Abstract
Legionella pneumophila persists for a long time in aquatic habitats, where the bacteria associate with biofilms and replicate within protozoan predators. While L. pneumophila serves as a paradigm for intracellular growth within protozoa, it is less clear whether the bacteria form or replicate within biofilms in the absence of protozoa. In this study, we analyzed surface adherence of and biofilm formation by L. pneumophila in a rich medium that supported axenic replication. Biofilm formation by the virulent L. pneumophila strain JR32 and by clinical and environmental isolates was analyzed by confocal microscopy and crystal violet staining. Strain JR32 formed biofilms on glass surfaces and upright polystyrene wells, as well as on pins of “inverse” microtiter plates, indicating that biofilm formation was not simply due to sedimentation of the bacteria. Biofilm formation by an L. pneumophila fliA mutant lacking the alternative sigma factor σ28 was reduced, which demonstrated that bacterial factors are required. Accumulation of biomass coincided with an increase in the optical density at 600 nm and ceased when the bacteria reached the stationary growth phase. L. pneumophila neither grew nor formed biofilms in the inverse system if the medium was exchanged twice a day. However, after addition of Acanthamoeba castellanii, the bacteria proliferated and adhered to surfaces. Sessile (surface-attached) and planktonic (free-swimming) L. pneumophila expressed β-galactosidase activity to similar extents, and therefore, the observed lack of proliferation of surface-attached bacteria was not due to impaired protein synthesis or metabolic activity. Cocultivation of green fluorescent protein (GFP)- and DsRed-labeled L. pneumophila led to randomly interspersed cells on the substratum and in aggregates, and no sizeable patches of clonally growing bacteria were observed. Our findings indicate that biofilm formation by L. pneumophila in a rich medium is due to growth of planktonic bacteria rather than to growth of sessile bacteria. In agreement with this conclusion, GFP-labeled L. pneumophila initially adhered in a continuous-flow chamber system but detached over time; the detachment correlated with the flow rate, and there was no accumulation of biomass. Under these conditions, L. pneumophila persisted in biofilms formed by Empedobacter breve or Microbacterium sp. but not in biofilms formed by Klebsiella pneumoniae or other environmental bacteria, suggesting that specific interactions between the bacteria modulate adherence.
Bacteria belonging to the genus Legionella are ubiquitous in natural and man-made aquatic habitats, where they colonize biofilms and replicate intracellularly in a wide range of protozoa (reviewed in references 7, 11, and 39). If inhaled via contaminated aerosols, Legionella spp. can cause the severe pneumonia Legionnaires' disease. The clinically most relevant species is Legionella pneumophila, which proliferates within alveolar macrophages of the human lung, causing disease (17, 24). The mechanisms of phagocytosis and intracellular replication of L. pneumophila in amoebae and macrophages are similar and are governed by the bacterial Icm/Dot type IV secretion system (T4SS) (13, 33, 36, 38, 43). Moreover, growth of L. pneumophila on the surface of agar plates in the presence of Acanthamoeba castellanii also requires the Icm/Dot T4SS (1).
While intracellular replication of L. pneumophila in phagocytes is increasingly well characterized on the molecular level, colonization of surfaces and biofilm formation by L. pneumophila are less well understood. Notably, it is not clear at present whether L. pneumophila replicates extracellularly in biofilms or merely persists in biofilms. To date, only a few studies have been conducted to determine the role and composition of biofilms in which L. pneumophila persists and proliferates. In these studies, tap water was used as the source of nutrients for biofilms composed of preselected mixtures of defined or undefined heterotrophic bacteria in continuous-flow reactors (23, 30, 41) or in a static system (16). The presence of microcolonies (31) or the growth of a noninfectious L. pneumophila strain in a complex biofilm that included cycloheximide-treated protozoa was interpreted as extracellular growth (42). However, in other studies L. pneumophila was found to persist in defined biofilms but not to replicate in the absence of amoebae. In a preformed biofilm composed of Pseudomonas aeruginosa, Klebsiella pneumoniae, and a Flavobacterium sp., L. pneumophila persisted in the absence or in the presence of the amoeba Hartmanella vermiformis, yet replication and release of planktonic (free-swimming) L. pneumophila into the bulk phase occurred only in the presence of amoebae (8, 23). A recent study in which an undefined microbial community was used confirmed that H. vermiformis amoebae were required for proliferation of L. pneumophila in a mixed-species static biofilm (16).
The identification of factors from bacterial metabolic networks that may promote growth of L. pneumophila is a challenge. When tap water is used as a source of nutrients, it does not seem to support growth of L. pneumophila in biofilms at a rate convenient for routine laboratory analysis. In the current study, we avoided these difficulties by using a rich medium which supports axenic growth of L. pneumophila in order to analyze adherence and biofilm formation under static, “quasi-static,” and continuous-flow conditions.
MATERIALS AND METHODS
Bacteria, plasmids, and amoebae.
The L. pneumophila strains and plasmids used in this study are listed in Table 1. L. pneumophila was routinely grown on charcoal yeast extract agar plates (6), in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract (AYE) medium, or occasionally in minimal medium (27, 28). Derivatives of the thymidine auxotroph strain Lp02 were grown in the presence of 100 μg/ml thymidine and 0.5% bovine serum albumin (14). To culture the conditional csrA mutant strain MB464, expression of csrA from plasmid p206-csrA was induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (22). Under the conditions used to test biofilm formation (see below), strain MB464 grew in the presence and in the absence of IPTG to the same optical density within 5 days even when the cells were washed prior to subcultivation. Chloramphenicol, kanamycin, and gentamicin were used at concentrations of 5 μg/ml, 50 μg/ml, and 10 μg/ml, respectively. Acanthamoeba castellanii ATCC 30234 was grown in proteose-yeast extract-glucose medium as described previously (19, 36).
TABLE 1.
