Abstract
The developing intestinal microbiota of breast-fed infants is considered to play an important role in the priming of the infants' mucosal and systemic immunity. Generally, Bifidobacterium and Lactobacillus predominate the microbiota of breast-fed infants. In intervention trials it has been shown that lactobacilli can exert beneficial effects on, for example, diarrhea and atopy. However, the Lactobacillus species distribution in breast-fed or formula-fed infants has not yet been determined in great detail. For accurate enumeration of different lactobacilli, duplex 5′ nuclease assays, targeted on rRNA intergenic spacer regions, were developed for Lactobacillus acidophilus, Lactobacillus casei, Lactobacillus delbrueckii, Lactobacillus fermentum, Lactobacillus paracasei, Lactobacillus plantarum, Lactobacillus reuteri, and Lactobacillus rhamnosus. The designed and validated assays were used to determine the amounts of different Lactobacillus species in fecal samples of infants receiving a standard formula (SF) or a standard formula supplemented with galacto- and fructo-oligosaccharides in a 9:1 ratio (OSF). A breast-fed group (BF) was studied in parallel as a reference. During the 6-week intervention period a significant increase was shown in total percentage of fecal lactobacilli in the BF group (0.8% ± 0.3% versus 4.1% ± 1.5%) and the OSF group (0.8% ± 0.3% versus 4.4% ± 1.4%). The Lactobacillus species distribution in the OSF group was comparable to breast-fed infants, with relatively high levels of L. acidophilus, L. paracasei, and L. casei. The SF-fed infants, on the other hand, contained more L. delbrueckii and less L. paracasei compared to breast-fed infants and OSF-fed infants. An infant milk formula containing a specific mixture of prebiotics is able to induce a microbiota that closely resembles the microbiota of BF infants.
The intestinal microbiota composition is regarded as an important factor for infant health and well-being (15, 32). A lower incidence of gastrointestinal and other infections has been found in breast-fed infants (43), which partly may be related to their microbiota composition. The intestinal microbiota of breast-fed infants is generally dominated by the genera Bifidobacterium and Lactobacillus (35), which are able to inhibit the growth of pathogens by lowering the pH, due to the production of lactic and acetic acid (1), or by competing for nutrients and epithelial adhesion sites (2). In contrast to breast-fed infants, formula-fed infants possess a more diverse microbiota which is mainly composed of Bacteroides, Bifidobacterium, Staphylococcus, Escherichia coli, and Clostridium spp. (19).
Several concepts are being used to modify the intestinal microbiota, such as nutritional changes or the consumption of pro- and/or prebiotics (10). Prebiotics are defined as nondigestible food ingredients that selectively stimulate the growth and/or activity of one or more bacteria in the colon and thereby beneficially affect the host (14). For infant formulas, a specific prebiotic mixture of galacto-oligosaccharides (GOS) and fructo-oligosaccharides (FOS) has been described that can stimulate the growth of bifidobacteria and lactobacilli similar to milk oligosaccharides in human breast milk (6, 8, 42). Several reports showed that the supplementation of infant formulas with this specific mixture of GOS and FOS increases the numbers of Bifidobacterium (7, 21, 36) and the total numbers of Lactobacillus (28), reduces the numbers of pathogens (20), and induces a short-chain fatty acid profile similar to that found in breast-fed infants (4, 21). Addition of the specific prebiotic mixture of GOS and FOS also results in a distribution of the different Bifidobacterium species similar to that found in breast-fed infants (16).
Although the supplementation of specific Lactobacillus strains, such as Lactobacillus rhamnosus, Lactobacillus reuteri, Lactobacillus acidophilus, and Lactobacillus fermentum, to infant formulas has been reported (2), the distribution of the different Lactobacillus species in breast-fed or formula-fed infants has not been studied in detail. To determine the composition of the different Lactobacillus species in breast-fed and formula-fed infants and to study the effects of nutritional interventions, it is relevant to quantitatively determine lactobacilli at the species level. For this purpose, species-specific duplex 5′ nuclease assays (quantitative real-time PCR) were developed for Lactobacillus acidophilus, Lactobacillus casei, Lactobacillus delbrueckii, Lactobacillus fermentum, Lactobacillus paracasei, Lactobacillus plantarum, Lactobacillus reuteri, and Lactobacillus rhamnosus. With these assays the different Lactobacillus species were quantified in breast-fed infants (BF) and infants receiving a standard formula (SF) or a standard formula supplemented with the specific prebiotic GOS-FOS mixture (OSF).
MATERIALS AND METHODS
Study design and sample collection.
Fecal samples were collected from an intervention trial with exclusively formula-fed infants, aged 28 to 90 days, receiving a standard formula (SF group; age, 60.3 ± 6.9 days [mean ± the standard error of the mean], ranging from 29.0 to 85.0 days) or a prebiotic formula containing 0.8 g of GOS-FOS/100 ml in a 9 to 1 ratio (OSF group; age, 51.9 ± 7.2 days, ranging from 30.0 to 86.0 days). A group of exclusively breast-fed infants was studied in parallel and used as a reference (BF group; age: 56.7 ± 7.4 days, ranging from 27.0 to 88.0 days). For study details, see also references 16 and 21.
Bacterial strains and culture conditions.
All bacterial strains used in the present study are listed in Table 1. All Bifidobacterium, Lactobacillus, Propionibacterium, Saccharomyces, Enterococcus, and Pediococcus strains were cultured in Mann Rogosa Sharp broth (Oxoid, Basingstoke, United Kingdom) at 37°C under anaerobic conditions.
TABLE 1.
