Abstract
The inclusion of antibiotic growth promoters, such as virginiamycin, at subtherapeutic levels in poultry feeds has a positive effect on health and growth characteristics, possibly due to beneficial effects on the host gastrointestinal microbiota. To improve our understanding of the chicken gastrointestinal microbiota and the effect of virginiamycin on its composition, we characterized the bacteria found in five different gastrointestinal tract locations (duodenal loop, mid-jejunum, proximal ileum, ileocecal junction, and cecum) in 47-day-old chickens that were fed diets excluding or including virginiamycin throughout the production cycle. Ten libraries (five gastrointestinal tract locations from two groups of birds) of approximately 555-bp chaperonin 60 PCR products were prepared, and 10,932 cloned sequences were analyzed. A total of 370 distinct cpn60 sequences were identified, which ranged in frequency of recovery from 1 to 2,872. The small intestinal libraries were dominated by sequences from the Lactobacillales (90% of sequences), while the cecum libraries were more diverse and included members of the Clostridiales (68%), Lactobacillales (25%), and Bacteroidetes (6%). To assess the effects of virginiamycin on the gastrointestinal microbiota, 15 bacterial targets were enumerated using quantitative, real-time PCR. Virginiamycin was associated with increased abundance of many of the targets in the proximal gastrointestinal tract (duodenal loop to proximal ileum), with fewer targets affected in the distal regions (ileocecal junction and cecum). These findings provide improved profiling of the composition of the chicken intestinal microbiota and indicate that microbial responses to virginiamycin are most significant in the proximal small intestine.
The animal gut is host to an abundant and diverse microbiota that plays an important role in the health and nutrition of the animal. Most of these organisms are considered commensal or symbiotic (4, 44), but the gastrointestinal microbiota can also have detrimental effects on host health and nutrition (15, 36). The relationship between the host animal and its gut microbiota can therefore be viewed as a balance between mutualism and pathogenicity (15). In agricultural animals, the routine inclusion of antibiotic growth promoters (AGPs) in diets has a beneficial effect on the growth and efficiency of feed conversion (17, 37), probably by beneficially modulating the gastrointestinal microbiota and suppressing the growth of pathogens (3, 18). The streptogramin antibiotic virginiamycin has been used for decades as an AGP in poultry feeds (5), but recent concerns over the possible selection for genes conferring resistance to the human therapeutic antibiotic quinupristin-dalfopristin (Synercid) and the detection of quinupristin-dalfopristin-resistant strains of Enterococcus faecium and other potentially pathogenic enterococci in retail poultry samples (8, 22, 28, 32) have led some to question the practice of using virginiamycin and other AGPs in commercial settings (10, 16, 29). In some European countries, certain AGPs have already been banned (39), and there is widespread anticipation of coming restrictions on their use in North America (41).
One approach to finding effective alternatives to AGPs is to understand both the normal gastrointestinal microbiota and the ways in which the microbiota is altered by the inclusion of AGPs in the diet. Much of the information that has accumulated on the composition of the gastrointestinal microbiota of poultry and other animals has come from culture-based studies (33). More recently, in an attempt to address the bias that culture studies impose, various PCR-based, culture-independent methods have been developed to characterize and quantify the gastrointestinal microbiota (38, 43). Commonly used methods include the amplification of 16S rRNA-encoding genes using universal primers and sequencing of randomly selected clones (2, 31), the development and application of species-specific 16S rRNA-targeted PCR primers (2), and denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA genes (26, 30). We have shown that an approximately 555-bp fragment of the chaperonin 60 (cpn60) gene can be amplified from essentially any genome using universal, degenerate PCR primers and is an effective gene target for molecular ecological studies of gastrointestinal microbiota using high-throughput sequencing of amplified products (23, 25). The cpn60 gene target provides data that are at least as phylogenetically informative as those provided by 16S rRNA-encoding DNA sequences (27) and can also in many cases offer improved interspecies discriminating power, especially between closely related strains (6). A reference database of cpn60 sequences is publicly available (24) which facilitates the identification of sequences generated from environmental templates. In addition, we have developed procedures for using real-time quantitative PCR (qPCR) that take advantage of the fact that cpn60 genes are normally present in a single copy to accurately and specifically enumerate target organisms within gastrointestinal tract samples using cpn60 sequences (12).
The objectives of the current study were twofold: to characterize the normal chicken gastrointestinal microbiota and to quantify the effects on this microbiota of the inclusion of subtherapeutic doses of virginiamycin throughout the broiler production cycle. We have used a combination of viable culture, high-throughput sequencing of cloned cpn60 gene fragments, and qPCR to achieve these objectives.
MATERIALS AND METHODS
Animal treatments.
Four hundred 1-day-old male broiler chickens (Cobb 500; Cobb Vantress Inc., Cleveland, GA) were distributed among 20 floor pens (20 birds/pen) containing used litter obtained from adult chickens not previously exposed to the antibiotic. Chickens were fed diets with (n = 10 pens) or without (n = 10 pens) added virginiamycin (20 g/tonne), formulated on corn and soybean meal to meet NRCC requirements (1994), in a four-phase feeding program to 50 days of age. At day 7, the number of birds per pen was adjusted to 18. Body weight, feed intake, feed conversion, and percent mortality were determined on days 15, 32, 42, and 50.
Sample collection.
At 47 days of age, two birds randomly selected from five pens per treatment were killed by cervical dislocation and the digestive tracts removed. Samples of intestinal contents were collected from a 10-cm segment from each of four small intestine locations: duodenal loop, mid-jejunum, proximal ileum, and ileocecal junction. In addition, the contents of one cecum per bird were collected. Contents from each of the sampled gut locations from each of two birds per pen were pooled into preweighed tubes and overlaid with nitrogen gas. A total of 50 samples (5 gut locations times 5 pairs of birds times 2 treatments) were collected. One aliquot of each sample was removed and frozen at −20°C for DNA extraction, and a second was suspended in prereduced peptone water with 0.05% cysteine-HCl and diluted for culture on selective medium.
