primers and represents the basic unit of another stDNA (called AgX367 hereafter) closely related to AgY477 but not Y-linked.
At high stringency, no AgY477-like sequences were detected from two other mosquitoes in the same subgenus, A. funestus and A. stephensi, nor from representatives of four other mosquito genera (Aedes aegypti, Armigeres subalbatus, Tripteroides bambusa, and Toxorhynchites amboinensis).
Copy number:
Dot blot analysis was carried out with a cloned AgY477 monomer probe. Because Southern analysis demonstrated cross-hybridization with another satellite(s), copy number was evaluated jointly. However, to distinguish the Y-linked satellites from others the hybridization experiment was performed using genomic DNA from males and females separately, from three different strains of A. gambiae. GA-M-Mali contains ∼1300 and ∼200 copies of AgY477-like sequences in males and females, respectively, whereas in the BKO and RSP strains these values are about twice as high. However, because of the limitations of the dot-blot technique, the values given above should be regarded only as crude estimates.
Physical mapping:
In situ hybridization of fluorescently labeled AgY477 probe to the ovarian nurse cell polytene chromosomes of A. gambiae revealed a strong signal in the centromeric region of the X chromosome (Figure 2). This result is consistent with PCR and Southern analyses indicating the presence of a cross-hybridizing 367-bp satellite (AgX367) in both sexes. Mapping of AgY477 on the Y was not attempted; due to its heterochromatic state, the Y does not polytenize in the only source of male polytene chromosomes—the larval salivary glands. Technical difficulties precluded use of larval mitotic chromosomes.
Figure 2.—
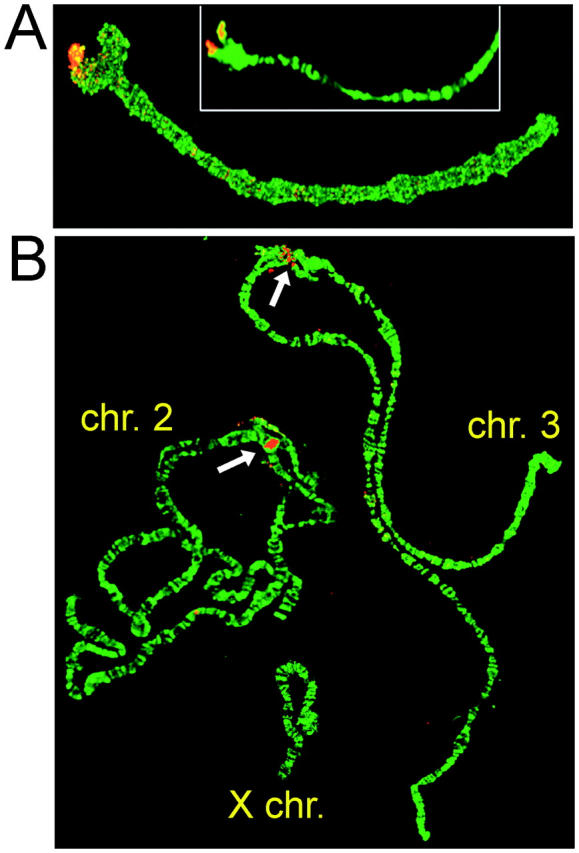
FISH of the AgY477 (A) and Ag53C (B) probes to A. gambiae ovarian nurse cell polytene chromosomes. The hybridization signal in B is indicated with arrows.
Sequence analysis:
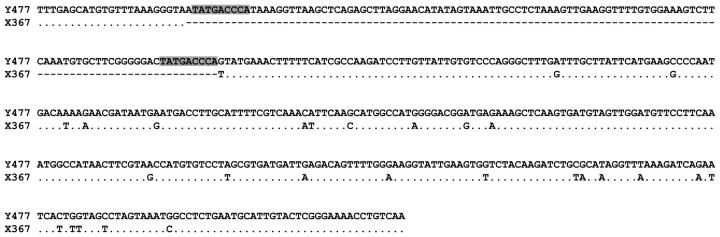
The level of intraspecific variation was assessed by comparison of monomer sequences of the Y-linked AgY477 and the X-linked AgX367 satellites from multiple clones (26 and 27, respectively) from five A. gambiae strains. A single 110-bp indel constituted the major difference between these satellites, which otherwise were highly similar (Figure 3). Although the full multiple sequence alignment revealed 91 segregating sites, only one polymorphism was fixed between the AgX367- and AgY477-bp satellites. Thirteen polymorphisms present in low frequency on the Y were fixed or nearly fixed on the X. These observations allowed us to recognize two recombinant 477-bp monomers from the GA-M-Mali strain that matched Y-linked monomers except for a contiguous tract of at least 240 bp that was identical to X-linked monomers (supplementary Figure 1 at http://www.genetics.org/supplemental/).
Figure 3.—
Alignment of AgY477 and AgX367 consensus monomer sequences. Dots, identical sites; dashes, missing sequences. Direct repeats flanking the Y-specific sequence are shaded.
Sequence analysis was extended to include four other species of the A. gambiae complex (four clones each). The multiple sequence alignment indicated length variation and substitutions diagnostic for a given species or a group of species. Relative to the X-linked stDNA from A. gambiae, monomers of all other species (except two in A. arabiensis) contained a 10-bp insertion located at one end of the 110-bp indel in A. gambiae. While entirely absent from the X-linked A. gambiae stDNA, 9 bp of this decamer were directly repeated at the ends of the 110-bp insertion of the Y-linked A. gambiae stDNA (see Figure 3). The average pairwise p-distances (excluding indels) between stDNA sequences from each species showed that, surprisingly, the A. gambiae Y and X stDNAs are not most similar (GAM-Y vs. GAM-X, 0.089; GAM-Y vs. ARA, MEL, MER or QUA, 0.060–0.084). Neighbor-joining phylogenetic analysis of all stDNA monomers indicated that A. gambiae X monomers were monophyletic, with A. arabiensis sequences inferred as their sister clade in the resulting tree (Figure 4). The two recombinant sequences from A. gambiae GA-M-Mali were inferred as basal to the (A. gambiae X + A. arabiensis) clade. All other Y-specific AgY477 stDNA monomers formed a single lineage, connected to an internal branch of the tree. Maximum-parsimony analyses gave similar results, regardless of gap information treatment.