Strains and plasmids used in this study
| Strain(s) or plasmid | Relevant features | Reference or source |
|---|---|---|
| L. pneumophila strains | ||
| AA100 | Virulent L. pneumophila serogroup 1 | 40 |
| BS100 | AA100 pilEL::Km | 40 |
| Corby | Virulent L. pneumophila serogroup 1 | 5 |
| Corby KH3 | Corby flaA::Km | 5 |
| GS28-K | JR32 lvh::Km | 34 |
| GS3011 | JR32 icmT3011::Km | 35 |
| JR32 | Salt-sensitive derivative of virulent L. pneumophila Philadelphia-1 serogroup 1 | 32 |
| LELA2883 | JR32 dotB2883::Tn903dIIlacZ | 32 |
| LELA2883-28 | JR32 dotB2883::Tn903dIIlacZ, lvh::Gm | 34 |
| LM1376 | JR32 rpoS4::Tn903dIIGm | 9 |
| MB110 | Lp02 wild type, Strr Thy− HsdR− | 10 |
| MB410 | Lp02 fliA 35::Km | 10 |
| MB413 | Lp02 letA 22-3::Km | 10 |
| MB464 | Lp02 csrA 5::Km, p206-csrA | 22 |
| Clinical isolates | Serogroup 1, except one strain each belonging to serogroups 4, 6, and 8-10 | Swiss National Reference Center for Legionella |
| Environmental isolates | Serogroup 1, except one strain each belonging to serogroups 4, 6, and 10 | Swiss National Reference Center for Legionella |
| Plasmids | ||
| pMMB207-Km14-GFPc | pMMB207-Km14, gfp, constitutive | This study |
| pMR119 | pMMB207, DsRed-Express, Ptac | 26 |
| pSW001 | pMMB207C, DsRed-Express, constitutive | This study |
| pTS13 | pMMB207C, lacZ, Ptac | This study |
To generate plasmid pMMB207-Km14-GFPc that constitutively produced enhanced green fluorescent protein (GFP), plasmid pMMB207-Km14 (36) was digested with EcoRV and MluI, blunted with the Klenow polymerase, and religated. This treatment partially deleted the lacIq gene, relieving repression of the Ptac promoter. The gfp gene was then released from plasmid pGS-GFP-04 (13) by digestion with EcoRI and SphI and ligated into the same restriction sites of pMMB207-Km14-ΔlacIq. To construct plasmid pTS13 expressing the β-galactosidase gene under control of the Ptac promoter, a 3.2-kb fragment was released from plasmid pBBR3535 (18) and cloned into the vector pMMB207C (3) using EcoRI and KpnI. Plasmid pSW001 constitutively expressing DsRed-Express was constructed by releasing the DsRed-Express gene from pMR119 (26) using SacI and Eco31I. The fragment was ligated into pMMB207C (3), which was digested with the same enzymes to obtain plasmid pSW002. The lacIq gene of pSW002 was deleted by digestion with MluI, treatment with the Klenow polymerase, and religation, which yielded pSW001.
Biofilm formation assay under static conditions.
Biofilm formation under static conditions was quantified by a crystal violet incorporation assay with 96-well plates, which was originally developed to analyze early steps of biofilm formation by Pseudomonas fluorescens (25). Unless indicated otherwise, we used upright, flat-bottom, 96-well polystyrene microtiter plates (no. 655180 Cellstar tissue culture plates; 34-mm2 base; surface area, 148 mm2; Greiner) containing 200 μl AYE medium per well. Twelve wells per sample were inoculated with 1:10 dilutions of L. pneumophila overnight cultures (mid-log to early stationary growth phase; optical density at 600 nm [OD600], 1.5 to 3.0) and incubated at 30°C for 5 days in a humid atmosphere (“wet box”) to prevent evaporation of liquid. The optical densities of planktonic cells in the 96-well plates were determined directly with a microplate reader (Spectramax Plus; Molecular Devices); attached cells did not contribute to the turbidity. Alternatively, attached cells together with planktonic cells were resuspended, and the optical density was determined with a photometer (Helios S; Thermo Spectronic). To quantify biofilm mass, planktonic bacteria were discarded, and surface-attached cells were fixed (10 min, 80°C) and stained for 15 min at room temperature with 200 μl/well of a filtered (pore size, 0.2 μm) 0.2% (wt/vol) crystal violet (Standard; Fluka) solution. Subsequently, the wells were washed twice with 340 μl (maximum well volume) and 200 μl water, and the dye was solubilized in 200 μl of 95% ethanol per well; 60 to 125 μl of the resulting solution was finally assayed to determine the absorbance at 600 nm with the microplate reader.
Inverse biofilm assay under static or quasi-static conditions.
Biofilm formation by L. pneumophila on inverse pins was quantified using transferable polystyrene transferable solid phase (TSP) plate lids (no. 445497; surface area, 95 mm2; Nunc). These plate lids resemble the “Calgary” biofilm device (2), yet they are used in the absence of a continuous flow. The plate lids were mounted on standard 96-well plates (no. 163320 Nunclon; Nunc) filled with 200 μl AYE medium, inoculated with L. pneumophila, and incubated for 5 days at 30°C in a “wet box” as described above. The growth of planktonic bacteria was assayed after resuspension by determining the OD600. To quantify adherent bacteria, the pins of the TSP plate lids were placed for 15 min in 96-well plates filled with 200 μl of a 0.2% crystal violet solution. The bound dye was then solubilized in 200 μl of 95% ethanol, and 125-μl aliquots were assayed to determined the absorbance at 600 nm.
To assay biofilm formation under “quasi-static” conditions, inverse microtiter plates were inoculated with L. pneumophila, and planktonic cells were allowed to attach for 2 h or 24 h. Subsequently, the lid of each plate was transferred to fresh medium once or twice a day (8 h and 16 h) for a total of 5 days. Where indicated below, A. castellanii (5 × 104 cells/200 μl) was present in the fresh medium. Wells filled with sterile AYE medium served as a negative control. Sessile (surface-attached) L. pneumophila was quantified by crystal violet staining or, to increase the sensitivity, counted by flow cytometry. In the latter assay, GFP-expressing L. pneumophila that had grown for 5 days was collected in SorC buffer (150 μl/well) (26) by vigorously vortexing the inverse 96-well plates twice for 1 min using the appropriate function of a “Spectramax” microplate reader. The contents of 12 wells per sample were combined, and the number of bacteria per 120 μl was determined with a FACScalibur flow cytometer (Becton Dickinson). Under these conditions, 81% of the planktonic L. pneumophila cells and 72% of the sessile L. pneumophila cells remained GFP positive after 5 days.
Flow chamber experiments.
In order to noninvasively monitor in real time the adherence of and biofilm formation by L. pneumophila under dynamic flow conditions, a continuous-flow chamber system was set up (20). Each channel (total volume, 160 μl) was equipped with bubble traps and flow breaks downstream of a peristaltic pump (IPC-N-16; Ismatec SA). The system was assembled using autoclaved silicon tubing (1 by 3 mm), sterilized with 0.5% sodium hypochlorite overnight, and rinsed excessively with sterile water. The routine operating conditions included a flow rate of 50 μl/min at 30°C and AYE medium diluted 1:10 (see Table S1 in the supplemental material).
Channels were inoculated by injection of 1:5 to 1:10 dilutions of virulent L. pneumophila strain JR32 constitutively expressing GFP that had been grown in AYE medium (30°C, overnight) to the mid-log or early stationary phase (OD600, 1.5 to 3.0) or with 1:50 to 1:100 dilutions of heterotrophic bacteria (see Table S2 in the supplemental material) that had been grown in AYE medium overnight. The bacteria were pumped into the flow chamber and allowed to attach for 45 to 60 min without flow. During the experiment, sterile AYE medium (diluted 1:10) was pumped through the system at the continuous-flow rates indicated below (5 μl/min to 50 μl/min). In AYE medium diluted 1:10 in Erlenmeyer flasks, L. pneumophila grew at a maximum growth rate (μmax) of 0.09 h−1, which corresponded to a doubling time of 8.2 h (data not shown). At the end of the experiment (usually 14 days), bacterial biomass was stained with SYTO62 (2 μM).