Culturing and viable counts.
The media (Becton Dickinson, Cockeysville, MD) and culture conditions used were as follows: MRS agar (anaerobic, 37°C for 24 to 48 h), bile esculin agar (aerobic, 37°C for 24 to 48 h), MacConkey agar (aerobic, 37°C for 24 to 48 h), and blood agar base with 5% sheep's blood (aerobic and anaerobic, 37°C for 24 to 48 h). Anaerobic conditions used an atmosphere of 10% CO2, 10% H2, and 80% N2. In addition, aliquots of each sample were plated on brilliant green agar as follows: 100 μl of a 1:10 dilution was spread directly (nonenriched), and 0.1 g of sample was added to 9.9 ml of cysteine selenite broth and incubated aerobically overnight at 37°C (enriched). An aliquot of 100 μl of the enriched culture was spread on a brilliant green plate, which was incubated overnight at 37°C.
Isolation and pooling of community DNA and generation of cpn60 PCR product libraries.
An aliquot of 0.2 g of each of the 50 digesta samples was removed, and total bacterial community DNA was isolated as described previously (23). The final DNA pellets were each dissolved in 200 μl of Tris-EDTA (TE) buffer. Equivalent volumes of DNA extracts corresponding to the same treatment group and intestinal location were pooled, resulting in a total of 10 DNA pools (5 intestinal locations times 2 treatment groups). Each pool was diluted 1:10 in TE buffer, and 1 μl was used as the template in a PCR with the cpn60 universal primers H279 and H280 at four annealing temperatures as described previously (23). Equal volumes of PCR products produced at each annealing temperature were mixed, agarose gel purified, and ligated into vector pGEM-T Easy (Invitrogen). Ligation mixtures were used to transform Escherichia coli JM109 (Invitrogen). Each of the 10 resulting libraries was plated on LB with ampicillin (50 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). A total of 1,344 white colonies were picked randomly from each of the 10 libraries into individual wells of 96-well plates containing 100 μl of LB-ampicillin. The plates were incubated overnight at 37°C, glycerol was added to 15% (vol/vol), and the cultures were stored at −80°C.
Sequencing and bioinformatics.
Template DNA for sequencing reactions was prepared directly from the frozen cultures using the TempliPhi system (Amersham Biosciences, Baie d'Urfé, QC, Canada). Single-pass sequences were generated using a BigDye Terminator cycle sequencing kit (Applied Biosystems) with T7 primer, and reactions were resolved on an ABI 3700 capillary sequencer. Raw sequences were trimmed for quality using Phred (14). The resulting sequences were clustered based on sequence identity using the d2_cluster (7). Clusters of identical sequences were then assembled into contigs or singletons using Phrap (21), incorporating the base quality information from Phred. Manual confirmation of contig assembly was done using Gap4 (version 4.6) in the Staden software package (release 2000.0; J. Bonfield, K. Beal, M. Betts, M. Jordan, and R. Staden, 2000). Sequence data, template information, and similarity results were placed in a MySQL database for storage and further analysis. Sequence manipulations, such as format changes and amino acid translations, were done using the EMBOSS software suite (35). Sequence alignments were done using CLUSTAL W (40). To determine the putative taxonomy of each contig and singleton arising from the assembly step, each sequence was compared to cpnDB, a reference set of cpn60 sequences (24), using FASTA (34).
Library sequences were deposited in GenBank (accession no. DQ106022 to DQ106393) and can also be retrieved from cpnDB (http://cpndb.cbr.nrc.ca).
Phylogenetic analysis was done using programs in the PHYLIP software package (J. Felsenstein, 1993, PHYLIP [Phylogeny Inference Package] version 3.5c, distributed by the author, Department of Genetics, University of Washington, Seattle). Specifically, distances were calculated with the maximum likelihood option of dnadist. Dendrograms were constructed from distance data by neighbor joining.
Quantitative PCR assays.
A summary of qPCR assay components is presented in Table S1 in the supplemental material. The locations of oligonucleotide “signatures” that differentiate the target cpn60 sequence from those of related organisms were identified using Signature Oligo software (LifeIntel Inc., Port Moody, BC, Canada). Signature regions were specified as potential primer landing sites, and compatible PCR primers were determined using Oligo6 (Molecular Biology Insights Inc., Cascade, CO) or Beacon Designer (Premier Biosoft International, Palo Alto, CA) software. In some cases, as specified in Table S1 in the supplemental material, compatible TaqMan probes (Sigma-Genosys Canada, Oakville, ON, Canada) were identified with Beacon Designer to detect the amplified products. The specificity of the PCR primers was initially verified by comparison to all of the other library sequences and reference sequences in cpnDB, using BLASTn (1) configured for short, nearly exact matches. Primer specificity was further validated by the lack of a detectable PCR product using template DNA derived from related strains as described previously (12). qPCR assays used Platinum quantitative PCR SuperMix-UDG or Platinum SYBR green qPCR SuperMix-UDG (Invitrogen) as appropriate in the presence of 3 mM MgCl2, 500 nM of each primer, and (for TaqMan assays) 200 nM TaqMan probe. Pooled DNA (50 μl from each of five extracts) from each gastrointestinal tract location and treatment (total of 10 pools) was purified on a QIAGEN genomic tip 20/G and dissolved in 100 μl of TE buffer. An aliquot of 1 μl of each purified genomic DNA pool was used as a template for the qPCR assays. All amplifications were preceded by the following steps: 50°C for 2 min (uracil DNA glycosylase activation) and then 95°C for 3 min (well factor collection); this was followed immediately by the PCR cycling conditions for each primer set indicated in Table S1 in the supplemental material. Data collection was set at the annealing or annealing/extension step. Amplifications were performed on an iCycler (Bio-Rad).
Statistical analysis.
The effect of treatment on production performance and number of bacteria determined by culture-based enumeration was determined using one-way analysis of variance (SPSS Inc., Chicago, IL).
RESULTS
Production response to inclusion of virginiamycin.