Figure 4.—
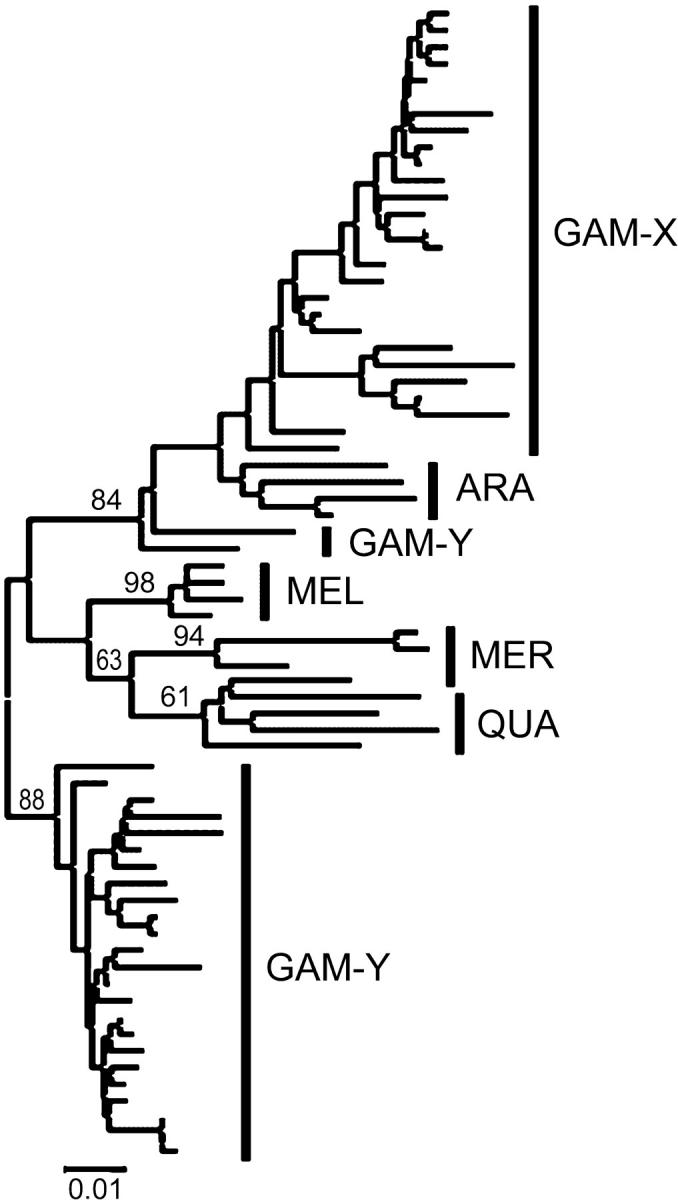
Phylogenetic relationships (neighbor-joining tree) between AgY477 and AgX367 monomers isolated from species of the A. gambiae complex. Numbers represent bootstrap values. Abbreviations are given in Figure 1.
AgY53A, AgY53B, and Ag53C satellites:
As predicted from the in silico analysis, male-specific PCR amplification of AgY53A and AgY53B stDNA was achieved from multiple A. gambiae strains and the other tested species (A. arabiensis), whereas Ag53C stDNA amplified from both sexes of A. gambiae and four other species (data not shown).
Southern analysis:
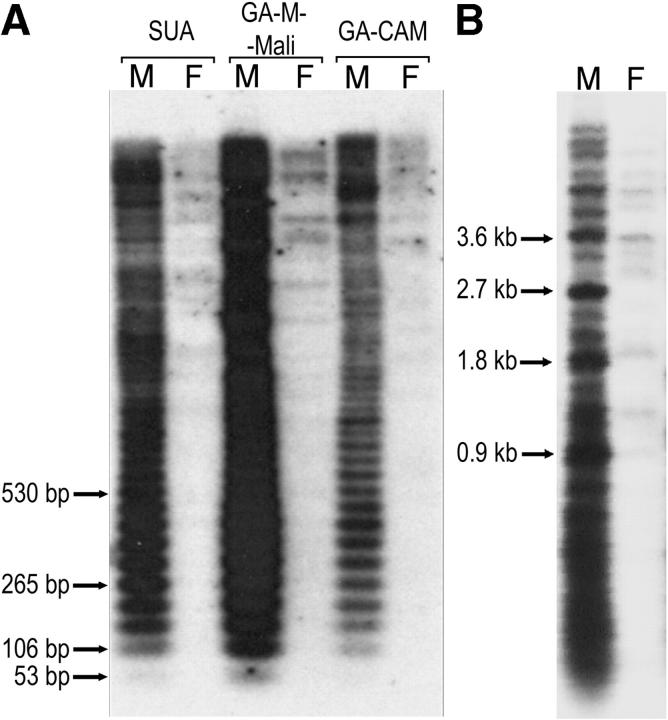
Southern blots of NsiI-digested DNA probed with a cloned AgY53A monomer revealed a strong 53-bp ladder only in male samples, indicating that the repeats are arrayed in tandem on the Y in each tested A. gambiae strain (Figure 5A). In addition, the probe hybridized to numerous male and a few female DNA fragments of higher molecular weight that could not be resolved in high-percentage agarose gels. Repeating the analysis with lower-percentage gels showed that in males of all strains except RSP, the strongest bands that could be resolved were evenly spaced by ∼900 bp (Figure 5B), suggesting a higher-order organization of tandemly arranged 17-monomer repeat units. Of the other four species tested, a faint 53-bp ladder was detected only in A. quadriannulatus males, although faint hybridization was detected to higher-molecular-weight fragments of both sexes (data not shown).
