Abstract
We have developed sediment-free anaerobic enrichment cultures that dechlorinate a broad spectrum of highly chlorinated polychlorinated biphenyls (PCBs). The cultures were developed from Aroclor 1260-contaminated sediment from the Housatonic River in Lenox, MA. Sediment slurries were primed with 2,6-dibromobiphenyl to stimulate Process N dechlorination (primarily meta dechlorination), and sediment was gradually removed by successive transfers (10%) to minimal medium. The cultures grow on pyruvate, butyrate, or acetate plus H2. Gas chromatography-electron capture detector analysis demonstrated that the cultures extensively dechlorinate 50 to 500 μg/ml of Aroclor 1260 at 22 to 24°C by Dechlorination Process N. Triplicate cultures of the eighth transfer without sediment dechlorinated 76% of the hexa- through nonachlorobiphenyls in Aroclor 1260 (250 μg/ml) to tri- through pentachlorobiphenyls in 110 days. At least 64 PCB congeners, all of which are chlorinated on both rings and 47 of which have six or more chlorines, were substrates for this dechlorination. To characterize the bacterial diversity in the enrichments, we used eubacterial primers to amplify and clone 16S rRNA genes from DNA extracted from cultures grown on acetate plus H2. Restriction fragment length polymorphism analysis of 107 clones demonstrated the presence of Thauera-like Betaproteobacteria, Geobacter-like Deltaproteobacteria, Pseudomonas species, various Clostridiales, Bacteroidetes, Dehalococcoides of the Chloroflexi group, and unclassified Eubacteria. Our development of highly enriched, robust, stable, sediment-free cultures that extensively dechlorinate a highly chlorinated commercial PCB mixture is a major and unprecedented breakthrough in the field. It will enable intensive study of the organisms and genes responsible for a major PCB dechlorination process that occurs in the environment and could also lead to effective remediation applications.
Polychlorinated biphenyls (PCBs) are priority pollutants that were used worldwide for a variety of applications for more than 50 years. PCB molecules are composed of a biphenyl backbone substituted with 1 to 10 chlorines. They were manufactured by catalytic chlorination of biphenyl to obtain specified weight percentages of chlorine for different applications. Consequently, they are complex mixtures, each composed of 60 to 90 PCB congeners. In the United States PCBs were produced under the trade name Aroclor. Several hundred million pounds of PCBs were released into the environment and pollute many rivers, lakes, and harbors worldwide. These PCBs persist in the sediments, accumulate in biota, and biomagnify in the food chain. Multiple adverse health effects have been attributed to them, and they are suspected human carcinogens (62). Microbial reductive PCB dechlorination provides a natural means of detoxifying PCBs in aquatic sediments because it reduces their persistence and increases their biodegradability and metabolism by other prokaryotes and by higher organisms (14, 16, 50, 51, 55).
There have been numerous reports of microbial dechlorination of Aroclors observed in situ and in laboratory experiments with sediment slurries (3, 9, 17, 18, 24, 34, 35, 47, 48, 53, 54, 66). Furthermore, eight microbial PCB dechlorination processes have been described (8, 9). Each of these dechlorination processes dechlorinates multiple PCB congeners in commercial PCB mixtures such as Aroclors, but they differ in the PCB congeners that are substrates, the PCB congeners that are products, and the chlorine positions targeted. However, until now no microbial PCB dechlorination process has ever been obtained in a sediment-free culture. Our objective was to develop and characterize a stable sediment-free culture that retained the ability to dechlorinate the same broad spectrum of PCBs that is dechlorinated in situ. The source of the sediment used to develop our cultures was one of the most highly PCB-contaminated rivers in the country, the upper Housatonic River in western Massachusetts. This river is contaminated with up to 668 mg/kg of Aroclor 1260 (15), a PCB mixture that is 60% chlorine by weight and is composed of congeners with 5 to 9 chlorines and an average chlorine number of 6.3.
Here we report the development and characterization of sediment-free enrichment cultures that extensively dechlorinate Aroclor 1260. The pattern of dechlorination is essentially identical to that of Process N, the major dechlorination activity observed in the Housatonic River. Hexa- through nonachlorobiphenyls were dechlorinated to less toxic and less persistent tri- through pentachlorobiphenyls. Our phylogenetic characterization of one sediment-free culture provides the best evidence to date of the bacteria that are actually involved in PCB dechlorination in the environment and suggests that Dehalococcoides spp. are involved.
MATERIALS AND METHODS
Sediment collection and storage.
Sediment samples were collected by repeated core sampling near the western shore of Woods Pond and transferred to 1-gallon glass jars which were then filled to the top with site water, sealed, and stored at 4°C until use.
Reagents.
PCB congeners (purity, 99.9%) were purchased from AccuStandard (New Haven, CT). 2,6-Dibromobiphenyl (26-BB) (purity, 99.9%) was a gift from GE Corporate Research and Development (Niskayuna, NY).
Microcosm preparation.
Microcosms were prepared in an anaerobic chamber (Coy Laboratories) in an atmosphere consisting of 95 to 97% N2 and up to 5% H2. Sediment was sieved to remove debris and then combined with anoxic sterile ultrapure H2O to form a slurry (60% wet sediment, 40% water). Thirty-milliliter aliquots of the slurry were dispensed into 60-ml serum bottles. We added sterile disodium malate (pH 7.0) to a final concentration of 10 mM and 26-BB to a final concentration of 350 μM from a 70 mM stock solution in gas chromatography (GC)-grade acetone (OmniSolv; EM Science). Triplicate microcosms were sealed with Teflon-lined butyl rubber septa (West Company) and aluminum crimp caps and were then incubated in the dark at 22 to 24°C. Sterile slurries for initial transfers were prepared by pasteurizing the microcosms at 75°C for 10 min, incubating them at 22 to 24°C for 24 h, and finally autoclaving them at 121°C for 3 h. The enrichments were transferred onto sterile sediment slurries three times and incubated under a nitrogen headspace. These subcultures were supplemented so that they contained each of the following vitamins at a final concentration of 50 μg/liter: p-aminobenzoic acid, d-biotin, folic acid, niacinamide, d-pantothenic acid, pyridoxal, pyridoxamine, pyridoxidine, riboflavin, thiamine, dl-6,8-thioctic acid, and vitamin B12.
Transfer onto medium without sediment.
We transferred an inoculum (final volume, 10%) from triplicate sediment slurries to triplicate bottles containing defined medium and incubated them under a headspace consisting of 20% CO2 and 80% N2. We used a sulfide-free bicarbonate-buffered minimal medium amended with a selenite-tungstate solution, vitamins (including vitamin B12), and a trace element solution (SL9) as described previously (2) and reduced with a solution of titanium(III) chloride (0.1 M)-citrate (0.2 M), pH 7, as described previously (2). However, after two transfers without sediment, we were unable to transfer a third time until we supplemented the medium with yeast extract. We eventually determined that 0.01% yeast extract maintained high dechlorination activity (see below). The enrichments were supplemented with pyruvate, butyrate, acetate plus formate, or acetate plus H2. Hydrogen was added by injecting 5 ml with a sterile syringe into a 30-ml culture to obtain a nominal concentration of 7.5 mM. Initially, each carbon source was added at a final concentration of 5 mM. After this, enrichments were fed every 3 to 4 weeks. Acetate or acetate-plus-formate enrichments were fed by adding each compound at a concentration of 5 mM, and pyruvate and butyrate enrichments were fed by adding the compound at a concentration of 3 mM. The medium was amended with Aroclor 1260 (lot 023-150B; AccuStandard, New Haven, CT) at the concentrations indicated below. The Aroclor 1260 (and 26-BB in some cases) was added as a concentrated acetone solution to 300 mg of sterile silica (∼240 mesh; Fisher Scientific). The bottoms and walls of the serum bottles were coated with the silica-PCB-acetone mixture by rotating the bottles on a vortex mixer, and then the acetone was gently evaporated with a stream of N2 gas. After the silica was totally dry, the medium was added, and the silica-PCB mixture was resuspended by vortexing. We believe that this procedure greatly increases the surface area for deposition of PCBs and thus makes these compounds more readily available to the bacteria. Furthermore, the fine silica-PCB mixture is easily suspended and allows highly reproducible samples to be taken. The first few transfers on sediment-free medium were also amended with 26-BB (350 μM).
