Abstract
Scanning of bacterial genomes to identify essential genes is of biological interest, for understanding the basic functions required for life, and of practical interest, for the identification of novel targets for new antimicrobial therapies. In particular, the lack of efficacious antimicrobial treatments for infections caused by the Burkholderia cepacia complex is causing high morbidity and mortality of cystic fibrosis patients and of patients with nosocomial infections. Here, we present a method based on delivery of the tightly regulated rhamnose-inducible promoter PrhaB for identifying essential genes and operons in Burkholderia cenocepacia. We demonstrate that different levels of gene expression can be achieved by using two vectors that deliver PrhaB at two different distances from the site of insertion. One of these vectors places PrhaB at the site of transposon insertion, while the other incorporates the enhanced green fluorescent protein gene (e-gfp) downstream from PrhaB. This system allows us to identify essential genes and operons in B. cenocepacia and provides a new tool for systematically identifying and functionally characterizing essential genes at the genomic level.
As the number of sequenced bacterial genomes rapidly expands, there is increased interest in learning how many and which of the annotated open reading frames (ORFs) fall into the category “essential.” Essential genes encode functions that are absolutely required for growth or viability (38). The discovery of novel essential genes not only contributes to the unraveling of previously unrecognized, essential cellular functions but also may help in identifying novel targets for new antibacterial molecules (16, 40).
Despite vast differences in size and gene repertories among bacterial genomes, a substantial number of essential genes appears to be conserved (24), suggesting that a core set of genes encodes key cellular functions (18). Methods of scanning microbial genomes for essential genes include direct gene disruption strategies such as random transposition (1, 17, 23, 43) and systematic gene-by-gene inactivation (26, 29, 46). This approach does not consider that many essential genes exist in operons (11, 35), and it has the potential to lead to incorrect classifications of nonessential genes as essential due to polar effects in operons containing a mixture of both essential and nonessential genes. This was experimentally assessed by Thanassi et al. (46), who found that 42% of the putative essential genes identified in Streptococcus pneumoniae were misidentified as such due to polar inactivation of true essential genes downstream. Furthermore, recent work has shown not only that essential genes are more likely to exist within operons than are nonessential genes (11, 35) but also that essential genes with related functions have a strong tendency to cluster even when they are not organized in operons (35).
Another general strategy for identifying essential genes is functional suppression either by antisense mRNA induction (15, 49) or by the transposon-based delivery of inducible promoters such as the arabinose-regulated promoter (PBAD) (25) and the tetracycline-inducible promoter (5, 13). When a conditionally lethal phenotype is obtained, the identified gene downstream of the inserted promoter is usually defined operationally as essential. Mutants with conditionally lethal phenotypes provide an opportunity for the functional characterization of essential genes. Growth conditions have a large impact on determining whether a particular gene is essential. However, growth on solid rich medium was commonly used in all of the above-mentioned methodologies.
The Burkholderia cepacia complex is a group of gram-negative bacteria comprising at least nine species (33, 48) which have emerged as multidrug-resistant nosocomial pathogens in immunocompromised patients, particularly in those with chronic granulomatous diseases and cystic fibrosis. B. cepacia complex isolates from patients with cystic fibrosis, particularly those from B. cenocepacia, can be transmitted from patient to patient, and the infection often results in rapid deterioration of the lung and a life-threatening pneumonia termed “cepacia syndrome” (22). Treatment of these infections is very difficult because of the intrinsic resistance of the B. cepacia complex to most clinically useful antibiotics (19). Thus, it becomes important to identify newer and improved antibacterial therapies for patients with cystic fibrosis. One potential approach is to identify in the B. cepacia complex novel essential genes whose products could become targets of new antibiotics.
The B. cepacia complex and Burkholderia in general are characterized by large genomes, possessing three to five chromosomes depending on the specific strain (31). Relative to other bacteria, very few molecular tools are available to genetically characterize and manipulate Burkholderia spp. (7, 12, 30, 47). We have previously reported the construction of an expression vector based on the Escherichia coli rhamnose-inducible promoter (PrhaB) (21), which provides tightly regulated gene expression in B. cenocepacia (4). In this study, we report the development of a transposon system that delivers an outwardly oriented PrhaB at two different distances from the point of insertion, based on incorporation of the enhanced green fluorescent protein gene (e-gfp) downstream from PrhaB near the insertion site. This system has allowed us to identify several essential genes and operons in B. cenocepacia.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. cenocepacia strain K56-2 (10) is a clinical isolate of the same clonal group as strain J2315, whose genome has recently been sequenced (http://www.sanger.ac.uk/Projects/B_cenocepacia/). E. coli K-12 strain DH5α [F− φ80lacZM15 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 ΔgyrA96 relA1 Δ(lacZYA-argF)U169] was used for the construction of plasposon derivatives and maintenance of the helper plasmid pRK2013 (see below). Bacteria were grown at 37°C in Luria-Bertani (LB) medium supplemented, as required, with 100 μg/ml trimethoprim (Tp) and 50 μg/ml gentamicin for B. cenocepacia and 50 μg/ml Tp or 40 μg/ml kanamycin for E. coli. All chemicals were purchased from Sigma Chemical Co., St. Louis, Mo., unless otherwise indicated.