Confocal laser scanning microscopy.
The architecture of GFP- and DsRed-labeled L. pneumophila biofilms was analyzed in glass-bottom microwell dishes (35-mm petri dishes; MatTek) under static conditions (no medium replacement) or quasi-static conditions (medium replaced twice a day). An inverted confocal microscope (Zeiss Axiovert 200 M) equipped with a ×100 oil objective (Plan Neofluar; Zeiss), an “Ultraview” confocal head (Perkin-Elmer), and a krypton argon laser (643-RYB-A01; Melles Griot) was used. To monitor the flow chambers, an inverted epifluorescence microscope (Zeiss Axioplan 2) equipped with an “apotome” device was used. Data processing and three-dimensional reconstruction were performed with the “Volocity” 2.6.1 software package (Improvision).
β-Galactosidase assay.
L. pneumophila JR32 harboring plasmid pTS13 (Table 1), which encodes β-galactosidase under control of the Ptac promoter, was cultivated in upright microtiter plates as described above. lacZ expression was induced either 3 days after inoculation by addition of 0.5 mM IPTG for 0.5 h and 3.5 h or throughout bacterial cultivation. For short-term induction, planktonic cells were collected, resuspended in AYE medium containing IPTG after centrifugation (13,000 × g, 1 min), and incubated in sterile microtiter plates. After removal of planktonic cells, surface-attached L. pneumophila was overlaid with AYE medium containing IPTG and incubated either immediately or after resuspension. β-Galactosidase activity was determined with o-nitrophenyl-β-d-galactopyranoside as a substrate.
16S rRNA determination.
Environmental bacterial isolates were identified by partial 16S rRNA gene amplification using primers fD1 and rP1 and subsequent sequencing of the PCR fragment (44).
RESULTS
L. pneumophila forms biofilms under static conditions in rich medium.
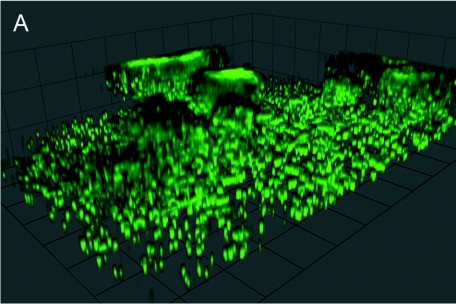
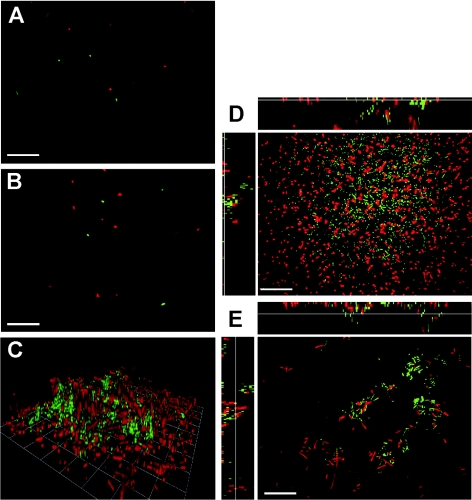
To investigate whether L. pneumophila forms three-dimensional structures under static conditions in rich medium, we used confocal laser scanning microscopy to analyze gfp-expressing wild-type strain JR32. After 3 days of growth in AYE medium, L. pneumophila formed irregular, rather fluffy aggregates that extended up to 20 μm into the medium (Fig. 1A). These aggregates formed in some areas of the glass surface, while in other areas single cells attached to the surface.
FIG.1.
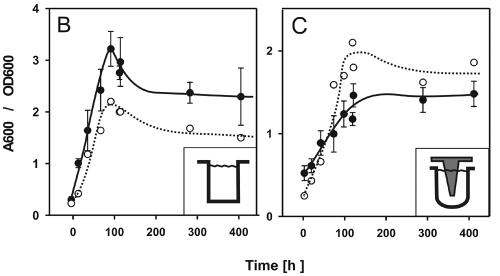
Biofilm formation by L. pneumophila under static conditions in rich medium. (A) Confocal laser scanning micrograph of a representative biofilm section formed in a glass-bottom dish by GFP-expressing L. pneumophila wild-type strain JR32. Unit cell, 13 μm. (B and C) Growth of planktonic L. pneumophila (OD600) (○) and crystal violet staining (A600) (•) of sessile bacteria in upright wells of polystyrene 96-well plates (B) or on polystyrene pins in an inverse biofilm assay (C). The assays were repeated at least three times with similar results.
Biofilm formation on abiotic surfaces can be quantified by crystal violet staining of the biomass attached to the surfaces of 96-well plates. In the present study, we tested microtiter plates made of polystyrene, microtiter plates made of polypropylene, and microtiter plates made of polyvinylchloride and found that flat-bottom polystyrene plates pretreated for cell culture yielded the largest amount of surface-attached biomass (data not shown). As the procedure originally described by O'Toole and Kolter (25) yielded only low maximum values for L. pneumophila (A600, <1.0), we optimized the crystal violet stain assay as described in Materials and Methods. In the optimized assay, biomass was formed at the bottom of the wells, as well as at the medium-air interface (data not shown), and the A600 increased 10-fold from 0.3 to 3.0 within 5 days (Fig. 1B). Under these conditions, L. pneumophila grew slowly with a doubling time of about 26 h (μmax, 0.036 h−1). The increase in crystal violet staining coincided with bacterial growth in the medium and ceased in the stationary growth phase. The presence of GFP or the growth phase of the inocula did not affect the ability of L. pneumophila to form a biofilm, as mid-log-phase cultures (OD600, 0.5) and stationary-phase cultures (OD600, 3.5) inoculated with the same initial dilution yielded similar amounts of sessile biomass after 5 days (data not shown).
The amount of biofilm formed determined by crystal violet staining may be overestimated in upright wells due to the contribution of cell sedimentation. To address this question, we tested the ability of L. pneumophila to adhere to pins of transferable microtiter plate lids (“inverse” assay) (Fig. 1C). In the inverse assay, the growth (doubling time, 26 h; μmax, 0.032 h−1) and the final cell densities of planktonic L. pneumophila were almost identical to the growth and final cell densities obtained with the upright system. Furthermore, as observed in the upright biofilm assay, the increase in crystal violet staining on the pins coincided with planktonic growth and ceased when the bacteria entered the stationary growth phase. The maximum values for crystal violet staining obtained with the inverse assay were about twofold lower than those obtained with the upright system, which roughly corresponded to the 1.5-fold-smaller surface area of the pins than of the upright wells. Therefore, the amounts of biomass per unit of surface area were similar in the upright and inverse systems, and adherence of bacteria due to sedimentation did not play a significant role.