Dietary inclusion of virginiamycin increased body weight and reduced the kilograms of feed required to achieve a kilogram of gain (feed efficiency) from day 0 to 15 of the study (Table 1). Chicken body weight, feed intake, and feed efficiency were not different for the remainder of the study. Cumulative mortality was reduced by more than 50% in chickens fed virginiamycin; however, this response was not significant due to high variation in the relatively small number of birds in the study.
TABLE 1.
Body weight, feed intake, mortality-corrected feed efficiency, and cumulative percent mortality observed in broiler chickens fed diet excluding or including virginiamycina
| Parameter | Day | −Vm | +Vm |
|---|---|---|---|
| Body wt (kg) | 15 | 0.304 ± 0.006b | 0.336 ± 0.004b |
| 32 | 1.314 ± 0.021 | 1.361 ± 0.012 | |
| 42 | 2.122 ± 0.025 | 2.130 ± 0.031 | |
| 50 | 2.723 ± 0.030 | 2.740 ± 0.048 | |
| Feed intake (g/day) | 15 | 0.447 ± 0.007 | 0.468 ± 0.006 |
| 32 | 2.046 ± 0.029 | 2.099 ± 0.018 | |
| 42 | 3.580 ± 0.054 | 3.587 ± 0.041 | |
| 50 | 4.964 ± 0.159 | 4.944 ± 0.067 | |
| Feed efficiency (F/G)c | 15 | 1.702 ± 0.076b | 1.587 ± 0.077b |
| 32 | 1.607 ± 0.025 | 1.590 ± 0.025 | |
| 42 | 1.720 ± 0.031 | 1.718 ± 0.043 | |
| 50 | 1.851 ± 0.044 | 1.833 ± 0.043 | |
| Mortality (%) | 15 | 1.68 ± 0.90 | 0.56 ± 0.59 |
| 32 | 2.23 ± 1.29 | 0.56 ± 0.59 | |
| 42 | 3.90 ± 1.52 | 0.56 ± 0.59 | |
| 50 | 6.12 ± 1.61 | 2.79 ± 1.31 |
Data are means ± standard errors. −Vm, without virginiamycin; +Vm, with virginiamycin.
Values in these rows are different (P < 0.05).
F/G, grams of feed per gram of body weight gain.
Characterization of the avian bacterial microbiota by plate counts.
Plate counts were performed for each gastrointestinal tract location that was sampled (see Table S3 in the supplemental material). For each of the selective media used, counts increased (P < 0.05) 1 to 3 orders of magnitude along the gastrointestinal tract from the proximal to distal small intestine and further increased (P < 0.05) 1 to 2 orders of magnitude from the distal ileum to the cecum. No significant effect of virginiamycin on plate counts determined at any intestinal location was observed, although in the proximal regions (duodenal loop, mid-jejunum, and proximal ileum), numeric increases of 0.4 to 1.2 log10 CFU/g were observed for total aerobes and lactobacilli in response to virginiamycin. The mid-jejunum showed the most marked increase. A few of the samples showed Salmonella in the absence of enrichment, but most were positive after enrichment. The inclusion of virginiamycin in the diet resulted in a decrease in Salmonella in the duodenal loop, but this effect was not noted in any other gastrointestinal tract location.
Identification of bacterial species resident in the avian gastrointestinal tract by chaperonin 60 gene sequencing.
Ten cpn60 PCR product libraries were generated, representing five gastrointestinal tract locations of broiler birds fed diets including or excluding virginiamycin. Each library was derived from the pooled contents from each gut location of 10 similarly treated birds. The library names, descriptions, and numbers of sequences that were analyzed from each library and the numbers of distinct sequences in each library are shown in Table 2. Only full-length cloned PCR product sequences were included in the analysis. While the small intestinal libraries were relatively homogenous, with only 29 to 58 distinct sequences per library, the cecum-derived libraries were comparatively diverse, with 110 (no virginiamycin) and 118 (with virginiamycin) different sequences recovered (Table 2). After all of the sequence data were pooled, the clustering process resulted in the identification of 370 different sequences that were recovered at various frequencies from each of the 10 libraries (see Table S5 in the supplemental material).
TABLE 2.
Number of cpn60 sequences determined in each poultry intestinal librarya
| Gut site | Treatmentb | No. of sequences | No. of distinct sequences |
|---|---|---|---|
| Duodenal loop | −Vm | 1,022 | 30 |
| Duodenal loop | +Vm | 1,087 | 53 |
| Mid-jejunum | −Vm | 1,208 | 38 |
| Mid-jejunum | +Vm | 1,103 | 29 |
| Proximal ileum | −Vm | 974 | 34 |
| Proximal ileum | +Vm | 1,158 | 58 |
| Ileocecal junction | −Vm | 1,183 | 35 |
| Ileocecal junction | +Vm | 1,193 | 35 |
| Cecum | −Vm | 1,016 | 110 |
| Cecum | +Vm | 988 | 118 |
A total of 1,344 clones (14 96-well plates) were picked from each library.
−Vm, without virginiamycin; +Vm, with virginiamycin.
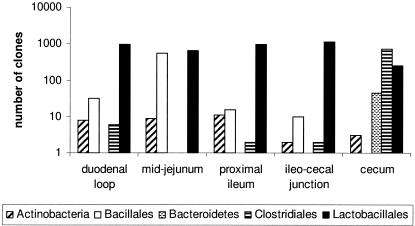
Individual sequences were identified by comparison to the cpnDB reference database and assigned to taxonomic groups (see Table S5 in the supplemental material). The distribution of taxa in each gut location for the control birds (no virginiamycin) is shown in Fig. 1. Organisms belonging to the Lactobacillales order dominated all small intestinal locations (duodenal loop, mid-jejunum, proximal ileum, and ileocecal junction) and were also detected in the cecum. However, the cecum microbiota composition was found to be almost completely distinct from the small intestine, with a predominance of sequences belonging to the Clostridiales order and Bacteroidetes phylum and relatively fewer examples of the Actinobacteria class and Bacillales order relative to the small intestinal populations examined. In fact, only four sequences (all lactobacilli) were found in both the small intestine and the cecum (see Table S5 in the supplemental material).