Recombinant DNA methods.
DNA extractions were performed with the DNeasy tissue kit from QIAGEN Inc., Canada. DNA ligations, restriction endonuclease digestions, and agarose gel electrophoresis were performed according to standard techniques (42). Restriction enzymes and T4 DNA ligase were purchased from Roche Diagnostics, Laval, Quebec, Canada. DNA transformation experiments with E. coli were carried out by the calcium chloride method (8). Plasmids were transferred into B. cenocepacia by triparental mating (9) using pRK2013 as a helper plasmid (14). PCRs were performed using the PTC-0200 or PTC-221 DNA engine (MJ Research, Incline Village, Nev.). Pwo polymerase (Roche) was used for cloning of the e-gfp gene. PCR amplifications larger than 3 kb were performed with the EXPAND High Fidelity PCR system (Roche). Colony PCR was performed with Taq polymerase using 0.5 M GC-rich resolution solution (Roche) when required. Amplification conditions were optimized for each primer pair. PCR products were separated on 0.7 to 1.0% (wt/vol) agarose gels, and the bands were purified with the QiaQuick gel extraction system (QIAGEN). Ligation mixtures were transformed into E. coli DH5α, and transformants were plated on LB agar plates with the appropriate antibiotic for selection. Resistant colonies were isolated and screened for the presence of plasmid.
Construction of vectors pSCrhaBout and pSCrhaBoutgfp.
Plasmids are listed in Table 1. pTnMod-OTp′ (12) was digested with KpnI and ligated to the arabinose-inducible system, amplified from the plasmid pBAD24 (20) by PCR. The resulting plasmid, pCM3, was used to clone the e-gfp gene under the control of the PBAD promoter. The e-gfp gene was amplified by PCR from pSCrhaB2-e-GFP using the primers 1045 and 1046 (Table 2). pCM3 and the amplified e-gfp gene fragment were digested with NsiI and SfiI, purified, and ligated, yielding pCM3gfp (Fig. 1). To generate pSCrhaBoutgfp, pCM3gfp was used as a template for inverse PCR amplification (34) with primers 776 and 1084 (Table 2), removing the arabinose-inducible system (Fig. 1). The rhamnose-inducible system was obtained from pSCrhaB2 (Table 1) by digestion with NsiI and NdeI and ligated to the amplified fragment from pCM3gfp. To obtain pSCrhaBout, the e-gfp gene from pSCrhaBoutgfp was removed by inverse PCR amplification of pSCrhaBoutgfp with primers 1512 and 1554, followed by purification and self-ligation (Fig. 1).
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pRK2013 | RK2 derivative, Kmrmob+tra+ ColE1 | 14 |
| pSCrhaB2 | oripBBR1rhaR rhaS PrhaB Tprmob+ | 4 |
| pSCrhaB2-e-GFP | pSCrhaB2, e-gfp | 4 |
| pCM3 | pTnMod-OTp′, araC PBAD | This work |
| pCM3gfp | pCM3, e-gfp | This work |
| pSCrhaBoutgfp | pTnMod-OTp′, rhaR rhaS PrhaB e-gfp | This work |
| pSCrhaBout | pTnMod-OTp′, rhaR rhaS PrhaB | This work |
Kmr, kanamycin resistance; Tpr, trimethoprim resistance.
TABLE 2.
Oligonucleotides used in this study
| Purpose and name | Oligonucleotide sequence, 5′-3′a |
|---|---|
| Vector construction | |
| 1045 | CCAATGCATATGGTGAGCAAGGGCGAG |
| 1046 | AATAATGGCCACCTAGGCCTTACTTGTA CAGCTCGTCC |
| 776 | ATTAGACCATATGGTGAGCAAGGGCG AGGA |
| 1084 | TTGATGCATTTGGTAACGAATCAGACA ATTGACG |
| 1512 | GCCCTTGCTCACCATATGTGATCCTGCT GAAT |
| 1554 | ATCACTCCATATGTGGACGAGCTGTAC AAGTAA |
| Sequencing of rhaBoutgfp and rhaBout mutants | |
| 824 | GCCCATTTTCCTGTCAGTAACGAGA |
| 1510 | CGATCACATGGTCCTGCTGGAG |
| RT-PCR | |
| nrfG2F | GAAGATCGTGTCGCCGCCGAAA |
| ppiA1R | TCGAGGAAGTTGGCGACGGATTT |
| ppiA2F | CGGTGTTCGGCAAGGTCGTGT |
| ppiB1R | CGAGTGGTTCAGGAAGTCGTTGT |
| ppiB2F | CGACAAGATCAAGGGCGTCAAGA |
| lpxH1R | GTGCAGATCGGAGAGAAACAGGAA |
| hisD1F | GCGTACCACGAGAAGCAGAAGAT |
| hisD2R | CCAGCACGAGGTCGTTCTTCA |
Restriction sites are underlined.
FIG. 1.