L. pneumophila clinical and environmental isolates form biofilms.
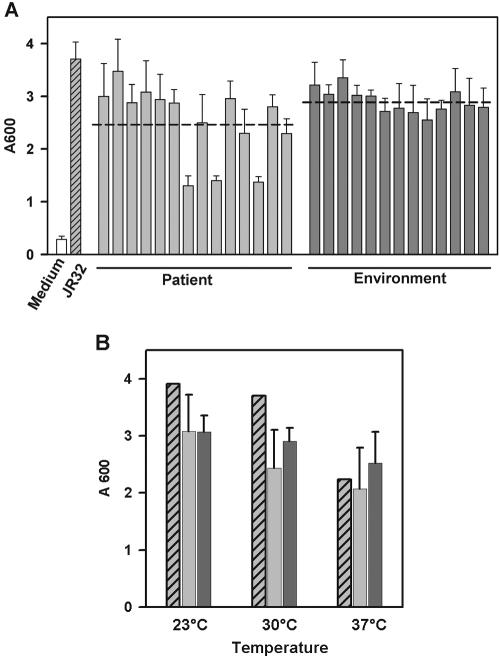
We used the upright assay to compare the biofilm formation by virulent laboratory strain JR32 with the biofilm formation by 27 L. pneumophila strains isolated from patients, cooling towers, or freshwater at different locations in Switzerland and France (Table 1). All of the environmental isolates formed biofilms comparable to those formed by strain JR32, demonstrating that the potential to form biofilms is (i) widespread among L. pneumophila strains, (ii) independent of the serogroup, and (iii) not affected by passage of JR32 over laboratory medium (Fig. 2A). In contrast, 3 of 14 clinical isolates produced about 50% less biomass on the surfaces of the wells. The three clinical strains that exhibited impaired biofilm formation, as well as other clinical strains, belong to serogroup 1, and therefore, the reduced biofilm formation was not due to a different lipopolysaccharide structure. Moreover, although they belong to different serogroups, the majority of the clinical and environmental isolates formed biofilms that were of similar sizes.
FIG. 2.
Biofilm formation by clinical and environmental L. pneumophila isolates. (A) Crystal violet staining of 5-day-old biofilms of L. pneumophila wild-type strain JR32, clinical and environmental L. pneumophila isolates, and medium. The dashed line indicates the average biomass accumulation. (B) Crystal violet staining of biofilms formed by L. pneumophila JR32 (cross-hatched bars), clinical isolates (average, standard deviation) (light gray bars), and environmental isolates (average, standard deviation) (dark gray bars) at different temperatures. The experiments were done in triplicate.
To determine the effect of temperature on biofilm formation by L. pneumophila, strain JR32 and the clinical and environmental isolates were cultivated until they reached the stationary growth phase in the upright static biofilm system at 23°C, 30°C, or 37°C, and this was followed by crystal violet staining. While the average level of biomass for the clinical and environmental strains decreased only marginally with increasing temperature, JR32 formed about 30% less biofilm at 37°C than at 23°C or 30°C (Fig. 2B). In general, biofilm formation by the environmental isolates appeared to be more robust and less influenced by temperature than biofilm formation by the clinical isolates. In light of these results, we routinely assayed biofilm formation at 30°C.
Biofilm formation by defined L. pneumophila mutants.
The static biofilm system allows identification and characterization of L. pneumophila genes involved in surface attachment and biofilm formation. We used the upright biofilm assay to test defined L. pneumophila mutants lacking genes potentially involved in surface attachment and biofilm formation (Table 2). The L. pneumophila mutants tested lacked pili, flagella, the Icm/Dot T4SS, the Lvh T4SS, both T4SSs, stationary-phase (RpoS) or alternative (FliA) sigma factors, the two-component response regulator LetA, or the global repressor CsrA. Of the mutant strains tested, only the fliA mutant was significantly affected in biofilm formation under the conditions tested, and it reproducibly produced 30% less biomass within 5 days. It is noteworthy that mutants that lacked rpoS or letA, both of which are required for expression of transmissive (virulence) traits of L. pneumophila, were not affected in biofilm formation.
TABLE 2.
Biofilm formation under static conditions by defined L. pneumophila mutant strains
| L. pneumophila mutant strain | Relevant characteristicsa | % of referenceb | SD (%) |
|---|---|---|---|
| BS100 | ΔpilEL, lacks type IV pili | 105 | 11 |
| Corby KH3 | ΔflaA, lacks flagella | 104 | 22 |
| GS28-K | Δlvh, lacks functional Lvh T4ASS | 106 | 15 |
| GS3011 | ΔicmT, lacks functional Icm/Dot T4BSS | 80 | 15 |
| LELA2883 | ΔdotB, lacks functional Icm/Dot T4BSS | 102 | 9 |
| LELA2883-28 | ΔdotBΔlvh, lacks Icm/Dot T4BSS and Lvh T4ASS | 104 | 10 |
| LM1376 | ΔrpoS, lacks stationary-phase sigma factor RpoS (σ38) | 110 | 15 |
| MB410 | ΔfliA, lacks flagellar sigma factor FliA (σ28) | 68 | 15 |
| MB413 | ΔletA, lacks two-component response regulator LetA (GacA) | 97 | 13 |
| MB464 | ΔcsrA/p206-csrA, mRNA-binding global repressor/essential activator of replication CsrA expressed under control of Ptac | 94, 122c | 11, 16c |
T4ASS, type IVA secretion system; T4BSS, type IVB secretion system.
The reference was the value for the corresponding isogenic wild-type strain (AA100, Corby, JR32, or Lp02). The experiments were done in triplicate and were reproduced at least twice with similar results.
The first value is the value obtained in the presence of IPTG, and the second value is the value obtained in the absence of IPTG.
Sessile L. pneumophila does not accumulate biomass under quasi-static conditions.