FIG. 1.
Taxonomic group assignments and clone frequencies of cpn60 sequences isolated from five gastrointestinal tract locations in control birds (no virginiamycin).
To identify differences in the microbial population compositions of birds in the two treatment groups and further refine the data set, all of the sequence and frequency data from all small intestinal libraries (173 sequences) were pooled and subjected to phylogenetic analysis. Based on the analysis, 20 sequences or clusters of virtually identical sequences (at least 95% pairwise identity) were identified, with total frequencies of occurrences of at least 10. As shown in Table 3, representatives of small intestinal groups A, B, C, E, F, H, and I were also detected in the cecum, while all other groups were detected only in the small intestine. Groups E, K, M, and N showed a weak but consistent trend of increased abundance in the proximal small intestine in the presence of virginiamycin and were chosen as targets for further investigation by qPCR. Group M was also chosen as a target due to its peculiar abundance in the library prepared from the mid-jejunum of birds fed diets without virginiamycin.
TABLE 3.
Abundance of groups in the small intestine
| Group | Description | No. of unique sequences in group | Clone frequency ina:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duodenal loop
|
Mid-jejunum
|
Proximal ileum
|
Ileocecal junction
|
Cecum
|
All locations | ||||||||
| −Vm | +Vm | −Vm | +Vm | −Vm | +Vm | −Vm | +Vm | −Vm | +Vm | ||||
| A | 97-100% identical to Lactobacillus gallinarum and L. crispatus (acidophilus group) | 31 | 372 | 589 | 23 | 601 | 711 | 528 | 544 | 1101 | 120 | 164 | 4,753 |
| B | 99% identical to Lactobacillus buchneri ATCC 4005 | 4 | 234 | 122 | 401 | 108 | 65 | 202 | 156 | 29 | 63 | 50 | 1,430 |
| C | 96-99% identical to Lactobacillus salivarius subsp. salivarius ATCC 11741 | 5 | 326 | 103 | 6 | 148 | 90 | 79 | 157 | 13 | 44 | 15 | 981 |
| D | 95-98% identical to Lactobacillus aviarius subsp. aviarius ATCC 43234 | 4 | 5 | 28 | 42 | 40 | 16 | 41 | 168 | 1 | 0 | 0 | 341 |
| E | 86% identical to Lactobacillus vaginalis ATCC 49540 | 1 | 5 | 32 | 8 | 34 | 4 | 31 | 4 | 1 | 0 | 5 | 124 |
| F | 99% identical to Lactobacillus vaginalis ATCC 49540 | 4 | 6 | 17 | 16 | 21 | 4 | 27 | 4 | 5 | 8 | 3 | 111 |
| G | 84% identical to Lactobacillus buchneri ATCC 4005 | 2 | 9 | 25 | 37 | 24 | 6 | 47 | 6 | 9 | 0 | 0 | 163 |
| H | 99% identical to Lactobacillus johnsonii ATCC 33200 | 6 | 0 | 18 | 2 | 11 | 23 | 19 | 60 | 10 | 16 | 15 | 174 |
| I | 84% identical to Lactobacillus buchneri ATCC 4005 | 4 | 1 | 10 | 3 | 2 | 0 | 9 | 1 | 2 | 0 | 2 | 30 |
| J | 84% identical to Pediococcus parvulus ATCC 43013 | 2 | 13 | 40 | 85 | 33 | 9 | 42 | 14 | 17 | 0 | 0 | 253 |
| K | 99% identical to Enterococcus cecorum ATCC 43198 | 4 | 3 | 5 | 0 | 6 | 4 | 6 | 31 | 0 | 1 | 0 | 56 |
| L | 95-98% identical to Globicatella sanguinis ATCC 51173 | 3 | 0 | 7 | 0 | 10 | 1 | 6 | 0 | 0 | 0 | 0 | 24 |
| M | 74% identical to Staphylococcus equorum ATCC 43958 | 3 | 5 | 4 | 423 | 1 | 5 | 4 | 0 | 0 | 0 | 0 | 442 |
| N | 77% identical to Staphylococcus sciuri ATCC 29060 | 1 | 2 | 8 | 0 | 9 | 0 | 11 | 0 | 0 | 0 | 0 | 30 |
| O | 75-76% identical to Staphylococcus pasteuri BM10426 | 7 | 4 | 14 | 2 | 13 | 2 | 13 | 2 | 0 | 0 | 0 | 50 |
| P | 76-77% identical to Staphylococcus intermedius ATCC 29663 | 3 | 6 | 11 | 2 | 10 | 0 | 13 | 0 | 0 | 0 | 0 | 42 |
| Q | 88-89% identical to Clostridium polysaccharolyticum ATCC 33142 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 2 | 0 | 0 | 21 |
| R | 74% identical to Macrococcus bovicus ATCC 51825 | 3 | 0 | 0 | 57 | 3 | 2 | 6 | 0 | 0 | 0 | 0 | 68 |
| S | 75-76% identical to Macrococcus carouselicus ATCC 51828 | 3 | 0 | 0 | 41 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 45 |
| T | 77-78% identical to Staphylococcus aureus subsp. aureus MRSA252 | 4 | 10 | 8 | 18 | 5 | 3 | 10 | 1 | 1 | 0 | 0 | 56 |
−Vm, without virginiamycin; +Vm, with virginiamycin.
qPCR analysis of the abundance of bacterial targets and changes in response to the inclusion of virginiamycin in feeds.