Bacterial strains used in this study
| Lactobacillus strains | Origina | Other strains | Origina | |
|---|---|---|---|---|
| Lactobacillus acidophilus | ATCC 4356 | Bacillus cereus | ATCC 11778 | |
| ATCC 521 | Bacteroides fragilis | LMG 10263T | ||
| DSM 20079T | Brevibacterium casei | ATCC 35513T | ||
| Lactobacillus amylovorus | DSM 20552 | Clostridium difficile | ATCC 9689T | |
| Lactobacillus bavaricus | JCM 1128 | Enterococcus faecalis | DSM 20478T | |
| Lactobacillus brevis | LMG 18022 | Escherichia coli | ATCC 35218 | |
| Lactobacillus bulgaricus | ATCC 11842T | Listeria monocytogenes | ATCC 7644 | |
| Lactobacillus casei | ATCC 393T | Pediococcus acidilactici | DSM 20284T | |
| DSM 20011T | Proplonibacterium avidum | DSM 4901 | ||
| Lactobacillus crispatus | ATCC 33820T | Pseudomonas aeruginosa | DSM 1117 | |
| Lactobacillus curvatus | ATCC 51436 | Saccharomyces cerevisiae | DSM 2548 | |
| Lactobacillus delbrueckii | JCM 1106 | Salmonella enterica serovar Typhimurium | ATCC 14028 | |
| JCM 1248T | Staphylococcus aureus | ATCC 29213 | ||
| Lactobacillus fermentum | DSM 20052T | Bifidobacterium adolescentis | ATCC 15703T | |
| Lactobacillus gasseri | LMG 11496T | Bifidobacterium angulatum | DSM 20098T | |
| Lactobacillus helveticus | CNRZ 3 | Bifidobacterium bifidum | DSM 20456T | |
| Lactobacillus johnsonii | ATCC 33200T | Bifidobacterium animalis | ATCC 25527T | |
| Lactobacillus kefir | LMG 11496 | Bifidobacterium gallicum | DSM 20093T | |
| Lactobacillus kefirgranum | DSM 10550T | Bifidobacterium dentium | ATCC 27534T | |
| Lactobacillus paracasei | ATCC 11582 | Bifidobacterium breve | ATCC 15700T | |
| ATCC 27216 | Bifidobacterium catenulatum | ATCC 27539T | ||
| Lactobacillus pentosus | JCM 8334 | Bifidobacterium infantis | LMG 8811T | |
| JCM8338 | Bifidobacterium longum | ATCC 15707T | ||
| Lactobacillus plantarum | DSM 20174T | |||
| NCIMB 8826 | ||||
| Lactobacillus reuteri | LMG 9213T | |||
| Lactobacillus rhamnosus | ATCC 53103 | |||
| ATCC 7469T | ||||
| Lactobacillus sake | DSM 6333 | |||
| Lactobacillus salivarius | ATCC 11741T |
ATCC, American Type Culture Collection, United States; CNRZ, Centre National de Recherches Zootechniques, France; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Germany; JCM, Japan Collection of Microorganisms, Japan; LMG, Laboratory for Microbiology, University of Gent, Belgium; NCIMB, National Collections of Industrial and Marine Bacteria, United Kingdom.
Gut commensals and pathogens, such as Bacteroides fragilis and Pseudomonas aeruginosa, were cultured in brain heart infusion broth (Oxoid, Basingstoke, United Kingdom) at 37°C, and Bacillus cereus, Brevibacterium casei, and Listeria monocytogenes were cultured at 30°C. Overnight cultures were stored at −20°C until further processing.
Qualitative PCR analysis.
For the species-specific qualitative PCR, DNA was isolated as described previously (16) earlier. PCRs were carried out as described previously (37, 40, 41) by using a PTC-200 Peltier Thermal Cycler (Biozym, Landgraaf, The Netherlands). Amplification products were checked by agarose gel electrophoresis and ethidium bromide staining.
Species-specific quantitative real-time PCR.
For the selection of primer and probe sequences, the 16S-23S intergenic spacer regions of the different Lactobacillus species were retrieved from the GenBank, EMBL, and DDBJ databases as follows: L. acidophilus (AB102855 [25], AF182726 [37], and U32971 [39]), L. alimentarius (AF500493 [33] and AF500492 [33]), L. amylovorus (AF182732 [37]), L. animalis (AY526616 and AY526614), L. brevis (AB102858 [25] and AF405353 [11]), L. bulgaricus (Z75475), L. casei (AB102854 [25], AF405352 [11], AF182729 [37], and AF121200 [38]), L. collinoides (AB117957 and AB117955), L. crispatus (AF182719 [37] and AF074857 [38]), L. curvatus (AF074858 [38], U97135 [5], and U97129 [5]), L. delbrueckii (]AB102856 [25], AB035485 [37], AB035484 [37], U32969 [39], U32968 [39], and U32967 [39]), L. farciminis (AF500491 [33] and AF500490 [33]), L. fermentum (AF182720 [37]), L. frumenti (AJ616011), L. gasseri (AB102860 [25], AF182721 [37], and AF074859 [38]), L. graminis (U97136 [5] and U97130 [5]), L. hamsteri (AF113601), L. helveticus (AF182728 [37]), L. jensenii (AB035486 [37] and U32970 [39]), L. johnsonii (AF074860 [38]), L. mindensis (AJ616016), L. panis (AJ616012), L. paracasei (AB035487 [37], AF182724 [37], and U32964 [39]), L. paralimentarius (AJ616014), L. paraplantarum (U97138 [5] and U97132 [5]), L. pentosus (U97141 [5], U97140 [5], and U97134 [5]), L. plantarum (AB102857 [25], AF405354 [11], AF182722 [37], U97139 [5], and U97133 [5]), L. sakei (U97137 [5] and U97131 [5]), L. salivarius (AB102859 [25], AB03488 [37], and AF182725 [37]), L. sharpeae (AF074861 [38]), L. reuteri (AF182723 [37]), L. rhamnosus (AF182730 [37], AF121201 [38], and U32966 [39]), L. ruminis (AF080103), L. vaginalis (AF182731), and L. zeae (AF074862). Sequences were aligned and the conserved regions were determined by using DNASIS for Windows V2.5 (Hitachi Software Engineering Co., Ltd., Wembley, United Kingdom). Using Primer Express 1.5a (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands), specific sequences were identified to design primers and probes for, respectively, all lactobacilli and the species: L. acidophilus, L. casei, L. delbrueckii, L. fermentum, L. paracasei, L. plantarum, L. rhamnosus, and L. reuteri. All primers and probes were tested for specificity using the basic local alignment search tool (BLAST) (3) and fulfilled the criteria described previously (16).