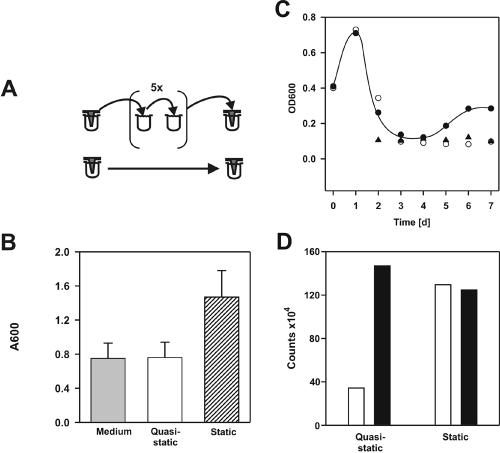
Biofilm formation by L. pneumophila under static conditions coincided with planktonic growth (Fig. 1B and C). To examine whether replication of planktonic or sessile bacteria accounted for the accumulation of sessile biomass, we performed the inverse assay under “quasi-static” conditions. L. pneumophila was allowed to adhere to pins of a TSP microtiter plate for 2 h. Subsequently, the lid was transferred for 5 days twice a day to a plate containing fresh medium, thereby exchanging the medium without disturbing the adherent bacteria (Fig. 3A). Even though under these conditions fresh medium was available for the bacteria at all times, no biomass accumulated on the pins (Fig. 3B), and no planktonic replication took place, as after transfer the OD600 decreased to and stayed at the level of the medium control (data not shown). In contrast, if after the first medium exchange the system was left static for 5 days, sessile L. pneumophila accumulated on the pins as described above, and the OD600 increased to 1.47. These findings suggest that biofilm formation by L. pneumophila under the conditions used is dependent on planktonic replication. Moreover, bacteria adhering to the pins after 2 h served as a reservoir for planktonic growth under static conditions.
FIG. 3.
L. pneumophila does not accumulate surface-attached biomass or grow under quasi-static conditions, but A. castellanii promotes biofilm formation. (A) Diagrams of static (no medium exchange) and quasi-static (medium exchanged twice a day) inverse biofilm assays. (B) Crystal violet staining of sessile L. pneumophila under static and quasi-static conditions and of noninoculated wells (medium). The results were reproduced in at least three independent experiments. (C) Growth of planktonic L. pneumophila (OD600) in the absence (○) and in the presence (•) of 5 × 104 A. castellanii cells per well and growth of A. castellanii alone (▴) in the inverse quasi-static biofilm assay. (D) Quantification of sessile, gfp-expressing L. pneumophila by flow cytometry in the absence (open bars) and in the presence (solid bars) of 5 × 104 A. castellanii cells per well under static (no medium exchange) or quasi-static (medium exchanged once a day) conditions in the inverse assay (5 days of growth).
Replication of L. pneumophila in A. castellanii promotes biofilm formation.
Rapid intracellular replication of L. pneumophila occurs in A. castellanii (∼100-fold increase in CFU in 24 h) (36). To determine whether L. pneumophila grown in amoebae contributes to biofilm formation, we used the inverse biofilm assay. After adherence of L. pneumophila to the pins for 24 h, lids were transferred twice a day to 96-well microtiter plates containing fresh medium with or without A. castellanii. In agreement with results described above, the OD600 decreased to the level of the medium control and no growth of L. pneumophila was observed in the planktonic phase in the absence of A. castellanii (Fig. 3C). In contrast, in the presence of 5 × 104 amoebae per well, the number of planktonic L. pneumophila cells increased after a lag phase of 4 days, as determined by the optical density (final OD600, 0.3), and the number of L. pneumophila cells attached to pins was comparable to the number of sessile bacteria observed under static conditions. Amoebae alone were not stained significantly with crystal violet. These results were confirmed by quantifying by flow cytometry L. pneumophila eluted from pins (Fig. 3D). After 5 days of growth under quasi-static conditions, three- to fourfold fewer adherent L. pneumophila cells were present in the absence of A. castellanii, and in the presence of amoebae similar numbers of bacteria were detected in quasistatic and static assays. These observations suggest that bacteria released from amoebae after intracellular replication accounted for the increases in both planktonic L. pneumophila and sessile biomass, supporting the notion that extrabiofilm bacterial replication is essential for biofilm formation.
Adherent L. pneumophila is metabolically active.
To test whether the lack of replication of surface-associated L. pneumophila correlates with a lack of metabolic activity, we quantified protein biosynthesis (β-galactosidase activity) in the upright static biofilm system. β-Galactosidase expression was induced by IPTG after 3 days (corresponding to the logarithmic growth phase [Fig. 1B]). After 0.5 h of induction, the β-galactosidase activity of sessile L. pneumophila or resuspended biofilm bacteria was similar to that of planktonic bacteria, while after 3.5 h of induction the enzyme activity was somewhat higher in planktonic bacteria (Fig. 4). Furthermore, if β-galactosidase was continuously expressed during cultivation, sessile L. pneumophila exhibited about one-half the activity that planktonic cells exhibited. As protein biosynthesis in biofilm-associated L. pneumophila is similar to protein biosynthesis in planktonic bacteria, the metabolic state of sessile L. pneumophila likely does not account for its lack of replication.
FIG. 4.
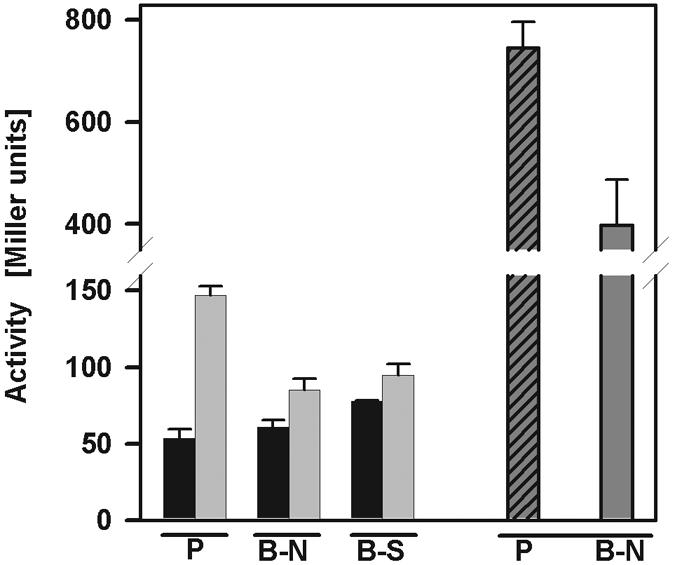
Expression of β-galactosidase activity by planktonic and sessile L. pneumophila. The β-galactosidase activities (in Miller units) of planktonic L. pneumophila, surface-attached, and resuspended biofilm bacteria were determined after induction with IPTG for 0.5 h (black bars) and 3.5 h (light gray bars) and after continuous induction with IPTG (cross-hatched bar and dark gray bar). P, planktonic bacteria; B-N, native biofilm; B-S, suspended biofilm. The data are representative of two independent experiments done in triplicate.
GFP-labeled L. pneumophila and DsRed-labeled L. pneumophila are randomly distributed on surfaces and in aggregates.