Construction of the transposon vectors pSCrhaBout and pSCrhaBoutgfp. The backbone of pCM3gfp was amplified by inverse PCR and ligated to the digested rhaR-rhaS-PrhaB region to form pSCrhaBoutgfp. pSCrhaBout was obtained by inverse PCR amplification of pSCrhaBoutgfp with divergent primers flanking the e-gfp gene. IR, inverted repeats; oriT, origin of transfer; dhfr, trimethoprim resistance cassette; pMB1 ori, origin of replication for E. coli; rhaR and rhaS, transcription regulator genes of the rhamnose system; PrhaB, rhamnose-inducible promoter. The numbers represent the primers used in inverse PCR, which are listed in Table 2.
Western blotting.
Bacterial cultures were grown in LB medium with 2% arabinose, or in rhamnose or glucose at the indicated levels, for 24 h. One milliliter of a bacterial culture of an optical density at 600 nm (OD600) of 3 was harvested by centrifugation and resuspended in loading dye (1 mg [wet weight]/μl), and the samples were incubated at 95°C for 5 min. Samples (5 ml) were loaded in a 15% sodium dodecyl sulfate-polyacrylamide gel, and the samples were separated by electrophoresis. Proteins were transferred to polyvinylidene difluoride membranes for 1 h at 250 mA, and GFP was reacted with rabbit anti-GFP antibody (Chemicon AB3080) as the primary antibody and horseradish peroxidase-linked sheep anti-rabbit immunoglobulin G IR dye 800 as the secondary antibody. Detection by chemiluminescence was performed with Chemiluminescence Blotting Substrate (Roche), as recommended by the manufacturer.
Generation of B. cenocepacia K56-2 rhamnose-dependent mutants.
Plasmid pSCrhaBoutgfp or pSCrhaBout was conjugated into B. cenocepacia K56-2 by triparental mating (9) using E. coli DH5α pRK2013 as a helper strain (14). Exconjugants were selected on LB agar plates supplemented with 100 μg/ml Tp, 50 μg/ml gentamicin, and 0.2% rhamnose. The resulting colonies were picked and arranged in LB plates supplemented with 100 μg/ml Tp and 0.2% rhamnose and then replica plated in LB plates with 100 μg/ml Tp and 0.2% rhamnose or 0.2% glucose. Transposon mutant clones that showed a rhamnose-dependent growth phenotype were picked and grown from an isolated colony. After reassessment of the conditional phenotype, clones were stored as glycerol stocks for further analysis.
Identification of transposon insertion sites.
The chromosomal sequences flanking the transposon insertions were identified by the self-cloning strategy described previously (18). Briefly, chromosomal DNA from B. cenocepacia transposon mutants was isolated and subjected to restriction endonuclease digestion by either NotI or XhoI. Digests were ligated under dilute conditions to favor intramolecular ligations with T4 DNA ligase and transformed into competent E. coli DH5α. Transformants were selected on LB agar supplemented with Tp at 50 mg/ml. Plasmids were isolated with the High Pure plasmid isolation kit (Roche Diagnostics) and sequenced at the Core Molecular Biology Facility (York University, Ontario, Canada) with primer 824 or 1510 (Table 2) for plasmid rhaBout or rhaBoutgfp, respectively. The DNA sequences were compared to the genome of B. cenocepacia J2315 by BLAST (32) to identify the precise insertion sites. These sequence data were produced by the B. cenocepacia J2315 Sequencing Group at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/bc/BC_chr1.dna. Analysis of the chromosomal region downstream of the transposon insertion was performed with Artemis software (41).
RNA isolation methods and RT-PCR analysis.
For RNA isolation, bacteria grown in liquid cultures were harvested and lysed in 10 mM Tris-Cl-1 mM EDTA, pH 8.0, containing 400 μg/ml lysozyme for 5 min at room temperature. RNA was recovered with the RNeasy Mini kit (QIAGEN) as instructed by the manufacturer. The integrity of the RNA was assessed by electrophoresis in a 1.0% agarose gel using Tris-borate-EDTA buffer. Residual DNA was removed by treatment with DNase I (30°C, 30 min) in DNase buffer (QIAGEN). The DNase was inactivated with 2.5 mM EDTA (65°C, 10 min). The RNA was then used as a template in reverse transcriptase (RT) PCR or aliquoted and stored at −80°C. Reverse transcription was performed with the Transcriptor Reverse Transcriptase kit (Roche) according to the manufacturer's instructions with 1.6 μM of the appropriate primers (Table 2). The resulting cDNA was subjected to PCR using Taq DNA polymerase (QIAGEN). The PCR amplification cycle consisted of 2 min at 94°C; followed by 24 three-step amplification cycles of 30 s at 94°C, 30 s at 55°C to 60°C, and 1 min at 72°C; and a final extension of 7 min at 72°C.
For each PCR, the appropriate controls with water and RNA in the absence of RT were included to ensure that the obtained amplifications were a result of cDNA and not of contaminating genomic DNA in the RNA preparation or in the reagents.
Semiquantitative RT-PCR.