The probe for the detection of the genus Lactobacillus is labeled with the 5′ reporter dye VIC and the 3′ quencher NFQ-MGB (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). The different lactobacilli species probes are labeled with the 5′ reporter dye 6-carboxy-fluorescein (FAM) and the 3′ quencher NFQ-MGB (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). To even further increase specificity and sensitivity, TaqMan minor groove binding probes were used (22).
For determination of the total bacterial load, an already-described probe and primer set was used (30). This universal oligonucleotide probe is labeled with the 5′ reporter dye FAM and the 3′ quencher dye 6-carboxy-tetramethyl-rhodamine (TAMRA).
The 5′ nuclease assays were performed as described earlier (16). Sequences of primers and probes are listed in Table 2. The optimized concentrations of the primers and probes are presented in Table 3.
TABLE 2.
Primers and probes used in the duplex 5′ nuclease assays
| Target | Primers and probes | Sequence (5′→3′) | Tm (°C) | % GC | BLAST ID number | Amplicon length (bp) |
|---|---|---|---|---|---|---|
| L. acidophilus | F_acid_IS | GAA AGA GCC CAA ACC AAG TGA TT | 59 | 43 | 1089017502-26171-202965955840 | 85 |
| R_acid_IS | CTT CCC AGA TAA TTC AAC TAT CGC TTA | 59 | 37 | 1089017571-27139-52545094772 | ||
| P_acid_IS | TAC CAC TTT GCA GTC CTA CA | 70 | 45 | 1089017717-29310-154296055415 | ||
| L. casei | F_case_IS | CTA TAA GTA AGC TTT GAT CCG GAG ATT T | 59 | 36 | 1037022798-023495-2136 | 132 |
| R_case_IS | CTT CCT GCG GGT ACT GAG ATG T | 59 | 55 | 1037022917-024843-29627 | ||
| P_case_IS | ACA AGC TAT GAA TTC ACT TGC | 70 | 38 | 1037022752-023005-20772 | ||
| L. delbrueckii | F_delb_IS | CAC TTG TAC GTT GAA AAC TGA ATA TCT TAAa | 58 | 30 | 1089018504-4206-64529811906 | 94 |
| R_delb_IS | CGA ACT CTC TCG GTC GCT TT | 58 | 55 | 1089018475-6841-166657768151 | ||
| P_delb_IS | CCG AGA ATC ATT GAG ATC | 68 | 44 | 1089018437-6309-163988227498 | ||
| L. fermentum | F_ferm_IS | AAC CGA GAA CAC CGC GTT AT | 58 | 50 | 1036676682-09669-23287 | 88 |
| R_ferm_IS | ACT TAA CCT TAC TGA TCG TAG ATC AGT CA | 58 | 38 | 1036676709-010209-2351 | ||
| P_ferm_IS | TAA TCG CAT ACT CAA CTA A | 68 | 32 | 1036676736-010547-20717 | ||
| L. paracasei | F_paca_IS | ACA TCA GTG TAT TGC TTG TCA GTG AAT AC | 60 | 38 | 1038306417-016220-23561 | 80 |
| R_paca_IS | CCT GCG GGT ACT GAG ATG TTT C | 60 | 55 | 1038306445-016796-3050 | ||
| P_paca_IS | TGC CGC CGG CCA G | 70 | 85 | 1038306524-018375-2626 | ||
| L. plantarum | F_plan_IS | TGG ATC ACC TCC TTT CTA AGG AAT | 58 | 42 | 1038305707-03107-18756 | 144 |
| R_plan_IS | TGT TCT CGG TTT CAT TAT GAA AAA ATAa | 58 | 26 | 1038305742-04177-12861 | ||
| P_plan_IS | ACA TTC TTC GAA ACT TTG T | 68 | 32 | 1038305778-04682-12880 | ||
| L. reuteri | F_reut_IS | ACC GAG AAC ACC GCG TTA TTT | 59 | 48 | 1089025339-29395-129280047216 | 93 |
| R_reut_IS | CAT AAC TTA ACC TAA ACA ATC AAA GAT TGT CT | 59 | 28 | 1089025385-30347-37558232754 | ||
| P_reut_IS | ATC GCT AAC TCA ATT AAT | 69 | 28 | 1089025413-30287-26112845854 | ||
| L. rhamnosus | F_rham_IS | CGG CTG GAT CAC CTC CTT T | 59 | 58 | 1023708254-09591-2284 | 97 |
| R_rham_IS | GCT TGA GGG TAA TCC CCT CAA | 59 | 52 | 1023708352-010389-16127 | ||
| P_rham_IS | CCT GCA CAC ACG AAA | 69 | 55 | 1023708453-011313-6655 | ||
| Lactobacillus spp. | F_alllact_IS | TGG ATG CCT TGG CAC TAG GA | 58 | 55 | 1024485925-024664-30598 | 92 |
| R_alllact_IS | AAA TCT CCG GAT CAA AGC TTA CTT AT | 58 | 35 | 1024478788-024701-16287 | ||
| P_alllact_IS | TAT TAG TTC CGT CCT TCA TC | 68 | 40 | 1024478009-017753-28422 | ||
| All bacteria | F_eub | TCC TAC GGG AGG CAG CAG T | 59 | -b | 466a | |
| R_eub | GGA CTA CCA GGG TAT CTA ATC CTG TT | 58 | ||||
| P_eub | CGT ATT ACC GCG GCT GCT GGC AC | 70 |
Concessions to the probe and primer design had to be made in these cases (more than three consecutive nucleotides are the same or an amplicon length greater than 150 bp).
