Abstract
Phenoxyalkanoic compounds are used worldwide as herbicides. Cupriavidus necator JMP134(pJP4) catabolizes 2,4-dichlorophenoxyacetate (2,4-D) and 4-chloro-2-methylphenoxyacetate (MCPA), using tfd functions carried on plasmid pJP4. TfdA cleaves the ether bonds of these herbicides to produce 2,4-dichlorophenol (2,4-DCP) and 4-chloro-2-methylphenol (MCP), respectively. These intermediates can be degraded by two chlorophenol hydroxylases encoded by the tfdBI and tfdBII genes to produce the respective chlorocatechols. We studied the specific contribution of each of the TfdB enzymes to the 2,4-D/MCPA degradation pathway. To accomplish this, the tfdBI and tfdBII genes were independently inactivated, and growth on each chlorophenoxyacetate and total chlorophenol hydroxylase activity were measured for the mutant strains. The phenotype of these mutants shows that both TfdB enzymes are used for growth on 2,4-D or MCPA but that TfdBI contributes to a significantly higher extent than TfdBII. Both enzymes showed similar specificity profiles, with 2,4-DCP, MCP, and 4-chlorophenol being the best substrates. An accumulation of chlorophenol was found to inhibit chlorophenoxyacetate degradation, and inactivation of the tfdB genes enhanced the toxic effect of 2,4-DCP on C. necator cells. Furthermore, increased chlorophenol production by overexpression of TfdA also had a negative effect on 2,4-D degradation by C. necator JMP134 and by a different host, Burkholderia xenovorans LB400, harboring plasmid pJP4. The results of this work indicate that codification and expression of the two tfdB genes in pJP4 are important to avoid toxic accumulations of chlorophenols during phenoxyacetic acid degradation and that a balance between chlorophenol-producing and chlorophenol-consuming reactions is necessary for growth on these compounds.
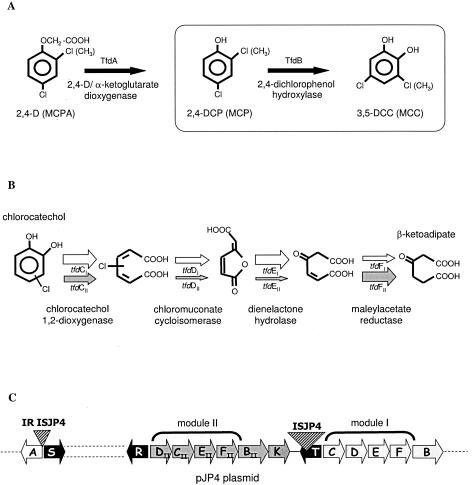
Bacteria play a fundamental role in the degradation of many chlorinated aromatic pollutants. Cupriavidus necator (formerly Ralstonia eutropha) JMP134(pJP4) was isolated for its ability to use 2,4-dichlorophenoxyacetate (2,4-D) as a sole carbon and energy source (5). This strain has been the focus of intensive studies because of its ability to degrade a wide spectrum of aromatic compounds, including 4-chloro-2-methylphenoxyacetate (MCPA) (29), 2,4,6-trichlorophenol (3), 4-fluorobenzoate (32), and 3-chlorobenzoate (5), among many others (D. Pérez-Pantoja, J. E. González-Pastor, D. H. Pieper, V. de Lorenzo, and B. González, unpublished data). The genes responsible for metabolism of 2,4-D/MCPA are localized in a 22-kb DNA region of plasmid pJP4 (37). The first step in 2,4-D/MCPA degradation involves cleavage of the ether bond by a 2,4-D/α-ketoglutarate dioxygenase encoded by the tfdA gene (8) to yield 2,4-dichlorophenol (2,4-DCP) and 4-chloro-2-methylphenol (MCP), respectively (Fig. 1A). These compounds are then transformed by a 2,4-DCP hydroxylase to produce 3,5-dichlorocatechol (3,5-DCC) or 5-chloro-3-methylcatechol (MCC) (6). A four-step modified ortho ring cleavage pathway consisting of a chlorocatechol 1,2-dioxygenase (TfdC), a chloromuconate cycloisomerase (TfdD), a dienelactone hydrolase (TfdE), and a maleylacetate reductase (TfdF) transforms 3,5-DCC into β-ketoadipate (Fig. 1B), which can be channeled to the central metabolism by two additional, chromosomally encoded enzymes (10). The pJP4 plasmid has two homologous, but nonidentical, gene clusters, tfdCIDIEIFI and tfdDIICIIEIIFII, encoding chlorocatechol degradation enzymes. Two different 2,4-dichlorophenol hydroxylases are encoded by the tfdB genes located immediately downstream of each of these clusters (Fig. 1C) (18, 37).
FIG. 1.
Genes involved in 2,4-D and MCPA degradation. (A) Chlorocatechol-producing peripheral reactions for 2,4-D and MCPA are carried out by enzymes encoded by the tfdA and tfdB genes in plasmid pJP4. (B) Chlorocatechol 1,2-dioxygenase (TfdC), chloromuconate cycloisomerase (TfdD), dienelactone hydrolase (TfdE), and maleylacetate reductase (TfdF) catalyze the conversion of chlorocatechols to chloromuconate, cis-dienelactone, maleylacetate, and β-ketoadipate, respectively. The arrow thicknesses indicate the relative specific activities of the enzymes encoded by each module (28, 30). (C) Organization of tfd genes in pJP4, including the tfdCIDIEIFIBI and tfdDIICIIEIIFIIBII gene clusters. The diagram is not drawn to scale.
Studies with C. necator JMP134 have demonstrated that each tfd gene module on pJP4 encodes functional enzymes, but the contribution of the enzymes encoded by each cluster to chlorocatechol degradation is not fully understood (18, 27, 30). Except for chloromuconate cycloisomerases, no great differences in substrate specificity between the enzymes encoded by each tfd module have been found, although it has been shown that the maintenance of both modules in pJP4 results in a higher expression of key catabolic activities that prevents the accumulation of toxic chlorocatechols (28, 30). An inactivation analysis of tfd functions was recently reported (19), suggesting that the tfdDIICIIEIIFIIBII genes are not essential for growth on 2,4-D but are relevant for growth on MCPA. Since no further information was given on intermediate accumulation during the degradation of chlorophenoxyacetates by these mutants, and since gene complementation was not reported, it is unclear if some of these phenotypes could be derived from polar effects on the chlorophenol hydroxylase genes. This is particularly relevant since chlorophenol substantially accumulates during 2,4-D degradation (22). Furthermore, no information on the contributions to total chlorophenol hydroxylase activity or differences in substrate specificity between TfdBI and TfdBII have been reported so far. In this work, we assess the contribution of each tfdB-encoded chlorophenol hydroxylase in this archetypal bacterial chlorophenoxyalkanoic acid degradation pathway.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. C. necator JMP134(pJP4), Burkholderia xenovorans LB400(pJP4), and 2,4-D/MCPA-mineralizing derivatives of these strains were grown at 30°C in liquid minimal medium (17), with 0.5 to 10 mM 2,4-D or MCPA or with 5 mM fructose as the sole carbon source. The plasmid pJP4-free derivative strain C. necator JMP222 was grown with 2 mM 2-methylphenol or with 5 mM fructose. Growth on each chlorophenoxyacetate was determined by measuring the increase in optical density at 600 nm (OD600). At least three replicates were used for each growth measurement. C. necator derivatives not able to proliferate on 2,4-D or MCPA were grown on 5 mM benzoate plus the appropriate antibiotic (Table 1) or on 5 mM fructose for induction experiments. Escherichia coli strains DH5α and BW25113 were maintained on Luria-Bertani (LB) agar plates. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml; and trimethoprim, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype and/or genotypea | Source or reference |
|---|---|---|
| C. necator strains | ||
| JMP134(pJP4) | 2,4-D+ MCPA+, pJP4 | DSMZb |
| JMP222 | pJP4-free derivative, 2,4-D− MCPA− Smr | H.-J. Knackmuss |
| JMP222(pJP4) | 2,4-D+ MCPA+, pJP4 | This study |
| B. xenovorans strains | ||
| LB400 | 2,4-D− MCPA− | DSMZb |
| LB400(pJP4) | 2,4-D+ MCPA+ | This study |
| E. coli strains | ||
| BW25113 | pKD46, red recombinase | 4 |
| BL21(DE3) | λDE3 lysogen, T7 RNA Pol | |
| Plasmids | ||
| pRK600 | Cmr | 11 |
| pGEM-T Easy | Apr | Promega Corp., Madison, WI |
| pGEMTFDB1 | Apr, tfdBI | This study |
| pGEMTFDB2 | Apr, tfdBII | This study |
| pGEMB3 | Apr, pheA | This study |
| pJP4-tfdBII mutant | tfdBI | This study |
| pJP4-tfdBI mutant | tfdBII | This study |
| pMLS7 | PS7 promoter, Tpr | 20 |
| pMLB1 | tfdBI, pMLS7 derivative | This study |
| pMLB3 | ORF4010, pMLS7 derivative | This study |
| pBBRMCS-2 | Kmr | 16 |
| pBTFDA | tfdA, pBBRMCS-2 derivative | Lab stock |
2,4-D+ and MCPA+, able to grow in 2,4-D and MCPA, respectively; tfd, catabolic genes from pJP4; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Smr, streptomycin resistance; Tpr, trimethoprim resistance; Kmr, kanamycin resistance.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany.