Primers hisD1F and hisD1R, which amplify an internal fragment of the hisD gene from B. cenocepacia, were used as the internal control for the quantification of gene expression. To determine that the PCR remained in the linear phase of amplification, aliquots from a 100-μl reaction were removed at different numbers of PCR cycles. The PCR amplification cycle consisted of 2 min at 94°C; followed by 29 three-step amplification cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C; and a final extension of 7 min at 72°C. The RT-PCR products were then analyzed by electrophoresis on a 1.2% agarose gel and visualized by a UV transilluminator after staining with ethidium bromide. Semiquantitative analysis of the RT-PCR products was performed by densitometry with Quantity One, version 4.50, software (Bio-Rad). The amplification product was normalized according to hisD expression.
Nucleotide sequence accession numbers.
The nucleotide sequences of plasmids pSCrhaBoutgfp and pSCrhaBout have been deposited in the GenBank database under accession no. DQ317694 and DQ317695, respectively.
RESULTS AND DISCUSSION
Rhamnose-inducible gene expression delivered into the chromosome of B. cenocepacia K56-2.
To identify essential B. cenocepacia K56-2 genes, we chose to deliver by transposition an outward arabinose-inducible promoter, as previously done by Judson and Mekalanos (25). Since the arabinose-regulated system in the plasmid pMLBAD provided inducible gene expression in B. cenocepacia (30), we predicted that insertion of the PBAD promoter upstream of an essential gene would result in the production of arabinose-dependent mutants. To test this hypothesis we constructed pCM3, a derivative of pTnMod-OTp′ carrying an outward arabinose-inducible promoter and the regulator gene araC in the opposite orientation (Table 1). After repeated attempts, we could not recover any transposon mutant with a conditionally lethal phenotype in the absence of arabinose (data not shown). To determine the level of chromosomal gene expression driven by the arabinose system in B. cenocepacia, we cloned the e-gfp gene within the inverted repeats and under the control of the PBAD promoter in pCM3. After conjugation of pCM3gfp into B. cenocepacia, several colonies were assayed by colony PCR to confirm the presence of the transposon insertion in the B. cenocepacia chromosome. Although the presence of an intact e-gfp gene in the chromosome was confirmed, we could not detect GFP by fluorescence microscopy or by Western blotting using anti-GFP antibodies under any of the arabinose concentrations examined, which ranged from 0.2% to 2% (wt/vol) (data not shown). The lack of e-gfp expression was not due to a mutation in this gene, since GFP was expressed from pCM3gfp in E. coli. We concluded that the chromosomal copy of PBAD could not drive detectable arabinose-regulated gene expression in B. cenocepacia. Therefore, the arabinose-inducible system is not appropriate for gene expression at chromosomal levels in B. cenocepacia.
Recently, we discovered that the E. coli rhamnose-inducible system (21) provides tight gene regulation in B. cenocepacia (4). Therefore, we developed a genetic system to deliver PrhaB into the B. cenocepacia chromosome by replacing the arabinose system in pCM3gfp with the E. coli rhamnose-inducible system, resulting in construction of the plasmid pSCrhaBoutgfp (Fig. 1). Also, the e-gfp gene was further removed from pSCrhaBoutgfp by inverse PCR and self-ligation to render the plasmid pSCrhaBout (Fig. 1). pSCrhaBoutgfp was conjugated into B. cenocepacia to verify that PrhaB expression can be detected after random insertion in the chromosome. Expression of GFP under the control of PrhaB was observed by Western blot analysis at different concentrations of rhamnose in the growth medium, while no GFP was detected in the absence of rhamnose or in the presence of glucose (Fig. 2a). These results demonstrate that PrhaB can be delivered to the B. cenocepacia chromosome, where it provides detectable expression levels under the control of rhamnose in the growth medium.
FIG. 2.
Identification of essential B. cenocepacia genes. (a) Western blot analysis of chromosomally located e-gfp expression under inducing and noninducing conditions. The arrow indicates the position of GFP. (b) Identification of rhamnose-dependent mutants by replica plating. (c) Rhamnose-dependent growth phenotype of mutant rhaBoutgfp 3. (d) Identification of the insertion site in rhamnose-dependent mutants by self-cloning. See the text for details.
Identification of essential genes and essential operons in B. cenocepacia K56-2.
Based on the previous results, B. cenocepacia K56-2 was mutagenized with the suicide plasposons pSCrhaBoutgfp and pSCrhaBout. Essential genes were screened and identified as depicted in Fig. 2b to d. Exconjugants obtained by triparental mating were arranged on LB agar plates containing rhamnose and were replica plated to plates with rhamnose or glucose (Fig. 2b). From 37,000 colonies screened in this manner, we identified 17 colonies that grew in plates with rhamnose but did not grow in the presence of glucose. These putative rhamnose-dependent colonies were repurified from the original plate and reexamined by plating on LB agar supplemented with either 0.2% rhamnose or 0.2% glucose (Fig. 2c). The rhamnose-dependent growth phenotype was confirmed for 15 of the 17 candidates. Figure 3a shows the growth of all mutants after 24 h of incubation with rhamnose or glucose. Most of the mutants reached levels of growth comparable to the wild type in the presence of saturating concentrations of rhamnose. Others showed comparatively less growth, probably due to a lack of optimal expression of the rhamnose-controlled genes at 0.2% rhamnose. Nevertheless, growth was repressed to different extents in all of the mutants. Induction ratios ranged from 1.5, in mutant rhaBout 13, to 18.7, in mutant rhaBoutgfp 9 (Fig. 3b).