Nadkami et al.
TABLE 3.
Optimized primer and probe concentrations for the duplex 5′ nuclease assays
| Target | 5′ Nuclease assay | Concn (nM)
|
||
|---|---|---|---|---|
| Forward primer | Reverse primer | Probe | ||
| L. acidophilus | L. acidophilus | 900 | 900 | 200 |
| All lactobacilli | 900 | 900 | 200 | |
| L. casei | L. casei | 900 | 900 | 200 |
| All lactobacilli | 300 | 300 | 50 | |
| L. delbrueckii | L. delbrueckii | 300 | 300 | 100 |
| All lactobacilli | 900 | 900 | 200 | |
| L. fermentum | L. fermentum | 300 | 300 | 100 |
| All lactobacilli | 300 | 300 | 100 | |
| L. paracasei | L. paracasei | 300 | 300 | 100 |
| All lactobacilli | 300 | 300 | 100 | |
| L. plantarum | L. plantarum | 300 | 300 | 100 |
| All lactobacilli | 300 | 300 | 100 | |
| L. reuteri | L. reuteri | 300 | 300 | 100 |
| All lactobacilli | 900 | 900 | 200 | |
| L. rhamnosus | L. rhamnosus | 900 | 450 | 200 |
| All lactobacilli | 150 | 100 | 100 | |
| Genus Lactobacillus | All lactobacilli | 600 | 600 | 100 |
| All bacteria | 300 | 300 | 100 | |
The relative amounts of the different Lactobacillus species in fecal samples were calculated after correction for differences in the amplification efficiencies of the duplex PCR as described previously (16, 24). The total counts of bacteria (cells per gram of feces) were determined by automated counting of microscopic images of fluorescently labeled cells. These counts, in combination with the percentages as determined with the duplex 5′ nuclease assays, were subsequently used to determine the numbers of lactobacilli per gram (wet weight) of feces (16).
The sensitivity of these duplex 5′ nuclease assays was compared to “conventional” PCR by testing dilution series of specific monocultures with both techniques. To determine the detection limit of the assay in CFU per milliliter, monocultures were also plated on Mann Rogosa Sharp agar and incubated under anaerobic conditions for 24 h at 37°C. The specificity of the assays was tested with the bacterial strains listed in Table 1.
The coefficients of variation (CV) within each duplex 5′ nuclease assay were determined by testing DNA isolated from feces spiked with a monoculture. This was performed 10 times for determination of the reproducibility and three times in quadruplicate for repeatability.
Data analyses.
For statistical analysis, the software package SPSS for Windows (version 12.0.1; SPSS, Inc., Chicago, Ill.) was used. All values were checked for normality by visual inspection of the normal probability plots. Differences were tested with paired sample t tests, and if P was <0.05 the difference was considered statistically significant. Although the breast-fed group is compared to the formula groups, it has to be kept in mind that no complete randomization was obtained because it is not possible to double blindly assign infants to a breast-fed group.
RESULTS
Species-specific quantitative real-time PCR.
The 5′ nuclease assay for detection of the genus Lactobacillus detected all Lactobacillus species tested, but no other closely related genera such as Enterococcus or Propionibacterium. The duplex 5′ nuclease assays for the detection of the different Lactobacillus species were specific as tested with the other (lactobacilli) strains.
Overall, the 5′nuclease assays were more sensitive than the conventional PCR assays (1,000- to 10,000-fold) and, by comparing conventional plating techniques with the duplex 5′ nuclease assays, the detection limits of the nuclease assays were found to range from 0.75 to 1.25 CFU/ml (Table 4). RNase-free and RNase-treated samples showed identical results demonstrating that contaminating RNA does not disturb the assays.
TABLE 4.
Detection limits and CV for reproducibility and repeatability of the duplex 5′ nuclease assays
| Target organism | Detection limit (CFU/ml) | CV
|
|
|---|---|---|---|
| Reproducibility | Repeatability | ||
| L. acidophilus | 0.75 | 0.11 | 0.13 |
| L. casei | 1.00 | 0.058 | 0.063 |
| L. delbrueckii | 1.15 | 0.059 | 0.035 |
| L. fermentum | 1.25 | 0.067 | 0.048 |
| L. paracasei | 1.25 | 0.052 | 0.11 |
| L. plantarum | 1.10 | 0.13 | 0.14 |
| L. reuteri | 1.00 | 0.062 | 0.11 |
| L. rhamnosus | 0.75 | 0.047 | 0.053 |
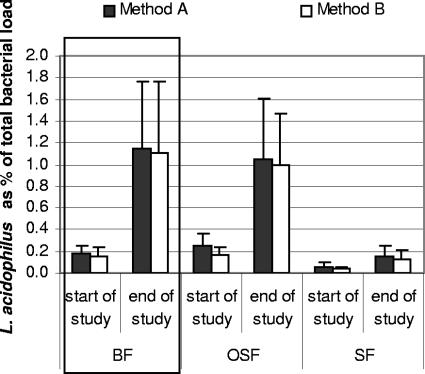
L. acidophilus as a percentage of the total bacterial load was determined directly, but also by combining the data for L. acidophilus as a percentage of the lactobacilli with the Lactobacillus data indicated as a percentage of the total bacterial load. There were no statistically significant differences between results obtained with the two methods (Fig. 1).