Extraction of PCBs and bromobiphenyls.
We sampled the microcosms at intervals of 7 to 10 days for PCB and bromobiphenyl extraction and analysis during the incubation periods (up to 152 days). For cultures containing significant amounts of sediment, the samples used for analysis (0.2 to 1 ml) were aseptically collected under a stream of sterile, O2-free N2 gas by using a micropipette with the end of the tip cut off. After the second transfer without sediment, samples were taken by syringe after the syringe was rinsed with 5 mM sulfide to remove O2. Samples were transferred to 8-ml glass vials fitted with Teflon-lined screw caps. Halogenated biphenyls were extracted with 4 to 5 ml of anhydrous diethyl ether (Mallinckrodt) by vigorous horizontal shaking on a platform shaker for a minimum of 16 h. Quantitative comparisons of samples extracted by this simple procedure and by a rigorous Soxhlet procedure (EPA 3540) (63) revealed no difference.
GC analysis and quantitation of halogenated biphenyls.
Dehalogenation of 26-BB was monitored by GC-mass spectrometry as previously described (12). Congener-specific PCB dechlorination was monitored by high-resolution capillary GC analysis with a 63Ni electron capture detector (ECD) as previously described (11). In brief, we used a Hewlett-Packard model 5890 GC-ECD operated in splitless mode and equipped with a DB-1 capillary column (length, 30 m; inside diameter, 0.25 mm; phase thickness, 0.25 μm; J & W Scientific, Inc., Folsom, CA). We used specially designed calibration standards prepared from Aroclor 1260 supplemented with all 43 of the PCB congeners previously identified as products of Dechlorination Process N (11, 58). The concentrations of the individual components of Aroclor 1260 in our standard were calculated from previously determined weight percent distributions of the congeners in Aroclor 1260 (25, 58). Our customized standard permits quantitation of 84 PCB peaks, including all significant peaks detected in our samples. The GC-ECD data were collected with Dionex AI-450 chromatography software (Dionex Corp., Sunnyvale, CA). PCBs were quantified with a five-point external calibration for the customized PCB standard (542 to 8,668 ng/ml) with a quadratic fit forced through zero. We calculated the mole percent value for each individual peak, the distribution of ortho, meta, and para chlorines per biphenyl, the total number of chlorines per biphenyl, and the PCB homolog distribution.
DNA extraction, cloning, and RFLP analysis.
DNA was extracted from triplicate cultures that had been transferred eight times on sediment-free medium. We used a BIO 101 Fast DNA Spin Kit for Soil (Q-Biogene), which worked equally well for DNA extraction from sediment slurries and from sediment-free cultures, with slight modifications of the manufacturer's protocol. A total of 1.5 ml of culture was transferred to an empty 2-ml screw-cap Fast Prep tube and centrifuged at 16,000 × g in a microcentrifuge for 15 min, and the supernatant was discarded. This procedure was repeated with a second 1.5 ml of culture. Lysing matrix E was added to the pellet. The second modification of the protocol was to process the preparation twice for 45 s at speed 5.5 with a Fast Prep (Savant). The DNA was diluted 1:10 with sterile PCR-quality water, and 1 μl was used as a template for PCR amplification in a 25-μl reaction mixture with eubacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (42, 43) and Eppendorf MasterTaq according to the manufacturer's instructions. We used the following hot start PCR program. PCR mixtures containing all reagents except Taq polymerase were incubated for 2 min at 94°C, and then 1 μl of Taq polymerase diluted 1:10 with 10× Taq buffer was added. Twenty-five cycles of 45 s at 94°C, 45 s at 45°C, and 45 s at 72°C were carried out, followed by final extension at 72°C for 7 min and a final hold at 4°C. Agarose gel electrophoresis revealed a single band that was the correct size (data not shown). We used the Topo TA Cloning Kit (Invitrogen) to clone 1.5 μl of the PCR product. Ligation was performed for 30 min. Cells were plated on Luria-Bertani (LB) plates with ampicillin (100 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (50 μg/ml). White colonies were picked onto fresh LB plates containing ampicillin and X-Gal, and 118 clones were checked for inserts by colony PCR with M13 primers as follows. PCR mixtures (25 μl) were prepared with no template, and then a sterile toothpick was used to gently touch a clone colony. The toothpick was inserted into the PCR mixture and stirred to transfer some cells to the reaction mixture. The amplification program was 7 min at 94°C and then 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, followed by a final extension at 72°C for 12 min and a final hold at 4°C. PCR products were analyzed on 1% agarose gels. All but two clones had inserts that were the proper length. The clones were then subjected to restriction fragment length polymorphism (RFLP) analysis as follows. Ten microliters of each PCR product was incubated overnight at 37°C with 2 U of HhaI and 2 U of MspI (TaKaRa) in 20 μl (total volume) of a solution containing (final concentrations) 33 mM Tris-acetate (pH 7.5), 10 mM magnesium acetate, 66 mM potassium acetate, 0.5 mM dithiothreitol, and 50 ng/μl bovine serum albumin. Following incubation, 2 μl of 10× loading dye was added to each reaction mixture, and the digests were electrophoresed on a 2% low-melting-point agarose gel (SFR; Amresco, Solon, Ohio) in 1× Tris-borate-EDTA at 170 V. RFLP patterns were analyzed visually.
Plasmid isolation and sequencing.
Clones that were representative of each RFLP group were grown, and the plasmids were isolated with a QIAGEN Spin Miniprep Kit used according to the manufacturer's instructions. Cloned 16S rRNA genes that were representative of all unique RFLP groups were sequenced from both ends using the M13F and M13R primers. The sequences were checked for chimeras by CHIMERA_CHECK (44) and then analyzed by the Basic Local Alignment Search Tool (BLAST) (4) and by the Sequence Match tool of Ribosomal Database Project II (RDP II), release 9.33 (updated 10 November 2005; http://rdp.cme.msu.edu) (20). The closest matching sequences and type strains were identified using the RDP II Sequence Match tool. When possible, the new sequences were classified according to Bergey's Taxonomic Outline of Prokaryotes (26), using the RDP II-release 9 Classification tool (20). The sequences were aligned by the Ribosomal Database Project staff by taking rRNA secondary structure into account. Phylogenetic trees were constructed using PAUP* 4.0b and were rooted using Aquifex pyrophilus as an outgroup. Maximum-likelihood trees were constructed using a heuristic search. Bootstrapped (1,000 bootstraps) neighbor-joining trees (using the Jukes-Cantor model) and maximum-parsimony trees were also constructed. Negative branches were not allowed, and zero-length branches were collapsed.
Nucleotide sequence accession numbers.
The nucleotide sequences of cloned 16S rRNA genes have been deposited in the GenBank database under accession numbers DQ168641 to DQ168658.
RESULTS
Establishment of sediment-free cultures.