DNA manipulation.
Restriction, ligation, and dephosphorylation reactions and the purification and electroporation of DNA were performed by standard procedures (1). Derivatives of the pJP4 plasmid were transferred from E. coli BW25113 to C. necator JMP222 by mating. Derivatives of the broad-host-range plasmid vector pMLS7 (pMLB1 and pMLB3) (20) were mobilized from E. coli DH5α to C. necator JMP222 by triparental mating with the helper strain E. coli HB101(pRK600), as previously described (27). Transconjugants were selected on minimal medium agar plates supplemented with 2 mM benzoate plus kanamycin or trimethoprim.
Inactivation of tfdBI and tfdBII genes.
The tfdBI and tfdBII genes were independently inactivated in E. coli BW25113 cells harboring the pJP4 plasmid by the method of Datsenko and Wanner (4). PCR primers FORTB1 (5′-AACAATTGACGGAGGAAGACATGGCATTGACGATCGAAACTGTAGGCTGGAGCTGC TTCG-3′) and REVTB1 (5′-ATAGATCGCGACGCCACTCCTTATATAGCTCGAGACGTAACAATGATATCCTCCTTAG-3′) (for tfdBI) and FORTB2 (5′-CCTATCGATAAGGAGACTTCATGACGAAAAACCAACACTGTAGGCTGGAGC TGCTTCG-3′) and REVTB2 (5′-CCGATCGGGTTTTTCTGGATCAGTGCGCGCAGAGCGCCCCATATGAATATCCTCCTTAG-3′) (for tfdBii), which contain 40-bp homology extensions of the tfdBI or tfdBII gene sequence (in bold) and 20-bp priming sequences for pKD4 (4), were synthesized. These primer pairs were used with pKD4 as the template to amplify the kanamycin resistance gene flanked by 40 bp of the tfdBI or tfdBII gene sequence. The following PCR program was used: 95°C for 5 min, 28 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 90 s, and then 72°C for 10 min. PCR products were used to inactivate the tfdBI or tfdBII gene in an E. coli BW25113(pJP4) strain harboring RecBCD recombinase by a previously described procedure (28). pJP4 derivatives containing an inactivated tfdBI or tfdBII gene were then transferred to strain JMP222 by biparental conjugation as previously described (27). Primer pair FORWB1 (5′-GGAGGAATTCATGGCATTGAC-3′; includes an EcoRI restriction site [in italics]) and REVWB1 (5′-GTTACGTCCGGTCCAGAAAA-3′) and primer pair FORWB2 (5′-TTCATATGTCTCCTTCGGGC-3′) and REVWB2 (5′-GCATTGCTGCGATTCACTT-3′) were used to verify correct recombinational insertion of the kanamycin resistance cassette in place of each tfdB gene. This was then confirmed by direct sequencing of the insertion region using these primers.
Constitutive and IPTG-induced expression of tfdB genes.
In order to clone the tfdBI gene into the pMLS7 constitutive expression vector, the FORWB1/REVWB1 primer pair was used to amplify a 1.9-kb DNA sequence corresponding to the complete tfdBI open reading frame (ORF) (the translation start site is included in the primer and is located immediately downstream of the EcoRI site). This fragment was cloned into pGEM-T Easy (Promega Corp., Madison, WI), and the resulting plasmid was digested with EcoRI to obtain a 2.0-kb fragment, which was inserted, in turn, into the pMLS7 plasmid (20) to produce pMLB1. The ORF4010 chromosomal gene was amplified from C. necator JMP134 genomic DNA using the primer pair FORWB3 (5′-GCGAATCTGGAGGAGAATTCCATGAAGACG-3′; includes an EcoRI restriction site [in italics]) and REVWB3 (5′-ATTTTCACGAGACCCAGACG-3′) to obtain a 1.8-kb DNA sequence. This PCR product was cloned into pGEM-T Easy, and the resulting plasmid, digested with EcoRI to release a 1.9-kb fragment, was then cloned into pMLS7 to produce pMLB3. The identities of the sequences cloned in pMLS7 with either the tfdBI or ORF4010 gene were confirmed by sequencing of the cloned regions in pMLB1 and pMLB3. For IPTG (isopropyl-β-d-thiogalactopyranoside)-induced expression of the tfdB genes in E. coli BL21(DE3), the primer pairs FORWB1L (5′-CTAGTCTGGTACCGCGAGATGAACAATCCCTGT-3′)/REVWB1 and FORWB2/REVWB2 were used for amplification of the tfdBI and tfdBII gene sequences, respectively. Each product was then cloned into pGEM-T Easy to give pGEMTFDB1 and pGEMTFDB2, respectively. The proper orientation of the insert was verified in each case by separate digestion with EcoRI, KpnI, and SacI. Selected plasmid clones were transferred to strain BL21(DE3) by electroporation. Expression of TfdBI and TfdBII was achieved by growing the resulting strains in LB broth, with 100 μg ml−1 of IPTG added at the exponential growth phase. Cells were harvested after 4 h of induction, and enzyme extracts were prepared as described below.
Toxicity tests for chlorophenols.
The MICs of 2,4-DCP and MCP were determined for strain JMP222(pJP4), the tfdBI mutant, the tfdBII mutant, and strain JMP222. Each strain was grown overnight in 5 mM fructose, and 10-μl aliquots of these cultures were transferred to fresh medium supplemented with fructose in the presence of different concentrations of 2,4-DCP or MCP (50 to 500 μM at increments of 25 μM). Growth was determined by the increase in OD600 after 24 h. Degradation of the added 2,4-DCP was also measured for each inoculum. At least three independent determinations were carried out for each reported MIC. The viability of cells growing on fructose and exposed to 2,4-DCP was studied by measuring the number of CFU on solid medium. Cells were grown on fructose, washed, resuspended in fresh medium without a carbon source to an OD600 of 0.5, and treated or not with 2 mM 2,4-DCP (time zero), and aliquots were taken every hour for up to 8 h. Appropriate dilutions were plated on agar supplemented with phenol as the sole carbon source. The reported CFU values are averages of three independent experiments.