FIG. 1.
Comparison of two methods to determine L. acidophilus as percentage of total bacterial load in breast-fed infants (BF) and infants receiving a standard formula supplemented with GOS-FOS (OSF) or a standard formula (SF). Bars represent the standard error. Method A shows a combination of the data of L. acidophilus as a percentage of the total lactobacilli and the genus Lactobacillus as a percentage of the total bacterial load. Method B shows L. acidophilus as a percentage of the total bacterial load.
The CV values for reproducibility and repeatability of the different assays ranged between 0.04 and 0.14 (Table 4).
Lactobacilli in fecal samples from the intervention study.
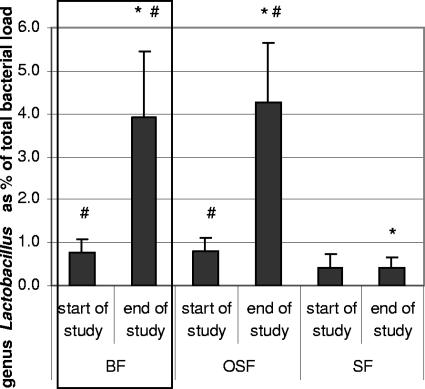
The levels of the different Lactobacillus species in fecal samples of breast-fed infants and infants receiving a standard formula or a standard formula supplemented with GOS/FOS were determined with the duplex 5′ nuclease assays. The number of lactobacilli as a percentage of the total bacteria is shown in Fig. 2. At the start of the study the percentages of lactobacilli in the OSF and SF group were not statistically different (0.8% ± 0.3% and 0.5% ± 0.3%, respectively). After 6 weeks of intervention, at the end of the study period, the percentage of lactobacilli in the OSF group (4.4% ± 1.4%) was significantly higher (P = 0.019) than in the SF group (0.4% ± 0.2%). Furthermore, there was a statistically significant increase in the percentages lactobacilli during the study period in the OSF group (0.8% ± 0.3% at the start versus 4.4% ± 1.4% at the end [P = 0.026]) and the BF group (0.8% ± 0.3% at start versus 4.1% ± 1.5% at the end [P = 0.034]).
FIG. 2.
Lactobacilli as a percentage of the total bacterial load in fecal samples of breast-fed infants (BF) and infants who received a standard formula supplemented with GOS-FOS (OSF) or a standard formula (SF). Bars represent SE. ✽, significant difference (P < 0.05) between the BF and SF group or the OSF and SF group at the end of the study; #, significant increase (P < 0.05) during the study period.
At the end of the study, breast-fed infants showed 3.0 ± 1.2 × 108 lactobacilli per g (wet weight) of feces, OSF-fed infants showed 3.3 ± 1.0 × 108 lactobacilli per g (wet weight) of feces, and SF-fed infants showed 5.4 ± 3.1 × 107 lactobacilli per g (wet weight) of feces.
The different Lactobacillus species expressed as a percentage of all lactobacilli are given in Table 5. In breast-fed infants L. acidophilus, L. paracasei, and L. casei were the most dominant species throughout the study period. The breast-fed infants also showed a significant increase during the study period of L. acidophilus (13.6% ± 3.4% versus 23.5% ± 4.5% [P = 0.017]), L. paracasei (7.2% ± 3.3% versus 22.1% ± 6.1% [P = 0.027]), and L. casei (4.0% ± 1.3% versus 6.0% ± 1.8% [P = 0.028]). At inclusion, the infants receiving OSF or SF showed a Lactobacillus distribution with relatively high proportions of L. acidophilus, L. casei, L. delbrueckii, and L. reuteri. During the intervention period a significant increase was shown for L. acidophilus (16.6% ± 3.3% versus 24.5% ± 3.9% [P = 0.001]), L. paracasei (0.8% ± 0.6% versus 16.8% ± 4.2% [P = 0.011]), and L. casei 5.6% ± 2.4% versus 10.7% ± 2.5% (P = 0.005)) as well as a significant decrease for L. delbrueckii (2.5% ± 1.1% versus 0.01% ± 0.01% [P = 0.045]) in infants receiving OSF. Consequently, the Lactobacillus distribution of the OSF group, at the end of the intervention study, mimics the distribution of breast-fed infants with L. acidophilus, L. paracasei, and L. casei as the predominant strains. In infants receiving SF a significant increase was seen in L. casei from 5.5% ± 1.5% to 8.3% ± 2.0% (P = 0.017) and in L. delbrueckii from 1.8% ± 0.7% to 6.9% ± 2.8% (P = 0.049). Also, a significant difference was found between the percentages of L. delbrueckii in infants receiving OSF and SF (0.01% ± 0.01% and 6.9% ± 2.8% [P = 0.033], respectively). At the end of the intervention period, the composition of the Lactobacillus microbiota in the SF-group represented more L. delbrueckii and L. reuteri and less L. acidophilus and L. paracasei compared to the BF and OSF groups.
TABLE 5.