Figure 5.—
Southern blots of genomic DNA from A. gambiae resolved on a 1.5% agarose gel (A), or 0.8% agarose gel (B), and probed with the AgY53A satellite monomer. Arrows indicate bands corresponding to a monomer and selected oligomers (A) and those corresponding to the higher-order units (B); the respective fragment sizes are shown.
Southern blots of FokI-digested DNA probed with the AgY53B monomer confirmed Y-linkage of the AgY53B array in A. gambiae and A. arabiensis (data not shown). Nearly perfect sequence homogenization of the repeats was observed in A. gambiae males, as the hybridization signal was essentially limited to the 53-bp monomer fragment rather than a 53-bp ladder. In contrast, a short and faint 53-bp ladder was detected in A. arabiensis males, with stronger signal originating from higher-molecular-weight DNA not obviously related to the ladder. No 53-bp monomer or ladder was detected in three other members of the A. gambiae complex.
As expected from in silico and PCR analyses, Southern analysis of NsiI-digested DNA using an Ag53C monomer probe revealed a 53-bp ladder in both sexes and in all five tested species in the A. gambiae complex (data not shown). A few additional larger DNA fragments were detected, among them an ∼1.7-kb fragment found in A. gambiae and A. melas. The Ag53C probe detected no related sequences in A. funestus.
Copy number:
A dot-blot experiment to estimate the abundance of AgY53A monomer repeats in the A. gambiae genome suggested ∼2000 copies in males. Although dot-blot experiments were not performed for AgY53B or Ag53C, minimum copy number was estimated from mate-pair sequence information on the basis of the assumption that a clone is composed entirely of satellite DNA if its ends bear the same satellite repeats. The longest identified clones containing the AgY53B or Ag53C repeats on each end were 50 kb (∼950 copies) and 15 kb (∼300 copies), respectively. These are very conservative estimates; actual copy number is likely much larger.
Physical mapping:
Despite repeated attempts, no hybridization of the AgY53A or AgY53B probes to the autosomes or the X was detected, consistent with a location on the Y chromosome. By contrast, Ag53C hybridized to the centromeric regions of chromosomes 2 and 3 (Figure 2).
Sequence analysis:
An alignment of monomer consensus sequences from each of the three 53-bp families in the A. gambiae PEST genome showed little overall similarity, with the distribution and proportion (32%) of conserved sites not exceeding that expected among unrelated sequences. However, higher sequence similarity observed in pairwise comparisons suggested that these satellites evolved from a common ancestor and that the two Y-linked families were more closely related to one another (62%) than to the autosomal family (AgY53A/Ag53C, 47%; AgY53B/Ag53C, 43%) (Figure 6).
Figure 6.—
An alignment of the monomer consensus sequences of the 53-bp stDNAs from A. gambiae PEST strain. Identical sites are denoted by dots, and missing sequences by dashes. Arrows indicate a tandemly repeated motif in AgY53A and AgY53B.
Using family-specific primers (supplementary Table 1), monomers and oligomers of each 53-bp family were amplified by PCR from multiple individuals of different strains and species. Cloned fragments selected for analysis contained 0.5–14 monomers. Hierarchical analysis of variance conducted on the fragments broken down into individual monomer sequences showed that most of the sequence variation within each data set (66–95%) was found within individuals, among units of an array. For autosomal Ag53C, between-species differences contributed little to the overall variation (5%), but, for the Y-linked families, usually more variation was partitioned between species (2–33% for internal monomers and 15% for junction monomers at the end of an array). No fixed interspecific substitutions were identified at the level of individual monomers of unknown position in the array; however, fixed differences were found between monomers at the end(s) of an array of AgY53B. Moreover, interspecific differences emerged if linked sequence changes from neighboring repeats were considered. Within clones containing AgY53A and AgY53B sequences, certain monomers from A. arabiensis are identical to A. gambiae monomers, but repeats adjacent to them possess several species-specific substitutions.
To evaluate patterns of variation within and among groups of adjacent monomer repeats within a strain, the A. gambiae Trace Archive (http://www.ncbi.nlm.nih.gov/blast/mmtrace.html) was searched for single-sequence reads (fragments) composed of ≥10 contiguous monomeric repeats from each 53-bp family. Ten high-quality sequence fragments of 10 monomers were randomly selected per family. Overall, monomer sizes were essentially constant, with only rare single-nucleotide deletions found (supplementary Figure 2 at http://www.genetics.org/supplemental/). Within the autosomal Ag53C family, sequences were more homogenous than those from the Y (38 haplotypes among 100 monomers; 5% variation among adjacent monomers on a fragment, 5% variation between fragments). Nearly half of the observed variability was due to singleton nucleotide substitutions. In the Y chromosome-linked stDNAs most of the variability was due to substitutions shared across a number of units (45–48 haplotypes among 100 monomers; 6–7% variation among adjacent monomers on a fragment, 6–8% variation between fragments). The shared substitutions fell into three categories: (1) rare and usually partitioned among fragments; (2) frequent (occurring in ≥15 monomers) and partitioned within fragments in adjacent monomers, but not correlated with other substitutions; and (3) frequent and partitioned within fragments, often in association with other specific substitutions (supplementary Figure 2). Substitutions in the second and third categories, typical of AgY53B and AgY53A, respectively, but absent from Ag53C, likely resulted from array homogenization. Because this pattern held when a larger set of 50 fragments per family was examined, it suggests that arrays on the Y and autosomes are shaped by evolutionary forces that differ in tempo or mode.