Enzyme assays.
For enzyme assays, cells were grown in minimal medium containing 2 mM 2,4-D. Strains not able to proliferate on 2,4-D were grown in minimal medium containing 5 mM fructose plus the corresponding antibiotic and were induced at late exponential growth phase with 0.5 mM 2,4-D for 3 h. One hundred milliliters of each culture was harvested at the end of the exponential growth phase (OD600 = 0.8 on fructose and 0.5 on 2,4-D), centrifuged, washed twice, and resuspended in 5 ml of a solution containing 50 mM Tris-HCl (pH 7.5) for chlorocatechol 1,2-dioxygenase activity measurements and 75 mM Na2HPO4/KH2PO4 buffer (pH 7.6), 1 mM dithiothreitol, 0.1 mM EDTA, 0.01 mM flavin adenine dinucleotide (FAD), and 0.1 mM 2,4-DCP for chlorophenol hydroxylase activity determinations. Cells were disrupted by sonication (Vibracell; Sonics & Materials, Inc., Newton, CT). The soluble protein fraction was obtained after 1 h of centrifugation at 130,000 × g in a Beckman L-80 ultracentrifuge. Cell extracts (0.1 to 5.0 mg of protein per ml) were used without further purification. Chlorocatechol 1,2-dioxygenase assays were performed as previously described (27). For chlorophenol hydroxylase activity, assay mixtures contained 50 mM Na2HPO4/KH2PO4 buffer (pH 7.6) and a volume of crude extract corresponding to 1 to 100 μg of protein (0.002 to 0.2 enzyme units). One unit of enzyme activity is the amount of crude extract that forms or consumes 1 μmol of product or substrate, respectively, per min. Protein determinations were performed using the Bradford reagent as previously described (27). Enzyme activities were determined in assays performed with a diode array Hewlett Packard HP 8452-A UV/Vis spectrophotometer. Chlorophenol hydroxylase activity was measured by the consumption of NADH, as indicated by the decrease in absorbance at 340 nm (ɛ340 = 6,300 M−1 cm−1). All assays were performed in the presence of 10 μM of FAD. Phenol-independent NADH oxidation was determined for each extract, and this contribution was subtracted from reported measurements. Each assay was started with the addition of 200 μM NADH to the reaction mixture containing fresh crude extract. Reported values are averages of at least three independent experiments. Transformation into the corresponding catechol was confirmed by high-performance liquid chromatography (HPLC) (see below) for every substituted phenol producing an NADH turnover rate higher than 5% of the rate produced by 2,4-DCP (see Table 2). Most chemicals were purchased from Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany; Baker Chemikalien, Griesheim, Germany; and Merck AG, Darmstadt, Germany. Chlorocatechols were obtained from Promochem, Wesel, Germany.
TABLE 2.
Chlorophenol hydroxylase activities against different phenolic substrates
| Substrate | Activity of C. necator strain against indicated substratea
|
|
|---|---|---|
| JMP222(pJP4-tfdBII) (TfdBI) | JMP222(pJP4-tfdBI) (TfdBII) | |
| 2,4-Dichloro | 100 (0.31) | 100 (0.13) |
| 2-Chloro | 11 | 31 |
| 3-Chloro | 3 | <2 |
| 4-Chloro | 29 | 68 |
| 2,3-/2,5-Dichlorob | 15 | <5 |
| 3,4-Dichloro | <2 | <2 |
| 2,3,5-/2,4,5-/2,4,6-Trichloro | <5 | <2 |
| 4-Chloro-2-methyl | 46 | 75 |
| 2-Methyl | 12 | 20 |
| 3-Methyl | <5 | <5 |
| 4-Methyl | 10 | 15 |
| 2,4-Dimethyl | 11 | <2 |
| 2-Fluoro/4-fluoro | <2 | <2 |
| 2-Nitro/4-nitro | <2 | <2 |
| 2,4-Dinitro | 4 | <2 |
| Phenol | 3 | <2 |
Relative values with respect to activity with 2,4-DCP. Absolute values (units mg of protein−1) are given in parenthesis.
Not transformed to chlorocatechols at detectable levels.
Detection of catabolic intermediates.
The accumulation of catabolic intermediates during the degradation of chloroaromatic compounds was determined by HPLC analysis. Cells were grown on 5 mM fructose and induced for at least 2 h with 0.5 mM 2,4-D. Experiments were designed to be performed under these conditions due to the slow growth of the tfdBI mutant on 2,4-D. Cells were pelleted, washed twice with minimal medium, and resuspended in this medium to an OD600 of 1.0. 2,4-D or MCPA (1 mM) was added, and the cells were incubated at 30°C in an orbital shaker. Two-hundred-microliter aliquots were taken at different times and cleared by centrifugation at 20,817 × g for 10 min, and supernatants were collected for HPLC. To detect catechols produced by chlorophenol hydroxylase, the enzyme reaction was stopped by acidification to pH 2 with HCl. Precipitated proteins were eliminated by centrifugation, and supernatants were collected. Samples (20 μl) from cell-free supernatants or enzyme reactions were injected directly into a Shimadzu LC-10AD liquid chromatograph system equipped with an SC125 Lichrospher 5-μm column (Bischoff, Leonberg, Germany). A methanol-H2O (60:40) mixture containing 0.1% (vol/vol) ortho-phosphoric acid was used as the solvent, at a flow rate of 1 ml min−1. The column effluent was monitored simultaneously at 210, 260, and 270 nm with an SPD-M10A diode array detector. The reported values are representative of at least two independent experiments. Retention volumes for relevant compounds were as follows: 3,5-dichlorocatechol, 1.7 ml; 3-methyl-5-chlorocatechol, 1.5 ml; 4-chlorocatechol, 1.3 ml; 3-chlorocatechol, 1.2 ml; 3-methylcatechol, 1 ml; 4-methylcatechol, 1.1 ml; 2,4-dichloromuconate, 0.7 ml; 2-methyl-4-chloromuconate, 0.6 ml; 2,4-D, 5.6 ml; MCPA, 6.0 ml; 2,4-DCP, 7.8 ml; and MCP, 7.1 ml.
RESULTS AND DISCUSSION
C. necator JMP222 derivatives containing tfdBI or tfdBII mutant pJP4 plasmid grow at different rates and extents on chlorophenoxyacetates.