Lactobacillus species as a percentage of the total Lactobacillus population in fecal samples of infants receiving breast milk, a standard formula supplemented with GOS and FOS, or a standard formulaa
| Lactobacillus sp. | % Lactobacillus spp. (SE) at:
|
|||||
|---|---|---|---|---|---|---|
| Start of the study (n = 10)
|
End of the study (n = 10)
|
|||||
| BF | OSF | SF | BF | OSF | SF | |
| L. acidophilus | 13.6 (3.4)A | 16.6 (3.3)A | 16.8 (4.1) | 23.5 (4.5)A | 24.5 (3.9)A | 19.2 (4.1) |
| L. casei | 4.0 (1.3)A | 5.6 (2.4)A | 5.5 (1.5)A | 6.0 (1.8)A | 10.7 (2.5)A | 8.3 (2.0)A |
| L. delbrueckii | 1.1 (0.8) | 2.5 (1.1)B | 1.8 (0.7)A | <0.001 (0.00)C | 0.01 (0.01)B,D | 6.9 (2.8)A,C,D |
| L. fermentum | <0.001 (0.00) | 0.2 (0.2) | 0.3 (0.3) | <0.001 (0.00) | <0.001 (0.00) | 0.05 (0.03) |
| L. paracasei | 7.2 (3.3)A | 0.8 (0.6)A | 0.9 (0.5) | 22.1 (6.1)A | 16.8 (4.2)A | 5.6 (3.3) |
| L. plantarum | <0.001 (0.00) | <0.001 (0.00) | <0.001 (0.00) | <0.001 (0.00) | <0.001 (0.00) | <0.001 (0.00) |
| L. reuteri | 2.2 (1.5) | 2.1 (0.8) | 1.9 (1.5) | 1.4 (0.6) | 1.3 (0.4) | 6.4 (3.2) |
| L. rhamnosus | <0.001 (0.00) | 0.2 (0.2) | 0.2 (0.2) | <0.001 (0.00) | <0.001 (0.00) | <0.001 (0.00) |
| Others | 71.9 (10.3)B | 72.1 (8.6)B | 72.5 (8.7)B | 47.0 (13.0)B | 46.8 (12.5)B | 53.5 (15.3)B |
BF, breast milk; OSF, standard formula supplemented with GOS and FOS; SF, standard formula. Superscripts: A, a significant increase (P < 0.05) during the study period; B, a significant decrease (P < 0.05) during the study period; C, a significant difference (P < 0.05) between the BF and SF groups; D, a significant difference (P < 0.05) between the OSF and SF groups.
L. fermentum, L. plantarum, and L. rhamnosus strains were present in very low percentages at the start of the intervention period, and these strains seemed to disappear completely during the intervention in all feeding groups.
DISCUSSION
Duplex 5′ nuclease assays were designed, optimized, validated, and used to study the distribution of Lactobacillus species in fecal samples of infants obtained from a nutritional intervention study. With these accurate assays, it was demonstrated that after an intervention with a mixture of galacto- and fructo-oligosaccharides the Lactobacillus species distribution in the feces of formula-fed infants closely resembles the distribution in breast-fed infants. Infants receiving SF showed a somewhat different pattern with relatively high levels of L. delbrueckii and lower levels of L. paracasei.
Species-specific quantitative real-time PCR.
Currently, traditional plating methods, conventional PCR, or fluorescent in situ hybridization (FISH) are used for the enumeration of lactobacilli. Traditional plating methods have some major disadvantages compared to modern molecular techniques, such as insufficient selectivity and the presence of “nonculturable” bacteria in fecal samples (31). The FISH technique is currently used to quantify the genus Lactobacillus in feces. However, with the commonly used FISH probe (S-G-Lab-0158-a-A20) for quantification of the genus Lactobacillus, genera such as Enterococcus, Pediococcus, Weissella, Vagococcus, Leuconostoc, and Oenococcus are also detected (17). In addition, the detection limit of FISH is rather high, which disables the quantification of very low bacterial numbers present in fecal samples of, for example, the different lactobacilli species. The conventional PCR is sufficiently sensitive for the detection of the genus Lactobacillus (40) and the different Lactobacillus species (37, 41). However, the conventional PCR can only be used for semiquantitative assessment, due to endpoint analyses limitations such as the plateau phase (29) and diminishing effects of differences in PCR product abundance (26). Contemporary quantitative real-time PCR allows the monitoring of the complete amplification and, as a consequence, overcomes the limitations correlated with endpoint analyses of the PCR process. To follow the PCR process, the use of specific fluorescently labeled probes or a minor-groove binding dye, like SYBR Green, can be utilized (9). A major disadvantage of the minor groove binding dyes is that these bind nonspecifically to all double-stranded DNA and may therefore reduce the specificity of a PCR.
For enumeration of the relatively small amounts of the different Lactobacillus species in fecal samples duplex 5′ nuclease assays were developed. These assays use a specific fluorescently labeled (TaqMan) probe during the amplification to ensure a high specificity and sensitivity.
The 16S-23S intergenic spacer rRNA gene sequences were used for the design of specific primers and probes for the duplex 5′ nuclease assays instead of the 16S rRNA gene, which is commonly used for the phylogenic analyses and specific detection of bacteria. Due to high similarities of the 16S rRNA gene sequences of the different Lactobacillus species, it is not feasible to develop highly specific primer and probe sets (23) for this gene. The intergenic spacer of 16S-23S rRNA gene can be used for a more detailed analysis of Lactobacillus species because sequences are less conserved than the 16S rRNA gene sequence (31).
The CV values (0.04 to 0.14) for the different species-specific duplex 5′ nuclease assays are acceptable and comparable to the CV values (0.09 to 0.28) reported earlier for determination of bacteria in fecal samples with the FISH technique (12, 18).
Lactobacilli in fecal samples from the intervention study.
In fecal samples of breast-fed infants, as well as in infants receiving a standard formula containing GOS-FOS, a significant increase in the percentage of lactobacilli was demonstrated during the study period. In contrast, the numbers in infants receiving a standard formula remained constant. The data presented here, obtained using quantitative molecular methods, support an earlier study in which traditional plating methods were used to show that GOS-FOS stimulates fecal lactobacilli (28). The sum of bifidobacteria and lactobacilli at the end of the study reaches ∼80% for the BF and OSF groups, whereas this percentage is ∼50% for the SF group. This is in correspondence with earlier findings, which state that the intestinal microbiota of breast-fed infants is generally dominated by the genera Bifidobacterium and Lactobacillus. Infants fed a standard formula are reported to have a more diverse microbiota with higher numbers of Bacteroides and Clostridium spp. (19, 35).