Satellite junctions and discovery of the AgY373 satellite:
To gain insight into the structure and organization of the boundaries between a given satellite and other sequences (hereafter, junctions), we identified 10 scaffolds from the A. gambiae genome assembly that contained sequences from one of the 53-bp satellites in juxtaposition with unrelated sequences. Because the scaffolds were short (1–11 kb), we utilized mate-pair information (mates are the sequences from the opposite ends of a clone) to extend the physical limits of the analysis. First, clone-end sequences spanning the junction regions were identified, using scaffold sequences as queries against the A. gambiae Trace Archive (unassembled clone-end sequences). Subsequently, their mates were retrieved and analyzed. The results are summarized in Table 2.
TABLE 2.
Satellite junction regions identified in theA. gambiae genome sequence
| Scaffold
|
Coordinates on scaffold
|
Mate pair information
|
||||||
|---|---|---|---|---|---|---|---|---|
| StDNA | Name | Length | Adjacent sequence similar to (expect value) |
53-bp satellite |
Adjacent sequence |
Junction-spanning fragment (orientation)a |
Insert size (kb) | Mate sequence similar to (expect value) |
| AgY53A | 4063 | 1330 | Repetitiveb; no similarity to known TEs | 1–351 | 352–1330 | 55433665 (+/+) | 10 | AgY53A |
| 6929 | 11068 | AgY373 stDNA | 675–11068c | 5–281 | 55794517 (+/+) | 10 | AgY53A | |
| 3901 | 2570 | ENSANGP00000018025, A. gambiae (8e-36)d | 2006–2570 | 10–660 | 56838282 (+/−) | 10 | AgY53B/AgY53A junction | |
| 1–456 | 56521932 (+/+) | 10 | AgY53A | |||||
| 4621 | 1276 | pygopus gene, D. melanogaster (2e-04)e | 355–1276 | 164–322 | 56037571 (+/−) | 10 | AgY53A | |
| 17–355 | 55582724 (+/+) | 10 | AgY53A | |||||
| AgY53B | 4339 | 1310 | AgY477 stDNA | 503–1310 | 1–497 | 59870233 (+/−) | 50 | AgY477 |
| 47452583 (+/+) | 2.5 | AgY53B | ||||||
| Ag53C | 3553 | 1113 | Transposon Tsessebe III; A. gambiae (0.0) | 623–1113 | 1–624 | 46844423 (+/+) | 2.5 | agCP7415, A. gambiae (e-140); blastopia |
| Polyprotein, D. melanogaster (1.2) | ||||||||
| 56602027 (+/−) | 10 | Ag53C | ||||||
| 4593 | 1340 | Repetitivef; no similarity to known TEs | 1–1137 | 1150–1340 | 55141577 (+/+) | 2.5 | Retrotransposon R2 gag polyprotein, Nasonia vitripennis (5e-05) |
|
| 7835 | 2991 | Repetitiveg; no similarity to known TEs | 692–2991 | 1–691 | 46910563 (+/+) | 2.5 | Ag53C | |
| 2370 | 1444 | Complex-B satellite | 1–289 | 303–1444 | 55518933 (+/−) | 10 | Ag53C | |
| 59577007h (+/+) | 15 | Complex-B | ||||||
| 5564 | 1216 | Complex-B satellite | 405–1216 | 1–403 | 117434796 (+/−) | 14 | Complex-B | |
| 56786386 (+/+) | 10 | Ag53C | ||||||
Fragment names are from the NCBI Trace Archive at http://www.ncbi.nlm.nih.gov/Traces/trace.fcgi; Orientation, +/+, forward, with mate sequence positioned downstream from the junction; +/−, reverse, with mate sequence upstream from the junction. Where available, mate-pair information provided for two clones to reveal the nature of sequences at both flanks of the junction.
At least 60 hits.
The 6829 scaffold encompasses a sequence gap between positions 2054 and 9833.
Similar to putative transposase from Oryza sativa (expected value 1.8).
Similar to the T1-1 transposon, A. gambiae (expected value 0.018).
Three hits, including the 4593 scaffold.
At least 20 hits.
The fragment shows slight sequence discrepancies when compared to the junction sequence on the scaffold and it may represent another, but nearly identical, junction.
Junctions were of two basic types: the interruption of one satellite sequence by the insertion of a known or putative transposable element, and the boundary between different satellite sequences. One scaffold (AAAB01003901; because all scaffold names differ only in the last four digits, they are given in an abridged form, e.g., 3901 hereafter), when extended using mate-pair information, exemplified both types: an insertion of a putative transposon into the AgY53A array only 10 kb downstream of a junction between AgY53A and AgY53B arrays. The latter array, spaced only 2 bp from the former, extends for at least 10 kb, as judged by further mate-pair analysis. Of the other four intersatellite junctions, three also could be shown by mate-pair analysis to represent the transition between long arrays (AgY53B/AgY477, Ag53C/complex-B; Table 2). Intersatellite junctions shared no obvious features. The AgY53A/AgY373 junction, spanning 395 bp, consisted of two short 53-bp tandem repeats dissimilar from each other and from the flanking AgY53A satellite (see below for information on AgY373). The first repeat contained a sequence stretch similar to the Ag53C satellite; the second showed no similarity to any of the 53-bp satellites described in this study, but was highly similar to a 53-bp sequence forming a short (decamer) array on chromosome 2 (scaffold 8960) and a trimer repeat on the X chromosome (scaffold 8846). The AgY53B/AgY477 junction comprised 10 bp, of which 7 bp matched the flanking AgY53B monomer. Of two Ag53C/complex-B junctions, one lacked intervening DNA (scaffold 5564) and the other consisted of 13 bp with no sequence similarity to the satellite units (scaffold 2370).