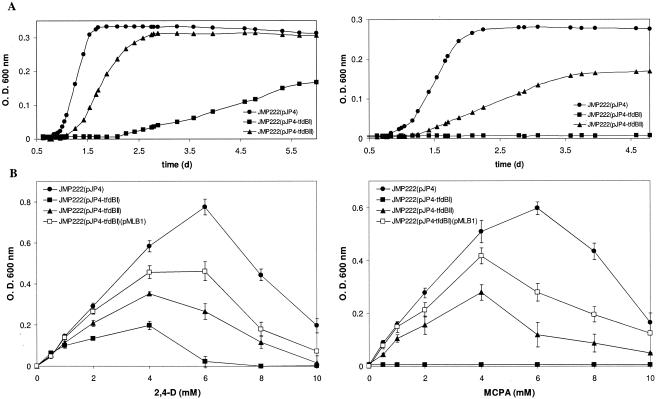
The specific contribution of each tfdB-encoded chlorophenol hydroxylase to growth on 2,4-D and MCPA was assessed by independently inactivating each tfdB gene in plasmid pJP4. Mutants of the pJP4 plasmid were constructed in E. coli BW25113 by allelic replacement of each of the chlorophenol hydroxylase-encoding genes (tfdBI or tfdBII) with a kanamycin resistance cassette. Recombination of the antibiotic resistance genes and disruption of each tfdB gene were verified by PCR amplification and direct sequencing of the target region (data not shown). The resulting pJP4 derivatives were then transferred by mating to the plasmid-free derivative strain JMP222. Growth of the tfdBI mutant and the tfdBII mutant on 2 mM 2,4-D or MCPA as the sole carbon and energy source was compared to the growth of C. necator JMP222 harboring the wild-type plasmid. The phenotype of the last strain is essentially equivalent to that of wild-type JMP134 (data not shown). Both tfdB mutant strains displayed slower growth on 2,4-D than strain JMP222(pJP4) (Fig. 2A). A much more pronounced effect was observed for the tfdBI mutant, which exhibited a growth rate of only 10% that of strain JMP222(pJP4), with a doubling time (Dt) of 40 h versus 4.5 h, and reached only 50% of the growth yield in stationary phase. In contrast, the growth rate of the tfdBII mutant was only slightly retarded (Dt = 7.7 h), and no reduction of the final growth yield was observed.
FIG. 2.
Growth of C. necator tfdBI and tfdBII mutants on chlorophenoxyacetates is differentially reduced. (A) Growth curves of strain JMP222(pJP4) (circles), the tfdBI mutant (squares), and the tfdBII mutant (triangles) on 2 mM 2,4-D (left) or MCPA (right). Representative OD600 values are shown for at least three independent experiments. (B) Growth of C. necator derivatives with different 2,4-D or MCPA concentrations. Data for the tfdBI mutant (pMLB1) constitutively expressing tfdBI are included. OD600 values were measured at stationary phase for all strains. Values shown are means of three independent experiments.
Similar patterns of growth were found when the mutant strains were incubated with MCPA as the carbon source (Fig. 2A). In this case, however, the change in phenotype was more severe, since growth was completely abolished in the tfdBI mutant, while the tfdBII mutant grew four times slower (Dt = 30.8 h) than strain JMP222(pJP4) (Dt = 8 h) and reached only 60% of the final growth yield. Differences between the growth of the tfdB mutants and that of strain JMP222(pJP4) could also be appreciated at higher substrate concentrations. As shown in Fig. 2B, the highest growth yield of strain JMP222(pJP4) was obtained at 6 mM 2,4-D (OD600 = 0.8), while both tfdB mutants reached lower yields at the same concentration. Accordingly, higher concentrations of MCPA as a substrate resulted in a strong impact on the growth of the tfdBII mutant strain, while the tfdBI mutant did not grow on this compound at any of the concentrations tested (Fig. 2B).
Growth measurements showed a differential contribution of each pJP4-encoded chlorophenol hydroxylase to the degradation of 2,4-D and MCPA. This observation correlates with total chlorophenol hydroxylase activity in cell extracts of the strain grown on fructose and induced with 0.5 mM 2,4-D. While total activity with 2,4-DCP was reduced from 0.38 ± 0.03 units mg of protein−1 in strain JMP222(pJP4) to 0.13 ± 0.05 units mg of protein−1 in the tfdBI mutant, the tfdBII mutant exhibited only a minor reduction in activity (0.31 ± 0.02 units mg of protein−1). Total hydroxylase activity with MCP was reduced to less than half the activity of strain JMP222(pJP4) in the tfdBI mutant (0.21 ± 0.04 units mg of protein−1 versus 0.10 ± 0.03 units mg of protein−1) and by about one-third in the tfdBII mutant (0.14 ± 0.02 units mg of protein−1). Chlorocatechol dioxygenase activities were compared as a reference and were found to be very similar in these three crude extracts (data not shown).
To ensure that the reduced growth on chlorophenoxyacetates by mutant strains was a result of tfdB gene inactivation, a plasmid for constitutive expression of the tfdBI gene was constructed to complement the tfdBI mutant strain, which shows the most severe phenotype. The pMLB1 plasmid, containing the tfdBI gene cloned downstream of the rps-7 promoter of Burkholderia cepacia (20), was introduced into the tfdBI mutant by mating. The resulting strain, JMP222(pJP4-tfdBI)(pMLB1), was able to grow on 2,4-D and MCPA, at concentrations up to 4 mM (Fig. 2B), but did not reach the same performance as strain JMP222(pJP4) at higher chlorophenoxyacetate concentrations. The chlorophenol hydroxylase activity in these cell extracts was 0.33 ± 0.03 units mg of protein−1 with 2,4-DCP as a substrate, which was higher than the activity of the tfdBI mutant but still lower than the activity of strain JMP222(pJP4). This may be due to a low level of expression of the tfdBI gene obtained with the exogenous PS7 promoter. Polar effects in the mutants were disregarded, since no catabolic function is encoded downstream of the tfdBI gene that could be affected by its inactivation, while the insertion in the tfdBII gene could only affect the expression of the tfdK gene, whose function as a 2,4-D uptake protein gene is only relevant for growth at very low concentrations of this compound (21).
The results showing that TfdBI is of major importance for 2,4-DCP or MCP transformation are in agreement with the inactivation analyses performed by Laemmli et al. (19) that indicated that the tfdBII gene is nonessential. However, we have shown that the tfdBII-encoded chlorophenol hydroxylase does make a contribution to the degradation of these compounds and that its function seems more relevant for the degradation of MCPA than for that of 2,4-D. In fact, C. necator JMP222(pJP4-tfdBI) is able to grow on 2,4-D, relying solely on the tfdBII-encoded chlorophenol hydroxylase.
The chlorophenol hydroxylases encoded by the pJP4 plasmid exhibit narrow substrate specificity profiles.
The differences in activity stated above for the TfdB enzymes with 2,4-DCP and MCP suggest that they have slightly different substrate specificities. Inactivation of each of the chlorophenol hydroxylase-encoding genes in pJP4 permitted the differentiation of each enzyme activity from its counterpart in a crude extract and allowed further investigation of the substrate preference of each of the TfdB enzymes. The results of the assays with crude extracts are summarized in Table 2. Both pJP4-encoded chlorophenol hydroxylases exhibited a very narrow specificity profile, being able to transform almost exclusively chlorinated phenols as long as they were substituted in the 2-, 4-, or 2,4- positions and allowing slow transformation of 2- and 4-methylphenol. The detection of catechols produced in the enzymatic assays by HPLC (not shown) allowed discrimination of true substrates, such as 2- and/or 4-chlorosubstituted phenols, from compounds such as 2,3- and 2,5-dichlorophenol, which elicited NADH oxidation but were not transformed to detectable levels. This has also been observed for other chlorophenol hydroxylases (2, 23) and described in detail for salicylate hydroxylase (36).