To further investigate whether L. pneumophila replicates on surfaces or in bacterial aggregates, we analyzed by confocal laser scanning microscopy the clonal distribution of L. pneumophila within biofilms formed by cocultures of GFP- and DsRed-labeled strains. A 1:1 mixture of GFP-tagged L. pneumophila and DsRed-tagged L. pneumophila was inoculated at a low density (OD600, 0.01) (Fig. 5A) and grown for 3 days in glass-bottom dishes without medium exchange. Under these conditions, L. pneumophila grew to an OD600 of about 1.0 and produced three-dimensional biofilm structures (Fig. 5C), in which individual green and red bacteria were interspersed homogeneously on the substratum (Fig. 5D) or in aggregates (Fig. 5E). We did not observe extended areas of single-color biofilm or microcolonies, suggesting that sessile L. pneumophila did not replicate at all or replicated only slowly. If after the initial attachment the medium was exchanged twice a day, hardly any growth took place in the medium (final OD600, 0.09) or on the substratum, and the few bacteria present were randomly distributed (Fig. 5B). Thus, under the conditions tested L. pneumophila biofilms seemed to be formed by random aggregation of planktonic bacteria rather than by clonal replication of sessile cells.
FIG. 5.
Distribution of GFP- and DsRed-labeled L. pneumophila in static and quasi-static cocultures. (A and B) Confocal laser scanning micrographs of GFP- and DsRed-expressing L. pneumophila JR32 on the surface of a glass-bottom dish at the time of inoculation (A) and after 3 days under quasi-static conditions (medium exchanged twice a day) (B). (C to E) Three-dimensional reconstruction (C) and sections at the surface (D) or through aggregates (E) of the biofilm formed after 3 days of growth under static conditions (no medium exchange). Bars, 10 μm; unit cell, 13 μm.
Adherence and persistence of L. pneumophila in a continuous-flow chamber system.
To monitor in real time adherence of and biofilm formation by L. pneumophila under dynamic flow conditions, we set up a flow chamber system. After inoculation (for about 1 h), a monolayer of single dispersed GFP-labeled L. pneumophila cells attached to the borosilicate glass (and polystyrene) substratum of the flow chambers in the absence and also in the presence of flow (Fig. 6 and 7A). The number of bacteria that initially attached depended on the density of the inoculum but not on the presence of GFP, the temperature (21°C to 37°C), the growth phase of the inoculum (mid-log or late log to early stationary), the source of the inoculum (liquid culture or agar plate), or the different media (see Table S1 in the supplemental material). Thus, L. pneumophila attaches to abiotic surfaces under a wide variety of conditions.
FIG. 6.
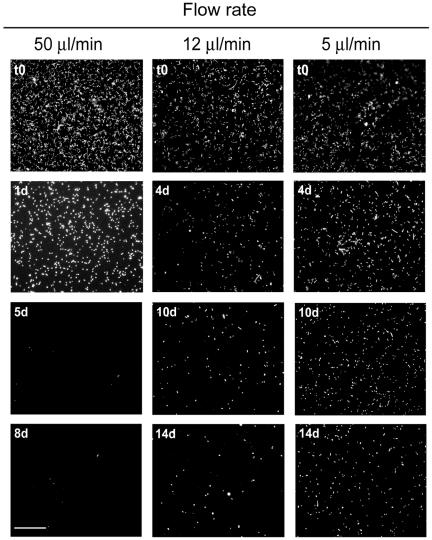
Adherence of L. pneumophila in a continuous-flow chamber system with a rich medium: fluorescence micrographs of GFP-expressing L. pneumophila in a continuous-flow chamber system operated at different flow rates with AYE medium (diluted 1:10). The bacteria initially adhered to surfaces and detached in a flow rate-dependent manner. The experiments were performed in triplicate and were independently reproduced at least twice with similar results. Bar, 50 μm.
FIG. 7.
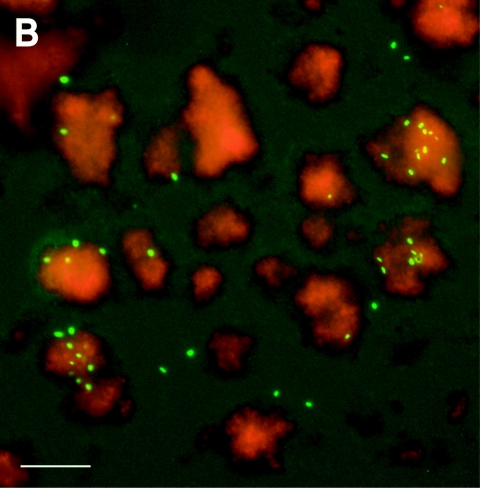
Effect of defined cocultures on persistence and localization of L. pneumophila in a continuous-flow chamber system. (A) Microbacterium sp. increased and K. pneumoniae decreased the persistence of GFP-expressing L. pneumophila in a continuous-flow chamber system operated with AYE medium (diluted 1:10) at a flow of 50 μl/min. Bar, 50 μm. (B) Fluorescence micrograph of a Microbacterium-Legionella coculture biofilm stained with the dye SYTO62 14 days after inoculation. Single L. pneumophila bacteria (green) predominantly colocalized with the Microbacterium sp. biofilm (red). Bar, 30 μm. Similar results were obtained in at least two independent experiments done in triplicate.
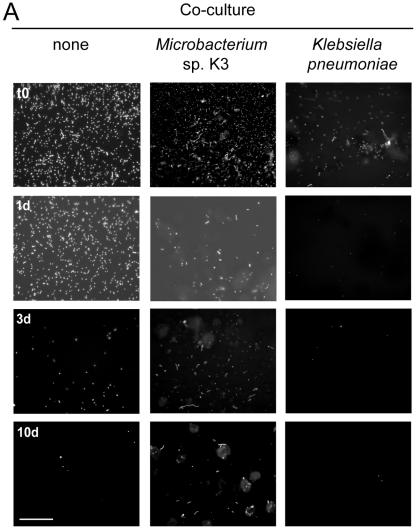
After the initial attachment, L. pneumophila JR32 did not form a biofilm for up to 14 days, despite a constant influx of nutrients (AYE medium diluted 1:10). Various conditions were tested, including different temperatures (21°C to 37°C), several media that support axenic growth in liquid culture, and sterile tap water with and without yeast extract (see Table S1 in the supplemental material). Essentially the same results were obtained for environmental and clinical L. pneumophila strains and other Legionella spp., which were inoculated either individually or as cocultures and monitored by bright-field microscopy (see Table S2 in the supplemental material).
Rather than accumulating biomass, L. pneumophila progressively detached from the surfaces of the flow chambers in a flow rate-dependent manner (Fig. 6 and 7A). While at a flow rate of 50 μl/min complete ablation of the bacteria was observed within 5 to 8 days, at flow rates of 12 μl/min and 5 μl/min increasing numbers of bacteria remained in the flow chamber 14 days after inoculation. Since the flow rate was the only parameter changed in this experiment, we concluded that mechanical forces acting on single dispersed Legionella cells attached to abiotic surfaces have a significant effect on the persistence of this bacterium in dynamic flow systems.