To extend the junction analysis, we identified 250 clone-end sequences from the Trace Archive with similarity to the AgY53A, AgY53B, Ag53C, AgY477, or complex-B satellites. The ∼100 longest clones (≥10 kb) were screened for mate pairs with dissimilar sequences, of which three were found. These three bore the following sequences at either end: Ag53C/complex-B, complex-B/93bp, and AgY477 with a previously undetected 373-bp tandem repeat (Trace Archive clone nos. 19600445662296, 19600445917325, and 19600446262512, respectively). A subsequent inspection of scaffolds containing the complex-B satellite sequences revealed that this and the 93-bp satellite arrays are directly juxtaposed, without any intervening sequence (cf. scaffold 7586 and the corresponding fragment 47552535 spanning the junction). Of special interest was the 373-bp tandem repeat, which appeared to be another satellite, not detected in our initial in silico screen for tandem repeats. The 373-bp monomer is highly similar to both AgX367 (73% sequence identity) and AgY477 (81% sequence identity), sharing the 110-bp insertion with the latter. Using the monomer sequence to search the Trace Archive, we detected numerous clones with insert sizes spanning 2.5–50 kb, having at both ends a tandemly organized sequence nearly perfectly matching the 373-bp query. The juxtaposition of Y-specific AgY477 and the 373-bp satellite indicated that the latter is present on the Y. PCR with primers specific to the 373-bp monomer (supplementary Table 1) provided evidence that this satellite is present exclusively on the Y chromosome (data not shown), leading us to designate it AgY373.
Internal structure:
Dot-matrix analysis of the satellite monomers failed to show clear evidence of internal substructure suggestive of their origin from shorter motifs. However, all satellites contained short inverted repeats potentially capable of forming dyad structures. In particular, two dyad symmetries that may constitute functional motifs were identified in the Ag53C and AgY477 satellites (Figure 7). Both share structural features resembling those proposed to define functional centromere DNA (Koch 2000).
Figure 7.—
Dyad symmetries (shaded in boldface type) identified within Ag53C and AgY477 monomers. Note identical pentanucleotide motifs (boxed) flanking both structures on the right. The GT motifs adjacent to the dyad structures are underlined.
DISCUSSION
Satellite DNA has remained essentially uncharacterized in mosquitoes. Only two articles regarding the subject have been published to date. In an early study of A. stephensi DNA using cesium chloride equilibrium centrifugation, four discrete satellite DNA bands distinct from the bulk of chromosomal DNA were found (Redfern 1981), although none of them was further characterized at the sequence level. The other account describes a short tandem-repeat array serendipitously discovered during characterization of a P-element construct integration site within the telomeric region of chromosome 2L in A. gambiae (Biessmann et al. 1996). A report describing the genome sequence of A. gambiae provided no further information regarding satellite sequences (Holt et al. 2002), beyond the suggestion that simple repeats are not more expanded in Anopheles than in the Drosophila genome. The implicit goal of the whole-genome shotgun sequencing approach is to determine the euchromatic segments of the genome (Myers et al. 2000). However, heterochromatic sequences, including satellites, are accumulated in large numbers as well, although, for technical reasons, most of them remain disregarded in the genome assembly, leaving large information gaps unresolved. The present study is the first step to close the gap in our understanding of the diversity, relative location, and evolution of satellite DNA in the A. gambiae genomic landscape.
A simple in silico strategy allowed us to successfully identify four satellite DNAs specific to the Y chromosome. They fall into two groups based on unit size and sequence similarity. The following discussion is focused on these satellites and their relatives, mapped to either the autosomal or the X-chromosomal centromeric regions.
The AgY477, AgY373, and AgX367 satellites share an extensive sequence similarity indicative of their origin from the same progenitor repeat unit. Another descendant satellite family that escaped in silico detection and experimental isolation was revealed by Southern blot in A. gambiae (shown in female lanes as the ladder corresponding to the AgY477; Figure 1). Unsuccessful attempts to amplify a 477-bp product from females using two pairs of primers designed from the AgY477 monomer sequence suggested that this enigmatic satellite does not have the target sites for the primers, although it shares a highly similar sequence to the AgY477 probe. The radiation of the AgY477-like satellites involved some rearrangements, including sequence insertions and deletions, although the sequence and chromosomal location of the ancestral array and the succession of evolutionary events remain unclear given the present data. Nevertheless, the discovered complexity suggests a rapid turnover of these stDNA sequences within the A. gambiae complex.
The phylogenetic relationships of the AgX367-like monomers from the A. gambiae complex (Figure 4) are identical with the phylogeny proposed for the complex on the basis of the rDNA sequences (Besansky et al. 1994). Although expected, this result should not to be taken for granted, because other studies showed that different portions of the Anopheles genome have strongly contrasting evolutionary histories resulting from apparently extensive interspecific introgression (Besansky et al. 2003). In consequence, true phylogenetic relationships among the members of the A. gambiae complex remain unknown. The rDNA locus, occupying the X chromosome heterochromatin, is closely linked to the centromeric region, to which the AgX367 was mapped. Concordant topologies of the phylogenetic trees inferred with these two markers attest to the same evolutionary fate of the involved segment of the X chromosome and to the presence of some phylogenetic signal in the AgX367-like stDNA sequences.