Apparent kinetic constants for 2,4-DCP calculated from double-reciprocal plots of activity versus substrate concentration were determined for the TfdBI and TfdBII hydroxylases by cloning and expression of the tfdBI and tfdBII genes in E. coli, using the pGEM-T Easy T7 promoter. Unfortunately, expression of the tfdB genes in E. coli was not high enough to allow quantification of each chlorophenol hydroxylase and a direct comparison of their Vmax values. The apparent Vmax of TfdBI under these conditions was 0.09 units mg of protein−1 (calculated with total protein), which was five times higher than the value obtained for TfdBII (0.02 units mg of protein−1). Assuming that both tfdB genes are similarly expressed from the IPTG-inducible T7 promoter in pGEM-T Easy, this difference would suggest that TfdBI is indeed more active than TfdBII. The apparent Km value determined for TfdBI was 11 μM, which is higher than the Km values reported for the TfdB proteins from B. cepacia 2A and Defluvibacter lusatiensis S1 (2, 24), which range from 3 to 4 μM, but is similar to the Km value of 14 μM reported for a hydroxylase purified from B. cepacia CSV90, another 2,4-D-degrading strain (31). The apparent Km for the TfdBII enzyme with 2,4-DCP was 2.5 μM, which is similar to the Km values of the reported hydroxylases and also similar to the Km previously described by Liu and Chapman for a strain JMP134 hydroxylase (23). The apparent kinetic constants described here suggest that the differences between the TfdBI and TfdBII hydroxylases are not merely a consequence of their differential expression but that the TfdBI enzyme is indeed more active than its counterpart, although TfdBII can be active at lower substrate concentrations.
A protein with chlorophenol hydroxylase activity from C. necator JMP134 was purified for an early report by Liu and Chapman (23) that suggested that dichlorophenol hydroxylase is a heterotetramer. However, using a different purification scheme, it was later shown that dichlorophenol hydroxylase is a homotetramer (6), and it was suggested that its identification as a heterotetramer was due to the presence of a copurified protein. No protein sequence data were provided by Liu and Chapman, and the amino acid composition of the copurified protein did not fit with either TfdBI or TfdBII sequence information or a mixture of both. Two other dichlorophenol hydroxylases have thus far been purified and characterized concerning substrate specificity, namely, the enzyme of B. cepacia 2a (formerly Acinetobacter) (2) and the enzyme from strain S1, a member of the α-2 subgroup of the Proteobacteria, recently identified as Defluvibacter lusatiensis (7, 24). Both enzymes have also been found to exhibit very narrow substrate preferences, being highly active with 2,4-dichloro- and 4-chloro-2-methylphenol but not able to transform phenol, and activity with 3-chlorophenol was either low or absent, in contrast to what is observed for multicomponent alkylphenol hydroxylases (13). Because TfdA, performing the first step in phenoxyalkanoic herbicide degradation, is an enzyme with relaxed substrate specificity (29), the strict substrate preference of TfdB enzymes may be the limiting factor in the range of chlorophenoxyacetates that can be degraded by C. necator JMP134. For the degradation of nonchlorinated substrates such as 2- and 4-methylphenoxyacetate, strain JMP134 can combine the use of TfdA with the recruitment of chromosomally encoded multicomponent phenol hydroxylases that degrade the corresponding phenols, probably via a meta cleavage pathway (29).
Although it is certain that the transformation of chlorophenols in C. necator JMP134 is achieved by the action of pJP4-encoded enzymes, a role of a chromosomally encoded chlorophenol hydroxylase activity in 2,4-DCP degradation has been suggested (15). This activity was only evident when the strain was grown on 2-methylphenol as a carbon source. C. necator carries a putative protein (ORF4010) with significant amino acid identity to either of the two plasmid-encoded chlorophenol hydroxylases (43% amino acid identity to TfdBI and 46% identity to TfdBII) and other phenol hydroxylases, which could indicate an enzyme capable of hydroxylating 2,4-DCP/MCP. In order to determine if ORF4010 codes for a functional chlorophenol hydroxylase, the corresponding DNA segment was cloned immediately downstream of the S7 promoter of pMLS7 to produce pMLB3. This plasmid was introduced into strain JMP222, and chlorophenol hydroxylase activity was measured with 2,4-DCP, MCP, and other substituted phenols in crude extracts of the strain growing on fructose. No activity against 2,4-DCP was found in strain JMP222(pMLB3), but some activity was detected with 2-methylphenol (0.05 ± 0.01 units mg of protein−1). The protein encoded by ORF4010 was more active with phenol (0.25 ± 0.02 units mg of protein−1) and 3-methylphenol (0.23 ± 0.02 units mg of protein−1), which contrasted with the negligible activities of the TfdBI and TfdBII enzymes for these substrates (Table 2). No phenol hydroxylase activity was found in crude extracts of strain JMP222 grown on fructose. These results indicate that the contribution of a chromosomally encoded putative chlorophenol hydroxylase to the degradation of phenoxyalkanoates seems to be negligible. Thus, despite the amino acid identity of the chromosomally encoded hydroxylase with the TfdB proteins, it exhibits a completely different substrate specificity which places it near other monocomponent flavin phenol hydroxylases, such as the PheA protein from Pseudomonas sp. strain 1001 (14), that can only transform phenol and 3-methylphenol (25).
Inactivation of the tfdBI or tfdBII gene results in the accumulation of toxic levels of chlorophenol during chlorophenoxyacetate degradation.
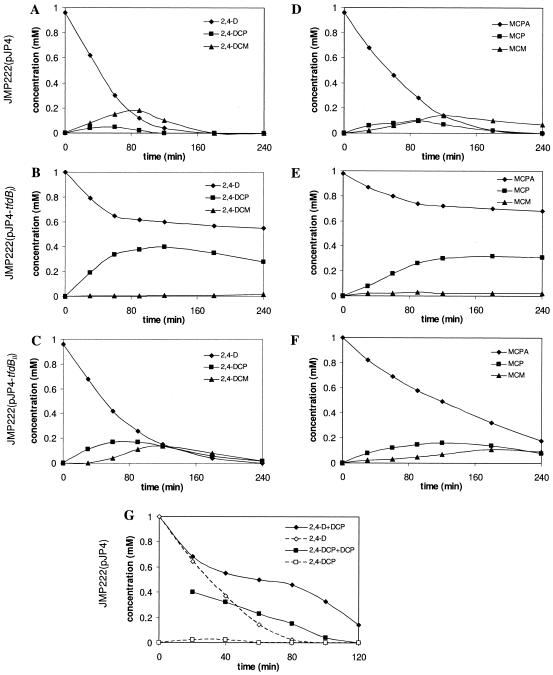
A discrete reduction in chlorophenol hydroxylase activity produced notorious changes in the ability of C. necator JMP134 derivatives to grow on chlorophenoxyacetates. The extent of reduction in growth of the tfdB mutants suggests that it was not merely due to kinetic pathway limitations but was probably raised from the toxic effects of both 2,4-DCP and MCP intermediates. Toxic effects of chlorophenols are mainly due to an increase in membrane fluidity due to their hydrophobicities and to their possible role as uncouplers of the electron transport chain (33, 35). Thus, relatively small changes in the levels of enzymes such as chlorophenol hydroxylase which are capable of transforming toxic intermediates can be responsible for substantial effects on growth rates and yields. In order to test if a higher level of 2,4-DCP accumulation during 2,4-D degradation could be detected in the mutant strains than in strain JMP222(pJP4), the presence of catabolic intermediates was analyzed in resting cells. As shown in Fig. 3A and D, strain JMP222(pJP4) accumulated low levels of the corresponding phenols and muconates when exposed to 2,4-D or MCPA. In contrast, 2,4-DCP was rapidly accumulated to a concentration of about 0.4 mM in the tfdBI mutant when 1 mM 2,4-D was given as the initial substrate, while 2,4-D degradation was almost completely stopped (to 7% of the initial rate) (Fig. 3B). A smaller chlorophenol accumulation (about 0.1 mM) was observed when the tfdBII mutant strain was incubated under the same conditions (Fig. 3C). This was associated with a reduction in the 2,4-D degradation rate to 41% of the initial value, while strain JMP222(pJP4) maintained a constant degradation rate throughout the same period (Fig. 3A).
FIG. 3.