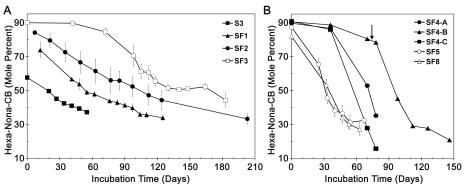
We previously determined that it is necessary to stimulate PCB dechlorination in the sediments studied by the addition of a high concentration (350 μM) of a single PCB congener or brominated biphenyl, a procedure that we called priming (6, 11, 13, 64). Hence, we established active PCB-dechlorinating cultures in sediment slurries carrying out Process N dechlorination by priming with 26-BB as previously reported (11). These cultures were transferred three times onto sterile sediment slurries when extensive dechlorination had occurred but before dechlorination had stopped. Malate, 26-BB, and vitamins were added at each transfer. In each case the 26-BB was completely dehalogenated to biphenyl, and the PCBs were dechlorinated without delay, as shown in Fig. 1, culture S3. We expressed the data as the decrease in hexa- through nonachlorobiphenyls over time because hexa- through nonachlorobiphenyls comprise ∼90% of Aroclor 1260. We then transferred the cultures onto sulfide-free bicarbonate-buffered minimal medium reduced with a Ti(III)-citrate solution; the final concentrations of Ti(III) and citrate were 0.8 mM and 1.6 mM, respectively. (We used sulfide-free medium because some organisms, including some sulfate reducers, are sensitive to sulfide.) Acetate and formate (5 mM each) were added as a carbon source and electron donor. PCB dechlorination was slow but steady for the first two transfers onto medium without sediment (Fig. 1, cultures SF1 and SF2), but three attempts to transfer a third time under the same conditions failed. After supplementing the medium with 0.1% yeast extract, we were able to transfer a third time without sediment, but there was a lag of about 2 months before PCB dechlorination began (Fig. 1, culture SF3). Since less than 5% of the 26-BB was dehalogenated after the third transfer without sediment, subsequent transfers were made without 26-BB. We subsequently decreased the yeast extract concentration to 0.01% and added 5 ml H2 per 30-ml culture as a potential electron donor. These changes reduced the lag time for the fourth transfer to about 35 days in two of the three triplicates (Fig. 1B). However, the third triplicate still showed little dechlorination after 70 days until we reinoculated the culture. Slow dechlorination and samples that showed little or no dechlorination were fairly common. Therefore, beginning with the fifth transfer without sediment, we increased the concentration of Aroclor 1260 from 5 μg/ml to 50 μg/ml (135 μM). This eliminated the lag time, greatly increased the rate of dechlorination (Fig. 1B, cultures SF5 and SF8), and resulted in far more reproducible dechlorination. We also eliminated the formate from subsequent transfers because cultures amended with formate, acetate, and H2 were no faster, and perhaps slightly slower, than cultures with acetate and H2 alone. Parallel transfers with pyruvate and butyrate were also carried out, and for several transfers PCB dechlorination was more rapid on these substrates (data not shown). However, by the eighth transfer without sediment, dechlorination was faster in the cultures transferred with acetate plus H2 (Fig. 1B, culture SF8); hence, the cultures grown on other substrates (data not shown) were discontinued.
FIG. 1.
Time course of dechlorination of Aroclor 1260 at various stages of the enrichment process. S3 is the third transfer on autoclaved sediment; SF1, SF2, SF3, SF4, SF5, and SF8 are the first, second, third, fourth, fifth, and eighth transfers without sediment, respectively. The Aroclor 1260 concentration was 5 μg/ml for SF1 to SF4 and 50 μg/ml for SF5 and SF8. For the S3 cultures, the sediment-associated Aroclor 1260 had been partially dechlorinated in situ. The concentration was approximately 45 μg/g (dry weight). Most data are averages for three replicates; for SF4 the data for three individual samples (SF4-A, SF4-B, and SF4-C) are shown. SF4-B showed minimal dechlorination by day 70 and was reinoculated from SF4-A and SF4-C on day 75 (indicated by an arrow). The error bars indicate the standard deviations of the means.
Effect of PCB concentration on dechlorination.
Increasing the concentration of Aroclor 1260 from 5 to 50 μg/ml had a profound impact on both the rate of dechlorination and the success of transfer. The maximum observed rate of dechlorination increased by a factor of more than 20 (Fig. 1 and Table 1). Also, transfers with 50 μg/ml had no lag time and were highly reproducible. Increasing the PCB concentration to 250 or 500 μg/ml further increased the rate of dechlorination (Table 1). These concentrations are 4 to 5 orders of magnitude greater than the solubility of Aroclor 1260, which is only 2.7 μg/liter (7.3 nM) (33, 61).
TABLE 1.
Maximum observed rates of dechlorination at different PCB concentrations
| No. of transfers without sediment | Expt | Aroclor 1260 concn
|
Maximum observed rate of dechlorination (nmol Cl/culture/day) (mean ± SD)a | |
|---|---|---|---|---|
| μg/ml | μM | |||
| 1 | JN2 | 5 | 13.5 | 4.71 ± 1.10 |
| 2 | JN3 | 5 | 13.5 | 3.53 ± 1.03 |
| 3 | JN7 | 5 | 13.5 | 4.88 ± 0.83 |
| 5 | JN12 | 50 | 135.0 | 169 ± 15 |
| 6 | JN13 | 50 | 135.0 | 99 ± 6 |
| 250 | 675.0 | 290 ± 51 | ||
| 500 | 1,350.0 | 594 ± 93 | ||
| 8 | JN18 | 50 | 135.0 | 100 ± 9 |
| 250 | 675.0 | 544 ± 44 | ||
The concentrations of Cl were calculated from the PCB analyses. Data were calculated per 30-ml culture.
Sediment-free cultures carry out extensive dechlorination of Aroclor 1260.
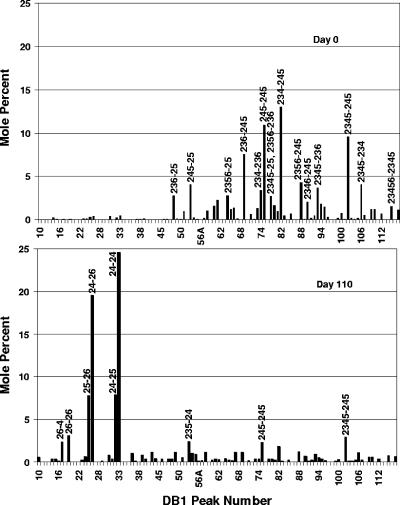
Figure 2 compares the PCB congener distribution for the eighth transfer without sediment at time zero and that after 110 days of incubation at 22 to 24°C. The data are the means for triplicate cultures incubated with 250 μg/ml of Aroclor 1260. Every peak for a congener with six or more chlorines was dramatically decreased. This analysis reveals an unprecedented breadth of substrate range in this sediment-free culture. Four major products were formed: 2,2′,4,4′-(tetra)chlorobiphenyl (24-24-CB), 24-26-CB, 24-25-CB, and 25-26-CB. (In this paper, we shall refer to PCB congeners by listing the substituted positions on each ring separated by a hyphen. Thus, 24-24-CB is the congener substituted at positions 2, 2′, 4, and 4′.) Significant amounts of 26-4-CB and 26-26-CB were also formed. This pattern of dechlorination is typical of Process N dechlorination, the dominant dechlorination activity in the Housatonic River (7, 10, 64). However, none of our previous investigations with sediment slurries have ever shown such extensive dechlorination.
FIG. 2.
Change in PCB congener distribution as a result of dechlorination in enrichment cultures transferred eight times without sediment. The data are the averages for three replicates. The concentration of Aroclor 1260 was 250 μg/ml (675 μM). A complete list of the congener assignments for all DB1 peaks is given in references 25 and 64.
Analysis of the PCB dechlorination.
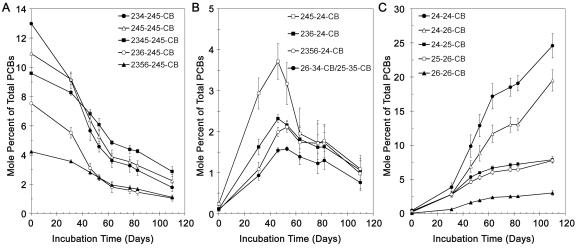
Analysis of the individual congener peaks over the course of the incubation showed that the congeners fell into three classes: substrates, intermediates, and terminal products. The results for several examples of each are shown in Fig. 3. All data are the averages for triplicate cultures incubated with 250 μg/ml of Aroclor 1260. Virtually all of the components of Aroclor 1260 were substrates. Hexachlorobiphenyls were dechlorinated at a higher rate than heptachlorobiphenyls (Fig. 3A), and heptachlorobiphenyls were dechlorinated more rapidly than octa- and nonachlorobiphenyls (data not shown).