At the start of the study a higher percentage of lactobacilli was expected in the breast-fed group compared to the OSF and SF group since earlier reports state that breast-fed infants have relatively high levels of lactobacilli (19, 35). The level of the genus Lactobacillus was, however, not elevated in breast-fed infants compared to infants receiving OSF or SF at the start of the present study, although they were exclusively breast-fed for 4 weeks before the start of the study. On the other hand, the Lactobacillus species distribution of breast-fed infants already differed from that of OSF- and SF-fed infants at study start and was mainly composed of L. acidophilus, L. casei, and L. paracasei.
A major finding of the present study is that GOS-FOS supplemented in a standard formula results in a Lactobacillus distribution with relatively high levels of L. acidophilus, L. casei, and L. paracasei, which is rather similar to that of breast-fed infants. Infants receiving a standard formula showed more L. delbrueckii and L. reuteri and less L. paracasei and L. acidophilus at the study end. In literature, it has only been described that L. acidophilus is one of the most common Lactobacillus species in infants (35) and also that L. reuteri, L. gasseri, L. paracasei, L. rhamnosus, and L. fermentum are commonly present (34, 44). In the present study, relatively high levels of L. acidophilus were also found in all of the infants. Conversely, no or very low levels of L. rhamnosus or L. fermentum were found in the feces of these infants. A large group of lactobacilli in the fecal samples of these infants (∼70% at the study start and ∼50% at the study end) is still unknown. This percentage of lactobacilli could consist partly of L. gasseri or other known human lactobacilli strains, such as L. crispatus, L. salivarius, L. johnsonii, L. ruminus, L. vitulinis, and L. brevis (13, 27, 34). The distribution of the unknown Lactobacillus species might still differ between the BF, OSF, and SF groups.
As previously shown for the Bifidobacterium population (16), an infant milk formula containing a specific mixture of prebiotics is also able to induce a Lactobacillus species distribution that mimics the distribution of breast-fed infants.
Acknowledgments
We thank the pediatric practitioners Sabine Gross, Klaus Helm, Malte Klarczyk, and Helmut Schöpfer for their collaboration and collection of the stool specimens. We thank Annelies Crienen for excellent technical assistance. We thank Günther Boehm for valuable advice throughout the study.
This study was partially supported by a grant from the European Commission through the PROEUHEALTH program (QLRT-2000-01273).
REFERENCES
- 1.Adams, M. R., and C. J. Hall. 1988. Growth inhibition of food-borne pathogens by lactic and acetic acids and their mixtures. Int. J. Food Sci. Technol. 23:287-292. [Google Scholar]
- 2.Agostoni, C., I. Axelsson, C. Braegger, O. Goulet, B. Koletzko, K. F. Michaelsen, J. Rigo, R. Shamir, H. Szajewska, D. Turck, and L. T. Weaver. 2004. Probiotic bacteria in dietetic products for infants: a commentary by the ESPGHAN Committee on Nutrition. J. Pediatr. Gastroenterol. Nutr. 38:365-374. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bakker-Zierikzee, A., M. S. Alles, J. Knol, F. Kok, J. J. M. Tolboom, and J. G. Bindels. 2005. Effects of infant formula containing a mixture of galacto- and fructooligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br. J. Nutr. 94:783-790. [DOI] [PubMed] [Google Scholar]
- 5.Berthier, F., and S. D. Ehrlich. 1998. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 161:97-106. [DOI] [PubMed] [Google Scholar]
- 6.Boehm, G., S. Fanaro, J. Jelinek, B. Stahl, and A. Marini. 2003. Prebiotic concept for infant nutrition. Acta Paediatr. 91(Suppl.):64-67. [DOI] [PubMed] [Google Scholar]
- 7.Boehm, G., M. Lidestri, P. Casetta, J. Jelinek, F. Negretti, B. Stahl, and A. Marini. 2002. Supplementation of a bovine milk formula with an oligosaccharide mixture increases counts of faecal bifidobacteria in preterm infants. Arch. Dis. Child Fet. Neonat. Ed. 86:F178-F181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehm, G., and B. Stahl. 2003. Oligosaccharides, p. 203-243. In T. Mattila-Sandholm and M. Saarela (ed.), Functional dairy products. Woodhead Publishing, Cambridge, United Kingdom.