Intermediate accumulation during chlorophenoxyacetate degradation by C. necator JMP134 derivatives. Chlorinated intermediates were detected by HPLC, using samples of supernatants after incubation of preinduced cell suspensions (OD600, 1.6) of strain JMP222(pJP4), the tfdBI mutant, and the tfdBII mutant with 1 mM 2,4-D (A to C) or MCPA (D to F). (G) Degradation of 2,4-D by strain JMP222(pJP4), as in panel A. 2,4-DCP (0.5 mM) was added to the cells after 20 min of incubation (+DCP). The discontinuous lines represent 2,4-D/2,4-DCP degradation in the absence of externally added 2,4-DCP. Symbols: diamonds, 2,4-D/MCPA; squares, 2,4-DCP/MCP; triangles, 2,4-dichloromuconate (2,4-DCM)/2-methyl-4-chloromuconate (MCM).
A similar scenario was observed for MCPA degradation (Fig. 3E and F), where the mutants also accumulated MCP to higher concentrations than did strain JMP222(pJP4), with 0.35 and 0.2 mM MCP in the tfdBI and tfdBII mutants, respectively, compared with 0.05 mM in the reference strain. When 0.4 mM 2,4-DCP was added directly to strain JMP222(pJP4) cells during 2,4-D degradation, a reduction in the rate of 2,4-D degradation was observed (to 14% of the initial rate), mimicking the phenotype of the tfdBI mutant. As the 2,4-DCP concentration decreased, the rate of 2,4-D degradation increased again, to 82% of the initial value (Fig. 3G). These observations confirmed that reductions in chlorophenol hydroxylase activity result in large increases in the concentration of chlorophenol, which can severely affect the growth of C. necator mutant strains on 2,4-D or MCPA.
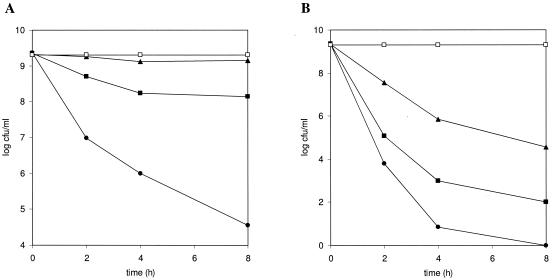
Since chlorophenols are accumulated during the degradation of chlorophenoxyacetates, the effect of direct exposure to 2,4-DCP on cell viability was compared for the strain JMP222(pJP4) and the mutant strains. Cells grown on fructose were washed and resuspended in fresh medium to an OD600 of 0.25 for treatment with different 2,4-DCP concentrations. Compared to that of strain JMP222(pJP4), survival of the tfdBI mutant was reduced by 2 and 3 orders of magnitude after 4 h and 6 h of 1 mM 2,4-DCP treatment, respectively (Fig. 4A). Survival of the tfdBII mutant was 1 order of magnitude lower than that of strain JMP222(pJP4) in the 8-h experiment. Treatment with 2 mM 2,4-DCP resulted in complete killing of the tfdBI mutant in 8 h, while the viability of the tfdBII mutant was decreased by 6 orders of magnitude (Fig. 4B). The viability of strain JMP222(pJP4) was reduced by 4 orders of magnitude at the end of the incubation period. By this time, strain JMP222(pJP4) degraded 15% of the initial 2,4-DCP, while the tfdBI mutant did not degrade any detectable amount of the compound. Therefore, chlorophenol disappearance alone does not explain the increased survival of strain JMP222(pJP4), but the metabolism of chlorophenol during this interval may allow energy-consuming adaptation mechanisms such as efflux pumps to function more efficiently (35). Despite these differences in survival, it is important that only a slight reduction in the viability of all strains tested was found when 0.5 mM 2,4-DCP was added to the cell suspensions (data not shown). The level of 2,4-DCP accumulated during 2,4-D degradation by the tfdBI mutant (Fig. 3B) is therefore unlikely to produce immediate cell death but is toxic enough to prevent 2,4-D degradation.
FIG. 4.
Effect of 2,4-DCP on cell viability of C. necator JMP134 derivatives. Cell suspensions of strain JMP222(pJP4) (triangles), the tfdBI mutant (circles), or the tfdBII mutant (squares) previously grown in 5 mM fructose were exposed to 1 mM (left) or 2 mM (right) 2,4-DCP, and samples were analyzed to determine the CFU at different times. Data for nonexposed tfdBI mutant cells (open squares) are included in each panel as a control of viability during the time of the assay. The values are averages based on two replicates.
In order to test if chlorophenol hydroxylase activity is relevant for the protection of C. necator directly exposed to chlorophenols, the MICs of 2,4-DCP and MCP for strain JMP222(pJP4) and tfdB mutant strains were compared. Strain JMP222(pJP4) was able to grow at concentrations of <0.4 or <0.3 mM of 2,4-DCP or MCP. The tfdBI mutant strain could only grow up to a concentration of 0.2 or 0.125 mM of 2,4-DCP or MCP, which was closer to the MICs for strain JMP222, at 0.075 mM and 0.05 mM, respectively. The elimination of tfdBII from pJP4 also reduced the MICs of chlorophenols, permitting growth of the mutant strain only at 2,4-DCP concentrations lower than 0.325 mM and resulting in a stronger effect on growth in MCP (MIC of 0.2 mM). These differences in the MICs between strain JMP222(pJP4) and the mutant strains can be explained because growth inhibition by 2,4-DCP is more effective in a strain that cannot degrade the compound. A strain JMP222(pJP4) inoculum completely degrades 2,4-DCP after 24 h of incubation in fructose plus 0.4 mM 2,4-DCP. In contrast, the tfdBI mutant can only degrade 16% of the total initial 2,4-DCP under the same incubation conditions. The ability to degrade 2,4-DCP thus seems intimately related to resistance to chlorophenol toxicity, explaining the differences in the above reported MICs.
Imbalance between production and degradation of chlorophenol affects growth on chlorophenoxyacetate by C. necator and B. xenovorans.
The results described so far show that chlorophenol turnover is a critical factor for chlorophenoxyacetate degradation by C. necator JMP134 due to the toxicity of this intermediate. If this is true, then a perturbation of this balance by an increase in the chlorophenol-producing enzyme TfdA should also have a negative effect on growth with these substrates. In order to test this possibility, the plasmid pBTFDA, harboring a constitutively expressed version of the tfdA gene, was introduced into strain JMP222(pJP4). Strain JMP222(pJP4)(pBTFDA) displayed reduced growth on 2,4-D compared with strain JMP222(pJP4), and this reduction became increasingly pronounced at higher 2,4-D concentrations (data not shown). Conversely, the introduction of plasmid pMLB1, harboring tfdBI, into strain JMP222(pJP4) resulted in a strain showing improved growth on the same substrate with respect to strain JMP222(pJP4) (data not shown). The total chlorophenol hydroxylase activity against 2,4-DCP measured in strain JMP222(pJP4)(pMLB1) reached up to 0.49 ± 0.04 units mg of protein−1, compared to 0.38 ± 0.03 units mg of protein−1 in strain JMP222(pJP4). These results imply that a moderate increase in chlorophenol hydroxylase activity can improve the efficiency of 2,4-D degradation in strain JMP222(pJP4). However, increased chlorophenol degradation can also result in intoxication of the strain because of chlorocatechol accumulation (28). This intermediate is highly toxic for bacteria, possibly due to oxidative damage of membranes and proteins (34). Nevertheless, the accumulation of chlorocatechols during 2,4-D degradation was negligible, even in strains harboring a constitutively expressed version of TfdBI (data not shown). This was probably due to the high activities of TfdC chlorocatechol dioxygenases towards 3,5-DCC, which is the best substrate for these enzymes (27).