FIG. 3.
Time course of changes in specific congeners during dechlorination of Aroclor 1260. (A) Dechlorination of five major components of Aroclor 1260. (B) Formation and subsequent dechlorination of intermediates. (C) Accumulation of major dechlorination products of Aroclor 1260. All data are averages for three replicates. The error bars indicate the standard deviations of the means. The concentration of Aroclor 1260 was 250 μg/ml (675 μM).
The time course of appearance of the five most abundant terminal products is shown in Fig. 3C. All products were formed throughout the entire incubation, although at different rates. To a large extent, the rate of formation of each product was apparently determined by the amount of available substrate that could be dechlorinated to that product. As shown clearly in Fig. 3C, the initial rates of formation of 24-25-CB, 24-26-CB, and 25-26-CB were identical and were only slightly lower than the initial rate of formation of 24-24-CB. This was confirmed by the nearly equal heights of the peaks on chromatograms at the earliest stages of dechlorination. Subsequently, 24-24-CB and 24-26-CB were formed at much higher rates. A likely explanation for this is that the pentachlorobiphenyls which were the immediate precursors of these dechlorination products are present at very low concentrations in Aroclor 1260. For example, Fig. 3B shows that 245-24-CB and 236-24-CB, which are precursors of 24-24-CB and 24-26-CB, respectively, comprise less than 0.25 mol% of the total PCBs in Aroclor 1260. However, the levels of these intermediates subsequently increased to 3.71 and 2.32 mol%, respectively, as they were formed through the dechlorination of hexa- and heptachlorobiphenyls.
A close examination of Fig. 2 shows that there are several additional peaks which were higher at day 110 than at zero time. These are the peaks for 26-34-CB, 236-24-CB, 245-24-CB, and 2356-24-CB (peaks 39, 49, 54, and 67). However, an analysis of these congeners over the entire time course (Fig. 3B) revealed that these congeners are all intermediates whose levels initially increased and which were subsequently further dechlorinated. Other congeners in this class are 246-34-CB/236-246-CB and 234-25-CB (peaks 55 and 58A, respectively).
Effect of dechlorination on PCB homolog distribution.
The dechlorination resulted in a shift in the homolog distribution from primarily hexa- and heptachlorobiphenyls to predominantly tetrachlorobiphenyls (Table 2). About one-half of the octa- and nonachlorobiphenyls were converted to less chlorinated congeners, but the experiment was terminated before dechlorination was complete and it is likely that the levels of these homologs would have been further decreased if the experiment had been continued. The level of pentachlorobiphenyls decreased only slightly, not because these compounds were not dechlorinated but because dechlorination was still in progress (Fig. 3B) and because new pentachlorobiphenyls, such as 246-24-CB, 246-25-CB, and 246-26-CB, were formed as terminal dechlorination products. Overall, the level of the hexa- through nonachlorobiphenyls decreased by 76%.
TABLE 2.
Effect of dechlorination on PCB homolog distribution
| PCB homologa | Amt (mol%) (avg ± SD)
|
% Decrease | |
|---|---|---|---|
| Day 0 | Day 110 | ||
| Trichlorobiphenyl | 0.41 ± 0.02 | 4.10 ± 0.47 | |
| Tetrachlorobiphenyl | 2.19 ± 0.08 | 65.23 ± 3.52 | |
| Pentachlorobiphenyl | 12.53 ± 0.06 | 10.46 ± 1.30 | 16.5 |
| Hexachlorobiphenyl | 47.33 ± 0.12 | 8.62 ± 1.49 | 81.8 |
| Heptachlorobiphenyl | 31.17 ± 0.10 | 8.43 ± 1.06 | 73.0 |
| Octachlorobiphenyl | 5.08 ± 0.08 | 2.38 ± 0.60 | 53.2 |
| Nonachlorobiphenyl | 1.09 ± 0.20 | 0.58 ± 0.03 | 46.5 |
No mono- or dichlorobiphenyls were detected.
The dechlorination was almost exclusively at the meta position, but 9.4% of the para chlorines were also removed (Table 3). The para dechlorination was most likely from 2,3,4,5- (2345-) and 23456-chlorophenyl rings, where the para chlorine is flanked by two meta chlorines. This hypothesis was supported by the fact that intermediates with 2356-chlorophenyl rings were formed (Fig. 3B). No ortho chlorines were removed.
TABLE 3.
Effect of dechlorination on chlorine position
| Chlorine position | No. of Cl/biphenyl (avg ± SD)
|
% Dechlorination | |
|---|---|---|---|
| Day 0 | Day 110 | ||
| ortho | 2.44 ± 0.00 | 2.49 ± 0.01 | 0 |
| meta | 2.47 ± 0.01 | 0.89 ± 0.09 | 63.9 |
| para | 1.37 ± 0.00 | 1.24 ± 0.00 | 9.4 |
| Total | 6.27 ± 0.01 | 4.61 ± 0.10 | 26.4 |
Specificity of dechlorination.
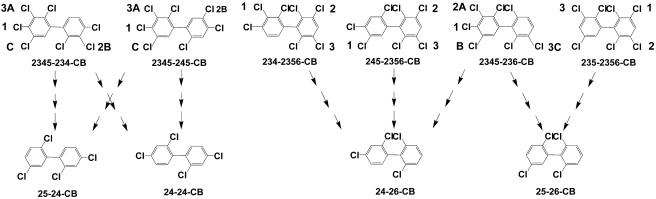
Figure 4 shows the proposed routes of dechlorination of six heptachlorobiphenyl components of Aroclor 1260 to the four major tetrachlorobiphenyl products. On the basis of the quantitative results, we propose that 2345-chlorophenyl rings may be dechlorinated by either of two pathways. It appears that the dominant mode of dechlorination, occurring approximately 60% of the time, is via loss of the para chlorine, followed by loss of the meta chlorine in position 3 to generate a 25-chlorophenyl ring. The second mode of dechlorination is via sequential loss of both meta chlorines, first from position 3 and then from position 5. Hence, we propose that 2345-234-CB is dechlorinated by two different pathways that lead to two different products. The dominant pathway is 2345-234-CB → 235-234-CB → 235-24-CB → 25-24-CB. The second pathway is 2345-234-CB → 245-234-CB → 245-24-CB → 24-24-CB. For each of the six heptachorobiphenyls we indicate the proposed order of chlorine removal in Fig. 4. Note that the chlorination pattern of both rings affects the order in which chlorines are removed. In general, it appears that the substrate preference for chlorophenyl rings is as follows: 23456 ≈ 2346 ≈ 2345 > 234 > 245 > 236 > 2356 > 235 > 34.
FIG. 4.
Proposed pathways for dechlorination of six heptachlorobiphenyls to the four major dechlorination products observed in the enrichment cultures. We propose that the first chlorine removed from 2345-chlorophenyl rings may be either the para chlorine (60% of the time) or the meta chlorine in position 3 (40% of the time). The proposed order of removal of the chlorines for each congener is also shown. For congeners with 2345-chlorophenyl rings we indicate the preferred order of attack by 1, 2, and 3 and the less frequent pattern of attack by A, B, and C. Note that the chlorination pattern of both rings affects the order in which chlorines are removed.