The evolutionary scenario appears relatively simple for the 53-bp satellites. Their ancestral array could have existed on the autosomes of the progenitor of the A. gambiae complex, from which some repeats were translocated onto the Y chromosome. Subsequently, these Y-linked sequences were split into two arrays and each assumed its own evolutionary trajectory. The Ag53C repeats, found in autosomal centromeric regions, are highly conserved between species, which may suggest that their sequences are functionally constrained, perhaps by the presence of the centromere-determining motif (see below). After translocation onto the Y, repeats presumably released from the constraint could freely vary, resulting in the observed divergence, which, remarkably, has been driven solely by nucleotide substitutions, rather than insertions and deletions. Despite compartmentalization into different genomic regions and substantial sequence differences, the arrays share the same monomer lengths, apparently maintained by nonneutral forces of molecular drive (Dover and Tautz 1986) or a structural constraint imposed by involvement of the satellites in nucleosome formation and positioning (Fitzgerald et al. 1994).
Apart from alterations in a monomer sequence, the evolution of a satellite can involve fluctuations in monomer copy number, sometimes leading to large array size differences over short periods of time (Lo et al. 1999). In the present study this phenomenon concerned the Y chromosome-linked AgY477-like arrays, which were found to differ in size among some A. gambiae strains. Because the Y chromosome in A. gambiae is not known to recombine during meiosis, two likely mechanisms causing a Y-linked array expansion are unequal sister chromatid exchange (Smith 1976) and rolling circle replication followed by DNA reintegration into the genome (Okumura et al. 1987). The latter mechanism, in particular, can cause extremely rapid, saltatory expansions of the arrays as exemplified in the genomes of South American rodents of the genus Ctenomys (Rossi et al. 1990; Slamovits et al. 2001). The first step in the rolling-circle model is the formation of an extrachromosomal circular structure, which appears to be common with satellites and other tandemly repeated sequences (Okumura et al. 1987; Cohen et al. 2003). The rolling-circle amplification leaves a footprint of periodically occurring patterns of substitutions. Small monomer sizes of the 53-bp satellites relative to the contiguous sequence stretches derived from the ends of the A. gambiae PEST strain genomic clones allowed us to investigate common sequence patterns in the closely linked units. The analysis revealed clear periodicity in the AgY53A satellite units and some signs of periodicity in the AgY53B satellite, indicating that the evolution of these arrays may have involved the rolling-circle replication mechanism. The erosion of this pattern in the AgY53B sequences may be due to subsequent substitutions and frequent slipped-strand mispairings (Levinson and Gutman 1987), as suggested by the presence of identical substitutions found in islands of adjacent units (pairs or triplets) and surrounded by units lacking the substitutions (supplementary Figure 2). It is conceivable that all the Y-linked satellites have been incorporated into the chromosome following rolling-circle amplification, although there is no evidence to support this notion. However, the observation of size differences among Y chromosomes in natural populations (Bonaccorsi et al. 1980) is consistent with the existence of rapid satellite array expansion mechanisms in A. gambiae.
Sequence comparisons of the Y chromosome-linked AgY477 and the X chromosome-linked AgX367 monomers from A. gambiae revealed two apparently recombinant monomers from the GA-M-Mali strain. Such sequences could have been generated either by natural recombination processes within the nucleus or in vitro during PCR. Pairing of all chromosomes, including sex chromosomes, during meiosis is necessary for their correct transmission to gametes. In Drosophila melanogaster pairing of the X with the Y occurs within the centromeric region and is effected via the rDNA sequences shared by both chromosomes (McKee and Karpen 1990). In Anopheles genetic evidence for crossing over between the long arms of the X and Y chromosomes has been obtained for A. culicifacies (Sakai et al. 1979). The AgY477 and AgX367 sequences may be responsible for pairing of the sex chromosomes in A. gambiae, which upon contact may occasionally recombine. This interpretation is tempered by the finding that PCR can produce recombinant DNA fragments when amplifying elements of repetitive families (Scharf et al. 1988). In vitro recombination occurs when the Taq polymerase incompletely extends the amplified DNA fragment by premature dissociation from the template during the extension step, and the partially amplified sequence acts as a primer during the subsequent amplification cycle. The possibility of generating chimeric molecules during PCR in the present study seems to be supported by the observation that some clones with the AgY53B and the Ag53C repeats terminated prematurely on one or the other end. Alternatively, shorter than expected products may have resulted from instability of repetitive sequences propagated in a plasmid vector. A similar problem concerning incomplete units, encountered during the analysis of satellites cloned directly from the digested genomic DNA of an ant, was interpreted as a potential cloning artifact (Lorite et al. 2002). Although potentially error prone, PCR is a very convenient strategy for rapid extraction of numerous satellite sequences from a large number of individuals. In addition, it remains the only feasible method for isolation of satellites present in a genome in a small copy number, prohibiting traditional direct cloning from genomic DNA.
Substantial differences were found between the overall nucleotide content of the A. gambiae genome and the nucleotide content of the satellites identified in this study (Table 1). Such disparity of various genomic DNA fractions influences their buoyant density, which is the underlying principle of the early studies of genomic DNA using cesium chloride gradient centrifugation. This strategy usually allows fractions of DNA differing in G + C content by >5% to be separated into different bands (Lewin 2000). Considering an average of 35.2% G + C in the A. gambiae genome this criterion is met for seven of the identified repeats (Table 1), which upon CsCl separation should theoretically form four distinct bands, in addition to the main chromosomal DNA fraction. However, the cesium chloride gradient centrifugation analyses of A. gambiae genomic DNA revealed no prominent satellite bands (Holt et al. 2002). Lack of such bands suggests that the repeats with divergent nucleotide composition may be represented in the genome by a small number of copies, below the threshold of visual detection.