- 9.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 10.Collins, M. D., and G. R. Gibson. 1999. Probiotics, prebiotics, and synbiotics: approaches for modulating the microbial ecology of the gut. Am. J. Clin. Nutr. 69:1052S-1057S. [DOI] [PubMed] [Google Scholar]
- 11.Dobson, C. M., H. Deneer, S. Lee, S. Hemmingsen, S. Glaze, and B. Ziola. 2002. Phylogenetic analysis of the genus Pediococcus, including Pediococcus claussenii sp. nov., a novel lactic acid bacterium isolated from beer. Int. J. Syst. Evol. Microbiol. 52:2003-2010. [DOI] [PubMed] [Google Scholar]
- 12.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujisawa, T., T. Yaeshima, and T. Mitsuoka. 1996. Lactobacilli in human feces. Biosci. Microflora 15:69-75. [Google Scholar]
- 14.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 15.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 16.Haarman, M., and J. Knol. 2005. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 71:2318-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmsen, H. J., P. Elfferich, F. Schut, and G. W. Welling. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb. Ecol. Health Dis. 11:3-12. [Google Scholar]
- 18.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen, H. J., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 20.Knol, J., G. Boehm, M. Lidestri, F. Negretti, J. Jelinek, M. Agosti, B. Stahl, A. Marini, and F. Mosca. 2005. Increase of faecal bifidobacteria due to dietary oligosaccharides induces a reduction of clinically relevant pathogen germs in the faeces of formula-fed preterm infants. Acta Paediatr. 94(Suppl.):31-33. [DOI] [PubMed] [Google Scholar]
- 21.Knol, J., P. Scholtens, C. Kafka, J. Steenbakkers, S. Gro, K. Helm, M. Klarczyk, H. Schopfer, H. M. Bockler, and J. Wells. 2005. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 40:36-42. [DOI] [PubMed] [Google Scholar]
- 22.Kutyavin, I. V., I. A. Afonina, A. Mills, V. V. Gorn, E. A. Lukhtanov, E. S. Belousov, M. J. Singer, D. K. Walburger, S. G. Lokhov, A. A. Gall, R. Dempcy, M. W. Reed, R. B. Meyer, and J. Hedgpeth. 2000. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leblond-Bourget, N., H. Philippe, I. Mangin, and B. Decaris. 1996. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int. J. Syst. Bacteriol. 46:102-111. [DOI] [PubMed] [Google Scholar]
- 24.Liu, W., and D. A. Saint. 2002. A new quantitative method of real-time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal. Biochem. 302:52-59. [DOI] [PubMed] [Google Scholar]
- 25.Massi, M., B. Vitali, F. Federici, D. Matteuzzi, and P. Brigidi. 2004. Identification method based on PCR combined with automated ribotyping for tracking probiotic Lactobacillus strains colonizing the human gut and vagina. J. Appl. Microbiol. 96:777-786. [DOI] [PubMed] [Google Scholar]
- 26.Mathieu-Daude, F., J. Welsh, T. Vogt, and M. McClelland. 1996. DNA rehybridization during PCR: the “Cot effect” and its consequences. Nucleic Acids Res. 24:2080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsuoka, T. 1992. The human gastrointestinal tract, p. 69-114. In B. J. B. Wood (ed.), The lactic acid bacteria, vol. 1. The lactic acid bacteria in health and disease. Elsevier Applied Science, New York, N.Y. [Google Scholar]
- 28.Moro, G., I. Minoli, M. Mosca, S. Fanaro, J. Jelinek, B. Stahl, and G. Boehm. 2002. Dosage-related bifidogenic effects of galacto- and fructooligosaccharides in formula-fed term infants. J. Pediatr. Gastroenterol. Nutr. 34:291-295. [DOI] [PubMed] [Google Scholar]
- 29.Morrison, C., and F. Gannon. 1994. The impact of the PCR plateau phase on quantitative PCR. Biochim. Biophys. Acta 1219:493-498. [DOI] [PubMed] [Google Scholar]
- 30.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 31.O'Sullivan, D. J. 2000. Methods for analysis of the intestinal microflora. Curr. Issues Intest. Microbiol. 1:39-50. [PubMed] [Google Scholar]
- 32.Ouwehand, A., E. Isolauri, and S. Salminen. 2002. The role of the intestinal microflora for the development of the immune system in early childhood. Eur. J. Nutr. 41:I32-I37. [DOI] [PubMed] [Google Scholar]
- 33.Rachman, C. N., P. Kabadjova, H. Prevost, and X. Dousset. 2003. Identification of Lactobacillus alimentarius and Lactobacillus farciminis with 16S-23S rDNA intergenic spacer region polymorphism and PCR amplification using species-specific oligonucleotide. J. Appl. Microbiol. 95:1207-1216. [DOI] [PubMed] [Google Scholar]
- 34.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 35.Satokari, R. M., E. E. Vaughan, C. F. Favier, J. Doré, C. A. Edwards, and W. M. de Vos. 2002. Diversity of Bifidobacterium and Lactobacillus spp. in breast-fed and formula fed infants as assessed by 16S rDNA sequence differences. Microb. Ecol. Health Dis. 14:97-105. [Google Scholar]
- 36.Schmelzle, H., S. Wirth, H. Skopnik, M. Radke, J. Knol, H. M. Bockler, A. Bronstrup, J. Wells, and C. Fusch. 2003. Randomized double-blind study of the nutritional efficacy and bifidogenicity of a new infant formula containing partially hydrolyzed protein, a high beta-palmitic acid level, and nondigestible oligosaccharides. J. Pediatr. Gastroenterol. Nutr. 36:343-351. [DOI] [PubMed] [Google Scholar]
- 37.Song, Y., N. Kato, C. Liu, Y. Matsumiya, H. Kato, and K. Watanabe. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 38.Tannock, G. W., A. Tilsala-Timisjarvi, S. Rodtong, J. Ng, K. Munro, and T. Alatossava. 1999. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yogurt by 16S-23S rRNA gene intergenic spacer region sequence comparisons. Appl. Environ. Microbiol. 65:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tilsala-Timisjarvi, A., and T. Alatossava. 1997. Development of oligonucleotide primers from the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int. J. Food Microbiol. 35:49-56. [DOI] [PubMed] [Google Scholar]
- 40.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weaver, L. T. 2003. Improving infant milk formulas: near the end of the trail for the holy grail? J. Pediatr. Gastroenterol. Nutr. 36:307-310. [DOI] [PubMed] [Google Scholar]
- 43.Wold, A. E., and I. Adlerberth. 2000. Breast feeding and the intestinal microflora of the infant: implications for protection against infectious diseases. Adv. Exp. Med. Biol. 478:77-93. [DOI] [PubMed] [Google Scholar]
- 44.Xanthopoulos, V., I. Ztaliou, W. Gaier, N. Tzanetakis, and E. Litopoulou-Tzanetaki. 1999. Differentiation of Lactobacillus isolates from infant faeces by SDS-PAGE and rRNA-targeted oligonucleotide probes. J. Appl. Microbiol. 87:743-749. [DOI] [PubMed] [Google Scholar]