Specific cocultures increase the persistence of L. pneumophila under dynamic flow conditions.
In natural biofilm habitats L. pneumophila associates with diverse heterotrophic bacteria that might promote its growth or prevent detachment. To address this question, the flow chamber system was inoculated with single species or mixed cultures of defined heterotrophic bacteria (see Table S2 in the supplemental material). Within 2 days, microcolonies of these bacteria became visible, as determined by bright-field microscopy, and the flow system was then inoculated with GFP-tagged L. pneumophila. In this capture assay, proliferation of L. pneumophila was not observed. However, L. pneumophila adhered to Empedobacter breve biofilms, to Microbacterium sp. biofilms, and, to a lesser extent, to Acinetobacter baumanii biofilms. After up to 14 days, only E. breve (data not shown) and Microbacterium sp. biofilms still retained L. pneumophila (Fig. 7A). Moreover, L. pneumophila colocalized in the flow chambers in regions where there were evident Microbacterium biofilms, suggesting that there were specific interactions between these two bacteria (Fig. 7B). In contrast, no attachment of L. pneumophila to Pseudomonas spp., Corynebacterium glutamicum, or K. pneumoniae biofilms was observed. The presence of K. pneumoniae even accelerated detachment and reduced adherence of L. pneumophila to abiotic surfaces. The latter findings raise the possibility that specific environmental bacteria could be used as biocontrol strains to prevent colonization of surfaces and biofilms by L. pneumophila.
DISCUSSION
L. pneumophila persists in aquatic biofilms under ambient conditions (7, 11, 39) and even at a low pH (pH 2.7) (37). Biofilms are complex communities consisting of prokaryotic and eukaryotic organisms (bacteria, algae, fungi, and protozoa) which are linked by manifold interactions, such as metabolic fluxes, adhesion, and ingestion. It is a matter of debate whether and how L. pneumophila proliferates in biofilms. The complex nutrient requirements of this facultative intracellular bacterium can be met by intracellular growth in protozoa and possibly also by extracellular growth in a biofilm network. Reproduction of such networks in the laboratory is a challenge, and to date it has not been possible to demonstrate extracellular replication of L. pneumophila in a defined biofilm under poor nutrient conditions. In one study, replication of a noninfectious L. pneumophila strain was observed in a biofilm containing protozoa treated with cycloheximide, an inhibitor of eukaryotic protein biosynthesis (42). However, the inoculum used in this study was a naturally occurring, not completely defined aquatic population that potentially contains Legionella. Furthermore, the possibility that the avirulent L. pneumophila strain replicated within cycloheximide-treated amoebae was not explored on a cellular level. Thus, L. pneumophila might persist rather than proliferate within mixed-species biofilms in the absence of protozoan hosts (8, 23).
To analyze extracellular growth on surfaces and biofilm formation by L. pneumophila under defined conditions, in this study we used a rich medium that supported axenic growth of L. pneumophila. Although L. pneumophila rarely encounters such nutrient-rich environments, in our study we explored the potential of this bacterium to form biofilms. We found that L. pneumophila formed biofilms in upright and inverse batch systems only when it grew in broth or within amoebae (Fig. 1, 3, and 5). No accumulation of biomass was observed in the inverse system under quasi-static conditions when the medium was exchanged twice a day (Fig. 3B). The transfer frequency was about twice the doubling time of L. pneumophila under these conditions (26 h) and prevented accumulation of planktonic bacteria in the medium. Furthermore, we confirmed by fluorescence microscopy that sessile L. pneumophila did not grow or grew only very slowly on the substratum or in aggregates (Fig. 5).
The inability of L. pneumophila to form biofilms under quasi-static conditions might be due to mechanical detachment of surface-attached bacteria or the failure of sessile cells to grow. While shear forces likely account for detachment of L. pneumophila in flow chambers (Fig. 6), in the quasi-static system only weak (if any) shear forces occur, and therefore, mechanical detachment of sessile bacteria is probably not the reason why sessile biomass does not accumulate in the latter system. Moreover, in the absence of shear forces (static conditions) sessile L. pneumophila also does not seem to grow (Fig. 5).
The failure of sessile L. pneumophila to grow might be due to decreased viability or metabolism of biofilm bacteria. We quantified β-galactosidase activity as a measure of viability and metabolic activity (Fig. 4). Sessile bacteria exhibited about one-half the β-galactosidase activity that planktonic L. pneumophila exhibited. This moderate reduction in protein biosynthesis activity is unlikely to account for the substantial decrease in proliferation. Alternatively, replication rather than metabolism might be specifically down-regulated in surface-attached bacteria. The mechanism by which this might happen is obscure. There is no evidence that accumulation of a bacterial factor is required for biofilm formation, since sessile biomass accumulation did not lag behind planktonic growth (Fig. 1B and C).
In agreement with our findings is a scenario in which planktonically growing L. pneumophila (Fig. 1 and 5) or bacteria released from amoebae after intracellular replication (Fig. 3) continuously aggregate on surfaces, forming biofilms. Similarly, the observation that surface-attached L. pneumophila in flow chambers does not form robust biofilms might be due to continuous removal of planktonic bacteria (Fig. 6 and 7). In summary, our experiments performed with an axenic, growth-promoting medium support the notion that extrabiofilm replication (e.g., growth in amoebae) is required for proliferation of L. pneumophila in biofilms (8, 16, 23).
There is increasing awareness that biofilm phenotypes depend on the specific growth conditions of bacteria and differ among bacterial strains. In the past, the concept of microbial biofilms was dominated by observations obtained with the model organism P. aeruginosa. When it is grown with glucose as a source of carbon and energy, P. aeruginosa develops complex three-dimensional “mushrooms” in flow chambers, and this architecture is most likely genetically controlled. In contrast, when it is grown in the presence of citrate, P. aeruginosa forms a flat biofilm and does not require some of the genes involved in the formation of mushrooms (15). Moreover, under continuous-flow conditions Escherichia coli K-12 autoaggregates and forms small irregular and unstable microcolonies that do not mature into biofilms (29). L. pneumophila seems to exhibit yet another autoaggregation and biofilm phenotype, which is observed only under static conditions (Fig. 1 and 5). The biofilm is characterized by rather delicate, fluffy extensions from the surface. Areas where these extensions are formed alternate with regions where single bacteria adhere to the surface. Cocultures of GFP-labeled L. pneumophila and DsRed-labeled L. pneumophila did not form extended clonal microcolonies.