Functions of satellite DNA remain unclear despite its ubiquitous presence in eukaryotic organisms. Invariant colocalization of satellite sequences with the cytologically defined primary constriction and site of centromere and kinetochore proteins points to the involvement of satellite DNA in the formation of the centromere complex (Henikoff et al. 2001; Sullivan et al. 2001; Talbert et al. 2004). However, the question of what constitutes the operational elements of a functioning centromere still remains unanswered. Lack of significant sequence similarity between centromeric satellite sequences from different species, or even among different chromosomes of an individual organism, clearly shows that a universal centromere sequence does not exist (Karpen and Allshire 1997; Choo 2000). In addition, satellite DNA is not even necessary for centromere function as illustrated by the analysis of human and Drosophila neocentromeres (Karpen and Allshire 1997; Choo 2000). These observations suggest that centromere formation might not depend solely on a specific primary DNA sequence, but might depend on such characteristics as sequence superstructures (e.g., secondary or tertiary structures). A sequence comparison of primate α-satellite and a human neocentromere revealed common structural features that include twofold (outer and inner) dyad symmetries and a short conserved GTGT nucleotide motif adjacent to the dyad symmetries (Koch 2000). A similar structure of the dyad symmetry and an identical GTGT motif were found in the Ag53C satellite, mapped to the centromeres of chromosomes 2 and 3 (Figure 7). Moreover, analogous dyad structure was identified in the Y-linked AgY477 and AgY373 stDNAs, although their exact chromosomal localization remains unknown. Remarkably, the right outer symmetry in the Y-linked satellites is flanked by the same pentanucleotide sequence (ATGTG) as the dyad symmetry in the Ag53C satellite. A different twofold dyad symmetry was also identified in the X-linked AgX367 satellite; however, it was not flanked by the pentanucleotide mentioned above nor was a GT motif found in the proximity of the dyad structure. It remains to be determined whether these sequences are indeed involved in the formation of the centromeres in A. gambiae; however, given the present scarcity of data on the DNA constituents of the centromere, all sequences present at the centromeric regions are potential candidates for functional centromeric DNA, and the mere suggestion of such structures might prove of value (Koch 2000).
Our study shed light on “the dark corners” of the A. gambiae genome and revealed unexpectedly complex patterns of satellites, some of them with interrelated evolutionary histories. We focused on satellites linked to the Y chromosome and attempted to find an explanation for the aspects of their structure and evolution; however, it is clear that a more complete understanding of these complex issues, including timing of stDNA transposition onto the Y, requires further research. Nevertheless, our strategies proved to be a successful step toward revealing the Y chromosome's secrets.
Acknowledgments
We thank Mathew Chrystal for providing computer support and two anonymous reviewers for comments on the manuscript. This work was supported by grant AI44003 from the National Institutes of Health.
References
- Besansky, N. J., and J. R. Powell, 1992. Reassociation kinetics of Anopheles gambiae (Diptera: Culicidae) DNA. J. Med. Entomol. 29: 125–128. [DOI] [PubMed] [Google Scholar]
- Besansky, N. J., J. R. Powell, A. Caccone, D. M. Hamm, J. A. Scott et al., 1994. Molecular phylogeny of the Anopheles gambiae complex suggests genetic introgression between principal malaria vectors. Proc. Natl. Acad. Sci. USA 91: 6885–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky, N. J., J. Krzywinski, T. Lehmann, F. Simard, M. Kern et al., 2003. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. USA 100: 10818–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann, H., J. Donath and M. F. Walter, 1996. Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol. Biol. 5: 11–20. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi, S., G. Santini, M. Gatti, S. Pimpinelli and M. Colluzzi, 1980. Intraspecific polymorphism of sex chromosome heterochromatin in two species of the Anopheles gambiae complex. Chromosoma 76: 57–64. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. B., M. D. Vibranovski, J. W. Carlson, S. E. Celniker, R. A. Hoskins et al., 2003. Y chromosome and other heterochromatic sequences of the Drosophila melanogaster genome: How far can we go? Genetica 117: 227–237. [DOI] [PubMed] [Google Scholar]
- Choo, K. H., 2000. Centromerization. Trends Cell Biol. 10: 182–188. [DOI] [PubMed] [Google Scholar]
- Clements, A. N., 1992 The Biology of Mosquitoes. Chapman & Hall, London.