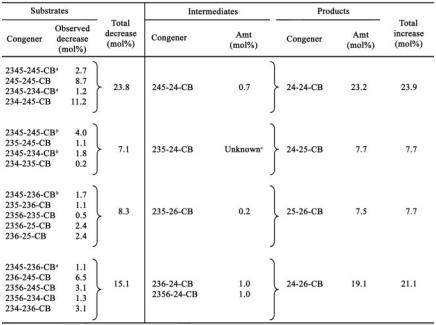
Table 4 lists the major congeners of Aroclor 1260 that are dechlorinated to each of the major tetrachlorobiphenyl products and shows the mole percent decrease of each in triplicate cultures incubated with 250 μg/ml of Aroclor 1260 for 110 days. The list of substrates includes some hexa- and pentachlorobiphenyls which not only are present in Aroclor 1260 but also are transient dechlorination products of higher congeners, such as 245-245-CB and 245-234-CB. The latter two congeners are major components of Aroclor 1260 and are also transient intermediates of 2345-245-CB and 2345-234-CB, respectively. Congeners which are proposed to be dechlorinated by two different pathways (initial para dechlorination or initial meta dechlorination) are present twice in Table 4, and the observed decreases for these congeners are apportioned as proposed above for each of the two pathways and terminal products. Table 4 also lists the observed mole percent increases for intermediates which accumulate (as shown in Fig. 3B) and for the terminal dechlorination products. These data illustrate why 24-24-CB and 24-26-CB accumulate to such high levels. They also permit a mass balance analysis. For 24-24-CB the mass balance is nearly perfect: an observed 23.9 mol% increase versus an observed 23.8 mol% decrease. For 24-25-CB and 25-26-CB the mass balances are also very good, approximately 108% and 94%, respectively. For 24-26-CB the mass balance is 139%. This could be due to an error in the calibration of 24-26-CB or to comigration of a contaminant with 24-26-CB, leading to an overestimate of this congener.
TABLE 4.
Mass balance of penta-, hexa-, and heptachlorobiphenyls dechlorinated to the four major tetrachlorobiphenyl products
The decrease for this congener was based on 40% meta dechlorination of the 2345-chlorophenyl ring to a 245-chlorophenyl ring.
The decrease for this congener was based on 60% para dechlorination of the 2345-chlorophenyl ring to a 235-chlorophenyl ring.
This congener coelutes with 245-25-CB and could not be resolved. It is a likely intermediate, but its contribution could not be determined.
Sixty-four PCB congeners were confirmed substrates, and 47 of these have six or more chlorines. Virtually all congeners containing 34-, 234-, 235-, 236-, 245-, 2345-, 2346-, and 2356-chlorophenyl rings and some congeners with 23456-chlorophenyl rings were substrates. However, as previously observed for Process N, the meta chlorines on 23- and 25-chlorophenyl groups were not substrates.
Analysis of sediment-free PCB-dechlorinating enrichment culture.
We amplified and cloned nearly complete 16S rRNA genes from genomic DNA extracted from our culture after eight transfers without sediment. RFLP analysis of 107 clones and subsequent sequencing of the 16S rRNA genes of all distinct RFLP groups revealed 11 operational taxonomic units (OTUs) (Table 5). Each OTU was phylogenetically classified using the naïve Bayesian rRNA classifier hosted on the RDP II site (20). This classifier is trained on all known type strain 16S rRNA sequences. The query sequence is assigned a taxonomic hierarchy, and the calculation is repeated for 100 trials to assign an estimate of the certainty of the taxonomic assignment. Table 5 shows the major phylogenetic group and the most precise taxonomic unit that could be assigned for each OTU and also shows the closest sequence match in the GenBank database as determined by using both BLAST and the RDP II Sequence Match tool (4, 20). All but one of the OTUs had close sequence matches (≥94 to >99% identity), but only four of those including Dehalococcoides matched isolated strains.
TABLE 5.
Phylogenetic analysis of sediment-free PCB-dechlorinating enrichment
| Clone distribution
|
Bergey's classification (RDP II) of OTU
|
No. of 16S rRNA genesc | Closest match in GenBank database as determined by RDP Sequence Match and BLAST tools | % Identity | |||
|---|---|---|---|---|---|---|---|
| Clone(s) | na | Normalized %b | Phylogenetic group | Closest classified relative (% certainty) | |||
| JN18 V62 Y, JN18 V2 A*, JN18 V17 A3 | 23 | 18-26 | Betaproteobacteria | Thauera (94) | 4d | Azoarcus strain LU1 (AJ007007) | 98-99 |
| JN18 A94 J | 2 | 3-4.5 | Deltaproteobacteria | Geobacter (96) | 2 | Uncultured bacterium clone W31 (AY770971) | 96 |
| JN18 A91 | 2 | 1.5-2 | Betaproteobacteria | Burkholderiales (95) | 3-6 | Uncultured bacterium clone B44 (AF407722) | 95 |
| JN18 A13 Q, JN18 A60 A4, JN18 A17R | 17 | 11-13 | Gammaproteobacteria | Pseudomonas (100) | 4-7 | Uncultured bacterium clone 69-7G (AY955095) | 99 |
| JN18 107 G | 42 | 27-32 | Bacteroidales | Porphyromonadacae (100) | 5-6 | Bacteroides sp. strain Z4 (AY949860) | >99 |
| JN18 A30 B, JN18 A96 B* | 4 | 13-18 | Chloroflexi | Dehalococcoides (100) | 1 | Dehalococcoides strains CBDB1 and FL2 (AF230641 and AF357918) | >99 |
| JN18 A14 H | 7 | 2.5-7 | Clostridiales | Sedimentibacter (100) | 3-12 | Uncultured bacterium clone PL-5B8 (AY570591) | 96 |
| JN18 V41 S | 3 | 1-3 | Clostridiales | Lachnospiracae (94) | 3-12 | Anaerotruncus colihominis HKU19 (DQ080188) | 94 |
| JN18 A56 K, JN18 A89 K* | 3 | 1-3 | Clostridiales | Clostridiacae (64) | 3-12 | Uncultured rumen bacterium clone 4C28d-15 (AB034128) | 87 |
| JN18 A24 M | 1 | 0.5-1 | Clostridiales | Syntrophomonadaceae (96) | 3-12 | Uncultured bacterium clone Vadin CA02 (UEU81706) | 97 |
| JN18 A7 F* | 3 | 2-10 | Eubacteria | Proteobacteria (38) | 1-7e | Uncultured bacterium clone SHA-53 (AJ249111) | 99 |
Number of clones.
Percentage of clones normalized for estimated rRNA copy number.
Number of 16S rRNA genes based on numbers reported for the same genus or closest relatives in reference 1 and in the online databases described in references 27 and 39 (see text).
Value for Azoarcus, the closest relative whose 16S rRNA copy number is known.
Value based on the range of rrn copy numbers for the Proteobacteria found in this study.
Table 5 shows the actual number of clones observed for each OTU for the 107 clones analyzed. However, due to potential PCR bias (5, 38, 41), cloning bias, and different numbers of rRNA gene copies for different bacteria (1, 27, 39), these numbers do not provide an accurate representation of the population distribution. PCR bias and artifact formation occur at higher rates during the last few cycles (38); therefore, PCR amplification was carried out for only 25 cycles in an effort to minimize such bias. Also, in order to minimize the rRNA copy number bias, we used data for 16S rRNA gene copy numbers from the closest phylogenetic relatives of our 11 OTUs for which such data are available to calculate percent distributions normalized by rRNA copy number. (The sources of these data were reference 1 and the online databases Ribosomal RNA Operon Copy Number Database, release 2.5 [posted 1 February 2004; http://rrndb/cme.msu.edu/rrndb/servlet/controller] and Genome Atlas Database [updated 8 November 2005; http://www.cbs.dtu.dk/services/GenomeAtlas/], as described in references 27 and 39.) Table 5 also shows the rRNA copy numbers for the genera most closely related to our OTUs, as well as the normalized percent distributions of the various OTUs.
The normalized data suggest that the most abundant bacteria in the PCB-dechlorinating culture were members of the Bacteroidales (27 to 32%) and Thauera-like Betaproteobacteria (18 to 26%), followed by Dehalococcoides spp. (13 to 18%) and Pseudomonas spp. (11 to 13%).