FIG. 3.
Growth of rhamnose-dependent mutants under inducing and noninducing conditions. Mutants were inoculated with toothpicks onto 96-well microtiter plates containing LB with rhamnose or glucose and incubated for 24 h at 37°C without shaking. (a) Growth was monitored by measuring the OD570. The numbers in the x axis correspond to the names of the mutants (Table 3); wt, B. cenocepacia K56-2 parental strain. Results are averages of 12 repetitions. (b) The induction ratio for each mutant was calculated as the OD570 under inducing conditions divided by the OD570 under repressing conditions, as shown in panel a.
The chromosomal insertion of the transposon sequence was demonstrated by colony PCR and Southern blot hybridization (data not shown). All of the mutants had only one transposon insertion. To identify the insertion site, the chromosomal DNA was extracted, digested, and self-ligated under dilute conditions, and the ligation mixture was transformed into E. coli (Fig. 2d). Recovered plasmids served as templates for sequencing reactions using primers homologous to the 3′ terminus of the transposon. Sequences were 100% identical to the corresponding sequences of the genome of B. cenocepacia J2315. In most cases, the insertions were in gene clusters, probably operons. Thus, the presence of at least one essential gene in a transcriptional unit would identify an “essential operon.” Two or more genes found downstream of a given transposon insertion site, in the same strand as PrhaB, and at a distance between genes equal to or less than 150 bp were considered part of the essential operon putatively controlled by the rhamnose-regulated promoter. The maximal 150-bp distance between genes for considering neighboring genes to be part of the same operon was adopted according to operon prediction studies (6). We tested this prediction experimentally by performing a transcriptional analysis of the nrfG-ppiA-ppiB-lpxC gene cluster by RT-PCR (Fig. 4). We prepared cDNA from RNA isolated from B. cenocepacia K56-2 and amplified it with primers that would allow detection of cotranscription (Fig. 4a). Amplification of DNA fragments of the expected size demonstrated that the contiguous genes in this region are organized into an operon structure (Fig. 4b).
FIG. 4.
Transcriptional analysis of the nrfG-ppiA-ppiB-lpxC cluster. (a) Schematic drawing of the putative operon (top) and location of the primers used in RT-PCR experiments and expected amplified band size for each pair of primers (bottom). (b) RT-PCR amplification of intergenic regions. The arrows indicate the positions of the amplified bands and the observed sizes.
DNA sequences of the regions flanking insertion sites revealed that all of the insertions occurred in the largest chromosome of B. cenocepacia and mainly in the leading strand, as has been described for essential genes in other studies (39). Only four mutants had rhamnose-controlled genes located in the lagging strand. The mutants and their characteristics are listed in Table 3. Some regions, such as the nrfG-ppiA-ppiB-lpxC cluster, were hit more than once in independent conjugation experiments. The insertion site was located upstream the nrfG gene and was identical for the mutants rhaBoutgfp 4 and rhaBout 12, even though conjugation was performed with the two different versions of the transposon (pSCrhaBoutgfp and pSCrhaBout). The rhaBoutgfp 7 and rhaBoutgfp 8 mutants were also identical, with the transposon insertion interrupting the ppiB gene. The rnc-era-recO operon (36) appears to be another hot spot, since two mutants, rhaBoutgfp 6 and rhaBoutgfp 11, were also obtained independently. Most rhamnose-dependent mutants had insertions upstream of genes whose homologues were shown in other studies to be essential (Table 3). There were only two exceptions: K56-2::rhaBoutgfp 3 and K56-2::rhaBoutgfp 5. In the former mutant, the transposon was inserted within the sensor kinase gene of a two-component regulator system, and presumably PrhaB controls the expression of the downstream response regulator gene atoC. It is not clear whether the essential phenotype is rescued by rhamnose-inducible expression of atoC or the downstream tRNA gene for phenylalanine. In K56-2::rhaBoutgfp 5, the transposon was inserted between two genes related to the biosynthesis of arginine and pyrimidine, carA and carB. In this mutant, PrhaB appears to control carB and, downstream, the genes greA and proP. GreA has been described as a transcription cleavage factor (3), and ProP is a transporter protein (37). None of these functions appear to be essential for growth of other bacteria, but one or more may be required for survival of B. cenocepacia. The rhaBout 14 and rhaBout 15 mutants have the transposon inserted in a position where there are no annotated downstream genes. The cause of the rhamnose-dependent phenotype in these mutants is unknown and currently under investigation.
TABLE 3.