To determine if chlorophenol turnover has a similar effect on a different catabolic strain, the pJP4 plasmid derivatives were transferred to B. xenovorans LB400. The resulting strain, B. xenovorans LB400(pJP4), had a growth yield on 2,4-D of about 50% that of strain JMP222(pJP4). The total chlorophenol hydroxylase activity for 2,4-DCP was 0.29 ± 0.02 units mg of protein−1, indicating that expression of the tfdB genes was not as high as that in C. necator, although TfdC activity reached similar levels (data not shown). When the pMLB1 plasmid harboring the constitutively expressed version of the tfdBI gene was introduced into strain LB400(pJP4), an increase in 2,4-DCP chlorophenol hydroxylase activity to twice (230%) the activity of strain LB400(pJP4) was produced. This resulted in a significant improvement (ranging from 1.5 to 2 times) in growth yields of strain LB400(pJP4)(pMLB1) in a wide range of 2,4-D concentrations (data not shown). Constitutive expression of the tfdBI gene in B. xenovorans LB400(pJP4) enabled the strain to grow faster on 2,4-D than strain LB400(pJP4), with a Dt of 3.2 h versus 5.9 h. On the other hand, the introduction of the tfdBI or tfdBII mutant pJP4 plasmid into strain LB400 resulted in a strain unable to grow on MCPA at all concentrations tested (data not shown), whereas only strain LB400(pJP4-tfdBII) could grow on 2,4-D, at concentrations below 4 mM. Increased expression of TfdA in strain LB400(pJP4)(pBTFDA) led to a reduction of growth on 2,4-D, which was more pronounced at higher 2,4-D concentrations. The results clearly indicate that avoiding chlorophenol accumulation is an important advantage for productive growth and for the degradation of chlorophenoxyacetates in β-proteobacteria. Intoxication due to unbalanced turnover of chlorophenols produces an immediate effect on chlorophenoxyacetate degradation and should be considered an important factor determining the extent and rate of growth of chlorophenoxyacetate-degrading strains on these xenobiotic compounds. Interestingly, toxicity by an imbalance between the production and degradation of initial intermediates in aromatic metabolism has also been reported for 3-chlorobenzoate (28), hydroxycinnamate (26), and protocatechuate (9).
TfdB enzyme sequences in pJP4 belong to different FAD monocomponent hydroxylase groups.
The results reported here have significant implications concerning the evolution of chloroaromatic compound metabolism pathways and also on the construction of chloroaromatic compound-degrading bacterial strains. Avoiding chlorophenol toxicity can be a selective pressure for an increased dosage of chlorophenol hydroxylase genes, produced either by gene duplication or by recruitment of different genes. A comparison of the two pJP4-encoded enzymes with other reported FAD monocomponent hydroxylases indicated that they may have been recruited from different pathways. The TfdBII protein sequence clusters together with homologues usually observed in isolates for which 2,4-D catabolic gene clusters have been analyzed (dendrogram not shown). Thus, TfdBII of strain JMP134 is highly related to TfdB enzymes from B. cepacia 2a, Delftia acidovorans P4a, Achromobacter xylosoxidans subsp. denitrificans EST4002, and Variovorax paradoxus TV1, sharing >86% amino acid sequence identity with them, while the TfdBI enzyme shares only approximately 60% amino acid sequence identity with members of this group. This supports the notion that the tfdBII gene is derived from a specialized 2,4-D degradation cluster. However, the TfdBII enzyme in C. necator JMP134 exhibits a low activity that is barely enough to allow growth of the strain on 2,4-D. This could indicate that tfdBII codes for a chlorophenol hydroxylase with intrinsically low activity or merely that this gene reaches a low level of expression in strain JMP134. This last possibility seems unlikely, since expression analyses have reported similar levels of transcription for the tfdBI and tfdBII genes when C. necator JMP134 was grown in continuous culture on fructose and 2,4-D was added as an inducer (22). In addition, transcriptomic preliminary data indicate that tfdBII transcripts are three to four times more abundant than tfdBI transcripts in cultures of C. necator JMP134 exposed to 2,4-D (D. Pérez-Pantoja, J. E. González-Pastor, D. H. Pieper, V. de Lorenzo, and B. González, unpublished data).
The TfdBI sequence belongs to a different group of FAD-dependent hydroxylases (data not shown). The absence of close relatives of tfdBI in all sequenced 2,4-D-degrading clusters suggests that this gene does not necessarily belong to a cluster specialized for 2,4-D degradation. However, partial amino acid sequences (covering approximately 12% of the protein) indicated that enzymes highly similar to TfdBI are present in 2,4-D-degrading Ralstonia and Burkholderia mallei strains (38), and a similar enzyme (DcpA) with 96% amino acid sequence identity with TfdBI has been reported for a Pseudomonas isolate (AY496436). The difference in chlorophenol hydroxylase activity of the TfdBI enzyme with respect to its TfdBII counterpart suggests that the function of TfdBI is not restricted to chlorophenoxyacetate degradation but that the enzyme also takes part in direct degradation of chlorophenols. This idea is supported by the significant identity of the TfdBI protein to ClpB (73% amino acid identity), a chlorophenol hydroxylase involved in 2,4-DCP degradation by D. lusatiensis, a strain that does not grow on 2,4-D (7). In fact, the tfdTCIDIEIFIBI gene cluster organization in pJP4 is different from the tfdRCEBKA organization of the 2,4-D-degrading gene clusters reported for strains where the tfd gene organization is known, i.e., V. paradoxus TV1(pTV1), B. cepacia 2a, A. xylosoxidans subsp. denitrificans EST4002, and D. acidovorans P4a (12).
The tfdA gene in strain JMP134 is separated by about 4 and 15 kb from tfdII and tfdI gene clusters, respectively, encoding enzymes for chlorophenol and chlorocatechol degradation, and it apparently forms a separate transcriptional unit (Fig. 1C). This may imply a different regulation for the tfdA gene and the rest of the tfd gene modules, making coordination between 2,4-DCP production and degradation difficult. Expression of these activities can be more balanced in tfdRCEBKA gene clusters, eliminating the need for an extra-high-performance chlorophenol hydroxylase like TfdBI. Accordingly, proteomic evidence obtained by our group shows that a larger amount of TfdA is produced with respect to both pJP4-encoded TfdB proteins when C. necator JMP134 is grown on 2,4-D (F. Flores-Aceituno, D. H. Pieper, and B. González, unpublished data).
Chlorophenols are highly toxic pollutants that can also be catabolic intermediates, as with phenoxyalkanoic herbicides. Due to this toxicity, the performance of catabolic bacteria is not dependent only on the presence of the characteristic enzymatic steps. This work clearly indicates that additional studies are required concerning the role in detoxification of reported chlorophenol and, possibly, phenol hydroxylases, focusing on their substrate specificities, activities, and regulation of gene expression.
Acknowledgments
We give special thanks to C. Lagos for plasmid pBTFDA.
This work was financed by FONDECYT 1030493, a MECESUP student travel grant, the collaboration grant CONICYT-Chile/BMBF-Germany, and the Millennium Scientific Initiative through Millennium Nucleus EMBA grant P/04-007-F. T.L. is a DIPUC Ph.D. fellow.
REFERENCES
- 1.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl (ed.). 1992. Short protocols in molecular biology, 2nd ed. Greene Publishing Associates, New York, N.Y.