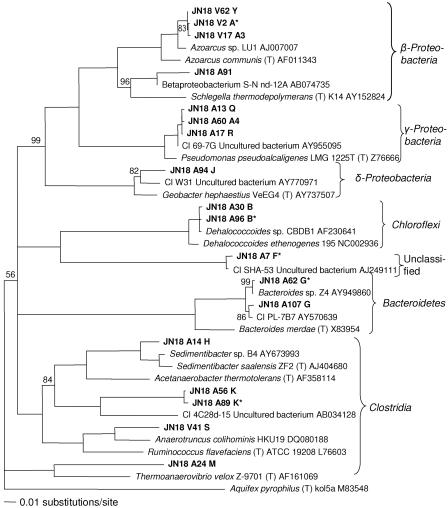
Figure 5 is a bootstrapped neighbor-joining tree showing the relationships of 18 sequences representing the 11 OTUs in the PCB-dechlorinating enrichment culture with their closest known type strains. The tree topologies were essentially identical for the neighbor-joining, maximum-parsimony, and maximum-likelihood trees.
FIG. 5.
Bootstrapped neighbor-joining tree of OTUs in sediment-free PCB-dechlorinating enrichment cultures and their closest relatives and type strains. The numbers at the nodes indicate the percentages of times that nodes appeared in 1,000 trials. Nodes that are not labeled appeared 100% of the time. Sequences cloned from DNA extracted from the enrichments after eight transfers without sediment are indicated by boldface type. JN18 refers to the experiment, and the additional letters and numbers indicate the clone name and RFLP designation where applicable. Type strains are indicated (T).
DISCUSSION
Establishment of sediment-free cultures that extensively dechlorinate Aroclor 1260.
Others have succeeded in sustaining PCB dechlorination activity in sediment-free media, but the reported activity of the cultures was restricted to a few PCB congeners that are chlorinated on only one ring and are not significant components of PCB mixtures found in the environment (31, 68). This is the first time that a sediment-free culture capable of degrading a complex Aroclor mixture has been developed. The cultures dechlorinate at least 64 PCB congeners, all of which are chlorinated on both rings and are present in Aroclor 1260 and 47 of which have six or more chlorines.
Our sediment-free enrichment cultures dechlorinated 76% of the hexa- through nonachlorobiphenyls in Aroclor 1260 (250 μg/ml) to tri- through pentachlorobiphenyls in 110 days at 22 to 24°C. These products are less toxic and less persistent in vertebrates (16). Nearly all congeners in Aroclor 1260 were substrates, including congeners with three or four ortho chlorines. The latter congeners exist as stable enantiomers, and we wondered whether their dechlorination would be limited. Stereoisomers of PCBs exist as racemic mixtures in commercial PCBs, and evidence for stereoselective dechlorination of some PCB congeners has been reported (52, 67). Our data show that there was well over 50% dechlorination of 236-236-CB, 236-245-CB, 2356-245-CB, and 2345-236-CB, demonstrating that both stereoisomers of each of these congeners are dechlorinated. However, since we did not separate the enantiomers, we do not know whether they were dechlorinated with the same efficiency. We thought that perhaps the presence of four ortho chlorines would sterically block access of dehalogenase enzymes, but the dechlorination of 236-236-CB shows that this clearly was not the case. This congener decreased from 1.56 mol% to 0.14 mol%, demonstrating that there was no steric hindrance or enantiomer selectivity.
Observed PCB dechlorination matches that seen in situ.
It is especially significant that the PCB dechlorination observed in our enrichment culture, Process N, is the same dechlorination process that occurs in situ in the Housatonic River (7, 10), the site from which the sediment used for inoculum was obtained. This means that we have retained all of the bacteria critical for Process N in our culture and are now positioned to begin to identify the particular bacteria and reductive dehalogenase gene(s) responsible for it. The breadth of the substrate specificity, 64 penta- through nonachlorobiphenyls and nine different chlorophenyl rings, indicates that the activity results from either multiple dechlorinators, multiple reductive dehalogenase genes in a single dechlorinator, or both. Raising the concentration of PCBs 10- to 100-fold increased the rate of dechlorination, eliminated the lag, and greatly improved the reproducibility of the transfers. This indicates that the dechlorinators are likely using the PCBs as terminal electron acceptors for halorespiration.
Process N dechlorination has also been observed in laboratory experiments with sediment slurries using microbial inocula from Silver Lake (Pittsfield, MA) (53), the Hudson River (New York) (9), and Baltimore Harbor (Maryland) (69); hence, the bacteria that carry out this process are apparently widespread in the environment.
Microbial community analysis with respect to halorespiration.
Even after normalization, Bacteroidales appeared to be the most abundant bacteria in our enrichment culture. No members of the Bacteroidales are known to be halorespirers, and the prominent presence of these bacteria in our culture may have more to do with the citrate used to chelate the Ti(III) reductant in our medium than with any role of these bacteria in dechlorination. Bacteroides spp. are known to carry out fermentation of citrate to formate, acetate, and bicarbonate (36; see Table 17.8 in reference 46). Clostridia have frequently been found in dechlorinating enrichments, and members of four clostridial families were present in our culture. These organisms are typically fermenters and may have been present due to the citrate and the yeast extract in our medium. Several clostridia are known to carry out halorespiration, although PCB dechlorination has not been demonstrated. The dechlorinating clostridia include Desulfitobacterium spp., Dehalobacter spp., and Clostridium bifermentans DPH1 (45, 57). The first two taxa belong to the family Peptococcaceae, which was not detected in our enrichment culture.
Beta-, Delta-, and Gammaproteobacteria were all present. No dehalogenating bacteria have been identified in the Beta- or Gammaproteobacteria, and the role of the Pseudomonas and Thauera-like bacteria in the enrichment culture is unclear. Diverse Azoarcus and Thauera spp. are known to degrade a wide variety of aromatic compounds using oxygen or nitrate as a terminal electron acceptor (49). There was probably still some biphenyl present in our culture from the dehalogenation of 26-BB in earlier transfers, but there was no nitrate in the medium. The prominent presence of Thauera-like bacteria in our enrichment culture suggests that they may have another means of anaerobic growth.
There are several halorespiring species in the Deltaproteobacteria. These include Desulfuromonas spp., Desulfomonile spp., Desulfovibrio dechloroacetovorans SF3, Anaeromyxobacter dehalogenans (45, 57), Geobacter lovleyi (60), and Trichlorobacter thiogenes (22), a very close relative of Geobacter (59). Clone JN18 A94 J in our enrichment culture is closely related to Geobacter; thus, we cannot rule out the possibility that it could be involved in the dechlorination.
We did not find any sequences that belong to the DF-1 or o-17 group of Chloroflexi (65). DF-1 and o-17 are distant relatives of Dehalococcoides (87 to 89% identity for 16S rRNA sequences) that are known to dechlorinate several individual PCB congeners (21, 70).
We cannot rule out the possibility that our culture contains previously unidentified dechlorinating bacteria. However, the most likely candidates for PCB dechlorinators in our enrichment culture are members of the genus Dehalococcoides. The two Dehalococcoides sequences that we found have 16S rRNA sequences that are nearly identical to those of Dehalococcoides spp. strains CBDB1, FL2, and BAV1, which belong to the Pinellas group of Dehalococcoides (30). Strain CBDB1 can use hexa- and pentachlorobenzenes as terminal electron acceptors (37) and can also dechlorinate 1,2,3,7,8-pentachloro-p-dibenzodioxin (19). Strain FL2 can use trichloroethene and cis-1,2-dichloroethene as electron acceptors, and strain BAV1 can use cis-1,2-dichloroethene and vinyl chloride as electron acceptors (28, 29). Our Dehalococcoides sequences are also closely related to the sequence of Dehalococcoides ethenogenes strain 195 (Fig. 5), which has been shown to dechlorinate 23456-CB to 2346-CB/2356-CB and 246-CB (23). Dehalococcoides strains 195, FL2, BAV1, and CBDB1 contain 18, 14, 7, and 32 nonidentical reductive dehalogenase genes (32, 40, 56). Together, these reductive dehalogenase genes comprise 42 different orthologous clusters. We found two different Dehalococcoides 16S rRNA gene sequences in our culture, and we expect that each of the corresponding strains also has multiple reductive dehalogenase genes. It is intriguing to speculate that two or more Dehalococcoides strains with different sets of reductive dehalogenase genes act together to achieve the broad PCB congener specificity characteristic of Process N dechlorination. However, definitive proof of this requires firm evidence that the dechlorination of Aroclor 1260 is linked to the growth of Dehalococcoides. Studies to determine if this is the case are in progress.