- Cohen, S., K. Yacobi and D. Segal, 2003. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 13: 1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, F. H., M. A. Mendez, M. O. Rasmussen, P. C. Mehaffey, N. J. Besansky et al., 1987. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am. J. Trop. Med. Hyg. 37: 37–41. [DOI] [PubMed] [Google Scholar]
- de la Herran, R., F. Fontana, M. Lanfredi, L. Congiu, M. Leis et al., 2001. Slow rates of evolution and sequence homogenization in an ancient satellite DNA family of sturgeons. Mol. Biol. Evol. 18: 432–436. [DOI] [PubMed] [Google Scholar]
- Dover, G. A., 1986. Molecular drive in multigene families; how biological novelties arise, spread and are assimilated. Trends Genet. 2: 159–165. [Google Scholar]
- Dover, G. A., and D. Tautz, 1986. Conservation and divergence in multigene families: alternatives to selection and drift. Philos. Trans. R. Soc. Lond. B Biol. Sci. 312: 275–289. [DOI] [PubMed] [Google Scholar]
- Elder, Jr., J. F., and B. J. Turner, 1994. Concerted evolution at the population level: pupfish HindIII satellite DNA sequences. Proc. Natl. Acad. Sci. USA 91: 994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, D. J., G. L. Dryden, E. C. Bronson, J. S. Williams and J. N. Anderson, 1994. Conserved patterns of bending in satellite and nucleosome positioning DNA. J. Biol. Chem. 269: 21303–21314. [PubMed] [Google Scholar]
- Heikkinen, E., V. Launonen, E. Muller and L. Bachmann, 1995. The pvB370 BamHI satellite DNA family of the Drosophila virilis group and its evolutionary relation to mobile dispersed genetic pDv elements. J. Mol. Evol. 41: 604–614. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Holt, R. A., G. M. Subramanian, A. Halpern, G. G. Sutton, R. Charlab et al., 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- Karpen, G. H., and R. C. Allshire, 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13: 489–496. [DOI] [PubMed] [Google Scholar]
- Koch, J., 2000. Neocentromeres and alpha satellite: a proposed structural code for functional human centromere DNA. Hum. Mol. Genet. 9: 149–154. [DOI] [PubMed] [Google Scholar]
- Krzywinski, J., D. R. Nusskern, M. K. Kern and N. J. Besansky, 2004. Isolation and characterization of Y chromosome sequences from the African malaria mosquito Anopheles gambiae. Genetics 166: 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V., and F. H. Collins, 1994. A technique for nucleic acid in situ hybridization to polytene chromosomes of mosquitoes in the Anopheles gambiae complex. Insect Mol. Biol. 3: 41–47. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Levinson, G., and G. A. Gutman, 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4: 203–221. [DOI] [PubMed] [Google Scholar]
- Lewin, B., 2000 Genes VII. Oxford University Press, Oxford.
- Lo, A. W., G. C. Liao, M. Rocchi and K. H. Choo, 1999. Extreme reduction of chromosome-specific alpha-satellite array is unusually common in human chromosome 21. Genome Res. 9: 895–908. [DOI] [PubMed] [Google Scholar]
- Lorite, P., J. A. Carrillo, A. Tinaut and T. Palomeque, 2002. Comparative study of satellite DNA in ants of the Messor genus. Gene 297: 113–122. [DOI] [PubMed] [Google Scholar]
- Mantovani, B., F. Tinti, L. Bachmann and V. Scali, 1997. The Bag320 satellite DNA family in Bacillus stick insects (Phasmodea): different rates of molecular evolution of highly repetitive DNA in bisexual and parthenogenetic taxa. Mol. Biol. Evol. 14: 1197–1205. [DOI] [PubMed] [Google Scholar]
- McKee, B. D., and G. H. Karpen, 1990. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell 61: 61–72. [DOI] [PubMed] [Google Scholar]
- Moore, D. P., and T. L. Orr-Weaver, 1998. Chromosome segregation during meiosis: building an unambivalent bivalent. Curr. Top. Dev. Biol. 37: 263–299. [DOI] [PubMed] [Google Scholar]
- Mravinac, B., M. Plohl, N. Mestrovic and D. Ugarkovic, 2002. Sequence of PRAT satellite DNA “frozen” in some Coleopteran species. J. Mol. Evol. 54: 774–783. [DOI] [PubMed] [Google Scholar]
- Myers, E. W., G. G. Sutton, A. L. Delcher, I. M. Dew, D. P. Fasulo et al., 2000. A whole-genome assembly of Drosophila. Science 287: 2196–2204. [DOI] [PubMed] [Google Scholar]
- Okumura, K., R. Kiyama and M. Oishi, 1987. Sequence analyses of extrachromosomal Sau3A and related family DNA: analysis of recombination in the excision event. Nucleic Acids Res. 15: 7477–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern, C. P., 1981. Satellite DNA of Anopheles stephensi Liston (Diptera: Culicidae). Chromosomal location and under-replication in polytene nuclei. Chromosoma 82: 561–581. [DOI] [PubMed] [Google Scholar]
- Rossi, M. S., O. A. Reig and J. Zorzopulos, 1990. Evidence for rolling-circle replication in a major satellite DNA from the South American rodents of the genus Ctenomys. Mol. Biol. Evol. 7: 340–350. [DOI] [PubMed] [Google Scholar]
- Sakai, R. K., R. H. Baker, K. Raana and M. Hassan, 1979. Crossing-over in the long arm of the X and Y chromosomes in Anopheles culicifacies. Chromosoma 74: 209–218. [Google Scholar]
- Scharf, S. J., C. M. Long and H. A. Erlich, 1988. Sequence analysis of the HLA-DR beta and HLA-DQ beta loci from three Pemphigus vulgaris patients. Hum. Immunol. 22: 61–69. [DOI] [PubMed] [Google Scholar]
- Sharakhov, I. V., A. C. Serazin, O. G. Grushko, A. Dana, N. F. Lobo et al., 2002. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science 298: 182–185. [DOI] [PubMed] [Google Scholar]
- Slamovits, C. H., J. A. Cook, E. P. Lessa and M. S. Rossi, 2001. Recurrent amplifications and deletions of satellite DNA accompanied chromosomal diversification in South American tuco-tucos (genus Ctenomys, Rodentia: Octodontidae): a phylogenetic approach. Mol. Biol. Evol. 18: 1708–1719. [DOI] [PubMed] [Google Scholar]
- Smith, G. P., 1976. Evolution of repeated DNA sequences by unequal crossover. Science 191: 528–535. [DOI] [PubMed] [Google Scholar]
- Sullivan, B. A., M. D. Blower and G. H. Karpen, 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2: 584–596. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2002 PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0 Beta. Sinauer Associates, Sunderland, MA.
- Talbert, P. B., T. D. Bryson and S. Henikoff, 2004. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkovic, D., and M. Plohl, 2002. Variation in satellite DNA profiles—causes and effects. EMBO J. 21: 5955–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]