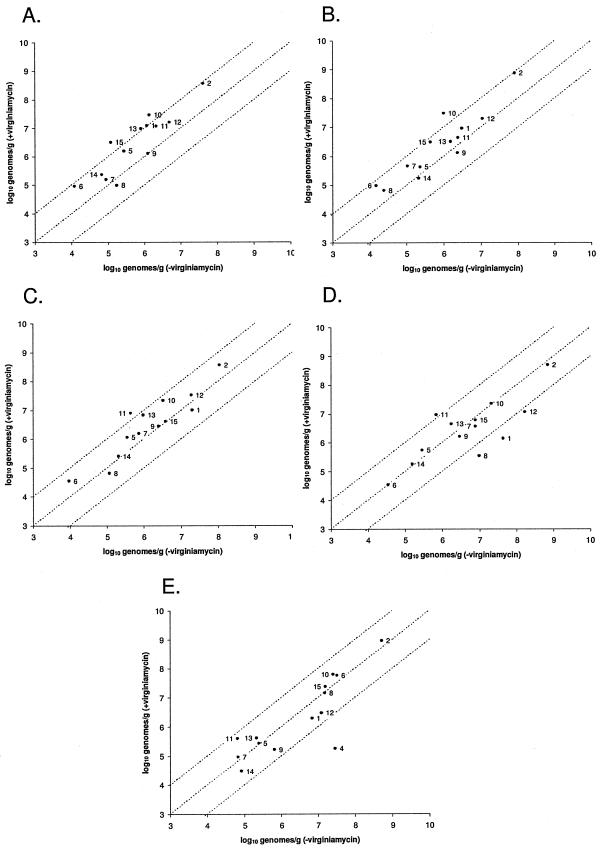
An examination of the frequency of clone recovery in each library suggested that the abundance of certain organisms may be affected by the inclusion of virginiamycin in the diet. These cpn60 sequences, which included both named (groups D, K, and L) and unnamed (groups M, E, and N) organisms, were therefore used to design specific PCR primers for qPCR analysis of the abundance of the target organisms (see Table S1 in the supplemental material). In addition to organisms chosen on the basis of clone frequency ratios, we targeted sequences that were recovered at low frequencies (fewer than 20 times) from the cecum (groups V, W, and X) and small intestine (Staphylococcus lentus) libraries. For these bacterial targets, no comparative clone frequency information was available, and these targets were chosen in order to probe the detection limits of the library sequencing method. We also included five targets for qPCR analysis that are organisms known to reside in the avian gastrointestinal tract (Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus johnsonii, Enterococcus faecalis, and Escherichia coli). A total of 15 bacteria were targeted for qPCR-based enumeration (see Table S1 in the supplemental material). DNA pools were prepared from each gastrointestinal tract location and diet (the same pools that were initially used to prepare the libraries) and were used as the template in a qPCR assay. We compared the results obtained from samples taken from the same gastrointestinal tract location from birds fed diets including or excluding virginiamycin and plotted this information for each target in each location (Fig. 2). In the duodenal loop, the abundance of 11 of the 13 targets that were detected increased in response to virginiamycin, many of them showing nearly or more than a 10-fold change in abundance (Fig. 2A). Four of the five targets from the lactobacilli increased in abundance in the duodenal loop: L. crispatus, library group D (Lactobacillus aviarius subsp. aviarius), L. johnsonii, and library group E (∼86% identical to Lactobacillus vaginalis). L. gasseri was not detected in the duodenal loop. Other bacterial targets showing a marked increase in response to virginiamycin were library group W (∼80% identical to Clostridium nexile), library group L (Globicatella sanguinis), library group K (Enterococcus cecorum), and library group X (∼85% identical to Corynebacterium glutamicum). Similarly, in the mid-jejunum, 11 of 13 targets that were detected increased in response to virginiamycin; only the abundances of library group M (∼74% identical to Staphylococcus equorum) and Staphylococcus lentus were unchanged or decreased in response to the antibiotic (Fig. 2B). However, only five of the bacterial targets increased at or near a 10-fold level (library group W, E. faecalis, L. johnsonii, library group E, and L. crispatus). In the proximal ileum, two targets (library groups N and E) increased at or near the 1-log level (Fig. 2C), and L. gasseri was detected at low levels (1.73 × 104 genomes/g digesta) uniquely in the proximal ileum in the presence of the antibiotic. In the ileocecal junction and cecum, only library group N increased substantially (Fig. 2D and E). In the distal locations, there were several targets that decreased markedly in abundance. For example, in the ileocecal junction, the abundances of three targets (E. coli, library group D, and library group K) were dramatically decreased in the presence of virginiamycin (Fig. 2D), while in the cecum, the numbers of library group V (∼80% identical to Eubacterium coprostanoligenes) were decreased by almost 2 orders of magnitude (Fig. 2E). The cecum was the only gastrointestinal tract location in which library group V was detected.
FIG.2.
Abundance of 15 bacterial targets in each gastrointestinal tract location in the presence and absence of virginiamycin as measured by qPCR. The dotted lines indicate a 10-fold increase (upper line), a 10-fold decrease (lower line), or no change (middle line). For all graphs, the data points are labeled as follows (see Table S1 in the supplemental material for library group identifications): 1, library group D; 2, L. crispatus; 3, L. gasseri; 4, library group V; 5, library group X; 6, library group W; 7, E. faecalis; 8, E. coli; 9, library group M; 10, library group E; 11, library group N; 12, library group K; 13, library group L; 14, S. lentus; and 15, L. johnsonii. Note that all primer sets were assayed in all gastrointestinal tract locations, and the absence of a target on a graph indicates that it was not detected in one or both treatment groups. (A) Duodenal loop. (B) Mid-jejunum. (C) Proximal ileum. (D) Ileocecal junction. (E) Cecum.
DISCUSSION
The sustained use of antibiotic growth promoters is a standard practice in the poultry industry. Although the beneficial effects on health and growth are believed to be mediated by effects on gastrointestinal microbiota, the underlying mechanisms remain to be discovered. The chicken gastrointestinal microbiota has been characterized by culture-based analysis (33) and, more recently, by molecular methods, including 16S rRNA gene sequencing (2, 19, 20, 31) and DGGE (26, 30, 42). We have used cpn60 gene sequencing to examine the microbiota in five locations in the chicken gastrointestinal tract in birds fed a control diet or a diet supplemented with virginiamycin. Some sequences were differentially recovered from cpn60 clone libraries, representing the control or virginiamycin-adapted microbiota. A number of these sequences were selected as targets for further analysis by qPCR.