Rhamnose-dependent mutants of B. cenocepacia K56-2a
| Mutant | IR-ATG (bp)b | Downstream gene(s) and putative operonsc | Predicted general functiond |
|---|---|---|---|
| RhaBoutgfp 1 | 68 | ygiH | Unknown |
| RhaBoutgfp 2 | 495 | ftsL ftsI murE murF mraY murD ftsW murG murC ddlA ftsQ ftsA ftsZ | Cell division |
| RhaBoutgfp 3 | 340 | atoC (CheY-like response regulator protein) | Signal transduction mechanism |
| RhaBoutgfp 4 | 25 | nrfG ppiA ppiB lpxH | Periplasmic protein folding and synthesis of lipid A |
| RhaBout 12 | 25 | nrfG ppiA ppiB lpxH | Periplasmic protein folding and synthesis of lipid A |
| RhaBoutgfp 5 | 422 | carB greA proP | Biosynthesis and transport of small molecules |
| RhaBoutgfp 6 | 587 | lepB rnc era recO pdxJ acpS bglX | Ribosome biogenesis |
| RhaBoutgfp 11 | 892 | lepB rnc era recO pdxJ acpS bglX | Ribosome biogenesis |
| RhaBoutgfp 7 | 244 | lpxH | Synthesis of lipid A |
| RhaBoutgfp 8 | 244 | lpxH | Synthesis of lipid A |
| RhaBoutgfp 9 | 120 | iscS iscU iscA djlA/hscB hscA fdx yfhJ | Housekeeping Fe-S cluster assembly |
| RhaBoutgfp 10 | 8 | hemE | Biosynthesis of heme |
| RhaBout 13 | 373 | purP/degA BCAL1801 BCAL1800 | Unknown |
| RhaBout 14 | Chromosome 1 base 202755 | Unknown | |
| RhaBout 15 | Chromosome 1 base 1988972 | Unknown |
Predicted B. cenocepacia proteins were matched with homologues by the COGnitor tool from the COGs database (45). When information was available, essential homologues in the genomes of E. coli (17), Haemophilus influenzae (1), Mycobacterium tuberculosis (43), and Pseudomonas aeruginosa (23) were identified.
Distance, in base pairs, between the inverted repeats (IR) of the transposon and the start codon (ATG) of the nearest downstream gene in the same strand as PrhaB.
Genes found to be essential in at least one of the previously mentioned genomes are shown in bold. Operons were arbitrarily defined as same-strand coding genes with intergenic distances of less than 150 bp.
General function was predicted based on information from the EcoCyc database (27).
The presence of the e-gfp gene downstream of PrhaB provides wild-type levels of expression of the nrfG-ppiA-ppiB-lpxC operon.
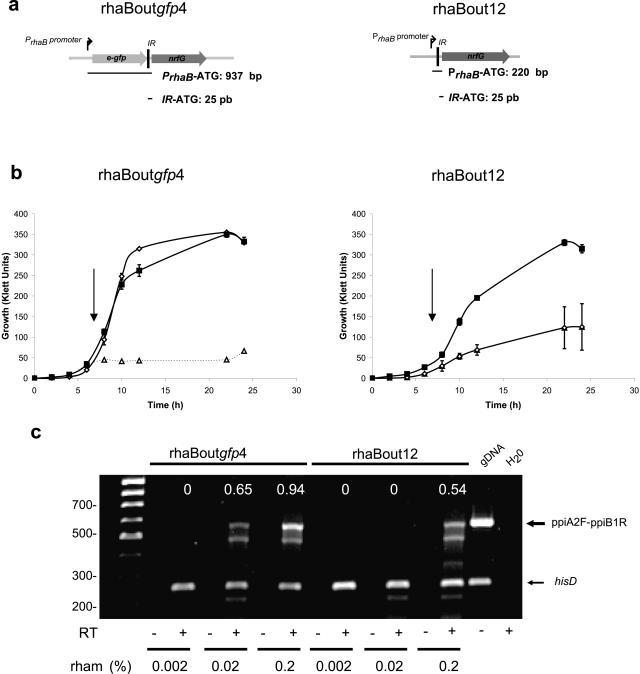
We hypothesized that transposons pSCrhaBoutgfp and pSCrhaBout would drive different levels of gene expression, given the different distances from PrhaB to the start codon of the putative essential gene, downstream of the insertion site (717 bp longer in pSCrhaBoutgfp than in pSCrhaBout). Also, we hypothesized that the presence or absence of the upstream e-gfp gene could influence the expression levels of the downstream genes. Isolation of the rhaBoutgfp 4 and rhaBout 12 mutants, which have identical insertion sites upstream of the nrfG gene (Fig. 5a), provided us an opportunity to address this idea experimentally. Figure 5b shows the growth curves of both mutants at three different concentrations of rhamnose. RhaBoutgfp 4 grew to wild-type levels at 0.02% and 0.2% rhamnose, while 0.002% rhamnose did not support growth. Conversely, neither 0.002% nor 0.02% rhamnose supported the growth of RhaBout 12 (Fig. 5b), while growth at 0.2% rhamnose was possible but to lower-than-wild-type levels. The differences in growth were not due to the expression of GFP itself, since growth of a B. cenocepacia K56-2 strain carrying a plasmid with a rhamnose-inducible e-gfp gene was identical at the three concentrations of rhamnose tested (data not shown). To demonstrate that the differences in growth in both strains were due to differences in the expression levels of the operon, a semiquantitative RT-PCR was performed (Fig. 5c). We normalized the transcription expression levels by amplifying a fragment of the hisD gene, as described before (2). Aliquots were taken during growth of rhaBoutgfp 4 and rhaBout 12 mutants at different levels of rhamnose. The RNA was extracted, and the relative levels of operon transcripts were analyzed. No transcription was detected at 0.002% rhamnose for rhaBoutgfp 4 or rhaBout 12 or at 0.02% rhamnose for rhaBout 12. In contrast, transcriptional expression was detected at 0.2% rhamnose for rhaBoutgfp 4 and rhaBout 12 and at 0.02% rhamnose for rhaBoutgfp 4. We conclude that the different expression levels of the essential operon in each mutant, which most likely reflect more-robust gene expression of the nrfG-ppiA-ppiB-lpxC operon in the presence of the intervening e-gfp gene, cause the growth differences between rhaBoutgfp 4 and rhaBout 12.