Under dynamic flow conditions L. pneumophila attaches but does not form robust biofilms (Fig. 6 and 7A). The inability of L. pneumophila to grow and form biofilms under dynamic flow conditions in a rich medium might be due to different reasons, including (i) mechanical detachment of single bacteria, (ii) dilution of a secreted bacterial factor required for growth on surfaces, (iii) removal of planktonically growing bacteria that may be required for biofilm formation, and (iv) suppression of processes relevant for colonization of surfaces, formation of biofilms, or persistence in aquatic ecosystems by the rich medium used in this study. However, under flow conditions L. pneumophila apparently colonizes biofilms formed by specific environmental bacteria, while other strains seem to compete with L. pneumophila for colonization of surfaces (Fig. 7). The flow chamber system described here should allow a detailed, noninvasive analysis of specific interactions between L. pneumophila and other environmental bacteria under continuous-flow conditions.
Surface attachment is the initial step in biofilm formation and is crucial for persistence and spread of L. pneumophila in the environment. The biofilm assays based on crystal violet staining allow identification of L. pneumophila genes involved in surface attachment and biofilm formation. Among the defined L. pneumophila mutants tested here, a strain lacking the flagellar sigma factor FliA (σ28) was found to be impaired for biomass accumulation (Table 2). FliA is required for expression of many traits associated with the transmissive phase of L. pneumophila, including flagella, contact-dependent cytotoxicity, phagocyte infection, and lysosome avoidance, as well as intracellular replication within the social amoeba Dictyostelium discoideum (10, 12, 21). In broth culture a fliA mutant strain bound more crystal violet than a wild-type control strain in the postexponential growth phase bound (21), indicating that the defect of the fliA mutant for biofilm formation is likely underestimated by using crystal violet binding as an assay. Only recently, L. pneumophila tatB and tatC mutant strains were found to be defective for sessile biomass accumulation, and therefore, the twin-arginine translocation (Tat) pathway seems to be another factor required for biofilm formation (4). It will be interesting to identify the targets and substrates of these systems involved in surface attachment and biofilm formation.
Supplementary Material
Acknowledgments
This work was supported by the Swiss Commission for Technology and Innovation (CTI/KTI grant 6629.2 BTS-LS), by the National Science Foundation (grant 631-065952), and by the Federal Office for Energy (BFE).
We thank Annette Oxenius for constructing plasmid pMMB207-Km14-GFPc, Wolf-Dietrich Hardt and his group for kind assistance with confocal microscopy, Klaus Heuner, Yousef Abu Kwaik, and Michele Swanson for providing L. pneumophila strains, and the Swiss National Reference Center for Legionella in Bellinzona for supplying clinical and environmental Legionella isolates. Margaret Clarke and Socorro Mesa kindly provided plasmids pMR119 and pBBR3535, respectively.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org.
REFERENCES
- 1.Albers, U., K. Reus, H. A. Shuman, and H. Hilbi. 2005. The amoebae plate test implicates a paralogue of lpxB in the interaction of Legionella pneumophila with Acanthamoeba castellanii. Microbiology 151:167-182. [DOI] [PubMed] [Google Scholar]
- 2.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358-1361. [DOI] [PubMed] [Google Scholar]
- 4.De Buck, E., L. Maes, E. Meyen, L. Van Mellaert, N. Geukens, J. Anne, and E. Lammertyn. 2005. Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem. Biophys. Res. Commun. 331:1413-1420. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 8.Fields, B. S. 2002. The social life of legionellae, p. 135-142. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Luck (ed.), Legionella. ASM Press, Washington, D.C.
- 9.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 11.Harb, O. S., L. Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 12.Heuner, K., C. Dietrich, C. Skriwan, M. Steinert, and J. Hacker. 2002. Influence of the alternative σ28 factor on virulence and flagellum expression of Legionella pneumophila. Infect. Immun. 70:1604-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilbi, H., G. Segal, and H. A. Shuman. 2001. icm/dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42:603-617. [DOI] [PubMed] [Google Scholar]
- 14.Horwitz, M. A., and S. C. Silverstein. 1983. Intracellular multiplication of Legionnaires' disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J. Clin. Investig. 71:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 16.Kuiper, M. W., B. A. Wullings, A. D. Akkermans, R. R. Beumer, and D. van der Kooij. 2004. Intracellular proliferation of Legionella pneumophila in Hartmannella vermiformis in aquatic biofilms grown on plasticized polyvinyl chloride. Appl. Environ. Microbiol. 70:6826-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDade, J. E., C. C. Shepard, D. W. Fraser, T. R. Tsai, M. A. Redus, and W. R. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory diseases. N. Engl. J. Med. 297:1197-1203. [DOI] [PubMed] [Google Scholar]
- 18.Mesa, S., Z. Ucurum, H. Hennecke, and H. M. Fischer. 2005. Transcription activation in vitro by the Bradyrhizobium japonicum regulatory protein FixK2. J. Bacteriol. 187:3329-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molofsky, A. B., L. M. Shetron-Rama, and M. S. Swanson. 2005. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect. Immun. 73:5720-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50:445-461. [DOI] [PubMed] [Google Scholar]
- 23.Murga, R., T. S. Forster, E. Brown, J. M. Pruckler, B. S. Fields, and R. M. Donlan. 2001. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147:3121-3126. [DOI] [PubMed] [Google Scholar]
- 24.Nash, T. W., D. M. Libby, and M. A. Horwitz. 1984. Interaction between the Legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J. Clin. Investig. 74:771-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 26.Otto, G. P., M. Y. Wu, M. Clarke, H. Lu, O. R. Anderson, H. Hilbi, H. A. Shuman, and R. H. Kessin. 2004. Macroautophagy is dispensable for intracellular replication of Legionella pneumophila in Dictyostelium discoideum. Mol. Microbiol. 51:63-72. [DOI] [PubMed] [Google Scholar]
- 27.Pine, L., J. R. George, M. W. Reeves, and W. K. Harrell. 1979. Development of a chemically defined liquid medium for growth of Legionella pneumophila. J. Clin. Microbiol. 9:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 30.Rogers, J., A. B. Dowsett, P. J. Dennis, J. V. Lee, and C. W. Keevil. 1994. Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabeling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58:2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 35.Segal, G., and H. A. Shuman. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 30:197-208. [DOI] [PubMed] [Google Scholar]
- 36.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheehan, K. B., J. M. Henson, and M. J. Ferris. 2005. Legionella species diversity in an acidic biofilm community in Yellowstone National Park. Appl. Environ. Microbiol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinert, M., U. Hentschel, and J. Hacker. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149-162. [DOI] [PubMed] [Google Scholar]
- 40.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Storey, M. V., J. Langmark, N. J. Ashbolt, and T. A. Stenstrom. 2004. The fate of legionellae within distribution pipe biofilms: measurement of their persistence, inactivation and detachment. Water Sci. Technol. 49:269-275. [PubMed] [Google Scholar]
- 42.Surman, S., G. Morton, B. Keevil, and R. B. Fitzgeorge. 2002. Legionella pneumophila proliferation is not dependent on intracellular replication, p. 86-89. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Luck (ed.), Legionella. ASM Press, Washington, D.C.
- 43.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 44.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.