Conclusion.
For the first time, this research has identified and developed robust, highly enriched, sediment-free cultures of the bacteria that carry out Dechlorination Process N, a major microbial PCB dechlorination activity which occurs naturally in the Housatonic River. This is a major and unprecedented breakthrough in the field which will, for the first time, make it possible to conclusively identify the bacterial strains and reductive dehalogenase genes responsible for a major dechlorination process that actually occurs in the environment. The insights that we gain about these bacteria and their gene expression should also be useful in the development of effective remediation applications.
Acknowledgments
We thank Lorenz Adrian, Frank Löffler, and Steve Zinder for helpful suggestions concerning cultivation of these organisms and Shawn Freitas for technical assistance. We thank the members of the Ribosomal Database Project for helpful discussions and for aligning our sequences.
This research was supported by National Science Foundation grant 0077837 and by a grant from GE Corporate Environmental Programs.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adrian, L., U. Szewzyk, and H. Görisch. 2000. Bacterial growth based on reductive dechlorination of trichlorobenzenes. Biodegradation 11:73-81. [DOI] [PubMed] [Google Scholar]
- 3.Alder, A. C., M. M. Häggblom, S. R. Oppenheimer, and L. Y. Young. 1993. Reductive dechlorination of polychlorinated biphenyls in anaerobic sediments. Environ. Sci. Technol. 27:530-538. [Google Scholar]
- 4.Altschul, S. F., Y. Ma, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Becker, S., P. Böger, R. Oehlmann, and A. Ernst. 2000. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl. Environ. Microbiol. 66:4945-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedard, D. L., S. C. Bunnell, and L. A. Smullen. 1996. Stimulation of microbial para-dechlorination of polychlorinated biphenyls that have persisted in Housatonic River sediment for decades. Environ. Sci. Technol. 30:687-694. [Google Scholar]
- 7.Bedard, D. L., and R. J. May. 1996. Characterization of the polychlorinated biphenyls in the sediments of Woods Pond: evidence for microbial dechlorination of Aroclor 1260 in situ. Environ. Sci. Technol. 30:237-245. [Google Scholar]
- 8.Bedard, D. L., E. A. Pohl, J. J. Bailey, and A. Murphy. 2005. Characterization of the PCB substrate range of microbial Dechlorination Process LP. Environ. Sci. Technol. 39:6831-6839. [DOI] [PubMed] [Google Scholar]
- 9.Bedard, D. L., and J. F. Quensen III. 1995. Microbial reductive dechlorination of polychlorinated biphenyls, p. 127-216. In L. Y. Young and C. E. Cerniglia (ed.), Microbial transformation and degradation of toxic organic chemicals. Wiley-Liss, New York, N.Y.
- 10.Bedard, D. L., L. A. Smullen, and R. J. May. 1996. Microbial dechlorination of highly chlorinated PCBs in the Housatonic River, p. 117. In Proceedings of the 1996 International Symposium on Subsurface Microbiology. Swiss Society of Microbiology and Institute of Plant Biology, University of Zurich, Davos, Switzerland.
- 11.Bedard, D. L., H. M. Van Dort, and K. A. DeWeerd. 1998. Brominated biphenyls prime extensive microbial reductive dehalogenation of Aroclor 1260 in Housatonic River sediment. Appl. Environ. Microbiol. 64:1786-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedard, D. L., and H. M. Van Dort. 1998. Complete reductive dehalogenation of brominated biphenyls by anaerobic microorganisms in sediment. Appl. Environ. Microbiol. 64:940-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedard, D. L., H. M. Van Dort, R. J. May, and L. A. Smullen. 1997. Enrichment of microorganisms that sequentially meta, para-dechlorinate the residue of Aroclor 1260 in Housatonic River sediment. Environ. Sci. Technol. 31:3308-3313. [Google Scholar]
- 14.Bedard, D. L., R. E. Wagner, M. J. Brennan, M. L. Haberl, and J. F. Brown, Jr. 1987. Extensive degradation of Aroclors and environmentally transformed polychlorinated biphenyls by Alcaligenes eutrophus H850. Appl. Environ. Microbiol. 53:1094-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasland, Bouck, & Lee, Inc. 2003. Housatonic River—rest of river RCRA facility investigation report. U.S. Environmental Protection Agency, Washington, D.C. [Online.] http://www.epa.gov/region1/ge/thesite/restofriver/reports/rcra_fir/200656.pdf.
- 16.Brown, J. F., Jr. 1994. Determination of PCB metabolic, excretion, and accumulation rates for use as indicators of biological response and relative risk. Environ. Sci. Technol. 28:2295-2305. [DOI] [PubMed] [Google Scholar]
- 17.Brown, J. F., Jr., D. L. Bedard, M. J. Brennan, J. C. Carnahan, H. Feng, and R. E. Wagner. 1987. Polychlorinated biphenyl dechlorination in aquatic sediments. Science 236:709-711. [DOI] [PubMed] [Google Scholar]
- 18.Brown, J. F., Jr., and R. E. Wagner. 1990. PCB movement, dechlorination, and detoxication in the Acushnet Estuary. Environ. Toxicol. Chem. 9:1215-1233. [Google Scholar]
- 19.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 20.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutter, L. A., J. E. M. Watts, K. R. Sowers, and H. D. May. 2001. Identification of a microorganism that links its growth to the reductive dechlorination of 2,3,5,6-chlorobiphenyl. Environ. Microbiol. 3:699-709. [DOI] [PubMed] [Google Scholar]
- 22.De Wever, H., J. R. Cole, M. R. Fettig, D. A. Hogan, and J. M. Tiedje. 2000. Reductive dehalogenation of trichloroacetic acid by Trichlorobacter thiogenes gen. nov., sp. nov. Appl. Environ. Microbiol. 66:2297-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fennell, D. E., I. Nijenhuis, S. F. Wilson, S. H. Zinder, and M. M. Häggblom. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38:2075-2081. [DOI] [PubMed] [Google Scholar]
- 24.Fish, K. M., and J. M. Principe. 1994. Biotransformations of Aroclor 1242 in Hudson River test tube microcosms. Appl. Environ. Microbiol. 60:4289-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frame, G. M., R. E. Wagner, J. C. Carnahan, J. F. Brown, Jr., R. J. May, L. A. Smullen, and D. L. Bedard. 1996. Comprehensive, quantitative, congener-specific analyses of eight Aroclors and complete PCB congener assignments on DB-1 capillary GC columns. Chemosphere 33:603-623. [Google Scholar]
- 26.Garrity, G. M. 2001. Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, N.Y.
- 27.Hallin, P. F., and D. Ussery. 2004. CBS Genome Atlas Database: a dynamic storage for bioinformatic results and sequence data. Bioinformatics 20:3682-3686. [DOI] [PubMed] [Google Scholar]
- 28.He, J., Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti, and F. E. Löffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 29.He, J. Z., K. M. Ritalahti, K. L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 30.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holoman, T. R., M. A. Elberson, L. A. Cutter, H. D. May, and K. R. Sowers. 1998. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl. Environ. Microbiol. 64:3359-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. von Wintzingerode, H. Görisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-rehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutzinger, O., S. Safe, and V. Zitco. 1974. The chemistry of PCBs. CRC Press, Inc., Cleveland, Ohio.