FIG. 5.
Comparative growth and gene expression analysis of rhaBoutgfp 4 and rhaBout12 mutants. (a) Distance of PrhaB from the start codon (ATG) of nrfG for both mutants. (b) Growth curves in LB medium. Black squares, 0.2% rhamnose; open diamonds, 0.02% rhamnose; open triangles and dotted lines, 0.002% rhamnose. Arrows represent the times at which aliquots were removed for RNA extraction. (c) Relative RT-PCR. Total RNA was extracted from mutants rhaBoutgfp 4 and rhaBout 12 at different levels of rhamnose (rham). The arrows indicate the positions of the internal control band hisD and the intergenic band amplified with primers ppiA2F and ppiB1R. gDNA, genomic DNA. The numbers at the top of the gel represent arbitrary levels of gene expression relative to the internal control hisD.
Concluding remarks.
We describe here a functional method for identifying essential genes and essential operons in B. cenocepacia by using the E. coli rhamnose-inducible promoter system. Delivery of an outward inducible promoter by transposition and identification of essential genes by screening for the conditional-growth phenotype were first developed by Judson and Mekalanos (25) using the arabinose-inducible promoter PBAD. These authors identified 16 arabinose-dependent-growth mutants in Vibrio cholerae. The same approach was further applied to E. coli (44). In this case, nine mutants were identified from over 25,000 colonies. However, the arabinose-regulated promoter did not provide lethal-conditional phenotypes in Salmonella, probably due to leakiness of the system (28). Together, these observations suggest that the PBAD promoter may not provide enough repression for the mutant to exhibit an arabinose-dependent growth phenotype. We show here that the PBAD promoter cannot provide gene expression at chromosomal levels in B. cenocepacia. Therefore, we turned to the rhamnose-inducible promoter system, which has a much tighter regulation than the arabinose systems in E. coli (21) and B. cenocepacia (4). Using pSCrhaBoutgfp and pSCrhaBout, we identified essential genes in B. cenocepacia K56-2 at frequencies of 1/2,600 and 1/2,900, respectively. The genome of B. cenocepacia J2315 comprises 8,128 predicted ORFs (the E. coli and V. cholerae genomes contain 4,409 ORFs and 3,890 ORFs, respectively). Thus, considering that the number of essential genes is not expected to increase with genome size, we predict that the rhamnose-inducible system will likely detect essential genes at a higher frequency than that previously described in other studies.
Regarding the identification of essential genes by conditional expression promoters, it has been argued that complete saturation of a genome would not be possible with a single vector because of the limited levels of basal and induced expression (25). We have demonstrated that in the case of the nrfG-ppiA-ppiB-lpxC operon, higher levels of regulated gene expression could be achieved by using pSCrhaBoutgfp, most likely because the levels of transcripts may remain more stable due to the transcription and/or translation of the e-gfp gene downstream of PrhaB.
In summary, the identification of 15 rhamnose-dependent B. cenocepacia mutants with the rhamnose-inducible promoter provides a starting point for developing a complete map of essential genes and essential operons in this bacterium, aiding the identification of novel targets for new antimicrobial drugs.
Acknowledgments
We thank Dobrinka Stoykova for technical assistance with replica plating and growth curves and Karen Keith for critical reading of the manuscript.