We analyzed the microbiota of the birds at day 47, which coincides with the end of the production cycle. Furthermore, the birds attain a mature microbiota by that time. The microbiota of the virginiamycin-fed birds therefore represented a virginiamycin-adapted microbiota that would be informative of the changes that are associated with the inclusion of virginiamycin in the diet. The results described here constitute one of the most comprehensive inventories of avian microbiota reported to date and also demonstrate the changes that occur in the abundance of specific organisms in response to the inclusion of dietary virginiamycin throughout the production cycle.
We observed a production performance response to dietary inclusion of virginiamycin in the early phase of this study. The lack of a significant performance response throughout the study is not surprising, given the small number of experimental units (pens in this case) employed and the fact that a growth promotion response to AGP is not always evident, particularly in highly sanitized research facility environments. Conduct of this study under commercial conditions may have provided a more consistent performance response to AGP; however, under such conditions, control of all other factors (such as diet composition, lighting, and vaccination program) is extremely difficult.
Examination of the cpn60 sequence information from the 10 libraries revealed that the most commonly recovered sequences were lactobacilli (library groups A, B, and C) and accounted for more than 65% of the total (Table 3). A predominance of lactobacilli, in particular L. crispatus (our group A) and Lactobacillus salivarius (our group C), was also noted by Lu et al. in the ileum of 49-day-old broilers in an analysis of 610 16S rRNA gene clones (31). These species were also identified as being abundant in the intestinal contents of 5- to 6-week-old broilers fed diets containing antibiotics (13, 30). L. crispatus, Lactobacillus buchneri, and L. salivarius may therefore be considered the most abundant microorganisms present in the gastrointestinal tract of 5- to 6-week-old broiler birds. Gong et al. (19) identified 15 bacterial species within a relatively small population of 51 16S rRNA gene clones, among which L. aviarius subsp. aviarius (our group D) and Enterococcus cecorum (our group K) were the most abundant.
The cecum microbiota was quite distinct from the small intestinal locations, with a predominance of sequences belonging to the Clostridiales order and Bacteroidetes phylum and relatively fewer examples of clones from the Actinobacteria class and Bacillales order than with sequences of the small intestinal populations (Fig. 1). Only three sequences (apparently from Lactobacillus crispatus, L. buchneri, and L. salivarius subsp. salivarius) were found in both the small intestine and the cecum (see Table S5 in the supplemental material). Although in younger birds the small intestinal and cecal bacterial populations are similar (37), several groups have observed that by 6 weeks of age, these populations are highly distinct from one another (2, 20, 26, 31, 42). This observation is hardly surprising given the different microenvironments provided by these gastrointestinal tract locations.
The analysis of cpn60 library clone frequencies provides information on the taxonomic composition of a sample, and comparative clone frequencies can be used as a means of identifying organisms whose abundances may change in response to perturbations, such as the inclusion of virginiamycin in the diet. Another molecular method for identifying organisms that change in abundance in response to environmental parameters is DGGE. Knarreborg et al. used DGGE to examine the effects of age, antibiotics, and dietary fats on the composition of the ileal bacterial community in broiler chickens and identified several species of Lactobacillus as well as Clostridium perfringens that responded to the treatments (30). In addition, Hume et al. monitored the changes in the gastrointestinal microbiota of developing and molting birds with DGGE (26); although they were able to detect changes in the bacterial composition of their samples, they also noted that the chosen method of analysis would likely not detect organisms present at fewer than 108 CFU/g in the intestinal samples. We have addressed the question of detection sensitivity in the current library sequencing study by choosing organisms that were detected at low frequencies as targets for qPCR analysis. For example, cpn60 sequences corresponding to S. lentus were identified only seven times in all libraries, and qPCR analysis showed that its abundance ranged from 104 to 105 genomes/g contents (Fig. 2). Furthermore, library group X (∼86% identical to Corynebacterium ammoniagenes; 21 clones identified in all libraries) was found at approximately 105 genomes/g in all locations, and library group W (∼80% identical to Clostridium nexile) was not detected in any of the small intestinal locations but ranged from 104 to 105 genomes/g in these locations (Fig. 2) (see Table S5 in the supplemental material). The library sequencing approach on the scale that we have used therefore appeared to be able to detect organisms whose abundance was near 105 genomes/g contents. This is comparable to the detection limit of the qPCR method in chicken gastrointestinal tract contents, which is approximately 104 genomes/g contents (12). Moreover, the clone frequencies that we identified were often a reasonably good indication of the change in abundance of the target organism in response to dietary virginiamycin: the clone frequency of library group W increased 1.95-fold in the cecal library with virginiamycin, and qPCR analysis showed that the increase was 1.77-fold. In some cases, however, clone frequencies were not an indication of true changes in response to virginiamycin; library group M (74% identical to Staphylococcus equorum) was apparently very abundant in the library prepared from the contents of the mid-jejunum in birds fed no antibiotic compared to birds fed virginiamycin, but qPCR analysis did not bear this out (Table 3 and Fig. 2). Thus, clone frequencies in the context of relatively large libraries as described here can be taken as a putative indication of the change in abundance of the target organism, but validation by qPCR analysis is necessary for confirmation.