- 2.Beadle, C. A., and A. R. W. Smith. 1982. The purification and properties of 2,4-dichlorophenol hydroxylase from a strain of Acinetobacter species. Eur. J. Biochem. 123:323-332. [DOI] [PubMed] [Google Scholar]
- 3.Clément, P., V. Matus, L. Cárdenas, and B. González. 1995. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol. Lett. 127:51-55. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farhana, L., and P. B. New. 1997. The 2,4-dichlorophenol hydroxylase of Alcaligenes eutrophus JMP134 is a homotetramer. Can. J. Microbiol. 43:202-205. [DOI] [PubMed] [Google Scholar]
- 7.Fritsche, K., G. Auling, J. R. Andreesen, and U. Lechner. 1999. Defluvibacter lusatiae gen. nov., sp. nov., a new chlorophenol-degrading member of the alpha-2 subgroup of proteobacteria. Syst. Appl. Microbiol. 22:197-204. [DOI] [PubMed] [Google Scholar]
- 8.Fukumori, F., and R. P. Hausinger. 1993. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase. J. Bacteriol. 175:2083-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerischer, U., and L. N. Ornston. 1995. Spontaneous mutations in pcaH and -G, structural genes for protocatechuate 3,4-dioxygenase in Acinetobacter calcoaceticus. J. Bacteriol. 177:1336-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gobel, M., K. Kassel-Cati, E. Schmidt, and W. Reineke. 2002. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: cloning, characterization, and analysis of sequences encoding 3-oxoadipate:succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase. J. Bacteriol. 184:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic selection markers for cloning and stable chromosomal insertion of foreign DNA in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann, D., S. Kleinsteuber, R. H. Muller, and W. Babel. 2003. A transposon encoding the complete 2,4-dichlorophenoxyacetic acid degradation pathway in the alkalitolerant strain Delftia acidovorans P4a. Microbiology 149:2545-2556. [DOI] [PubMed] [Google Scholar]
- 13.Jeong, J. J., J. H. Kim, C. K. Kim, I. Hwang, and K. Lee. 2003. 3- and 4-alkylphenol degradation pathway in Pseudomonas sp. strain KL28: genetic organization of the lap gene cluster and substrate specificities of phenol hydroxylase and catechol 2,3-dioxygenase. Microbiology 149:3265-3277. [DOI] [PubMed] [Google Scholar]
- 14.Kivisaar, M., R. Horak, L. Kasak, A. Heinaru, and J. Habicht. 1990. Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid 24:25-36. [DOI] [PubMed] [Google Scholar]
- 15.Koh, S. C., M. V. McCullar, and D. D. Focht. 1997. Biodegradation of 2,4-dichlorophenol through a distal meta-fission pathway. Appl. Environ. Microbiol. 58:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 17.Kröckel, L., and D. D. Focht. 1987. Construction of chlorobenzene-utilizing recombinants by progenitive manifestation of a rare event. Appl. Environ. Microbiol. 53:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, C. M., J. H. Leveau, A. J. B. Zehnder, and J. R. van der Meer. 2000. Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134 (pJP4). J. Bacteriol. 182:4165-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli, C. M., C. Werlen, and J. R. van der Meer. 2004. Mutation analysis of the different tfd genes for degradation of chloroaromatic compounds in Ralstonia eutropha JMP134. Arch. Microbiol. 181:112-121. [DOI] [PubMed] [Google Scholar]
- 20.Lefebre, M. D., and M. Valvano. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leveau, J. H., A. J. B. Zehnder, and J. R. van der Meer. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134 (pJP4). J. Bacteriol. 180:2237-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leveau, J. H., F. Konig, H. Fuchslin, C. Werlen, and J. R. van der Meer. 1999. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol. Microbiol. 33:396-406. [DOI] [PubMed] [Google Scholar]
- 23.Liu, T., and P. J. Chapman. 1984. Purification and properties of a plasmid-encoded 2,4-dichlorophenol hydroxylase. FEBS Lett. 173:314-318. [DOI] [PubMed] [Google Scholar]
- 24.Makdessi, K., and U. Lechner. 1997. Purification and characterization of 2,4-dichlorophenol hydroxylase isolated from a bacterium of the alpha-2 subgroup of the Proteobacteria. FEMS Microbiol. Lett. 157:95-101. [DOI] [PubMed] [Google Scholar]
- 25.Nurk, A., L. Kasak, and M. Kivisaar. 1991. Sequence of the gene (pheA) encoding phenol monooxygenase from Pseudomonas sp. EST1001: expression in Escherichia coli and Pseudomonas putida. Gene 102:13-18. [DOI] [PubMed] [Google Scholar]
- 26.Parke, D., and L. N. Ornston. 2004. Toxicity caused by hydroxycinnamoyl-coenzyme A thioester accumulation in mutants of Acinetobacter sp. strain ADP1. Appl. Environ. Microbiol. 70:2974-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Pantoja, D., L. Guzmán, M. Manzano, D. H. Pieper, and B. González. 2000. Role of tfdCIDIEIFI and tfdDIICIIEIIFII gene modules in catabolism of 3-chlorobenzoate by Ralstonia eutropha JMP134 (pJP4). Appl. Environ. Microbiol. 66:1602-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Pantoja, D., T. Ledger, D. H. Pieper, and B. González. 2003. Efficient turnover of chlorocatechols is essential for growth of Ralstonia eutropha JMP134 (pJP4) in 3-chlorobenzoic acid. J. Bacteriol. 185:1534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pieper, D. H., W. Reineke, K. H. Engesser, and H.-J. Knackmuss. 1988. Metabolism of 2,4-dichlorophenoxyacetic acid, 4-chloro-2-methylphenoxy-acetic acid and 2-methylphenoxyacetic acid by Alcaligenes eutrophus JMP134. Arch. Microbiol. 150:95-102. [Google Scholar]
- 30.Plumeier, I., D. Pérez-Pantoja, S. Heim, B. González, and D. H. Pieper. 2002. The importance of different tfd genes for the degradation of chloroaromatics by Ralstonia eutropha JMP134. J. Bacteriol. 184:4054-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radjendirane, V., M. A. Bhat, and C. S. Vaidyanathan. 1991. Affinity purification and characterization of 2,4-dichlorophenol hydroxylase from Pseudomonas cepacia. Arch. Biochem. Biophys. 288:169-176. [DOI] [PubMed] [Google Scholar]
- 32.Schlömann, M., E. Schmidt, and H.-J. Knackmuss. 1990. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J. Bacteriol. 172:5112-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schüürmann, G., H. Segner, and K. Jung. 1997. Multivariate mode of action analysis of acute toxicity of phenols. Aquat. Toxicol. 38:277-296. [Google Scholar]
- 34.Schweigert, N., A. J. B. Zehnder, and R. I. L. Eggen. 2001. Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ. Microbiol. 3:81-91. [DOI] [PubMed] [Google Scholar]
- 35.Sikkema, J., J. A. M. de Bont, and B. Poolman. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takemori, S., M. Kakamura, K. Suzuki, and M. Katagiri. 1970. The kinetics of salicylate hydroxylase reaction. FEBS Lett. 6:305-308. [DOI] [PubMed] [Google Scholar]
- 37.Trefault, N., R. De la Iglesia, A. M. Molina, M. Manzano, T. Ledger, D. Pérez-Pantoja, M. A. Sánchez, M. Stuardo, and B. González. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6:655-668. [DOI] [PubMed] [Google Scholar]
- 38.Vallaeys, T., L. Courde, C. McGowan, A. D. Wright, and R. R. Fulthorpe. 1999. Phylogenetic analyses indicate independent recruitment of diverse gene cassettes during assemblage of the 2,4-D catabolic pathway. FEMS Microbiol. Ecol. 28:373-382. [Google Scholar]