S.T.C. was supported by a postdoctoral fellowship from the Canadian Cystic Fibrosis Foundation. C.L.M. was supported by two consecutive undergraduate summer research awards from the Natural Sciences and Engineering Research Council of Canada. This study was supported by the special initiative in cystic fibrosis research “In Memory of Michael O'Reilly” from the Canadian Cystic Fibrosis Foundation and the Institute of Respiratory and Circulatory Health of the Canadian Institutes of Health Research. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Footnotes
Dedicated to the memory of Lucía Mondéjar de Cardona.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bittner, M., S. Saldias, C. Estevez, M. Zaldivar, C. L. Marolda, M. A. Valvano, and I. Contreras. 2002. O-antigen expression in Salmonella enterica serovar Typhi is regulated by nitrogen availability through RpoN-mediated transcriptional control of the rfaH gene. Microbiology 148:3789-3799. [DOI] [PubMed] [Google Scholar]
- 3.Borukhov, S., O. Laptenko, and J. Lee. 2001. Escherichia coli transcript cleavage factors GreA and GreB: functions and mechanisms of action. Methods Enzymol. 342:64-76. [DOI] [PubMed] [Google Scholar]
- 4.Cardona, S. T., and M. A. Valvano. 2005. An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid 54:219-228. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, P., D. G. Muttucumaru, and T. Parish. 2005. Use of a tetracycline-inducible system for conditional expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Appl. Environ. Microbiol. 71:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, X., Z. Su, P. Dam, B. Palenik, Y. Xu, and T. Jiang. 2004. Operon prediction by comparative genomics: an application to the Synechococcus sp. WH8102 genome. Nucleic Acids Res. 32:2147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, K. H., J. B. Gaynor, K. G. White, C. Lopez, C. M. Bosio, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443-448. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig, F. F., J. G. Coote, R. Parton, J. H. Freer, and N. J. Gilmour. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885-2890. [DOI] [PubMed] [Google Scholar]
- 10.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Daruvar, A., J. Collado-Vides, and A. Valencia. 2002. Analysis of the cellular functions of Escherichia coli operons and their conservation in Bacillus subtilis. J. Mol. Evol. 55:211-221. [DOI] [PubMed] [Google Scholar]
- 12.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, K. G. C., P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 16.Galperin, M. Y., and E. V. Koonin. 2004. “Conserved hypothetical” proteins: prioritization of targets for experimental study. Nucleic Acids Res. 32:5452-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil, R., F. J. Silva, J. Pereto, and A. Moya. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68:518-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govan, J. R., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldimann, A., L. L. Daniels, and B. L. Wanner. 1998. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J. Bacteriol. 180:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan, I. K., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin. 2002. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12:962-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Judson, N., and J. J. Mekalanos. 2000. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18:740-745. [DOI] [PubMed] [Google Scholar]
- 26.Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knuth, K., H. Niesalla, C. J. Hueck, and T. M. Fuchs. 2004. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol. Microbiol. 51:1729-1744. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefebre, M. D., and M. A. Valvano. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 32.Madden, T. L., R. L. Tatusov, and J. Zhang. 1996. Applications of network BLAST server. Methods Enzymol. 266:131-141. [DOI] [PubMed] [Google Scholar]
- 33.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 34.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal, C., and L. D. Hurst. 2004. Evidence against the selfish operon theory. Trends Genet. 20:232-234. [DOI] [PubMed] [Google Scholar]
- 36.Powell, B., H. K. Peters III, Y. Nakamura, and D. Court. 1999. Cloning and analysis of the rnc-era-recO operon from Pseudomonas aeruginosa. J. Bacteriol. 181:5111-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Racher, K. I., R. T. Voegele, E. V. Marshall, D. E. Culham, J. M. Wood, H. Jung, M. Bacon, M. T. Cairns, S. M. Ferguson, W. J. Liang, P. J. Henderson, G. White, and F. R. Hallett. 1999. Purification and reconstitution of an osmosensor: transporter ProP of Escherichia coli senses and responds to osmotic shifts. Biochemistry 38:1676-1684. [DOI] [PubMed] [Google Scholar]
- 38.Reich, K. A. 2000. The search for essential genes. Res. Microbiol. 151:319-324. [DOI] [PubMed] [Google Scholar]
- 39.Rocha, E. P., and A. Danchin. 2003. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 31:6570-6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosamond, J., and A. Allsop. 2000. Harnessing the power of the genome in the search for new antibiotics. Science 287:1973-1976. [DOI] [PubMed] [Google Scholar]
- 41.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 44.Serina, S., F. Nozza, G. Nicastro, F. Faggioni, H. Mottl, G. Deho, and A. Polissi. 2004. Scanning the Escherichia coli chromosome by random transposon mutagenesis and multiple phenotypic screening. Res. Microbiol. 155:692-701. [DOI] [PubMed] [Google Scholar]
- 45.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thanassi, J. A., S. L. Hartman-Neumann, T. J. Dougherty, B. A. Dougherty, and M. J. Pucci. 2002. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 30:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomlin, K. L., S. R. Clark, and H. Ceri. 2004. Green and red fluorescent protein vectors for use in biofilm studies of the intrinsically resistant Burkholderia cepacia complex. J. Microbiol. Methods 57:95-106. [DOI] [PubMed] [Google Scholar]
- 48.Valvano, M. A., K. E. Keith, and S. T. Cardona. 2005. Survival and persistence of opportunistic Burkholderia species in host cells. Curr. Opin. Microbiol. 8:99-105. [DOI] [PubMed] [Google Scholar]
- 49.Wang, B., and H. K. Kuramitsu. 2005. Inducible antisense RNA expression in the characterization of gene functions in Streptococcus mutans. Infect. Immun. 73:3568-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]