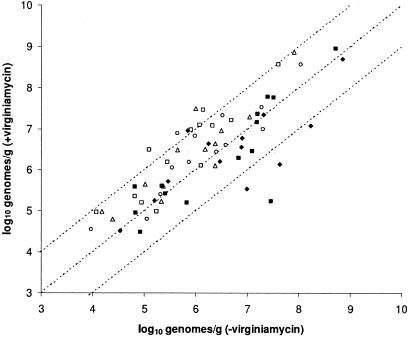
We used qPCR to examine the adundance of selected organisms in each gastrointestinal location in the presence and absence of virginiamycin. In general terms, dietary virginiamycin appeared to increase the abundance of the bacterial targets that we analyzed in the proximal gastrointestinal tract locations (duodenal loop, mid-jejunum, and proximal ileum), with no effect or a decrease in their abundances toward the distal end (ileocecal junction and cecum) (Fig. 3). Since culture-based analysis indicated that total bacterial numbers did not change significantly when virginiamycin was included in the diet, we suggest that the increase in the abundance of certain species may be explained by a virginiamycin-induced decrease in unenumerated competing species, which opens up a niche into which bacteria may grow. These bacteria may be beneficial to host health and nutrition, for example, by competitively excluding the colonization of the gastrointestinal tract by pathogenic bacteria, aiding in the digestion of certain feed components, producing essential metabolites, or degrading potentially antinutritional dietary components (11). The introduction of a reasonably broad-spectrum antibiotic at subtherapeutic doses into a complex and varied intestinal microbiota appeared to alter the balance of the ecosystem and ultimately resulted in both increases and decreases in the abundances of various species in different gastrointestinal locations. For example, we observed that the abundance of most of the Lactobacillus targets increased in the presence of virginiamycin in the proximal locations but did not change substantially or decreased in the distal locations. An increase in lactobacilli and other targets in the proximal locations is in contrast to the findings of Knarreborg et al., who reported significant decreases in the viable counts of lactobacilli and C. perfringens in the ileum of 14- to 21-day-old broiler chickens fed diets containing avilamycin and salinomycin (30). Although the effects of the antibiotics on the total viable counts of lactobacilli were not significant at 35 days of age, L. salivarius was observed by DGGE to be absent only in the antibiotic-fed birds at this age (30). Similar effects on the populations of C. perfringens and L. salivarius were reported in the gastrointestinal tracts of 5-week-old broilers fed diets containing zinc bacitracin and salinomycin (13). In contrast, Collier et al. (9) reported that tylosin increased the abundances of lactobacilli, especially L. gasseri, in the ileum of antibiotic-fed pigs compared with that of controls. These results are consistent with our observations in the proximal gastrointestinal tract of virginiamycin-fed chickens.
FIG. 3.
Abundance of each of 15 bacterial targets in the presence and absence of dietary virginiamycin grouped according to gastrointestinal tract location. Each gastrointestinal tract location is plotted separately: duodenal loop (□), mid-jejunum (▵), proximal ileum (○), ileocecal junction (♦), and cecum (▪). Note that the proximal locations are represented by open symbols while the distal locations are represented by closed symbols.
In conclusion, we provide here significant further characterization of microbial colonization in the chicken intestinal tract. The cpn60 sequence information will be valuable in assessing species colonization variation in response to nutritional, environmental, and genetic factors and ultimately linking specific organisms with nutritional or health benefits. In the case of virginiamycin, bacteria colonizing the proximal gastrointestinal tract, including regions proximal to those examined here, may be very sensitive to antibiotic inclusion and should be examined more closely for contribution to the beneficial effect of AGP, especially in a time course study and ideally in a commercial environment. We have identified a number of lactobacilli that were increased in proximal locations and may have probiotic potential. Similarly, bacterial species reduced by dietary virginiamycin, which were primarily identified in distal locations, may be considered potentially harmful organisms with respect to bird health and performance and could be targets for reduction by nonantibiotic approaches.
Supplementary Material
Acknowledgments
We thank Jason Marshall, Jennifer Town, Jodi Roach, Theresa L'Heureux, and Lisa Hrycan for providing technical assistance, along with Bill Crosby and Matt Links for bioinformatics support. Animal performance data and digesta samples were provided by Cargill Animal Nutrition.
This work was funded by Cargill Animal Nutrition.
Footnotes
Supplemental material for this article may be found at http://www.aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amit-Romach, E., D. Sklan, and Z. Uni. 2004. Microflora ecology of the chicken intestine using 16S ribosomal DNA primers. Poult. Sci. 83:1093-1098. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. B., V. J. McCracken, R. I. Aminov, J. M. Simpson, R. I. Mackie, M. W. A. Verstegen, and H. R. Gaskins. 1999. Gut microbiology and growth-promoting antibiotics in swine. Nutr. Abstr. Rev. 70:101-188. [Google Scholar]
- 4.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 5.Barrow, P. A. 1998. Use of virginiamycin as a growth promoter. Vet. Rec. 143:483-484. [PubMed] [Google Scholar]
- 6.Brousseau, R., J. E. Hill, G. Prefontaine, S. H. Goh, J. Harel, and S. M. Hemmingsen. 2001. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Appl. Env. Microbiol. 67:4828-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, J., D. Davison, and W. Hide. 1999. d2_cluster: a validated method for clustering EST and full-length cDNA sequences. Genome Res. 9:1135-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butaye, P., K. Van Damme, L. A. Devriese, L. Van Damme, M. Bael, S. Lauwers, and F. Haesebrouck. 2000. In vitro susceptibility of Enterococcus faecium isolated from food to growth-promoting and therapeutic antibiotics. Int. J. Food Microbiol. 54:181-187. [DOI] [PubMed] [Google Scholar]
- 9.Collier, C. T., M. R. Smiricky-Tjardes, D. M. Albin, J. E. Wubben, V. M. Gabert, B. Deplancke, D. Bane, D. B. Anderson, and H. R. Gaskins. 2003. Molecular ecological analysis of porcine ileal microbiota responses to antimicrobial growth promoters. J. Anim. Sci. 81:3035-3045. [DOI] [PubMed] [Google Scholar]
- 10.Cox, L. A., Jr., and D. A. Popken. 2004. Quantifying human health risks from virginiamycin used in chickens. Risk Anal. 24:271-288. [DOI] [PubMed] [Google Scholar]
- 11.Dibner, J. J., and J. D. Richards. 2005. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 84:634-643. [DOI] [PubMed] [Google Scholar]
- 12.Dumonceaux, T. J., J. E. Hill, S. A. Briggs, K. K. Amoako, S. M. Hemmingsen, and A. G. Van Kessel. 2006. Enumeration of specific bacterial populations in complex intestinal communities using quantitative PCR based on the chaperonin-60 target. J. Microbiol. Methods 64:46-62. [DOI] [PubMed] [Google Scholar]
- 13.Engberg, R. M., M. S. Hedemann, T. D. Leser, and B. B. Jensen. 2000. Effect of zinc bacitracin and salinomycin on intestinal microflora and performance of broilers. Poult. Sci. 79:1311-1319. [DOI] [PubMed] [Google Scholar]
- 14.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 15.Farthing, M. J. 2004. Bugs and the gut: an unstable marriage. Best Pract. Res. Clin. Gastroenterol. 18:233-239. [DOI] [PubMed] [Google Scholar]
- 16.Ferber, D. 2002. Antibiotic resistance. Livestock feed ban preserves drugs' power. Science 295:27-28. [DOI] [PubMed] [Google Scholar]
- 17.Frost, A. J., and J. B. Woolcock. 1991. Antibiotics and animal production, p. 181-194. In J. B. Woolcock (ed.), Microbiology of animals and animal products. Elsevier, New York, N.Y.