- 34.Imamoglu, I., K. Li, and E. R. Christensen. 2002. Modeling polychlorinated biphenyl congener patterns and dechlorination in dated sediments from the Ashtabula River, Ohio, USA. Environ. Toxicol. Chem. 21:2283-2291. [PubMed] [Google Scholar]
- 35.Imamoglu, I., K. Li, E. R. Christensen, and J. K. McMullin. 2004. Sources and dechlorination of polychlorinated biphenyl congeners in the sediments of Fox River, Wisconsin. Environ. Sci. Technol. 38:2574-2583. [DOI] [PubMed] [Google Scholar]
- 36.Janssen, P. H. 1991. Fermentation of l-tartrate by a newly isolated Gram-negative glycolytic bacterium. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 59:191-198. [DOI] [PubMed] [Google Scholar]
- 37.Jayachandran, G., H. Görisch, and L. Adrian. 2003. Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 180:411-416. [DOI] [PubMed] [Google Scholar]
- 38.Kanagawa, T. 2003. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96:317-323. [DOI] [PubMed] [Google Scholar]
- 39.Klappenbach, J. A., P. R. Saxman, J. T. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kube, M., A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian. 2005. Genome sequence of the chlorinated compound respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269-1273. [DOI] [PubMed] [Google Scholar]
- 41.Kurata, S., T. Kanagawa, Y. Magariyama, K. Takatsu, K. Yamada, T. Yokomaku, and Y. Kamagata. 2004. Reevaluation and reduction of a PCR bias caused by reannealing of templates. Appl. Environ. Microbiol. 70:7545-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 43.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1991. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, and C. R. Woese. 1993. The Ribosomal Database Project. Nucleic Acids Res. 21:3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Löffler, F. E., J. R. Cole, K. M. Ritalahti, and J. M. Tiedje. 2003. Diversity of dechlorinating bacteria, p. 53-87. In M. M. Häggblom and I. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Press, Boston, Mass.
- 46.Madigan, M. T., and J. M. Martinko. 2006. Brock biology of microorganisms, 11th ed. Pearson Prentice Hall, Upper Saddle River, N.J.
- 47.Magar, V. S., R. C. Brenner, G. W. Johnson, and J. F. Quensen III. 2005. Long-term recovery of PCB-contaminated sediments at the Lake Hartwell superfund site: PCB dechlorination. 2. Rates and extent. Environ. Sci. Technol. 39:3548-3554. [DOI] [PubMed] [Google Scholar]
- 48.Magar, V. S., G. W. Johnson, R. C. Brenner, J. F. Quensen III, E. A. Foote, G. Durell, J. A. Ickes, and C. Peven-McCarthy. 2005. Long-term recovery of PCB-contaminated sediments at the Lake Hartwell superfund site: PCB dechlorination. 1. End-member characterization. Environ. Sci. Technol. 39:3538-3547. [DOI] [PubMed] [Google Scholar]
- 49.Mechichi, T., E. Stackebrandt, N. Gad'on, and G. Fuchs. 2002. Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch. Microbiol. 178:26-35. [DOI] [PubMed] [Google Scholar]
- 50.Mousa, M. A., P. E. Ganey, J. F. Quensen III, B. V. Madhukar, K. Chou, J. P. Giesy, L. J. Fischer, and S. A. Boyd. 1998. Altered biologic activities of commercial polychlorinated biphenyl mixtures after microbial reductive dechlorination. Environ. Health Perspect. 106(Suppl. 6):1409-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mousa, M. A., J. F. Quensen III, K. Chou, and S. A. Boyd. 1996. Microbial dechlorination alleviates inhibitory effects of PCBs on mouse gamete fertilization in vitro. Environ. Sci. Technol. 30:2087-2092. [Google Scholar]
- 52.Pakdeesusuk, U., W. J. Jones, C. M. Lee, A. W. Garrison, W. L. O'Niell, D. L. Freedman, J. T. Coates, and C. S. Wong. 2003. Changes in enantiomeric fractions during microbial reductive dechlorination of PCB132, PCB149, and Aroclor 1254 in Lake Hartwell sediment microcosms. Environ. Sci. Technol. 37:1100-1107. [DOI] [PubMed] [Google Scholar]
- 53.Quensen, J. F., III, S. A. Boyd, and J. M. Tiedje. 1990. Dechlorination of four commercial polychlorinated biphenyl mixtures (Aroclors) by anaerobic microorganisms from sediments. Appl. Environ. Microbiol. 56:2360-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quensen, J. F., III, J. M. Tiedje, and S. A. Boyd. 1988. Reductive dechlorination of polychlorinated biphenyls by anaerobic microorganisms from sediments. Science 242:752-754. [DOI] [PubMed] [Google Scholar]
- 55.Quensen, J. F., III, M. Mousa, S. A. Boyd, J. T. Sanderson, K. L. Froese, and J. P. Giesy. 1998. Reduction of Ah receptor mediated activity of PCB mixtures due to anaerobic dechlorination. Environ. Toxicol. Chem. 17:806-813. [Google Scholar]
- 56.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. T. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 57.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 58.Smullen, L. A., K. A. DeWeerd, D. L. Bedard, W. A. Fessler, J. C. Carnahan, and R. E. Wagner. 1993. Development of a customized congener specific PCB standard for quantification of Woods Pond sediment PCBs, p. 45-59. In Research and development program for the destruction of PCBs: twelfth progress report. General Electric Company, Corporate Research and Development, Schenectady, N.Y.
- 59.Snoeyenbos-West, O., C. G. Van Praagh, and D. R. Lovley. 2001. Trichlorobacter thiogenes should be renamed as a Geobacter species. Appl. Environ. Microbiol. 67:1020-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sung, Y., K. E. Fletcher, K. M. Ritalahti, N. Ramos-Hernández, R. A. Sanford, N. H. Mesbah, and F. E. Löffler. 2006. Geobacter lovleyi sp. nov. strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Appl. Environ. Microbiol. 72:2775-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.United States Environmental Protection Agency. 1980. Ambient water quality criteria for PCBs. EPA-440/5-80-068. U.S. Environmental Protection Agency Criteria and Standards Division, Washington, D.C.
- 62.United States Environmental Protection Agency. 9 November 2005, posting date. Health effects of PCBs. U.S. Environmental Protection Agency, Washington, D.C. [Online.]. http://www.epa.gov/pcb/effects.html.
- 63.United States Environmental Protection Agency. 1986. EPA method 3540. Soxhlet extraction, chapter 4, section 4.2.1. In Test methods for evaluating solid waste: physical/chemical methods, 3rd ed., vol. 1B. Environmental Protection Agency, Washington, D.C.
- 64.Van Dort, H. M., L. A. Smullen, R. J. May, and D. L. Bedard. 1997. Priming microbial meta-dechlorination of polychlorinated biphenyls that have persisted in Housatonic River sediments for decades. Environ. Sci. Technol. 31:3300-3307. [Google Scholar]
- 65.Watts, J. E. M., S. K. Fagervold, H. D. May, and K. R. Sowers. 2005. A PCR-based specific assay reveals a population of bacteria within the Chloroflexi associated with the reductive dehalogenation of polychlorinated biphenyls. Microbiology 151:2039-2046. [DOI] [PubMed] [Google Scholar]
- 66.Wiegel, J., and Q. Wu. 2000. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol. Ecol. 32:1-15. [DOI] [PubMed] [Google Scholar]
- 67.Wong, C. S., A. W. Garrison, and W. T. Foreman. 2001. Enantiomeric composition of chiral polychlorinated biphenyl atropisomers in aquatic bed sediment. Environ. Sci. Technol. 35:33-39. [DOI] [PubMed] [Google Scholar]
- 68.Wu, Q., K. R. Sowers, and H. D. May. 2000. Establishment of a polychlorinated biphenyl-dechlorinating microbial consortium, specific for doubly flanked chlorines, in a defined, sediment-free medium. Appl. Environ. Microbiol. 66:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, Q., K. R. Sowers, and H. D. May. 1998. Microbial reductive dechlorination of Aroclor 1260 in anaerobic slurries of estuarine sediments. Appl. Environ. Microbiol. 64:1052-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, Q., J. E. M. Watts, K. R. Sowers, and H. D. May. 2002. Identification of a bacterium that specifically catalyzes the reductive dechlorination of polychlorinated biphenyls with doubly flanked chlorines. Appl. Environ. Microbiol. 68:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]