- 18.Gaskins, H. R., C. T. Collier, and D. B. Anderson. 2002. Antibiotics as growth promotants: mode of action. Anim. Biotechnol. 13:29-42. [DOI] [PubMed] [Google Scholar]
- 19.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, R. Wheatcroft, P. M. Sabour, and S. Chen. 2002. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol. 41:171-179. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 22.Hayes, J. R., L. L. English, P. J. Carter, T. Proescholdt, K. Y. Lee, D. D. Wagner, and D. G. White. 2003. Prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Env. Microbiol. 69:7153-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, J. E., S. M. Hemmingsen, B. G. Goldade, T. J. Dumonceaux, J. Klassen, R. T. Zijlstra, S. H. Goh, and A. G. Van Kessel. 2005. Comparison of ileum microflora of pigs fed corn-, wheat-, or barley-based diets by chaperonin-60 sequencing and quantitative PCR. Appl. Env. Microbiol. 71:867-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, J. E., S. L. Penny, K. G. Crowell, S. H. Goh, and S. M. Hemmingsen. 2004. cpnDB: a chaperonin sequence database. Genome Res. 14:1669-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill, J. E., R. P. Seipp, M. Betts, L. Hawkins, A. G. Van Kessel, W. L. Crosby, and S. M. Hemmingsen. 2002. Extensive profiling of a complex microbial community by high-throughput sequencing. Appl. Env. Microbiol. 68:3055-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hume, M. E., L. F. Kubena, T. S. Edrington, C. J. Donskey, R. W. Moore, S. C. Ricke, and D. J. Nisbet. 2003. Poultry digestive microflora biodiversity as indicated by denaturing gradient gel electrophoresis. Poult. Sci. 82:1100-1107. [DOI] [PubMed] [Google Scholar]
- 27.Jian, W., L. Zhu, and X. Dong. 2001. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 51:1633-1638. [DOI] [PubMed] [Google Scholar]
- 28.Joseph, S. W., J. R. Hayes, L. L. English, L. E. Carr, and D. D. Wagner. 2001. Implications of multiple antimicrobial-resistant enterococci associated with the poultry environment. Food Addit. Contam. 18:1118-1123. [DOI] [PubMed] [Google Scholar]
- 29.Kelly, L., D. L. Smith, E. L. Snary, J. A. Johnson, A. D. Harris, M. Wooldridge, and J. G. Morris, Jr. 2004. Animal growth promoters: to ban or not to ban? A risk assessment approach. Int. J. Antimicrob. Agents 24:205-212. [DOI] [PubMed] [Google Scholar]
- 30.Knarreborg, A., M. A. Simon, R. M. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Env. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Env. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald, L. C., S. Rossiter, C. Mackinson, Y. Y. Wang, S. Johnson, M. Sullivan, R. Sokolow, E. DeBess, L. Gilbert, J. A. Benson, B. Hill, and F. J. Angulo. 2001. Quinupristin-dalfopristin-resistant Enterococcus faecium on chicken and in human stool specimens. N. Engl. J. Med. 345:1155-1160. [DOI] [PubMed] [Google Scholar]
- 33.Mead, G. C. 1997. Bacteria in the gastrointestinal tract of birds, p. 216-240. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology: gastrointestinal microbes and host interactions, vol. 2. Chapman and Hall, New York, N.Y. [Google Scholar]
- 34.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 36.Schauer, D. B. 1997. Paving the way for pathogens? Curr. Biol. 7:R75-R77. [DOI] [PubMed] [Google Scholar]
- 37.Stutz, M. W., and G. C. Lawton. 1984. Effects of diet and antimicrobials on growth, feed efficiency, intestinal Clostridium perfringens, and ileal weight of broiler chicks. Poult. Sci. 63:2036-2042. [DOI] [PubMed] [Google Scholar]
- 38.Tannock, G. W. 2001. Molecular assessment of intestinal microflora. Am. J. Clin. Nutr. 73:410S-414S. [DOI] [PubMed] [Google Scholar]
- 39.Taylor, D. J. 1999. EU ban on four antibiotic growth promoters. Vet. Rec. 144:158. [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnidge, J. 2004. Antibiotic use in animals-prejudices, perceptions and realities. J. Antimicrob. Chemother. 53:26-27. [DOI] [PubMed] [Google Scholar]
- 42.van der Wielen, P. W., D. A. Keuzenkamp, L. J. Lipman, F. van Knapen, and S. Biesterveld. 2002. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 44:286-293. [DOI] [PubMed] [Google Scholar]
- 43.Vaughan, E. E., F. Schut, H. G. Heilig, E. G. Zoetendal, W. M. de Vos, and A. D. Akkermans. 2000. A molecular view of the intestinal ecosystem. Curr. Issues Intest. Microbiol. 1:1-12. [PubMed] [Google Scholar]
- 44.Xu, J., and J. I. Gordon. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci. USA 100:10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.